Abstract
Purpose
Long non-coding RNA H19 was highly expressed in the latent period of epilepsy, contributing to apoptosis of hippocampal neurons by targeting let-7b. Transforming growth factor beta receptor 1 (TGFBR1), a target of let-7b, is located on the susceptibility locus for epilepsy. In this context, we investigated the association between tagSNPs in long non-coding RNA H19 and transforming growth factor beta receptor 1 (TGFBR1) rs6478974 and the risk of epilepsy.
Patients and Methods
The present study consisted of 302 patients with epilepsy and 612 age- and gender-matched controls. The polymorphisms were analyzed using a TaqMan allelic genotyping assay. H19 and TGFBR1 mRNA levels were determined using quantitative real-time polymerase chain reaction.
Results
The TGFBR1 AT and TT genotypes emerged as a protective factor for the risk of epilepsy (AT vs AA: adjusted OR = 0.59, 95% CI: 0.39–0.89, P = 0.01; TT vs AA: adjusted OR = 0.53, 95% CI: 0.35–0.80, P = 0.002, respectively). The protective effect was also observed in recessive genetic model (adjusted OR = 0.56, 95% CI: 0.38–0.82, P = 0.003). Individuals carrying the rs6478974 TT genotype had lower levels of TGFBR1 mRNA. Moreover, the TCTAT and TCCAA haplotypes emerged as a risk factor for epilepsy and the rs3741219-rs2839698-rs6478974 was associated with an interactive effect on the risk of epilepsy.
Conclusion
The current study provides evidence of the rs6478974 TT genotype decreasing the susceptibility to epilepsy by reducing the levels of TGFBR1 mRNA.
Keywords: long non-coding RNA H19, transforming growth factor beta receptor 1, genetic susceptibility, quantitative PCR
Introduction
Epilepsy is a neurological disorder that is characterized by recurrent epileptic seizures, affecting about 39 million people worldwide in 20151 and resulting in direct economic costs of about $1 billion annually in the United States.2 Current knowledge of the exact reason for epilepsy remains unclear. Established acquired causes include traumatic brain injury,3 stroke,4 brain tumors5 and infective lesions of the brain.6 Besides the acquired factors, genetic factors have been demonstrated to play crucial roles in most cases.6,7 Twin studies showed that concordance rates for epilepsy in monozygotic twins were four times higher than those in dizygotic twins.8 Close relatives of a patient with epilepsy had a five-fold higher risk than those of the general population.9 Moreover, a series of genes, such as sodium voltage-gated channel alpha subunit 1, cholinergic receptor nicotinic alpha 4 subunit and potassium voltage-gated channel subfamily Q members 2 and 3 have been identified to contribute to epileptogenesis.10–13
Apart from protein-coding RNAs mentioned above, some non-coding RNAs have been reported to be involved in the development and progression of epilepsy.14–17 Although the study of microRNAs (miRNAs) has dominated the field of non-coding RNAs’ biology over the past years, long non-coding RNAs (lncRNAs) have attracted growing attention in recent years.14,15,18 LncRNAs, defined as non-coding RNAs with lengths exceeding 200 nucleotides, are found to execute multiple biological functions, including regulating gene transcription and/or post-transcriptional processing.14,19,20 In both human mesial temporal lobe epilepsy and animal model of temporal lobe epilepsy, amounts of lncRNAs were observed to be differentially expressed.14,17,21 Among them, lncRNA H19 was reported to be highly expressed in the latent period of epilepsy, contributing to apoptosis of hippocampal neurons by targeting let-7b and hippocampal glial cell activation via JAK/STAT signaling.14,15 Transforming growth factor beta receptor 1 (TGFBR1), as a target gene of let-7b, was found to be up-regulated in patients with temporal lobe epilepsy.22–25
It is evident that chromosome 9q21-q22 is a susceptibility locus for epilepsy.26,27 TGFBR1, located in the region of 9q22.33 in human genome, has been identified to be related to the pathogenesis of epilepsy.25,28 And thus we hypothesized that single nucleotide polymorphisms (SNPs) in TGFBR1 may be associated with the risk of epilepsy. Due to rs6478974 in TGFBR1 affecting expression level of miRNAs,29 we investigated in this study the association between the potential functional SNP rs6478974 and risk of epilepsy in a Chinese population. Since epilepsy is a complex disease that is triggered by more than one gene, tagSNPs in lncRNA H19 were also examined. We found that the TGFBR1 rs6478974, H19 rs3741219 and rs2839698 may have an interactive effect on the development of epilepsy.
Patients and Methods
Study Population
A hospital-based case control study was conducted in the Northeast of China. A total of 302 patients with epilepsy were recruited from the China-Japan Union Hospital of Jilin University between January 2012 and June 2019. Meanwhile, 612 control blood samples were obtained from healthy volunteers who lived in the same area during the same period. Patients with epilepsy were diagnosed according to the criteria based on the International League Against Epilepsy.30 Among the patients, 186 suffered from drug-responsive epilepsy and 116 suffered from drug-resistant epilepsy. Drug-responsive patients were defined as those with more than 50% reduction of seizure frequency or seizure free after treatment with antiepileptic drugs, and drug-resistant patients were defined as those with failure to achieve sustained seizure freedom after treatment with two established antiepileptic drugs.31 Exclusion criteria were as follows: (a) patients with psychiatric comorbidity; (b) a family history of epilepsy; (c) history of pseudoseizures; (d) alcohol and/or drug addiction; (e) not Chinese Han ethnicity; (f) patients with combined tumor. The study protocol was reviewed and approved by the Institutional Ethical Committee of the China-Japan Union Hospital of Jilin University (Approved number: 0034), and written informed consent was signed by all subjects or their relatives.
SNPs Selection
We selected tagSNPs in H19 with minor allele frequency (MAF) more than 10% in Chinese Han population. Moreover, functional SNP in TGFBR1 was also selected according to the following criteria: (a) MAF > 10% in Chinese Han population; (b) affecting TGFBR1 expression based on data from expression Quantitative Trait Loci (eQTL, https://www.gtexportal.org/).
DNA and RNA Extraction
For each subject, 3–5 mL of anticoagulation peripheral blood sample was collected. Genomic DNA was extracted using the isolation kit according to the manufacturer’s instruction (Tiangen, Beijing, China). Total RNA was extracted using the RNAprep pure Blood Kit (Tiangen, Beijing, China). DNA and RNA concentration and purity were determined using the NanoDrop ND-1000 spectrophotometer from NanoDrop Technologies (Rockland, DE). The 260/280 ratio for DNA ranging between 1.7 and 1.9 and the 260/280 ratio for RNA > 1.9 were considered acceptable.
Genotyping
H19 polymorphisms (ie, rs3741219, rs2839698, rs217727 and rs3741216) and TGFBP1 rs6478974 were genotyped by using the ABI 7500 real-time PCR System (Applied Biosystems, Foster City, CA, USA). For quality control, about 5% of the subjects were randomly selected for repeat analysis, and inconsistent results were resolved by validation with Sanger sequencing.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
H19 and TGFBR1 mRNA levels in patients with epilepsy and controls were examined by using qRT-PCR. Isolated RNA was converted to synthesize cDNA using the First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instruction. Amplification was performed on the ABI 7500 qRT-PCR System (Applied Biosystems) using a SYBR Green kit. Primer sequences used were as follows:32–34 GAPDH forward, CTCTCTGCTCCTCCTGTTCGAC and GAPDH reverse, TGAGCGATGTGGCTCGGCT; H19 forward, TGCTGCACTTTACAACCACTG and H19 reverse, ATGGTGTCTTTGATGTTGGGC; TGFBR1 forward, GAGGAAAGTGGCGGGGAG and TGFBR1 reverse, CCAACCAGAGCTGAGT CCAAGTA. The thermocycling conditions were set as follows: initial preincubation at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 5 sec and annealing/extension at 60°C for 30 sec. The relative expression levels of H19 and TGFBR1 mRNA were calculated using the 2 −ΔCt method, with GAPDH as an internal control.35
Statistical Analysis
Quanto software version 1.2 was performed for evaluation of the statistical power. The genotype distributions of the selected SNPs were tested for Hardy-Weinberg equilibrium (HWE) using a goodness-of-fit χ2 test, with p > 0.05 indicating agreement with HWE. The association between H19 and TGFBR1 polymorphisms and epilepsy risk was compared using chi-square test. Adjusted logistic regression analysis based on age and gender was used to compute odds ratios (ORs) and 95% confidence intervals (CIs). Haplotype analyses for the SNPs were carried out using online SHEsis software, and Bonferroni correction was used for multiple comparisons. Multifactor dimensionality reduction (MDR) platform was used to evaluate H19-TGFBR1 interaction.36 Data of qRT-PCR were analyzed using Mann–Whitney U-test. A p value of <0.05 was considered to be statistically significant. All data were analyzed using the SPSS software version 19.0 (SPSS, Chicago, IL, USA).
Results
Characteristics of Study Population
Table 1 shows the demographic and clinical data of the study population that was used for SNPs analysis and qRT-PCR. The mean age of patients with epilepsy was not significantly different from that of healthy controls (P = 0.69). Additionally, no significant difference of gender distribution was observed between epilepsy patients and controls (P = 0.33). Among the 302 patients enrolled in this study, 164 (54.3%) had generalized epilepsy and 138 (45.7%) had focal epilepsy, with 97 (32.1%) cryptogenic, 92 (30.5%) idiopathic and 113 (37.4%) symptomatic epilepsy; 186 (61.6%) were diagnosed drug-responsive and 116 (38.4%) had drug-resistant epilepsy.
Table 1.
Demographics of Controls and Patients with Epilepsy
| Subjects for SNPs Analysis | Subjects for qRT-PCR | |||||
|---|---|---|---|---|---|---|
| Patients with Epilepsy | Controls | P value | Patients with Epilepsy | Controls | P value | |
| N | 302 | 612 | 108 | 108 | ||
| Age, mean ± SD (years) | 34.00 ± 15.85 | 34.00 ± 11.96 | 0.69 | 32.00 ± 13.7 | 32.00 ± 12.8 | 0.80 |
| Age of onset, mean ± SD (years) | 24.00 ± 17.68 | 20.4 ± 13.9 | ||||
| Gender, n (%) | ||||||
| Male | 192 (63.6) | 409 (66.8) | 0.33 | 64 (59.3) | 73 (67.6) | 0.20 |
| Female | 110 (36.4) | 203 (33.2) | 44 (40.7) | 35 (32.4) | ||
| Seizure type, n (%) | ||||||
| Generalized | 164 (54.3) | 60 (55.6) | ||||
| Focal | 138 (45.7) | 48 (44.4) | ||||
| Epilepsy syndrome, n (%) | ||||||
| Cryptogenic | 97 (32.1) | 37 (34.3) | ||||
| Idiopathic | 92 (30.5) | 36 (32.4) | ||||
| Symptomatic | 113 (37.4) | 35 (33.3) | ||||
| Antiepileptic drug therapy, n (%) | ||||||
| Drug-responsive | 186 (61.6) | 68 (63.0) | ||||
| Drug-resistant | 116 (38.4) | 40 (37.0) | ||||
Abbreviations: SNPs, single nucleotide polymorphisms; qRT-PCR, quantitative real-time polymerase chain reaction; SD, standard deviation.
Association Between H19 and TGFBR1 Polymorphisms and the Risk of Epilepsy
The genotype frequencies of H19 and TGFBR1 polymorphisms (ie, rs3741219, rs2839698, rs217727, rs3741216 and rs6478974) among epilepsy patients and controls are shown in Table 2. None of the genotype distributions in controls deviated from HWE. Compared to the TGFBR1 rs6478974 AA genotype, the AT and TT genotypes emerged as a protective factor for the risk of epilepsy (AT vs AA: adjusted OR = 0.59, 95% CI: 0.39–0.89, P = 0.01; TT vs AA: adjusted OR = 0.53, 95% CI: 0.35–0.80, P = 0.002, respectively). The protective effect was also observed in recessive genetic model (adjusted OR = 0.56, 95% CI: 0.38–0.82, P = 0.003). However, we failed to find any association between tagSNPs in H19 (ie, rs3741219, rs2839698, rs217727 and rs3741216) and epilepsy risk. Stratification analysis also showed no significant association between the 5 selected SNPs and antiepileptic drug therapy (drug-resistant vs drug-responsive) (Table 3). When stratified analysis was performed based on age of onset, gender, seizure type and epilepsy syndrome, no significant association was found (data not shown).
Table 2.
Association Between H19 and TGFBR1 Polymorphisms and the Risk of Epilepsy
| Polymorphisms | Controls, n = 612, n (%)† | Patients, n = 302, n (%)† | Adjusted OR (95% CI)‡ | P value |
|---|---|---|---|---|
| H19 rs3741219 | ||||
| TT | 339 (55.4) | 173 (57.3) | Reference | |
| CT | 228 (37.3) | 104 (34.4) | 0.88 (0.66–1.19) | 0.41 |
| CC | 45 (7.4) | 25 (8.3) | 1.08 (0.64–1.82) | 0.78 |
| Dominant | 273 (44.6) | 129 (42.7) | 0.92 (0.69–1.21) | 0.54 |
| Recessive | 567 (92.6) | 277 (91.7) | 1.14 (0.68–1.90) | 0.62 |
| H19 rs2839698 | ||||
| CC | 343 (56.1) | 164 (54.3) | Reference | |
| CT | 221 (36.1) | 120 (39.7) | 1.12 (0.83–1.50) | 0.45 |
| TT | 48 (7.8) | 18 (6.0) | 0.78 (0.44–1.39) | 0.40 |
| Dominant | 269 (43.9) | 138 (45.7) | 1.07 (0.81–1.41) | 0.65 |
| Recessive | 564 (92.2) | 284 (94.0) | 0.74 (0.42–1.30) | 0.29 |
| H19 rs217727 | ||||
| CC | 265 (43.3) | 123 (40.7) | Reference | |
| CT | 261 (42.6) | 129 (42.7) | 1.08 (0.80–1.45) | 0.63 |
| TT | 86 (14.1) | 50 (16.6) | 1.25 (0.83–1.88) | 0.29 |
| Dominant | 347 (56.7) | 179 (59.3) | 1.12 (0.84–1.48) | 0.44 |
| Recessive | 526 (85.9) | 252 (83.4) | 1.22 (0.83–1.78) | 0.32 |
| H19 rs3741216 | ||||
| AA | 468 (76.5) | 223 (73.8) | Reference | |
| AT | 130 (21.2) | 71 (23.5) | 1.14 (0.82–1.59) | 0.45 |
| TT | 14 (2.3) | 8 (2.6) | 1.19 (0.49–2.90) | 0.70 |
| Dominant | 144 (23.5) | 79 (26.2) | 1.14 (0.83–1.57) | 0.41 |
| Recessive | 598 (97.7) | 294 (97.4) | 1.19 (0.49–2.87) | 0.70 |
| TGFBR1 rs6478974 | ||||
| AA | 72 (11.8) | 58 (19.2) | Reference | |
| AT | 250 (40.8) | 120 (39.7) | 0.59 (0.39–0.89) | 0.01 |
| TT | 290 (47.4) | 124 (41.1) | 0.53 (0.35–0.80) | 0.002 |
| Dominant | 322 (52.6) | 178 (58.9) | 0.78 (0.59–1.03) | 0.08 |
| Recessive | 540 (88.2) | 244 (80.8) | 0.56 (0.38–0.82) | 0.003 |
Notes: †The percentage is not always 100 due to rounding. ‡Adjusted by age and gender.
Abbreviations: TGFBR1, transforming growth factor beta receptor 1; OR, odds ratio; CI, confidence interval.
Table 3.
Distribution of H19 and TGFBR1 Polymorphisms in Drug-Responsive and -Resistant Patients with Epilepsy
| Drug-Responsive, n = 186, n (%) | Drug-Resistant, n = 116, n (%) | Adjusted OR (95% CI)† | P value | |
|---|---|---|---|---|
| H19 rs3741219 | ||||
| TT | 105 (56.5) | 68 (58.6) | Reference | |
| CT/CC | 81 (43.5) | 48 (41.4) | 0.93 (0.58–1.49) | 0.76 |
| H19 rs2839698 | ||||
| CC | 104 (55.9) | 60 (51.7) | Reference | |
| CT/TT | 82 (44.1) | 56 (48.3) | 1.16 (0.73–1.85) | 0.53 |
| H19 rs217727 | ||||
| CC | 75 (40.3) | 48 (41.4) | Reference | |
| CT/TT | 111 (59.7) | 68 (58.6) | 0.95 (0.59–1.52) | 0.82 |
| H19 rs3741216 | ||||
| AA | 141 (75.8) | 82 (70.7) | Reference | |
| AT/TT | 45 (24.2) | 34 (29.3) | 1.33 (0.79–2.24) | 0.29 |
| TGFBR1 rs6478974 | ||||
| AA/AT | 113 (60.8) | 65 (56.0) | Reference | |
| TT | 73 (39.2) | 51 (44.0) | 0.83 (0.52–1.33) | 0.45 |
Note: †Adjusted by age and gender.
Abbreviations: TGFBR1, transforming growth factor beta receptor 1; OR, odds ratio; CI, confidence interval.
Haplotype Analysis and Interaction Analysis
Compared to the TCCAT haplotype, the TCTAT and TCCAA haplotypes emerged as a risk factor for epilepsy (OR = 1.63, 95% CI: 1.13–2.35, P = 0.008; OR = 1.81, 95% CI: 1.26–2.62, P = 0.001, respectively) (Table 4).
Table 4.
Haplotype Analyses of H19 and TGFBR1 Polymorphisms with the Risk of Epilepsy
| Haplotype† | Controls, n (%) | Patients, n (%) | OR (95% CI) | P value |
|---|---|---|---|---|
| TCCAT | 260 (21.2) | 94 (15.6) | Reference | |
| TCTAT | 134 (10.9) | 79 (13.1) | 1.63 (1.13–2.35) | 0.008 |
| TCCAA | 122 (10.0) | 80 (13.2) | 1.81 (1.26–2.62) | 0.001 |
| TTCAT | 96 (7.8) | 36 (6.0) | 1.04 (0.66–1.63) | 0.87 |
| CCCAT | 90 (7.4) | 51 (8.4) | 1.57 (1.03–2.38) | 0.03 |
| TCTAA | 68 (5.6) | 40 (6.6) | 1.63 (1.03–2.57) | 0.04 |
| TTTAT | 59 (4.8) | 22 (3.6) | 1.03 (0.60–1.78) | 0.91 |
| TTCAA | 41 (3.3) | 27 (4.5) | 1.82 (1.06–3.13) | 0.03 |
| CCCAA | 39 (3.2) | 18 (3.0) | 1.28 (0.70–2.34) | 0.43 |
| CCTAT | 36 (2.9) | 8 (1.3) | 0.62 (0.28–1.37) | 0.23 |
| TCTTT | 30 (2.4) | 10 (1.7) | 0.92 (0.43–1.96) | 0.83 |
| TCCTT | 29 (2.4) | 16 (2.6) | 1.53 (0.79–2.94) | 0.20 |
| CTTAT | 27 (2.2) | 13 (2.2) | 1.33 (0.66–2.69) | 0.42 |
| CCTAA | 26 (2.1) | 17 (2.8) | 1.81 (0.94–3.48) | 0.07 |
| TTTAA | 24 (2.0) | 9 (1.5) | 1.04 (0.47–2.31) | 0.93 |
| CTCAT | 22 (1.8) | 10 (1.7) | 1.26 (0.57–2.75) | 0.57 |
| CTCAA | 12 (1.0) | 7 (1.2) | 1.61 (0.62–4.22) | 0.33 |
Note: †Only the frequency more than 1% was presented.
Abbreviations: TGFBR1, transforming growth factor beta receptor 1; OR, odds ratio; CI, confidence interval.
Gene–gene interaction analysis showed that the rs3741219-rs2839698-rs6478974 was the best candidate model, with the accuracy of 0.60 and cross-validation consistency of 9/10 (OR = 2.00, 95% CI: 1.51–2.64, P < 0.001) (Table 5).
Table 5.
Interaction Analysis of H19 and TGFBR1 Polymorphisms with the Risk of Epilepsy
| Best Candidate Models | Accuracy | Cross-Validation Consistency | Sensitivity | Specificity | OR (95% CI) | P value |
|---|---|---|---|---|---|---|
| rs3741219-rs6478974 | 0.57 | 4/10 | 0.48 | 0.62 | 1.47 (1.11–1.95) | 0.006 |
| rs3741219-rs2839698-rs6478974 | 0.60 | 9/10 | 0.53 | 0.64 | 2.00 (1.51–2.64) | <0.001 |
Abbreviations: TGFBR1, transforming growth factor beta receptor 1; OR, odds ratio; CI, confidence interval.
The rs6478974 TT Genotype Associated to Lower Levels of TGFBR1 mRNA
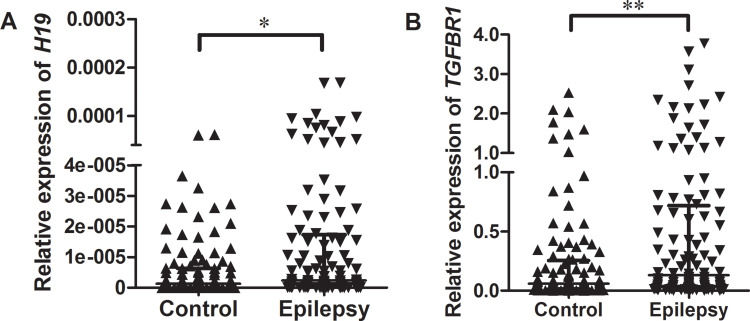
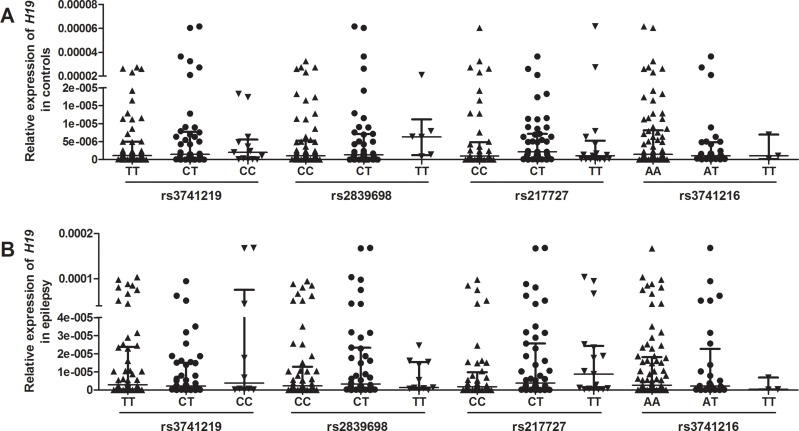
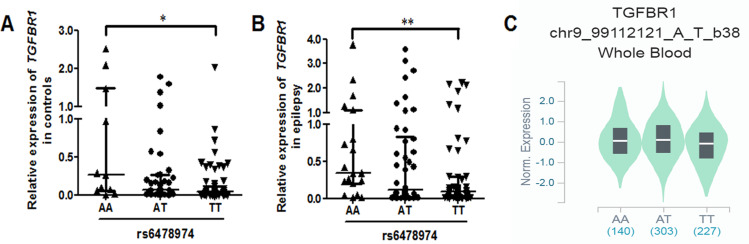
Relative expression of H19 and TGFBR1 in epilepsy patients and controls was examined using qRT-PCR (n = 108). As shown in Figure 1, both H19 and TGFBR1 mRNA levels were significantly higher in epilepsy patients than those in controls. Genotype-phenotype analysis showed that the rs3741219, rs2839698, rs217727 and rs3741216 did not influence H19 expression (Figure 2). However, compared to carriers with the rs6478974 AA genotype, carriers with the rs6478974 TT genotype had lower levels of TGFBR1 mRNA in both epilepsy patients (Figure 3A) and controls (Figure 3B), which was confirmed by data from eQTL (P = 9.8 X 10−14) (Figure 3C). When the patients were classified into cryptogenic, idiopathic and symptomatic groups, no relevant data were found regarding TGFBR1 mRNA levels to the rs6478974.
Figure 1.
Relative expression of H19 and TGFBR1 mRNA in epilepsy patients and controls. RNA was extracted from blood samples and qRT-PCR was used to examine the expression levels of H19 (A) and TGFBR1 mRNA (B) in epilepsy patients and controls. GAPDH was used as an internal control. Data are presented as median with interquartile range (*P < 0.05, **P < 0.01).
Figure 2.
Association between tagSNPs in H19 and its expression. The relationship between tagSNPs in H19 (ie, rs3741219, rs2839698, rs217727 and rs3741216) and H19 expression in controls (A) and patients with epilepsy (B).
Figure 3.
The rs6478974 TT carriers exhibited lower levels of TGFBR1. The relationship between the rs6478974 AA, AT and TT genotypes and TGFBR1 mRNA levels in controls (A) and patients with epilepsy (B) (*P < 0.05, **P < 0.01). Data from eQTL showed that the rs6478974 TT genotype was associated with lower expression of TGFBR1 (P = 9.8 X 10−14) (C).
Discussion
In this study, we for the first time investigated the association between tagSNPs in H19 and TGFBR1 rs6478974 and susceptibility to epilepsy in the Chinese Han population. Our study of 302 patients with epilepsy and 612 controls found significant differences in genotypic and allelic frequencies of the rs6478974 between cases and controls. Haplotype analysis showed that the frequencies of the TCTAT and TCCAA haplotypes were higher in epilepsy patients than those in controls. MDR analysis revealed that a three-loci model of rs3741219-rs2839698-rs6478974 was the best for predicting the risk of epilepsy. Additionally, our study found that carriers with the rs6478974 TT genotype displayed lower levels of TGFBR1 mRNA. Our study had 80.3% power to evaluate the effect of H19-TGFBR1 SNPs on the risk of epilepsy when setting the relative risk of 1.6 under a dominant genetic model. These findings indicate that the rs6478974 may be a susceptibility locus for the occurrence of epilepsy.
Growing evidence has shown that brain inflammation is a cause or a consequence of epilepsy.37 Transforming growth factor-β1 (TGF-β1), an important regulator in the brain’s responses to injury and inflammation, has been reported to be implicated in the pathophysiology of epilepsy.37,38 By binding to TGF-β, TGFBR1 mediates the induction of several genes involved in brain disorder, such as epilepsy/seizure.28 In patients with temporal lobe epilepsy, TGFBR1 protein was found to be up-regulated, acting as a therapeutic target for preventing status epilepticus.25,28 TGFBR1 is located on the region of 9q22.33 that has been identified to be a susceptibility locus for epilepsy.26,27 We speculated therefore that SNP in TGFBR1 may affect the occurrence of epilepsy. We in this study genotyped a functional SNP rs6478974 in TGFBR1 and found that the rs6478974AT and TT genotypes emerged as a protective factor for the risk of epilepsy. To determine the reason for TGFBR1 rs6478974 decreasing epilepsy risk, we analyzed the expression levels of TGFBR1 mRNA in both patients with epilepsy and controls. We found that TGFBR1 mRNA was higher in patients than that in controls. More importantly, we found that the presence of rs6478974 TT genotype resulted in lower levels of TGFBR1 mRNA. The impact of the rs6478974 on TGFBR1 expression levels was also evident in eQTL analysis of RNA-seq data of blood cells. Taken together, a conclusion might be made that the rs6478974 TT genotype exerted a protective effect on epileptogenesis by decreasing TGFBR1 expression at the transcriptional level.
Epilepsy is not a single gene disorder but verified existence of a series of susceptibility genes.13 LncRNAs can modulate gene expression via multiple modes, participating in the pathogenesis of epilepsy.20 H19, a type of lncRNA, was highly expressed in the latent period of epilepsy, contributing to apoptosis of hippocampal neurons by targeting let-7b and hippocampal glial cell activation via JAK/STAT signaling.14,15 Therefore, in this study, we genotyped tagSNPs in H19 (ie, rs3741219, rs2839698, rs217727 and rs3741216) and performed H19-TGFBR1 interaction analysis to clarify the effect of gene–gene interaction on epilepsy risk. Although no significant association between the SNPs and epilepsy risk was found in single site comparison, haplotype analysis revealed the TCTAT and TCCAA haplotypes had a 1.63- and 1.81-fold increased risk of epilepsy, respectively. Notably, a significant three-loci interaction model of rs3741219-rs2839698-rs6478974 was identified to increase the risk of epilepsy. Our results were consistent with some previous reports in central nervous system diseases, which found that the Grs217727Ars2839698Grs3741219 haplotype carriers were less likely to develop glioma39 and the 3-loci model of rs2280543-rs217727-rs2839698 conferred the risk of intracranial aneurysm.40 With regard to the association between H19 polymorphisms and risk ischemic stroke (IS), conflicting results were obtained. Zhu et al reported the H19 rs217727 increasing the susceptibility of small vessel IS,41 whereas Huang et al reported no significant association between SNPs in H19 and IS risk.42 Discrepancies of the results may arise from diversities of genetic background in different diseases, affection of environmental factors and limited sample sizes. Further analyses of gene–environment interaction based on larger sample sizes will be a benefit for the better understanding of the effect of H19 and TGFBR1 on epilepsy risk.
In this study, we have to acknowledge some limitations. The major concern of a hospital-based case-control study is selection bias. Although HWE was present in the current study, population-based case cohort studies are still valuable to confirm our results. Additionally, China has multiple ethnic populations encompassing 56 ethnicities. To avoid the heterogeneity, only Chinese Han was enrolled in this study, and thus the data cannot directly extend to other ethnic groups. Intra-ethnic comparative studies are necessary to support our findings.
In conclusion, the current study provides direct evidence of the rs6478974 TT genotype decreasing the susceptibility to epilepsy and the rs6478974 TT being associated with lower levels of TGFBR1 mRNA. Given the important biological role of TGFBR1 in the pathogenesis of epilepsy, the rs6478974 may be potentially used as a biomarker for the development of epilepsy. Extension of current findings to other neurological diseases will be necessary in determining whether the genetic marker is specific to epilepsy. Further studies are needed to understand how the rs6478974 predisposes to epilepsy and affects the expression of TGFBR1 mRNA. Once accomplished, it will help to predict the potential therapeutic value of the rs6478974 in the treatment of epilepsy.
Funding Statement
This study was supported by the Project of International Cooperation of Jilin Province in China [20180414062GH] and Natural Science Foundation of Jilin Province in China [20180101300JC].
Ethics Approval and Informed Consent
All procedures performed in studies involving human participants were in accordance with the ethical standards of China-Japan Union Hospital of Jilin University and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Disease GBD, Injury I, Prevalence C. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388(10053):1545–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilden JA, Cohen-Gadol AA. Evaluation of first nonfebrile seizures. Am Fam Physician. 2012;86(4):334–340. [PubMed] [Google Scholar]
- 3.Lucke-Wold BP, Nguyen L, Turner RC, et al. Traumatic brain injury and epilepsy: underlying mechanisms leading to seizure. Seizure. 2015;33:13–23. doi: 10.1016/j.seizure.2015.10.002 [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y, Li X, Zhang K, Tong T, Cui R. The progress of epilepsy after stroke. Curr Neuropharmacol. 2018;16(1):71–78. doi: 10.2174/1570159X15666170613083253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Politsky JM. Brain tumor-related epilepsy: a current review of the etiologic basis and diagnostic and treatment approaches. Curr Neurol Neurosci Rep. 2017;17(9):70. doi: 10.1007/s11910-017-0777-3 [DOI] [PubMed] [Google Scholar]
- 6.Berkovic SF, Mulley JC, Scheffer IE, Petrou S. Human epilepsies: interaction of genetic and acquired factors. Trends Neurosci. 2006;29(7):391–397. doi: 10.1016/j.tins.2006.05.009 [DOI] [PubMed] [Google Scholar]
- 7.Pandolfo M. Genetics of epilepsy. Semin Neurol. 2011;31(5):506–518. doi: 10.1055/s-0031-1299789 [DOI] [PubMed] [Google Scholar]
- 8.Berkovic SF, Howell RA, Hay DA, Hopper JL. Epilepsies in twins: genetics of the major epilepsy syndromes. Ann Neurol. 1998;43(4):435–445. doi: 10.1002/ana.410430405 [DOI] [PubMed] [Google Scholar]
- 9.Bhalla D, Godet B, Druet-Cabanac M, Preux PM. Etiologies of epilepsy: a comprehensive review. Expert Rev Neurother. 2011;11(6):861–876. doi: 10.1586/ern.11.51 [DOI] [PubMed] [Google Scholar]
- 10.Steinlein OK. Genetics and epilepsy. Dialogues Clin Neurosci. 2008;10(1):29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang N, Huang HL, Zhou H. Study of candidate gene cHRNA4 for familial epilepsy syndrome. Eur Rev Med Pharmacol Sci. 2018;22(6):1765–1769. doi: 10.26355/eurrev_201803_14594 [DOI] [PubMed] [Google Scholar]
- 12.Goto A, Ishii A, Shibata M, Ihara Y, Cooper EC, Hirose S. Characteristics of KCNQ2 variants causing either benign neonatal epilepsy or developmental and epileptic encephalopathy. Epilepsia. 2019;60(9):1870–1880. doi: 10.1111/epi.16314 [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Lin ZJ, Liu L, et al. Epilepsy-associated genes. Seizure. 2017;44:11–20. doi: 10.1016/j.seizure.2016.11.030 [DOI] [PubMed] [Google Scholar]
- 14.Han CL, Ge M, Liu YP, et al. Long non-coding RNA H19 contributes to apoptosis of hippocampal neurons by inhibiting let-7b in a rat model of temporal lobe epilepsy. Cell Death Dis. 2018;9(6):617. doi: 10.1038/s41419-018-0496-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han CL, Ge M, Liu YP, et al. LncRNA H19 contributes to hippocampal glial cell activation via JAK/STAT signaling in a rat model of temporal lobe epilepsy. J Neuroinflammation. 2018;15(1):103. doi: 10.1186/s12974-018-1139-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao Y, Chen Y. Pathophysiology and clinical utility of non-coding RNAs in epilepsy. Front Mol Neurosci. 2017;10:249. doi: 10.3389/fnmol.2017.00249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han CL, Liu YP, Zhao XM, et al. Whole-transcriptome screening reveals the regulatory targets and functions of long non-coding RNA H19 in epileptic rats. Biochem Biophys Res Commun. 2017;489(2):262–269. doi: 10.1016/j.bbrc.2017.05.161 [DOI] [PubMed] [Google Scholar]
- 18.Henshall DC, Hamer HM, Pasterkamp RJ, et al. MicroRNAs in epilepsy: pathophysiology and clinical utility. Lancet Neurol. 2016;15(13):1368–1376. doi: 10.1016/S1474-4422(16)30246-0 [DOI] [PubMed] [Google Scholar]
- 19.Gou Q, Gao L, Nie X, et al. Long noncoding RNA AB074169 inhibits cell proliferation via modulation of KHSRP-mediated CDKN1a expression in papillary thyroid carcinoma. Cancer Res. 2018;78(15):4163–4174. doi: 10.1158/0008-5472.CAN-17-3766 [DOI] [PubMed] [Google Scholar]
- 20.Villa C, Lavitrano M, Combi R. Long non-coding RNAs and related molecular pathways in the pathogenesis of epilepsy. Int J Mol Sci. 2019;20(19):4898. doi: 10.3390/ijms20194898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui Z, Zhang X, Song H, et al. Differential long non-coding RNA (lncRNA) profiles associated with hippocampal sclerosis in human mesial temporal lobe epilepsy. Int J Clin Exp Pathol. 2019;12(1):259–266. [PMC free article] [PubMed] [Google Scholar]
- 22.Yan S, Yu Z, Ning L, et al. Let-7b promotes alpaca hair growth via transcriptional repression of TGFbetaR I. Gene. 2016;577(1):32–36. doi: 10.1016/j.gene.2015.11.022 [DOI] [PubMed] [Google Scholar]
- 23.Wang B, Jha JC, Hagiwara S, et al. Transforming growth factor-beta1-mediated renal fibrosis is dependent on the regulation of transforming growth factor receptor 1 expression by let-7b. Kidney Int. 2014;85(2):352–361. doi: 10.1038/ki.2013.372 [DOI] [PubMed] [Google Scholar]
- 24.Xicola RM, Bontu S, Doyle BJ, et al. Association of a let-7 miRNA binding region of TGFBR1 with hereditary mismatch repair proficient colorectal cancer (MSS HNPCC). Carcinogenesis. 2016;37(8):751–758. doi: 10.1093/carcin/bgw064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Y, Xue T, Yuan J, et al. Increased expression of TGFbeta type I receptor in brain tissues of patients with temporal lobe epilepsy. Clin Sci. 2009;117(1):17–22. doi: 10.1042/CS20080347 [DOI] [PubMed] [Google Scholar]
- 26.Tikka-Kleemola P, Artto V, Vepsalainen S, et al. A visual migraine aura locus maps to 9q21-q22. Neurology. 2010;74(15):1171–1177. doi: 10.1212/WNL.0b013e3181d8ffcb [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deprez L, Peeters K, Van Paesschen W, et al. Familial occipitotemporal lobe epilepsy and migraine with visual aura: linkage to chromosome 9q. Neurology. 2007;68(23):1995–2002. doi: 10.1212/01.wnl.0000262764.78511.17 [DOI] [PubMed] [Google Scholar]
- 28.Mercado-Gomez O, Landgrave-Gomez J, Arriaga-Avila V, Nebreda-Corona A, Guevara-Guzman R. Role of TGF-beta signaling pathway on Tenascin C protein upregulation in a pilocarpine seizure model. Epilepsy Res. 2014;108(10):1694–1704. doi: 10.1016/j.eplepsyres.2014.09.019 [DOI] [PubMed] [Google Scholar]
- 29.Slattery ML, Trivellas A, Pellatt AJ, et al. Genetic variants in the TGFbeta-signaling pathway influence expression of miRNAs in colon and rectal normal mucosa and tumor tissue. Oncotarget. 2017;8(10):16765–16783. doi: 10.18632/oncotarget.14508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher RS, Cross JH, French JA, et al. Operational classification of seizure types by the international league against epilepsy: position paper of the ILAE commission for classification and terminology. Epilepsia. 2017;58(4):522–530. doi: 10.1111/epi.13670 [DOI] [PubMed] [Google Scholar]
- 31.Kwan P, Arzimanoglou A, Berg AT, et al. Definition of drug resistant epilepsy: consensus proposal by the ad hoc Task Force of the ILAE Commission on Therapeutic Strategies. Epilepsia. 2010;51(6):1069–1077. doi: 10.1111/j.1528-1167.2009.02397.x [DOI] [PubMed] [Google Scholar]
- 32.Han X, Wang C, Tang D, Shi Y, Gao M. Association of genetic polymorphisms in chromosome 9p21 with risk of ischemic stroke. Cytokine. 2020;127:154921. doi: 10.1016/j.cyto.2019.154921 [DOI] [PubMed] [Google Scholar]
- 33.Wang S, Huang M, Wang Z, et al. MicroRNA133b targets TGFbeta receptor I to inhibit TGFbetainduced epithelialtomesenchymal transition and metastasis by suppressing the TGFbeta/SMAD pathway in breast cancer. Int J Oncol. 2019;55(5):1097–1109. doi: 10.3892/ijo.2019.4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bitarafan S, Yari M, Broumand MA, et al. Association of increased levels of lncRNA H19 in PBMCs with risk of coronary artery disease. Cell J. 2019;20(4):564–568. doi: 10.22074/cellj.2019.5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 36.Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19(3):376–382. doi: 10.1093/bioinformatics/btf869 [DOI] [PubMed] [Google Scholar]
- 37.Vezzani A, French J, Bartfai T, Baram TZ. The role of inflammation in epilepsy. Nat Rev Neurol. 2011;7(1):31–40. doi: 10.1038/nrneurol.2010.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eftekhari S, Mehrabi S, Karimzadeh F, et al. Brain derived neurotrophic factor modification of epileptiform burst discharges in a temporal lobe epilepsy model. Basic Clin Neurosci. 2016;7(2):115–120. doi: 10.15412/J.BCN.03070205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng Y, Zhou L, Yao J, et al. Associations of lncRNA H19 polymorphisms at microrna binding sites with glioma susceptibility and prognosis. Mol Ther Nucleic Acids. 2020;20:86–96. doi: 10.1016/j.omtn.2020.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Sima X. Replication of GWAS loci revealed an increased risk of BET1L and H19 polymorphisms with intracranial aneurysm. Dis Markers. 2019;2019:9490639. doi: 10.1155/2019/9490639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu R, Liu X, He Z. Long non-coding RNA H19 and MALAT1 gene variants in patients with ischemic stroke in a northern Chinese Han population. Mol Brain. 2018;11(1):58. doi: 10.1186/s13041-018-0402-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang J, Yang J, Li J, et al. Association of long noncoding RNA H19 polymorphisms with the susceptibility and clinical features of ischemic stroke in southern Chinese Han population. Metab Brain Dis. 2019;34(4):1011–1021. doi: 10.1007/s11011-019-00417-0 [DOI] [PubMed] [Google Scholar]