Abstract
The adipokine leptin regulates energy homeostasis through ubiquitously expressed leptin receptors. Leptin has a number of major signaling targets in the brain, including cells of the anterior pituitary (AP). We have previously reported that mice lacking leptin receptors in AP somatotropes display growth hormone (GH) deficiency, metabolic dysfunction, and adult-onset obesity. Among other targets, leptin signaling promotes increased levels of the pituitary transcription factor POU1F1, which in turn regulates the specification of somatotrope, lactotrope, and thyrotrope cell lineages within the AP. Leptin’s mechanism of action on somatotropes is sex dependent, with females demonstrating posttranscriptional control of Pou1f1 messenger RNA (mRNA) translation. Here, we report that the stem cell marker and mRNA translational control protein, Musashi1, exerts repression of the Pou1f1 mRNA. In female somatotropes, Msi1 mRNA and protein levels are increased in the mouse model that lacks leptin signaling (Gh-CRE Lepr-null), coincident with lack of POU1f1 protein, despite normal levels of Pou1f1 mRNA. Single-cell RNA sequencing of pituitary cells from control female animals indicates that both Msi1 and Pou1f1 mRNAs are expressed in Gh-expressing somatotropes, and immunocytochemistry confirms that Musashi1 protein is present in the somatotrope cell population. We demonstrate that Musashi interacts directly with the Pou1f1 mRNA 3′ untranslated region and exerts translational repression of a Pou1f1 mRNA translation reporter in a leptin-sensitive manner. Musashi immunoprecipitation from whole pituitary reveals coassociated Pou1f1 mRNA. These findings suggest a mechanism in which leptin stimulation is required to reverse Musashi-mediated Pou1f1 mRNA translational control to coordinate AP somatotrope function with metabolic status.
Keywords: MSI1, MSI2, POU1F1, somatotrope, pituitary, plasticity
The anterior pituitary (AP) coordinates metabolism, growth, homeostasis, and reproduction through controlled secretion of signaling hormones from distinct AP cell populations: somatotropes, growth hormone (GH); lactotropes, prolactin (PRL); thyrotropes, thyroid stimulating hormone (TSH); gonadotropes, luteinizing hormone (LH), and follicle-stimulating hormone (FSH); and corticotropes, adrenocorticotropin (ACTH). The AP demonstrates a high level of cell population plasticity to meet changing metabolic and reproductive needs (1, 2). The mechanisms controlling AP cell plasticity are unknown but may parallel those used during pituitary embryonic development (3). During pituitary development, Sox2-expressing stem cells arise early (mouse embryonic day 9.5) followed by the appearance of cells expressing the Prop1 transcription factor (embryonic day 11.5), which is the earliest marker of pituitary tissue identity (4, 5). Prop1 activates expression of the Pou1f1 transcription factor that is required for maturation of the somatotrope, thyrotrope, and lactotrope AP cell populations (6). AP cell plasticity in the adult tissue may be mediated through a similar maturation mechanism in response to metabolic signals.
The adipokine leptin is produced in proportion to available fat stores and signals via leptin receptors (LEPRs) to most tissues, including the pituitary (7-11). We have previously shown that loss of LEPR signaling in somatotropes (Gh-CRE Lepr-null) results in GH deficiency, subfertility, and a disrupted metabolic phenotype including adult-onset obesity, thus establishing a requirement for leptin in the maturation of somatotrope function (12-15). Notably, female somatotropes from these mutant mice had significant reductions in POU1F1 protein levels, suggesting that leptin signaling impinges on pituitary cell maturation (14, 16). Moreover, the reduced levels of POU1F1 protein in the female Lepr-null somatotrope model occurred in the absence of reduced levels of Pou1f1 messenger RNA (mRNA), suggesting posttranscriptional control of Pou1f1 mRNA (16). Here, we have investigated the mechanism governing leptin-dependent control of somatotrope Pou1f1 mRNA translation in females. An in silico analysis of the regulatory 3′ untranslated region (UTR) of the Pou1f1 mRNA identified multiple binding sites for the stem/progenitor cell fate regulatory protein, Musashi. The Musashi family of RNA binding proteins (Musashi1 and Musashi2) bind to target mRNAs in a sequence-specific manner and are thought to promote stem and progenitor cell self-renewal and to oppose their differentiation (17). Using RNA binding and RNA reporter assays, we determined that Musashi can directly bind to, and repress translation of, the Pou1f1 mRNA in a leptin-sensitive manner. Use of single-cell RNA sequencing (scRNAseq) unexpectedly revealed expression of both isoforms (Msi1 and Msi2 mRNA) in a significant proportion of the nonstem/progenitor cell populations in adult pituitary, including the Pou1f1 mRNA-expressing somatotrope, lactotrope, and thyrotrope lineages. Immunolabeling confirmed that Musashi proteins are present within all AP hormone-producing cell lineages including somatotropes. Immunoprecipitation (IP) of Musashi1 from the pituitary revealed coassociated Pou1f1 mRNA in vivo. These findings indicate that the stem/progenitor cell regulatory protein Musashi can exert translational control over the Pou1f1 mRNA to coordinate somatotrope function with metabolic status.
Materials and Methods
Animals
All animals belonged to the sighted FVB.129P hybrid strain, from Jackson Laboratories (FVB.129P2‐Pde6b+ Tyrc‐ch/Ant) and are referred to as FVB mice. Development of our somatotrope Lepr Exon 1-null line (rGhp-Cre; Lepr exon 1–/–) has been described previously (13) as has the introduction of the Cre-reporter tdTomato-eGFP transgene (Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo) used for purification of somatotropes from Cre-bearing controls and mutants by fluorescence-activated cell sorting (FACS) (16). Primers and genotyping for floxed Lepr (Lepr exon 1LoxP/LoxP) are described in our recent study (13). The primers and genotyping protocol for floxed tdTomato-eGFP are found at the Jackson Laboratories website (18). Control, female mice used for scRNAseq come from a line of mice bearing 2 alleles each of floxed Msi1 and floxed Msi2 (19) (Msi1LoxP/LoxP; Msi2LoxP/LoxP), but do not express the Cre recombinase. In addition, some experiments used the sighted FVB.129P hybrid mice that carried no transgenes as controls. All animals were humanely killed before 9 am. The use of animals was approved by the University of Arkansas for Medical Sciences Animal Care and Use Committee.
Single-cell RNA sequencing
Pituitaries from 8-week-old, control, diestrous FVB females bearing 2 alleles each of floxed MSI1 and MSI2 (n = 6) were collected and individually dispersed as previously described (15). Following mechanical dispersion, cells were pelleted and washed once with Dulbecco’s Modified Eagle’s Medium (DMEM). Cells were pelleted again and fixed with methanol according to the 10x Genomics methanol fixation protocol (20). The morning of sample submission, cells were rehydrated and pooled into 2 pools of 3 pituitaries each. Each pool was then counted and submitted for processing to the Genomics Core at the Arkansas Children’s Hospital Center for Translational Pediatric Research. Single-cell 3′ library generation was performed using a 10x Genomics Chromium Controller and the v3 reagent kit (10x Genomics) according to the manufacturer’s protocol. Cell suspensions were loaded onto the middle 4 channels of a chromium single-cell A chip according to the manufacturer’s instructions, aiming for 10 000 cells per channel. Following generation of single-cell gel bead-in-emulsions, reverse transcription, fragmentation, and complementary DNA (cDNA) amplification, library preparations and bar coding were performed using the Illumina TruSeq Stranded mRNA Sample Preparation Kit v2 as per the manufacturer’s instructions. Libraries were validated with a fragment analyzer for fragment size and quantified by use of a Qubit fluorometer. A total of 200 pM of each library was pooled for sequencing on the Illumina NextSeq 500 platform using a high output flow cell (100 cycles) to generate approximately 25 million 75 base reads per sample. 10x Genomics Cell Ranger 3.1.0 mkfastq wrapper and was used to perform sample demultiplexing and generate fastq files.
Bioinformatic analyses
Demultiplexed fastq files were analyzed with the 10x Genomics Cell Ranger 3.1.0 count function for sequence alignment and gene counting. The reads were aligned to a custom reference using STAR and transcript counts were generated (21, 22). The University of California, Santa Cruz (UCSC) mm10 reference mouse transcriptome was amended by annotating chromosome 11: 50 948 572-50 949 192 in the gene transfer file as an artificial single exon gene on the negative strand named “Prop1L” as described by Cheung et al (23). A custom reference was then compiled using the cellranger mkref command, and the custom reference was used for Cell Ranger/STAR alignment instead of the original UCSC mm10 transcriptome. Seurat (24) was used to preprocess the raw counts generated by cellranger count to filter cells with low-quality cells, cell doublets, and cells with a high percentage of mitochondrial genes. We filtered cells that have unique feature counts over more than the 75th percentile plus 1.5 times the interquartile range or less than the 25th percentile minus 1.5 times the interquartile range and cells with mitochondrial counts outside the same range with respect to mitochondrial gene percentage. Counts were normalized using the LogNormalize method, and the 2000 highest variable features were selected and data were scaled by linear transformation and principal component analysis performed on the scaled data. A JackStraw procedure determined the significant principal component analysis components that have a strong enrichment of low P value features. A graph-based clustering approach embedded cells in a graph structure by gene expression patterns and into interconnected “quasi-cliques” or “communities” with Uniform Manifold Approximation and Projection (UMAP) (25), which visually groups cells with similar gene expression signatures. Seurat FindNeighbors and FindClusters functions were optimized to label clusters based on the visual clustering in the projections and cell types assigned (23, 26). The top 10 genes expressed in each cell type cluster are presented in Supplementary Table 1 (27), and share considerable overlap with the genes identified as enriched in the same pituitary cell types from the scRNAseq data sets of Ho et al (28). Differential expression analysis was performed using MAST, a generalized linear model framework that treats cellular detection rate as a covariate (29). Data were evaluated from each pool separately and revealed a high level of reproducibility between the samples, and the data from each pool (9230 and 9071 cells, respectively) were normalized for read depth and aggregated into one data set containing 18 301 cells. The aggregate UMAP plot was very similar to the individual UMAP plots, indicating that there was very limited biological or technical variability between the duplicate sample pools. After clustering, we found highly expressed hormone genes (Gh, Prl, and Pomc) could be detected in blood cells. This most likely arose as a consequence of cell lysis during cell preparation, resulting in a cross contamination of the released mRNAs. This issue is a common problem for scRNAseq protocols (28, 30). Since these hormone genes would not be expected to be expressed in blood cells, they represent background noise signals in the analysis. Consequently, we employed an expression cutoff threshold for the UMAP plots in Fig. 1 and dot plot (Fig. 2B) to remove the background cross-contamination signal.
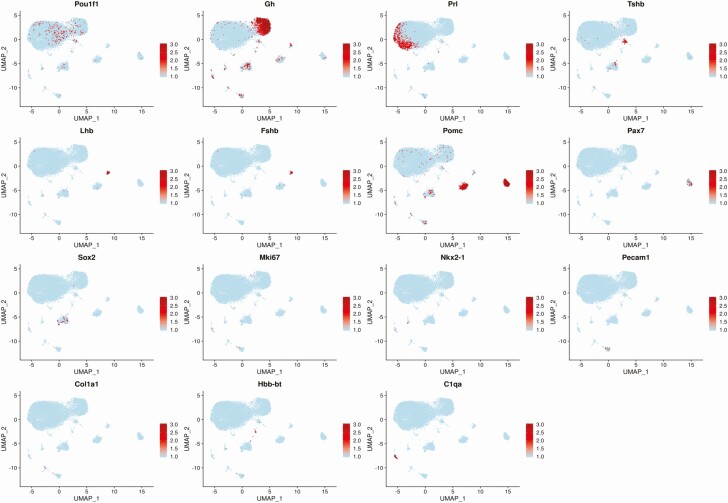
Figure 1.
Pituitary endocrine and cell type markers detected by single-cell RNA sequencing (scRNAseq). The normalized expression from the 85th percentile to the maximum of each gene is indicated by a gradient of light to dark red. Expression below the 85th percentile for a given gene is displayed as light blue. Pou1f1, Gh, Prl, Tshb, Lhb, Fshb, Pomc, and Pax7 are hormonal cell markers, whereas Sox2 marks stem cells. Nkx2-1, Pecam1, Co1a1, Hbb-bt, and C1qa are markers of nonhormonal cell types, and Mki67 is a marker of cell proliferation (see main text).
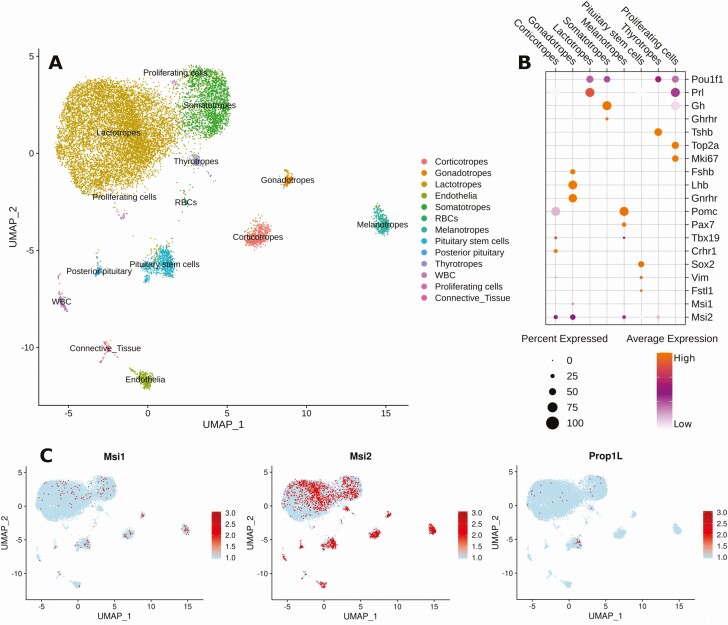
Figure 2.
Expression of Prop1, Msi1, and Msi2 within the pituitary cell populations. A, Cell type clusters were manually assigned based on the expression of known marker gene set within 13 original clusters determined by shared nearest neighbor modularity optimization. Cells in each cluster are marked with the same color. B, Dot plot of normalized gene expression level of representative markers of each cluster. C, Expression of Msi1, Msi2, and Prop1L genes are displayed by a gradient of light to dark red from minimum to maximum normalized expression for each gene. A light blue cell indicates zero transcripts for the gene were detected in the cell.
Cell dispersion and fluorescence-activated cell sorting
Pituitary cells were dispersed into single cells using the previously published protocol (16), resuspended in FACS running buffer (PBS containing 15 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethane sulfonic acid], 1 mM EDTA, and 1% bovine serum albumin [BSA]) and stored on ice. Prior to sorting, cells were redispersed using 26-gauge syringe and filtered to remove cell clumps (BD Falcon, 352235). The pituitary cells were sorted with a FACSAria machine (BD Biosciences) into 5-mL polystyrene tubes (coated with 4% BSA in 1 × PBS) containing 200 μL of DMEM + ITS supplement (insulin, transferrin, and sodium selenite; Sigma). Following sorting, cells were stored on ice until centrifugation at 1400 revolutions per minute for 20 minutes at 4 °C. The buffer was removed and samples stored until processed for protein or RNA. For studies of responses to leptin, a subset of purified somatotrope fractions were stimulated at 37 °C immediately following sorting with mild agitation for 3 hours with vehicle or 10 nM leptin (Sigma L3772), followed by protein extraction and enzyme immunoassay (EIA) analysis for MSI1.
Pituisphere culture
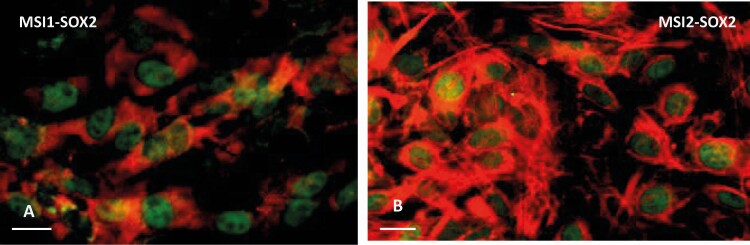
Pituitary stem cell–dependent, floating colonies (pituispheres) were prepared from dispersed whole control female pituitaries (FVB.129P) as previously described (31). After growth as floating colonies on low-adherent multiwell plates for 7 days, pituispheres were fixed and dual-labeled for immunocytochemistry. The immunolabeling involved the use of 1:1000 rabbit anti-SOX2 (green fluorescence, Alexa-Fluor 488 conjugate anti Sox2-Millipore AB5603A4; RRID: AB_11205731 [32]) and goat anti-Musashi1 (RRID: AB_2147926 [33]) or anti-Musashi2 (RRID: AB_2147942 [34]). The 2 primary antibodies were added together for 30 minutes and after washing, horse anti-immunoglobulin G (IgG)-linked to Dylight 594 (RRID: AB_2336777 (35)) was added to detect the Musashi by red fluorescence.
Immunocytochemistry
Dispersed pituitary cells from control FVB.129P mice were fixed in 4% paraformaldehyde as previously described (16). The cells were then immunolabeled for GH and Musashi1 or Musashi2 with the use of rabbit antibody to GH (Hormone Distribution Program, AF Parlow; RRID AB_2629219 [36]). The full validation of this antibody has been published and described in detail in a recent publication (37). We used the antibody at 1:80 000 to 1:100 000. The Musashi1 and Musashi2 antibodies were made in goats and purchased from R&D Systems (AF3255 and AF2628). They were diluted to 1:5000 to 1:10 000 and validated by absorption controls in which 100 ng/mL Musashi1 or Musashi2 peptides (ABCAM, ab178003; ab136164) neutralized labeling with the anti-Musashi1 or anti-Musashi2, respectively. The 2 diluted primary antibodies were added together and cells were incubated for 30 minutes at 37 °C. After washing in Tris buffer, the cells were incubated in horse antirabbit IgG linked to Dylight 594 (red) (RRID: AB_2336414 [38]) and horse antigoat IgG linked to Dylight 488 (green) (RRID: AB_2336776 [39]). GH-expressing cells were thus labeled with red fluorescence and anti-Musashi labeling used green fluorescence. We mounted the cells on coverslips in 4′,6-diamidino-2-phenylindole (DAPI)-containing mounting media so the nuclei were labeled blue.
For the detection of MSI1 or MSI2 expression by corticotropes, thyrotropes, and lactotropes, direct dual immunolabeling was applied to freshly plated mixed cultures of whole pituitary cells on coverslips from diestrous female mice. The cells had been fixed in 4% paraformaldehyde for 30 minutes, followed by 4 washes in PBS. The protocol involved 3 washes in 0.05 M Tris buffer, pH 7.6 followed by 5 minutes in 0.3% Triton X and another 3 Tris buffer washes. The cells were then treated with diluent buffer containing 10% normal horse serum and 0.1% BSA for 15 minutes and then were exposed to a solution containing diluted primary antibody conjugates for 30 minutes at 37 °C in a hybridization incubator with gentle rotation. After this exposure, the cells were washed with Tris buffer, mounted on slides in Vectashield vibrance antifade mounting media with DAPI (Vector Laboratories), and viewed in a fluorescence microscope.
The antibodies used were as follows: Thyrotropes were immunolabeled with 1:400 to 1:500 polyclonal rabbit antimouse TSHB antibody conjugated to Cy3 produced against amino acids 21 to 138 with recombinant TSHb (Phe21-Val138) (LSBio-LS C703313 RRID:AB_2884004 [40]). Prolactin cells were immunolabeled with 1:400 to 1:500 polyclonal rabbit antimouse PRL antibody conjugated to Cy3, which was made against mouse prolactin (LSBio-LS-C699468; RRID:AB_2884005 [41]). Corticotropes were immunolabeled with 1:2000 to 1:10 000 polyclonal rabbit antimouse proopiomelanocortin (POMC) conjugated to Cy3, which was made against amino acids 27 to 235 with recombinant POMC (Trp27-Gln235) as immunogen (LSBio-LS-C700398. RRID:AB_2884003 [42]). Each cell type was labeled with Cy3 or orange-red fluorescence with its respective antibody. Specificity tests for anti-TSHB or PRL involved Western blots (LS-Bio) and reduction of immunolabeling following liquid-phase immunoabsorption with 2 to 4 µg of antigen used to make the antibody for 24 hours at 4 °C before application. Specificity tests for Pomc involved Western blot analysis (LS-Bio) and immunoabsorption tests.
For dual labeling, we added anti-MSI1 or anti MSI2 to the diluted antihormone antibodies so they could be applied in one step. These antibodies were conjugated to a green fluorescent fluorophore. MSI1 was detected by 1:500 to 1:1000 polyclonal goat antihuman MSI1 antibody conjugated to Alexa Fluor 488 (R&D Systems AF2628, RRID:AB_2147926 [33]). Its specificity had been tested in pituitary cells and tumor cell lines, as previously reported (43) and by Western blots (Novus Biologicals). MSI2 was detected by 1:500 to 1:1000 polyclonal rabbit anti MSI2 antibody conjugated to DyLight 488 (Novus Biologicals, RRID:AB_2884006 [44]). Specificity was ascertained by Western blots (Novus Biologicals) and neutralization of immunolabeling following the addition of 1 to 10 µg/mL of MSI2 to the antiserum.
Musashi1 enzyme immunoassay
Following FACS, purified somatotropes were lysed in radioimmunoprecipitation assay buffer (RIPA; Sigma Aldrich, R0278) with 10 mL/mL protease inhibitor cocktail (Fisher Scientific, PI78425) on ice. The extracts were then centrifuged at 4 °C at 14 000 revolutions per minute for 20 minutes. Supernatant was removed and stored at –20 °C. Somatotrope protein extracts were assayed for Musashi1 protein content using an enzyme-linked immunosorbent assay (MyBiosource.com; MBS9353445; RRID: AB_2864367 [45]) and normalized to total protein levels (DC Protein Assay Kit, BioRad, 5000112).
Pou1f1 3′ untranslated region cloning
Polymerase chain reaction (PCR) primers were designed to amplify the entire Pou1f1 3′ UTR (RefSeq Accession NM_008849), which contains 8 consensus Musashi binding elements (MBEs) from murine whole pituitary poly[A]+ RNA preparations after 3′ RNA ligation to an oligonucleotide primer P1 and first-strand cDNA synthesis using a primer complementary to P1 (46). The Pou1f1 3′ UTR was amplified using primers to add a 5′ Nhe1 (fwd: 5′-GCGGCTAGCGGCTGGCGTGTGATAGCCATGTGTGG-3′) and a 3′ Sal1 (rev: 5′-GCGGTCGACGTTTATAGATTCATTTATTTTGTTTCATTTTATTTTATGGG-3′) and subcloned into Nhe1/Sal1-digested pmiRGLO (Promega). The resulting 552-base pair (bp) 3′ UTR was sequenced and found to be considerably shorter than the 1030-bp 3′ UTR predicted in the database (NM_008849). The difference in length of the cloned vs predicted Pou1f1 3′ UTR was a consequence of diversity of tetranucleotide microsatellite repeat sequences within the middle region of the Pou1f1 3′ UTR. The predicted 1030-bp 3′ UTR contains 92 perfect copies of the CTTT repeat and 24 copies of the CCTT repeat within this region. By contrast, our cloned 552-bp Pou1f1 3′ UTR contains 18 CTTT repeats and no CCTT repeats. In all other respects, the cloned 3′ UTR matched the database Pou1f1 sequence, including the 8 consensus MBEs. The resultant clone placed the Pou1f1 3′ UTR downstream of the Firefly Luciferase (FLuc) open reading frame and was designated pmiRGLO 552bp Pou1f1 3′ UTR.
Electrophoretic mobility shift assay detection of Musashi binding to the Pou1f1 messenger RNA 3′ untranslated region
The human cDNAs encoding MSI1 and MSI2 were purchased from Open BioSystems. The hMSI1 and hMSI2 open reading frames were subcloned into the pXen2 vector (47) using 5′ Cla1 and 3′ BamH1 PCR primers (hMSI1 fwd: 5′-GCCCATCGATATGGAGACTGACGCGCCCCAG-3′; hMSI1 rev: 5′-GCGGGATCCTCAGTGGTACCCATTGGTGAAGGCTGTGGCAATCAAAGGG-3′; hMSI2 fwd: 5′-GCGATCGATATGGCCCGGGGACTGCCTTACACCATG-3′; hMSI2 rev: 5′-CCGGGATCCTCAATGGTATCCATTTGTAAAGGC-3′). The resulting pXen hMSI1 and pXen hMSI2 constructs encode the Musashi proteins linked with an in-frame N-terminal glutathione S-transferase (GST) epitope tag. Quikchange II (Agilent) PCR mutagenesis was used to mutate alanine 184 to valine in hMSI1 (fwd: 5′-CAAAATGGTGGAATGTAAGAAAGTTCAGCCAAAGGAGGTGATGTCG-3′ and rev: 5′-CGACATCACCTCCTTTGGCTGAACTTTCTTACATTCCACCATTTTG-3′) and separately, alanine 185 to valine in hMSI2 (fwd: 5′-ATGGTAGAATGTAAGAAAGTTCAGCCGAAAGAAGTCATG-3′ and rev: 5′-CATGACTTCTTTCGGCTGAACTTTCTTACATTCTACCAT-3′). To generate constructs expressing just the N-terminal 199 amino acids of hMSI1 and hMSI2, we employed Quikchange mutagenesis to introduce a STOP codon after amino acid 199 and a BamH1 site (hMSI1 fwd: 5′-GGCTCAGCCCGGGGGAGGTGAGGATCCTCTCGAGTCATGCCCTAC-3′ and rev: 5′-GTAGGGCATGACTCGAGAGGATCCTCACCTCCCCCGGGCTGAGCC-3′; hMSI2 fwd: 5′-CCTGGGACAAGAGGCCGGTGAGGATCCGCCCGGGGACTGCCTTAC-3′ and rev: 5′-GTAAGGCAGTCCCCGGGCGGATCCTCACCGGCCTCTTGTCCCAGG-3′). Subsequent BamH1 digestion released the C-terminal fragment, which was discarded, and the remaining plasmid sequence was religated. The resulting constructs, pXen hNMSI1 and pXen hNMSI2, encode just the first 199 amino acids including both RNA recognition motif domains. A similar strategy was employed to generate pXen hNMSI1 A184V and pXen hNMSI2 A185V, where the N-terminal RNA binding domain of each isoform carries either the A184V or A185V mutation. All constructs were sequence verified. Prior to in vitro RNA transcription/translation, the pXen hMSI1-based plasmids were linearized with Pst1, while the pXen hMSI2-based plasmids were linearized with EcoR1.
GST fusion proteins were in vitro transcribed/translated using TNT SP6-coupled Reticulocyte Lysate System (Promega) from pXen1 (47) (for GST), or pXen plasmids encoding wild-type, N-terminal domain or RNA-binding mutant forms of Musashi1 or Musashi2 as described earlier. A 5′ biotin-labeled RNA oligonucleotide probe was synthesized by Integrated DNA Technologies corresponding to 44 nucleotides of the murine Pou1f1 3′ UTR, which contains the 3′ most consensus Musashi-binding element (5′-BioCUAGCCAUGCAAGUGGUGCACAGAUAUACAUGUAGGCAAAACAC-3′), and 80 fmol of labeled probe was incubated with 1 μL of reticulocyte lysate in binding buffer (50 mM Tris pH 7.5, 20 mM KCl, 150 mM NaCl, 2 mM EGTA [ethylene glycol tetraacetic acid], 0.05% NP-40, 6 mM DTT [dithiothreitol], 8U RNase OUT; [48]) in a final volume of 20 μL. The binding reaction was incubated at room temperature for 20 minutes, then 0.5 μL of 200 mg/mL heparin was added (to reduce nonspecific binding) and incubated for a further 20 minutes. A 5-μL volume of the binding reaction was run on a 6% DNA retardation gel (Thermo Fisher Scientific/Invitrogen) and transferred to Biodyne B membranes (Pierce) according to the manufacturer’s instructions. After UV crosslinking, biotinylated RNA was detected using Chemiluminescent Nucleic Acid Detection Module (Pierce) and an AlphaInnotech ChemiImager as previously described (49). An additional 5 μL volume of each binding reaction was processed for Western blotting to assess levels of the expressed tagged proteins in the reticulocyte lysates.
Western blotting
Protein lysates or electrophoretic mobility shift assay (EMSA) binding reaction were added to NuPAGE (Thermo Fisher Scientific/Invitrogen) sample loading buffer and electrophoresed through a 10% NuPAGE gel (Thermo Fisher Scientific/Invitrogen) and transferred to a 0.2-µm pore size nitrocellulose membrane (Protran; Midwest Scientific). The membrane was blocked with 5% nonfat dried milk TBST (20 mM Tris pH 7.5, 150 mM NaCl, 0.05% Tween20) for 60 minutes at room temperature, or overnight at 4 °C and incubated with primary antibody overnight at 4 °C. Filters were washed 3 times for 10 minutes in TBST, incubated with horseradish peroxidase–conjugated secondary antibody, or Protein A HRP-conjugate (No. 12291, Cell Signaling) then washed 3 times for 10 minutes in TBST. Blots were developed using enhanced chemiluminescence in a Fluorchem 8000 Advanced Imager (ProteinSimple). Western blots were quantified using Fluorchem FC2 software. Antibodies used for immunodetection were anti-GST (1:5000, No. SC-138, Santa Cruz; RRID: AB_627677 [50]), anti-Musashi1 (1:1000, No. ab21628, AbCam; RRID: AB_2144988 [51]), anti-Musashi2 (1:000, No. ab50829, AbCam; RRID: AB_880995 [52]), and anti-Tubulin (1:5000, No. ab7291, AbCam; RRID: AB_2241126 [53]), diluted in TBST + 0.5% nonfat milk.
Cell transfection and luciferase assays
NIH3T3 cells (ATCC CRL-1658) were cotransfected with the pmiRGLO 552bp Pou1f1 3′ UTR plasmid along with either Msi1-eGFP, an RNA binding mutant of Msi1 (Msi1-bm-eGFP) or eGFP control plasmids as described previously (54, 55). The Msi1-bm construct is mutated at 3 positions within the first RNA recognition motif and has attenuated target RNA association (56). Expression of the Musashi1-eGFP, Musashi1-bm-eGFP, and eGFP proteins was confirmed by fluorescence microscopy. Where indicated, leptin (Sigma L3772) was added to cell media at a final concentration of 100 nM immediately after cotransfection of the pmiRGLO 552bp Pou1f1 3′ UTR reporter plasmid and the plasmid expressing Msi1-eGFP. Luciferase activity was determined in quadruplicate after 24 hours, using the Dual-Luciferase Reporter Assay System (Promega, E2920) and Turner Biosystems luminometer (Promega) according to the supplier’s protocol. Data are expressed as relative luciferase activity (FLuc/Renilla luciferase) in arbitrary units. The experiments were repeated on 4 separate occasions.
RNA immunoprecipitation of endogenous Musashi1-Pou1f1 messenger RNA complexes and quantitative polymerase chain reaction
Musashi1 was immunoprecipitated from lysate from 6 adult control mice pituitaries equally split between Musashi1 antibody (Abcam, ab52865; RRID: AB_881168 [57]) and control rabbit IgG (Millipore 12-370; RRID: AB_145841 [58]) as per the kit supplier’s protocol (Millipore Magna-RIP kit (Millipore Sigma, 17-700). The protein was degraded, and RNA was precipitated and analyzed by quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) for Pou1f1 mRNA content relative to the housekeeping transcript peptidylprolyl isomerase A/cyclophilin A (Ppia) with the QuantStudio 12k Flex system (Applied Biosystems) (59). Musashi1 immunoprecipitation (IP) was confirmed by Western blot (data not shown). The IP experiments were performed twice, and each qRT-PCR repeated in triplicate.
Statistics
Five or more animals were used for each test, and in vitro tests were repeated at least 3 times. Cell counts and assay values were analyzed with Prism statistical software with analysis of variance, followed by Sidak, Tukey, or Bonferroni post hoc tests, as described previously (13, 16, 60). When 2 groups were compared, a t test was run. P less than .05 was considered significant. The results of the statistical analyses are included in the text of the figure legends.
Results
Musashi is widely expressed in the adult anterior pituitary
To determine whether Musashi is a potential regulator of Pou1f1 mRNA translation in somatotropes, we first needed to elucidate which cells of the adult AP express Musashi mRNA (Msi1 and/or Msi2). To this end, we performed scRNAseq on pooled pituitary samples from 8-week-old, virgin, diestrous female mice. Pituitaries from 3 animals were pooled on 2 separate occasions to generate 2 sample sets for scRNAseq. After sequencing and bioinformatic clustering of cells with similar gene signatures using the 10x Genomics cellular barcodes and unique molecular identifier counts (see “Materials and Methods”), the enrichment of established pituitary endocrine expression markers (Pou1f1, Gh, Prl, Tshb, Lhb, Fshb, Pomc, and Pax7) was examined and used to guide identification of the predominant pituitary cell types (somatotropes, lactotropes, thyrotropes, gonadotropes, corticotropes, and melanotropes). Fig. 1 shows the UMAP feature plots from normalizing the 2 samples to generate an aggregate data set that includes all 6 animals to visualize the indicated cell type markers. The highest levels of Pou1f1 did not occur in cells with the highest levels of Prl expression, Gh expression, or Tshb expression, suggesting an inverse relationship between expression of the Pou1f1 lineage determinant and cells expressing the highest levels of downstream hormone gene expression. A small cluster of Tshb-expressing thyrotropes was observed. Lh- and Fshb-expressing cells showed a high degree of overlap and represent the gonadotrope population. Two major Pomc-expressing cell clusters can be distinguished, representing corticotropes and melanotropes (which showed Pomc/Pax7coexpression). Five percent of the cells in our samples express Sox2, a marker of adult pituitary stem cells (61). As observed by Cheung et al in their characterization of male pituitaries by scRNAseq (23), marker genes for a number of other cell types were also detected in our female whole-pituitary samples: Nkx2.1 (posterior pituitary), Pecam1 (platelets and endothelial cells), Col1a1 (connective tissue), Hbb-bt (red blood cells), and C1qa (white blood cells). Because whole isolated pituitaries were harvested, these additional cell types were expected. We observed very few cells with expression of the cell proliferation marker Mki67, reflecting the mature, postmitotic status of this adult tissue. Notably, the cells that were enriched for Mki67 also expressed Pou1f1 and were uniquely enriched for genes associated with cell division, chromosome organization, and the mitotic cell cycle (see Supplementary Table 1 [27]). They were consequently designated proliferating cells and occupy 2 positions on the UMAP plot (see also Fig. 2A).
For the aggregated data set, we manually assigned cell types based on gene enrichment within individual cells (see Supplementary Table 1 [27] for the top 10 genes in each cluster) with the cells of each cluster designated by a shared color (see Fig. 2A). We separately generated a dot plot (Fig. 2B) to summarize at the population level the proportion of those cells that express or coexpress key genes. Prl-expressing lactotropes and Gh-expressing somatotropes were the most abundant cell types (see Fig. 2A). We found Pou1f1 expression in a significant proportion of somatotropes, lactotropes, and thyrotropes, consistent with its role as a lineage determinant (Fig. 2B). Similarly to Cheung et al (23), we did not identify the separate multihormonal cluster seen by Ho and colleagues (28). However, the proliferating cell cluster (which is shown as 2 locations on the UMAP plot; see Fig. 2A) showed significant coexpression of Pou1f1, Gh, and Prl and so may represent a progenitor pool in our analyses (see Fig. 2B).
Expression of the pituitary progenitor cell marker Prop1 is found predominantly within a subset of the Sox2-expressing stem cell population (Fig. 2C), although some Prop1 expression is observed in more mature cells of the Pou1f1-dependent lineages (somatotropes and lactotropes). Expression of the stem/progenitor cell markers Msi1 and Msi2 was observed in Sox2- and Sox2/Prop1-expressing stem and progenitor cell populations (see Fig. 2C) as expected given the characterized role of Musashi1 and Musashi2 in adult stem and progenitor cells of other tissues (17). Interestingly, Msi1 and Msi2 were also found to be expressed throughout all 5 of the differentiated, hormone-expressing anterior pituitary cell lineages, with Msi2 showing consistently higher expression than Msi1 (see Fig. 2C). We conclude that in addition to stem and progenitor cell expression, Msi1 and Msi2 are also expressed in the more mature, hormone-expressing cell lineages of the adult AP. The expression of Musashi (Msi1 and Msi2) in somatotropes is consistent with it being a potential regulator of Pou1f1 mRNA translation.
Musashi protein is expressed in the somatotrope population
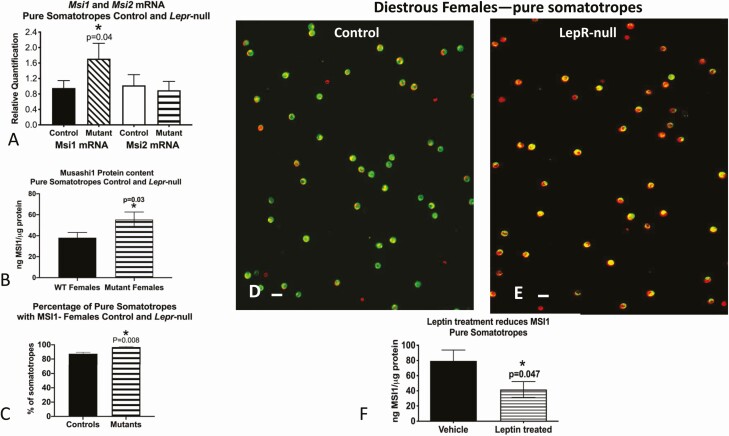
To identify the relationship between Musashi and the AP cell maturation stimulus leptin, we assessed Musashi protein levels and Msi1 and Msi2 mRNA levels in FACS-purified somatotropes from control female mice (littermate controls; see “Materials and Methods”) and from female somatotrope-specific, Lepr-null mice. Msi1 mRNA and Musashi1 protein (MSI1) levels were increased in the purified Lepr-null somatotropes (Fig. 3A and 3B). Musashi1 immunolabeling detected a slight increase in somatotropes expressing Musashi1 proteins in Lepr-null somatotropes from 87% with a range of 83% to 93% of the control fields to 97% with a range of 92% to 99% of the deletion mutant fields (MSI1, Fig. 3C). What is more striking however is the increase in density of labeling for Musashi1 in the Lepr-null mutant somatotropes, although we did not quantify this increase (Fig. 3D and 3E). To directly assess the relationship between leptin signaling and Musashi1 protein levels, somatotropes from control FVB.129P females were stimulated with 100nM leptin for 3 hours and Musashi1 protein levels assessed by EIA, revealing an approximate 50% decrease in somatotrope Musashi1 protein levels (MSI1, Fig. 3F). Taken together, these data confirm Musashi expression in purified adult pituitary somatotropes and suggest that leptin signaling through the LEPR leads to the downregulation of Musashi1 in somatotrope populations.
Figure 3.
Analysis of MSI1 and MSI2 expression in control and somatotrope Lepr-null mice. A, Levels of Msi1 and Msi2 messenger RNA (mRNA); B, levels of MSI1 proteins by enzyme immunoassay (EIA); C, cell counts of somatotropes with MSI1 proteins detected by immunolabeling; D, somatotropes identified by green fluorescence (eGFP-Cre-recombinase reporter) and immunolabeled for MSI1 with red fluorescence (Dylight 594); E, increased MSI1 expression in Lepr-null somatotropes; F, treatment of pure somatotropes with leptin reduces MSI1 protein expression as assessed by EIA. Bar = 10 μm. Statistics; t test A, MSI1 mRNA control vs mutant: t = 1.91, degree of freedom (df) = 8, P = .04; B, t = 2.01, df = 9, P = .03; C, t = 3.09; df = 7.23, P = .04; F, t = 1.897, df = 8, P = .047.
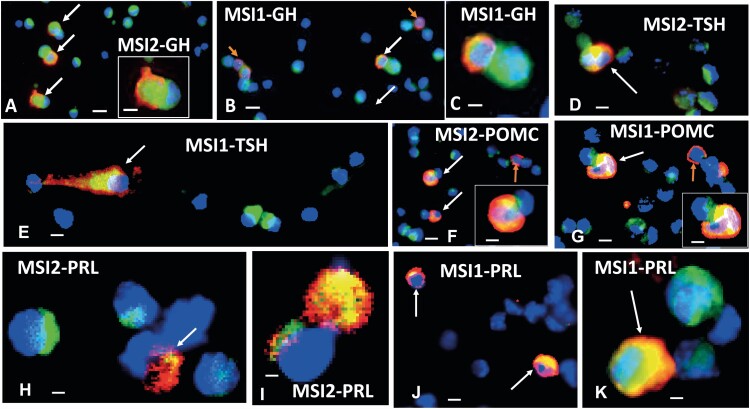
To further validate our scRNAseq data (see Fig. 2) and determine whether Musashi proteins were expressed in other hormone-producing cell lineages, we sought to characterize MSI1 and MSI2 protein expression in relation to various AP hormones using immunocytochemistry on dispersed whole pituitary cells from wild-type diestrous female mice. Consistent with the FACS data (see Fig. 3), female mice showed coimmunolabeling for MSI2 (Fig. 4A) or MSI1 (Fig. 4B) and GH. More GH cells express MSI2 than MSI1. These images show that the MSI proteins are near the nucleus in patches, whereas the GH stores are in the periphery of the cells. There is almost no overlap in the labeling, so the green and red fluorescence can be distinguished in most areas of the same cells (Fig. 4A inset and Fig. 4C). Fig. 4D and 4E illustrate that all the cells immunolabeled for TSHB also express MSI1 or MSI2, and that the Musashi proteins are centrally located near the nucleus. Nearby are cells labeled green only for MSI1 or MSI2. In contrast, immunolabeling for Pomc detected MSI1 or MSI2 in only 66% to 70% of the POMC-labeled corticotropes. Fig. 4F and 4G illustrate fields showing 2 dual-labeled corticotropes near corticotropes that are labeled red only for Pomc. Prolactin cells also show Musashi coexpression, although there is variability in the level of expression of MSI1 or MSI2 (Fig. 4H-4K). Fig. 4H and 4I show 2 PRL cells that are also weakly labeled for MSI2, while very strong labeling is evident in one of the cells in Fig. 4I. This cell is so strongly labeled that the nucleus can be visualized only at the top of the upper cell in Fig. 4I. A similar variability of expression of MSI1 is seen in PRL cells (compare the 3 cells in Fig. 4J and 4K). These whole-cell images show some overlap in red and green fluorescence, which produces a yellow color because the antigens may lie above one another in different focal planes. However, MSI1 and MSI2 tend to be more centrally located near the nucleus, and a layer of green fluorescence over the nucleus turns the region a cyan color. Fig. 4K shows a good example of this layering in the lactotrope. Nearby is a cell that produces only MSI1, and one can see the overlay of patches of MSI1 over the DAPI-stained deep-blue nucleus. Given our previous report showing Musashi protein expression in gonadotropes (43), our data indicate that MSI1 and MSI2 proteins are expressed within all 5 of the differentiated, hormone-expressing anterior pituitary cell lineages.
Figure 4.
Musashi expression in dispersed anterior pituitary (AP) cell lineages. Immunolabeling for growth hormone (GH), thyroid stimulating hormone-β (TSH-β), proopiomelanocortin (POMC), or prolactin (Prl; red fluorescence, Dylight 594 or Cy3) and MSI2 or MSI1 (green fluorescence Dylight or Alexa Fluor 488). Blue is DAPI (4′,6-diamidino-2-phenylindole) fluorescence, indicating nuclei. A number of cells are labeled only for MSI2 or MSI1 (green patches near blue nuclei). White arrows show dual-labeled cells. A shows multiple GH cells with MSI2, and one of them is shown in a higher magnification inset. GH is cytoplasmic and MSI is more centrally located, nearer the blue nucleus. B shows some GH cells without MSI1 (orange arrows). C is a higher magnification of 2 cells shown in B, one showing MSI1 and GH (red and green) and the other showing only MSI1. D shows 2 cells immunolabeled for TSH-β and MSI2 next to several cells labeled only for MSI2. E shows a cell dual-labeled for MSI1 and TSH-β, with MSI1 more centrally located. F shows a field with 3 POMC cells, 2 of which are labeled for POMC and MSI2. A third cell (upper right) contains only Pomc (orange arrow). G shows 3 POMC cells, 2 of which are strongly labeled for MSI1. One of the POMC cells is shown in higher magnification in the inset in F and G. H and I show Prl cells dual-labeled for Prl and MSI2 and illustrates the variety in the labeling patterns. J and K illustrate Prl cells dual-labeled for Prl and MSI1. Bar = 10 μm (A, B, D, F, G, and J); 20 μm (insets and C, E, H, I, and K).
Musashi1 and Musashi2 proteins are expressed in pituitary stem cells
Given the coexpression of Msi1 and Msi2 mRNA in the Sox-expressing cell cluster in the scRNAseq data set (see Fig. 2C), we wanted to confirm that Musashi1 and Musashi2 proteins were also expressed in the Sox2+ pituitary stem cell population. Stem cell-dependent, floating pituisphere colonies were prepared from dispersed whole pituitaries, and nuclear Sox2 protein (labeled green) coexpression with cytoplasmic Musashi1 or Musashi2 proteins (MSI1 and MSI2, labeled red) was readily identified (Fig. 5A and 5B). Of note, some Musashi1 and Musashi2 protein–containing cells did not contain Sox2, indicating the presence of Musashi in non–stem cell populations within the 7-day-old pituispheres.
Figure 5.
Musashi expression in pituitary stem cells. Duel immunofluorescent labeling for SOX2 (green fluorescence) and A, MSI1, red fluorescence or B, MSI2, red fluorescence in pituispheres. MSI1/2 are localized in the cytoplasm and SOX2 shows nuclear localization. Bar = 10 μm.
Musashi binds to the Pou1f1 3′ untranslated region and represses translation
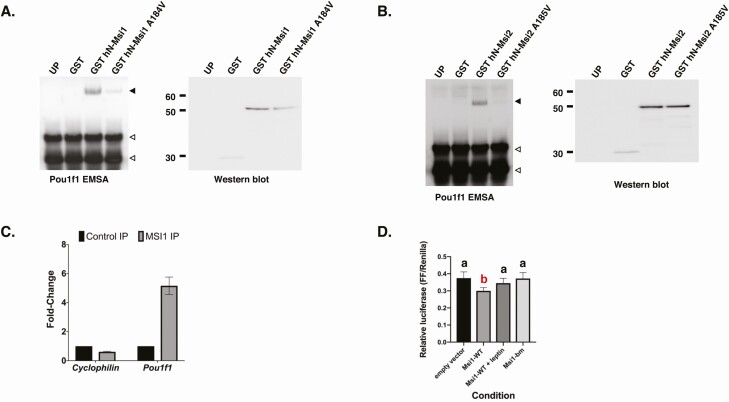
We next explored a possible direct role for Musashi in the regulation of Pou1f1 mRNA translation. The Pou1f1 mRNA regulatory 3′ UTR contains 8 potential MBEs (consensus sequence [A/G]U1-3G (56). We used an RNA EMSA to determine if the Pou1f1 mRNA 3′ UTR associates with Musashi protein, using the N-terminal RNA binding domain of the human Musashi1 and Musashi2 proteins. Both Musashi1 and Musashi2 proteins bound to the Pou1f1 mRNA 3′ UTR probe (Fig. 6A and 6B). Pou1f1 3′ UTR RNA binding to Musashi1 was dramatically attenuated with a pathologically occurring, single-nucleotide mutation of Musashi1 (Musashi1 A184V) that has been previously shown to attenuate interaction with target mRNAs (Fig. 6A) (62). The corresponding mutation in the Musashi2 protein (Musashi2 A185V) similarly resulted in disrupted association (see Fig. 6B), indicating that the Pou1f1 mRNA specifically associates with Musashi1 and Musashi2 in vitro. The full-length Musashi1 and Musashi2 also interacted specifically with the Pou1f1 3′ UTR, although less strongly than the individual N-terminal RNA binding domains (Supplementary Fig. 1 [27]).
Figure 6.
Musashi binding to the Pou1f1 3′ untranslated region (UTR) and translational control of reporter messenger RNAs (mRNAs) fused to the Pou1f1 3′ UTR. A, Left panel: RNA electrophoretic mobility shift assay (EMSA) using biotinylated Pou1f1 3′ UTR incubated with control unprogrammed reticulocyte lysate (UP) or reticulocyte lysate programmed to express glutathione S-transferase (GST), GST–N-terminal Musashi1 (hN-Msi1), or an RNA binding mutant form of Musashi1 (GST-hN_Msi1 A184V). The Musashi1-specific complex is indicated by a solid arrowhead; free probe is marked by open arrowheads. Right panel, Western blot of the reticulocyte lysates used in RNA EMSA detected through anti-GST antibodies to assess the relative expression level of the programmed proteins. The position of molecular weight standards (kDa) are shown. B, Right and left panels as described in A, but with reticulocytes programed with GST–N-terminal Musashi2 (hN-Msi2) or the RNA binding mutant of MSi2 (GST-hN-Msi2 A185V). The Musashi2-specific complex is indicated by a solid arrowhead, whereas a nonspecific band is indicated by an asterisk. C, Immunoprecipitation of whole-pituitary lysate from pooled control female mice. Results are expressed as fold-enrichment calculated from quantitative reverse-transcriptase–polymerase chain reaction of target mRNAs from control rabbit immunoglobulin or anti-Musashi1 immunoprecipitation p = 0.0456, n = 2, 1-tailed t-test. No enrichment of a control gene, Cyclophilin/peptidylprolyl isomerase A/cyclophilin (PPIA) was observed. D, NIH3T3 cells cotransfected with the pmiRGLO 552bp Pou1f1 3′ UTR Firefly Luciferase (FLuc) reporter plasmid and either peGFPN1 (encoding the eGFP moiety alone), peGFP Msi1 in the absence or presence of leptin (Msi1 + leptin) or control, peGFP Msi1-bm. FLuc values were normalized to the expression of a control Renilla luciferase expressed from the same plasmid (FF/Renilla). Summary graph of 4 separate experiments, in which each condition was measured in triplicate. Statistics: one-way analysis of variance (ANOVA) with Sidak multiple comparison test, F(3,44) = 15.4, P < .0001. Values that do not significantly differ from each other are represented by “a,” whereas “b” indicates a value that differs significantly after one-way ANOVA from all “a” values. Specifically, empty vector vs wild-type (WT) Msi1, P < .0001; WT Msi1 vs WT Msi1 + leptin, P < .004; and WT Msi1 vs Msi1-bm, P < .0001.
To determine whether the Pou1f1 mRNA is associated with endogenous Musashi in the pituitary, Musashi1 was immunoprecipitated from lysates of whole pituitaries from adult female mice, and coassociated Pou1f1 mRNA was assessed by qRT-PCR. Pou1f1 mRNA was enriched approximately 5-fold in the Musashi1 IPs over the level in control rabbit IgG IP (Fig. 6C). No enrichment of the housekeeping gene Ppia mRNA (encoding peptidylprolyl isomerase A/cyclophilin A) was observed.
To assess the functional role of the Musashi-Pou1f1 mRNA interaction, we employed a reporter mRNA assay, in which the Pou1f1 mRNA 3′ UTR was cloned downstream of the FLuc open reading frame and cotransfected into NIH3T3 cells with a Musashi protein expression plasmid (43, 56). Fig. 6D is a summary graph of 4 separate experiments in which each condition was measured in triplicate. Musashi expression resulted in a significant repression of the FLuc reporter (see Fig. 6D, 19.7%, P < .0001). Treatment with leptin attenuated the ability of Musashi1 to exert repression (see Fig. 6D). The RNA binding mutant form of Musashi protein (Msi1-bm) did not show significant repression. The individual experiments are presented in Supplementary Fig. 2 (27). Taken together, these data indicate that the Pou1f1 mRNA binds Musashi directly both in vitro and in the whole pituitary, and Musashi1 can exert translational repression of the Pou1f1 mRNA in a leptin-regulated manner.
Discussion
The Musashi RNA binding proteins support stem and/or progenitor cell populations in physiological and pathological tissues by promoting cell self-renewal and opposing cell differentiation and maturation (17). An examination of mRNA expression from the Genotype-Tissue Expression project (63) indicates expression of Msi2 in all normal human tissues tested, with pituitary ranking as the second highest for expression after the ovary (Supplementary Fig. 3 (27)). Expression of Msi1 showed a more restricted pattern of tissue expression, with Msi1 expression levels consistently lower than that of Msi2 (Supplementary Fig. 3 [27]). Interestingly, in males the pituitary also ranked as the second highest for Msi1 expression after the testis. Moreover, with the exclusion of sex-specific ovary and testis, the pituitary ranks as the tissue with highest expression both of Msi1 and Msi2.
In this study, we were interested in examining the expression of Msi1 and Msi2 in the mouse pituitary. Our scRNAseq analyses show that in the mouse pituitary, Msi1 and Msi2 are both expressed in the Sox2+ adult pituitary stem cell population, as expected, and also identify Msi1 and Msi2 expression in a subset of all hormone-expressing cell lineages of the anterior pituitary (see Fig. 2). Further, we confirmed expression of Musashi1 and Musashi2 protein in both adult stem cells (see Fig. 5) and AP hormone-producing cell lineages (see Fig. 4). The expression of Musashi within hormone-producing cells of the adult mouse pituitary suggests a level of immaturity or flexibility of fate that may contribute to the plasticity of these cells. Reanalysis of published scRNAseq data from 2 murine pituitary scRNAseq experiments (23, 28) shows a similar distribution of Msi1 and Msi2 both in the Sox2+ stem cells and also in a subset of each distinct hormone-producing cell lineage (Supplementary Figs. 4-6 [27]). The role of Msi1 and Msi2 expression in these more mature, non–stem cell populations remains an open question. We hypothesize that Musashi may serve to control production of hormones and consequently maintain the plasticity of the pituitary. Consistent with this idea, we have previously reported that Musashi1 exerts repression of translation of the gonadotropin releasing hormone receptor (Gnrhr) mRNA in adult pituitary gonadotropes, and that repression is reversed in response to leptin signaling (43). Plasticity of the pituitary cell populations has been documented to allow remodeling of the hormone-producing cell lineages to respond to the requirements of postnatal lactation (28, 64), hypothyroidism (65-67), or due to lack of negative feedback from the adrenal (68, 69) or gonads (70-72). Our findings position Musashi to contribute to maintenance of pituitary functional plasticity.
We used a recently available resource, the Tabula Muris (73), to determine whether the expression of Msi1 and Msi2 mRNA both in stem cells and in differentiated, non–stem cell populations was peculiar to the pituitary. The Tabula Muris is a searchable scRNAseq data set repository representing 20 organ tissues harvested from the same animals, generated by either Dropseq from FACS-enriched cells or scRNAseq using cell dispersion and the 10x Genomics platform. Although the pituitary is not one of the characterized tissues, many other organ and tissue systems are represented. Querying this repository revealed that Msi2 mRNA is broadly expressed both in stem and non–stem cell types across all 20 sampled tissues. Msi1 mRNA expression, by contrast, is much more restricted and is detectable only within limited stem cell populations. A composite summary of Msi1 and Msi2 mRNA tissue expression is shown in Supplementary Fig. 7 (FACS/Dropseq) (27) and Supplementary Fig. 8 (10X Genomics) (27). From this analysis, it appears that the broad expression of Msi2 may not necessarily be an indicator of flexibility in the cell fate of differentiated tissue. By contrast, the expression of Msi1 and/or coexpression of both Msi1 and Msi2 may more accurately predict cell plasticity. Consistent with this idea and as noted earlier, the human pituitary has the highest expression of both Msi1 and Msi2 of all the tissues examined. However, we cannot rule out an alternate interpretation of Msi2 expression in the Tabula Muris findings that many tissue cell types retain a marked level of plasticity. Further experiments targeting the role of the Musashi isoforms in specific AP populations will be necessary to elucidate the precise role and function of Msi1 and Msi2 expression in the adult pituitary gland.
A prior study from our laboratory had suggested that leptin acts at a posttranscriptional level to promote Pou1f1 mRNA translation in adult female pituitary somatotropes (16). The Pou1f1 mRNA 3′ UTR contains multiple consensus MBEs, and our scRNAseq data showing overlap of Msi1 and Msi2 with Pou1f1 expression in POU1F1-dependent lineages (somatotropes, lactotropes, and thyrotropes) supports a role for Musashi as a regulator of Pou1f1 mRNA translation (see Figs. 1 and 2). EIA and immunocytochemistry confirm Musashi protein expression in somatotrope populations (see Figs. 3 and 4). Interestingly, there does appear to be heterogeneity of Msi1 and Msi2 protein expression within the somatotrope population as assessed by immunocytochemistry (see Fig. 3D). Whether this indicates an underlying spatial gradient of Musashi protein levels within the AP parenchyma prior to cell dispersal remains to be determined. We also note that there is a discrepancy between the number of somatotropes with detectable Msi1 protein (see Figs. 3 and 4) and with detectable Msi1 mRNA expression (see Fig. 2). There are several possible explanations for this discrepancy, including technical limitations on the level of sensitivity of Msi1 transcript detection. As others have noted (74), not all transcripts are efficiently reverse transcribed and, given the stochastic nature of mRNA capture prior to library generation, more highly expressed mRNAs are favored over those of lower expression. Consequently, some mRNAs may be underrepresented or absent (a phenomenon termed dropout). Thus, the absence of a certain gene in a cell’s scRNAseq library does not always mean it is not expressed. Because Msi1 mRNA is expressed at lower levels than the Msi2 mRNA in the mouse pituitary (75), the discrepancy between Msi1 protein levels and Msi1 mRNA that we observe is most likely a consequence of dropout.
At this juncture, the molecular basis for the sex-specific regulation of Pou1f1 mRNA translation (16) is unclear. We note that males and females both express Msi1 and Msi2 within the AP (Supplementary Figs. 5 and 6 [27]) and FACS-sorted somatotropes express comparable levels of MSI1 protein (Supplemental Fig. 9 [27]), so differences in expression of MSI1 alone cannot account for the sex-specific posttranscriptional control of POU1F1 levels.
With regard to regulation by leptin, Musashi1 mRNA and protein levels are increased in somatotropes that are blocked from leptin signaling (Lepr-null mutants) (see Fig. 3A and 3B). Consistent with these findings, Musashi1 protein levels decrease dramatically after leptin stimulation in purified somatotropes (see Fig. 3F). Of note, we do not see a similar change in Msi2 mRNA levels indicating isoform-specific regulation of Musashi in the pituitary. As expected for these characterized stem cell regulatory proteins, Musashi1 and Musashi2 mRNAs and proteins were also detected in Sox2+ pituitary stem cells (see Figs. 2 and 5).
Musashi1 and Musashi2 association with the MBE-containing Pou1f1 mRNA 3′ UTR was confirmed both in the in vitro EMSA assay and through co-IP from the adult pituitary (see Fig. 6). We further demonstrated that Musashi1 exerts repression of translation of the Pou1f1 reporter mRNA in a heterologous cell assay and that this repression is attenuated after leptin treatment (see Fig. 6). Together, these findings are consistent with pituitary-expressed Musashi proteins exerting translational repression of the Pou1f1 mRNA until exposure to leptin signaling. The data parallel our earlier findings that Musashi controls translation of the Gnrhr mRNA in gonadotropes (43) and suggest that Musashi may target and regulate multiple mRNAs that encode key pituitary cell fate determinants to control hormone production within the anterior pituitary. We have recently shown that Musashi can also exert translational regulation of the Tsh and Prl mRNAs, indicating that Musashi can impinge directly on translation of specific AP hormone mRNAs (37). An important future direction will be to selectively ablate Musashi in individual AP cell lineages to assess specific in vivo requirements for Musashi-dependent mRNA translational regulation to modulate pituitary function. Taken together, our findings suggest a critical role for Musashi in mediating AP cell plasticity in response to metabolic cues, both as a direct regulator of specific AP hormone mRNAs and a regulator of the AP cell lineage determinant POU1F1.
Acknowledgments
We thank Drs Leonard Cheung and Yugong Ho for their help accessing their respective scRNAseq data sets, Drs Fanni Gergely and Trevor Sweeney for helpful discussions on A184V RNA binding–deficient forms of Musashi, and Allen Gies for assistance with the GEO data repository submission.
Financial Support: This work was supported by the National Institutes of Health (grant No. R01HD093461 to A.M.M., G.V.C., and M.C.M.), (grant No. R01DK113776-01 to G.V.C., A.M.M., and M.C.M.), and (grant No. R01HD087057 to G.V.C. and A.M.M.); and the National Institute of General Medical Sciences (grant Nos. P20 GM103425 and P30GM11070 to Dr Edgar Garcia-Rill).
Glossary
Abbreviations
- ACTH
adrenocorticotropin
- AP
anterior pituitary
- BSA
bovine serum albumin
- cDNA
complementary DNA
- EIA
enzyme immunoassay
- EMSA
electrophoretic mobility shift assay
- FACS
fluorescence-activated cell sorting
- FSH
follicle-stimulating hormone
- GH
growth hormone
- GST
glutathione S-transferase
- IP
immunoprecipitation
- LEPR
leptin receptor
- LH
luteinizing hormone
- MBE
Musashi binding element
- mRNA
messenger RNA
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- POMC
proopiomelanocortin
- PRL
prolactin
- qRT
quantitative reverse transcriptase
- scRNAseq
single-cell RNA sequencing
- TSH
thyroid stimulating hormone
- UTR
untranslated region
Additional Information
Disclosures: The authors have nothing to disclose.
Data Availability
The scRNAseq data described in this study have been deposited in the Gene Expression Omnibus under accession number GSE153045.
References
- 1. Prevot V. Plasticity of neuroendocrine systems. Eur J Neurosci. 2010;32(12):1987-1988. [DOI] [PubMed] [Google Scholar]
- 2. Childs GV, MacNicol AM, MacNicol MC. Molecular mechanisms of pituitary cell plasticity. Front Endocrinol (Lausanne). 2020;11:656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vankelecom H. Pituitary stem/progenitor cells: embryonic players in the adult gland? Eur J Neurosci. 2010;32(12): 2063-2081. [DOI] [PubMed] [Google Scholar]
- 4. Kelberman D, Rizzoti K, Lovell-Badge R, Robinson IC, Dattani MT. Genetic regulation of pituitary gland development in human and mouse. Endocr Rev. 2009;30(7):790-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Youngblood JL, Coleman TF, Davis SW. Regulation of pituitary progenitor differentiation by β-Catenin. Endocrinology. 2018;159(9):3287-3305. [DOI] [PubMed] [Google Scholar]
- 6. Pérez Millán MI, Brinkmeier ML, Mortensen AH, Camper SA. PROP1 triggers epithelial-mesenchymal transition-like process in pituitary stem cells. eLife. 2016;5:e14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iqbal J, Pompolo S, Considine RV, Clarke IJ. Localization of leptin receptor-like immunoreactivity in the corticotropes, somatotropes, and gonadotropes in the ovine anterior pituitary. Endocrinology. 2000;141(4):1515-1520. [DOI] [PubMed] [Google Scholar]
- 8. Jin L, Burguera BG, Couce ME, et al. Leptin and leptin receptor expression in normal and neoplastic human pituitary: evidence of a regulatory role for leptin on pituitary cell proliferation. J Clin Endocrinol Metab. 1999;84(8):2903-2911. [DOI] [PubMed] [Google Scholar]
- 9. Jin L, Zhang S, Burguera BG, et al. Leptin and leptin receptor expression in rat and mouse pituitary cells. Endocrinology. 2000;141(1):333-339. [DOI] [PubMed] [Google Scholar]
- 10. Sone M, Nagata H, Takekoshi S, Osamura RY. Expression and localization of leptin receptor in the normal rat pituitary gland. Cell Tissue Res. 2001;305(3):351-356. [DOI] [PubMed] [Google Scholar]
- 11. Shimon I, Yan X, Magoffin DA, Friedman TC, Melmed S. Intact leptin receptor is selectively expressed in human fetal pituitary and pituitary adenomas and signals human fetal pituitary growth hormone secretion. J Clin Endocrinol Metab. 1998;83(11):4059-4064. [DOI] [PubMed] [Google Scholar]
- 12. Akhter N, Odle AK, Allensworth-James ML, et al. Ablation of leptin signaling to somatotropes: changes in metabolic factors that cause obesity. Endocrinology. 2012;153(10):4705-4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Allensworth-James ML, Odle A, Haney A, Childs G. Sex differences in somatotrope dependency on leptin receptors in young mice: ablation of LEPR causes severe growth hormone deficiency and abdominal obesity in males. Endocrinology. 2015;156(9):3253-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Allensworth-James ML, Odle A, Haney A, MacNicol M, MacNicol A, Childs G. Sex-specific changes in postnatal GH and PRL secretion in somatotrope LEPR-null mice. J Endocrinol. 2018;238(3):221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Childs GV, Akhter N, Haney A, et al. The somatotrope as a metabolic sensor: deletion of leptin receptors causes obesity. Endocrinology. 2011;152(1):69-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Odle AK, Allensworth-James ML, Akhter N, et al. A sex-dependent, tropic role for leptin in the somatotrope as a regulator of POU1F1 and POU1F1-dependent hormones. Endocrinology. 2016;157(10):3958-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fox RG, Park FD, Koechlein CS, Kritzik M, Reya T. Musashi signaling in stem cells and cancer. Annu Rev Cell Dev Biol. 2015;31:249-267. [DOI] [PubMed] [Google Scholar]
- 18. JAX genotyping resources. https://www.jax.org/jax-mice-and-services/customer-support/technical-support/genotyping-resources [Google Scholar]
- 19. Li N, Yousefi M, Nakauka-Ddamba A, et al. The Msi family of RNA-binding proteins function redundantly as intestinal oncoproteins. Cell Rep. 2015;13(11):2440-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. 10x Genomics methanol fixation protocol for scRNAseq. https://support.10xgenomics.com/single-cell-gene-expression/sample-prep/doc/demonstrated-protocol-methanol-fixation-of-cells-for-single-cell-rna-sequencing. Accessed 2020. [Google Scholar]
- 21. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng GX, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheung LYM, George AS, McGee SR, et al. Single-cell RNA sequencing reveals novel markers of male pituitary stem cells and hormone-producing cell types. Endocrinology. 2018;159(12):3910-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888-1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McInnes L, Healy J, Melville J. UMAP: uniform manifold approximation and projection for dimension reduction. ArXiv. 2018;arXiv:1802.03426. [Google Scholar]
- 26. Franzén O, Gan LM, Björkegren JLM. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database (Oxford). 2019;2019:baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Supplemental Data. 10.5061/dryad.51c59zw5s [DOI] [Google Scholar]
- 28. Ho Y, Hu P, Peel MT, et al. Single-cell transcriptomic analysis of adult mouse pituitary reveals sexual dimorphism and physiologic demand-induced cellular plasticity. Protein Cell. 2020;11(8):565-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Finak G, McDavid A, Yajima M, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fletcher PA, Smiljanic K, Maso Prévide R, et al. Cell type- and sex-dependent transcriptome profiles of rat anterior pituitary cells. Front Endocrinol (Lausanne). 2019;10:623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen J, Hersmus N, Van Duppen V, Caesens P, Denef C, Vankelecom H. The adult pituitary contains a cell population displaying stem/progenitor cell and early embryonic characteristics. Endocrinology. 2005;146(9): 3985-3998. [DOI] [PubMed] [Google Scholar]
- 32. RRID:AB_11205731. https://antibodyregistry.org/search?q=RRID:AB_11205731 [Google Scholar]
- 33. RRID:AB_2147926. https://antibodyregistry.org/search?q=RRID:%20AB_2147926 [Google Scholar]
- 34. RRID:AB_2147942. https://antibodyregistry.org/search?q=RRID:%20AB_2147942RRID:%20AB_2147942 [Google Scholar]
- 35. RRID:AB_2336777. https://antibodyregistry.org/search?q=RRID:%20AB_2336777RRID:%20AB_2336777 [Google Scholar]
- 36. RRID:AB_2629219. https://antibodyregistry.org/search?q=RRID%20AB_2629219RRID%20AB_2629219 [Google Scholar]
- 37. Allensworth-James ML, Odle AK, Lim J, et al. Metabolic signalling to somatotrophs: transcriptional and post-transcriptional mediators. J Neuroendocrinol. 2020;32(11):e12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. RRID:AB_2336414. https://antibodyregistry.org/search?q=RRID:%20AB_2336414RRID:%20AB_2336414 [Google Scholar]
- 39. RRID:AB_2336776. https://antibodyregistry.org/search?q=RRID:%20AB_2336776RRID:%20AB_2336776 [Google Scholar]
- 40. RRID:AB_2884004. https://antibodyregistry.org/search?q=RRID:AB_2884004RRID:AB_2884004 [Google Scholar]
- 41. RRID:AB_2884005. https://antibodyregistry.org/search?q=RRID:AB_2884005RRID:AB_2884005 [Google Scholar]
- 42. RRID:AB_2884003. https://antibodyregistry.org/search?q=RRID:AB_2884003RRID:AB_2884003 [Google Scholar]
- 43. Odle AK, Beneš H, Melgar Castillo A, et al. Association of Gnrhr mRNA with the stem cell determinant Musashi: a mechanism for leptin-mediated modulation of GnRHR expression. Endocrinology. 2018;159(2):883-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. RRID:AB_2884006. https://antibodyregistry.org/search?q=RRID:AB_2884006RRID:AB_2884006 [Google Scholar]
- 45. RRID:AB_2864367. https://antibodyregistry.org/search?q=RRID:%20AB_2864367RRID:%20AB_2864367 [Google Scholar]
- 46. Rassa JC, Wilson GM, Brewer GA, Parks GD. Spacing constraints on reinitiation of paramyxovirus transcription: the gene end U tract acts as a spacer to separate gene end from gene start sites. Virology. 2000;274(2):438-449. [DOI] [PubMed] [Google Scholar]
- 47. MacNicol MC, Pot D, MacNicol AM. pXen, a utility vector for the expression of GST-fusion proteins in Xenopus laevis oocytes and embryos. Gene. 1997;196(1-2):25-29. [DOI] [PubMed] [Google Scholar]
- 48. Okabe M, Imai T, Kurusu M, Hiromi Y, Okano H. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature. 2001;411(6833): 94-98. [DOI] [PubMed] [Google Scholar]
- 49. Charlesworth A, Wilczynska A, Thampi P, Cox LL, MacNicol AM. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 2006;25(12):2792-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. RRID:AB_627677. https://antibodyregistry.org/search?q=RRID:%20AB_627677RRID:%20AB_627677 [Google Scholar]
- 51. RRID:AB_2144988. https://antibodyregistry.org/search?q=RRID:%20AB_2144988RRID:%20AB_2144988 [Google Scholar]
- 52. RRID:AB_880995. https://antibodyregistry.org/search?q=RRID:%20AB_880995RRID:%20AB_880995 [Google Scholar]
- 53. RRID:AB_2241126. https://antibodyregistry.org/search?q=RRID:%20AB_2241126RRID:%20AB_2241126 [Google Scholar]
- 54. MacNicol AM, Hardy LL, Spencer HJ, MacNicol MC. Neural stem and progenitor cell fate transition requires regulation of Musashi1 function. BMC Dev Biol. 2015;15:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. MacNicol MC, Cragle CE, MacNicol AM. Context-dependent regulation of Musashi-mediated mRNA translation and cell cycle regulation. Cell Cycle. 2011;10(1):39-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Imai T, Tokunaga A, Yoshida T, et al. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001;21(12):3888-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. RRID:AB_881168. https://antibodyregistry.org/search?q=RRID:%20AB_881168RRID:%20AB_881168 [Google Scholar]
- 58. RRID:AB_145841. https://antibodyregistry.org/search?q=RRID:%20AB_145841RRID:%20AB_145841 [Google Scholar]
- 59. Odle AK, Haney A, Allensworth-James M, Akhter N, Childs GV. Adipocyte versus pituitary leptin in the regulation of pituitary hormones: somatotropes develop normally in the absence of circulating leptin. Endocrinology. 2014;155(11): 4316-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Akhter N, CarlLee T, Syed MM, et al. Selective deletion of leptin receptors in gonadotropes reveals activin and GnRH-binding sites as leptin targets in support of fertility. Endocrinology. 2014;155(10):4027-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vankelecom H, Gremeaux L. Stem cells in the pituitary gland: a burgeoning field. Gen Comp Endocrinol. 2010;166(3):478-488. [DOI] [PubMed] [Google Scholar]
- 62. Chavali PL, Stojic L, Meredith LW, et al. Neurodevelopmental protein Musashi-1 interacts with the Zika genome and promotes viral replication. Science. 2017;357(6346):83-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Carithers LJ, Ardlie K, Barcus M, et al. ; GTEx Consortium . A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv Biobank. 2015;13(5):311-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Le Tissier PR, Hodson DJ, Martin AO, Romanò N, Mollard P. Plasticity of the prolactin (PRL) axis: mechanisms underlying regulation of output in female mice. Adv Exp Med Biol. 2015;846:139-162. [DOI] [PubMed] [Google Scholar]
- 65. Jobin M, Delgado A, Fortier C. Interactions between the pituitary, thyroid and adrenal cortex during acute exposure to cold or to electric shocks in the rat. Horm Res. 1975;6(4): 199-212. [DOI] [PubMed] [Google Scholar]
- 66. Childs GV, Taub K, Jones KE, Chin WW. Triiodothyronine receptor beta-2 messenger ribonucleic acid expression by somatotropes and thyrotropes: effect of propylthiouracil-induced hypothyroidism in rats. Endocrinology. 1991;129(5): 2767-2773. [DOI] [PubMed] [Google Scholar]
- 67. Vidal S, Horvath E, Kovacs K, Cohen SM, Lloyd RV, Scheithauer BW. Transdifferentiation of somatotrophs to thyrotrophs in the pituitary of patients with protracted primary hypothyroidism. Virchows Arch. 2000;436(1):43-51. [DOI] [PubMed] [Google Scholar]
- 68. Childs GV, Ellison DG, Lorenzen JR, Collins TJ, Schwartz NB. Retarded development of castration cells after adrenalectomy or sham adrenalectomy. Endocrinology. 1983;113(1):166-177. [DOI] [PubMed] [Google Scholar]
- 69. Lorenzen JR, Schlepphorst C, Schwartz NB. The interaction of castration and adrenalectomy on pituitary responses to loss of target gland negative feedback in the male rat. Endocrinology. 1980;106(2):592-599. [DOI] [PubMed] [Google Scholar]
- 70. Childs GV, Lloyd JM, Unabia G, Gharib SD, Wierman ME, Chin WW. Detection of luteinizing hormone β messenger ribonucleic acid (RNA) in individual gonadotropes after castration: use of a new in situ hybridization method with a photobiotinylated complementary RNA probe. Mol Endocrinol. 1987;1(12):926-932. [DOI] [PubMed] [Google Scholar]
- 71. Childs GV, Unabia G, Wierman ME, Gharib SD, Chin WW. Castration induces time-dependent changes in the follicle-stimulating hormone beta-subunit messenger ribonucleic acid-containing gonadotrope cell population. Endocrinology. 1990;126(4):2205-2213. [DOI] [PubMed] [Google Scholar]
- 72. Moriarty GC. Adenohypophysis: ultrastructural cytochemistry. a review. J Histochem Cytochem. 1973;21(10):855-894. [DOI] [PubMed] [Google Scholar]
- 73. Tabula Muris Consortium; Overall coordination; Logistical coordination; Organ collection and processing; Library preparation and sequencing; Computational data analysis; Cell type annotation; Writing group; Supplemental text writing group; Principal investigators. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562(7727): 367-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cheung LYM, Rizzoti K. Cell population characterization and discovery using single-cell technologies in endocrine systems. J Mol Endocrinol. 2020;65(2):R35-R51. [DOI] [PubMed] [Google Scholar]
- 75. Szabat M, Kalynyak TB, Lim GE, et al. Musashi expression in β-cells coordinates insulin expression, apoptosis and proliferation in response to endoplasmic reticulum stress in diabetes. Cell Death Dis. 2011;2:e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The scRNAseq data described in this study have been deposited in the Gene Expression Omnibus under accession number GSE153045.