Treating multiple cells from a single animal as independent for statistical analysis is pseudoreplication and can result in bogus estimates of significance.
Abstract
This article reviews how to analyze data from experiments designed to compare the cellular physiology of two or more groups of animals or people. This is commonly done by measuring data from several cells from each animal and using simple t tests or ANOVA to compare between groups. I use simulations to illustrate that this method can give erroneous positive results by assuming that the cells from each animal are independent of each other. This problem, which may be responsible for much of the lack of reproducibility in the literature, can be easily avoided by using a hierarchical, nested statistics approach.
Readers will be aware of concerns about the lack of reproducibility of scientific research (Ioannidis, 2005). Perhaps these issues should not be a surprise: research is performed by humans and will never be perfect. The problem is serious, however, and a variety of factors contribute, as reviewed previously (Brown and Ramaswamy, 2007; Loscalzo, 2012; Arrowsmith et al., 2015; Enserink, 2017; Eisner, 2018). These include fraud, carelessness, and uncontrolled issues relating to cell lines and animals. Problems with experimental design and statistical analysis are also a major concern.
The purpose of this tutorial is to concentrate on one statistical issue that, although widely discussed (Lazic, 2010; Sikkel et al., 2017; Lazic et al., 2018), is still a major problem: this is the subject of pseudoreplication (Hurlbert, 1984) in which data points are treated as independent biological estimates when they are really technical replicates. A common example arises in physiology experiments comparing tissues or cells that come from two or more groups in order to investigate whether the groups differ. Often, the groups are different animals. For example, a comparison may be between wild type and transgenic, control and heart failure, or naive and conditioned. Tissue may also be taken from human subjects: for example, diabetic versus healthy or pregnant versus control. In tissue culture experiments, comparisons can be made between cells transfected with active and scrambled siRNAs. In many such projects, the question is whether the properties of cells or tissues are different between the two groups of animals or people. For example, is Ca2+ handling or ion channel kinetics or density different in the two conditions?
Because of cost and practical issues, it can be difficult to obtain large numbers of animals or subjects, and therefore many cells are studied from each of a small number of animals. Consider a typical case in which there are “N” animals in each of two groups, with “n” cells (or tissues) being studied from each animal. Both groups therefore contain “N * n” cells, and such experiments are often incorrectly analyzed by performing statistical tests, such as t tests or ANOVA, taking the number of samples in each group as N * n. One of the critical assumptions of t tests and ANOVA is that each observation in a dataset is independent of other observations. Violating this independence assumption results in an inflated type I error rate (i.e., thinking you have a difference between conditions when, in fact, no difference occurs—in other words, a false positive).
The flaw can be seen by considering the limiting case in which n = 1; a single animal is used in each group. Imagine that 100 cells are studied from each of two animals. The standard error of the mean is (SD / √number of cells), here equal to 0.1 SD. This very small value will mean that even a modest difference in the average value of the two animals can result in an apparently statistically significant difference. This would be equivalent to addressing the question of whether blood pressure is different in people who live in London and New York by studying one individual from each city and measuring her blood pressure 100 times.
While I am sure that most readers need no convincing that it is invalid to study a single animal, the literature contains many studies where, say, three animals are used and five cells are studied from each animal. There are two sources of variation in such an experiment: (i) variation among animals and (ii) variation among cells isolated from an individual animal. These can be represented by their SDs, SDanimal and SDcell, respectively. The variation represented by SDanimal includes not only factors present in the animals but also those resulting from differences between different cell isolations. An extreme example can be considered whereby SDcell is 0. Under these conditions, the five cells studied will give identical values. The 15 cells will be made up of five identical replicates of three different values. By chance, these three values (from the three animals) may be different. While a t test based on n = 3 animals finds no significant difference, use of n = 15 cells may suggest a spurious significant difference.
The problem can be appreciated from the results of the simulation shown below. Here, we assume that there is no real difference between two groups of animals and that each has a mean value of 1.0. The value for a given animal will be normally distributed with SDanimal. The circles in Fig. 1 A give the values for the three animals selected randomly in each of groups 1 and 2. An unpaired t test gave a nonsignificant (P = 0.15) value. For each animal, the program then selected five cells randomly from another normal distribution with the mean equal to the animal mean using SDcell. The values for these cells are shown as the smaller open circles, distributed around the animal mean (Fig. 1 B). An unpaired t test was then performed comparing the 15 cells in condition 1 with those in condition 2. In this particular trial, there was a highly significant difference between the cells in the two conditions. On average, with SDanimal and SDmean both equal to 0.3, the simulation found that P was <0.05 in 29% of trials. Since there were no real differences in this simulation, these 29% are all false positives.
Figure 1.
Simulation of the effects of comparing 15 cells drawn from 3 animals. (A) Values of three selected animals in each group. In each group, three animal values were drawn from a normal distribution: mean 1.0; SDanimal, 0.3. The means of the three points are shown by the horizontal black lines. A t test shows no significant difference. (B) The same three animals have been selected, and from each animal five cells (open circles of the same color as the animal in A) were drawn using a normal distribution, with a mean given by the animal value and SDcell of 0.3. The mean values of the 15 cells are denoted by the horizontal black lines. A t test shows a significant difference.
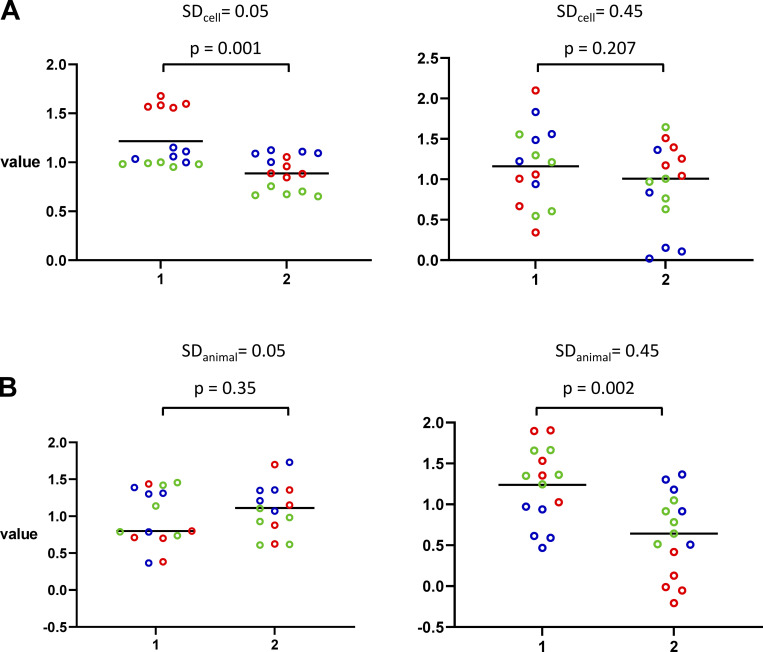
The likelihood of obtaining a false positive depends on the animal and cell SDs. Further simulations show that increasing SDcell decreases the probability of finding a significant difference (Fig. 2 A). In both panels, SDanimal is 0.3. SDcell is 0.05 in the left graph and 0.45 in the right. The lower SDcell in the left panel results in a small spread of cell values and an apparent significant difference, while the greater spread in the right means that no significant difference is seen. On average, an SDcell of 0.05 produces 47% false positives, whereas an SDcell of 0.45 only 21%. As demonstrated in Fig. 2 B, increasing SDanimal makes it more likely that an apparent significant difference will be noted. This is because the increased variance among the animals increases the chance of a large difference between the three animals in one case compared with the other. With a fixed SDcell of 0.3, increasing SDanimal from 0.05 to 0.45 increases the false-positive rate from 6% to 37%.
Figure 2.
Effects of animal and cell SD on apparent significance. (A) Effects of SDcell. In both panels, SDanimal is 0.3. SDcell is 0.05 (left) and 0.45 (right), giving P values of 0.001 and 0.207, respectively. (B) Effects of SDanimal. In both panels, SDcell is 0.3. SDanimal is 0.05 (left) and 0.45 (right), giving P values of 0.35 and 0.002, respectively. In both panels, the black horizontal lines denote the mean value of the 15 cells.
Fig. 3 A shows a more complete analysis; 200,000 trials were performed for each condition with values of SDcell and SDanimal between 0 and 0.5. The P value for a t test was calculated for each trial and the fraction giving a value of P < 0.05, the false-positive rate, was recorded.
Figure 3.
Dependence of apparent significance on SD and sample numbers. (A) Effects of SD. SDcell and SDanimal were varied from 0 to 0.5 in steps of 0.025. 200,000 trials were done for each condition. The colored areas indicate the percentage of trials which gave a P value of <0.05. Points A–D are referred to in the text. (B) Effects of numbers of animals and cells. The plots show the effects of altering animal number (x axis) and number of cells per animal (y axis). The contours and colors indicate the percentage of trials giving P < 0.05. In all simulations, SDanimal and SDcell were 0.3. 20,000 trials were performed at each condition.
Given that the animals and cells were drawn from identical populations, one might expect P < 0.05 in 5% of the trials. This is, however, only seen for a combination of low SDanimal and high SDcell (point A in Fig. 3 A). Under these conditions, the t test is influenced by a very large variation in the values from the cells. As SDanimal increases and SDcell decreases, the fraction of trials for which P < 0.05 increases to values of 45%, a very high false-positive rate (point B). Points C and D show equal SDs for cell and animal, with both being low for C and high for D. In both cases, the simulation predicts a false discovery rate of ∼35%.
The effects of SDcell were previously highlighted by considering the intraclass correlation coefficient, or ICC (Sikkel et al., 2017). A very low SDcell corresponds to a maximum (1.0) value of ICC (i.e., the values of all the cells from a given animal are identical). In other words, no information is provided by these multiple cells compared with a single measurement and the error using a simple t test is large. At the other extreme (when SDcell is high compared with SDanimal), the ICC is 0 and there is no clustering, so the error is much less. Sikkel et al. (2017) provide a means to calculate the effective sample size in an experiment in which n cells are studied from each of N animals. This value is [N × n] / [1 + (n − 1) × ICC]. Only in the trivial case where ICC = 0 (SDcell is large) does this equal the value of n × N used for a simple t test. As ICC approaches 1 (SDcell = 0), the effective sample size falls to a value of N, the number of animals, indicating the futility of considering multiple cells from each animal.
The simulation of Fig. 3 B was designed to investigate how the number of cells used per animal affects the error. Only when a single cell is used from each animal does the false-positive rate equal 5%; as the number of cells per animal is increased, the false-positive rate increases. Even when only two cells are used from each animal, the false-positive rate is >10%. It may seem counterintuitive that increasing the number of cells increases the false-positive rate. This occurs because the higher the number of cells, the greater the level of pseudoreplication and the more flawed the t test is (the same holds for ANOVA). As also shown by Fig. 3 B, the number of animals used has very little effect on the error. In summary, irrespective of the number of animals used, studying more than one cell per animal and assuming that cells are independent can dramatically increase the false-positive rate. In these simulations, each animal provides the same number of cells, whereas in most papers, the number of cells studied per animal varies from day to day, further complicating the issue.
I should make four points clear. (1) There is nothing wrong with basing statistics on the number of cells in experiments when one is not comparing between animals. For example, when studying the effects of a drug on an ion channel, n can be the number of cells. There is, of course, a different discussion to be had about how many animals should be used to ensure that the sampled population is representative. (2) I have focused above on studies that base statistical analysis on the number of cells rather than the number of animals. A further complexity comes when one makes several measurements from each cell. One example might be measuring the size of cellular organelles. Another comes in studies analyzing the properties of calcium sparks and whether they differ between different groups of animals. Here, hundreds of sparks are often recorded from each cell, and the degree of pseudoreplication produced by treating sparks as independent is enormous when it comes to addressing questions such as whether the amplitude or spatial spread of these sparks is altered. It is therefore important to examine the possibility that many of the reported differences between animals in, for example, spark amplitude may be artifactual (Sikkel et al., 2017). (3) Related problems arise in tissue culture experiments (Lazic et al., 2018). The above discussion has been couched in terms of animals and cells, but similar problems arise if wells or dishes taken from the same culture are treated as independent. (4) Finally, it is important to note that variations in the properties of cells may reflect not only experimental variation but also real heterogeneity within the animal. In the latter case, it is important to quantify this heterogeneity and how it changes rather than simply assessing the mean value.
Given the above, it is clearly inappropriate to use an analysis that assumes that different cells from the same animal provide independent measures. So, what can be done? One simple solution is to average the values from all the cells taken from a given animal and do a t test (or ANOVA) with N being the number of animals. As discussed previously (Sikkel et al., 2017), the disadvantage of this method is that it makes it likely that real effects will be missed and also takes no account of the fact that some animals provide more cells than others. A better approach is to use hierarchical (nested) analysis or linear mixed modeling, which takes explicit account of the structure of the data, specifically how many cells come from each animal. The reader is referred to a full explanation of this approach as commonly applied to cell physiology (Sikkel et al., 2017). In brief, the method makes use of the structure of the data. At one extreme (corresponding to a low SDcell in the simulations above), the data provided by all the cells from a particular animal are clustered together and the error produced by using a simple t test is large (Fig. 3 A). In this case, the hierarchical approach uses this clustering to make a large correction. At the other extreme, when there is less clustering within an animal (high SDcell), less correction is needed. If such analysis is applied to the simulations above, the erroneous false positives disappear.
In the past, the required software was not as readily available as that for performing t tests, although the major commercial programs (including GraphPad Prism, SPSS, Stata, and SAD) as well as open-source data analysis software such as R, Stan, and Julia, provide it. Data S1 shows, for example, how nested analysis of the data in Fig. 1 can be performed in GraphPad Prism. Finally, the use of different-colored symbols in Figs. 1 and 2 makes it clear how the values from cells from a single animal are clustered. This identification of cells is not normally performed in the literature and is worth considering to give a graphical impression of the degree of clustering.
As suggested previously, pseudoreplication and inappropriate statistical analysis likely account for considerable lack of reproducibility in a variety of fields in physiology, including neuroscience (Lazic, 2010) and cardiac calcium signaling (Sikkel et al., 2017). Rectifying this will require not only the use of proper analysis but also often the use of more animals and human subjects. While this will obviously make research both more expensive and slower, these steps need to be taken to ensure that pseudoreplication does not continue to cast a shadow over physiology.
Online supplemental material
Data S1 provides a guide to performing hierarchical analysis using GraphPad Prism.
Supplementary Material
provides a guide to performing hierarchical analysis using GraphPad Prism.
Acknowledgments
Olaf S. Andersen served as editor.
I am grateful to the following colleagues for very useful discussions: Alan Batterham, Henk Granzier, Chris Lingle, Joe Mindell, Jeanne Nerbonne, Crina Nimigean, Eduardo Rios, Néstor Saiz, Godfrey Smith, Andrew Stewart, Andrew Trafford, and Susan Wray.
The author was supported by The British Heart Foundation (grant CH/2000004/12801).
The author declares no competing financial interests.
References
- Arrowsmith, C.H., Audia J.E., Austin C., Baell J., Bennett J., Blagg J., Bountra C., Brennan P.E., Brown P.J., Bunnage M.E., et al. . 2015. The promise and peril of chemical probes. Nat. Chem. Biol. 11:536–541. 10.1038/nchembio.1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, E.N., and Ramaswamy S.. 2007. Quality of protein crystal structures. Acta Crystallogr. D Biol. Crystallogr. 63:941–950. 10.1107/S0907444907033847 [DOI] [PubMed] [Google Scholar]
- Eisner, D.A. 2018. Reproducibility of science: Fraud, impact factors and carelessness. J. Mol. Cell. Cardiol. 114:364–368. 10.1016/j.yjmcc.2017.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink, M. 2017. Sloppy reporting on animal studies proves hard to change. Science. 357:1337–1338. 10.1126/science.357.6358.1337 [DOI] [PubMed] [Google Scholar]
- Hurlbert, S.H. 1984. Pseudoreplication and the Design of Ecological Field Experiments. Ecol. Monogr. 54:187–211. 10.2307/1942661 [DOI] [Google Scholar]
- Ioannidis, J.P.A. 2005. Why most published research findings are false. PLoS Med. 2:e124 10.1371/journal.pmed.0020124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazic, S.E. 2010. The problem of pseudoreplication in neuroscientific studies: is it affecting your analysis? BMC Neurosci. 11:5 10.1186/1471-2202-11-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazic, S.E., Clarke-Williams C.J., and Munafò M.R.. 2018. What exactly is ‘N’ in cell culture and animal experiments? PLoS Biol. 16:e2005282 10.1371/journal.pbio.2005282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscalzo, J. 2012. Irreproducible experimental results: causes, (mis)interpretations, and consequences. Circulation. 125:1211–1214. 10.1161/CIRCULATIONAHA.112.098244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikkel, M.B., Francis D.P., Howard J., Gordon F., Rowlands C., Peters N.S., Lyon A.R., Harding S.E., and MacLeod K.T.. 2017. Hierarchical statistical techniques are necessary to draw reliable conclusions from analysis of isolated cardiomyocyte studies. Cardiovasc. Res. 113:1743–1752. 10.1093/cvr/cvx151 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
provides a guide to performing hierarchical analysis using GraphPad Prism.