Microtubule attachments to spindle poles in yeast are flexible, which allows the microtubules to pivot. Fong et al. directly measure the pivoting flexibility of the microtubule–pole interface and show that this flexibility is important for timely pole separation during mitosis.
Abstract
To assemble a bipolar spindle, microtubules emanating from two poles must bundle into an antiparallel midzone, where plus end–directed motors generate outward pushing forces to drive pole separation. Midzone cross-linkers and motors display only modest preferences for antiparallel filaments, and duplicated poles are initially tethered together, an arrangement that instead favors parallel interactions. Pivoting of microtubules around spindle poles might help overcome this geometric bias, but the intrinsic pivoting flexibility of the microtubule–pole interface has not been directly measured, nor has its importance during early spindle assembly been tested. By measuring the pivoting of microtubules around isolated yeast spindle poles, we show that pivoting flexibility can be modified by mutating a microtubule-anchoring pole component, Spc110. By engineering mutants with different flexibilities, we establish the importance of pivoting in vivo for timely pole separation. Our results suggest that passive thermal pivoting can bring microtubules from side-by-side poles into initial contact, but active minus end–directed force generation will be needed to achieve antiparallel alignment.
Introduction
Duplicated spindle poles are usually tethered side by side when they begin nucleating the microtubules that will make up the spindle (Moens and Rapport, 1971; Byers and Goetsch, 1975; O’Toole et al., 1999; Winey and O’Toole, 2001). The microtubules have structural polarity and assemble with their fast-growing plus ends pointing outward from the poles and their minus ends anchored at the poles (Dammermann et al., 2003; Byers et al., 1978; Winey et al., 1995; McIntosh and Euteneuer, 1984). Motor proteins and cross-linkers then actively push the newly duplicated poles apart by bundling some of the microtubules from each pole together to form a force-generating “midzone” (Hoyt et al., 1992; Lim et al., 2009; Kapitein et al., 2005; Kashina et al., 1997; Ding et al., 1993). The plus end–directed activity of midzone motors (e.g., kinesin-5s) will only drive pole separation when the motors act on overlapping, antiparallel microtubules.
The initial side-by-side configuration of the poles and the importance of midzone pushing for pole separation are conserved features of mitosis (Leary et al., 2019; Crasta and Surana, 2006; Bettencourt-Dias and Glover, 2007; van den Wildenberg et al., 2008; Kapitein et al., 2005), and they raise a general question: How are antiparallel microtubule overlaps ensured? Building a spindle requires enough antiparallel interactions for plus end–directed motors to push the poles apart. But the arrangement of microtubules emanating from two nearby poles will disfavor antiparallel interactions, instead favoring parallel interactions. Midzone cross-linkers and motors bind preferentially to antiparallel microtubules (Kapitein et al., 2005; van den Wildenberg et al., 2008), but their approximately threefold preference for this arrangement seems inadequate to completely avoid formation of parallel bundles, which, if they were numerous enough, would inhibit pole separation. This problem is common across eukaryotes but seems particularly challenging in the budding yeast Saccharomyces cerevisiae. The spindle pole bodies (SPBs) of budding yeast are puck-shaped discs embedded in the nuclear envelope (Jaspersen and Winey, 2004; O’Toole et al., 1999; Byers and Goetsch, 1975). When SPBs are side by side, the surfaces from which they grow spindle microtubules face in the same direction. This geometry will strongly favor the creation of parallel microtubule arrays, the opposite of what is needed to support a midzone-pushing mechanism. Yet, like all eukaryotes, budding yeast reliably separate their poles and build a bipolar spindle with opposing, antiparallel microtubules.
If midzone pushing is required for separating SPBs from their side-by-side configuration, then pole-attached microtubules probably need to pivot so they can form the antiparallel overlaps the motors need to push the poles apart (Fig. 1 A). EM tomograms of side-by-side SPBs in S. cerevisiae show microtubules emanating at a wide variety of angles, with some filaments interdigitating over the tether between the two poles (O’Toole et al., 1999; Bullitt et al., 1997; Leary et al., 2019). In live Schizosaccharomyces pombe cells, dynamic pivoting of microtubules around spindle poles is visible by fluorescence microscopy and is implicated in kinetochore capture and spindle reassembly after cold stress (Kalinina et al., 2013; Blackwell et al., 2017; Winters et al., 2019). These observations suggest that pivoting might also be crucial for the initial conversion from side-by-side poles into a bipolar arrangement. But close proximity of the poles and the relatively short lengths of pole-anchored microtubules preclude direct observation of pivoting in vivo during this fundamental stage of spindle assembly. Moreover, while microtubules in S. pombe appear to pivot freely (Kalinina et al., 2013; Prelogović et al., 2019; Winters et al., 2019; Cojoc et al., 2016), mechanical flexibility at the microtubule–pole interface has not been directly measured or modified in any organism. Thus, the importance of pivoting specifically during initial establishment of spindle bipolarity remains uncertain.
Figure 1.
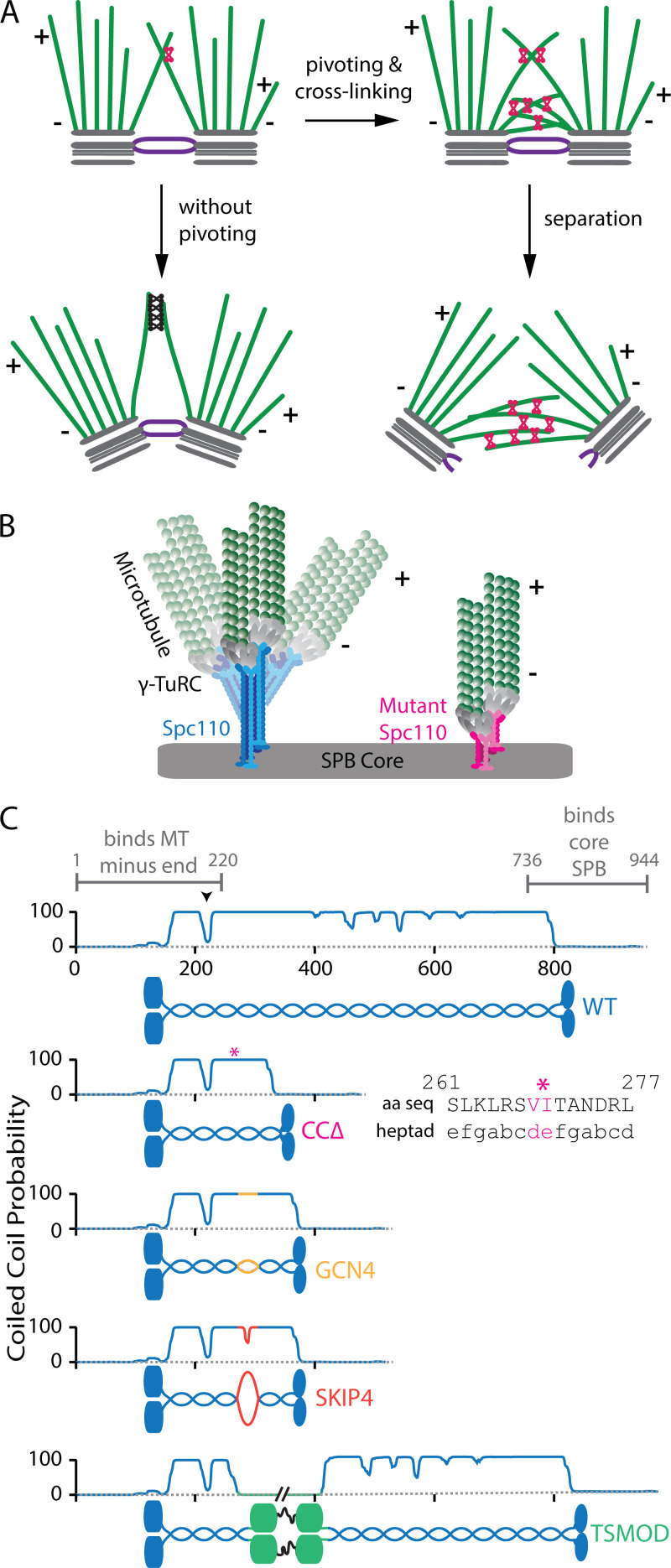
Hypothesized role of pivoting during spindle assembly and design of mutants to alter pivoting flexibility. (A) Pivoting of microtubules about their spindle pole attachments is a prerequisite for establishing the antiparallel interactions (red linkers) needed to drive pole separation. Pivoting might also help avoid an overabundance of parallel interactions (black linkers), which would inhibit pole separation. (B) Pivoting could occur via bending of the Spc110 tethering molecule, which links microtubule (MT) minus end–capping complexes (i.e., the γ-tubulin ring complexes, or γ-TuRCs) into the core of the spindle pole. (C) Four constructs designed to alter pivoting flexibility: CCΔ removes much of the WT coiled coil while preserving known protein-binding regions. GCN4 and SKIP4 are nearly identical in length, with short ectopic insertions. Predicted coiled coil propensity remains high across the GCN4 insertion but drops markedly in the SKIP4 insertion, which includes a distinct flexible hinge. TSMOD adds a large insertion, with spider silk sequence (black) flanked by fluorescent proteins (CFP and YFP, green globular domains). A break in the coiled coil required for binding the γ-TuRC (black arrowhead) was preserved in all mutants. Asterisks (magenta) mark the junction in CCΔ. Inset shows preserved heptad registration in the amino acid sequence (aa seq) surrounding the junction. Coiled coil probabilities were calculated using MARCOIL (Delorenzi and Speed, 2002; Zimmermann et al., 2018).
Here, by mutating a tethering protein, Spc110, which links microtubule minus ends to the SPB core, we altered the flexibility of pivoting around the S. cerevisiae SPB. Pivoting flexibility was measured directly by tracking thermal movements of individual microtubules nucleated in vitro from purified SPBs (Fong et al., 2016, 2017). A series of Spc110 constructs, engineered to have higher and lower flexibility than WT, illustrate that a long coiled coil within Spc110 is a key determinant of microtubule pivoting flexibility. The relative timing of SPB separation in cells expressing these constructs correlates with pivoting flexibilities measured in vitro. We conclude that microtubule pivoting is specifically important during early spindle assembly, when SPBs separate from their side-by-side configuration. Our results also suggest that thermal pivoting is sufficient to bring microtubules from side-by-side poles into contact, but probably insufficient to bring them all the way into antiparallel alignment. Thus, formation of antiparallel bundles probably occurs in two stages, with passive thermal pivoting bringing microtubules into initial contact and active minus end–directed force generators subsequently driving them into an antiparallel arrangement.
Results and discussion
Design of mutants to alter flexibility of microtubule-pole attachments
As the only known linker between the minus ends of nuclear microtubules and the core of the SPB, Spc110 seemed likely to confer flexibility to the SPB–microtubule interface (Fig. 1 B). Spc110 is a dimeric protein with globular N- and C-termini connected by a coiled coil stalk (Mirzayan et al., 1992; Kilmartin et al., 1993; Muller et al., 2005; Lyon et al., 2016). Its N-terminus (residues 1–220) binds the γ-tubulin small complex, which assembles into oligomeric caps on microtubule minus ends (Nguyen et al., 1998; Knop and Schiebel, 1998; Lyon et al., 2016; Kollman et al., 2010). Its C-terminus (residues 736–944) interacts with the core of the SPB (Viswanath et al., 2017; Muller et al., 2005; Adams and Kilmartin, 1999; Fig. 1 C). The stalk that connects its N- and C-termini forms a nearly continuous coiled coil ∼80 nm in length, with a few interruptions (Mirzayan et al., 1992; Kilmartin et al., 1993). Previous work showed that the length of this Spc110 coiled coil dictates the separation between the SPB core and the minus ends of attached microtubules (i.e., between “central” and “inner” plaques) and that large regions of the coiled coil are dispensable for viability (Kilmartin et al., 1993). Hypothesizing that bending within Spc110 might govern the degree to which microtubules can pivot about their SPB attachments, we designed a series of mutations in the coiled coil to either increase or decrease its mechanical flexibility.
For one mutant, we removed most of the coiled coil but retained all previously described interaction sites within its N- and C-termini (Fig. 1 C, magenta). Heptad repeats were carefully preserved across the junction. By shortening the coiled coil and removing the intermittent breaks, we reasoned that this coiled coil delete (CCΔ) construct would be less flexible than full-length WT Spc110. At the other extreme, we designed a construct that we hypothesized would be more flexible than WT. A 124-residue portion of the coiled coil was replaced with the tension sensor module (TSMOD; Grashoff et al., 2010; Brenner et al., 2016), which consists of 40 residues of spider silk protein sequence flanked by fluorescent proteins YFP and CFP (Fig. 1 C, green). Insertion of the stretchy spider silk sequence, the globular fluorescent proteins, and the unstructured linkers all seemed likely to confer extra flexibility. Because our CCΔ and TSMOD constructs had different total lengths, we designed two additional constructs predicted to differ in flexibility but with nearly identical lengths. Starting with our truncated CCΔ construct, we inserted into the junction site 33 amino acids of well-characterized, leucine zipper coiled coil from the transcription factor GCN4 (Harbury et al., 1993) or 34 amino acids of the SKIP4 motif from the stalk of myosin, which includes a distinct, flexible hinge (Taylor et al., 2015). Predicted coiled coil propensity for the GCN4 construct was continuously high across the insertion (Fig. 1 C, gold), suggesting that it would be relatively stiff. In contrast, predicted coiled coil propensity for the SKIP4 construct dropped markedly at the insertion due to the extra hinge (Fig. 1 C, orange), suggesting that this construct would be more flexible than GCN4.
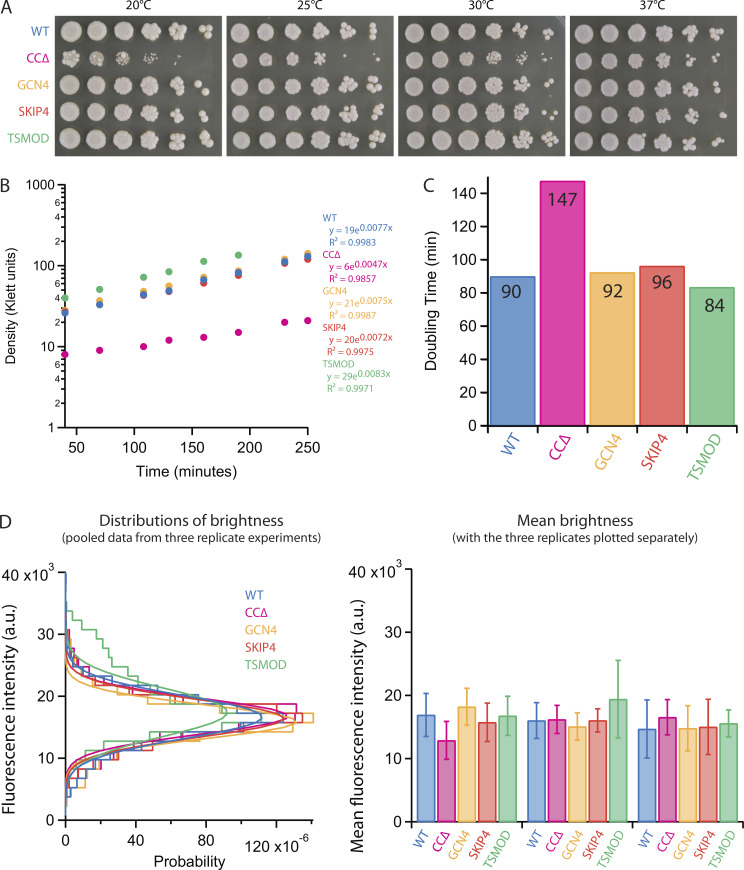
When transformed into yeast, each of the four mutant constructs supported cell viability as the sole copy of the SPC110 gene (Fig. S1 A). Growth rates for three of the four mutant strains, TSMOD, GCN4, and SKIP4, were indistinguishable from those of WT (Fig. S1, B and C). The CCΔ strain grew 1.6-fold more slowly and was cold sensitive, indicating that the CCΔ construct was not as well tolerated as the others. Nevertheless, SPBs in all the strains recruited nearly identical levels of Spc97 (Fig. S1 D), confirming that the engineered mutations did not interfere with assembly of Spc110 into the SPBs nor with its ability to recruit the γ-tubulin small complex. Moreover, all four strains grew sufficiently well for purifying SPBs and measuring the timing of SPB separation.
Figure S1.
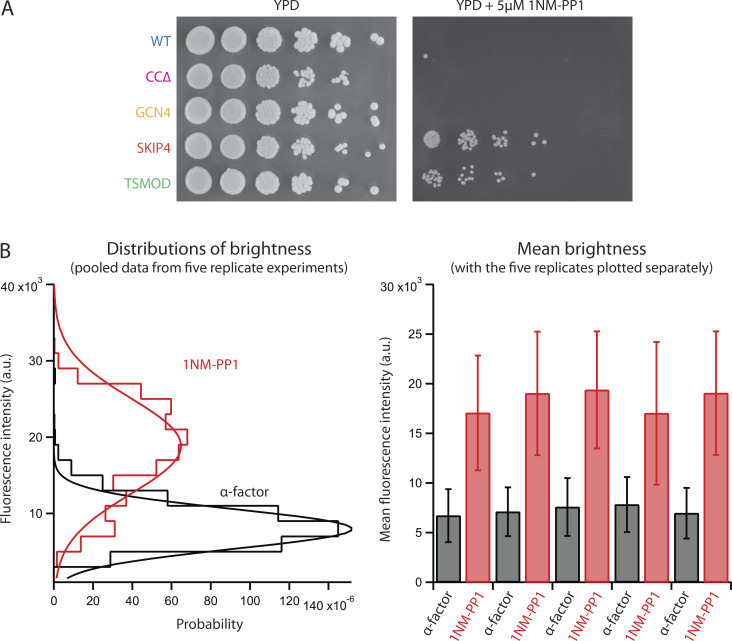
Growth data for WT and Spc110 mutant strains. (A) Serial dilutions of indicated strains grown at indicated temperatures. CCΔ cells exhibited cold sensitivity with reduced growth below 37°C. (B) Growth curves and parameters for best-fit lines for indicated strains grown at 30°C. (C) Doubling times calculated from the growth curves in B. (D) SPBs in all the mutant strains recruit similar levels of γ-tubulin small complex. Cells expressing a fluorescent-tagged component of the γ-tubulin small complex, Spc97-GFP, together with either WT or mutant Spc110 were imaged in asynchronous cell culture. Fluorescence intensities of individual green spots within the cells were then measured. At left: brightness distributions and Gaussian fits from pools of three replicate experiments. At right: average brightness from each replicate experiment (mean ± SD, obtained from Gaussian fits to brightness distributions, each with between 228 and 9,073 spots).
Engineered mutants affect microtubule pivoting flexibility in vitro
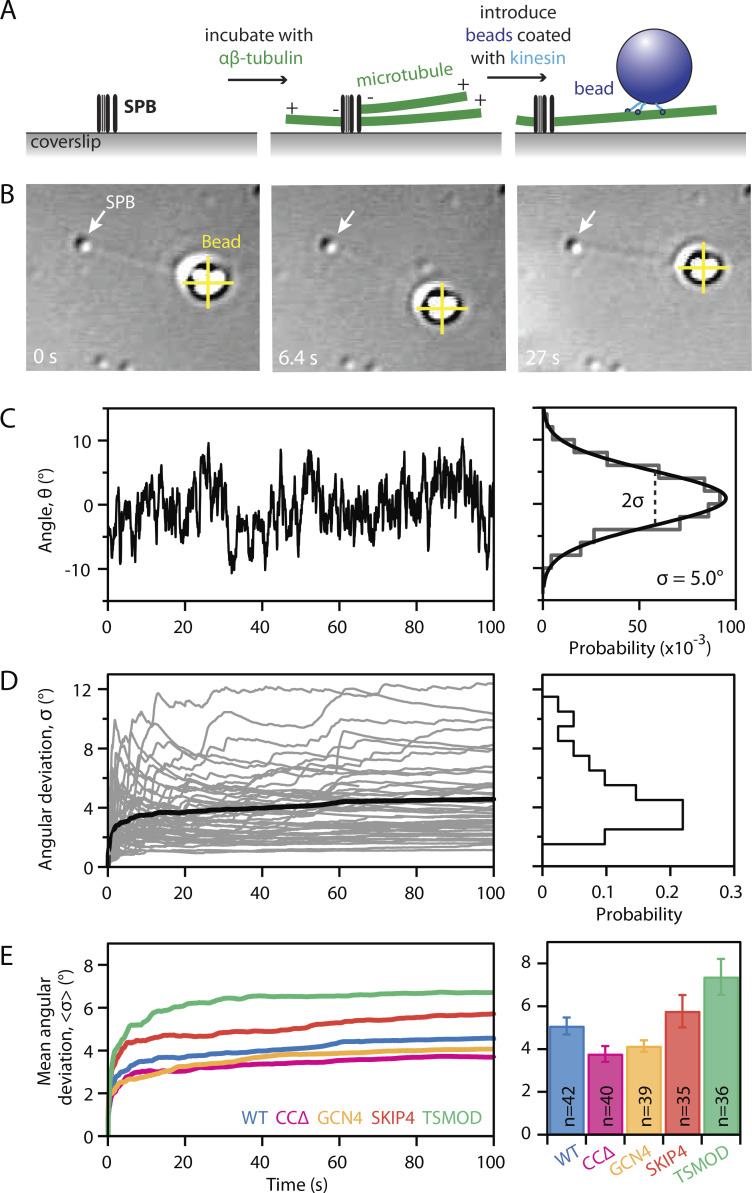
To assess the flexibility of SPB-microtubule attachments, we measured thermal pivoting of individual microtubules nucleated from isolated SPBs in vitro. The amount of pivoting in vitro provides a quantitative measure of intrinsic flexibility at the SPB–microtubule interface, avoiding possible contributions from microtubule cross-linkers, motor proteins, or other active processes that might affect pivoting in vivo (Hepperla et al., 2014; Winters et al., 2019). SPBs were isolated from cells expressing either WT Spc110 or one of the mutants via affinity purification followed by velocity sedimentation (Fong et al., 2016, 2017). The isolated SPBs were adhered to glass coverslips, microtubules were nucleated from them and stabilized with Taxol, and then free tubulin was washed out while kinesin-coated beads were introduced (Fig. 2 A). When viewed by video-enhanced differential interference contrast microscopy, the individual coverslip-anchored SPBs were clearly visible and the nucleated microtubules pivoted about their points of attachment to the SPBs (Fong et al., 2017). Beads bound to the microtubules served as fiducials, enabling automated tracking of the angular motion of the filaments (Fig. 2, B and C, left). The distribution of angles for each microtubule was fit with a Gaussian, and the standard deviation, , was used as a measure of pivoting flexibility (Fig. 2 C). After ∼100 s, the angular deviations equilibrated to a steady value (Fig. 2 D), indicating that the microtubules did not swivel freely but instead pivoted around a stable mean orientation. For each type of SPB, the average angular deviation, , was computed from many individual SPB-microtubule attachments (n ≥ 35; Fig. 2 E).
Figure 2.
Mutating Spc110 alters flexibility of microtubule pivoting about purified spindle poles. (A) Schematic of the assay. SPBs nonspecifically adsorbed to a coverslip (left panel) nucleate microtubules, which are stabilized with Taxol (middle). Kinesin-coated beads bound to the microtubules (right) allow automated tracking of microtubule orientation. (B) Time-lapse images of microtubule pivoting observed via video-enhanced differential interference contrast microscopy. A surface-adsorbed SPB appears as a small punctum (white arrows). Bead motion in two dimensions is tracked (yellow crosses) relative to the SPB, and angular deflection of the microtubule is computed. (C) Angular deflection, θ, of an SPB-microtubule attachment plotted against time (left) and corresponding distribution of angles (right) fit with a Gaussian. The standard deviation of the fit, σ, provides a measure of pivoting flexibility. (D) Evolution of the angular deviation, σ, for 42 WT SPB-microtubule attachments (gray curves, at left). The deviation typically reached a steady value after 100 s, indicating that the microtubules pivoted around a stable mean orientation. Black curve shows average of all 42 individual curves, which plateaus at <σ> = 5.1° ± 0.4° (mean ± SEM; n = 42 SPB-microtubule attachments). The distribution of angular deviations after 100 s is shown as a histogram (at right). (E) Mean angular deviation <σ> for microtubules pivoting around WT SPBs and indicated mutants. Evolution of <σ> is plotted versus time (at left). Bars (at right) show final, cumulative estimates computed from Gaussian fits (mean ± SEM; n = 35–42 SPB-microtubule attachments).
In general, the measured flexibilities accorded well with predictions based on the Spc110 coiled coil structures. The average angular deviation for microtubules emanating from WT SPBs was = 5.1 ± 0.4° (mean ± SEM; n = 42), whereas the average for mutant CCΔ SPBs, = 3.8 ± 0.4° (n = 40), was lower than for WT (Fig. 2 E), confirming that truncation of the Spc110 coiled coil reduces pivoting flexibility. Conversely, the average deviation for microtubules emanating from TSMOD SPBs, = 7.4 ± 0.8° (n = 36), was higher than for WT, confirming that the large insertion added into the Spc110 coiled coil increased its flexibility. The two mutant SPB types with isometric Spc110 constructs had intermediate flexibilities. SKIP4 ( = 5.8 ± 0.8°; n = 35) was more flexible than GCN4 ( = 4.1 ± 0.3°; n = 39), as expected given the hinge in the former construct. (P values for all pairwise combinations can be found in Table S1). Altogether, these in vitro data show that pivoting flexibility at the SPB–microtubule interface is determined in part by properties of the Spc110 coiled coil and that mutants can be engineered with increased and decreased flexibility relative to WT.
Flexibility mutants affect timing of pole separation in vivo
Separation of duplicated poles is fundamental to the formation of a bipolar spindle and is driven by motor proteins (e.g., kinesin-5s), which slide antiparallel microtubules to push the poles apart (Blangy et al., 1995; Kapitein et al., 2005; Scholey et al., 2016; Hoyt et al., 1992; Roof et al., 1992). Following duplication, SPBs are tethered side by side via a “bridge,” and their microtubule-nucleating surfaces are cooriented (Bullitt et al., 1997; O’Toole et al., 1999). Thus, some microtubules presumably must pivot to interdigitate over the bridge in an antiparallel fashion before motors can drive pole separation (Kilmartin et al., 1993; O’Toole et al., 1999; Hepperla et al., 2014; Leary et al., 2019). Considering this likely sequence of events, we hypothesized that altering the flexibility of pivoting might affect the timing of pole separation in vivo.
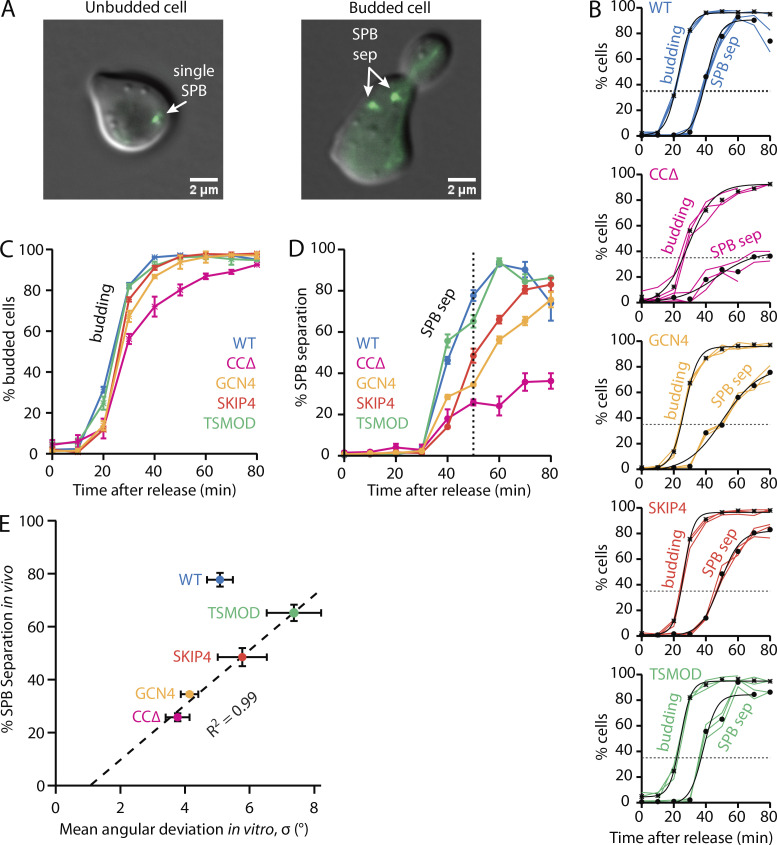
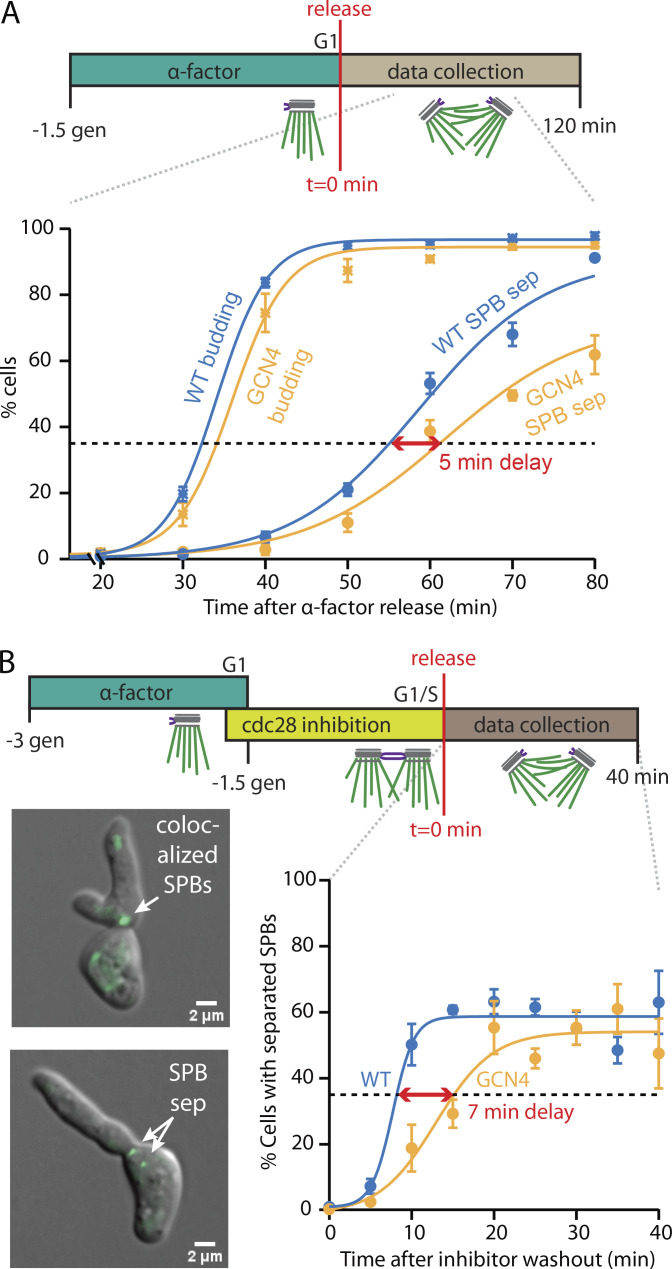
To monitor the timing of SPB separation in cells, we tagged their SPBs with Spc72-GFP. The cells were initially synchronized by treatment with α-factor, which arrests them in G1 of the cell cycle before duplication of their SPBs (Winey and O’Toole, 2001; Winey and Byers, 1993; Byers and Goetsch, 1975). After release from α-factor, the cells grow buds while their SPBs are duplicated and separated to form a bipolar spindle (Fig. 3 A). We counted cells with two visible poles and cells with buds at 10-min intervals to follow the progress of each population over time (Fig. 3 B). The delay between 35% cell budding and 35% SPB separation, interpolated from the fitted curves, provided a measure of SPB separation timing for each cell type. Cells with WT Spc110 separated their SPBs 18 min after bud emergence (Fig. 3 B, blue), whereas the delay was much longer, 44 min, for cells with the least flexible mutant, CCΔ (Fig. 3 B, magenta). Cells with the isometric Spc110 mutants, GCN4 or SKIP4, were also delayed relative to wild type, separating their SPBs 24 or 23 min after bud emergence, respectively (Fig. 3 B, gold and orange). Cells with the most flexible Spc110 construct, TSMOD, exhibited the briefest delay, 15 min (Fig. 3 B, green).
Figure 3.
Flexibility mutants affect timing of pole separation in vivo. (A) Representative images of WT cells after synchronous release from α-factor. Immediately after release (t = 0 min, left), the cells are unbudded and their unduplicated SPB appears as a single green spot. Later (t = 50 min, right), the cells have buds and their SPBs are duplicated and separated (sep). (B) Timing of bud emergence and SPB separation. For each time point, the percentages of cells with buds or with separated SPBs were counted. Each colored line represents a biological replicate of the α-factor arrest. Mean percentages of cells with buds (crosses) or with separated SPBs (closed circles) were fit with sigmoidal curves. Horizontal dotted lines indicate 35% thresholds used to compare SPB separation timing relative to bud emergence. (C and D) Overlays of bud emergence (C) and SPB separation (D) versus time. Mean percentages from B are replotted here for comparison. Error bars represent SD (from n = 3 cultures). (E) SPB separation timing in vivo correlates with pivoting flexibility at the SPB–microtubule interface measured in vitro. The percentages of cells with separated SPBs at t = 50 min after α-factor release (indicated by dotted vertical line in D) are plotted against angular deviation (from Fig. 2 E). Mutant data are fitted with a line. Vertical error bars represent SD (from n = 3 cultures). Horizontal error bars represent SEM (from n = 35–42 SPB-microtubule attachments).
To compare WT and mutant data directly, without curve fitting or interpolation, the fractions of cells measured at each time point were averaged across three independent cultures and overlaid (Fig. 3, C and D). Bud emergence followed a similar time course for all cell types (Fig. 3 C). At 50 min after release from α-factor, the fractions of mutant GCN4, SKIP4, and TSMOD cells with buds were very high (>90%) and were indistinguishable from the fraction of WT cells with buds (97% ± 1%; mean ± SD from n = 3 cultures). The mutant CCΔ cells exhibited a modest delay in bud emergence, but most (80% ± 5%) had nevertheless grown buds at 50 min after release, and they reached WT levels of budding at 80 min. These data show that all the cultures were well synchronized and suggest that the Spc110 coiled coil mutations had little effect on the budding process. In contrast, the fractions of cells with separated SPBs varied more widely (Fig. 3 D). Most WT cells, 78% ± 5%, had separated their SPBs by 50 min after release, but significantly fewer of the mutant cells had done so. Cells with the least flexible Spc110 mutant, CCΔ, exhibited the most severe delay, with only 26% ± 3% separating their SPBs after 50 min. Mutant GCN4 and SKIP4 cells also exhibited delays, with 34% ± 1% and 48% ± 6%, respectively, separating their SPBs by 50 min. Cells with the most flexible TSMOD construct exhibited only a slight delay compared with WT, with 65% ± 5% of the mutant TSMOD cells separating their SPBs by 50 min.
To examine the relationship between SPB separation and pivoting flexibility, we plotted the separation data measured in vivo versus the pivoting data measured in vitro (Fig. 3 E). Results from both sets of experiments were correlated, with data from the four mutants falling along a single line. These observations show that reduced pivoting at the SPB–microtubule interface in vitro is associated with delayed SPB separation in vivo.
Notably, cells expressing WT Spc110 fall distinctly above the line in Fig. 3 E. This observation suggests that, relative to the mutants, WT Spc110 confers a slight advantage in SPB separation efficiency that is independent of pivoting flexibility. A short segment of the Spc110 coiled coil with no known function (residues 267–387) is missing in all the mutants. We speculate that this region might be involved in unknown interactions or processes that help promote SPB separation.
Low-flexibility mutant specifically delays pole separation
Our hypothesis was that reduced pivoting would specifically delay the SPB separation process. However, because α-factor arrests cells before SPB duplication, every cell after release from α-factor must first duplicate its SPB and then insert the new copy into the nuclear envelope before SPB separation can begin (Crasta and Surana, 2006; Jaspersen and Winey, 2004). To isolate the separation process from the duplication and insertion processes, we reversibly arrested cells by inhibiting Cdc28, the main cyclin-dependent kinase in budding yeast. Cells carrying an analogue-sensitive cdc28-as1 allele can be arrested by the small-molecule chemical inhibitor 1NM-PP1, which selectively inhibits the modified Cdc28-as1 enzyme (Bishop et al., 2000). Inhibiting Cdc28-as1 in asynchronous cultures arrests the cells at two distinct, Cdc28-dependent transition points in the cell cycle, G1/S and G2/M. However, by initially synchronizing the cells in G1 using α-factor and subsequently adding 1NM-PP1 to inhibit Cdc28-as1, the population can be uniformly synchronized at G1/S (Bishop et al., 2000), with duplicated SPBs inserted into the nuclear envelope but not yet separated (Crasta et al., 2006; Fig. S2). Synchronizing cells at this later stage allowed us to measure the effect of a low-flexibility Spc110 mutant specifically on SPB separation.
Figure S2.
Characterization of cdc28-as1 strains expressing either WT Spc110 or the flexibility mutants. (A) Serial dilution of cdc28-as1 cells expressing the indicated Spc110 constructs grown in the absence (left) or presence (right) of 5 µM 1NM-PP1, which arrests the cells by specifically inhibiting the modified enzyme Cdc28-as1. Notably, cells expressing SKIP4 or TSMOD were able to escape the 1NM-PP1 arrest. Previous work indicates that Cdc28 inhibition blocks SPB separation due to reduced levels of kinesin-5 motors and cross-linkers (Crasta et al., 2006). We therefore speculate that our SKIP4 and TSMOD strains escaped this arrest because the increased flexibility at their SPB–microtubule interfaces meant that fewer motors and cross-linkers were needed. (B) Confirmation that when cdc28-as1 cells are synchronized in G1/S, their SPBs are duplicated but unseparated. Populations of cells expressing Spc110-GFP were either arrested in G1 using α-factor alone or synchronized at G1/S using the double-synchronization protocol shown in Fig. 4 B, with α-factor followed by 500 nM 1NM-PP1. Fluorescence intensities of individual green spots within the cells were then measured during the α-factor (G1) or 1NM-PP1 (G1/S) arrest. Spot brightness roughly tripled during the 1NM-PP1 arrest. This tripling indicates that the spots in 1NM-PP1 (G1/S) contained duplicated but unseparated SPBs, tethered too closely together to resolve by conventional light microscopy, given that individual SPBs during α-factor (G1) arrest contain two thirds the amount of Spc110 found at SPBs in cycling cells (Yoder et al., 2003). At left: brightness distributions from pools of five replicate experiments. At right: average brightness from each replicate experiment (mean ± SD, obtained from Gaussian fits to brightness distributions, each with between 62 and 886 spots).
We constructed five strains expressing either WT Spc110 or each of the four flexibility mutants together with the cdc28-as1 allele. Unfortunately, cells with CCΔ and cdc28-as1 grew extremely slowly, making synchronization impractical. Moreover, the two most flexible mutants, SKIP4 and TSMOD, both escaped the arrest (Fig. S2 A), precluding their synchronization. However, cells with either WT Spc110 or with the low-flexibility GCN4 mutant in combination with cdc28-as1 grew and arrested sufficiently well to be initially synchronized with α-factor, subsequently arrested with 1NM-PP1, and then finally released from G1/S for examination of SPB separation by fluorescence microscopy. As a control, we also examined SPB separation for these same two strains after release from α-factor alone (i.e., from G1, without 1NM-PP1).
After release from G1 using α-factor alone, the cdc28-as1 cells with WT Spc110 separated their SPBs 22 min after budding, and those with GCN4 separated their SPBs 27 min after budding, a delay 5 min longer than that of WT (Fig. 4 A). This relative timing recapitulated our earlier measurements with strains lacking cdc28-as1, confirming that the modified Cdc28-as1 kinase retains sufficient activity to support normal SPB duplication, insertion, and separation. When the two strains were instead released from G1/S using the double-synchronization protocol with α-factor followed by 1NM-PP1, SPB separation occurred much more quickly, presumably because SPB duplication and insertion had already been completed before the release. The GCN4 cells still separated their SPBs 7 min later than those with WT Spc110 (Fig. 4 B). This observation, that cells carrying the low-flexibility GCN4 mutant consistently separate their SPBs 5–7 min later than cells with WT Spc110, irrespective of whether they are synchronized before or after SPB duplication/insertion, indicates that the mutant specifically delays the SPB separation process.
Figure 4.
Microtubule pivoting flexibility specifically affects rate of pole separation. (A) Timing of bud emergence and SPB separation for cdc28-as1 cells expressing GCN4 (gold) versus WT (blue) Spc110 after release from G1 using α-factor alone. Cells were synchronized in G1 with single, unduplicated SPBs using α-factor for 1.5 generation (gen)-times. For each time point after release, percentages of cells with buds or with separated SPBs were counted. SPB separation (sep) was delayed in GCN4 cells by 5 min relative to WT cells. (B) Timing of bud emergence and SPB separation for cdc28-as1 cells expressing GCN4 (gold) versus WT (blue) Spc110 after release from G1/S using the double-synchronization protocol. Cells were initially synchronized with single SPBs using α-factor alone and then transferred into 1NM-PP1 alone to allow SPB duplication and insertion. For each time point after release, percentages of cells with separated SPBs were counted. SPB separation was delayed in GCN4 cells by 7 min relative to WT cells. Inset: Representative images of cdc28-as1 cells expressing WT Spc110. Immediately after release from G1/S, the duplicated SPBs are colocalized at the bud neck (t = 0 min, top) and appear as a single green spot. Later (t = 20 min, bottom), the SPBs are separated (sep). Hyperpolarized cell shape is characteristic of Cdc28-as1 inhibition (Bishop et al., 2000; Crasta et al., 2006). Symbols and error bars in A and B represent mean ± SD (from n = 3 cultures). Horizontal dotted lines indicate 35% thresholds used to compare SPB separation timing across strains.
Implications for the mechanisms underlying pole separation
Many studies have shown the importance of plus end–directed motility, generated by kinesin-5 motors acting on antiparallel midzone microtubules, for pushing spindle poles apart (Leary et al., 2019; Gerson-Gurwitz et al., 2011; Saunders and Hoyt, 1992; Saunders et al., 1995; Hoyt et al., 1992; Hagan and Yanagida, 1990; van den Wildenberg et al., 2008; Heck et al., 1993; Kapitein et al., 2005; Shimamoto et al., 2015; Sawin et al., 1992; Enos and Morris, 1990; Blangy et al., 1995; Singh et al., 2018; Kashina et al., 1997). However, few have focused specifically on the earliest separation events, when the poles transition from closely tethered together to apart. This transition is fundamental but difficult to study in vivo owing to the close initial proximity of the poles and the high density of microtubules in their vicinity, so the precise mechanisms remain uncertain. Nevertheless, our direct measurements of flexibility at the SPB–microtubule interface have implications for how thermal fluctuations, passive cross-linkers, and minus end–directed force generators might create the antiparallel midzone required for plus end–directed motors to drive pole separation.
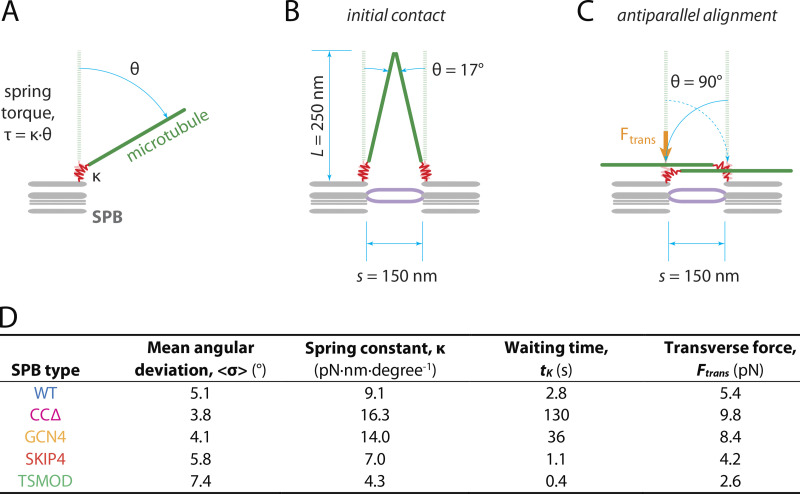
Assuming ideal “Hookean” spring behavior and applying the equipartition principle (Howard, 2001), the 5.1° angular deviation we measured for microtubules pivoting around WT SPBs implies a torsional spring constant of κ = 9.1 pN·nm·degree−1 (Fig. 5, A and D). Working against this torsional spring, thermal pivoting is expected to bring the short microtubules typically found on side-by-side poles (O’Toole et al., 1999) into frequent contact; for example, spontaneous deflections of = 17° (Fig. 5, B and D) should occur every few seconds. But complete deflection to = 90° (Fig. 5 C) would essentially never occur via thermal pivoting alone, implying that antiparallel alignment probably cannot be achieved by a purely passive mechanism (see Materials and methods for calculations underlying these estimates). However, once a pair of microtubules from the two SPBs makes contact, microtubule cross-linkers can bind them together, trapping them in an arrangement where active, minus end–directed force generators could drive them into antiparallel alignment. Only about 5 pN of minus end–directed transverse force would suffice to overcome the torsional springs and deflect the filaments completely to = 90°. This modest force could be supplied in S. cerevisiae by several Cin8 motors, which under certain circumstances can generate minus end–directed force (Roostalu et al., 2011; Gerson-Gurwitz et al., 2011; Fallesen et al., 2017), or in S. pombe by Cut7 or Pkl1 (Winters et al., 2019; Yukawa et al., 2018). In principle, it could also be supplied by a disassembling microtubule tip, which can generate substantial pulling force (Driver et al., 2017; Volkov et al., 2013; Grishchuk et al., 2005; Lombillo et al., 1995) and might explain how S. pombe and S. cerevisiae cells can sometimes build spindles even without mitotic kinesin activity (Rincon et al., 2017; Yukawa et al., 2020; Leary et al., 2019).
Figure 5.
Estimated torsional spring constants, waiting times for thermal pivoting, and transverse forces for antiparallel alignment. (A) The SPB–microtubule interface was assumed to behave as a Hookean spring (red), whose angular deflection, θ = τ/κ, varies linearly with applied torque, τ, where κ represents the torsional spring constant. (B) The waiting time for thermal pivoting to bring microtubules from adjacent SPBs into contact was estimated from the first-passage time for deflection to θ = 17°, the minimum angle needed to bring the tips of two microtubules of length L = 250 nm, separated laterally by distance s = 150 nm, into contact. (C) The transverse force required for deflection to θ = 90° was estimated by assuming a force acting perpendicularly to the microtubule at distance s = 150 nm from its pivot point (Ftrans, orange arrow). Values of L and s were chosen to match the physiological arrangement of SPBs when they are tethered together by a bridge (purple), after duplication and insertion into the nuclear envelope (O’Toole et al., 1999). (D) Torsional spring constants, κ, and transverse forces required to bring microtubules from adjacent, side-by-side SPBs into antiparallel alignment, Ftrans, estimated from the measured angular deviations, <σ>. Approximate waiting times for thermal pivoting to bring microtubules from adjacent SPBs into initial contact, tK, were estimated using Kramer’s rate theory (Howard, 2001). See Materials and methods for details.
The variation in SPB separation timing across our mutant strains can be explained by the different flexibilities at their SPB–microtubule interfaces. Angular deviations for microtubules pivoting around the mutant SPBs ranged from 3.8° to 7.4°, which corresponds to a fourfold range of torsional spring constants (from = 4.3 to 16 pN·nm·degree−1) and roughly a 300-fold range in the frequency at which microtubules from side-by-side poles will thermally pivot into contact (Fig. 5 D). A short microtubule attached via the most flexible mutant, TSMOD, for example, is predicted to deflect by = 17° two or three times per second, whereas the same microtubule attached via the stiffest mutant, CCΔ, would do so only about once every 2 min. This substantial delay might explain the slow growth and cold sensitivity of CCΔ cells. Overall, the mutants span a wide range of flexibilities, and the stiffer ones should significantly increase the time required for a cell to build an antiparallel bundle between side-by-side SPBs.
Concluding remarks
Seeking to engineer mutants with different pivoting flexibilities, we focused on the tethering protein Spc110, which contains a long coiled coil that links the minus ends of spindle microtubules into the core of the SPB. Our design strategy assumed that shorter coiled coils with fewer putative “hinges” (i.e., fewer breaks in predicted coiled coil propensity) would be stiffer. The angular deviations we measured for microtubules pivoting around SPBs in vitro confirm our strategy was successful and establish that the coiled coil of Spc110 is indeed a key determinant of pivoting flexibility. The relative timing of SPB separation in cells expressing the various Spc110 mutants correlates strongly with pivoting flexibility in vitro, indicating that intrinsic, mechanical resistance to pivoting at the SPB–microtubule interface poses a significant barrier that S. cerevisiae cells must overcome before separating their SPBs.
Recent work implicated microtubule pivoting in reassembly of spindles in S. pombe after release from cold stress, which triggers spindle disassembly and creates a situation with the two SPBs already far apart from one another (Winters et al., 2019). This approach enabled direct observation of microtubules pivoting randomly around the two poles, making initial contact, and then being driven into antiparallel alignment by minus end–directed movement of the S. pombe kinesin-5, Cut7. However, the role of pivoting during normal spindle assembly, beginning with side-by-side poles, could not be examined. Our work now establishes that pivoting is important for separation of poles from the normal, initially side-by-side arrangement.
We took advantage of the uniquely well-understood molecular architecture of S. cerevisiae SPBs, but the question of how dividing cells avoid parallel microtubule interactions and create instead the antiparallel bundles needed to drive pole separation is common across eukaryotes. Duplicated spindle poles are generally close together at the onset of mitosis, when their microtubule nucleation activity dramatically increases. Thus, microtubule pivoting flexibility could facilitate the formation of antiparallel bundles between side-by-side poles in a variety of cell types. In metazoan cells, inward pulling forces generated by kinesin-14s help keep duplicated spindle poles close together until mitosis (Decarreau et al., 2017; Hata et al., 2019). In principle, these same minus end–directed motors could also drive pivoting of microtubules into antiparallel alignment between the neighboring poles, thereby enabling plus end–directed kinesin-5 motors to subsequently push the poles apart. As molecular understanding of microtubule–centrosome linkages improves, it should become possible to engineer linkages with altered mechanical properties in metazoan cells and test the role of microtubule pivoting flexibility more broadly.
Materials and methods
Mutant design and plasmids
All plasmids used in this study are listed in Table S2. The TSMOD was inserted into the coiled coil of Spc110, replacing 372 base pairs (base pairs 267–387) of endogenous Spc110 sequence. The TSMOD consisted of 40 amino acids of spider silk protein sequence flanked N-terminally by YFP and C-terminally by CFP. The yeast-integrating plasmid (pAZ127) containing the TSMOD in Spc110 was verified by sequencing.
The CCΔ mutant was designed to contain the minimal coiled coil that would maintain the interaction sites between the γ-tubulin small complex and the N-terminus of Spc110 and the interactions of the SPB core proteins with the C-terminus of Spc110. The sequence of the CCΔ was analyzed by MARCOIL (Delorenzi and Speed, 2002; Zimmermann et al., 2018) to verify high coiled coil probability across the junction site, such that the heptad repeats were maintained. The GCN4 and SKIP4 mutant alleles were similarly designed and analyzed by MARCOIL. To insert the GCN4 domain and the SKIP4 domain, DNA fragments spanning the coiled coil deletion between restriction sites BglII and BseRI were synthesized (GenScript), digested by restriction enzymes, and ligated into an integrating plasmid containing the CCΔ allele. The constructs were verified by sequencing.
Strains and media
All yeast strains used in this study were derived from W303 and are listed in Table S3. In strains used for SPB purification, Spc97 was C-terminally tandem affinity purification (TAP)–tagged by PCR amplification of the TAP-kanMX6 cassette from pFA6a-CTAP-kanMX6-2XPA using primers that shared homology with the flanking sequences of the SPC97 stop codon. Spc72 was C-terminally tagged with GFP by PCR amplification of the GFP-HIS3MX6 cassette from pFA6a-GFP(S65T)-HIS3MX6 (Wach et al., 1997) using primers that shared homology with the flanking sequences of the SPC72 stop codon. Spc110 was C-terminally tagged with GFP by PCR amplification of the GFP-kanMX6 cassette from pFA6a-GFP(S65T)-kanMX6 (Wach et al., 1997) using primers that shared homology with the flanking sequences of the SPC110 stop codon.
Integrating plasmids containing the Spc110 mutant alleles were linearized with MluI and transformed into HSY2-1C for a plasmid shuffle. Briefly, the endogenous copy of SPC110 was deleted by replacement with TRP1. The haploid carried a plasmid, pHS26, with a WT copy of SPC110 and ADE3. The presence of ADE3 results in a buildup of red pigment in the yeast cells. When the mutant allele of SPC110 was integrated into yeast cells, pHS26 was stochastically lost, resulting in white colonies. These white colonies were isolated, and the presence of the mutant allele of Spc110 was verified by PCR and sequencing (Widlund and Davis, 2005).
Yeast extract–Peptone–Dextrose (YPD) medium is as described (Burke et al., 2000). YPD 3X Ade medium is YPD medium with 15 mg/ml adenine. α-Factor (GenScript) was stored in methanol at 10 mg/ml and was used in experiments at a final concentration of 6 µM. InSolution PP1 Analogue II (1NM-PP1; EMD Millipore) was used at a final concentration of 500 nM.
Cell synchronization
To study SPB separation in vivo, strains were constructed with WT or mutant Spc110 and with Spc72-GFP to label the poles with a fluorescent marker. To study SPB separation relative to bud emergence, cells were arrested with α-factor and imaged after release from arrest. Cells were grown in YPD culture to 30 Klett units, and α-factor was added to 6 µM final concentration. Cells were arrested for 1.5 generations at 30°C. Cells were sonicated, filtered, and resuspended in fresh prewarmed YPD. Samples were taken from the culture at 10-min intervals for 2 h for formaldehyde fixation. Cells were fixed in 3.7% formaldehyde for 5 min before pelleting and resuspending in PBS.
To study SPB separation relative to SPB duplication and insertion into the nuclear envelope, cells were first synchronized with α-factor before they were arrested with 1NM-PP1. Cells were grown in YPD 3X Ade culture to 30 Klett units. α-Factor was added to a final concentration of 6 µM, and cells were arrested for 1.5 generations at 30°C. For the final 20 min of α-factor arrest, 1NM-PP1 was added to a final concentration of 500 nM. Cells were sonicated, filtered, and resuspended in fresh prewarmed YPD 3X Ade. 1NM-PP1 was added to a final concentration of 500 nM. Cells were arrested for 1.5 generations at 30°C. Cells were sonicated, filtered, and resuspended in fresh prewarmed YPD 3X Ade, and samples were fixed with formaldehyde every 5 min for 40 min after release. Cells were fixed in 3.7% formaldehyde for 5 min before pelleting and resuspension in PBS. For each strain, arrests were completed in technical triplicates.
Fluorescence microscopy
Images were acquired using an Axio Observer 7 inverted microscope (Zeiss), a Plan-Apochromat 63×/1.46 Oil Korr objective (Zeiss), a SPECTRA light engine Generation III (Lumencor), and an ORCA-Flash4.0 V3 digital complementary metal oxide semiconductor (CMOS) camera (Hamamatsu). Exposures were 500 ms for GFP and 60 ms for differential interference contrast (DIC) imaging. Three z-stacks were taken for each field of view. For each of the formaldehyde-fixed samples, cells were mounted on an agarose pad as previously described (Muller et al., 2005). Images were processed using ZEN software (Zeiss) and exported as 16-bit uncompressed Tagged Image File Format (TIFF) files. Fluorescence intensity measurements were made with Fluorcal, custom MATLAB software described previously (Shimogawa et al., 2010).
SPB purification
SPBs were purified by a C-terminal TAP-tag on Spc97, as previously described (Fong et al., 2016). Cells were grown in YPD and harvested at 150 Klett units. Yeast cells were pelleted and washed two times with dH2O before resuspension in buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [Hepes], pH 7.4, 1.2% polyvinylpyrrolidone [average molecular weight 40,000], 1 mM DTT, 1 mM PMSF, 4 µg/ml aprotinin, 4 µg/ml chymostatin, 4 µg/ml leupeptin, 4 µg/ml pepstatin, 10 mM sodium fluoride, 1 mM sodium pyrophosphate, and 1 mM β-glycerophosphate). Cells were pelleted and flash frozen after extrusion through a syringe into liquid nitrogen. Cells were lysed by cryogrinding in a PM100 ball mill grinder (Retsch). Lysed cells were resuspended and homogenized in lysis buffer (20 mM Hepes buffer, pH 7.4, 0.5% Triton X-100, 2 mM MgCl2, 100 µM GTP, 1 mM ATP, 1 mM DTT, 1 mM PMSF, 4 µg/ml aprotinin, 4 µg/ml chymostatin, 4 µg/ml leupeptin, 4 µg/ml pepstatin, 10 mM sodium fluoride, 1 mM sodium pyrophosphate, 1 mM β-glycerophosphate, and 5% glycerol) with 300 mM NaCl. The lysate was cleared at 2,000× g for 10 min at 4°C. Dynabeads (Invitrogen) conjugated to rabbit immunoglobulin G (according to manufacturer’s protocols) were incubated with the cleared lysate for 30 min at 4°C. The Dynabeads were magnetized and washed three times with lysis buffer with 200 mM NaCl. To elute the SPBs, the Dynabeads were resuspended in tobacco etch virus (TEV) cleavage buffer (40 mM Hepes buffer, pH 7.4, 200 mM NaCl, 2 mM MgCl2, 1 mM GTP, 1 mM ATP, 1 mM EDTA, 1 mM DTT, and 5% glycerol) and incubated with 1 µg of TEV for 2 h at 4°C.
Sucrose gradients were made by allowing five steps of sucrose solutions (10%, 20%, 30%, 40%, 2.5 M sucrose in 10 mM Bis-Tris buffer, pH 6.5, and 0.1 mM MgCl2) to equilibrate for 2 h at 4°C. The TEV eluate was applied to the sucrose gradient and spun at 50,000 rpm for 5 h at 4°C in a TLS55 rotor (Beckman Coulter). Fractions were removed from the top of the gradient with wide-bore tips. The presence of SPBs was determined by Western blot analysis, probing for Spc110 and Spc97. Aliquots of SPBs were flash frozen and stored at −80°C.
Preparation of kinesin-coated beads
An N-terminal derivative of the Drosophila melanogaster kinesin heavy chain, DmK401, was purified as previously described (Asbury et al., 2003). DmK401 is a homodimer of the N-terminal 401 amino acids of the kinesin heavy chain with a C-terminal hexahistidine tag. DmK401 was expressed in BL21 Star (DE3) cells (Invitrogen). Cultures were induced with 1 mM IPTG for 2 h at 20°C. Rifampicin was added to a final concentration of 200 µM, and cultures were grown overnight at 20°C. Cells were pelleted and resuspended in an equivalent volume of lysis buffer (250 mM potassium phosphate buffer, pH 7.6, 300 mM KCl, 10 mM imidazole, pH 7.0, 1 mM β-mercaptoethanol, 4 mM MgCl2, and Complete protease inhibitors [Roche]), lysed with a French press, and clarified at 18,000 rpm for 20 min at 4°C. The cleared lysate was added to 5 ml of nickel-nitrilotriacetic acid (Ni-NTA) resin and incubated for 1 h at 4°C on a nutator. The resin was washed twice with wash buffer (250 mM potassium phosphate buffer, pH 7.6, 300 mM KCl, 25 mM imidazole, pH 7.0, 1 mM β-mercaptoethanol, 4 mM MgCl2, and Complete protease inhibitors). The kinesin was eluted from the resin with four elutions of increasing imidazole concentrations (50 mM, 100 mM, 200 mM, and 300 mM imidazole in 250 mM potassium phosphate buffer, pH 7.6, 300 mM KCl, 1 mM β-mercaptoethanol, 4 mM MgCl2, and Complete protease inhibitors). Aliquots of purified kinesin were flash frozen and stored at −80°C.
Streptavidin-coated polystyrene beads (0.56 µm in diameter; Spherotech) were functionalized with biotinylated anti-His5 antibodies (Qiagen) and stored with continuous rotation at 4°C in BRB80 (80 mM Pipes buffer, 1 mM MgCl2, and 1 mM EGTA, pH 6.9) supplemented with 8 mg/ml BSA. Before each experiment, 6 pM beads was incubated with purified kinesin for 1 h at 4°C in assay buffer (BRB80, 5 mg/ml BSA, 11.5 µM Taxol, and 1 mM DTT). Beads were sonicated for 2 min to reduce clumping before adding to the flow chamber.
Thermal fluctuation measurements
To measure SPB-microtubule attachment flexibility in vitro, SPBs were imaged in a flow chamber (∼10 µl volume). The flow chamber was constructed with a glass slide, double-stick tape, and a KOH-cleaned coverslip, as previously described (Fong et al., 2017). SPBs were diluted with 5× BRB80 (400 mM Pipes buffer, pH 6.8, 5 mM MgCl2, and 5 mM EGTA), 40 mg/ml BSA, and 2.67 M KCl to a final concentration of 1× BRB80, 8 mg/ml BSA, and 500 mM KCl. The diluted SPBs were flowed into the chamber and allowed to nonspecifically adhere to the coverslip for 30 min at room temperature. The adhered SPBs were washed thoroughly with BRB80. Bovine brain tubulin, purified as previously described (Castoldi and Popov, 2003), was cleared of aggregates by ultracentrifugation at 90,000 rpm for 10 min at 4°C in a TLA100 (Beckman Coulter). The tubulin polymerization buffer (1× BRB80, 1 mg/ml κ-casein, 5 mg/ml BSA, 2 µM Taxol, 1 mM GTP, 1 mM DTT, and 20 µM cleared tubulin) was flowed into the chamber. Taxol-stabilized microtubules nucleated from SPBs at room temperature for 9 min. Free tubulin was washed out with wash buffer (1× BRB80, 1 mg/ml κ-casein, 5 mg/ml BSA, 11.5 µM Taxol, and 1 mM DTT). Kinesin-coated beads (described above) were flowed in with an oxygen-scavenging system (500 µg/ml glucose oxidase, 60 µg/ml catalase, and 25 mM glucose). The ends of the flow chamber were sealed to prevent evaporation.
The laser trap was described previously (Akiyoshi et al., 2010; Franck et al., 2010; Sarangapani and Asbury, 2014; Fong et al., 2017). All laser trap experiments were performed in a temperature-controlled room maintained at 23°C. SPBs with nucleated microtubules were identified using the DIC imaging module (part of the laser trap setup). At the beginning of each experiment, some microtubules already had a bead attached, while others were bare. For bare microtubules, beads were trapped in solution and brought close to the microtubule to promote bead-microtubule attachment. The thermal fluctuations of the bead attached to the SPB-nucleated microtubule were recorded for at least 100 s.
Thermal fluctuation analysis
Custom MATLAB (MathWorks) software was used to convert DVD recordings to AVI files, which were subsequently broken up into separate files for individual SPB-microtubule attachments. Using custom LabVIEW (National Instruments) software, the positions of the SPB and the bead were tracked throughout the recorded data. Custom MATLAB software then used the position tracking files to calculate the angle of the microtubule based on the positions of the SPB and bead. For a small number of cases, the SPB could not be tracked through the whole video, and a reference marker was instead tracked to correct for drift. The angle of the microtubule was calculated throughout the recording, and the distribution of angles was fit with a Gaussian curve. The standard deviation of this Gaussian fit, , was used as a measure of the flexibility of the SPB-microtubule attachment, as seen in the bar graph on the right in Fig. 2 E. To monitor the progression of σ over time, as seen in the plot on the left in Fig. 2 E, σ was calculated arithmetically rather than using the Gaussian fitting method. The arithmetic method was computationally simpler but resulted in slightly different mean values in the σ-over-time plot on the left in Fig. 2 E versus the bar graph on the right.
The torsional stiffness of the SPB–microtubule interface was estimated by assuming that the angular deflection of an SPB-attached microtubule, , varies linearly with applied torque, (Fig. 5 A). In this case, the torsional spring stiffness is given by , where = 4.1 pN·nm represents thermal energy (i.e., Boltzmann’s constant, , multiplied by absolute temperature, ) and represents the mean-squared angular deviation of the microtubule (i.e., the angular variance) measured at thermal equilibrium. To estimate the transverse force needed to bring microtubules emanating from adjacent, side-by-side SPBs into antiparallel alignment, we calculated the magnitude of force, applied perpendicular to a microtubule at distance, , from its pivot point, that would be needed to deflect it to = 90° (Fig. 5 C). The distance, = 150 nm, was chosen to match the separation between adjacent SPBs when they are side by side, after duplication and insertion into the nuclear envelope (O’Toole et al., 1999). To estimate the waiting time for thermal pivoting to bring microtubules from adjacent SPBs into contact, we calculated Kramer’s first-passage time for rotational diffusion to deflect the microtubule to = 17°,
where is the potential energy barrier due to the spring (Howard, 2001). We assumed a rotational drag coefficient,
which approximates the microtubule as a slender rod of length, , and radius, , pivoting around one end. The radius of a microtubule is = 12.5 nm (Amos and Klug, 1974). The length, = 250 nm, was chosen to match the typical lengths of microtubules emanating from side-by-side SPBs (O’Toole et al., 1999). The angle, = 17°, represents the minimum needed to bring the tips of two microtubules, separated laterally by distance = 150 nm, into contact (Fig. 5 B). Because the first-passage time increases very steeply with angle, deflections all the way to = 90° are expected to take far longer than the generation time of a yeast cell. Thus, thermal pivoting alone is unlikely to achieve antiparallel alignment. Step-by-step calculations of the torsional stiffnesses, transverse forces, and waiting times for the WT and mutant SPBs are provided in an online spreadsheet file (Table S4).
Online supplemental material
Fig. S1 summarizes strain growth and SPB fluorescence in cells expressing either wild-type or mutant Spc110. Fig. S2 summarizes strain growth and SPB fluorescence in cells expressing cdc28-as1 and either WT or mutant Spc110 under 1NM-PP1 inhibition. Table S1 contains the P values from pairwise comparisons of mean angular deviations related to Fig. 2 E. Table S2 presents the plasmids used in this study. Table S3 lists the strains used in this study. Table S4 presents calculations of torsional spring constants and transverse forces from the experimentally measured angular deviations.
Supplementary Material
lists P values from pairwise comparisons of mean angular deviations related to Fig. 2 E.
lists plasmids used in this study.
lists strains used in this study.
presents calculations of torsional spring constants and transverse forces from the experimentally measured angular deviations.
Acknowledgments
We thank Aida Llauró for developing the method to track pivoting by video-enhanced differential interference contrast; David Agard, Andrew Lyon, Ivan Rayment, and Michael Andreas for advice on mutant designs; Mark Winey and Sue Jaspersen for advice on cdc28-as1 experiments; Sue Jaspersen (Stowers Institute for Medical Research, Kansas City, MO) for providing the cdc28-as1 strain; the spindle pole body P01 group for feedback and suggestions; and Mark Winey, Sue Jaspersen, Josh Larson, Lucas Murray, and Grace Hamilton for comments on our manuscript.
This work was supported by National Institutes of Health grants R35GM134842 (to C.L. Asbury), R35GM130293 (to T.N. Davis), and P01GM105537 (Principal Investigator, Mark Winey; sub-Principal Investigators, C.L. Asbury and T.N. Davis) and by a Packard Fellowship (to C.L. Asbury).
The authors declare no competing financial interests.
Author contributions: K.K. Fong and C.L. Asbury conceived and designed the research and wrote the manuscript. C.L. Asbury supervised the research. K.K. Fong conducted the experiments and analyzed the data. T.N. Davis contributed materials, designed experiments, and edited the manuscript.
References
- Adams, I.R., and Kilmartin J.V.. 1999. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 145:809–823. 10.1083/jcb.145.4.809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi, B., Sarangapani K.K., Powers A.F., Nelson C.R., Reichow S.L., Arellano-Santoyo H., Gonen T., Ranish J.A., Asbury C.L., and Biggins S.. 2010. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 468:576–579. 10.1038/nature09594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos, L., and Klug A.. 1974. Arrangement of subunits in flagellar microtubules. J. Cell Sci. 14:523–549. [DOI] [PubMed] [Google Scholar]
- Asbury, C.L., Fehr A.N., and Block S.M.. 2003. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 302:2130–2134. 10.1126/science.1092985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias, M., and Glover D.M.. 2007. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 8:451–463. 10.1038/nrm2180 [DOI] [PubMed] [Google Scholar]
- Bishop, A.C., Ubersax J.A., Petsch D.T., Matheos D.P., Gray N.S., Blethrow J., Shimizu E., Tsien J.Z., Schultz P.G., Rose M.D., et al. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 407:395–401. 10.1038/35030148 [DOI] [PubMed] [Google Scholar]
- Blackwell, R., Sweezy-Schindler O., Edelmaier C., Gergely Z.R., Flynn P.J., Montes S., Crapo A., Doostan A., McIntosh J.R., Glaser M.A., and Betterton M.D.. 2017. Contributions of Microtubule Dynamic Instability and Rotational Diffusion to Kinetochore Capture. Biophys. J. 112:552–563. 10.1016/j.bpj.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy, A., Lane H.A., d’Hérin P., Harper M., Kress M., and Nigg E.A.. 1995. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 83:1159–1169. 10.1016/0092-8674(95)90142-6 [DOI] [PubMed] [Google Scholar]
- Brenner, M.D., Zhou R., Conway D.E., Lanzano L., Gratton E., Schwartz M.A., and Ha T.. 2016. Spider Silk Peptide Is a Compact, Linear Nanospring Ideal for Intracellular Tension Sensing. Nano Lett. 16:2096–2102. 10.1021/acs.nanolett.6b00305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullitt, E., Rout M.P., Kilmartin J.V., and Akey C.W.. 1997. The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell. 89:1077–1086. 10.1016/S0092-8674(00)80295-0 [DOI] [PubMed] [Google Scholar]
- Burke, D., Dawson D., and Stearns T.. 2000. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 205 pp. [Google Scholar]
- Byers, B., and Goetsch L.. 1975. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124:511–523. 10.1128/JB.124.1.511-523.1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, B., Shriver K., and Goetsch L.. 1978. The role of spindle pole bodies and modified microtubule ends in the initiation of microtubule assembly in Saccharomyces cerevisiae. J. Cell Sci. 30:331–352. [DOI] [PubMed] [Google Scholar]
- Castoldi, M., and Popov A.V.. 2003. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr. Purif. 32:83–88. 10.1016/S1046-5928(03)00218-3 [DOI] [PubMed] [Google Scholar]
- Cojoc, G., Florescu A.-M., Krull A., Klemm A.H., Pavin N., Jülicher F., and Tolić I.M.. 2016. Paired arrangement of kinetochores together with microtubule pivoting and dynamics drive kinetochore capture in meiosis I. Sci. Rep. 6:25736. 10.1038/srep25736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta, K., and Surana U.. 2006. Disjunction of conjoined twins: Cdk1, Cdh1 and separation of centrosomes. Cell Div. 1:12. 10.1186/1747-1028-1-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta, K., Huang P., Morgan G., Winey M., and Surana U.. 2006. Cdk1 regulates centrosome separation by restraining proteolysis of microtubule-associated proteins. EMBO J. 25:2551–2563. 10.1038/sj.emboj.7601136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann, A., Desai A., and Oegema K.. 2003. The minus end in sight. Curr. Biol. 13:R614–R624. 10.1016/S0960-9822(03)00530-X [DOI] [PubMed] [Google Scholar]
- Decarreau, J., Wagenbach M., Lynch E., Halpern A.R., Vaughan J.C., Kollman J., and Wordeman L.. 2017. The tetrameric kinesin Kif25 suppresses pre-mitotic centrosome separation to establish proper spindle orientation. Nat. Cell Biol. 19:384–390. 10.1038/ncb3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorenzi, M., and Speed T.. 2002. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics. 18:617–625. 10.1093/bioinformatics/18.4.617 [DOI] [PubMed] [Google Scholar]
- Ding, R., McDonald K.L., and McIntosh J.R.. 1993. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell Biol. 120:141–151. 10.1083/jcb.120.1.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver, J.W., Geyer E.A., Bailey M.E., Rice L.M., and Asbury C.L.. 2017. Direct measurement of conformational strain energy in protofilaments curling outward from disassembling microtubule tips. eLife. 6:e28433. 10.7554/eLife.28433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enos, A.P., and Morris N.R.. 1990. Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans. Cell. 60:1019–1027. 10.1016/0092-8674(90)90350-N [DOI] [PubMed] [Google Scholar]
- Fallesen, T., Roostalu J., Duellberg C., Pruessner G., and Surrey T.. 2017. Ensembles of Bidirectional Kinesin Cin8 Produce Additive Forces in Both Directions of Movement. Biophys. J. 113:2055–2067. 10.1016/j.bpj.2017.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, K.K., Graczyk B., and Davis T.N.. 2016. Purification of fluorescently labeled Saccharomyces cerevisiae spindle pole bodies. Methods Mol. Biol. 1413:189–195. 10.1007/978-1-4939-3542-0_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, K.K., Sarangapani K.K., Yusko E.C., Riffle M., Llauró A., Graczyk B., Davis T.N., and Asbury C.L.. 2017. Direct measurement of microtubule attachment strength to yeast centrosomes. Mol. Biol. Cell. 28:1853–1861. 10.1091/mbc.E17-01-0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck, A.D., Powers A.F., Gestaut D.R., Davis T.N., and Asbury C.L.. 2010. Direct physical study of kinetochore-microtubule interactions by reconstitution and interrogation with an optical force clamp. Methods. 51:242–250. 10.1016/j.ymeth.2010.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerson-Gurwitz, A., Thiede C., Movshovich N., Fridman V., Podolskaya M., Danieli T., Lakämper S., Klopfenstein D.R., Schmidt C.F., and Gheber L.. 2011. Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. EMBO J. 30:4942–4954. 10.1038/emboj.2011.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grashoff, C., Hoffman B.D., Brenner M.D., Zhou R., Parsons M., Yang M.T., McLean M.A., Sligar S.G., Chen C.S., Ha T., and Schwartz M.A.. 2010. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 466:263–266. 10.1038/nature09198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk, E.L., Molodtsov M.I., Ataullakhanov F.I., and McIntosh J.R.. 2005. Force production by disassembling microtubules. Nature. 438:384–388. 10.1038/nature04132 [DOI] [PubMed] [Google Scholar]
- Hagan, I., and Yanagida M.. 1990. Novel potential mitotic motor protein encoded by the fission yeast cut7+ gene. Nature. 347:563–566. 10.1038/347563a0 [DOI] [PubMed] [Google Scholar]
- Harbury, P.B., Zhang T., Kim P.S., and Alber T.. 1993. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 262:1401–1407. 10.1126/science.8248779 [DOI] [PubMed] [Google Scholar]
- Hata, S., Pastor Peidro A., Panic M., Liu P., Atorino E., Funaya C., Jäkle U., Pereira G., and Schiebel E.. 2019. The balance between KIFC3 and EG5 tetrameric kinesins controls the onset of mitotic spindle assembly. Nat. Cell Biol. 21:1138–1151. 10.1038/s41556-019-0382-6 [DOI] [PubMed] [Google Scholar]
- Heck, M.M.S., Pereira A., Pesavento P., Yannoni Y., Spradling A.C., and Goldstein L.S.B.. 1993. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J. Cell Biol. 123:665–679. 10.1083/jcb.123.3.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepperla, A.J., Willey P.T., Coombes C.E., Schuster B.M., Gerami-Nejad M., McClellan M., Mukherjee S., Fox J., Winey M., Odde D.J., et al. 2014. Minus-end-directed Kinesin-14 motors align antiparallel microtubules to control metaphase spindle length. Dev. Cell. 31:61–72. 10.1016/j.devcel.2014.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, J. 2001. Mechanics of Motor Proteins and the Cytoskeleton. Sinauer Associates, Inc., Sunderland, MA. [Google Scholar]
- Hoyt, M.A., He L., Loo K.K., and Saunders W.S.. 1992. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J. Cell Biol. 118:109–120. 10.1083/jcb.118.1.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen, S.L., and Winey M.. 2004. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 20:1–28. 10.1146/annurev.cellbio.20.022003.114106 [DOI] [PubMed] [Google Scholar]
- Kalinina, I., Nandi A., Delivani P., Chacón M.R., Klemm A.H., Ramunno-Johnson D., Krull A., Lindner B., Pavin N., and Tolić-Nørrelykke I.M.. 2013. Pivoting of microtubules around the spindle pole accelerates kinetochore capture. Nat. Cell Biol. 15:82–87. 10.1038/ncb2640 [DOI] [PubMed] [Google Scholar]
- Kapitein, L.C., Peterman E.J.G., Kwok B.H., Kim J.H., Kapoor T.M., and Schmidt C.F.. 2005. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 435:114–118. 10.1038/nature03503 [DOI] [PubMed] [Google Scholar]
- Kashina, A.S., Rogers G.C., and Scholey J.M.. 1997. The bimC family of kinesins: essential bipolar mitotic motors driving centrosome separation. Biochim. Biophys. Acta. 1357:257–271. 10.1016/S0167-4889(97)00037-2 [DOI] [PubMed] [Google Scholar]
- Kilmartin, J.V., Dyos S.L., Kershaw D., and Finch J.T.. 1993. A spacer protein in the Saccharomyces cerevisiae spindle poly body whose transcript is cell cycle-regulated. J. Cell Biol. 123:1175–1184. 10.1083/jcb.123.5.1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M., and Schiebel E.. 1998. Receptors determine the cellular localization of a γ-tubulin complex and thereby the site of microtubule formation. EMBO J. 17:3952–3967. 10.1093/emboj/17.14.3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman, J.M., Polka J.K., Zelter A., Davis T.N., and Agard D.A.. 2010. Microtubule nucleating γ-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 466:879–882. 10.1038/nature09207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary, A., Sim S., Nazarova E., Shulist K., Genthial R., Yang S.K., Bui K.H., Francois P., and Vogel J.. 2019. Successive Kinesin-5 Microtubule Crosslinking and Sliding Promote Fast, Irreversible Formation of a Stereotyped Bipolar Spindle. Curr. Biol. 29:3825–3837.e3. 10.1016/j.cub.2019.09.030 [DOI] [PubMed] [Google Scholar]
- Lim, H.H., Zhang T., and Surana U.. 2009. Regulation of centrosome separation in yeast and vertebrates: common threads. Trends Cell Biol. 19:325–333. 10.1016/j.tcb.2009.03.008 [DOI] [PubMed] [Google Scholar]
- Lombillo, V.A., Stewart R.J., and McIntosh J.R.. 1995. Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature. 373:161–164. 10.1038/373161a0 [DOI] [PubMed] [Google Scholar]
- Lyon, A.S., Morin G., Moritz M., Yabut K.C.B., Vojnar T., Zelter A., Muller E., Davis T.N., and Agard D.A.. 2016. Higher-order oligomerization of Spc110p drives γ-tubulin ring complex assembly. Mol. Biol. Cell. 27:2245–2258. 10.1091/mbc.E16-02-0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh, J.R., and Euteneuer U.. 1984. Tubulin hooks as probes for microtubule polarity: an analysis of the method and an evaluation of data on microtubule polarity in the mitotic spindle. J. Cell Biol. 98:525–533. 10.1083/jcb.98.2.525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzayan, C., Copeland C.S., and Snyder M.. 1992. The NUF1 gene encodes an essential coiled-coil related protein that is a potential component of the yeast nucleoskeleton. J. Cell Biol. 116:1319–1332. 10.1083/jcb.116.6.1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens, P.B., and Rapport E.. 1971. Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae (Hansen). J. Cell Biol. 50:344–361. 10.1083/jcb.50.2.344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, E.G.D., Snydsman B.E., Novik I., Hailey D.W., Gestaut D.R., Niemann C.A., O’Toole E.T., Giddings T.H. Jr., Sundin B.A., and Davis T.N.. 2005. The organization of the core proteins of the yeast spindle pole body. Mol. Biol. Cell. 16:3341–3352. 10.1091/mbc.e05-03-0214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, T., Vinh D.B.N., Crawford D.K., and Davis T.N.. 1998. A genetic analysis of interactions with Spc110p reveals distinct functions of Spc97p and Spc98p, components of the yeast γ-tubulin complex. Mol. Biol. Cell. 9:2201–2216. 10.1091/mbc.9.8.2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole, E.T., Winey M., and McIntosh J.R.. 1999. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 10:2017–2031. 10.1091/mbc.10.6.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelogović, M., Winters L., Milas A., Tolić I.M., and Pavin N.. 2019. Pivot-and-bond model explains microtubule bundle formation. Phys. Rev. E. 100:012403. 10.1103/PhysRevE.100.012403 [DOI] [PubMed] [Google Scholar]
- Rincon, S.A., Lamson A., Blackwell R., Syrovatkina V., Fraisier V., Paoletti A., Betterton M.D., and Tran P.T.. 2017. Kinesin-5-independent mitotic spindle assembly requires the antiparallel microtubule crosslinker Ase1 in fission yeast. Nat. Commun. 8:15286. 10.1038/ncomms15286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof, D.M., Meluh P.B., and Rose M.D.. 1992. Kinesin-related proteins required for assembly of the mitotic spindle. J. Cell Biol. 118:95–108. 10.1083/jcb.118.1.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu, J., Hentrich C., Bieling P., Telley I.A., Schiebel E., and Surrey T.. 2011. Directional switching of the kinesin Cin8 through motor coupling. Science. 332:94–99. 10.1126/science.1199945 [DOI] [PubMed] [Google Scholar]
- Sarangapani, K.K., and Asbury C.L.. 2014. Catch and release: how do kinetochores hook the right microtubules during mitosis? Trends Genet. 30:150–159. 10.1016/j.tig.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, W.S., and Hoyt M.A.. 1992. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 70:451–458. 10.1016/0092-8674(92)90169-D [DOI] [PubMed] [Google Scholar]
- Saunders, W.S., Koshland D., Eshel D., Gibbons I.R., and Hoyt M.A.. 1995. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J. Cell Biol. 128:617–624. 10.1083/jcb.128.4.617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin, K.E., LeGuellec K., Philippe M., and Mitchison T.J.. 1992. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 359:540–543. 10.1038/359540a0 [DOI] [PubMed] [Google Scholar]
- Scholey, J.M., Civelekoglu-Scholey G., and Brust-Mascher I.. 2016. Anaphase B. Biology (Basel). 5:51. 10.3390/biology5040051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto, Y., Forth S., and Kapoor T.M.. 2015. Measuring Pushing and Braking Forces Generated by Ensembles of Kinesin-5 Crosslinking Two Microtubules. Dev. Cell. 34:669–681. 10.1016/j.devcel.2015.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawa, M.M., Wargacki M.M., Muller E.G., and Davis T.N.. 2010. Laterally attached kinetochores recruit the checkpoint protein Bub1, but satisfy the spindle checkpoint. Cell Cycle. 9:3619–3628. 10.4161/cc.9.17.12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S.K., Pandey H., Al-Bassam J., and Gheber L.. 2018. Bidirectional motility of kinesin-5 motor proteins: structural determinants, cumulative functions and physiological roles. Cell. Mol. Life Sci. 75:1757–1771. 10.1007/s00018-018-2754-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, K.C., Buvoli M., Korkmaz E.N., Buvoli A., Zheng Y., Heinze N.T., Cui Q., Leinwand L.A., and Rayment I.. 2015. Skip residues modulate the structural properties of the myosin rod and guide thick filament assembly. Proc. Natl. Acad. Sci. USA. 112:E3806–E3815. 10.1073/pnas.1505813112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Wildenberg, S.M.J.L., Tao L., Kapitein L.C., Schmidt C.F., Scholey J.M., and Peterman E.J.G.. 2008. The homotetrameric kinesin-5 KLP61F preferentially crosslinks microtubules into antiparallel orientations. Curr. Biol. 18:1860–1864. 10.1016/j.cub.2008.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanath, S., Bonomi M., Kim S.J., Klenchin V.A., Taylor K.C., Yabut K.C., Umbreit N.T., Van Epps H.A., Meehl J., Jones M.H., et al. 2017. The molecular architecture of the yeast spindle pole body core determined by Bayesian integrative modeling. Mol. Biol. Cell. 28:3298–3314. 10.1091/mbc.e17-06-0397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov, V.A., Zaytsev A.V., Gudimchuk N., Grissom P.M., Gintsburg A.L., Ataullakhanov F.I., McIntosh J.R., and Grishchuk E.L.. 2013. Long tethers provide high-force coupling of the Dam1 ring to shortening microtubules. Proc. Natl. Acad. Sci. USA. 110:7708–7713. 10.1073/pnas.1305821110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., Brachat A., Alberti-Segui C., Rebischung C., and Philippsen P.. 1997. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 13:1065–1075. [DOI] [PubMed] [Google Scholar]
- Widlund, P.O., and Davis T.N.. 2005. A high-efficiency method to replace essential genes with mutant alleles in yeast. Yeast. 22:769–774. 10.1002/yea.1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey, M., and Byers B.. 1993. Assembly and functions of the spindle pole body in budding yeast. Trends Genet. 9:300–304. 10.1016/0168-9525(93)90247-F [DOI] [PubMed] [Google Scholar]
- Winey, M., and O’Toole E.T.. 2001. The spindle cycle in budding yeast. Nat. Cell Biol. 3:E23–E27. 10.1038/35050663 [DOI] [PubMed] [Google Scholar]
- Winey, M., Mamay C.L., O’Toole E.T., Mastronarde D.N., Giddings T.H. Jr., McDonald K.L., and McIntosh J.R.. 1995. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 129:1601–1615. 10.1083/jcb.129.6.1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winters, L., Ban I., Prelogović M., Kalinina I., Pavin N., and Tolić I.M.. 2019. Pivoting of microtubules driven by minus-end-directed motors leads to spindle assembly. BMC Biol. 17:42. 10.1186/s12915-019-0656-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder, Tennessee J, Pearson Chad G, Bloom Kerry, and Davis Trisha N. 2003. The Saccharomyces cerevisiae spindle pole body is a dynamic structure. Mol Biol Cell. 14(8):3494–3505. . 10.1091/mbc.e02-10-0655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukawa, M., Yamada Y., Yamauchi T., and Toda T.. 2018. Two spatially distinct kinesin-14 proteins, Pkl1 and Klp2, generate collaborative inward forces against kinesin-5 Cut7 in S. pombe. J. Cell Sci. 131:jcs210740. 10.1242/jcs.210740 [DOI] [PubMed] [Google Scholar]
- Yukawa, M., Teratani Y., and Toda T.. 2020. How Essential Kinesin-5 Becomes Non-Essential in Fission Yeast: Force Balance and Microtubule Dynamics Matter. Cells. 9:1154. 10.3390/cells9051154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, L., Stephens A., Nam S.Z., Rau D., Kübler J., Lozajic M., Gabler F., Söding J., Lupas A.N., and Alva V.. 2018. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 430:2237–2243. 10.1016/j.jmb.2017.12.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists P values from pairwise comparisons of mean angular deviations related to Fig. 2 E.
lists plasmids used in this study.
lists strains used in this study.
presents calculations of torsional spring constants and transverse forces from the experimentally measured angular deviations.