Abstract
We present here the second complete genome of anaerobic ammonium oxidation (anammox) bacterium, Candidatus (Ca.) Brocadia pituitae, along with those of a nitrite oxidizer and two incomplete denitrifiers from the anammox bacterial community (ABC) metagenome. Although reduction to NO is considered to be the first step in anammox, Ca. B. pituitae lacks nitrite reductase genes (nirK and nirS) responsible for this reaction. Comparative genomics of Ca. B. pituitae with Ca. Kuenenia stuttgartiensis and six other anammox bacteria with nearly complete genomes revealed that their core genome structure contains 1,152 syntenic orthologues. But nitrite reductase genes were absent from the core, whereas two other Brocadia species possess nirK and these genes were horizontally acquired from multiple lineages. In contrast, at least five paralogous hydroxylamine oxidoreductase genes containing candidate ones (hao2 and hao3) encoding another nitrite reductase were observed in the core. Indeed, these two genes were also significantly expressed in Ca. B. pituitae as in other anammox bacteria. Because many nirS and nirK genes have been detected in the ABC metagenome, Ca. B. pituitae presumably utilises not only NO supplied by the ABC members but also NO and/or NH2OH by self-production for anammox metabolism.
Keywords: anammox, metagenomics, physiological potential, core genome structure
1. Introduction
Anaerobic ammonium oxidation (anammox) bacteria couple nitrite reduction to ammonium oxidation, with nitric oxide (NO) and hydrazine as intermediates, ultimately producing dinitrogen gas and nitrate.1–3 NO, a key intermediate in anammox bacteria, is commonly produced by the reduction of nitrite to NO. This reaction is catalysed by two different types of enzymes—copper-containing (NirK) and cd1 (NirS) nitrite reductases.4 However, anammox bacteria do not necessarily possess the genes encoding these enzymes. Interestingly, the nirK gene has been detected in the Candidatus (Ca.) Brocadia sp. UTAMX25 and Ca. Brocadia caroliniensis6 genomes despite their low completeness; however, neither nirK nor nirS genes have been detected in other nearly complete Brocadia genomes. Ca. Brocadia sinica, which is one of the species with nearly complete genomes, lacks genes encoding NO-forming nitrite reductase (i.e. nirK and nirS).7 But, 15N-tracer experiments have demonstrated that Ca. B. sinica could reduce to NH2OH, instead of NO, with as yet unidentified nitrite reductase. On the other hand, anammox genomes contain 10 or 11 different types of paralogous genes encoding octaheme hydroxylamine oxidoreductase (Hao).8,9 Hao-like proteins are thought to be the most likely candidate enzymes catalysing nitrite reduction to NO or NH2OH in anammox bacteria. Indeed, among the Hao-like protein genes of Ca. Kuenenia stuttgartiensis,10KSMBR1_2163 (formerly kustc0458) and KSMBR1_3792 (formerly kuste4574) have been postulated to function in nitrite reduction based on their gene expression profile and protein sequence analyses, making them candidates for the elusive nitrite reductases producing NO.8,11–13 Nevertheless, the physiological functions of Hao-like proteins remain enigmatic. Considering the published literature, a number of questions arise. First, what is the significance of the presence or absence of the nirS or nirK genes in anammox bacteria, and do any other members of an anammox bacterial community (ABC) have such genes? Second, do all anammox bacteria have the potential to reduce nitrite with Hao-like proteins? To answer these questions, we attempted to reconstruct the genomes of anammox and other predominant bacteria, from the ABC metagenome in the anammox bioreactor fed with nitrite and ammonium.14 We then performed comparative genomics on the anammox bacteria to determine their core genome structure, which was vertically inherited from a common ancestor, and examined the expression profiles of the key genes related to nitrogen metabolism. We also characterized the physiological and metabolic potential of not only anammox bacteria but also the predominant ABC members, especially for nitrite oxidizers, to determine their lifestyle in the anammox bioreactor.
2. Materials and methods
2.1. Anammox bacterial consortium
Anammox population was enriched in a small up-flow reactor, in which nonwoven fabric supports were placed in15,16 and a synthetic medium was continuously supplied. A plastic cylinder (38-mm inner diameter, 400-mm length), of which bottom end was plugged with a rubber stopper, was set up in vertical position. Two sheets of porous polyester nonwoven fabric support (3-mm thick, 30 × 200 mm) were submerged to activated sludge, obtained from a municipal sewage treatment plant in central Japan, and placed in a plastic cylinder as a fixed-bed for the microbial population. Void volume of the reactor was 410 ml excluding the void volume of fabric supports, because supplied medium may not freely exchange inside and outside of support. The synthetic medium consisted of (per litre of tap water): NH4Cl, 103–480 mg; NaNO2, 104–570 mg; NaHCO3, 500 mg; KH2PO4, 27 mg; MgSO4·7H2O, 300 mg; and CaCl2·7H2O, 18 mg. Dissolved oxygen in the medium was not eliminated and pH was controlled at 7.2–7.5. Concentrations of NH4Cl and NaNO2 were increased stepwise after anammox activity was detected. Samples were obtained from the medium reservoir and the effluent two to three times a week, and , , and were determined using ion chromatograph (IC-2010, Tosoh Corp. Ltd., Tokyo, Japan).14 In the anammox enrichment culture, neither nitrite nor ammonium was consumed for 106 days after starting to supply the medium to the reactor, but consumption of nitrite and ammonium as well as production of nitrate was found on 107 days of operation, indicating that anammox organisms were growing in the reactor. Temperature of the reactor was maintained at 32.1 ± 1.0°C during 254 days of operation. Because the temperature unexpectedly raised and sometimes reached to around 35°C, the temperature was lowered to 30°C at the day 327, which was stably controlled until the 421 days, when the biomass was sampled. In addition to the measurement of anammox activity, we confirmed the 16S rDNA sequence of anammox bacteria amplified by PCR.
2.2. DNA and RNA isolation
The biomass of the anammox bacterial consortium was filtered through a 53-μm mesh to make it uniform (final wet weight, 3.6 g). DNA was extracted using the ISOIL for Bead-Beating kit (Nippon Gene) according to the manufacturer’s protocol, except for an additional treatment with a lytic enzyme.17 Briefly, a biomass sample was suspended in 30 ml of TE buffer and vortexed vigorously. The biomass suspension was evenly divided among 20 tubes containing zirconia/silica beads (0.1 mm, 0.47 g; 0.5 mm, 0.23 g). After the addition of lysozyme solution (0.71 mg/ml as a final concentration) to the tube, the suspension was incubated for 15 min at 37 °C with gentle shaking, and then incubated for 15 min at 37 °C after the addition of 20 µl of proteinase K solution (12 U). The lytic enzyme–treated biomass was bead-beaten by vortexing at power range 5 for 7 min. The solution was then centrifuged at 12,000 × g for 5 min at room temperature and the DNA in the supernatant was purified.
To concentrate DNA from Ca. Brocadia species used for genome reconstruction, we tested the beating conditions with 0.5-mm zirconia/silica beads alone by changing the beating power and time. The DNA concentration was evaluated based on the amount of PCR product using a primer set for the 16S rRNA gene (Bro5F, GGGTATGATCTTGGCTCAGAACGA; Bro232R, CGCACGTTTGACTATCATCACC) of Ca. B. pituitae. For RNA isolation, the biomass of the anammox bacterial consortium (2.5 g wet weight) was filtered as described above for DNA isolation and transferred to 50-ml tubes containing 5 ml TE buffer (pH 8.0). Ten millilitres of Bacterial RNA protect reagent (Qiagen) was added to the biomass suspension and vortexed vigorously. After the biomass suspension was allowed to stand for 5 min at room temperature, it was centrifuged at 5,000 × g for 10 min at 4°C to collect the biomass, and the pellet was washed with TE buffer. Lysozyme (1.36 mg/ml as a final concentration) was added to biomass suspended in 5 ml TE buffer. After a 10-min incubation at 37°C, 12 U of Proteinase K was added for another 10-min incubation. To increase the disruption efficiency, the lytic enzyme-treated biomass was transferred to a new tube containing 2.7 g zirconia silica beads (0.1 mm, 1.8 g; 0.5 mm, 0.9 g) and then vortexed at maximum power for 10 min. RNA was extracted and purified using the RNeasy Protect Bacteria Mini kit and QIAzol Lysis Reagent (Qiagen). DNase I-treated RNA was re-purified using the RNeasy Mini Clean-up Kit (Qiagen), and rRNA was removed using the Ribo-Zero rRNA Removal Kit (Gram-negative bacteria; Epicentre).
2.3. Sequencing and raw data treatment
Genomic DNA was sheared using a Focused ultrasonicator (Covaris). A paired-end library with an insert size of 450 bp was prepared using a TruSeq DNA PCR-free LT Sample Prep Kit (Illumina) and sequenced on an Illumina MiSeq platform, generating 6 million reads with read lengths of 300 bp. The PEAR software (v0.9.6) was used to merge these paired-end reads after filtering the low-quality scores of the sequence using the FASTX toolkit and eliminating duplicates using PRINSEQ. Gene prediction from the merged sequences was performed using MetaGeneAnnotator.18 Until the final creation of a multi-FASTA file of amino acid sequences including these multiple processes, it was pipelined as MAPLE Submission Data Maker (MSDM).19 MSDM is available from https://maple.jamstec.go.jp/maple/maple-2.3.1/softdownload/MSDM.html. The multi-FASTA file consisting of 3 million amino acid sequences was subjected to metagenomic analysis using GenomapleTM (formerly MAPLE) ver. 2.3.2. RNA-seq libraries were prepared from the anammox bacterial consortium according to the standard Illumina protocol, and cDNA libraries were checked for quality and quantity using the DNA-100 kit (Agilent Technologies) and a 2100 Bioanalyzer. Each library was sequenced with the Illumina Sequencing Kit v2 on one lane of a MiSeq desktop sequencer (Illumina) to obtain 150-bp average paired-end reads. Reads Per Kilobase per Million mapped reads (RPKM) values were calculated according to the standard method. The RPKM ratio, calculated by dividing the RPKM of each gene by the mean RPKM of all ribosomal proteins, was used to determine relative gene expression levels. Amino acid sequences used for GenomapleTM analysis are available at https://zenodo.org/record/3491404#.XigFVhP7RTY. BioSample accession numbers: SAMD00057694 for metagenomic sequence, AP021856 for Ca. B. pituitae, AP021857 for Ca. Desulfobacillus denitrificans, AP021858 for Ca. Nitrosymbiomonas proteolyticus, BLAA01000001- BLAA01000004 for 4 contigs of Ca. D. symbiosum, and DRA009157 for RNA sequence RNA-seq, of the ABC.
2.4. Community structure analysis
We calculated the proportional representation of bacteria in the metagenome based on the mapping pattern of module M00178 for bacterial ribosomes.20,21 We calculated the proportion of bacteria at the individual taxonomic rank as defined by the Kyoto Encyclopaedia of Genes and Genomes (KEGG; i.e. phylum, order, and class level). For a more detailed analysis of bacterial communities, we searched all sequences assigned to ribosomal proteins by GenomapleTM using the NCBI non-redundant protein database.
2.5. Reconstruction of genomes and data analyses
In addition to the Illumina platform, a PacBio RS II DNA sequencer was also used for metagenomic sequencing. Paired-end reads (400-bp insertion size on average) and mate-pair reads (3- and 5-kb insertion sizes on average) generated on a HiSeq 2500 (Illumina) were assembled using Platanus ver. 1.2.122 to reconstruct the genome of predominant species in Ca. genus Brocadia. Reads from PacBio RS II were assembled using HGAP3. In the finishing step, a fosmid library was constructed according to the previously published method23 and sequenced by Sanger and Illumina Technologies to fill gaps and increase sequence quality. In addition to Ca. Brocadia species, the genomes of three other predominant species in the phylum Armatimonadetes, class Betaproteobacteria (β-proteobacteria), and family Anaerolineae were reconstructed in the same manner. Gene identification and annotation were initially performed using DFAST.24 We then manually curated the output results by carefully comparing the BLAST search results with the NCBI protein database as the genome database of DFAST is too small for annotating the genomes of uncultured microbes. A circular map of each reconstructed genome was constructed using the CGView Server.25
2.6. Phylogenetic and genomic similarity analyses
To determine the phylogenetic position of Ca. Brocadia species whose genome was reconstructed in this study among anammox bacteria, we retrieved ribosomal proteins from each complete or draft genome registered in the NCBI database. Subsequently, we selected 20 commonly conserved ribosomal proteins (rplA, rplA, rplN, rplQ, rplV, rplW, rplX, rplC, rplE, rplF, rplI, rpsK, rpsM, rpsP, rpsS, rpsB, rpsD, rpsE, rpsH, and rpsI) among the anammox bacteria. These concatenated protein sequences were aligned with those of Rhodopirellula baltica, used as an outgroup. We used the LG+G model in MEGA 6.06 to construct a phylogenetic tree using the maximum likelihood method.26 We tested the average nucleotide identity among the genomes used for phylogenetic analysis to identify species in the genus Ca. Brocadia using JSpeciesWS.27 We also determined the phylogenetic positions of the nitrite reductases (NirK and NirS) horizontally acquired in the anammox bacterial genomes using the LG+G + I model in MEGA 6.06.
2.7. Orthologous analysis and estimation of the core genome structure
Orthologous groups (OGs) for Ca. Brocadia species, Ca. K. stuttgartiensis, and six other anammox bacteria were constructed using the rapid classification programme DomClust28 on the Microbial Genome Database (MBGD) server.29 A core genome is defined as a set of genes (OGs) syntenically conserved in at least half of the compared strains. In our study, a set of genes in the syntenic regions shared by at least five species was defined as the core genome for the eight analysed anammox species. A set of syntenic regions and the consensus order of the OGs in these regions, designated as the core genome structure, were created using the CoreAligner programme30 based on conserved linkages between orthologous genes in each chromosome. Comparative analysis of gene organization around the nirK gene was performed using GenomeMatcher.31
2.8. Evaluation of the metabolic and physiological potential
The patterns of metabolic and physiological potential of Ca. Brocadia species and three other predominant species were investigated using GenomapleTM (formerly MAPLE) ver. 3.2.21,32 GenomapleTM is available through a web interface (https://maple.jamstec.go.jp/maple/maple-2.3.1/) and as a stand-alone package from Docker Hub (https://hub.docker.com/r/genomaple/genomaple). Genes were mapped to 795 functional modules defined by the KEGG (pathways, 305; complexes, 294; functional sets, 157; and signatures, 40), and the module completion ratio (MCR) was calculated according to a Boolean algebra–like equation described previously.20 To evaluate MCR, Q-value suggesting working probability of the modules was also calculated by GenomapleTM. The Q-value near zero indicates high working probability of the module.21 The MCR and Q-value patterns of Ca. Brocadia species were compared with those of three other anammox species with genomes reconstructed to fewer than five contigs.
2.9. Determination of anammox activity using a 15N-tracer
The potential anammox and denitrification activities of biomass samples were determined using a 15N-tracer technique based on a method described previously.33 To determine the activities of the samples, reactive substrates for anammox were added to the vials under the following three combinations: (i) 1 mM unlabelled NH4Cl and 1 mM Na15NO2 or 1 mM 15NH2OH (SI Science, Tokyo, Japan; 99.9 atom% 15N), (ii) 1 mM 15NH4Cl (SI Science; 99.9 atom% 15N) and 1 mM unlabelled NaNO2 or NH2OH, and (iii) 0.4 mM 15NH4Cl without nitrite. During anaerobic incubation with substrate combinations (i) and (ii), - or NH2OH-dependent anammox could be detected by the production of 14N15N (29N2), because anammox uses 1 mol each of and or NH2OH. Substrate combination (iii) was a negative control to determine whether anammox occurs without nitrite or NH2OH. The biomass granule of the anammox bacterial consortium, which was sieved through a 53-μm mesh, was used for measurement of anammox activity. When NO was used as a reactive substrate instead of , we added 2500 ppm of 15NO gas (SI Science; 99.6 atom% 15N, +98%) to the headspace of the vial containing the ABC biomass suspended in 5-ml substrate solution. The concentration of dissolved NO was ∼7 µM. The following standard gas was used for GC/MS analysis. A small amount of 15N-labelled N2 gas (53.4 15N atom%; SI science, Co. Ltd., Tokyo, Japan) was added in a vial filled with ultrapure He (>99.99995%) as background to prepare standard to demine 29N2, and 30N2. Concentration of 29N2 and 30N2 in the preparation was, 386 and 221 ppmv, respectively. Other GC/MS analytical conditions and subsequent calculations were performed as described previously,14,33 except that the ABC biomass was not sieved. After the GC/MS measurement, the biomass was recovered by filtration and dried at 100 °C for 12 h to calculate the dry weight. To disperse the ABC granules, the sample was agitated using an SW-M6000 stirrer at 130 rpm before measurement of anammox activity.
3. Results and discussion
3.1. Metagenomic analysis of anammox bioreactor
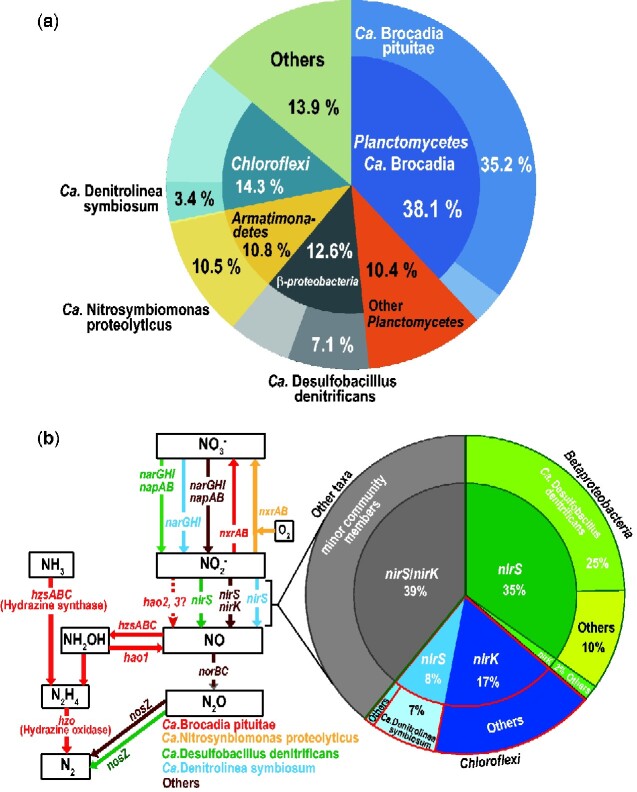
The ABC comprised members of four major taxa, Planctomycetes (48.5%), Chloroflexi (14.3%), Armatimonadetes (10.8%), and β-proteobacteria (12.6%); Ca. Brocadia was the most predominant member in Planctomycetes, with a relative abundance of 38.1% (Fig. 1a). Accordingly, we performed genome reconstruction of Ca. Brocadia species and representative species of three other major taxa using sequenced fosmid clones and shotgun reads. We successfully reconstructed the genomes and determined the whole-genome sequences of three currently non-isolatable species in genus Ca. Brocadia, phylum Armatimonadetes, and class β-proteobacteria. Similarly, we were able to assemble four contigs of Anaerolineaceae species in phylum Chloroflexi. We designated this anammox bacteria as Ca. Brocadia pituitae (‘pituitae’ is ‘sludge’ in Latin). Ca. B. pituitae was shown to be a new species by phylogenetic analysis based on the sequences of concatenated ribosomal proteins and genomic identity analysis among 12 Brocadia strains including Ca. B. pituitae (Supplementary Fig. S1 and Supplementary Table S1). We also designated 3 other predominant species as Ca. N. proteolyticus (Armatimonadetes), Ca. D. denitrificans (β-proteobacteria), and Ca. D. symbiosum (Anaerolineaceae) based on their genomic features and metabolic and physiological potential deduced by the Genomaple. The description of Ca. N. symbiomonas is as follows: Nitrosymbiomonas (ni.tro.sym.bi.o’mo.nas. L. n. nitrum nitrate; Gr. n. symbios a companion; Gr. n. monas a unit, monad; M.L. fem. n. Nitrosymbiomonas, nitrate producing symbiotic monad.), proteolyticus (pro.te.o.ly’ti.cus. Ger. protein from Gr. protos first; adj. lyticus dissolving; M.L. adj. proteolyticus protein-dissolving). We also designated β-proteobacteria and Anaerolineae phylotypes as Ca. D. denitrificans and Ca. D. symbiosum. The description of Ca. D. denitrificans and Ca. D. symbiosum are as follows: Desulfobacillus (de.sul.fo.ba.cil’lus. L. pref. de from; L. n. sulphur; L. dim. n. bacillus, a small rod; M.L. masc. Desulfobacillus a bacillus that reduces sulphur compound), denitrificans (de.ni.tri’fi.cans. L. prep. de away from; L. n. nitrum soda; M.L., nitrum nitrate; M.L. v. denitrifico. denitrify; and M.L. part. adj. denitrificans denitrifying). Denitrolinea (de.ni.tro.li.ne’a. L. pref. de from; L. n. nitrum nitrate; L. fem. n. linea line; N.L. fem. n. Denitrolinea line-shaped nitrate reducing), symbiosum (sym.bi.o’sum. Gr. n. symbios a companion; N.L. neut. adj. symbiosum living together with, symbiotic).
Figure 1.
Metagenomic analysis of anammox reactor. (a) Community structure analysis based on ribosomal proteins identified in the metagenome. The genomes of unisolable Ca. B. pituitae, Ca. N. proteolyticus, and Ca. D. denitrificans were reconstructed in this study, although the genome of Ca. D. symbiosum is still divided into four contigs. Taxa representing <2% of the total were classified as ‘Other’. (b) Schematic representation of transformation steps of nitrogen compounds and the genes associated with each reaction step in the ABC. Dashed line indicates the predicted reaction.
3.2. Genomic features of predominant ABC members
The genome of Ca. B. pituitae consists of a single circular chromosome (4,075,302 bp) with mean G + C content of 43.4% (Table 1 and Supplementary Fig. S2). We identified 3,593 protein coding sequences (CDSs), 47 tRNA genes and 3 rRNA genes. Out of the 3,593 CDSs in the Ca. B. pituitae genome, 1,138 were included in the core genome structure, comprising 1,152 OGs (Supplementary Fig. S3 and Table S2). The genomes of two other predominant species, Ca. N. proteolyticus and Ca. D. denitrificans, are also single circular chromosomes consisting of 2,809,316 and 3,145,360 bp with a higher G + C content of 61.1% and 66.7%, respectively. Although the genome of Ca. D. symbiosum is still divided into 4 contigs, this genome is thought to be nearly completed because 50 ribosomal protein genes were identified in the 4 contigs. In general, a bacterial ribosome is composed of 52 ribosomal proteins with 1 or 2 of them missing depending on the bacterial strain. Total length of the contigs is 3,705,798 bp and their mean G + C content is also as high (59.5%) as the genomes from two other predominant species (Table 1).
Table 1.
General features of the reconstructed genomes of four major members in the anammox reactor
| General features | Ca. B. pituitae | Ca. N. proteolyticus | Ca. D. denitrificans | Ca. D. symbiosum |
|---|---|---|---|---|
| Size (bp) | 4,075,302 | 2,809,316 | 3,145,360 | 3,705,798 |
| contig | 1 | 1 | 1 | 4 |
| G + C (%) | 43.4 | 61.1 | 66.7 | 59.5 |
| protein coding genes | 3,593 | 2,561 | 3,104 | 3,442 |
| function assigned | 2,015 | 1,626 | 2,378 | 1,811 |
| conserved hypothetical | 1,418 | 869 | 684 | 1,520 |
| hypothetical | 160 | 66 | 42 | 111 |
| rRNA | 3 | 3 | 4 | 3 |
| tRNA | 47 | 47 | 51 | 49 |
| ribosomal protein (KO-assigned) | 51 | 54 | 53 | 50 |
Genomic analysis of the reconstructed genomes revealed that Ca. B. pituitae possesses no nitrite reductase genes, such as nirS and nirK, and these genes were not included in the core structure of the anammox bacterial genome. In the anammox bioreactor, however, those genes were complemented by those from other ABC members (i.e. NO producers from nitrite) in the bioreactor. Of all nitrite reductases detected in the ABC bioreactor, the nirS genes derived from Ca. D. denitrificans and Ca. D. symbiosum occupied 25% and 7%, respectively (Fig. 1b). These two ABC members also possessed nitrate reductase genes (narGHI and/or napAB) but not NO reductase genes (norBC), indicating that they are imperfect denitrifiers, although norBC genes are present in other ABC members. Indeed, when a small portion of biomass granules was anaerobically incubated with ammonium and nitrite as reactive substrates, 84% of the total nitrogen emission was due to the anammox reaction, and the remaining 16% was due to denitrification. Thus, it is likely that ABC members capable of producing NO were supplying it to Ca. B. pituitae, which is able to use nitrite. Indeed, in the case of NO, 69% of the total nitrogen emission was derived from the anammox reaction (Supplementary Table S3). Expression of the nirS genes was confirmed by metatranscriptomic analysis, which showed that the gene expression level from Ca. D. denitrificans and Ca. D. symbiosum was 5- to 10-fold lower than the average level for ribosomal proteins (Supplementary Table S4). Thus, although predominant ABC members including imperfect denitrifiers such as Ca. D. denitrificans and Ca. D. symbiosum have the ability to produce and supply NO to Ca. B. pituitae, Ca. B. pituitae does not necessarily depend solely on other ABC members for NO production. If an anaerobic incubation experiment, in which is not added but only 15NO2− is added to anammox population, is performed, it is expected that 15 reduction (to NO) rate would be similar to anammox rate, when anammox bacteria depend on NO supplied by cross feeding. In this experimental system, because anammox reaction consuming NO is suppressed, NO would be accumulated in the incubation vial and it has been reported that NO could enhance reduction activity.34,35 Thus, the fair comparison of reduction rate between only 15 and 15 + is thought to be difficult.
On the other hand, the genome of Ca. K. stuttgartiensis possesses the nirS gene, but this gene is hardly expressed at the transcriptional and protein levels compared with those encoding other key catabolic genes.12,13 In contrast, nirS is one of the highest expressed genes in the marine anammox species, Ca. Scalindua brodae.36 On the other hand, Ca. Jettenia caeni KSU1, as well as Ca. B. fulgida and Ca. B. caroliniensis, possess nirK instead of nirS, although other Brocadia species possess no such nitrite reductase (Supplementary Fig. S1). Ca. B. pituitae does not possess these genes although this species utilises nitrite, NO, and NH2OH as the substrate for anammox metabolism.
Ca. N. proteolyticus was shown to be a nitrite oxidizer through the detection of nxrAB genes encoding nitrite oxidoreductase. Ca. N. proteolyticus possesses 78 genes encoding peptidases and 27 of them were predicted to be extracellular enzymes by the SOSUI program (http://harrier.nagahama-i-bio.ac.jp/sosui/). Although the expression level varied depending on the individual gene, almost all peptidase genes were expressed (Supplementary Table S5). Such a large number of peptidase genes were not observed in other reconstructed genomes. In addition, this species possesses 32 genes encoding cell wall-associated enzymes such as glycosyl transferase, lytic transglycosidase, and cell wall-associated hydrolase assisting in adherence, autoaggregation, turnover of cell wall and autolysis.37–39 Out of those genes, 22 genes containing 6 secretory soluble enzyme genes were expressed with various expression levels (Supplementary Table S6). Since Ca. N. proteolyticus also possesses multiple expressed genes related to Type II secretory pathway (Supplementary Table S7), many of the peptidase and cell wall-associated enzymes are presumably secreted via Type II secretory pathway.40 It is not yet clear how ABC members are selected and maintained, but it seems that the members of anammox bioreactors using non-woven fabric as a carrier share some common features. In fact, because 16S rRNA genes with more than 99% identity to that of Ca. N. proteolyticus have been detected in many other anammox bioreactors,41,42 this microbe was inevitably suggested to be an ABC member responsible for nitrite oxidation via consumption of O2 in the anammox bioreactor. Ca. N. proteolyticus is one of nitrite oxidizing bacteria (NOBs), but it has no potential for carbon fixation unlike the other NOBs (Supplementary Table S10) although the bioreactor is maintained through feeding with a synthetic inorganic medium containing sodium bicarbonate as the sole carbon source. Given such operating conditions, the immediate question of how Ca. N. proteolyticus cells acquire nutrients and are keeping their population in the bioreactor arises.
Since Ca. N. proteolyticus possesses multiple secretory peptidases and lytic transglycosidases (which are expected to be attractive new targets for the development of broad-spectrum antibiotics38) and also Type II secretion systems,40 it is possible that proteolysis of biomass from autolysed old cells and also the lysis of active ABC members sensitive to these enzymes may supply nutrients to Ca. N. proteolyticus itself and to other heterotrophic predominant species such as Ca. D. denitrificans and Ca. D. symbiosum, for maintaining their populations. In fact, ABC members in the anammox bioreactor repeat cell metabolism, but cannot maintain an active cell population in the reactor unless biomass from old cells is properly reused. In addition, keeping the population balance of ABC members is considered to be an important factor in maintaining stable anammox activity. In that sense, Ca. N. proteolyticus possessing high proteolysis potential must be one of the important key species necessary for keeping balanced population of ABC members and the stable anammox activity in the bioreactor.
On the other hand, imperfect denitrifiers, Ca. D. denitrificans and Ca. D. symbiosum, commonly appeared even in the ABC of anaerobically controlled anammox bioreactor. In fact, the genes with 98–99% identities to Ca. D. denitrificans (bin ID: PRO2, β-proteobacteria) and Ca. D. symbiosum (bin ID: CFX3, Chloroflexi) were detected in the metagenomic sequences from this bioreactor although their abundance is very low (1%>).5 Therefore, the involvement of these common ABC members in anammox metabolism may be related to the difficulty of isolating anammox bacteria.
3.3. Placement of nitrite reductase gene on the anammox bacterial genome and its phylogeny
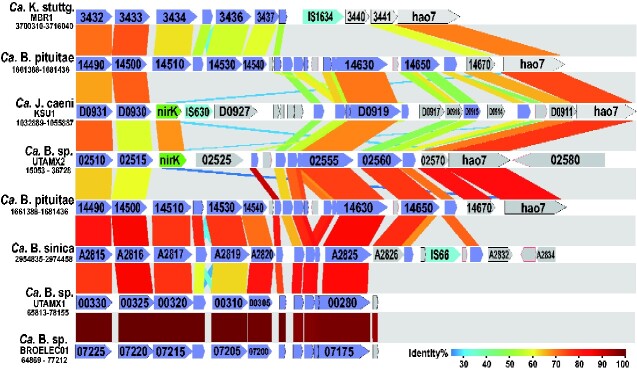
As mentioned earlier, anammox bacteria do not necessarily possess NirK and NirS-type nitrite reductase genes but where and how their genes are located on the anammox genome is still a question. In fact, Ca. Brocadia sp. UTAMX2, (identified as Ca. B. fulgida based on the results of phylogenetic and genomic identity analyses; Supplementary Fig. S2 and Table S1), and J. caeni KSU1, possess the nirK gene unlike Ca. B. pituitae. In contrast, Ca. K. stuttgartiensis possesses the nirS gene, and Ca. Scalindua rubra BSI-1 possesses both genes. A comparison of the genome structure near nirK with those of other anammox bacteria that lack the gene revealed that the nirK in the KSU1 genome is positioned between the core gene set together with an insertion sequence (IS) belonging to the IS630 family43 (Fig. 2). In addition, the nirK gene was incidentally inserted at the same position of the UTAMX2 genome, even though there was no IS element nearby. This mobile gene-like behaviour makes it clear that the nirK gene was acquired by horizontal gene transfer. Different types of ISs have also been found in this region of Ca. K. stuttgartiensis and of Ca. B. sinica,44 which lacks the nirK gene. Indeed, Ca. K. stuttgartiensis and Ca. B. pituitae genomes contain 74 and 68 transposase genes for ISs, respectively, belonging to IS families IS630, IS4, and ISL3, which are widely disseminated throughout various bacterial genomes.43,45 These foreign nirK genes exhibit 67% identity with each other, but their ancestral host organism is unknown. The nirK gene has also been detected in Ca. Scalindua draft genomes with low completeness, but whether it was acquired by the same route as the other two genes remains unknown.
Figure 2.
Comparison of gene organization around the nirK gene among anammox bacteria. Gene organization was compared among six anammox bacteria with and without the nirK gene. Genomic fragments containing nirK or the corresponding region were extracted from four nearly completed draft genomes and two completed genomes, and then aligned. Purple arrows show the core gene set among anammox bacteria, as shown in Supplementary Fig. S3. K. stuttg: Kuenenia stuttgartiensis, B: Brocadia, J: Jettenia.
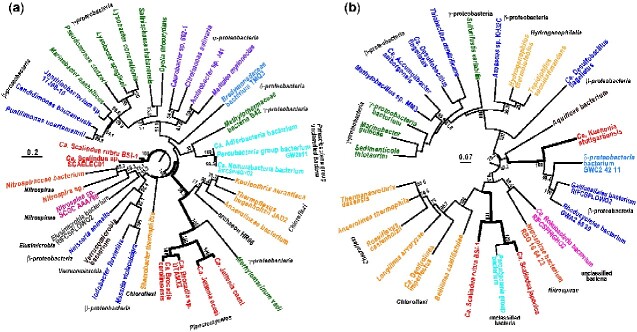
To identify the ancestral host organism, we constructed an unrooted phylogenetic tree based on the amino acid sequences of NirK protein. Ca. Brocadia5,6 and Ca. Jettenia46,47 species formed a cluster together with Methylomarinum vadi in γ-proteobacteria, and two Ca. Scalindua species independently formed their own cluster, although all anammox bacteria were classified into the family Brocadiaceae (Fig. 3a). Proteobacteria formed a large cluster comprising four subdivisions (α, β, γ, and δ), but some of the species in β- and γ-proteobacteria formed another small cluster with other taxa. Given the complicated phylogenetic relationships among NirK protein clusters, except for the major Proteobacteria cluster, it appears that anammox bacteria did not necessarily acquire the nirK gene from monophyly. In addition, the nirS genes of Ca. Kuenenia and Ca. Scalindua species did not form a cluster, but Ca. Kuenenia formed a cluster with subsets of β-proteobacteria and δ-proteobacteria, whereas Ca. Scalindua formed a cluster with the Parcubacteria within the unclassified bacteria group (Fig. 3b). Because the nirS gene, like the nirK gene, was also not horizontally acquired from monophyly (Fig. 3b), it was not possible to determine the original taxonomic affiliation of the nitrite reductase gene acquired in the anammox bacteria genome.
Figure 3.
Phylogenetic position of NirK and NirS proteins identified in anammox bacteria. (a) A maximum likelihood tree based on NirK proteins. (b) A maximum likelihood tree based on NirS proteins. The numbers indicate the percentages of bootstrap support. Bootstrap values <50% were omitted from this figure.
3.4. Metabolic and physiological potential harboured in the reconstructed genomes
To examine metabolic and physiological potential harboured in the anammox bacterial genomes, we applied Ca. J. caeni and Ca. B. sinica genomes reconstructed to less than five contigs together with the two complete genomes (Ca. B. pituitae and Ca. K. stuttgartiensis) to GenomapleTM and compared the patterns of MCR and Q-value among the four anammox bacteria (Supplementary Table S9). We found that their pattern is quite similar among the four species through all KEGG modules. In the pathway modules, when completed modules or modules with a Q-value of < 0.5 (i.e. suggesting high working probability of the module) were picked up in each species, only Brocadia species had the module for undecaprenyl phosphate alpha-L-Ara4N biosynthesis (M00761) involved in amino and nucleotide sugar metabolism despite the fact that this is γ-proteobacteria-specific rare module (Supplementary Fig. S4). In contrast, Euryarchaeota/Planctomyces-specific rare module for haem biosynthesis (M00847) was incomplete only in Ca. B. sinica. Since there was no noticeable difference regarding the other pathway modules, we inferred that there is no difference in the basic anabolic potential of the four major anammox bacteria.
In general, NOB have an ability to fix carbon through Calvin cycle,48 but Ca. N. proteolyticus has only enzymes involved in 5 reaction steps of the module for Calvin cycle (M00165), which comprises 11 reaction steps (i.e. MCR: 54.5%). On the other hand, a complete ammonia oxidizer (Comammox) can use reductive citrate cycle for carbon fixation,49 but Ca. N. proteolyticus has no ATP citrate lyase and citryl-CoA synthetase, which are key enzymes for this cycle. So far, seven carbon fixation pathways including these two are known, but Ca. N. proteolyticus does not have any of them and thus it seems to lack carbon fixation ability unlike the other NOBs (Supplementary Table S10). On the other hand, one of the incomplete denitrifiers, Ca. D. denitrificans, was found to complete the module for dissimilatory sulphur reduction (M00596) unlike Ca. D. symbiosum.
3.5. Orthologous analysis of Hao-like protein genes in anammox bacteria
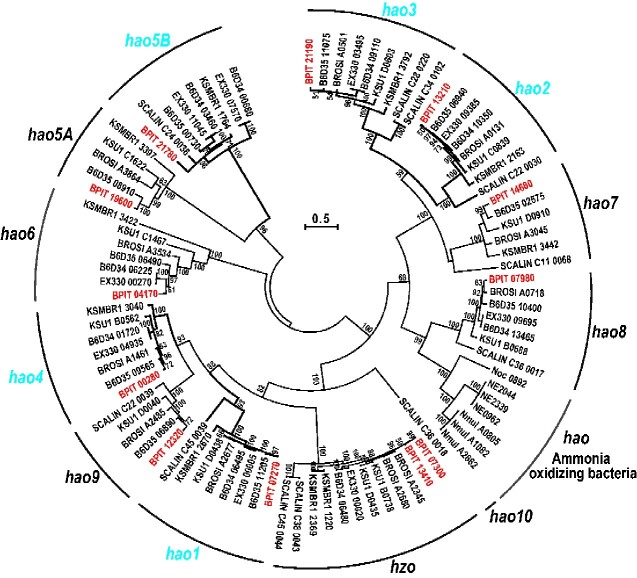
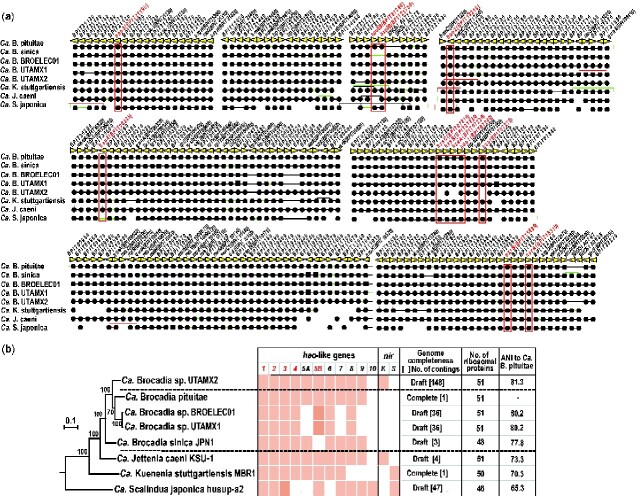
Hao catalyses oxidation of NH2OH to NO2− or NO in the nitrification process of ammonia oxidizing bacteria (AOB).9 On the other hand, Hao-like proteins are predicted to be the most likely candidate enzymes catalysing reduction to NO in Ca. K. stuttgartiensis10,12 and Ca. Scalindua profunda40 and to NH2OH in Ca. B. sinica.7Ca. B. pituitae possesses nine paralogous genes encoding Hao-like proteins, similar to other anammox bacteria. Thus, we first classified hao- and hao-like genes identified in typical AOB and all available anammox bacterial genomes through clustering analysis. We designated them as hao1–hao10 and one of the large independent clusters, hao5, was divided into two groups, hao5A and hao5B and the hao8 cluster constructed a large one with the AOB hao cluster (Fig. 4). As a result, Ca. B. pituitae was found to possess all genes categorized into hao1–hao 9 except for hao10, which is specific to Ca. Scalindua. Among those genes, at least 5 (hao1–hao4 and hao5B) were found in the core genome whereas Ca. B. sinica is missing hao5B gene (Fig. 5a and b). The genes encoding hydrazine synthase subunit A, B, and C (hzsABC) and hydrazine oxidoreductase (hzo), which are responsible for the main anammox reaction (Fig. 1), and nitrite oxidase (nxrAB) are also included in the core genome (Fig. 5a and Supplementary Table S10). Ca. B. fulgida UTMX2 and Ca. J. caeni KSU1 possessed the same hao-like gene repertoire as Ca. B. pituitae, but Ca. K. stuttgartiensis were missing hao8 and hao9 genes.
Figure 4.
Phylogenetic tree of Hao-like proteins. KSU1 C0486, which was identified as Hao5B by MBGD 27, is not shown in this figure because of its short partial sequence. Hao proteins from AOB were also included in this figure as the reference proteins. BPIT: Ca. B. pituitae, BROSI: Ca. B. sinica JPN1, EX330: Ca. Brocadia sp. BROELEC01, B6D34; Ca. Brocadia sp. UTAMX1, B6D35: Ca. Brocadia sp. UTAMX2, KSU1: Ca. J. caeni KSU1, KSMBR1: Ca. K. stuttgartiensis MBR1, SCALIN: Ca. Scalindua japonica husup-a2, Noc: Nitrosococcus oceani ATCC19707, NE: Nitrosomonas europaea ATCC 19718, Nmul: Nitrosospira multiformis ATCC 25196. Amino acid sequences of hao-like gene products were aligned by MUSCLE. Phylorgenetic relationships were inferred by Maximum Likelihood method with LG+G + I model using MEGA6.06 24.
Figure 5.
Phylogenetic relationships and core gene set among anammox bacteria. (a) Phylogenetic tree based on amino acid sequences of the concatenated ribosomal proteins of anammox bacteria and retention patterns of hao-like and nitrite reductase nirS and nirK, genes. Two complete genomes Ca. B. pituitae and Ca. K. stuttgartiensis, and six nearly completed draft genomes were used for this study. (b) Core gene alignment of anammox bacteria. Rows and columns represent genomes and OGs, respectively. Black lines represent direct adjacency, green lines represent non-adjacent neighbourhoods indicating the existence of insertions, and red lines represent inversions. Closed circles and squares indicate single and paralogous genes, respectively. Dashed lines show the connection to the divided gene. Red characters show genes associated with anammox reactions. ANI: average nucleotide identity. The ANI values show identity % of Ca. B. pituitae genome to other anammox bacterial with nearly complete genome.
The expression of KSMBR1_2670 gene (formerly kustc1061) corresponding to hao1 (Fig. 5), and its gene products have been experimentally confirmed as hydroxylamine oxidase in Ca. K. stuttgartiensis. In addition, the expression of KSMBR1_2163 (formerly kustc0458) and KSMBR1_3792 (formerly kustc4574) genes corresponding to hao2 and hao3, respectively, has also been confirmed along with their gene products.8 Based on protein sequence analyses, it has been postulated that the KSMBR1_2163 (hao2) and KSMBR1_3792 (hao3) genes encode the elusive nitrite reductases reducing nitrite to NO in Ca. K. stuttgartiensis. However, Ca. B. sinica JPN1 as well as Ca. B. pituitae lack the genes encoding canonical NO-producing nitrite reductases (i.e. nirS and nirK; Fig. 5b). Reduction of nitrite to NH2OH and not NO was confirmed by 15N-tracer experiments in Ca. B. sinica although the enzyme responsible for this reaction is still unidentified. It was also confirmed that this species utilises NH2OH and , but not NO and NH4+ for N2H4 synthesis, which is downstream of nitrite reduction, demonstrating that the anammox metabolism of Ca. B. sinica is NH2OH-dependent.7 We also confirmed that Ca. B. pituitae can utilise NH2OH and for anammox metabolism similar to Ca. B. sinica (Supplementary Fig. S5). Oshiki et al.7 speculated that Hao-like proteins are the most likely candidate enzymes catalysing nitrite reduction to NH2OH because -forming pentahaem cytochrome c nitrite reductase has evolved to octahaem cytochrome c protein (Hao)50–52 and this Hao protein is capable of reduction of nitrite to NH2OH using electrons shuffled from quinone pools by a membrane-anchored cytochrome c protein that appeared during the evolutionary process.53,54 High expression of genes corresponding to hao1–hao3 and hao5A (BROSI_A2677, BROSI_A0131, BROSI_A0501, and BROSI_A3864) was observed in Ca. B. sinica JPN1, and the expression levels of those genes were lower under conditions unfavourable for anammox metabolism.54 Through a series of experimental results, they arrived at BROSI_A0501 (hao3) as the most plausible candidate gene responsible for nitrite reduction to NH2OH. In addition, it was suggested that NH2OH-forming nitrite reductase is involved in nitrite reduction by Ca. B. sinica by difference in nitrogen isotope fractionation in comparison with anammox bacteria possessing NO-forming enzymes.55 Out of the five Brocadia species, only Ca. B. sinica lacks hao5B gene included in the core genome. Ca. B. fulgida lacks hao5A, whereas hao5B is duplicated in its genome (Fig. 4 and Supplementary Fig. S2). Although the function of hao5B is still unknown, since both Ca. K. stuttgartiensis and Ca. B. pituitae possess hao5B together with hao2 and hao3 and can utilize NO for N2H4 synthesis, there could be some relationship between the presence of hao5B and the availability of NO.
On the other hand, all hao-like genes in Ca. B. pituitae genome are expressed at various expression levels under regular operational conditions of an anammox bioreactor. High RPKM ratio of more than 10 was observed in several core genes, hao1, hao2, and hao5B, and the hao3 gene was also significantly expressed with the RPKM ratio of 3.09 (Supplementary Table S4). As predicted in Ca. K. stuttgartiensis and Ca. B. sinica, assuming hao2 and hao3 encode NO- and NH2OH-forming nitrite reductase, respectively, it is possible that Ca. B. pituitae reduces nitrite to NO and NH2OH using these enzymes and utilize them for N2H4 biosynthesis. Although details are unclear as yet, the expression of these two genes may be regulated in response to the environment in the anammox bioreactor. Therefore, Ca. B. pituitae presumably utilizes not only NO supplied by the ABC members, but also NO and NH2OH by self-production for anammox metabolism.
Supplementary data
Supplementary data are available at DNARES online.
Supplementary Material
Acknowledgements
We thank Profs K. Kurokawa and H. Mori of the National Institute of Genetics for treatment of the raw data and data registration and Prof T. Sumino of Toyo University for his useful suggestion. We also thank W. Arai of JAMSTEC, Dr C. Katsuyama of Hiroshima University, J. Okada and T. Ide of Chuo University and the members of Comparative Genomics Laboratory at NIG for technical and computational assistance. This work was supported by KAKENHI Grants-in-Aid for Scientific Research to H.T. [17H00793 and 15KT0039]; by KAKENHI Grants-in-Aid for Scientific Research to Y.S. [25241021] and M.K [19H04241]; and by a grant from the collaborative research programme of the National Institute for Basic Biology NIBB, to H.T. and I.U. [No. 19-450]; Finally, the sequencing work was supported by MEXT KAKENHI grant No. 221S0002 to Y.S.
Conflict of interest
None declared.
References
- 1. Kartal B., van Niftrik L., Keltjens J.T., Op den Camp H.J., Jetten M.S.M.. 2012, Anammox-Growth physiology, cell biology, and metabolism, Adv. Microb. Physiol., 60, 211–62. [DOI] [PubMed] [Google Scholar]
- 2. van Niftrik L., Jetten M.S.M.. 2012, Anaerobic ammonium-oxidizing bacteria: unique microorganisms with exceptional properties. Microbiol, Mol. Biol. Rev., 76, 585–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Katal B., de Almeida N.M., Maalcke W.J., Op den Camp H.J.M., Jetten M.S.M., Keltjens J.T.. 2013, How to make a living from anaerobic ammonium oxidation, FEMS Microbiol. Rev., 37, 428–61. [DOI] [PubMed] [Google Scholar]
- 4. Vlaemick S.E., Hay A.G., Maignien L., Verstraete W.. 2011, In quest of the nitrogen oxidizing prokaryotes of the early Earth, Environ. Microbiol., 13, 283–95. [DOI] [PubMed] [Google Scholar]
- 5. Lawson C.E., Wu S., Bhattacharjee A.S.. 2017, Metabolic network analysis reveals microbial community interactions in anammox granules, Nat. Commun., 10, 15416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Park H., Brotto A.C., van Loosdrecht M.C.M., Chandran K.. 2017, Discovery and metagenomic analysis of an anammox bacterial enrichment related to Candidatus “Brocadia caroliniensis” in a full-scale glycerol-fed nitritation-denitritation separate centrate treatment process, Water Res., 111, 265–73. [DOI] [PubMed] [Google Scholar]
- 7. Oshiki M., Ali M., Shinyako-Hata K., Satoh H., Okabe S.. 2016, Hydroxylamine-dependent anaerobic ammonium oxidation anammox, by “Candidatus Brocadia sinica”, Environ. Microbiol., 18, 3133–43. [DOI] [PubMed] [Google Scholar]
- 8. Kartal B., Keltjens J.T.. 2016, Anammox biochemistry: a tale of heme c proteins, Trends Biochem. Sci., 41, 998–1011. [DOI] [PubMed] [Google Scholar]
- 9. Caranto J.D., Lancaster K.M.. 2017, Nitric oxide is an obligate bacterial nitrification intermediate produced by hydroxylamine oxidereductase, Proc. Natl. Acad. Sci. USA, 114, 8217–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frank J., Lücker S., Vossen R.H.A.M., et al. 2018, Resolving the complete genome of Kuenenia stuttgartiensis from a membrane bioreactor enrichment using single-molecule real-time sequencing, Sci. Rep., 8, 4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu Z., Wessels H.J.C.T., van Alen T., Jetten M.S.M., Kartal B.. 2019, Nitric oxide-dependent anaerobic ammonium oxidation, Nat. Commun., 10, 1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Strous M., Pelletier E., Mangenot S., et al. 2006, Deciphering the evolution and metabolism of an anammox bacterium from a community genome, Nature, 440, 790–4. [DOI] [PubMed] [Google Scholar]
- 13. Kartal B., Maalcke W.J., de Almeida N.M., et al. 2011, Molecular mechanism of anaerobic ammonium oxidation, Nature, 479, 127–30. [DOI] [PubMed] [Google Scholar]
- 14. Nakamura T., Harigaya Y., Kimura Y., et al. 2017, Quantitative evaluation of inhibitory effect of various substances on anaerobic ammonia oxidation anammox, J. Biosci. Bioeng., 124, 333–8. [DOI] [PubMed] [Google Scholar]
- 15. Furukawa K., Rouse J.D., Imajo U., Nakamura K., Ishida H.. 2002, Anaerobic oxidation of ammonium confirmed in continuous flow treatment using a non-woven biomass carrier, Japanese J. Wat. Treat. Biol., 38, 87–94. [Google Scholar]
- 16. Isaka K., Sumino T., Tsuneda S.. 2007, High nitrogen removal performance at moderately low temperature utilizing anaerobic ammonium oxidation reactions, J. Biosci. Bioeng., 103, 486–90. [DOI] [PubMed] [Google Scholar]
- 17. Morita H., Kuwahara T., Ohshima K., et al. 2007, An improved DNA isolation method for metagenomic analysis of the microbial flora of the human intestine, Microb. Environ., 22, 214–22. [Google Scholar]
- 18. Noguchi H., Taniguchi T., Itoh T.. 2008, MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes, DNA Res., 15, 387–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takami H. 2019, MAPLE enables functional assessment of microbiota in various environments. In: Gojobori T., Wada T., Kobayashi T., Mineta K. eds, Marine Metagenomics-Technological Aspects and Applications. Singapore: Springer, pp. 85–119. [Google Scholar]
- 20. Takami H., Taniguchi T., Moriya Y., Kuwahara T., Kanehisa M., Goto S.. 2012, Evaluation method for the potential functionome harbored in the genome and metagenome, BMC Genomics, 13, 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takami H., Taniguchi T., Arai W., Takemoto K., Moriya Y., Goto S.. 2016, An automated system for evaluation of the potential functionome: MAPLE version 2.1.0, DNA Res., 23, 467–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kajitani R., Toshimoto K., Noguchi H., et al. 2014, Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads, Genome Res., 24, 1384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nunoura T., Hirayama H., Takami H., et al. 2005, Genetic and functional properties of uncultivated thermophilic crenarchaeotes from a subsurface gold mine as revealed by analysis of genome fragments, Environ. Microbiol., 7, 1967–84. [DOI] [PubMed] [Google Scholar]
- 24. Tanizawa Y., Fujisawa T., Nakamura Y.. 2018, DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication, Bioinformatics., 34, 1037–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grant J.R., Stothard P.. 2008, The CGView Server: a comparative genomics tool for circular genomes, Nucleic Acids Res., 36, W181–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.. 2013, MEGA6: Molecular evolutionary genetics analysis version 6.0, Mol. Biol. Evol., 30, 2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Richter M., Rosselló-Móra R., Glöckner F.O., Peplies J.. 2016, JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison, Bioinformatics, 32, 929–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Uchiyama I. 2006, Hierarchical clustering algorithm for comprehensive orthologous-domain classification in multiple genomes, Nucleic Acids Res., 34, 647–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Uchiyama I., Mihara M., Nishide H., Chiba H.. 2015, MBGD update 2015: microbial genome database for flexible ortholog analysis utilizing a diverse set of genomic data, Nucleic Acids Res., 43, D270–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Uchiyama I. 2008, Multiple genome alignment for identifying the core structure among moderately related microbial genomes, BMC Genomics, 9, 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ohtsubo Y., Ikeda-Ohtsubo W., Nagata Y., Tsuda M.. 2008, GenomeMatcher: a graphical user interface for DNA sequence comparison, BMC Bioinfomatics, 9, 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arai W., Taniguchi T., Goto S., et al. 2018, MAPLE 2.3.0: an improved system for evaluating the functionomes of genomes and metagenomes, Biosci. Biotech. Biochem., 82, 1515–7. [DOI] [PubMed] [Google Scholar]
- 33. Yoshinaga I., Amano T., Yamagishi T., et al. 2011, Distribution and diversity of anaerobic ammonium oxidation Anammox, bacteria the sediment of a eutrophic freshwater lake, Lake Kitaura, Microbes Environ., 26, 189–97. [DOI] [PubMed] [Google Scholar]
- 34. Spiro S. 2012, Nitrous oxide production and consumption: regulation of gene expression by gas-sensitive transcription factors, Philos. Trans. R Soc. B, 367, 1213–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nadeem S., Dörsch P., Bakken L.R.. 2013, The significance of early accumulation of nanomolar concentrations of NO as an inducer of denitrification, FEMS Microbiol. Ecol., 83, 672–84. [DOI] [PubMed] [Google Scholar]
- 36. van de Vossenberg J., Woebken D., Maalcke S.W.J., et al. 2013, The metagenome of the marine anammox bacterium ‘Candidatus Scalindua profunda’ illustrates the versatility of this globally important nitrogen cycle bacterium, Environ. Microbiol., 15, 1275–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yakovlieva L., Walvoort M.T.C.. 2020, Processivity in bacterial glycosyltransferases, ACS Chem. Biol., 15, 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scheurwater E., Reid C.W., Clarke A.J.. 2008, Lytic transglycosylases: bacterial space-making autolysis, Int. J. Biochem. Cell Biol., 40, 586–91. [DOI] [PubMed] [Google Scholar]
- 39. Vermassen A., Leroy S., Talon R., Provot C., Popowska M., Desvaux M.. 2019, Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan, Front. Microbiol., 10, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ciancitto N.P. 2005, Type II secretion: a protein secretion system for all seasons, Trends Microbiol., 13, 581–8. [DOI] [PubMed] [Google Scholar]
- 41. Liu C., Yamamoto T., Nishiyama T., Fujii T., Furukawa K.. 2009, Effect of salt concentration in anammox treatment using non-woven biomass carrier, J. Biosci. Bioeng., 107, 519–23. [DOI] [PubMed] [Google Scholar]
- 42. Park G., Takekawa M., Soda S., Ike M., Furukawa K.. 2017, Temperature dependence of nitrogen removal activity by anammox bacteria enriched at low temperatures, J. Biosci. Bioeng., 123, 505–11. [DOI] [PubMed] [Google Scholar]
- 43. Chandler M., Mahilion J.. 2002, Insertion sequences revisited. In: Craig N.L., Gellert R., Lambowitz M., Craigie A.M. eds. Mobile DNA II. Washington: ASM Press, pp. 305–366. [Google Scholar]
- 44. Oshiki M., Shinyako-Hata K., Satoh H., Okabe S.. 2015, Draft genome sequence of an anaerobic ammonium-oxidizing bacterium, “Candidatus Brocadia sinica”, Genome Announc., 3, e00267-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Takami H., Han C.G., Takaki Y., Ohtsubo E.. 2001, Identification and distribution of new insertion sequences in the genome of alkaliphilic Bacillus halodurans C-125, J. Bacteriol., 183, 4345–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hira D., Toh H., Migita C.T., et al. 2012, Anammox organism KSU-1 expresses a NirK-type copper-containing nitrite reductase instead of a NirS-type with cytochrome cd1, FEBS Lett., 586, 1658–63. [DOI] [PubMed] [Google Scholar]
- 47. Mardanov A.V., Beletsky A.V., Ravin N.V., Botchkova E.A., Litti Y.V., Nozhevnikova A.N.. 2019, Genome of a Novel Bacterium “Candidatus Jettenia ecosi” reconstructed from the metagenome of an anammox bioreactor, Front. Microbiol., 10, 2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bergmann D.J., Hooper A.B., Klotz M.G.. 2005, Structure and sequence conservation of hao cluster genes of autotrophic ammonia-oxidizing bacteria: evidence for their evolutionary history, Appl. Environ Microbiol., 71, 5371–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Palomo A., Pedersen A.G., Fowler S.J., Dechesne A., Sicheritz-Pontén T., Smets B.F.. 2018, Comparative genomics sheds light on niche differentiation and the evolutionary history of commamox Nitrospira, ISME J., 12, 1779–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Klotz M.G., Schmid M.C., Strous M., op den Camp H.J.M., Jetten M.S.M., Hooper A.B.. 2008, Evolution of an octahaem cytochrome c protein family that is key to aerobic and anaerobic ammonia oxidation by bacteria, Environ. Microbiol., 10, 3150–63. [DOI] [PubMed] [Google Scholar]
- 51. Klotz M.G., Stein L.Y. 2008, Nitrifier genomics and evolution of the nitrogen cycle, FEMS Microbiol. Lett., 278, 146–56. [DOI] [PubMed] [Google Scholar]
- 52. Campbell B.J., Smith J.L., Hanson T.E., et al. 2009, Adaptations to submarine hydrothermal environments exemplified by the genome of Nautilia profundicola, PLoS Genet., 5, e1000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hanson T.E., Campbell B.J., Kalis K.M., Campbell M.A., Klotz M.G.. 2013, Nitrate ammonification by Nautilia profundicola AmH: experimental evidence consistent with a free hydroxylamine intermediate, Front. Microbiol., 4, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rathnayake R.M.L.D., Oshiki M., Ishii S., Segawa T., Satoh H., Okabe S.. 2018, Experimental evidence for in situ nitric oxide production in anaerobic ammonia-oxidizing bacterial granules, Environ. Sci. Technol., 52, 5744–52. [DOI] [PubMed] [Google Scholar]
- 55. Kobayashi K., Makabe A., Yano M., et al. 2019, Dual nitrogen and oxygen isotope fractionation during anaerobic ammonium oxidation by anammox bacteria, ISME J., 13, 2426–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.