Abstract
BACKGROUND
Recent studies have demonstrated that circular RNA AKT3 (circAKT3) plays a crucial role in regulating the malignant phenotypes of tumor cells. However, the potential effects of circAKT3 on esophageal cancer have not been investigated.
AIM
To illuminate the role of circAKT3 in malignant behaviors of esophageal cancer cells and its underlying mechanism.
METHODS
Clinical samples were collected to detect the expression of circAKT3. The role of circAKT3 in proliferation, migration, invasion, and apoptosis of esophageal cancer cells was evaluated using Cell Counting Kit-8, wound healing assays, Transwell assays, and fluorescence analysis, respectively. The target of circAKT3 was screened and identified using an online database and luciferase reporter assay. A xenograft nude mouse model was established to investigate the role of circAKT3 in vivo.
RESULTS
In vitro assays showed that proliferative, migratory, and invasive capacities of esophageal cancer cells were significantly enhanced by circAKT3 overexpression. Furthermore, miR-17-5p was screened as the target of circAKT3, and miR-17-5p antagonized the effects of circAKT3 on esophageal cancer cells. Moreover, we identified RHOC and STAT3 as the direct target molecules of miR-17-5p, and circAKT3 facilitated expression of RHOC and STAT3 by inhibiting miR-17-5p. In vivo assays showed circAKT3 knockdown inhibited growth of esophageal cancer.
CONCLUSION
CircAKT3 contributed to the malignant behaviors of esophageal cancer in vitro and in vivo by sponging miR-17-5p thus providing a potential target for treatment of esophageal cancer.
Keywords: Esophageal cancer, Circular RNA AKT3, miR-17-5p, Proliferation, Migration, Invasion
Core Tip: Circular RNA AKT3 was overexpressed in esophageal cancer and enhanced proliferative, migratory, and invasive capacities of esophageal cancer cells by sponging miR-17-5p to exert protumor effects. Downregulation of circular RNA AKT3 inhibited xenograft tumor growth of esophageal cancer in vivo. This research provided a potential target for better understanding the underlying mechanism of esophageal cancer.
INTRODUCTION
As one of the most malignant tumors, esophageal cancer ranks in the top ten in morbidity and mortality of the tumors worldwide[1]. However, the exact mechanism is still in discussion. Currently, surgery is the predominant approach for treatment of esophageal cancer in combination with some other alternatives[2,3]. Although great progress has been made in the diagnosis and treatment of esophageal cancer, the prognosis has not been improved satisfactorily with a 5-year survival rate of less than 30%[4]. Thus, investigating the underlying mechanism is conducive to deepening the pathogenesis of esophageal cancer and developing targeted therapies.
Circular RNAs (circRNAs) belong to endogenous noncoding RNAs, which could be expressed in eukaryotic cells[5]. Unlike other RNA molecules, circRNAs possess the structure of a closed loop and cannot be degraded by RNase R[6]. Accumulating evidence has demonstrated that circRNAs participate in the modulation of protein expression by sponging microRNAs (miRNAs) or interacting with RNA binding proteins[7,8]. In tumor tissues and cell lines, the aberrant expression of circRNAs is involved in the regulation of malignant phenotypes of tumor cells. Wang et al[9] reported that circLMTK2 is overexpressed in gastric cancer, which could also be a predictor for a poor prognosis in patients with gastric cancer. In colorectal cancer, circFBXW7 is significantly downregulated, and circFBXW1 overexpression suppresses proliferative, migratory, and invasive abilities of colorectal cancer cells[10]. Therefore, differential expression of circRNAs exerts the opposite effects on tumor cells. However, only a few studies have concentrated on the effects of circRNAs on esophageal cancer.
Circular RNA AKT3 (circAKT3) is a novel molecule, which is derived from the AKT3 gene and identified in gastric cancer with cisplatin resistance[11]. The in vitro and in vivo assays have revealed that circAKT3 is overexpressed in cisplatin resistant-gastric cancer and leads to the increased expression of PIK3R1 to mediate resistance of gastric cancer cells to cisplatin[11]. However, Xia et al[12] demonstrated that glioblastoma expressed a low level of circAKT3, and circAKT3 overexpression suppressed proliferation and promoted sensitivity to radiation of glioblastoma cells. The contradictory findings remind us of the potential role of circAKT3 in esophageal cancer.
MATERIALS AND METHODS
Collection of clinical samples
The clinical tissue samples were acquired from patients with esophageal cancer (n = 82) who underwent surgery at China-Japan Union Hospital of Jilin University. The relative expression of circAKT3 less than the mean value was classified into the circAKT3 low group (n = 43), and the relative expression of circAKT3 more than the mean value was classified into the circAKT3 high group (n = 39).
In vivo assay
The nude mice (n = 12) were purchased from Experimental Animal Center of China-Japan Union Hospital of Jilin University, housed in cages under the condition of the 12-h shift of light-dark cycles, and randomly divided into two groups. Negative control short hairpin RNA and short hairpin RNA against circAKT3, purchased from Integrated Biotech Solutions Co. (China), were transfected into TE-1 cells in the presence of puromycin (3 mg/mL; Thermo Fisher Scientific, United States) to establish the circAKT3 knockdown cell lines. The mice injected with the negative control short hairpin RNA-transfected TE-1 cells were designated as the negative control short hairpin RNA group, and mice injected with the short hairpin RNA against circAKT3-transfected TE-1 cells were designated as the short hairpin RNA against circAKT3 group. After feeding for 30 d, the mice were anesthetized with pentobarbital sodium (25 mg/kg; Sigma-Aldrich, United States) intraperitoneally and killed by decapitation. The xenografts were isolated carefully and weighed.
Cell culture
Human esophageal cancer cell lines (KYSE-150, TE-10, TE-1) and human embryonic kidney cell line (HEK-293T) were cultured in Dulbecco’s Modified Eagle Medium containing 10% fetal bovine serum (Gibco, United States). Human normal esophageal epithelial cell line (HEEC) was cultured in Dulbecco’s Modified Eagle Medium with high glucose containing 10% fetal bovine serum (Gibco, United States). All of the cells were cultured in a CO2 incubator chamber (Thermo Fisher Scientific, United States).
Quantitative real time-polymerase chain reaction
TRIzol (Ambion, United States) was utilized to extract the total RNA. SuperScript III (Invitrogen, United States) was used to obtain cDNA. SYBR Premix Ex Taq II (Takara, Japan) was utilized to perform quantitative real time(qRT)-PCR. The primer sequences of circAKT3 and U6 were adopted as previously reported[11]. The primer sequences of miR-17-5p were obtained from recent research[13]. The primer sequences of AKT3, RHOC, STAT3, and GAPDH were obtained from the NCBI online database (https://www.ncbi.nlm.nih.gov/gene/). CircAKT3 (forward, 5’-TCCAAATAAACGCCT TGGTGG-3’; reverse, 5’-CCTCAGAGAACACCCGCTCT-3’); miR-17-5p (forward, 5’-CGGCGGCAAAGTGCTTACAG-3’; reverse, 5’-GTGCAGGGTCCGAGGT-3’); U6 (forward, 5’-CTCGCTTCGGCAGCACA-3’; reverse, 5’-AACGCTTCACGA ATTTGCGT-3’); AKT3 (forward, 5’-CGGCTTTCTCACGGATCACA-3’; reverse, 5’-CGGGACACTTTCCTTCCTCC-3’); RHOC (forward, 5’-AAGTTCCCTTTGCCCG TCTG-3’; reverse, 5’-ACAGTGGCAACTCAAGGGTC-3’); STAT3 (forward, 5’-CCAGGTACCGTGTGTCAAGC-3’; reverse, 5’-CAGACCTGACACCTGTGTTG-3’); GAPDH (forward, 5’-TGAAATGTGCACGCACCAAG-3’; reverse, 5’-GGGAAGCAGCATTCAGGTCT-3’). All of the primers were synthesized by Sangon (China).
Cell transfection
The siRNAs against circAKT3, miR-17-5p mimics, miR-17-5p inhibitor, indicated negative control, and plasmids were synthesized and purchased from Sangon (China). Lipofectamine 3000 (Invitrogen, United States) was used to conduct the transfection. RNase R (Epicentre, United States) was used to digest linear RNAs (but not circRNAs) in line with the manufacturer’s instruction.
Fluorescence analysis
The Hoechst Staining Kit (Beyotime, China) was used to evaluate apoptotic cells. In brief, TE-1 cells were seeded at a density of 2.0 × 105 cells per well. After cell transfection with indicated vectors, 4% paraformaldehyde was used to fix the cultured TE-1 cells, followed by using Hoechst 33258 to stain the cells. A fluorescence microscope (Olympus, Japan) was utilized to observe the cells.
Measurement of cell viability
The Cell Counting Kit-8 (CCK-8; Beyotime, China) was utilized to measure cell viability. After transfection, TE-1 cells were seeded at a density of 1 × 104 cells per well for the indicated time. CCK-8 solution was added into each well for 2 h, and the Microplate Reader (Bio-Rad, United States) was used to record the value at 450 nm.
Wound healing assay
The wound healing assay was used to measure the migratory ability. After transfection, TE-1 cells were seeded into a 6-well plate for 24 h. The cells were scratched by a 10 mL Finntip and cultured in serum-free medium. Images were captured at 0 h and 24 h and analyzed by ImageJ software.
Measurement of invasive ability
The Transwell assay was conducted to evaluate the invasive ability. TE-1 cells with indicated transfection were cultured in Millicell Standing Cell Culture 24 well (Millipore, United States). After incubation for 24 h, glutaraldehyde solution was used to fix the cells, and crystal violet was used to stain the cells.
Luciferase reporter assay
ENCORI database was used to acquire the predicted binding sites between circAKT3 and miR-17-5p. TargetScanHuman database was utilized to acquire the potential binding sites of miR-17-5p with RHOC or STAT3. The procedures of the luciferase reporter assay were conducted in line with the previous study[11]. In our research, HEK-293T cells were utilized to detect the binding ability by cotransfecting the indicated vectors (Sangon, China) with Lipofectamine 3000 (Invitrogen, United States). The luciferase activity was evaluated by Bio-GloTM Luciferase Assay System (Promega, United States).
Western blot
Western blot was carried out in line with the previous report[14]. In brief, the total protein was lysed by RIPA lysis buffer (Beyotime, China), and the BCA Kit (Thermal Fisher Scientific, United States) was used to detect the precise concentration. The total protein was added into SDS-PAGE gel (Beyotime, China). Primary antibodies [STAT3 rabbit monoclonal antibody (Cell Signaling Technology, United States); RHOC rabbit monoclonal antibody (Cell Signaling Technology, United States); GAPDH mouse monoclonal antibody (Cell Signaling Technology, United States)] were used overnight at 4 °C. Secondary antibodies with horseradish peroxidase (Beyotime, China) were applied for 2 h, and the indicated proteins were monitored by ChemDocTM XRS+ System (Bio-Rad, United States).
Statistical analysis
The data in our research were displayed as mean ± standard deviation and analyzed by GraphPad Prism 8.0 Software (GraphPad, United States). The t-test, two-way ANOVA or Chi-squared test was used. Statistical significance was set as the P value less than 0.05.
RESULTS
CircAKT3 was overexpressed in esophageal cancer
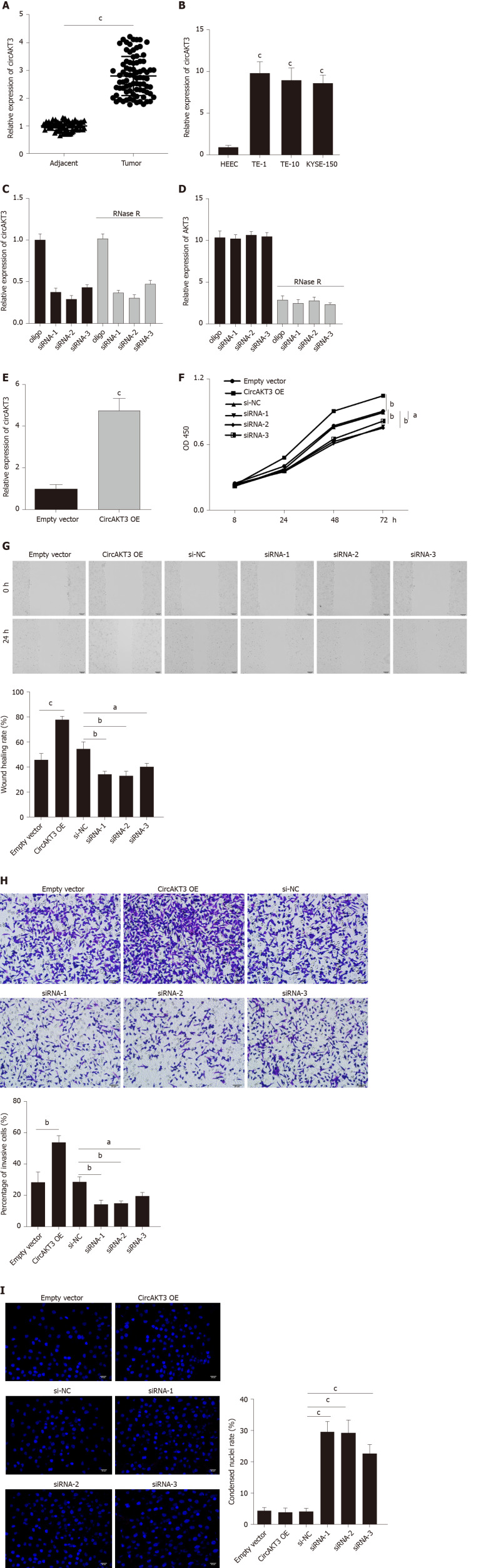
First, we detected the expression of circAKT3 in the esophageal cancer tissue samples and cell lines. The qRT-PCR results showed that circAKT3 was significantly upregulated in esophageal cancer tissue samples (Figure 1A). The characteristic analysis of the patients with esophageal cancer indicated that the tumor size, lymphatic metastasis, and clinical tumor node metastasis staging were positively related to the upregulation of circAKT3 in the tissue samples (Table 1). We also found that human esophageal cancer cell lines TE-1 cells, TE-10 cells, and KYSE-150 cells expressed a significantly higher level of circAKT3 compared to human normal esophageal epithelial cell line HEEC cells (Figure 1B).
Figure 1.
Circular RNA AKT3 promoted malignant phenotypes of TE-1 cells. A: The expression of circular RNA AKT3 (circAKT3) in tissue samples; B: The expression of circAKT3 human normal esophageal epithelial cell line (HEEC) and human esophageal cancer cell lines; C and D: TE-1 cells were transfected with control oligo and circAKT3 siRNAs in the presence or absence of RNase R to detect the expression of circAKT3 and AKT3; E: The expression of circAKT3 in TE-1 cells with circAKT3 overexpression (OE); F: Cell viability of TE-1 cells as detected by the Cell Counting Kit-8 assay was performed to detect cell viability of TE-1 cells with indicated transfection; G: Migratory capacity of TE-1 cells with circAKT3 OE and knockdown; H: Invasive ability of TE-1 cells with circAKT3 OE and knockdown; I: Apoptosis of TE-1 cells with circAKT3 OE and knockdown by Hoechst staining. aP < 0.05; bP < 0.01; cP < 0.001. NC: Negative control; OD: Optical density.
Table 1.
Characteristics of patients with esophageal cancer
|
Characteristic
|
CircAKT3 low
|
CircAKT3 high
|
P
value
|
| Age in yr | 0.824 | ||
| < 50 | 19 | 19 | |
| ≥ 50 | 24 | 20 | |
| Tumor diameter in cm | 0.026 | ||
| < 3 | 27 | 14 | |
| ≥ 3 | 16 | 25 | |
| Lymphatic metastasis | 0.027 | ||
| No | 24 | 12 | |
| Yes | 19 | 27 | |
| TNM staging | 0.016 | ||
| I-II | 26 | 13 | |
| III-IV | 17 | 26 |
CircAKT3: Circular RNA AKT3; TNM: Tumor node metastasis.
CircAKT3 promoted malignant behaviors of TE-1 cells
Next, siRNAs against circAKT3 were transfected into TE-1 cells, and qRT-PCR analysis revealed a marked efficiency of circAKT3 knockdown (Figure 1C). As the differential structure between circAKT3 and AKT3 mRNA, circAKT3 was resistant to RNase R, while AKT3 mRNA was sensitive to RNase R (Figure 1C and 1D), which confirmed the circular structure of circAKT3. Moreover, the circAKT3 overexpression plasmids were transfected into TE-1 cells and could significantly increase the expression of circAKT3 (Figure 1E). CCK-8 assay showed that circAKT3 siRNAs inhibited proliferation of TE-1 cells, while circAKT3 overexpression plasmids facilitated proliferation of TE-1 cells (Figure 1F). We also found that circAKT3 knockdown suppressed migration and invasion of TE-1 cells, while circAKT3 overexpression exerted the opposite role by conducting the wound healing assay and Transwell assay, respectively (Figure 1G and 1H). We also measured the effect of circAKT3 on apoptosis. The fluorescence analysis indicated that circAKT3 siRNAs increased the number of condensed nuclei in TE-1 cells; however, circAKT3 overexpression had no effect on the apoptotic rate of TE-1 cells (Figure 1I). These findings suggested that circAKT3 facilitated proliferative, migratory, and invasive capacities of esophageal cancer cells.
miR-17-5p acted as the downstream molecule of circAKT3 to antagonize the effects of circAKT3 on TE-1 cells
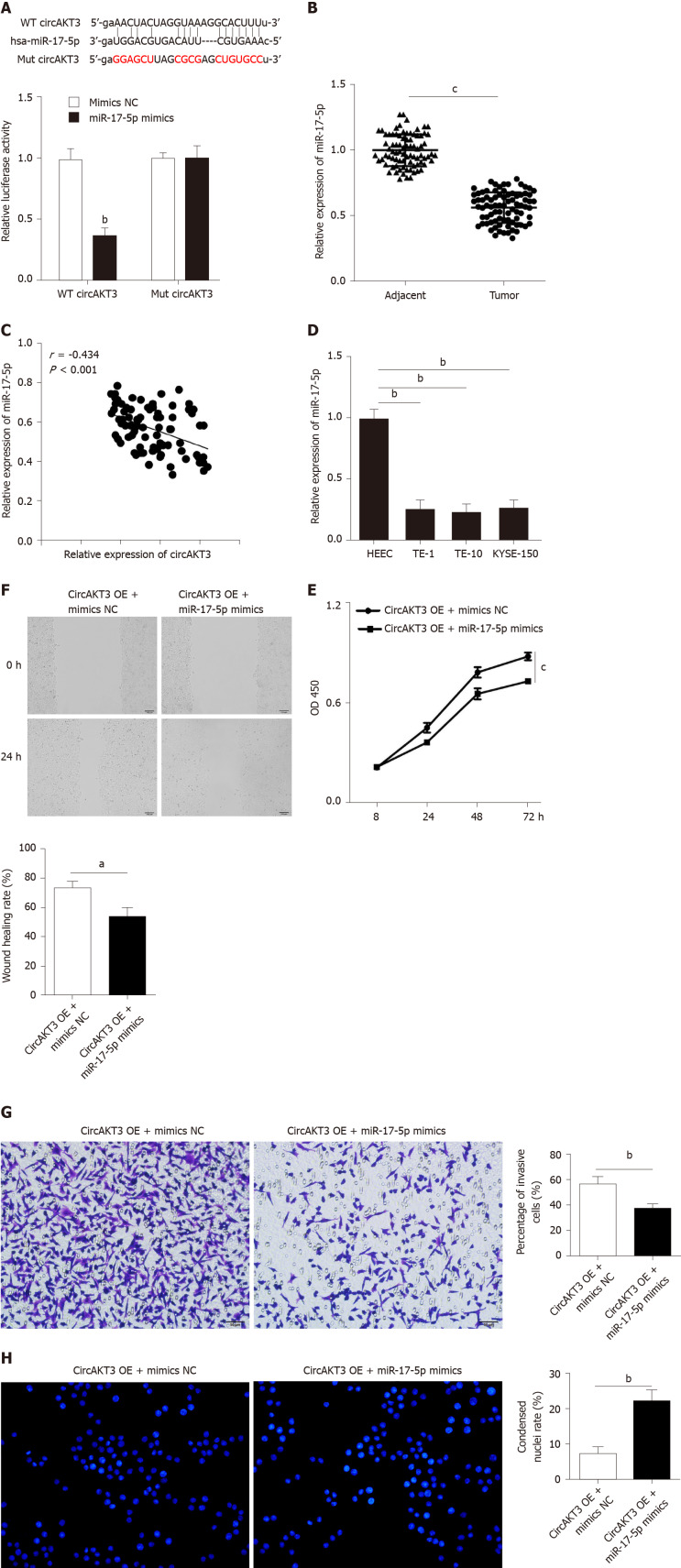
Accumulating evidence has reported that circRNAs regulate cell functions by sponging miRNAs[7]. The ENCORI database (http://starbase.sysu.edu.cn/index.php) was utilized to predict the potential target of circAKT3, and we identified miR-17-5p as the target of circAKT3. We carried out a luciferase reporter assay and found that TE-1 cells with wild-type circAKT3 and miR-17-5p mimics cotransfection presented a remarkably decreased luciferase activity (Figure 2A). Next, we detected the expression of miR-17-5p and found a lower level of miR-17-5p in esophageal cancer tissue samples (Figure 2B). Moreover, miR-17-5p expression was inversely associated with the expression of circAKT3 in clinical tissue samples (Figure 2C). Meanwhile, esophageal cancer cell lines showed decreased expression of miR-17-5p (Figure 2D). These results indicated that miR-17-5p was inhibited by circAKT3. We then explored the role of miR-17-5p functioning as a downstream molecule of circAKT3 in TE-1 cells. CCK-8 assay showed that cell viability of TE-1 cells with cotransfection of circAKT3 overexpression plasmids and miR-17-5p mimics was decreased compared to that with circAKT3 overexpression plasmids transfection (Figure 2E). Moreover, miR-17-5p mimics suppressed migratory and invasive abilities of circAKT3 overexpression plasmids-transfected TE-1 cells by the wound healing assay and Transwell assay, respectively (Figure 2F and 2G). In parallel, miR-17-5p mimics increased the apoptotic rate of circAKT3 overexpression plasmids-transfected TE-1 cells (Figure 2H).
Figure 2.
miR-17-5p acted as the downstream molecule of circular RNA AKT3 to antagonize the effects of circular RNA AKT3 on TE-1 cells. A: Schematic of the miR-17-5p binding sites in wild-type (WT) circular RNA AKT3 (circAKT3) and the mutant (Mut) circAKT3 (up). Luciferase reporter assay was performed in HEK-293T cells to determine the binding ability between miR-17-5p and circAKT3 (down); B: The expression of miR-17-5p in tissue samples; C: CircAKT3 was negatively correlated with miR-17-5p in esophageal cancer tissue samples; D: miR-17-5p expression in indicated cell lines; E-H: Cell viability (E), migratory ability (F), invasive ability (G), and apoptosis (H) of circAKT3 overexpression (OE)-transfected TE-1 cells in the presence or absence of miR-17-5p mimics. aP < 0.05; bP < 0.01; cP < 0.001. HEEC: Human normal esophageal epithelial cell line; NC: Negative control; OD: Optical density.
RHOC and STAT3 were modulated by the circAKT3/miR-17-5p axis
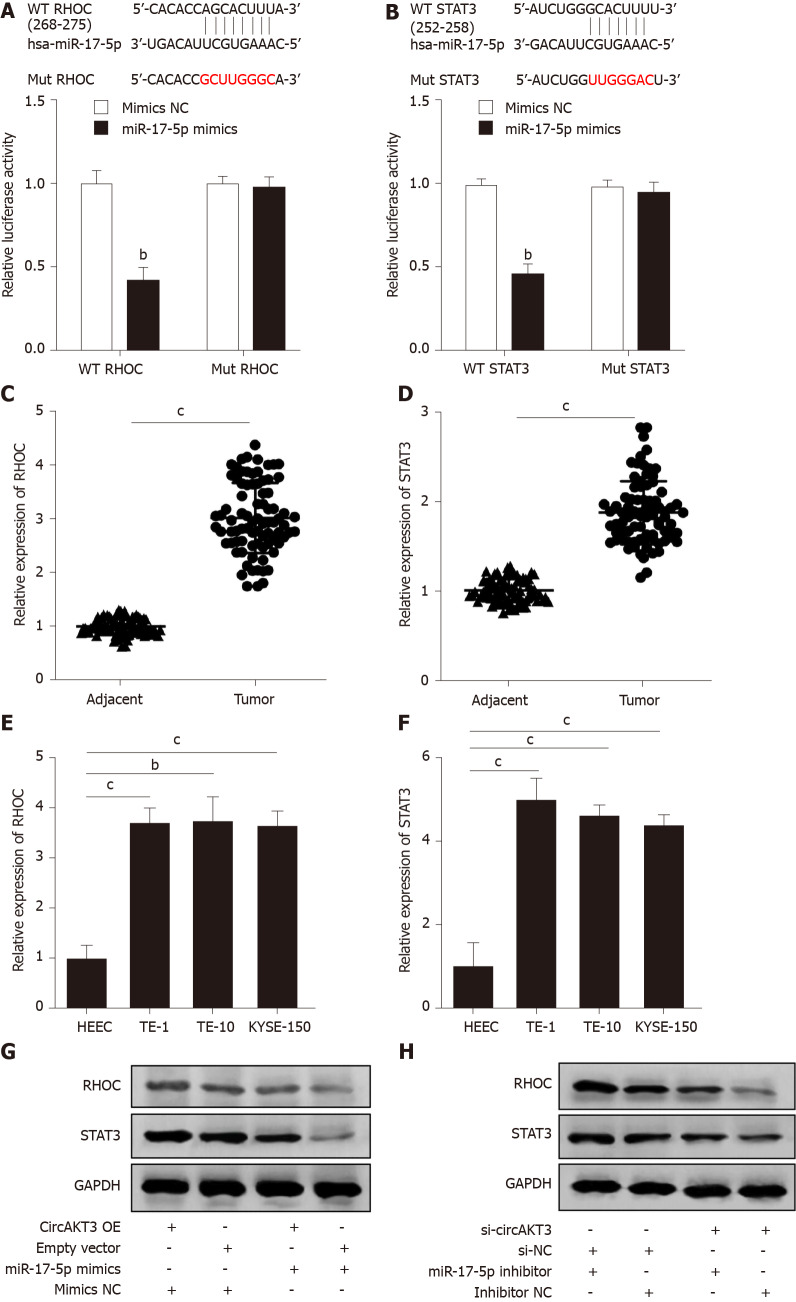
We then identified RHOC and STAT3 as the potential targets of miR-17-5p by using TargetScanHuman database (http://www.targetscan.org/vert_72/). The luciferase reporter assay showed that TE-1 cells with cotransfection of wild-type RHOC and miR-17-5p mimics presented a lower luciferase activity (Figure 3A). In addition, the decreased luciferase activity was also shown in TE-1 cells with cotransfection of wild-type STAT3 and miR-17-5p mimics (Figure 3B). Next, we detected the mRNA level of RHOC and STAT3 in clinical samples and found that both RHOC and STAT3 were upregulated in esophageal cancer tissue samples (Figure 3C and 3D). Moreover, the expression of RHOC and STAT3 was increased in TE-1 cells, TE-10 cells, and KYSE-150 cells (Figure 3E and 3F). Western blot analysis showed that circAKT3 promoted the expression of RHOC and STAT3 in TE-1 cells, and miR-17-5p led to the reduced expression of RHOC and STAT3, which was reversed by circAKT3 overexpression plasmids cotransfection (Figure 3G). Conversely, circAKT3 siRNA inhibited the expression of RHOC and STAT3 in TE-1 cells, and a miR-17-5p inhibitor promoted the expression of RHOC and STAT3, which was reverted by circAKT3 siRNA cotransfection (Figure 3H). In consequence, circAKT3 facilitated the expression of RHOC and STAT3 by sponging miR-17-5p in esophageal cancer cells.
Figure 3.
RHOC and STAT3 were modulated by the circular RNA AKT3/miR-17-5p axis. A: Schematic of the miR-17-5p binding sites in wild-type (WT) RHOC and the mutant (Mut) RHOC (up). Luciferase reporter assay was performed in HEK-293T cells to determine the binding ability between miR-17-5p and RHOC (down); B: Schematic of the miR-17-5p binding site in WT STAT3 and the Mut STAT3 (up). Luciferase reporter assay was performed in HEK-293T cells to determine the binding ability between miR-17-5p and STAT3 (down); C and D: The expression of RHOC (C) and STAT3 (D) in tissue samples; E and F: The expression of RHOC (E) and STAT3 (F) in indicated cell lines; G and H: Protein level of RHOC and STAT3 in TE-1 cells with indicated transfection. bP < 0.01; cP < 0.001. circAKT3: Circular RNA AKT3; HEEC: Human normal esophageal epithelial cell line; NC: Negative control; OE: Overexpression.
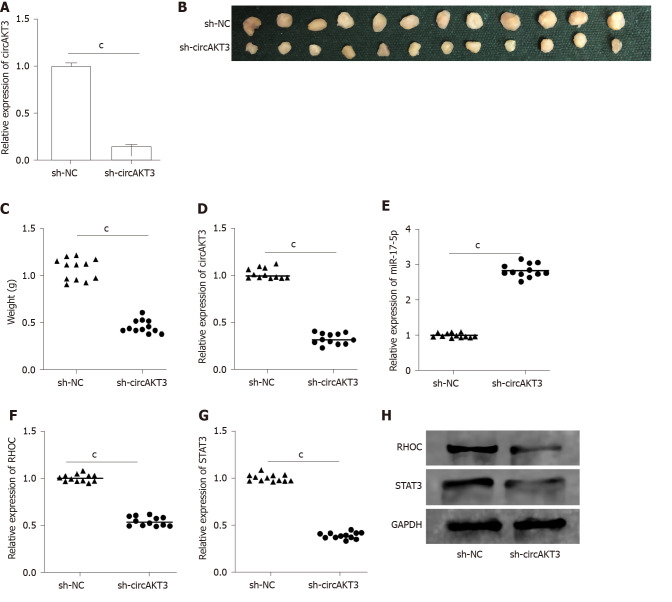
CircAKT3 knockdown suppressed growth of esophageal cancer in vivo
CircAKT3 shRNA was transfected into TE-1 cells to establish the stable circAKT3 knockdown cell line (Figure 4A). The in vivo assay showed that the size and weight of the xenograft tumor injected with circAKT3 shRNA-transfected TE-1 cells were smaller and lighter (Figure 4B and 4C). The qRT-PCR analysis indicated that the xenograft tumor injected with circAKT3 shRNA-transfected TE-1 cells expressed a lower level of circAKT3, RHOC, and STAT3 and a higher level of miR-17-5p (Figure 4D-G). In parallel, the protein levels of RHOC and STAT3 were decreased in the xenograft tumor injected with circAKT3 shRNA-transfected TE-1 cells (Figure 4H).
Figure 4.
Circular RNA AKT3 knockdown suppressed growth of esophageal cancer in vivo. A: The expression of circular RNA AKT3 (circAKT3) in TE-1 cells with circAKT3 short hairpin (sh)RNA transfection; B and C: The size (B) and weight (C) of the xenograft tumor injected with circAKT3 shRNA-transfected TE-1 cells; D-G: The expression of circAKT3 (D), miR-17-5p (E), RHOC (F) and STAT3 (G) in the xenograft tumor injected with circAKT3 shRNA-transfected TE-1 cells by quantitative real time-polymerase chain reaction; H: The protein levels of RHOC and STAT3 in the xenograft tumor injected with circAKT3 shRNA-transfected TE-1 cells. cP < 0.001. sh-NC: Negative control shRNA; sh-circAKT3: Circular RNA AKT3 shRNA.
DISCUSSION
Increased proliferative, migratory, and invasive abilities are hallmarks of tumor cells[15]. Thus, exploring the exact mechanism of malignant phenotypes of tumor cells contributes to clarifying the pathogenesis of tumorigenesis. Herein, we reported the protumor role of circAKT3 in esophageal cancer cells by measuring the proliferative, migratory, and invasive abilities. In addition, miR-17-5p acted as a sponge molecule to inhibit the effects of circAKT3 on esophageal cancer cells. Furthermore, two protumor molecules, RHOC and STAT3, were the direct downstream targets of miR-17-5p, and circAKT3 upregulated RHOC and STAT3 by inhibiting miR-17-5p in esophageal cancer cells. Finally, we found circAKT3 knockdown inhibited growth of esophageal cancer in vivo.
CircRNAs function among the complex molecular network and are implicated in the expression of functional proteins[5,7]. Mechanistically, the inhibitory effect of circRNAs on miRNAs is one of the predominant mechanisms involving regulation of proteins[7]. Increasing evidence has focused on the effects of circRNAs on the modulation of malignant phenotypes of tumor cells; however, only a few studies explore the relationship between circRNAs and esophageal cancer. A recent study reported that circZNF292 is upregulated in the esophageal cancer cell line Eca-109, and inhibition of circZNF292 enhanced the expression of miR-206 to limit the proliferative, migratory, and invasive ability of Eca-109 cells[16]. In addition, Shi et al[17] demonstrated that overexpressed circPRKCI promoted the expression of AKT3 by inhibiting miR-3680-3p to facilitate proliferation and migration of esophageal cancer cells. In addition, circSMAD7[18] and circFOXO3[19] could sponge different miRNAs to suppress malignant phenotypes of esophageal cancer cells. Therefore, it can be inferred that circRNAs exert the opposite roles in development and progression of esophageal cancer.
Our research identified circAKT3 as a motivator in development and progression of esophageal cancer cells. The gene symbol of circAKT3 is AKT3, which is a key member of the PI3K/AKT signaling pathway[11,20]. Previously, Huang et al[11] reported that circAKT3 overexpression in gastric cancer participated in cisplatin resistance by restraining miR-198. However, the contradiction about the effects of circAKT3 on tumors is that circAKT3 could suppress development of glioblastoma[12] and progression of clear cell renal cell carcinoma[20]. These findings, combined with our results, indicate a tumor specificity of circAKT3. Moreover, we identified miR-17-5p as a downstream molecule of circAKT3 to suppress the effects of circAKT3 on esophageal cancer cells by evaluating proliferation, migration, and invasion of TE-1 cells and found that esophageal cancer tissue samples and cell lines showed a significantly lower level of miR-17-5p. It has been reported that the expression of miR-17-5p decreased esophageal cancer stem-like cells and mediated radiation resistance[13], which further reinforced our findings.
Additionally, we screened two protumor molecules RHOC and STAT3 as the direct downstream targets of miR-17-5p. RHOC is one of the Ras homolog family members and functions as a protumor molecule in multiple tumors[21,22]. Clinical data shows that RHOC is positively expressed in esophageal cancer tissue samples and could be regarded as a predictor for patients with a poor prognosis of esophageal cancer[23,24]. The in vitro and in vivo experiments manifest that RHOC enhanced the proliferative and invasive capacity of esophageal cancer cells[25]. Similar to RHOC, STAT3 is a pivotal molecule that accelerates development and progression of malignant tumors[26,27]. In esophageal cancer cells, activation of STAT3 dramatically reduced apoptotic rate and contributed to increased proliferative ability[28]. Moreover, STAT3 promoted the expression of programmed death ligand 1 to mediate immune evasion of esophageal cancer cells[29].
CONCLUSION
Taken together, our research discovered that circAKT3 enhanced proliferative, migratory, and invasive capacities of esophageal cancer cells by sponging miR-17-5p to exert protumor effects, and downregulation of circAKT3 inhibited xenograft tumor growth of esophageal cancer in vivo, providing a potential target for better understanding the underlying mechanism of esophageal cancer.
ARTICLE HIGHLIGHTS
Research background
Esophageal cancer is one of the most malignant tumors in the digestive system and exploring the underlying mechanism is a key issue that needs to be addressed. Circular RNA AKT3 (circAKT3) is a novel noncoding RNA participating in the development and progression of multiple tumors.
Research motivation
To identify biomarkers for the diagnosis and treatment of esophageal cancer.
Research objectives
To explore the mechanism of the role of circAKT3 in the development of esophageal cancer.
Research methods
Cell Counting Kit-8, wound healing assay, Transwell assay, and fluorescence analysis were used to evaluate the circAKT3 effect on proliferation, migration, invasion, and apoptosis of esophageal cancer cells.
Research results
In vitro assays results revealed that proliferative, migratory, and invasive capacities of esophageal cancer cells were significantly enhanced by circAKT3 overexpression. miR-17-5p was screened as the target of circAKT3, and miR-17-5p antagonized the effects of circAKT3 on esophageal cancer cells. Moreover, we identified RHOC and STAT3 as the direct target molecules of miR-17-5p, and circAKT3 facilitated expression of RHOC and STAT3 by inhibiting miR-17-5p. In vivo assays showed circAKT3 knockdown inhibited growth of esophageal cancer.
Research conclusions
CircAKT3 enhanced proliferative, migratory, and invasive capacities of esophageal cancer cells by sponging miR-17-5p to exert protumor effects, and downregulation of circAKT3 inhibited xenograft tumor growth of esophageal cancer in vivo, providing a potential target for better understanding the underlying mechanism of esophageal cancer.
Research perspectives
The role of circAKT3 in other tumors may be uncovered in the future, and its application in anticancer therapy will be promoted.
Footnotes
Institutional review board statement: This research was reviewed and approved by the Ethics Committee of China-Japan Union Hospital of Jilin University and complied with the guidelines of Declaration of Helsinki.
Institutional animal care and use committee statement: The procedures were in accordance with guidelines of Animal Use and Care Committee of China-Japan Union Hospital of Jilin University.
Conflict-of-interest statement: All the authors have no conflict of interest related to the manuscript.
ARRIVE guidelines statement: The authors have read the ARRIVE guidelines, and the manuscript was prepared and revised according to the ARRIVE guidelines.
Manuscript source: Unsolicited manuscript
Peer-review started: October 30, 2020
First decision: November 30, 2020
Article in press: December 16, 2020
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kravtsov V S-Editor: Chen XF L-Editor: Filipodia P-Editor: Ma YJ
Contributor Information
Hong-Liang Zang, Department of Hepatobiliary and Pancreatic Surgery, China-Japan Union Hospital of Jilin University, Changchun 130033, Jilin Province, China.
Fu-Jian Ji, Department of Colorectal Surgery, China-Japan Union Hospital of Jilin University, Changchun 130033, Jilin Province, China.
Hai-Ying Ju, Department of Hematology, Jilin Province Blood Center, Changchun 130000, Jilin Province, China.
Xiao-Feng Tian, Department of Hepatobiliary and Pancreatic Surgery, China-Japan Union Hospital of Jilin University, Changchun 130033, Jilin Province, China. fengwykctut4210@163.com.
Data sharing statement
No additional data are available.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Lagergren J, Smyth E, Cunningham D, Lagergren P. Oesophageal cancer. Lancet. 2017;390:2383–2396. doi: 10.1016/S0140-6736(17)31462-9. [DOI] [PubMed] [Google Scholar]
- 3.van Rossum PSN, Mohammad NH, Vleggaar FP, van Hillegersberg R. Treatment for unresectable or metastatic oesophageal cancer: current evidence and trends. Nat Rev Gastroenterol Hepatol. 2018;15:235–249. doi: 10.1038/nrgastro.2017.162. [DOI] [PubMed] [Google Scholar]
- 4.Ohashi S, Miyamoto S, Kikuchi O, Goto T, Amanuma Y, Muto M. Recent Advances From Basic and Clinical Studies of Esophageal Squamous Cell Carcinoma. Gastroenterology. 2015;149:1700–1715. doi: 10.1053/j.gastro.2015.08.054. [DOI] [PubMed] [Google Scholar]
- 5.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 6.Xia L, Song M, Sun M, Wang F, Yang C. Circular RNAs as Biomarkers for Cancer. Adv Exp Med Biol. 2018;1087:171–187. doi: 10.1007/978-981-13-1426-1_14. [DOI] [PubMed] [Google Scholar]
- 7.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J Neurosci Res. 2020;98:87–97. doi: 10.1002/jnr.24356. [DOI] [PubMed] [Google Scholar]
- 9.Wang S, Tang D, Wang W, Yang Y, Wu X, Wang L, Wang D. circLMTK2 acts as a sponge of miR-150-5p and promotes proliferation and metastasis in gastric cancer. Mol Cancer. 2019;18:162. doi: 10.1186/s12943-019-1081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu H, Yao B, Wen X, Jia B. FBXW7 circular RNA regulates proliferation, migration and invasion of colorectal carcinoma through NEK2, mTOR, and PTEN signaling pathways in vitro and in vivo. BMC Cancer. 2019;19:918. doi: 10.1186/s12885-019-6028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang X, Li Z, Zhang Q, Wang W, Li B, Wang L, Xu Z, Zeng A, Zhang X, Zhang X, He Z, Li Q, Sun G, Wang S, Li Q, Wang L, Zhang L, Xu H, Xu Z. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18:71. doi: 10.1186/s12943-019-0969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia X, Li X, Li F, Wu X, Zhang M, Zhou H, Huang N, Yang X, Xiao F, Liu D, Yang L, Zhang N. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer. 2019;18:131. doi: 10.1186/s12943-019-1056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynam-Lennon N, Heavey S, Sommerville G, Bibby BA, Ffrench B, Quinn J, Gasch C, O'Leary JJ, Gallagher MF, Reynolds JV, Maher SG. MicroRNA-17 is downregulated in esophageal adenocarcinoma cancer stem-like cells and promotes a radioresistant phenotype. Oncotarget. 2017;8:11400–11413. doi: 10.18632/oncotarget.13940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhan N, Li B, Xu X, Xu J, Hu S. Inhibition of FASN expression enhances radiosensitivity in human non-small cell lung cancer. Oncol Lett. 2018;15:4578–4584. doi: 10.3892/ol.2018.7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pastushenko I, Blanpain C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Hu G, Zhao Y, Xiao Z, Yan M, Ren M. Silence of cZNF292 suppresses the growth, migration, and invasion of human esophageal cancer Eca-109 cells via upregulating miR-206. J Cell Biochem. 2020;121:2354–2362. doi: 10.1002/jcb.29458. [DOI] [PubMed] [Google Scholar]
- 17.Shi N, Shan B, Gu B, Song Y, Chu H, Qian L. Circular RNA circ-PRKCI functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-3680-3p in esophageal squamous cell carcinoma. J Cell Biochem. 2019;120:10021–10030. doi: 10.1002/jcb.28285. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Wang Q, Zhu D, Rong J, Shi W, Cao X. Up-regulation of circ-SMAD7 inhibits tumor proliferation and migration in esophageal squamous cell carcinoma. Biomed Pharmacother. 2019;111:596–601. doi: 10.1016/j.biopha.2018.12.116. [DOI] [PubMed] [Google Scholar]
- 19.Xing Y, Zha WJ, Li XM, Li H, Gao F, Ye T, Du WQ, Liu YC. Circular RNA circ-Foxo3 inhibits esophageal squamous cell cancer progression via the miR-23a/PTEN axis. J Cell Biochem. 2020;121:2595–2605. doi: 10.1002/jcb.29481. [DOI] [PubMed] [Google Scholar]
- 20.Xue D, Wang H, Chen Y, Shen D, Lu J, Wang M, Zebibula A, Xu L, Wu H, Li G, Xia L. Circ-AKT3 inhibits clear cell renal cell carcinoma metastasis via altering miR-296-3p/E-cadherin signals. Mol Cancer. 2019;18:151. doi: 10.1186/s12943-019-1072-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guan X, Chen S, Zhao Y. The role of RhoC in malignant tumor invasion, metastasis and targeted therapy. Histol Histopathol. 2018;33:255–260. doi: 10.14670/HH-11-915. [DOI] [PubMed] [Google Scholar]
- 22.Thomas P, Pranatharthi A, Ross C, Srivastava S. RhoC: a fascinating journey from a cytoskeletal organizer to a Cancer stem cell therapeutic target. J Exp Clin Cancer Res. 2019;38:328. doi: 10.1186/s13046-019-1327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faried A, Faried LS, Usman N, Kato H, Kuwano H. Clinical and prognostic significance of RhoA and RhoC gene expression in esophageal squamous cell carcinoma. Ann Surg Oncol. 2007;14:3593–3601. doi: 10.1245/s10434-007-9562-x. [DOI] [PubMed] [Google Scholar]
- 24.Zhao ZH, Tian Y, Yang JP, Zhou J, Chen KS. RhoC, vascular endothelial growth factor and microvascular density in esophageal squamous cell carcinoma. World J Gastroenterol. 2015;21:905–912. doi: 10.3748/wjg.v21.i3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faried A, Faried LS, Kimura H, Nakajima M, Sohda M, Miyazaki T, Kato H, Usman N, Kuwano H. RhoA and RhoC proteins promote both cell proliferation and cell invasion of human oesophageal squamous cell carcinoma cell lines in vitro and in vivo. Eur J Cancer. 2006;42:1455–1465. doi: 10.1016/j.ejca.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 26.Huynh J, Chand A, Gough D, Ernst M. Therapeutically exploiting STAT3 activity in cancer - using tissue repair as a road map. Nat Rev Cancer. 2019;19:82–96. doi: 10.1038/s41568-018-0090-8. [DOI] [PubMed] [Google Scholar]
- 27.Thilakasiri PS, Dmello RS, Nero TL, Parker MW, Ernst M, Chand AL. Repurposing of drugs as STAT3 inhibitors for cancer therapy. Semin Cancer Biol. 2019 doi: 10.1016/j.semcancer.2019.09.022. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Zhi Y, Song H, Zong M, Yi J, Mao G, Chen L, Huang G. S1PR1 promotes proliferation and inhibits apoptosis of esophageal squamous cell carcinoma through activating STAT3 pathway. J Exp Clin Cancer Res. 2019;38:369. doi: 10.1186/s13046-019-1369-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang W, Pang Q, Yan C, Wang Q, Yang J, Yu S, Liu X, Yuan Z, Wang P, Xiao Z. Induction of PD-L1 expression by epidermal growth factor receptor-mediated signaling in esophageal squamous cell carcinoma. Onco Targets Ther. 2017;10:763–771. doi: 10.2147/OTT.S118982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data are available.