Abstract
The new Coronavirus disease 2019 (COVID-19) is rapidly affecting the world population with statistics quickly falling out of date. Due to the limited availability of annotated Coronavirus X-ray and CT images, the detection of COVID-19 remains the biggest challenge in diagnosing this disease. This paper provides a promising solution by proposing a COVID-19 detection system based on deep learning. The proposed deep learning modalities are based on convolutional neural network (CNN) and convolutional long short-term memory (ConvLSTM). Two different datasets are adopted for the simulation of the proposed modalities. The first dataset includes a set of CT images, while the second dataset includes a set of X-ray images. Both of these datasets consist of two categories: COVID-19 and normal. In addition, COVID-19 and pneumonia image categories are classified in order to validate the proposed modalities. The proposed deep learning modalities are tested on both X-ray and CT images as well as a combined dataset that includes both types of images. They achieved an accuracy of 100% and an F1 score of 100% in some cases. The simulation results reveal that the proposed deep learning modalities can be considered and adopted for quick COVID-19 screening.
Keywords: Deep learning, COVID-19, Coronavirus, Analysis, Medical images, Convolutional neural networks
Introduction
Coronaviruses are a large group of viruses known to cause comorbidities that range from colds to more severe diseases, such as middle east respiratory syndrome (MERS) and severe acute respiratory syndrome (SARS). A new Coronavirus has emerged in 2019, in Wuhan, China. This virus is a new strain that has not been previously identified in humans [1]. The infection is transmitted by droplets when a person contacts someone with respiratory symptoms (such as coughing or sneezing) with close contact (within a distance of 1 m), which makes this person at risk of exposing his mucous membranes (mouth and nose) or conjunctiva (eye) to potential respiratory droplets. To be contagious, the infection may also be transmitted by contaminated devices found in the environment surrounding the infected person [8]. Therefore, infection with the virus that causes COVID-19 disease can be transmitted either by direct contact with infected individuals or indirect contact with surfaces in the surrounding environment, or tools used by the infected person (such as a stethoscope or thermometer). According to Medical News today [9], as the COVID-19 pandemic continues to claim victims around the world, the need for early diagnosis of infections is increasing. Also, the limited image data sources with expert labeled data for COVID-19 detection are insufficient. In addition, manual detection of COVID-19 is time consuming. Several medical image processing techniques have been involved in several applications in recent years [2–7]. Therefore, many computer-aided diagnostic (CAD) systems that depend on by automated artificial intelligence (AI) methods have been designed to detect and differentiate COVID-19 [2, 10–15].
At present, recent advances in machine learning, particularly data-driven deep learning (DL) methods using CNNs and ConvLSTM have shown promising performance in identifying, classifying, and quantifying disease patterns in medical images [3, 16–21], especially in the detection of COVID-19 [11, 12]. For instance, Rajaraman et al. [10] presented a deep learning algorithm using a CNN for COVID‐19 detection. They carried out their algorithm on a type of X-ray images called CXR images. They obtained accuracies of 0.5555 and 0.6536 in classifying Twitter and Montreal COVID‐19 CXR test data, respectively. Asif and Wenhui [11] developed a method for automatic prediction of COVID-19 using a deep convolutional neural network (DCNN) based on Inception V3 model and chest X-ray images. They obtained more than 96% accuracy for the detection of COVID-19. Moura et al. [12] presented a deep learning method for the classification of COVID-19, pneumonia and healthy chest X-ray radiographs. They obtained the best average accuracy for validation as 0.9725. He et al. [22] presented a deep transfer learning method for COVID-19 diagnosis based on CT scans. They achieved an F1 score of 0.85 and an AUC of 0.94 in diagnosing COVID-19 from CT scans. Jelodar et al. [23] introduced a method for automated detection of COVID-19 using the LSTM recurrent neural network approach. Also, Yan et al. [24] presented an LSTM-based COVID-19 confirmed cases prediction model. However, most of the previous deep learning approaches have the problem of data irregularity, which usually results in mis-calibration between classes in the dataset. Also, most of these studies worked on a limited number of images [7, 25], which achieves low accuracy for detection of COVID-19. In addition, these studies have overfitting problems and regularization errors. The work in [26] presented a deep learning approach for COVID-19 detection. This approach is based on a CNN and a ConvLSTM. The presented deep learning models are tested on both X-ray and CT biomedical images. The main idea in this work is to investigate the effect of performing several data augmentation techniques such as rotation, flipping and resizing as well as data augmentation based on conditional generative adversarial networks (CGANs). This work achieved an accuracy of 99% for X-ray images and 96% for CT images.
In this paper, we present an effective and robust solution for the detection of COVID-19 cases from small data based on CNN and ConvLSTM. Moreover, the proposed method reduces the overfitting and regularization errors through the utilization of an effective augmentation technique for COVID-19 recognition. The main contributions of this paper are as follows:
We propose novel and efficient modalities for the detection of COVID-19 based on CNN and ConvLSTM.
We introduce a novel dataset, which is a combination of CT and X-ray images in normal and COVID-19 cases.
We get a high accuracy of classification on small data based on CNN and ConvLSTM.
We present an effective augmentation technique for COVID-19 detection to reduce overfitting and regularization errors.
The rest of this paper is structured as follows. Section 2 describes in detail the proposed modalities. Section 3 presents experimentation, dataset and performance evaluation of the proposed modalities. Section 4 gives a discussion of the results. Section 5 gives the concluding remarks and the future work.
Deep learning modalities for augmented detection of COVID-19
This paper presents a diagnosis system for COVID-19 disease with deep learning. The proposed deep learning modalities are based on CNN and ConvLSTM. The objective is to design deep learning modalities that can extract feature maps from the input images and enroll these feature maps into a classification network to discriminate between the normal and abnormal states. The performance of these modalities is evaluated by the capability of discrimination of the normal and abnormal states with low false-alarm rates. So, the main contribution is to design an efficient deep learning architecture. This architecture consists of a hierarchy of convolutional layers, pooling layers, and ConvLSTM layers. In addition, the classification network handles the feature maps generated from the deep learning architecture to obtain a decision whether the input images are normal or not.
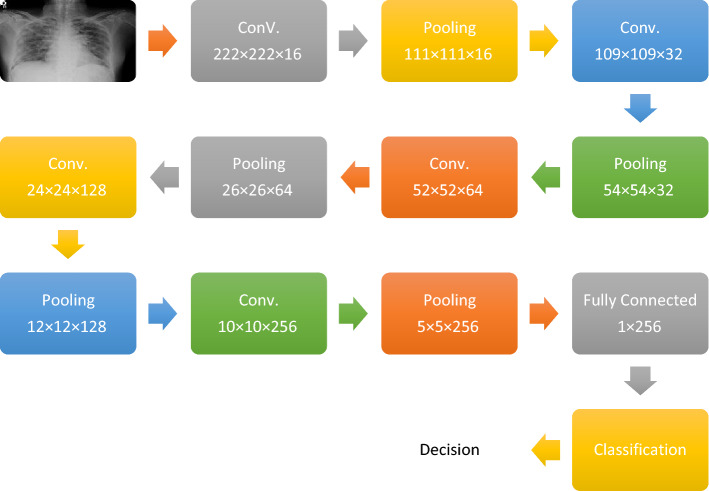
The first proposed deep learning modality is based on CNN. This modality consists of five convolutional (CNV) layers followed by five pooling (PL) layers. This hierarchy is implemented to extract features from the input images and generate feature maps that are enrolled to the classification network. Each CNV layer in the deep learning architecture generates a feature map whose depth equals the number of digital filters included in this layer. In addition, the PL layer is implemented for feature reduction. It can be implemented with two methodologies (max pooling and mean pooling). The feature map is segmented into square windows with a certain size. The max pooling technique is based on extracting the maximum value from each window, while the mean pooling extracts the mean value of each window. Furthermore, the classification network consists of two layers: fully-connected layer and classification layer. The fully-connected layer handles the feature map, which is generated from the hierarchy of CNV and PL layers. This layer converts the 3D feature map into a feature vector that is enrolled into the classification layer that determines whether the input image belongs to normal or abnormal classes. Figure 1 shows the proposed CNN model.
Fig. 1.
Architecture of proposed CNN modality
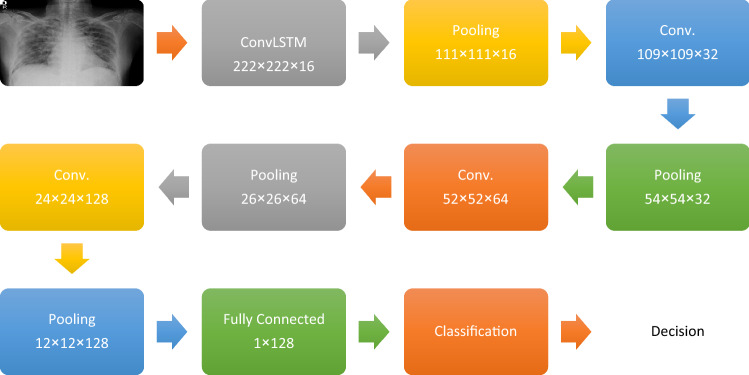
Another deep learning modality is proposed in this paper. It is a hybrid one that contains both ConvLSTM and CNN modalities. The ConvLSTM can be considered as the 2D version of the LSTM. The main idea of the LSTM is to remember the previous states and construct the current state. This modality is a double-edged technique, because the current state depends on the whole previous states. So, the drop in such a state would lead to a damaging performance in the next states. Hence, this type of deep learning modalities needs to be handled carefully and observed in the training phase to correct any disorder that may occur. This deep learning modality consists of ten layers. The ConvLSTM layer is followed by a PL layer. Then, a set of three CNV layers followed by three PL layers is used. The classification network is the same as in the first deep learning modality. It can be observed that this model consists of less layers than those of the first deep learning modality based on CNNs. This modality is designed to reduce the complexity of the designed deep learning structure (Fig. 2).
Fig. 2.
Architecture of hybrid ConvLSTM-CNN modality
Both the first and second proposed modalities consist of three phases (training, validation, and testing). The training needs an optimization methodology to update the values of weights. Adam optimizer is used to minimize the cross-entropy loss function [27–30].
Backpropagation algorithm is implemented to minimize the error between the real and estimated targets. This error is minimized based on the cross-entropy loss function. This type of loss function mainly depends on the difference between the exponential log of both real and estimated targets, as shown in Eq. 1.
Experimentation, dataset and performance evaluation
This section describes the datasets used in our study and gives a discussion of the experimentation, datasets and performance evaluation of the proposed modalities on these datasets. The proposed deep learning modalities are implemented on both CT and X-ray images. In addition, they are implemented on an augmented dataset. The proposed modalities are implemented and evaluated by the accuracy, F1 score and MCC. In addition, the sensitivity, specificity, PPV and NPV are considered in the evaluation process. These metrics help in indicating whether the true infection case is estimated as an infection or misclassified as a normal case. These metrics may be useful for technicians.
Dataset description
The proposed deep learning modalities are tested on three datasets. The first dataset includes CT images [31]. This dataset consists of 288 COVID-19 images and 288 normal images. This dataset is augmented by several rotations and scaling operations. This augmentation process generates 2880 COVID-19 and 2880 normal images. The second dataset includes X-ray images [32]. This dataset consists of two different augmented subsets. Each subset consists of 304 COVID-19 images and 304 normal images.
The third dataset is the COVID-19 radiography dataset [33]. This dataset includes X-ray images. In addition, the COVID-19 radiography dataset consists of three categories: COVID-19, normal and viral pneumonia. The proposed modalities are tested on this dataset to discriminate between the cases of COVID-19 and viral pneumonia. Furthermore, the proposed modalities are tested on a combined dataset [34] including the first and second datasets in order to combine both CT and X-ray images in normal and COVID-19 cases. Table 1 shows a description of each dataset.
Table 1.
Brief description of the datasets
| Dataset | Imaging technology | Target categories | No. of COVID-19 images | No. of Non-COVID images | Splitting ratios |
|---|---|---|---|---|---|
| COVID CT scans [31] | CT scan | COVID-19 and Normal | 288 | 288 | 70–30 |
| Augmented COVID dataset [32] | X-ray | COVID-19 and Normal | 304 | 304 | 70–30 |
| X-ray | COVID-19 and Normal | 304 | 304 | 70–30 | |
| COVID-19 radiography [33] | X-ray | COVID-19 and Pneumonia | 912 | 1345 | 70–30 |
| Combined dataset [34] | CT scan and X-ray | COVID-19 and Normal | 3065 | 3065 | 70–30 |
Results on CT dataset
The proposed deep learning modalities are tested on CT images. The dataset is split into 70% for training and 30% for testing. In addition, in the training phase, the dataset is fragmented into batches, where the batch size is 20. Furthermore, the number of training epochs is 40.
CNN modality
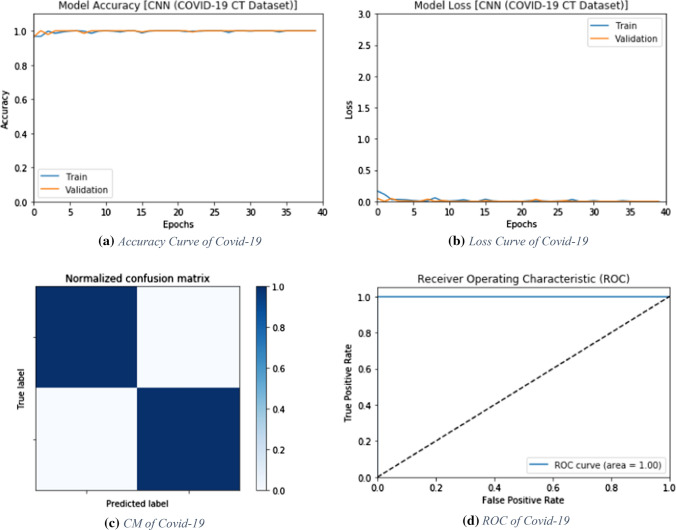
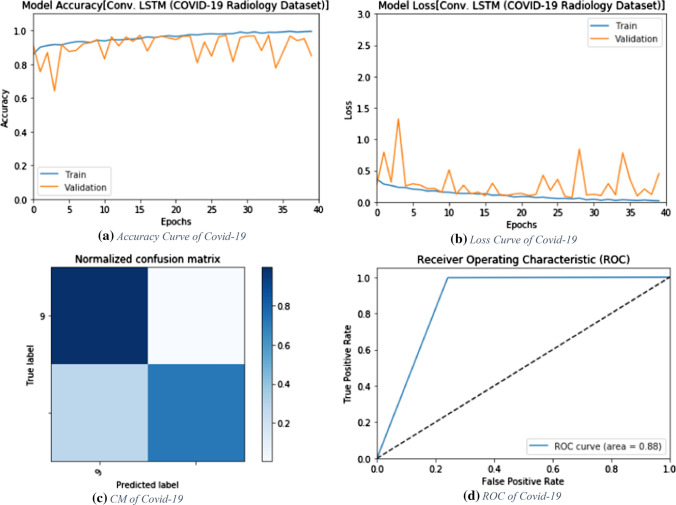
The proposed CNN deep learning modality is tested on the CT images for COVID-19 detection. The simulation results are evaluated by accuracy, F1 score and MCC. Figure 3 shows the evaluation metrics of the proposed CNN modality. In addition, Fig. 4a and b show the accuracy and loss curves of both training and validation phases. Furthermore, Fig. 4c and d show the confusion matrix and the ROC curve for the testing phase.
Fig. 3.
Evaluation metrics of the proposed CNN modality on CT images
Fig. 4.
Accuracy, loss, confusion matrix and ROC curve for the proposed CNN modality on CT images
ConvLSTM modality
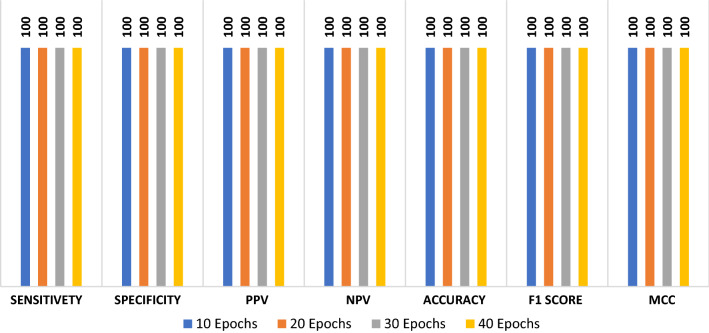
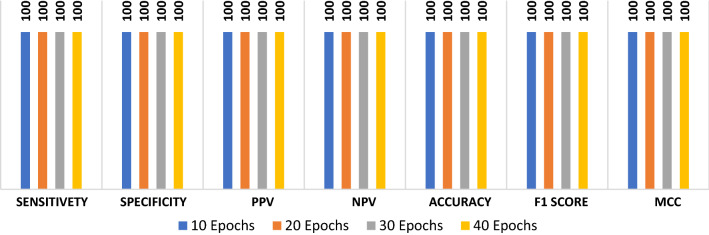
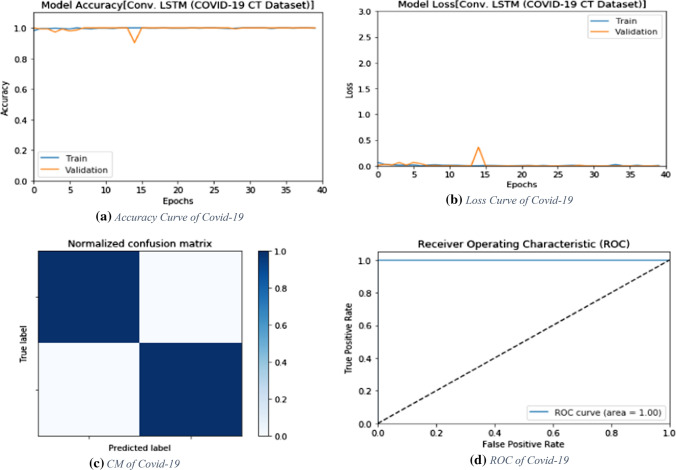
The proposed ConvLSTM deep learning modality is tested on the CT dataset. Like the CNN modality, it is evaluated with accuracy of detection, F1 score and MCC. Figure 5 shows the values of evaluation metrics at different numbers of epochs. The proposed modality achieves an accuracy of 100%. This is attributed to the nature of the CT images and the efficient design of the deep learning modality for sufficient feature extraction. In addition, Fig. 6a and b show the accuracy and loss curves of the training and validation data, while Fig. 6c and d show the confusion matrix and ROC curve of the testing data.
Fig. 5.
Evaluation metrics of the ConvLSTM modality
Fig. 6.
Accuracy, loss, confusion matrix and ROC curve for the proposed ConvLSTM modality on CT images
Results on augmented datasets
Both of the proposed deep learning modalities are tested on the augmented COVID-19 dataset. This dataset includes different X-ray chest images. The augmentation process is performed by rotations with several rotation angles. This dataset includes two categories: COVID-19 and normal. The target of the proposed modalities is to discriminate between COVID-19 and normal cases.
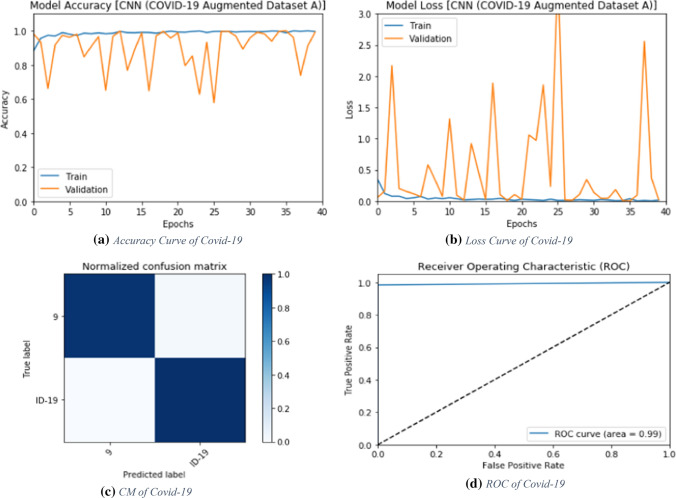
Results on augmented dataset A
The proposed deep learning modalities are tested on X-ray images, which are included in the first augmented dataset (Dataset A). Figures 7 and 9 show the evaluation metrics for the CNN and the ConvLSTM modalities, respectively. In addition, Figs. 9, 10, 13, and 14 show the accuracy and loss curves for these modalities. Furthermore, Figs. 11, 12, 15, and 16 show the confusion matrix and ROC curves.
Fig. 7.
Evaluation metrics of the proposed CNN modality on augmented dataset A
Fig. 9.
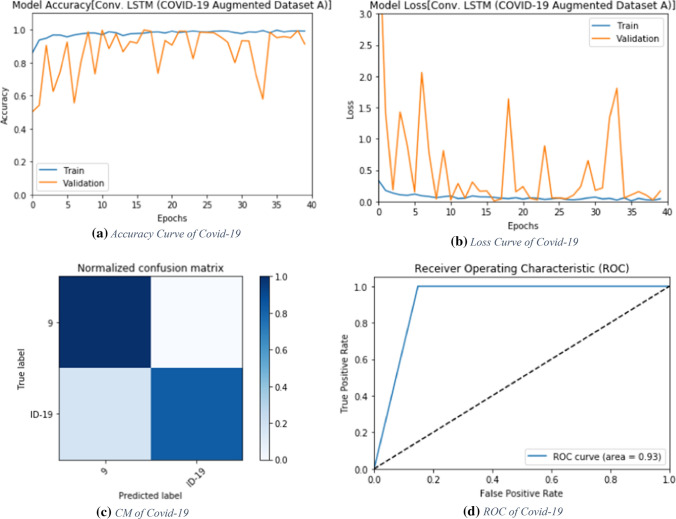
Evaluation metrics of the proposed ConvLSTM modality for augmented dataset A
Fig. 10.
Accuracy, loss, confusion matrix and ROC curve for the proposed ConvLSTM modality for augmented dataset A
Fig. 13.
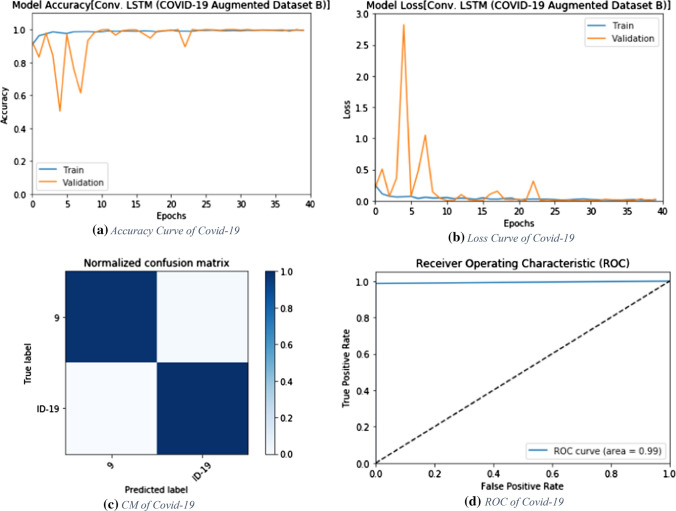
Evaluation metrics of the proposed ConvLSTM modality on the augmented dataset B
Fig. 14.
Accuracy, loss, confusion matrix and ROC curve for the proposed ConvLSTM modality on the augmented dataset B
Fig. 11.
Evaluation metrics of the proposed CNN modality for augmented dataset B
Fig. 12.
Accuracy, loss, confusion matrix and ROC curve for the proposed CNN modality for augmented dataset B
Fig. 15.
Evaluation of the proposed CNN modality on the combined dataset
Fig. 16.
Accuracy, loss, confusion matrix and ROC curve for the proposed CNN modality on the combined dataset
CNN modality
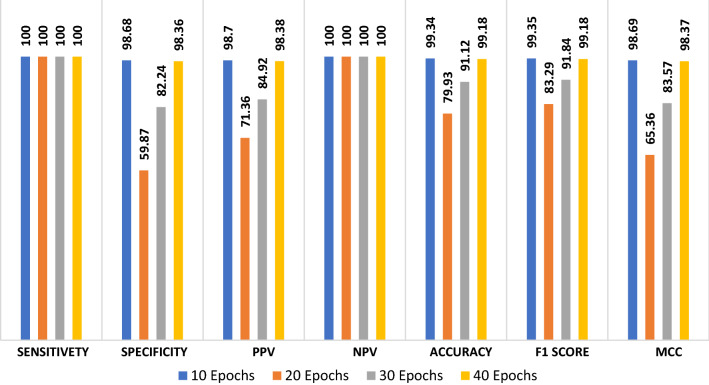
The proposed CNN modality is tested on the first X-ray augmented dataset. Figure 7 shows the evaluation metrics of the proposed CNN modality with different numbers of epochs. The simulation results reveal that the proposed CNN modality achieves an accuracy of 99.18%, an F1 score of 99.18%, and MCC of 98.37%. Figure 8a and b show the accuracy and loss curves of the training and validation data. In addition, Figs. 11 and 12 show the confusion matrix and ROC curve for the testing data. The testing accuracy is 99%.
Fig. 8.
Accuracy, loss, confusion matrix and ROC curve for the proposed ConvLSTM modality on CT images
ConvLSTM modality
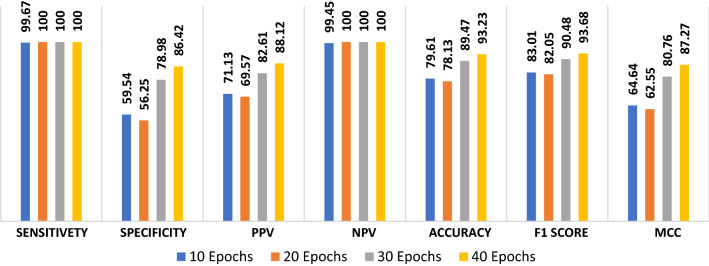
The proposed ConvLSTM modality is tested on the X-ray dataset. Figure 9 shows the values of the evaluation metrics with different numbers of epochs. The proposed ConvLSTM modality achieves an accuracy of 93.23 with 40 epochs. In addition, the proposed modality achieves an F1 score of 93.68% and MCC of 87.27%. Figure 10a and b show the accuracy and loss curves of training and validation data. Furthermore, Fig. 10c and d illustrate the confusion matrix and ROC curve of the proposed modality.
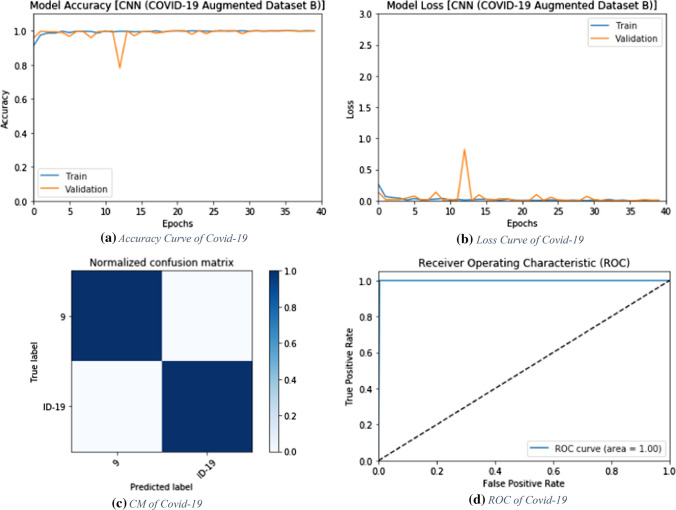
Results on augmented dataset B
The proposed deep learning modalities are tested on the second augmented dataset (Augmented Dataset B) including X-ray images. Both the CNN and ConvLSTM modalities are tested on 608 X-ray images. This dataset is split into 304 COVID-19 images and 304 normal images.
CNN modality
The proposed CNN modality is tested on the second augmented dataset. Figure 11 shows the values of the evaluation metrics of the proposed CNN modality with different numbers of epochs. The simulation results reveal that the proposed CNN modality achieves an accuracy of 100% with 20, 30 and 40 epochs. Figure 12a and b show the accuracy and loss curves of the training and validation data. In addition, Fig. 12c and d show the confusion matrix and ROC curve of the testing data. The testing accuracy of the proposed modality is 100%.
ConvLSTM modality
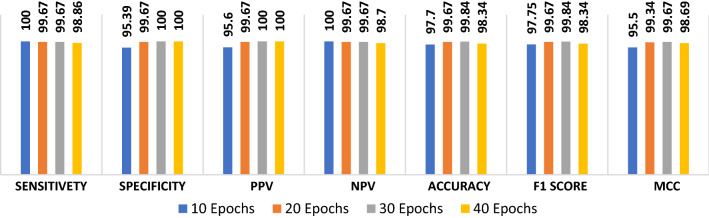
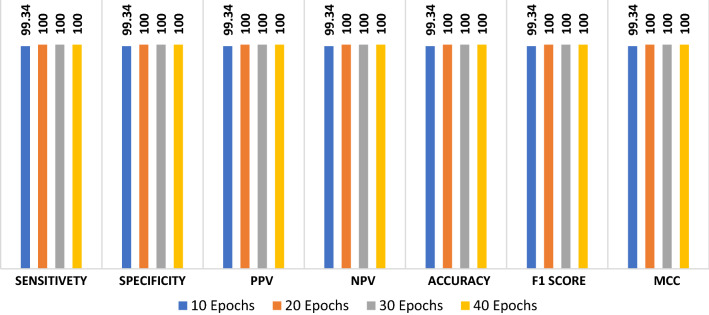
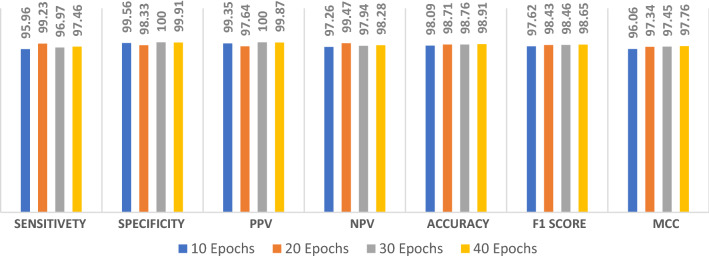
The proposed ConvLSTM modality is tested on the second augmented dataset. Figure 13 shows the simulation results with different numbers of epochs. The simulation results reveal that the proposed ConvLSTM modality achieves accuracies of 97.7%, 99.67%, 99.84% and 98.34% with of 10, 20, 30 and 40 epochs, respectively. In addition, Fig. 14a and b show the accuracy and loss curves of the training and validation data. Furthermore, Fig. 14b and d show the confusion matrix and ROC curve of the testing data. The proposed ConvLSTM modality achieves a testing accuracy of 99%.
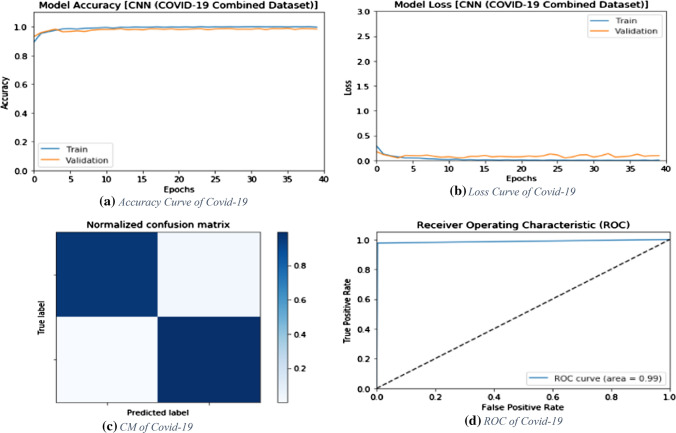
Results of the combined dataset
Another scenario that is proposed in this paper is based on CT and X-Ray images. The proposed deep learning modalities are tested on a combined dataset including both CT and X-Ray images.
CNN modality
The proposed CNN modality is tested on the combined dataset. Figure 15 shows the simulation results of the proposed CNN modality with different numbers of epochs. The simulation results reveal that the proposed CNN modality achieves an accuracy of 98.97% with 40 epochs, while it achieves an F1 score of 98.65%. In addition, the proposed modality achieves an MCC of 97.76%. Figure 16a and b show the accuracy and loss curves of the training and validation data, while Fig. 16c and d show the confusion matrix and ROC curve of the proposed CNN modality. The proposed CNN modality achieves a testing accuracy of 99%.
ConvLSTM modality
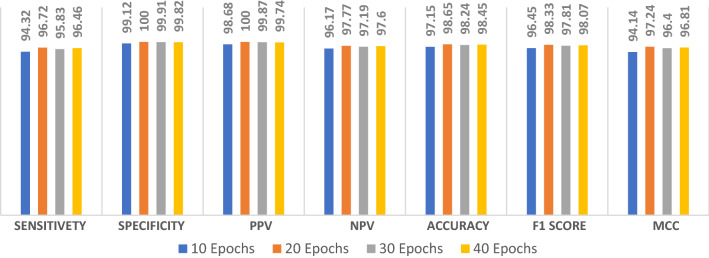
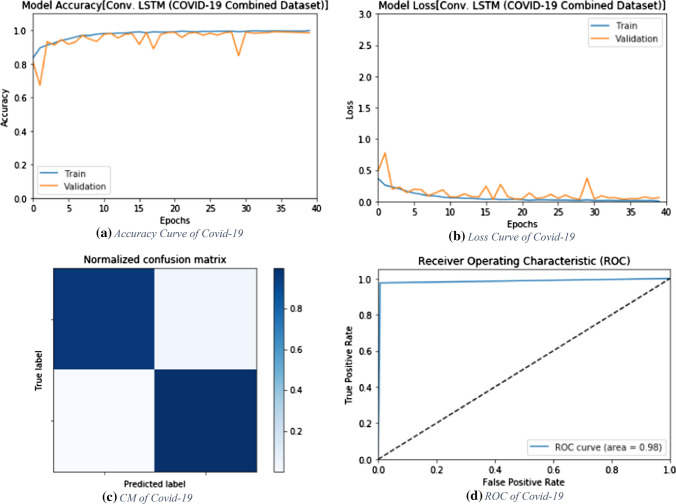
The proposed ConvLSTM modality is tested on the combined dataset. Figure 17 shows the simulation results of the proposed ConvLSTM modality with different numbers of epochs. The simulation results reveal that the proposed ConvLSTM modality achieves an accuracy of 98.45% at epoch 40, while it achieved an F1 score of 98.07%. In addition, the proposed modality achieves an MCC of 96.81%. Figure 18a and b show the accuracy and loss curves of the training and validation data, while Fig. 18c and d show the confusion matrix and ROC curve of the proposed ConvLSTM modality. The proposed ConvLSTM modality achieves a testing accuracy of 98%.
Fig. 17.
Evaluation metrics of the proposed ConvLSTM modality on the combined dataset
Fig. 18.
Accuracy, loss, confusion matrix and ROC curve for the proposed ConvLSTM modality on the combined dataset
Results on COVID-19 radiography dataset
This paper presents a different scenario of COVID-19 detection. The detection process is based on discrimination between COVID-19 and pneumonia diseases. This scenario is a challenging one, because it is required to discriminate between two diseases with high similarity in features instead of normal and abnormal states like the previous scenarios. Both of the proposed modalities are tested on the radiography dataset to discriminate between viral pneumonia and COVID-19 X-Ray images.
CNN modality
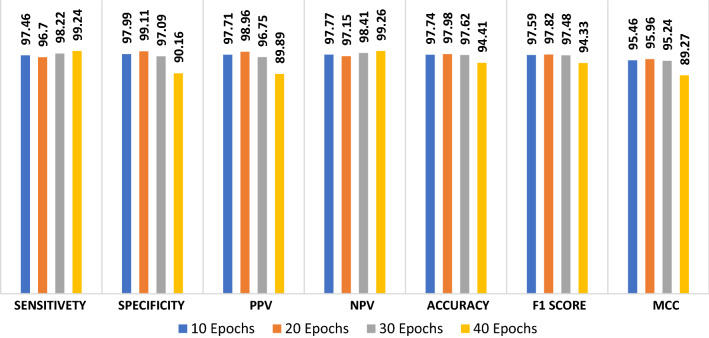
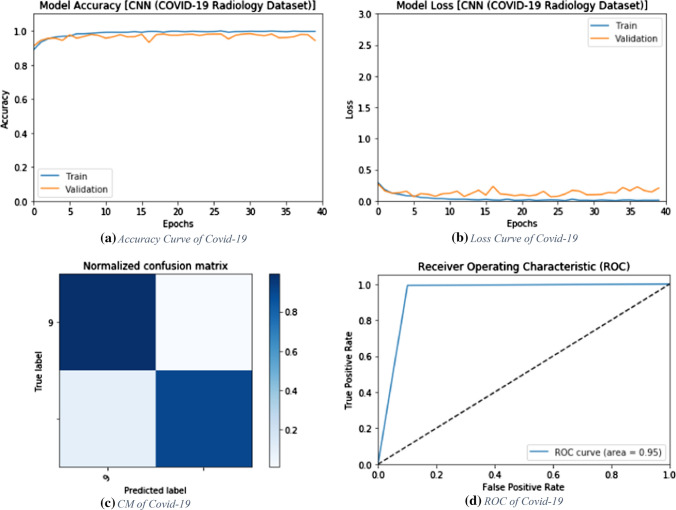
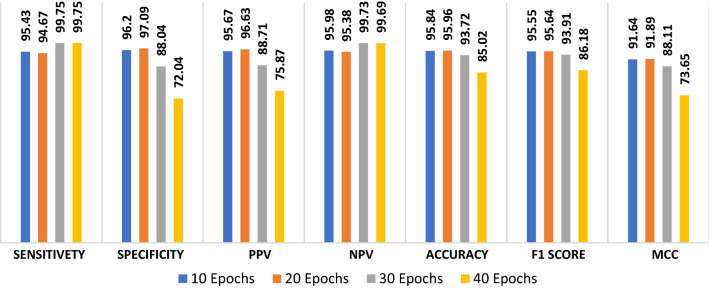
The proposed CNN modality is tested on the radiography dataset in order to discriminate between viral pneumonia and COVID-19 states. Figure 19 shows the simulation results of the proposed modality with a number of epochs of 10, 20, 30, and 40. The simulation results show that the proposed CNN modality achieves an accuracy of 97.74%, 97.98%, 97.62%, and 94.41% with the number of epochs of 10, 20, 30, and 40, respectively. In addition, the proposed modality achieves an F1 score of 97.59%, 97.82%, 97.48%, and 94.33% for the number of epochs of 10, 20, 30, and 40, respectively. Furthermore, it achieves an MCC of 95.46%, 95.46%, 95.96% and 89.27% with 10, 20, 30 and 40 epochs, respectively. Figure 20a and b show the accuracy and loss curves of the training and validation data. In addition, Fig. 20c and d show the confusion matrix and ROC curve of the testing data. The proposed modality achieves a testing accuracy of 95%.
Fig. 19.
Evaluation metrics of the proposed CNN modality on radiography dataset
Fig. 20.
Accuracy, loss, confusion matrix and ROC curve for the proposed CNN modality on radiography dataset
ConvLSTM modality
The proposed ConvLSTM modality is tested on the radiography dataset in order to discriminate between viral pneumonia and COVID-19 states. Figure 21 shows the simulation results of the proposed modality at a number of epochs of 10, 20, 30, and 40. The simulation results show that the proposed ConvLSTM modality achieves an accuracy of 95.84%, 95.96%, 93.72%, and 85.02.41% at the number of epochs of 10, 20, 30 and 40, respectively. In addition, the proposed modality achieves an F1 score of 95.55%, 95.64%, 93.91%, and 86.18% for the number of epochs of 10, 20, 30, and 40, respectively. Furthermore, it achieves an MCC of 91.64%, 91.89%, 88.11% and 73.65% for number of epochs of 10, 20, 30 and 40, respectively. Figure 22a and b show the accuracy and loss curves of the training and validation data, while Fig. 22c and d show the confusion matrix and ROC curve of the testing data. The proposed modality achieves a testing accuracy of 88%.
Fig. 21.
Evaluation metrics of the proposed ConvLSTM modality on radiography dataset
Fig. 22.
Accuracy, loss, confusion matrix and ROC curve for the proposed ConvLSTM model for radiography dataset
Results and discussion
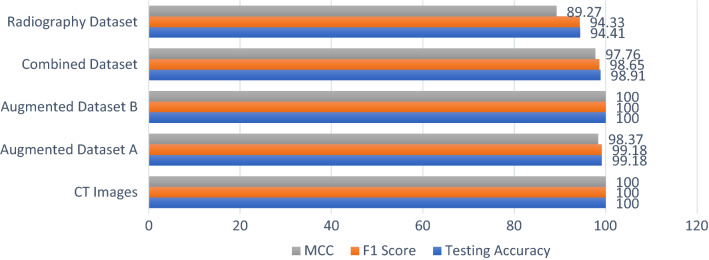
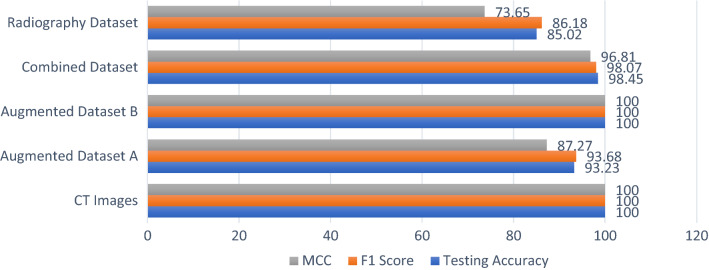
This paper includes a study of a COVID-19 diagnosis system using deep learning modalities. The proposed deep learning modalities are based on CNN and ConvLSTM. In addition, the proposed modalities are tested on both CT and X-ray images of COVID-19 and normal cases. The proposed CNN modality achieves accuracies of 100%, 99,18%, 100%, 98.91%, and 94.33% for CT images, augmented dataset (A), augmented dataset (B), combined dataset, and COVID-19 radiography dataset, respectively. On the other hand, the ConvLSTM modality achieved accuracies of 100%, 93.23%, 100%, 98.45%, and 85.02% for CT images, augmented dataset (A), augmented dataset (B), combined dataset and COVID-19 radiography dataset, respectively. Furthermore, F1 score and MCC evaluation metrics are calculated in order to present a fair evaluation of the proposed deep learning modalities. Although the proposed modalities achieve high performance in some cases, they achieve poor performance in others, especially for the ConvLSTM modality. The reason for this poor performance is the dependency on the previous states in the structure of the ConvLSTM modality. Generally, the proposed modalities reveal high performance in the detection of COVID-19. So, the proposed modalities can be considered in an efficient diagnosis system for COVID-19. Figure 23 shows the evaluation metrics of the proposed CNN modality for all datasets, while Fig. 24 shows the evaluation metrics of the proposed ConvLSTM modality.
Fig. 23.
Evaluation metrics of the training phase for CNN model
Fig. 24.
Evaluation metrics of the training phase for ConvLSTM modality
In addition, the proposed deep learning modalities are evaluated in the testing stage using the area under ROC curve (AUC). The proposed CNN modality achieves testing accuracies of 100%, 99%, 100%, 999% and 95% for CT images, augmented dataset (A), augmented dataset (B), combined dataset and COVID-19 radiography dataset, respectively. On the other hand, the ConvLSTM modality achieves testing accuracies of 100%, 93%, 99%, 98% and 88% for CT images, augmented dataset (A), augmented dataset (B), combined dataset and COVID-19 radiography dataset, respectively.
Table 2 shows a brief comparison between the proposed modalities and the work presented in the literature. This comparison is performed in two directions. The first direction depends on the works, which were tested on X-ray images. In this direction, the proposed deep learning modalities achieve an accuracy of 99%, while the previous works achieved a range of 95 to 98%. On the other hand, another direction is considered for the comparison between the proposed modalities and the previous works, which were tested on CT images. The works in literature achieved an accuracy of 83% to 90.1%, while the proposed modalities achieve an accuracy of 99%. So, the proposed deep learning modalities can be considered in an efficient diagnosis systems for the detection of COVID-19.
Table 2.
Brief comparison between the proposed modalities and the traditional works
| Work | Year | Data type | Modality | Accuracy (%) |
|---|---|---|---|---|
| [11] | 2020 | X-ray | Deep CNN | 96 |
| [12] | 2020 | X-ray | CNN | 98 |
| [16] | 2020 | X-ray | DeTraC Deep Learning CNN | 95.12 |
| [17] | 2020 | X-ray | ResNet | 96.23 |
| [18] | 2020 | X-ray | Deep Learning Feature Extraction + Machine Learning | 98 |
| Proposed Modalities | 2020 | X-ray | CNN | 99 |
| 2020 | X-ray | ConvLSTM | 99 | |
| [22] | 2020 | CT | Pre-trained Deep Learning Modality | 83 |
| [35] | 2020 | CT | DeCovNet | 90.1 |
| Proposed Modalities | 2020 | CT | CNN | 99 |
| 2020 | CT | ConvLSTM | 99 |
Conclusion and future work
This paper presented an effective and robust solution for the detection of the COVID-19 cases based on two efficient deep learning modalities: CNN and ConvLSTM. Two different augmentation techniques have been utilized for COVID-19 detection in order to reduce overfitting and regularization errors. We introduced a novel dataset, which is a combination of CT and X-ray images with normal and COVID-19 cases. The proposed modalities have been tested on both CT and X-ray images of COVID-19 and normal cases. The proposed CNN modality achieved a high classification performance with accuracies of 100%, 99,18%, 100%, 98.91% and 94.33% for CT images, augmented dataset (A), augmented dataset (B), combined dataset and COVID-19 radiography dataset, respectively. On the other hand, the ConvLSTM model achieved accuracies of 100%, 93.23%, 100%, 98.45% and 85.02% for CT images, augmented dataset (A), augmented dataset (B), combined dataset and COVID-19 radiography dataset, respectively. Hence, the proposed modalities can be considered in an efficient diagnosis system for the detection of COVID-19 and other relevant infections.
Acknowledgements
The authors are acknowledged the support of Menoufia University.
Author’s contribution
All authors contributed equally.
Compliance with ethical standards
Conflict of interest
None.
Ethics approval
Confirm.
Consent to participate
Confirm.
Consent for publication
Confirm.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Coronavirus Disease (COVID-2019) (2020) Situation reports, [Online]. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. Accessed 15 Oct 2020
- 2.Dorgham O, Al-Rahamneh B, Almomani A, Khatatneh KF. Enhancing the security of exchanging and storing DICOM medical images on the cloud. Int J Cloud Appl Comput IJCAC. 2018;8(1):154–172. [Google Scholar]
- 3.Gudivada A, Philips J, Tabrizi N. Developing concept enriched models for big data processing within the medical domain. Int J Softw Sci Comput Intell IJSSCI. 2020;12(3):55–71. doi: 10.4018/IJSSCI.2020070105. [DOI] [Google Scholar]
- 4.Qureshi B. An affordable hybrid cloud based cluster for secure health informatics research. Int J Cloud Appl Comput IJCAC. 2018;8(2):27–46. [Google Scholar]
- 5.Sarivougioukas J, Vagelatos A. Modeling deep learning neural networks with denotational mathematics in UbiHealth environment. Int J Softw Sci Comput Intell IJSSCI. 2020;12(3):14–27. doi: 10.4018/IJSSCI.2020070102. [DOI] [Google Scholar]
- 6.Goléa NEH, Melkemi KE. ROI-based fragile watermarking for medical image tamper detection. Int J High Perform Comput Netw. 2019;13(2):199–210. doi: 10.1504/IJHPCN.2019.097508. [DOI] [Google Scholar]
- 7.Ghoneim A, Muhammad G, Amin SU, Gupta B. Medical image forgery detection for smart healthcare. IEEE Commun Mag. 2018;56(4):33–37. doi: 10.1109/MCOM.2018.1700817. [DOI] [Google Scholar]
- 8.Bai X, et al. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. 2020 doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raoofi A, Takian A, Sari AA, Olyaeemanesh A, Haghighi H, Aarabi M. COVID-19 pandemic and comparative health policy learning in Iran. Arch Iran Med. 2020;23(4):220–234. doi: 10.34172/aim.2020.02. [DOI] [PubMed] [Google Scholar]
- 10.Rajaraman S, Antani SK (2020) Training deep learning algorithms with weakly labeled pneumonia chest X-ray data for COVID-19 detection. medRxiv
- 11.Asif S, Wenhui Y (2020) Automatic detection of COVID-19 using X-ray images with deep convolutional neural networks and machine learning. medRxiv
- 12.de Moura J, Novo J, Ortega M (2020) Fully automatic deep convolutional approaches for the analysis of COVID-19 using chest X-ray images. medRxiv [DOI] [PMC free article] [PubMed]
- 13.Naseer A, Rani M, Naz S, Razzak MI, Imran M, Xu G. Refining Parkinson’s neurological disorder identification through deep transfer learning. Neural Comput Appl. 2020;32(3):839–854. doi: 10.1007/s00521-019-04069-0. [DOI] [Google Scholar]
- 14.Razzak MI, Imran M, Xu G. Big data analytics for preventive medicine. Neural Comput Appl. 2020;32(9):4417–4451. doi: 10.1007/s00521-019-04095-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li D, Deng L, et al. A novel CNN based security guaranteed image watermarking generation scenario for smart city applications. Inf Sci. 2019;479:432–447. doi: 10.1016/j.ins.2018.02.060. [DOI] [Google Scholar]
- 16.Abbas A, Abdelsamea MM, Gaber MM (2020) Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. arXiv preprint arXiv:2003.13815 [DOI] [PMC free article] [PubMed]
- 17.Farooq M, Hafeez A (2020) COVID-resnet: a deep learning framework for screening of COVID-19 from radiographs. arXiv preprint arXiv:2003.14395
- 18.Hall LO, Paul R, Goldgof DB, Goldgof GM (2020) Finding COVID-19 from chest x-rays using deep learning on a small dataset. arXiv preprint arXiv:2004.02060
- 19.Gozes O, Frid-Adar M, Sagie N, Zhang H, Ji W, Greenspan H (2020) Coronavirus detection and analysis on chest ct with deep learning. arXiv preprint arXiv:2004.02640
- 20.AlZu’bi S, Shehab M, Al-Ayyoub M, Jararweh Y, et al. Parallel implementation for 3d medical volume fuzzy segmentation. Pattern Recogn Lett. 2020;130:312–318. doi: 10.1016/j.patrec.2018.07.026. [DOI] [Google Scholar]
- 21.Wang H, Li Z, Li Y, Gupta BB, Choi C. Visual saliency guided complex image retrieval. Pattern Recogn Lett. 2020;130:64–72. doi: 10.1016/j.patrec.2018.08.010. [DOI] [Google Scholar]
- 22.He X, Yang X, Zhang S, Zhao J, Zhang Y, Xing E, Xie P (2020) Sample-efficient deep learning for COVID-19 diagnosis based on CT scans. medRxiv
- 23.Jelodar H, Wang Y, Orji R, Huang H (2020) Deep Sentiment classification and topic discovery on novel coronavirus or COVID-19 online discussions: NLP using LSTM recurrent neural network approach. arXiv preprint arXiv:2004.11695 [DOI] [PubMed]
- 24.Yan B, Tang X, Liu B, Wang J, Zhou Y, Zheng G, Zou Q, Lu Y, Tu W (2020) An improved method of COVID-19 case fitting and prediction based on LSTM. arXiv preprint arXiv:2005.03446
- 25.Wang L, Li L, Li J, Li J, Gupta BB, Liu X. Compressive sensing of medical images with confidentially homomorphic aggregations. IEEE Internet Things J. 2018;6(2):1402–1409. doi: 10.1109/JIOT.2018.2844727. [DOI] [Google Scholar]
- 26.Sedik A, Iliyasu AM, El-Rahiem A, Abdel Samea ME, Abdel-Raheem A, Hammad M, Peng J, El-Samie A, Fathi E, El-Latif AAA. Deploying machine and deep learning models for efficient data-augmented detection of COVID-19 infections. Viruses. 2020;12(7):769. doi: 10.3390/v12070769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z (2018) Improved adam optimizer for deep neural networks. In: 2018 IEEE/ACM 26th international symposium on quality of service (IWQoS), pp 1–2. IEEE
- 28.El-Ashkar AM, Sedik A, Shendy H, Taha TES, El-Fishawy AS, El-Nabi A, Khalaf AA, El-Banby GH, El-Samie A, Fathi E. Classification of reconstructed SAR images based on convolutional neural network. Menoufia J Electron Eng Res. 2019;28(ICEEM2019-Special Issue):122–125. doi: 10.21608/mjeer.2019.76897. [DOI] [Google Scholar]
- 29.Al-Azrak FM, Sedik A, Dessowky MI, El Banby GM, Khalaf AA, Elkorany AS, El-Samie FEA. An efficient method for image forgery detection based on trigonometric transforms and deep learning. Multimed Tools Appl. 2020;79:1–23. doi: 10.1007/s11042-019-7523-6. [DOI] [Google Scholar]
- 30.Abd El-Rahiem B, Sedik A, El Banby GM, Ibrahem HM, Amin M, Song OY, Khalaf AAM, Abd El-Samie FE (2020) An efficient deep learning model for classification of thermal face images. J Enterp Inf Manag
- 31.COVID-CT-Dataset (2020) A CT scan dataset about COVID-19. https://github.com/UCSD-AI4H/COVID-CT. Accessed 15 April 2020
- 32.Alqudah AM, Qazan S (2020) Augmented COVID-19 x-ray images dataset, vol 4. 10.17632/2FXZ4PX6D8.4
- 33.https://www.kaggle.com/tawsifurrahman/covid19-radiography-database. Accessed 15 Oct 2020
- 34.Sedik A, El-Samie A, Fathi E, El-Latif A, Ahmed A, Hammad M. Combined COVID-19 dataset. Mendeley Data. 2020 doi: 10.17632/3pxjb8knp7.3. [DOI] [Google Scholar]
- 35.Zheng C, Deng X, Fu Q, Zhou Q, Feng J, Ma H, Liu W, Wang X (2020) Deep learning-based detection for COVID-19 from chest CT using weak label. medRxiv
- 36.Ghoshal B, Tucker A (2020) Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. arXiv preprint arXiv:2003.10769
- 37.Alom MZ, Rahman MM, Nasrin MS, Taha TM, Asari VK (2020) COVID_MTNet: COVID-19 detection with multi-task deep learning approaches. arXiv preprint arXiv:2004.03747
- 38.Amrani M, Hammad M, Jiang F, Wang K, Amrani A. Very deep feature extraction and fusion for arrhythmias detection. Neural Comput Appl. 2018;30(7):2047–2057. doi: 10.1007/s00521-018-3616-9. [DOI] [Google Scholar]
- 39.Alghamdi A, Hammad M, Ugail H, Abdel-Raheem A, Muhammad K, Khalifa HS, Abd El-Latif AA (2020) Detection of myocardial infarction based on novel deep transfer learning methods for urban healthcare in smart cities. Multimed Tools Appl 1–22
- 40.Khatami A, Nazari A, Khosravi A, Lim CP, Nahavandi S. A weight perturbation-based regularisation technique for convolutional neural networks and the application in medical imaging. Expert Syst Appl. 2020;149:113196. doi: 10.1016/j.eswa.2020.113196. [DOI] [Google Scholar]
- 41.Hajabdollahi M, Esfandiarpoor R, Sabeti E, Karimi N, Soroushmehr SMR, Samavi S. Multiple abnormality detection for automatic medical image diagnosis using bifurcated convolutional neural network. Biomed Sig Process Control. 2020;57:101792. doi: 10.1016/j.bspc.2019.101792. [DOI] [Google Scholar]