Abstract
The effect of somatic mutations and the gene expression profiles on the prognosis is well documented in cancer research. This study was conducted to evaluate the association of GATA3 somatic mutations with tumor features, survival, and expression profiles in breast cancer. Clinicopathological information was compared between TCGA-BRCA patients with GATA3-mutant and non-mutant tumors in all patients as well as in ER-positive subgroup. Cox-regression method was used to evaluate the association of the GATA3 mutation status with overall survival time. Differential gene expression, functional annotation, and protein–protein interaction analyses were performed using edgeR, Metascape, DAVID, STRING and CytoNCA. GATA3-mutant and non-mutant samples had significantly different clinicopathological features (p < 0.05). While GATA3 mutation status was not associated with the overall survival in the entire cohort (padj = 0.52), the GATA3-wild type ER-positive cases had a better prognosis than mutant ones (padj = 0.04). GATA3 expression was higher in tumors than normal tissues. Several pathways were different between mutant and non-mutant groups (p < 0.05). Interleukin-6 was found as the highest scored gene in both comparisons (normal vs. mutant and normal vs. non-mutant groups) in the entire patient and in the ER-positive subgroup, suggesting the association of IL6 with breast tumorigenesis. These findings suggest that GATA3 mutations can be associated with several tumor characteristics and influence the pattern of gene expression. However, GATA3 mutation status seems to be a prognostic factor for the disease only in ER-positive patients.
Subject terms: Cancer, Computational biology and bioinformatics, Genetics, Molecular biology, Biomarkers, Oncology, Risk factors
Introduction
Breast cancer, the most common type of cancer in women worldwide, is a heterogeneous disease with different pathological and molecular features and subtypes1. The disease is caused by both environmental and genetic factors2. In this regard, numerous genetic risk factors have been identified for tumor development and progression3. Except for the genes with highly penetrant and hereditary mutations, such as BRCA1 and BRCA24, the genetic basis of breast cancer and the role of genetic variations and their effects on malignant transformation are currently complex and requires further investigations. Several studies have demonstrated that somatic mutations in oncogenes and tumor suppressor genes are major drivers of different types of breast tumors and correlate with clinicopathological characteristics of the disease, response to therapy, or prognosis5–7. GATA binding protein 3 (GATA3) is one of the important genes involved in breast cancer development8.
GATA binding protein 3 is a transcription factor that encodes a protein member of the GATA family. GATA family members have two conserved Zinc-finger DNA binding domains. This transcription factor binds to promoters of target genes through the consensus (A/T)GATA(A/G) motifs9. Previous studies have demonstrated that GATA3 protein has crucial roles in cell development and differentiation in different types of cells, including mammary tissue10. Therefore, variations in its expression can affect downstream pathways and result in changes in cellular characteristics as its higher expression has been identified in hormone receptor-positive breast cancer patients11. While some data have pointed out that the GATA3 expression level is not an independent prognostic factor11, several researchers have reported that it was associated with better survival in breast cancer patients12,13. Also, it has been reported that breast tumors expressing low levels of GATA3 were correlated with larger tumors14. A literature report suggests that related pathways may be the reason for the association of this gene with some clinical features of breast cancer15,16. In light of these findings, GATA3 has been considered as an important gene in breast development and cancer17. However, the role of GATA3 somatic mutations in the development of breast tumor characteristics, patient survival outcomes, and its impact on tumor gene expression profiles is poorly understood.
In this study, we evaluated the genomic alterations of GATA3 in breast tumors, using the data collected by TCGA18, and analyzed the associations of GATA3 somatic mutations with tumor features, patient survival, and tumor gene expression profiles to highlight the clinical importance of this gene in breast cancer.
Results
GATA3 somatic mutation status and association with clinicopathological features
In the TCGA-BRCA cohort, tumors of 975/1085 female patients were evaluated for somatic mutations. Among these patients, a total of 103 different GATA3 mutations were identified in 138 patients (14.15%). Insertions constituted the largest type of mutations (50.5%), followed by deletions (29.1%) and substitutions (20.4%). A large portion of the mutations (74.7%) resulted in frame-shifts and variant effect predictor (VEP)19 has indicated 96.3% of all mutations were predicted to have a high or moderate impact. The most frequent mutation was X309, which is a two-base pairs (CA) deletion/splice site mutation (chr10:g.8069470delCA, annotated as GATA3 X309_splice in the GDC portal). This mutation was detected in tumors from 21 patients (15.22% of patients with GATA3 mutations). There were 11 additional recurrent mutations identified in more than one patient (n = 2–8), while the rest of the mutations were detected only in one patient.
The average diagnosis age was 45.66 ± 13.65 and 58.77 ± 12.97 in patients with and without GATA3 mutations, respectively (p = 0.001; Table 1). We compared the GATA3 mutation status in patients with different age categories. This analysis showed that the proportion of the patients with GATA3 mutated tumors was higher in the patients diagnosed under 40 years of age compared to those who were diagnosed after 40 years of age [20 of 89 patients under 40 years old (22.5%) and 118 of 885 patients above 40 years old (13.3%), respectively; p = 0.02]. In addition to age at diagnosis, menopausal status was significantly different between patients with and without GATA3 mutations (p = 0.00004; Table 1). Other clinicopathological characteristics that were associated with GATA3 mutation status in this patient cohort (Table 1) are the following: pathologic tumor size was significantly different between patients with GATA3 mutant tumors compared to patients with wild-type GATA3 tumors (p = 0.01). A significant difference was also seen with tumor histological types. There was a strong relationship between the GATA3 mutation and ER/PR status; almost none of the tumors with GATA3 mutations were ER-negative (Table 1). Additionally, in the multivariable logistic regression analysis, age at diagnosis, tumor size (pT), PR status, and histological tumor type were found to be independently associated factors of GATA3 mutation status in breast cancer (Table 2).
Table 1.
Results of univariate logistic regression analysis examining the association between GATA3 mutation status and clinical features.
| Variables | All patients | ER-Positive patients | ||||||
|---|---|---|---|---|---|---|---|---|
| Categories | Wild n (%) | Mutant n (%) | p valuea | OR (95% CI) | Wild n (%) | Mutant n (%) | p valuea | OR (95% CI) |
| Age | ||||||||
| Mean | 58.77 ± 12.97 | 45.66 ± 13.65 | 0.001 | 0.98 (0.96–0.99) | 60.03 ± 13.02 | 54.81 ± 13.68 | 0.00005 |
0.09 (0.95–0.98) |
| Age ≤ 35 | 25 (3%) | 6 (4.3%) | 16 (2.8) | 5 (3.9) | ||||
| Age > 35 | 811 (97%) | 14 (95.7%) | 0.40 | 0.68 (0.27–1.68) | 565 (97.2) | 123 (96.1) | 0.489 |
1.43 (0.52–3.99) |
| Age ≤ 40 | 69 (8.3%) | 20 (14.5%) | 39 (6.7) | 19 (14.8) | ||||
| Age > 40 | 767 (91.7%) | 118 (85.5%) | 0.02 | 0.53 (0.31–0.91) | 542 (93.3) | 109 (85.2) | 0.003 |
2.42 (1.35–4.35) |
| Menopause status | ||||||||
| Peri and Pre | 193 (25.6%) | 53 (43.8%) | 123 (23.3) | 49 (43.4%) | ||||
| Post | 562 (74.4%) | 68 (56.2%) | 0.00004 | 0.44 (0.30–0.65) | 405 (76.7) | 64 (56.6) | 0.00002 |
2.52 (1.61–3.85) |
| Race | ||||||||
| White | 577 (75.8%) | 95 (73.1%) | 418 (81.2) | 85 (70.8) | ||||
| Black/African-American | 138 (18.1%) | 22 (16.9%) | 0.90 | 0.97 (0.59–1.60) | 75 (14.5) | 22 (18.3) | 0.175 |
1.44 (0.85–2.45) |
| Asian | 46 (6.1%) | 13 (10%) | 0.10 | 1.72 (0.89–3.30) | 22 (4.3) | 13 (10.8) | 0.004 |
2.91 (1.41–5.99) |
| History of other malignancy | ||||||||
| No | 783 (93.7%) | 132 (95.7%) | 537 (92.4) | 122 (95.3) | ||||
| Yes | 53 (6.3%) | 6 (4.3%) | 0.37 | 0.67 (0.28–1.59) | 44 (7.6) | 6 (4.7) | 0.253 |
0.60 (0.25–1.44) |
| History of neoadjuvant therapy | ||||||||
| No | 826 (98.8%) | 135 (98.5%) | 572 (98.3) | 125 (98.4) | ||||
| Yes | 10 (1.2%) | 2 (1.5%) | 0.80 | 1.22 (0.27–5.65) | 10 (1.7) | 2 (1.6) | 0.910 |
0.91 (0.20–4.23) |
| Margin status | ||||||||
| Negative | 698 (89.9%) | 117 (88%) | 482 (89.1) | 108 (87.1) | ||||
| Positive/Close | 78 (10.1%) | 16 (12%) | 0.49 | 1.22 (0.69–2.17) | 59 (10.9) | 16 (12.9) | 0.526 |
1.21 (0.67–2.18) |
| Number of involved lymph node | ||||||||
| Median (Q1–Q3) | 2.28 ± 4.46 | 1.95 ± 3.31 | 0.45 | 0.98 (0.93–1.03) | 1 (0–3) | 1 (0–3) | 0.284 |
0.97 (0.91–1.03) |
| Lymph node ratio | ||||||||
| Median (Q1–Q3) | 0.16 ± 0.26 | 0.18 ± 0.27 | 0.56 | 1.25 (0.61–2.57) | 0.06 (0.0–0.25) | 0.06 (0.0–0.24) | 0.756 |
0.88 (0.39–1.98) |
| Lymph node ratio category | ||||||||
| Negative = 0 | 349 (49.4%) | 53 (46.5%) | 218 (44.8) | 48 (45.7) | ||||
| Low (> 0–0.2) | 179 (25.3%) | 29 (25.4%) | 0.79 | 1.07 (0.66–1.74) | 136 (27.9) | 29 (27.6) | 0.902 |
0.97 (0.58–1.61) |
| Intermediate (> 0.2–0.65) | 118 (16.7%) | 23 (20.2%) | 0.36 | 1.28 (0.75–2.19) | 89 (18.3) | 21 (20.0) | 0.812 |
1.07 (0.61–1.79) |
| High (> 0.65) | 61 (8.6%) | 9 (7.9%) | 0.94 | 0.97 (0.46–2.07) | 11 (9.0) | 7 (6.7) | 0.457 |
0.71 (0.31–1.70) |
| AJCC pT | ||||||||
| T1 and T2 | 717 (85.9%) | 106 (77.4%) | 496 (85.4) | 99 (78.0) | ||||
| T3 and T4 | 118 (14.1%) | 31 (22.6%) | 0.01 | 1.78 (1.14–2.77) | 85 (14.6) | 28 (22.0) | 0.040 |
1.65 (1.02–2.66) |
| AJCC pN | ||||||||
| Negative | 398 (48.4%) | 62 (46.6%) | 256 (44.8) | 57 (46.0) | ||||
| Positive | 424 (51.6%) | 71 (53.4%) | 0.70 | 1.07 (0.74–1.55) | 315 (55.2) | 67 (54.0) | 0.818 |
0.95 (0.65–1.41) |
| AJCC pM | ||||||||
| Negative | 702 (97.2%) | 115 (97.5%) | 492 (98.6) | 105 (97.2) | ||||
| Positive | 15 (2.1%) | 3 (2.5%) | 0.76 | 1.22 (0.35–4.28) | 7 (1.4) | 3 (2.8) | 0.318 |
2.01 (0.51–7.89) |
| AJCC stage | ||||||||
| Stage 1 and 2 | 627 (76.7%) | 96 (71.1%) | 428 (75.1) | 90 (72.0) | ||||
| Stage 3 and 4 | 191 (23.3%) | 39 (28.9) | 0.16 | 1.33 (0.89–2.00) | 142 (24.9) | 35 (28.0) | 0.473 |
1.17 (0.76–1.81) |
| ER status by IHC | ||||||||
| Negative | 216 (27.1%) | 1 (0.8%) | – | – | – | – | ||
| Positive | 582 (72.9%) | 128 (99.2%) | 0.0001 | 47.51 (6.60–341.94) | – | – | – | – |
| PR status by IHC | ||||||||
| Negative | 282 (35.5%) | 24 (18.5%) | 83 (14.3) | 23 (18.0) | ||||
| Positive | 513 (64.5%) | 106 (81.5%) | 0.0002 | 2.43 (1.52–3.87) | 497 (85.7) | 105 (82.0) | 0.295 |
0.76 (0.46–1.27) |
| HER2 statusb | ||||||||
| Negative | 589 (81.5%) | 91 (85%) | 431 (81.6) | 91 (85.0) | ||||
| Positive | 134 (18.5%) | 16 (15%) | 0.37 | 0.77 (0.44–1.36) | 97 (18.4) | 16 (15.0) | 0.400 |
0.78 (0.4401.39) |
| Receptor statusc | ||||||||
| ER and/or PR positive | 598 (77.3%) | 129 (100%) | – | – | – | – | ||
| HER2 overexpressed | 32 (4.1%) | 0 (0%) | 1.00 | 0.00 (–) | – | – | – | – |
| TNBC | 144 (18.6%) | 0 (0%) | 1.00 | 0.00 (–) | – | – | – | – |
| Anatomic neoplasm subdivision | ||||||||
| Left | 432 (51.6%) | 75 (54.3%) | 299 (51.4) | 69 (53.9) | ||||
| Right | 405 (48.4%) | 63 (45.7%) | 0.55 | 0.90 (0.62–1.29) | 283 (48.6) | 59 (46.1) | 0.604 |
0.90 (0.62–1.33) |
| Histological type of tumord | ||||||||
| IDC | 617 (73.8%) | 103 (74.6%) | 400 (68.7) | 96 (75.0) | ||||
| ILC | 148 (17.7%) | 14 (10.1%) | 0.06 | 0.57 (0.32–1.02) | 136 (23.4) | 14 (10.9) | 0.005 |
0.43 (0.24–0.78) |
| Other | 71 (8.5%) | 21 (15.2%) | 0.03 | 1.77 (1.04–3.01) | 46 (7.9) | 18 (14.1) | 0.104 |
1.63 (0.91–2.94) |
AJCC American Joint Committee on Cancer, CI confidence interval, ER estrogen receptor, IDC invasive ductal carcinoma, IHC immunohistochemistry, ILC invasive lobular carcinoma, ISH in situ hybridization, OR odds ratio, PR progesterone receptor, TNBC triple negative breast cancer.
aSignificant p values are shown in bold.
bAccording to ISH/IHC results.
cAssociation between the receptor status and GATA3 mutation status cannot be estimated because all GATA3 mutant tumors are also ER and/or PR positive. Significant p values are shown in bold.
dOther category includes rare types of tumors (e.g. Metaplastic, Medullary tumors).
Table 2.
Results of the multivariable logistic regression analysis.
| All patients | ER-Positive patients | ||||
|---|---|---|---|---|---|
| Variable | p value | OR (95% CI) | Variable | p value | OR (95% CI) |
| Age at diagnosis | 0.00040 | 0.97 (0.96–0.99) | Age at diagnosis | 0.001 | 0.97 (0.96–0.99) |
| Tumor size (T3 and T4 vs. T1 and T2) | 0.00262 | 2.09 (1.29–3.38) | Tumor size (T3 and T4 vs. T1 and T2) | 0.195 | 1.45 (0.83–2.45) |
| Histological typea | 0.00513 | Histological type | |||
| ILC versus IDC | 0.00968 | 0.44 (0.23–0.82) | ILC versus IDC | 0.011 | 0.42 (0.21–0.81) |
| Other type versus IDC | 0.12139 | 1.59 (0.88–2.86) | Other type versus IDC | 0.075 | 1.76 (0.94–3.30) |
| PR status (positive vs. negative) | 0.00001 | 2.92 (1.81–4.73) | Race | ||
| - | - | - | Black/African American versus White | 0.285 | 1.35 (0.78–2.34) |
| - | - | - | Asian versus White | 0.031 | 2.29 (1.08–4.86) |
Significant p values are shown in bold.
CI confidence interval, IDC invasive ductal carcinoma, ILC invasive lobular carcinoma, OR odds ratio, PR progesterone receptor.
aOther category includes rare histological types, such as metaplastic and medullary tumors.
We repeated these analyses in the ER-positive subgroup (Tables 1, 2). Overall, the results in this subgroup analysis were similar to that of the entire patient cohort. An interesting finding in the ER-positive subgroup analysis was that the mutant cases were more frequently presented than non-mutants in the Asian population (Table 1).
GATA3 somatic mutations and prognosis
The median overall survival was 10.80 ± 0.7 years (11.69 ± 3.63 and 10.61 ± 2.19 years in patients without GATA3 mutation compared with patients with GATA3 mutation, respectively; p = 0.73). There was no significant difference between the two groups in terms of median survival time. This finding was also similar in the ER-positive subgroup (Table 3).
Table 3.
Results of the univariate and multivariable Cox regression analysis for GATA3 mutation status.
| Cox regression | All patients | ER-Positive patients | ||||
|---|---|---|---|---|---|---|
| Variable | p value | HR (95% CI) | Variable | p value | HR (95% CI) | |
| Univariate | GATA3 mutation status (yes vs. no) | 0.73 | 1.09 (0.66–1.80) | GATA3 mutation status (yes vs. no) | 0.40 | 1.26 (0.74–2.15) |
| Multivariable | GATA3 mutation status (yes vs. no) | 0.52 | 1.22 (0.66–2.26) | GATA3 mutation status (yes vs. no) | 0.040 | 1.84 (1.03–3.27) |
| Age at diagnosis | 0.0001 | 1.03 (1.02–1.05) | Age at diagnosis | 2.411E−8 | 1.05 (1.03–1.07) | |
| Stage category (S3 and S4 vs. S1 and S2) | 7.461E−10 | 4.11 (2.62–6.44) | Stage category (S3 and S4 vs. S1 and S2) | 0.026 | 1.90 (1.09–3.33) | |
| Radiation therapy status (yes vs. no) | 0.003 | 0.49 (0.31–0.79) | Lymph node status category (positive vs. negative) | 0.104 | 1.63 (0.90–2.96) | |
CI confidence interval, HR hazards ratio.
Univariate Cox proportional hazard analysis indicated age at diagnosis, menopause status, lymph node ratio, history of neoadjuvant therapy and adjuvant radiation therapy to be associated with survival times in the patients. Also, several tumor characteristics including margin status, pathologic tumor size (pT), lymph node (pN), and stage were associated with overall survival (Table S2).
While Multivariable Cox regression model adjusting for prognostic factors revealed that GATA3 somatic mutation status was not an independent prognostic factor for all patients (padj = 0.52), wild type samples indicated better prognosis in the ER-positive subgroup (padj = 0.04) (Table 3). However, age (padj = 0.0001), stage (padj = 7.461E−10) and radiation therapy (p = 0.003) were significantly and independently associated with overall survival time in the entire patient cohort. Analysis of the ER-positive cases indicated age (padj = 2.411E−8) and stage (padj = 0.026) as independent factors associated with overall survival time.
Gene expression analysis
According to the TCGA expression data, GATA3 expression level was higher in GATA3-mutant (log FC = 2.78, p = 4.38E−34 in all patients and log FC = 2.66, p = 2.07E−57 in ER-positive subgroup) and non-mutant (log FC = 1.76, p = 2.11E−21 in all patients and log FC = 1.96, p = 3.24E−46 in ER-positive subgroup) tumors than normal tissues. While mutant tumors had a higher level than non-mutants (log FC = 1.02; p = 1.15E−12), this was not detected in the analysis of the ER-positive breast cancer patients.
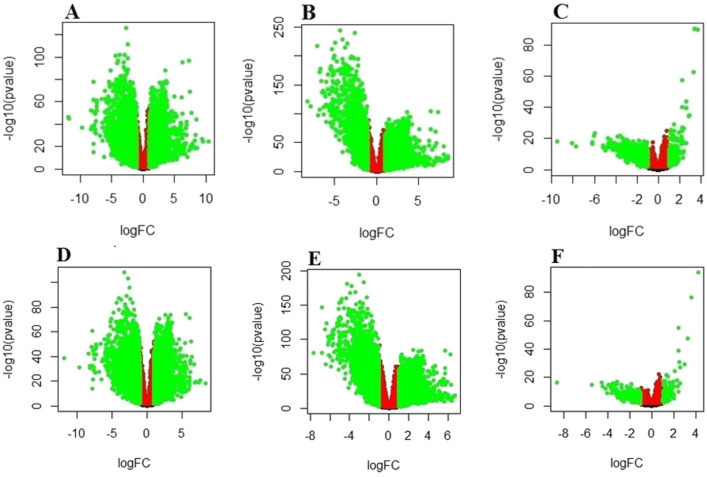
A total of 4816 differentially expressed genes (DEGs) were observed between the GATA3-mutant and normal tissues (2476 up-regulated and 2340 down-regulated genes). Additionally, there were a total of 4308 DEGs between the GATA3-non-mutant and normal tissues (2593 up-regulated and 1715 down-regulated genes). Finally, 907 DEGs between the non-mutant and mutant tumors were found: 169 genes were up-regulated and 738 genes were down-regulated at an FDR < 0.05 and log fold change (log FC) > 1. In the ER-positive subgroup, 4522 (2143 up-regulated and 2379 down-regulated genes), 4066 (2055 up-regulated and 2011 down-regulated genes) and 480 genes (103 up-regulated and 377 down-regulated genes) were found in the comparison between mutant versus normal, non-mutant versus normal and non-mutant versus mutant tumors, respectively. Volcano plots are shown in Fig. 1.
Figure 1.
Volcano plats showed analysis of differential expressed genes (DEGs) between the normal compared with the tumors (GATA3-mutant and non-mutant). (A) Log2-fold change mutant and normal; (B) non-mutant and normal; (C) non-mutant and mutant; (D) Log2-fold change mutant and normal in ER-positive patients; (E) non-mutant and normal in ER-positive patients; (F) non-mutant and mutant in ER-positive patients. Green dots represent significantly DEGs (FDR < 0.05 and log FC > 1).
The most up and down-regulated DEGs in three categories of comparison are listed in Table 4. MYH2 and CKM in mutant versus normal and non-mutant versus normal and SMR3B in mutant versus non-mutant were the top down-regulated genes. The top up-regulated genes were MUC2, S100A7A and ALDOB in mutant versus normal, non-mutant versus normal and mutant versus non-mutant, respectively. The ER-positive subgroup analysis showed MUC2, CST5 and ALDOB as the top up-regulated genes and MYH2 as the top down-regulated gene between mutant versus normal, non-mutant versus normal, and CSN1S1 as the top down-regulated gene between mutants versus non-mutant samples.
Table 4.
Differentially expressed genes (DEGs) between GATA3-mutant versus normal and GATA3-non-mutant versus normal and GATA3-mutant versus non-mutant tissues according to the TCGA data.
| Gene name | Fold change | FDR | Gene name | Fold change | FDR | |
|---|---|---|---|---|---|---|
| GATA3 mutant versus normal | All patients: 4816 (2476 up-regulated and 2340 down-regulated genes) | ER-Positive patients: 4522 (2143 up-regulated and 2379 down-regulated genes) | ||||
| Up-regulated DEGs | MUC2 | 10.43879 | 3.33E−24 | MUC2 | 8.329625376 | 2.62 E−18 |
| CGA | 9.492646 | 3.97 E−24 | CHRNA9 | 7.628457572 | 7.85 E−21 | |
| CHRNA9 | 9.389171 | 1.25E−26 | CGA | 7.332730083 | 1.31E−18 | |
| CPLX2 | 8.522081 | 5.63E−19 | PCSK1 | 6.922966106 | 1.79E−19 | |
| CST4 | 8.506592 | 2.09E−43 | TRH | 6.85432771 | 8.50E−19 | |
| Down-regulated DEGs | MYH2 | − 11.8785 | 1.83E−45 | MYH2 | − 11.8811779 | 6.71E−38 |
| CKM | − 11.8163 | 3.02E−44 | NRAP | − 9.667105996 | 8.76E−31 | |
| NRAP | − 9.63614 | 4.01E−36 | TNNC2 | − 8.329242401 | 9.99E−42 | |
| TNNC2 | − 8.32733 | 3.04E−51 | TCAP | − 8.199816299 | 5.68E−32 | |
| ACTA1 | − 8.21929 | 4.09E−35 | ACTA1 | − 8.118831848 | 4.32E−30 | |
| GATA3 non-mutant versus normal | All patients: 4308 (2593 up-regulated and 1715 down-regulated genes) | ER-Positive patients: 4066 (2055 up-regulated and 2011 down-regulated genes) | ||||
| Up-regulated DEGs | S100A7A | 8.467232 | 9.82E−22 | CST5 | 6.658392275 | 1.67E−18 |
| CSAG1 | 8.423582 | 3.50E−26 | S100A7A | 6.546948974 | 1.96E−16 | |
| CST5 | 8.403968 | 2.03E−22 | CGA | 6.525469762 | 1.16E−17 | |
| CGA | 8.399469 | 2.10E−22 | CARTPT | 6.349247937 | 1.00E−10 | |
| MAGEA12 | 8.173768 | 5.55E−22 | CST4 | 6.32237147 | 7.11E−34 | |
| Down-regulated DEGs | MYH2 | − 8.18882 | 1.49E−120 | MYH2 | − 7.688190533 | 6.83E−80 |
| CKM | − 7.80643 | 4.34E−112 | PYGM | − 6.809240454 | 7.50E−145 | |
| PYGM | − 7.07911 | 1.61E−215 | ACTA1 | − 6.782578399 | 4.19E−79 | |
| NRAP | − 6.89436 | 1.09E−95 | ATP2A1 | − 6.539245722 | 2.40E−113 | |
| ATP2A1 | − 6.75453 | 8.90E−171 | NRAP | − 6.423415223 | 1.04E−63 | |
| GATA3 mutant versus non-mutant | All patients: 907 (169 up-regulated and 738 down-regulated genes) | ER-Positive patients: 480 (103 up-regulated and 377 down-regulated genes) | ||||
| Up-regulated DEGs | ALDOB | 3.731806 | 9.25E−87 | ALDOB | 4.193275263 | 1.63E−90 |
| AMY2A | 3.414509 | 9.25E−87 | AMY2A | 3.62447513 | 3.97E−73 | |
| C8orf34 | 3.308972 | 2.16E−59 | C8orf34 | 3.23103563 | 1.77E−44 | |
| ZPLD1 | 2.907425 | 4.75E−32 | ZPLD1 | 2.950533096 | 9.72E−27 | |
| LOC284749 | 2.889682 | 1.42E−31 | ||||
| Down-regulated DEGs | SMR3B | − 9.48864 | 3.11E−16 | CSN1S1 | − 8.610397883 | 1.76E−14 |
| CSN1S1 | − 8.13224 | 3.62E−15 | MYOC | − 5.400766258 | 8.60E−13 | |
| CSN3 | − 7.72831 | 1.09E−13 | MSLN | − 4.537413853 | 3.42E−14 | |
| C4orf7 | − 6.20695 | 1.00E−14 | DMBT1 | − 4.476329021 | 4.97E−12 | |
| FABP7 | − 6.20416 | 7.96E−16 | MYH2 | − 4.192987371 | 2.92E−10 | |
| SMR3B | − 4.029963607 | 1.15E−07 | ||||
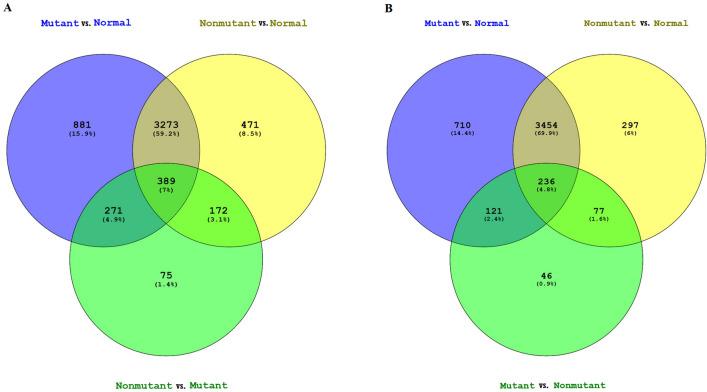
Venn diagram shows the common and specific genes in every group. As it can be seen in Fig. 2, 389 and 236 genes are common in the three groups of all and ER-positive patients, respectively, that might be involved in breast carcinogenesis and also be influenced by GATA3 mutations.
Figure2.
Venn diagram indicating differentially expressed genes overlapping between the samples in (A) the entire patient and (B) ER-positive subgroup. Blue: GATA3-Mutant versus Normal; Yellow: GATA3-Non-mutant versus Normal; Green: GATA3-Mutant versus Non-mutant.
Functional annotation analysis of differentially expressed genes
To gain an insight into the functionality of the DEGs between normal and tumor (mutant and non-mutant) samples, gene set enrichment analysis was performed using the Metascape and DAVID functional enrichment tool. According to DAVID outputs, 36 pathways found to be significantly different between GATA3-mutant and normal samples, 7 pathways had been previously reported as the most important pathways related to breast cancer (p ≤ 0.05)20–22. Evaluation of non-mutant tumors against normal tissue samples indicated 37 significantly different. Also, 3 different pathways (protein digestion and absorption; Wnt signalling; and cell adhesion molecules) were significantly different between mutant and non-mutant tumor tissues. Analysis of ER-positive patients indicated 37 and 36 significantly different pathways in normal samples in comparison with mutant and non-mutant tumors, respectively. Furthermore, pancreatic secretion pathway was different between mutant and non-mutant tumors. These results are shown in the supplementary information file, Table S3.
PPI network of module analysis
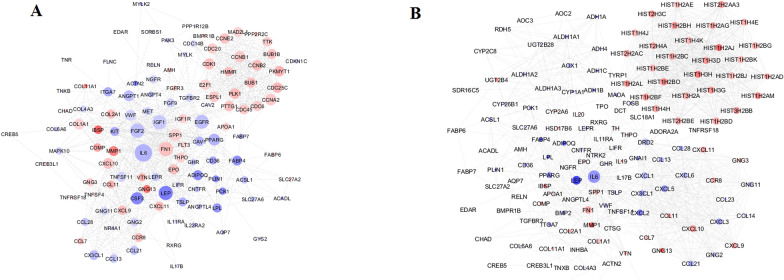
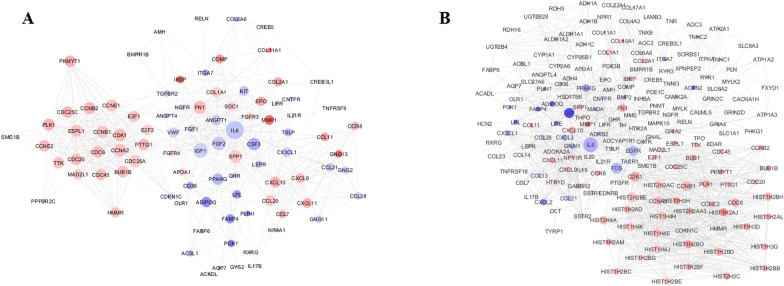
To gain a better understanding of the biological relationships between breast cancer-related genes, the genes that share the same GO term related to breast cancer were examined in the STRING database. Results indicated that 116 and 95 genes (proteins) for all patients and 142 and 191 for ER-positive subgroup matched the database and were used to construct the PPI network between GATA3 mutant tumor and normal tissues (Fig. 3) and between GATA3 non-mutant tumor and normal tissues, respectively (Fig. 4).
Figure 3.
PPI network of breast cancer differentially expressed genes (DEGs) between normal and GATA3 mutant samples in (A) the entire patient and (B) ER-positive subgroup. The node size is proportional to the degree value as the bigger size means the larger degree value. The color of the node is related to the expression of genes: up regulated genes are shown in Red and down regulated genes are shown in Blue.
Figure 4.
PPI network of breast cancer differentially expressed genes DEGs between normal and GATA3-non-mutant in (A) the entire patient and (B) ER-positive subgroup. The node size is proportional to the degree value as the bigger size means the larger degree value. The color of the node is related to the expression of genes: up regulated genes are shown in Red and down regulated genes are shown in Blue.
The top nodes with high topology score that were calculated by three centrality methods, were considered as hub nodes. Interleukin 6 (IL6) had the highest scores in three centrality methods in both comparisons between normal and mutant and normal and non-mutant groups in all patients as well as ER-positive subgroup. FN1, IGF1, FGF2 and LEP in all patients and, LEP and FN1 genes in the ER-positive subgroup could be considered as hub nodes in normal and mutant. Moreover, IGF1, FGF2, FN1 and SPP1 genes in all patients and FOS, FGFR, LEP and CDK1 genes in ER-positive subgroup could be considered as hub nodes in normal and non-mutant groups. PPI analysis did not find any prominent network when the two mutant and non-mutant groups were compared, which may be due to the limited number of identified gene sets.
Discussion
Cancer, as a multifactorial disease with complex pathological features, is influenced by genetic factors. However, somatic mutations are amongst the most important well-known genetic factors involved in cancer. The role of somatic mutations in tumor development and progression of cancer has been confirmed through advances in technology and increasing knowledge about mutation characteristics. In this study, we focused on the analysis of a gene with known roles in breast cancer, GATA38,16, using the large-scale data obtained by the TCGA project18. In this cohort, the frequency of somatic mutations in GATA3 was 14.15%. As previously reported, this gene is one of the three genes representing more than 10% somatic mutations in all breast cancer patients23. The analysis of clinical factors in relationship with the GATA3 somatic mutations reported in TCGA-BRCA project revealed that GATA3 mutations were associated with several clinical features and pathological subtypes of breast cancer. Also, differential gene expression analysis has identified different patterns of expression in normal samples, GATA3 mutant and non-mutant tumor tissues in the entire cohort as well as in the ER-positive cases. Furthermore, our results also showed three pathways were significantly different between GATA3 mutant and non-mutant tumors.
Our results suggested that patients with GATA3 mutant tumors were significantly younger than those patients without GATA3 mutations. A previous report has indicated that younger luminal B cases had GATA3 mutations more frequently than older patients24. This finding has also been validated in metastatic breast cancer patients27. Since ER-positive younger patients indicated poorer prognosis28, a higher rate of GATA3 mutations may have clinical importance.
Our results suggested the importance of GATA3 in tumor size in the TCGA dataset. It has been previously reported that mutational load is correlated with the size of tumor in breast cancer patients29. Therefore, it is expected to observe a higher rate of GATA3 mutation in larger tumors. Furthermore, a higher rate of rare types of tumor (Mixed Histology, Mucinous Carcinoma and Medullary Carcinoma) was observed in association with GATA3 mutations (Table 1). Conversely, after adjustment and also in the ER-positive group, we found a significant difference in mutation status between ILC and IDC, but not in rare types of breast cancer (Table 2). These results may be affected by the small number of rare types in comparison with ductal carcinoma of the breast. However, this may highlight the impact of mutations on different features of breast tumors (Table 2). In addition, the results of our analysis showed ER-positive tumors harbored almost all GATA3 somatic mutations detected in the patient cohort. This finding confirms previous reports showing an association of GATA3 with ER-positive status and luminal differentiation, which may reflect its role in response to chemotherapy30. Also, a study has shown that GATA3 up-regulates and stabilizes ER mRNA transcription31. In contrast, GATA3 expression is down-regulated by progestin-induced PR activation32. It may explain the association of GATA3 mutations with the luminal type of breast cancer as a hormone receptor-positive type.
As the two aspects of GATA3 have been studied, i.e. a difference in expression between mutant and non-mutant or normal tissues and the impact of its mutations on tumor properties, it can be postulated that in agreement with previous studies, our data support the higher level of expression in tumor tissues than normal samples16,33 and the lack of importance of GATA3 somatic mutations as an independent factor in patient survival11,34. However, non-mutant samples showed better survival than others in ER-positive patients. METABRIC data indicated the prognostic value of GATA3 X308_Splice mutation, as the mutant samples had better survival than wild-type ones both in all patients and ER-positive patients35. On the other hand, in samples representing a high expression of GATA3, mutant patients had longer survival than wild-types, and mutations in the second GATA3 zinc-finger (ZnFn2) was associated with lower survival time than other mutations36. Another study has also reported that a significant association of GATA3 mutations with hormone receptor-positive situation may reflect the better prognosis of the disease17. All of these different findings suggest the importance of mutation type and co-consideration of other related factors in the association of GATA3 somatic mutation with overall survival. However, different factors including the number of mutant samples and the study settings may cause this variation. We acknowledge such variation in these findings can make it more difficult to come to a straightforward conclusion. Regarding to the higher level of expression in tumor samples (GATA3 mutant and non-mutant) than normal ones, a meta-analysis study confirmed the relation between GATA3 overexpression and favorable phenotypes including ER-positive status14. On the other hand, a cell line study indicated the active GATA3 transcription factors cause proliferative phenotypes and promote the growth of ER-positive breast cancer cell lines37. In addition to the impact of the mutation on expression level, somatic mutations may affect the binding site and influence the rate of downstream genes expression and result in a changed transcriptional network36,38. Furthermore, it has been observed that higher rate of GATA3 mutations in ER-positive patients may lead to resistance to endocrine therapy27. Therefore, all of these findings indicate diverse activities of GATA3 protein which affect the luminal breast epithelial cells via different pathways can neutralize the impact of this gene on the prognosis of the disease.
MYH2, as a down-regulated gene in GATA3-mutant and non-mutant samples, encodes an Actin-based motor protein with the skeletal muscle contraction activity. According to the Human Protein Atlas18, MYH2 protein was not detected in breast tissues, however, low amount of RNA has been observed39. Since GATA3 mutants compared with non-mutant tumor samples did not indicate any difference in expression of MYH2, its lower expression in tumor samples may be resulted due to the tumor environment. Similar to MYH2, CKM (Muscle type of CK) is down-regulated in tumor (GATA3 mutant and non-mutant) tissues. Expression of this gene in mRNA level has previously been shown in breast samples39. Furthermore, a decreased level of serum CK has been specified in breast cancer patients40. Moreover, SMR3B gene (submaxillary gland androgen-regulated protein 3B) was identified to have differential expression between GATA3 mutant and non-mutant tumor tissues as mutant samples indicate a lower level of expression. Previously, it has been predicted SMR3B has GATA3 transcription factor binding site motif41 and is expressed more in triple-negative breast cancer patients with poor prognosis compared to the low-risk patients42. As GATA3 protein has a role in expression regulation, lower level of SMR3B expression in tumor carrying GATA3 mutations can be explained by this fact. CSN1S1 (Casein Alpha S1), is another top down-regulated gene in GATA3 mutant samples compared to non-mutants in the ER-positive subgroup. Its RNA expression has been identified in breast tissue, however, the protein has only been detected in lactating breast. Because of significantly different protein expression in benign prostate hyperplasia compared with normal and tumor prostate tissues, CSN1S1 has been reported as a potential biomarker for early identification of benign prostate hyperplasia patients43. Moreover, CSN1S1 has identified as a tumor suppressor that controls breast tumor growth and metastasis44. According to our finding, GATA3-mutants had larger tumor size that it may be due to the down-regulation of CSN1S1.
We found MUC2 over-expression in GATA3-mutant tumor than normal samples. MUC2 is up-regulated in mucinous carcinomas45, and have higher expression in invasive breast tumors than adjacent normal tissues. A significantly higher level of serum MUC2 has also been found in breast cancer patients compared with healthy people46. Furthermore, as a prognostic effector, MUC2 protein is associated with shorter disease-free survival47. Evaluation of a cell line with the limited expression of MUC2 indicated a decreased rate of proliferation and better response to chemotherapy by efficiently induced apoptosis48. These findings confirmed the potential prominent role of MUC2 expression as the prognostic marker in breast cancer. However, the relationship between GATA3 and MUC2 remains to be evaluated. Another up-regulated gene, S100A15, is a calcium-binding protein with higher expression in non-mutant tumors than normal ones. While there is evidence which indicates elevated S100A15 transcripts in ER/PR negative breast cancers49, the association of this gene with breast cancer prognosis has not been confirmed50. In the ER-positive subgroup, CST5 (Cystatin D), was the first top differentially up-regulated gene between non-mutants and normal. This gene has been down-regulated in colon cancer51, and its induction by calcitriol can also prevent the breast cancer cells growth52. The mutant and non-mutant comparison showed Aldolase B (ALDOB), a glycolytic enzyme, to be up-regulated in GATA3 mutant samples. However, tumor samples did not show differential expression in comparison with normal ones. Previous studies indicated a decreased level of ALDOB in several cancers53,54. Therefore, the higher expression of ALDOB in GATA3 mutant breast cancer tumors may be caused by involved common regulatory pathways that need to be confirmed by functional and gene–gene interaction analyses. Furthermore, according to the Venn diagram, 75 genes in the entire patient group and 46 in ER-positive subgroup, were differentially expressed between GATA3-mutant and non-mutant tumors that may indicate the impact of GATA3 in the expression profile of the tumor cells.
Considering the differently expressed pathways, previously indicated to be associated with breast cancer, protein digestion and absorption pathway was different between all categories55. Other pathways were specifically different between mutant and non-mutant tumors. Wnt/β-catenin signaling pathway is a modulating factor of mammary gland morphogenesis and cell properties20 and mediates the increase of GATA3 expression21. Consistent with our finding, a previous study indicated WNT/β-catenin signaling as an enriched gene set in GATA3 X308_Splice mutant breast tumor35. Cell adhesion molecules (CAMs) was another different gene set between tumor samples. The role of this pathway has been recognized in the carcinogenesis and metastasis of breast cancer. Therefore, evaluation of the involved genes can be diagnostic, prognostic and therapeutic targets22,56. Besides, the regulatory role of GATA3 in adhesion molecules expression has been identified in cell culture analysis57. Hence, expression variation of these genes can happen in association with GATA3 situation induced by mutations. These findings may reflect the interactions between GATA3 and genes involved in WNT and cell adhesion molecules pathways in the pathogenesis of breast cancer. Furthermore, we found that systemic lupus (SLE) erythematosus pathway is differentially expressed between ER-positive breast tumor and normal tissues. It has been shown that SLE is influenced by estrogen-estrogen receptor-mediated signaling through the modulation of cytokine production58. There are also reports indicating a lower rate of hormone-dependent cancers in SLE patients although they may tend for a higher incidence of triple-negative breast cancer compared to general population59. As the main finding of the protein–protein interaction analysis, IL6 was identified to be an important hub node in the comparison between tumor and normal samples. In line with our results indicating the contribution of this gene in different pathways, IL6 overexpression has been previously described in breast cancer60. Many cellular functions including oncogenesis are influenced by IL661. These findings suggest the crucial role of IL6 in the pathogenesis of breast cancer and the importance of targeting this gene in the treatment of the disease.
Conclusion
In conclusion, our results suggest that GATA3 mutation status is associated with a number of clinicopathological features, as well as with overall survival time only in ER-positive breast cancer. Our results also indicate a possible common biological process involving GATA3 mutations and ER/PR status, which needs to be confirmed by functional analyses. The GATA3 mutations may influence the expression profile of the tumor cells via impact on expression and activity rate of the GATA3 gene. These findings should also be confirmed using gene–gene interaction analyses and homogenous samples.
Methods
Patients and data files
The study population has consisted of female breast cancer patients in the TCGA-BRCA cohort. Information on the GATA3 mutations in the tumors was retrieved from https://portal.gdc.cancer.gov. This information was available for 975 of the patients. The tumor mRNA expression data (level 3 data; including raw count data) was extracted from Illuminahiseq_rnaseqV2-exon_quantification (MD5) data file at https://gdac.broadinstitute.org/. This data was available for 771 tumor (671 non-mutant and 100 mutant) and 99 normal tissues of the patients. Demographic and clinical data were obtained from the file rendered by the Legacy Archive of the GDC portal at https://portal.gdc.cancer.gov/legacy-archive/files/735bc5ff-86d1-421a-8693-6e6f92055563. Categorization of the study population was performed according to standard protocols62–68. All analyses were also replicated in ER-positive samples including 482 non-mutant and 92 mutant tumor tissues.
Computational analysis of expression profile
The edgeR program (http://bioconductor.org/packages/release/bioc/html/edgeR.html) is a Bioconductor software package for examining the differential expression of replicated count data using an over-dispersed Poisson model and Empirical Bayes methods to account for both biological and technical variability and moderate the degree of over-dispersion across transcripts69. This program was used to determine the DEGs in the normal tissues when compared to the tumors (GATA3 mutant and non-mutant). The probabilistic methods were used by edgeR to evaluate the differential expression. The affected genes determined based on a false discovery rate (FDR) < 0.05 and a log Fold change (FC) > 1.
Functional annotation of differentially expressed genes (DEGs)
The proteins encoded by DEGs were analyzed, and annotated using Metascape, “A Gene Annotation and Analysis Resource”, which can be used to analyze multi-platform OMICs data (http://metascape.org/gp/index.html), DAVID “Database for Annotation, Visualization and Integrated Discovery” (https://david.ncifcrf.gov/)70–72 to test for gene set enrichment analysis, Gene Ontology (GO) terms and pathways. According to the database, DAVID pathways output is based on KEGG (Kyoto Encyclopedia of Genes and Genomes). Only terms with modified Fisher Exact p value ≤ 0.05 were considered significant. Metascape is a web-based portal, and is useful for functional annotations of genes73.
Protein–protein interaction (PPI) network
DEGs (corrected p values ≤ 0.05) were imported to the search tool of STRING (v10.0, http://string-db.org/) for the retrieval of interacting genes/proteins by selecting Homo sapiens as the organism. STRING can identify a network of close interactions among this set of genes based on information on experimental as well as predicted protein interactions. The three methods including degree centrality, betweenness centrality, and closeness centrality were used to calculate the topology scores of nodes in the PPI network using the CytoNCA74.
Statistical analysis
Demographic and clinical/molecular data that were examined during statistical analyses are shown in the supplementary information file, Table S1. Comparison between variables between the two groups (mutant vs. non-mutant) was examined using Pearson's Chi-squared test for categorical variables and independent sample t test for continuous variables. Univariate logistic regression analysis was used to examine the associations of GATA3 somatic mutation status with different variables, and the odds ratios (OR) and 95% confidence intervals (CIs) were presented. Multivariate logistic regression analysis was used to assess the variables that were independently predictive of the GATA3 mutation status. For this purpose, covariates with p values ≤ 0.05 in the univariate analysis were entered into a multivariable model, excluding the rare variables (ER status and hormone receptor status). In addition, menopause status and age at diagnosis were highly associated, thus, menopausal status (which had more missing data than the age at diagnosis) was excluded from the multivariable model.
Overall survival (OS) time is defined as the time from diagnosis till the time of death or last contact. Associations between variables and OS were examined using the Kaplan–Meier plots/Log-rank test and Cox proportional hazards regression methods. Results of the univariate Cox regression analysis was used to select the variables to be entered into the multivariable Cox regression models. For this purpose, covariates with p values less than 0.05 in the univariate analysis were entered into a covariate selection method (Backward-LR), excluding the rare variables, such as metastasis status (pM) and history of neoadjuvant therapy, and highly correlated variables. Highly correlated variables included menopausal status (excluded) and age at diagnosis, tumor size (pT) (excluded) and stage, and lymph node ratio (excluded) and lymph node status (pN). As a result, age, stage, and radiation therapy status were selected for the analysis of the entire cohort. Association of the GATA3 mutation status with OS was then examined in a multivariable Cox model after adjusting for these clinical factors. Similar to this process, OS analysis was done for the ER-positive subgroup. After excluding the rare variables, such as metastasis status and history of neoadjuvant therapy, and highly correlated variables including menopausal status, tumor size and lymph node ratio, the variables including age, stage, and lymph node status were selected for the assessment of the GATA3 mutations’ association with OS in a multivariable Cox model. The hazard rate ratio (HR) and 95% CIs were calculated by the Cox models.
A p value < 0.05 was considered significant. All statistical analyses were performed using SPSS 16.0 (IBM, USA).
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Supplementary Information
Acknowledgments
This work was mainly conducted in a sabbatical period at the Discipline of Genetics, Faculty of Medicine, Memorial University, St. John's, Canada (FA), with the financial support from the Mashhad University of Medical Sciences, Mashhad, Iran. Authors gratefully acknowledge the use of data generated by TCGA Research Network (https://www.cancer.gov/tcga).
Author contributions
F. A. and S. S. contributed to the design of the work. F. A., S. S., A. S. and A. P. contributed to data analysis and interpretation. F. A. and S. S. contributed to drafting and editing the article. A. P. contributed to critical revision of the article and approving the final version of this paper. All authors also participated in the finalization of the manuscript and approved the final draft.
Funding
This work was financially supported by Mashhad University of Medical Sciences under grant 931185.
Data availability
The Data belongs to TCGA Research Network and is available in https://www.cancer.gov/tcga.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Fahimeh Afzaljavan, Ayeh Sadat Sadr and Sevtap Savas.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-80680-9.
References
- 1.Ferlay J, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Rudolph A, Chang-Claude J, Schmidt MK. Gene–environment interaction and risk of breast cancer. Br. J. Cancer. 2016;114:125–133. doi: 10.1038/bjc.2015.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Encinas G, et al. Somatic mutations in breast and serous ovarian cancer young patients: a systematic review and meta-analysis. Rev. Assoc. Med. Bras. 2015;61:474–483. doi: 10.1590/1806-9282.61.05.474. [DOI] [PubMed] [Google Scholar]
- 4.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pereira B, et al. Erratum: The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat. Commun. 2016;7:11908. doi: 10.1038/ncomms11908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Usary J, et al. Mutation of GATA3 in human breast tumors. Oncogene. 2004;23:7669–7678. doi: 10.1038/sj.onc.1207966. [DOI] [PubMed] [Google Scholar]
- 8.Takaku M, Grimm SA, Wade PA. GATA3 in breast cancer: tumor suppressor or oncogene? Gene Expr. 2015;16:163–168. doi: 10.3727/105221615x14399878166113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou J, Provot S, Werb Z. GATA3 in development and cancer differentiation: cells GATA have it! J. Cell. Physiol. 2010;222:42–49. doi: 10.1002/jcp.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miettinen M, et al. GATA3: a multispecific but potentially useful marker in surgical pathology: a systematic analysis of 2500 epithelial and nonepithelial tumors. Am. J. Surg. Pathol. 2014;38:13–22. doi: 10.1097/PAS.0b013e3182a0218f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voduc D, Cheang M, Nielsen T. GATA-3 expression in breast cancer has a strong association with estrogen receptor but lacks independent prognostic value. Cancer Epidemiol. Biomark. Prevent. 2008;17:365–373. doi: 10.1158/1055-9965.epi-06-1090. [DOI] [PubMed] [Google Scholar]
- 12.Cakir A, et al. GATA3 expression and its relationship with clinicopathological parameters in invasive breast carcinomas. Pathol. Res. Pract. 2017;213:227–234. doi: 10.1016/j.prp.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez RS, et al. GATA-3 expression in male and female breast cancers: comparison of clinicopathologic parameters and prognostic relevance. Hum. Pathol. 2013;44:1065–1070. doi: 10.1016/j.humpath.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Guo Y, et al. Prognostic and clinicopathological value of GATA binding protein 3 in breast cancer: a systematic review and meta-analysis. PLoS ONE. 2017;12:e0174843. doi: 10.1371/journal.pone.0174843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albergaria A, et al. Expression of FOXA1 and GATA-3 in breast cancer: the prognostic significance in hormone receptor-negative tumours. Breast Cancer Res. 2009;11:R40. doi: 10.1186/bcr2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoon NK, et al. Higher levels of GATA3 predict better survival in women with breast cancer. Hum. Pathol. 2010;41:1794–1801. doi: 10.1016/j.humpath.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gustin JP, et al. GATA3 frameshift mutation promotes tumor growth in human luminal breast cancer cells and induces transcriptional changes seen in primary GATA3 mutant breast cancers. Oncotarget. 2017;8:103415–103427. doi: 10.18632/oncotarget.21910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinstein JN, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLaren W, et al. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rangel MC, et al. Developmental signaling pathways regulating mammary stem cells and contributing to the etiology of triple-negative breast cancer. Breast Cancer Res. Treat. 2016;156:211–226. doi: 10.1007/s10549-016-3746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Di L-J. Wnt/β-catenin mediates AICAR effect to increase GATA3 expression and inhibit adipogenesis. J. Biol. Chem. 2015;290:19458–19468. doi: 10.1074/jbc.M115.641332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossetti C, et al. Adhesion molecules in breast carcinoma: a challenge to the pathologist. Rev. Assoc. Méd. Bras. 2015;61:81–85. doi: 10.1590/1806-9282.61.01.081. [DOI] [PubMed] [Google Scholar]
- 23.Koboldt DC, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffith OL, et al. The prognostic effects of somatic mutations in ER-positive breast cancer. Nat. Commun. 2018;9:3476. doi: 10.1038/s41467-018-05914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang M-X, Ren J-T, Tang L-Y, Ren Z-F. Molecular features in young vs elderly breast cancer patients and the impacts on survival disparities by age at diagnosis. Cancer Med. 2018;7:3269–3277. doi: 10.1002/cam4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Y-Z, Yu K-D, Zuo W-J, Peng W-T, Shao Z-M. GATA3 mutations define a unique subtype of luminal-like breast cancer with improved survival. Cancer. 2014;120:1329–1337. doi: 10.1002/cncr.28566. [DOI] [PubMed] [Google Scholar]
- 27.Azim HA, Jr, Nguyen B, Brohée S, Zoppoli G, Sotiriou C. Genomic aberrations in young and elderly breast cancer patients. BMC Med. 2015;13:266. doi: 10.1186/s12916-015-0504-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancello G, et al. Prognosis and adjuvant treatment effects in selected breast cancer subtypes of very young women (<35 years) with operable breast cancer. Ann. Oncol. 2010;21:1974–1981. doi: 10.1093/annonc/mdq072. [DOI] [PubMed] [Google Scholar]
- 29.Budczies J, et al. Classical pathology and mutational load of breast cancer—integration of two worlds. J. Pathol. Clin. Res. 2015;1:225–238. doi: 10.1002/cjp2.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tominaga N, et al. Clinicopathological analysis of GATA3-positive breast cancers with special reference to response to neoadjuvant chemotherapy. Ann. Oncol. 2012;23:3051–3057. doi: 10.1093/annonc/mds120. [DOI] [PubMed] [Google Scholar]
- 31.Hostetter C, Licata L, Keen J. A role for GATA-3 in control of estrogen receptor alpha expression. Can. Res. 2009;69:3050. doi: 10.1158/0008-5472.sabcs-3050. [DOI] [Google Scholar]
- 32.Izzo F, et al. Progesterone receptor activation downregulates GATA3 by transcriptional repression and increased protein turnover promoting breast tumor growth. Breast Cancer Res. 2014;16:491. doi: 10.1186/s13058-014-0491-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H, Wilkerson ML, Lin F, Shi J. Immunohistochemical evaluation of GATA3 expression in tumors and normal tissues: a useful immunomarker for breast and urothelial carcinomas. Am. J. Clin. Pathol. 2012;138:57–64. doi: 10.1309/ajcp5uafmsa9zqbz. [DOI] [PubMed] [Google Scholar]
- 34.McCleskey BC, et al. GATA3 expression in advanced breast cancer: prognostic value and organ-specific relapse. Am. J. Clin. Pathol. 2015;144:756–763. doi: 10.1309/ajcp5mmr1fjvvtpk. [DOI] [PubMed] [Google Scholar]
- 35.Hruschka N, et al. The GATA3 X308_Splice breast cancer mutation is a hormone context-dependent oncogenic driver. bioRxiv. 2019 doi: 10.1101/664367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takaku M, et al. GATA3 zinc finger 2 mutations reprogram the breast cancer transcriptional network. Nat. Commun. 2018;9:1059. doi: 10.1038/s41467-018-03478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emmanuel N, et al. Mutant GATA3 actively promotes the growth of normal and malignant mammary cells. Anticancer Res. 2018;38:4435–4441. doi: 10.21873/anticanres.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mair B, Konopka T. Gain- and loss-of-function mutations in the breast cancer gene GATA3 result in differential drug sensitivity. PLoS Genet. 2016;12:e1006279. doi: 10.1371/journal.pgen.1006279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uhlen M, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Pan H, et al. Low serum creatine kinase levels in breast cancer patients: a case-control study. PLoS ONE. 2013;8:e62112–e62112. doi: 10.1371/journal.pone.0062112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rouillard AD, et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database. 2016;7:8. doi: 10.1093/database/baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lv X, et al. Identification of potential key genes and pathways predicting pathogenesis and prognosis for triple-negative breast cancer. Cancer Cell Int. 2019;19:172. doi: 10.1186/s12935-019-0884-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu K, Ling MT, Wang X, Wong YC. Evidence of a novel biomarker, αs1-Casein, a milk protein, in benign prostate hyperplasia. Prostate Cancer Prostatic Dis. 2006;9:293–297. doi: 10.1038/sj.pcan.4500872. [DOI] [PubMed] [Google Scholar]
- 44.Bonuccelli G, et al. The milk protein α-casein functions as a tumor suppressor via activation of STAT1 signaling, effectively preventing breast cancer tumor growth and metastasis. Cell Cycle. 2012;11:3972–3982. doi: 10.4161/cc.22227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rakha EA, et al. Expression of mucins (MUC1, MUC2, MUC3, MUC4, MUC5AC and MUC6) and their prognostic significance in human breast cancer. Mod. Pathol. 2005;18:1295–1304. doi: 10.1038/modpathol.3800445. [DOI] [PubMed] [Google Scholar]
- 46.Bademler S, et al. Clinical significance of serum membrane-bound mucin-2 levels in breast cancer. Biomolecules. 2019;9:40. doi: 10.3390/biom9020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walsh MD, McGuckin MA, Devine PL, Hohn BG, Wright RG. Expression of MUC2 epithelial mucin in breast carcinoma. J. Clin. Pathol. 1993;46:922–925. doi: 10.1136/jcp.46.10.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Astashchanka A, Shroka TM, Jacobsen BM. Mucin 2 (MUC2) modulates the aggressiveness of breast cancer. Breast Cancer Res. Treat. 2019;173:289–299. doi: 10.1007/s10549-018-4989-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolf R, et al. Highly homologous hS100A15 and hS100A7 proteins are distinctly expressed in normal breast tissue and breast cancer. Cancer Lett. 2009;277:101–107. doi: 10.1016/j.canlet.2008.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cancemi P, et al. A multiomics analysis of S100 protein family in breast cancer. Oncotarget. 2018;9:29064–29081. doi: 10.18632/oncotarget.25561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alvarez-Díaz S, et al. Cystatin D is a candidate tumor suppressor gene induced by vitamin D in human colon cancer cells. J. Clin. Investig. 2009;119:2343–2358. doi: 10.1172/jci37205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.University of Medicine and Dentistry of New Jersey (UMDNJ). Vitamin D Found To Stimulate A Protein That Inhibits The Growth Of Breast Cancer Cells. https://www.sciencedaily.com/releases/2009/02/090204172437.htm. Accessed 21 Feb 2018.
- 53.Asaka M, et al. Alteration of aldolase isozymes in serum and tissues of patients with cancer and other diseases. J. Clin. Lab. Anal. 1994;8:144–148. doi: 10.1002/jcla.1860080306. [DOI] [PubMed] [Google Scholar]
- 54.He J, et al. Downregulation of ALDOB is associated with poor prognosis of patients with gastric cancer. Onco Targets Ther. 2016;9:6099–6109. doi: 10.2147/OTT.S110203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akkiprik M, et al. Identification of differentially expressed IGFBP5-related genes in breast cancer tumor tissues using cDNA microarray experiments. Genes (Basel) 2015;6:1201–1214. doi: 10.3390/genes6041201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saadatmand S, et al. Expression of cell adhesion molecules and prognosis in breast cancer. Br. J. Surg. 2013;100:252–260. doi: 10.1002/bjs.8980. [DOI] [PubMed] [Google Scholar]
- 57.Kim KS, Kim J, Oh N, Kim MY, Park KS. ELK3-GATA3 axis modulates MDA-MB-231 metastasis by regulating cell-cell adhesion-related genes. Biochem. Biophys. Res. Commun. 2018;498:509–515. doi: 10.1016/j.bbrc.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 58.Kassi E, Moutsatsou P. Estrogen receptor signaling and its relationship to cytokines in systemic lupus erythematosus. J. Biomed. Biotechnol. 2010;2010:317452. doi: 10.1155/2010/317452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan K, et al. Breast cancer in systemic lupus erythematosus (SLE): receptor status and treatment. Lupus. 2018;27:120–123. doi: 10.1177/0961203317713146. [DOI] [PubMed] [Google Scholar]
- 60.Kozlowski L, Zakrzewska I, Tokajuk P, Wojtukiewicz MZ. Concentration of interleukin-6 (IL-6), interleukin-8 (IL-8) and interleukin-10 (IL-10) in blood serum of breast cancer patients. Rocz. Akad. Med. Bialymst. 2003;1995(48):82–84. [PubMed] [Google Scholar]
- 61.Dethlefsen C, Hojfeldt G, Hojman P. The role of intratumoral and systemic IL-6 in breast cancer. Breast Cancer Res. Treat. 2013;138:657–664. doi: 10.1007/s10549-013-2488-z. [DOI] [PubMed] [Google Scholar]
- 62.Health, N. I. O. Racial and Ethnic Categories and Definitions for NIH Diversity Programs and for Other reporting Purposes. https://grants.nih.gov/grants/guide/notice-files/not-od-15-089.html. Accessed 21 Feb 2018 (2015).
- 63.Breast Cancer Rates by Race and Ethnicity. Center for Disease Control and Prevention. https://www.cdc.gov/cancer/breast/statistics/race.htm. Accessed 21 Feb 2018.
- 64.Houssami N, Macaskill P, Marinovich ML, Morrow M. The association of surgical margins and local recurrence in women with early-stage invasive breast cancer treated with breast-conserving therapy: a meta-analysis. Ann. Surg. Oncol. 2014;21:717–730. doi: 10.1245/s10434-014-3480-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lakhani S, Ellis I, Schnitt S, &, , et al. WHO Classification of Tumours of the Breast. 4. Lyon: IARC Press; 2012. [Google Scholar]
- 66.Reeves GK, Pirie K, Green J, Bull D, Beral V. Reproductive factors and specific histological types of breast cancer: prospective study and meta-analysis. Br. J. Cancer. 2009;100:538–544. doi: 10.1038/sj.bjc.6604853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tseng LA, et al. The association of menopausal status with physical function: the Study of Women’s Health Across the Nation (SWAN): menopausal status and physical function. Menopause (New York, N.Y.) 2012;19:1186–1192. doi: 10.1097/gme.0b013e3182565740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vinh-Hung V, et al. Lymph node ratio as an alternative to pN staging in node-positive breast cancer. J. Clin. Oncol. 2009;27:1062–1068. doi: 10.1200/jco.2008.18.6965. [DOI] [PubMed] [Google Scholar]
- 69.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 71.da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dennis G, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:R60. doi: 10.1186/gb-2003-4-9-r60. [DOI] [PubMed] [Google Scholar]
- 73.Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Data belongs to TCGA Research Network and is available in https://www.cancer.gov/tcga.