Abstract
Background
Dengue and Chikungunya viruses can cause large-scale epidemics, with attack rates of up to 80%. In Tanzania, there have been repeated outbreaks of dengue fever, the most recent in 2018 and 2019, mostly affecting the coastal areas. Despite the importance of these viruses, there is limited knowledge on the epidemiology of dengue (DENV) and Chikungunya (CHIKV) in Tanzania. This study was conducted to investigate the prevalence of DENV and CHIKV in Kilombero Valley, Tanzania.
Methods
A cross-sectional study was conducted at Kibaoni Health Center in Kilombero Valley, Southeastern Tanzania, in the rainy and dry seasons of 2018. Febrile patients of any age and gender were enrolled from the outpatient department. Blood samples were taken and screened for DENV and CHIKV viral RNA by real-time reverse transcription polymerase chain reaction assays.
Results
Overall, 294 patients were recruited. Most were females (65%), and one-third of patients were aged 14–25 years. DENV and CHIKV were detected in 29 (9.9%) and 3 (1.0%) patients, respectively. DENV was detected across all age groups during both the dry and rainy seasons. Although all 4 DENV serotypes were detected, serotypes 1 and 3 dominated and were present in 14 patients (42.4%) each. Additionally, the study showed DENV-1 and DENV-3 co-infections.
Conclusions
This study reveals the co-circulation of all 4 DENV serotypes and CHIKV in Kilombero. Importantly, we report the first occurrence of DENV-4 in Tanzania. Unlike previous DENV outbreaks caused by DENV-2, the 2018 outbreak was dominated by DENV-1 and DENV-3. The occurrence of all serotypes suggests the possibility of severe clinical outcomes in future DENV epidemics in Tanzania.
Keywords: Chikungunya virus, dengue serotypes, dengue virus, Kilombero district, Tanzania
Dengue (DENV) and Chikungunya (CHIKV) viruses pose a significant public health threat, with increasing geographic range and severity. There have been several acute-onset large-scale epidemics with attack rates of DENV and CHIKV reaching 90% and 85%, respectively [1–3]. Annually, DENV causes up to 400 million infections [4], while CHIKV has been reported in nearly 40 countries around the world with an estimate of 2 million cases [5]. Numerous outbreaks have been noted in Africa, including in East African countries such as Kenya, where outbreaks have been reported in 2016, 2019, and 2020 [6, 7]. The clinical manifestations of both diseases are similar to malaria and thus could lead to misdiagnosis and underreporting of such infections in the absence of specific laboratory diagnostic testing in malaria-endemic areas [8].
In Africa, out of the 4 immunogenically and genetically distinct DENV serotypes (DENV 1–4), DENV-2 is the most frequently documented [9–12]. Importantly, exposure to 1 DENV serotype does not confer immunity to other serotypes [13], and secondary infections by another serotype or mixed infections increase the risk of developing dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS), both being potentially lethal manifestations of dengue [14]. The 3 genotypes of CHIKV are Asian, West African, and East/Central/South African (ECSA) [15]. Since its discovery in 1952 in Tanzania, there have been several reports from studies mostly conducted in Northern and Central Tanzania [10, 16, 17].
In Tanzania, DENV outbreaks were reported in 2010, 2012–2014 [9, 18–20], and recently in 2018 and 2019 [21]. The 2014 outbreak resulted in 1018 confirmed cases and 4 deaths, mainly in Dar es Salaam [22], with a few confirmed cases in Kilosa district [10]. However, the worst documented dengue outbreak occurred in 2019 in Dar es Salaam and later in Tanga [21], with 6873 cases and 13 deaths [21]. Severe dengue infection is uncommon in Tanzania, reported in 3%–5% of dengue-infected patients in the Dar es Salaam region [23, 24]. DENV-2 is the most widely reported [23], even though co-circulation of DENV-1 and DENV-3 was demonstrated in the recent outbreak in 2019 [25]. In addition, there is evidence of DENV-1 and DENV-3 in travelers who returned to Japan from Tanzania [26, 27].
CHIKV infections, on the other hand, have been documented mostly in a few districts in Northern and Central Tanzania, with a prevalence ranging from 4% to 8% [10, 17, 28]. Currently, there is little information available regarding the prevalence of the circulating DENV serotypes and CHIKV in several areas of Tanzania. Unfortunately, there is also a lack of appropriate pathogen diagnosis in febrile diseases, and no surveillance programs have been conducted so far. Therefore, sporadic cases may have occurred but may have been undetected or misdiagnosed as malaria or other viral diseases because of similar clinical symptoms. The aim of this study was to determine the prevalence of DENV and CHIKV, including identification of DENV serotypes, in Kilombero Valley, Southeastern Tanzania.
METHODS
Study Setting
This study was conducted in Ifakara Town Council (ITC), located in Kilombero Valley, Southeastern Tanzania. The Kilombero Valley is surrounded by wildlife management areas, rainforests, and savannahs. The Kilombero River runs through the valley, creating one of the largest flood plains in Africa. The area has a dry season lasting from June to November and 2 rainy seasons, with heavy rains from March to May and short rains in December and January. Administratively, ITC has 1 division subdivided into 9 wards and 11 villages. Five wards are in urban areas, while 4 are in rural areas. This study was conducted at Kibaoni Health Center (KHC) located within ITC. KHC serves a population of about 106 424 people mainly from ITC and other parts of the Kilombero and Ulanga districts.
Study Design and Population
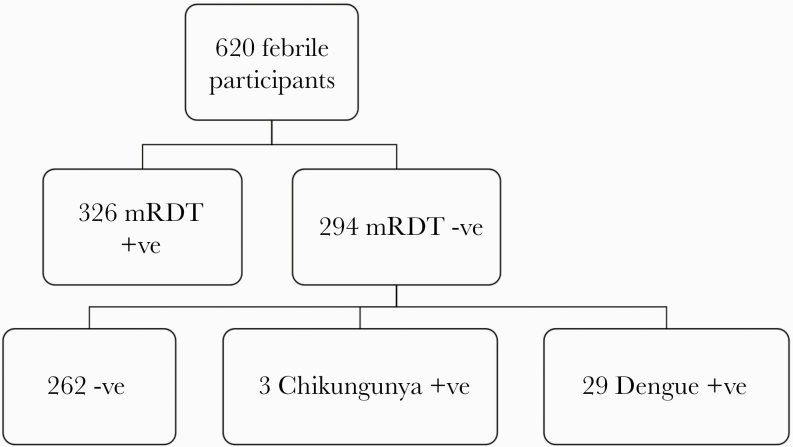
A cross-sectional study of patients presenting to the KHC with fever was conducted from March to May 2018 (rainy season) and from June to October 2018 (dry season). Eligible criteria were febrile patients of any age and gender with a measured axillary or rectal temperature of ≥37.5°C or ≥38°C, respectively. Patients were excluded from the study if they presented with severe illnesses that required immediate medical care, and individuals with positive malaria results by rapid test were excluded in the subsequent laboratory analysis, Figure 1.
Figure 1.
Flowchart of study participants’ enrollment and specimen processing. Abbreviations: +ve, individuals who tested positive; –ve, individuals who tested negative; mRDT, malaria rapid diagnostic test.
Sample Size
The sample size was estimated to be 334 and was calculated with the formula reported in Naing et al. (2006), using a prevalence of 29% based on a study conducted in febrile patients in Kilosa district, located 180 km from the study area [10, 29]. The assumptions for the confidence level and margin of error were 95% (1.96) and 5% (0.05), respectively, while recording error, as well as other such contingencies, was 5%.
Clinical Examination and Sample Collection
A trained physician at KHC obtained informed consent from eligible febrile patients. Information on demographics was collected using a standardized questionnaire. A physical examination was performed for each patient. The provisional clinical diagnosis and patient management were done by the hospital clinical team according to the local standard of care. Venous blood (5 mL from children and adults, or 2–3 mL from infants) was collected aseptically into EDTA vacutainer tubes for viral diagnostics. The blood was immediately transported to the Ifakara Health Institute (IHI) laboratory for phase separation. Plasma fractions were obtained and stored at –80°C until analyzed.
Laboratory Analysis
All enrolled patients were initially screened by malaria rapid diagnostic test (mRDT) against malaria parasites using SD BIOLINE Malaria Ag Pf/Pan (Standard Diagnostics, Inc.) at KHC. Samples whose mRDT results were negative were further analyzed by DENV and CHIKV real-time reverse transcriptase polymerase chain reaction (RT-PCR) assays at the IHI laboratory, Ifakara, Tanzania.
Real-time RT-PCR for Detection of DENV and CHIKV
RNA was extracted from plasma samples using the QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany). Real-time RT-PCR kits were used for detection and differentiation of CHIKV and DENV 1–4 serotypes (Infectious Diseases Horizontal Technology Centre, A*STAR). The kits were comprised of (i) Mix 1, which had specific oligonucleotide primers and Taqman probes for multiplex detection of DENV-1 and DENV-3, (ii) Mix 2, with specific oligonucleotide primers and probes for detection of DENV-2, DENV-4, and CHIKV, and (iii) internal control (IC) targeting β-actin protein in each respective mix and positive controls for all 4 DENV serotypes and CHIKV. The real-time RT-PCR for both Mix 1 (DENV1 and DENV3) and Mix 2 (DENV2, DENV4, and CHIKV) was carried out in a total reaction volume of 25 µL, with primer and probe concentrations as indicated in Supplementary Table 1. Thermal cycler conditions were used as follows: reverse transcription at 50°C for 20 minutes, initial denaturation at 95°C for 2 minutes, and 45 cycles of repeated denaturation at 95°C for 45 seconds and extension at 60°C for 1 minute 15 seconds. One-step real-time RT-PCR was performed in a Rotor Gene Q thermocycler (Qiagen).
Patient Consent Statement
A written informed consent was obtained from each participant. For children aged <12 years, a written informed consent was sought from a parent or guardian. In addition, verbal assent was also obtained from children aged 7–12 years, whereas those aged 12 – 18 years provided their own written consent in addition to consent of a parent or guardian. This study was approved by Institutional Review Board of Ifakara Health Institute (IHI/IRB/No. 12–2017) and the Medical Research Coordinating Committee of Tanzania’s National Institute for Medical Research (NIMR/HQ/R.8a/Vol.1X/2565). Each patient was assigned a study identification number to ensure confidentiality, which were applied to all patient specimens and data. All data were stored on a password-locked computer.
RESULTS
A total of 294 febrile patients were recruited into this study. One-third (33.0%) of patients were aged 14–25 years, and another third (30%) were aged 26–45 years (Table 1). Most participants were female (65.0%), and half (51.7%) were recruited during the dry season. One hundred eighty-four patients (62.6%) presented at KHC with a fever duration of 1–4 days, and 101 (34.4%) had a duration of 5–7 days. About half of the participants were subsistence farmers (52%), while 12.6% were engaging in business. The rest of the participants were students and children. Administratively, the majority of patients (88.1%) who visited KHC were from ITC, and very few came from Kilombero and Ulanga districts. The main presenting symptoms were headache (136, 46.3%), coughing (131, 44.6%), malaise (66, 22.4%), diarrhea (42, 14.3%), and vomiting (9, 0.3%).
Table 1.
Demographic Information of the Study Participants
| Category | Subcategory | No. | % |
|---|---|---|---|
| Age | ≤5 y | 25 | 8.5 |
| 6–13 y | 24 | 8.2 | |
| 14–25 | 97 | 33.0 | |
| 26–45 | 88 | 29.9 | |
| >45 | 60 | 20.4 | |
| Gender | Male | 103 | 35.0 |
| Female | 191 | 65.0 | |
| Season | Rainy | 142 | 48.3 |
| Dry | 152 | 51.7 | |
| Occupation | Farmer | 153 | 52.0 |
| Employed | 2 | 0.7 | |
| Students | 80 | 27.2 | |
| Business | 37 | 12.6 | |
| Children | 22 | 7.5 | |
| Location (by administration) | Ifakara Town Council | 268 | 91.2 |
| Kilombero district | 9 | 3.1 | |
| Ulanga district | 17 | 5.8 | |
| Location (urban vs rural) | Urban | 196 | 66.7 |
| Rural | 98 | 33.3 | |
| Duration of fever | 1–4 d | 184 | 62.5 |
| 5–7 d | 101 | 34.4 | |
| Undetermined | 9 | 3.1 |
Prevalence of DENV and CHIKV
The internal and positive controls for both DENV and CHIKV were amplified as shown in Supplementary Figures 1–6. All patients’ samples were successfully tested against dengue and Chikungunya viruses. Of 294 febrile patients, 29 (9.9%) tested positive for DENV by real-time RT-PCR. DENV was detected across all age groups, although the majority was observed in participants aged 14–25 years and >45 years. Among 29 DENV-positive samples, 19 were females and 10 were males (Table 2). DENV was detected in both seasons, that is, 16 and 13 cases in the rainy and dry seasons, respectively. DENV viral RNA copies were detected mostly in patients with a fever duration of 1–4 days. In terms of location (by administration), most of the DENV patients came from ITC. In this study, all DENV serotypes were detected, but DENV-1 and DENV-3 were more frequent (14 cases each), as summarized in Table 3. DENV-2 was detected in 2 patients, while DENV-4 was observed in 1 patient. Furthermore, our findings revealed co-infection between DENV-1 and DENV-3 in 2 patients as indicated in Table 3.
Table 2.
Characteristics of Patients With DENV by Real-time RT-PCR
| Category | Subcategory | DENV+ (%) | Odds Ratio | 95% CI |
|---|---|---|---|---|
| Overall | All patients | 29 (9.9) | N/A | N/A |
| Age | ≤5 y | 3 (1.0) | 1 | N/A |
| 6–13 y | 1 (0.3) | 0.26 | 0.02–2.78 | |
| 14–25 | 10 (3.4) | 0.74 | 0.18–2.94 | |
| 26–45 | 6 (2.0) | 0.55 | 0.12–2.39 | |
| >45 | 9 (3.1) | 1.18 | 0.29–4.82 | |
| Gender | Male | 10 (3.4) | 1 | N/A |
| Female | 19 (6.5) | 1.01 | 0.45–2.27 | |
| Season | Dry | 13 (4.4) | 1 | N/A |
| Rainy | 16 (5.4) | 1.40 | 0.65–3.03 | |
| Location | Ifakara Town Council | 26 (8.8) | 1 | N/A |
| Kilombero district | 2 (0.7) | 2.71 | 0.53–13.75 | |
| Ulanga district | 1 (0.3) | 0.59 | 0.07–4.65 | |
| Location (urban vs rural) | Rural | 9 (31.0) | 1 | N/A |
| Urban | 20 (69.0) | 1.14 | 0.50–2.62 | |
| Duration of fever | 1–4 d | 27 (9.2) | 1 | N/A |
| 5–7 d | 2 (0.7) | 0.90 | 0.20–4.09 | |
| Symptoms | Headache | 13 (44.8) | N/A | N/A |
| Cough | 10 (34.5) | N/A | N/A | |
| General body pain | 5 (17.2) | N/A | N/A | |
| Diarrhea | 3 (10.3) | N/A | N/A | |
| Skin rash | 1 (3.4) | N/A | N/A |
Abbreviations: DENV, dengue virus; RT-PCR, reverse transcriptase polymerase chain reaction.
Table 3.
Proportion of DENV Serotypes Among PCR-Positive Samples (n = 29)
| Dengue Serotype | No. (%) |
|---|---|
| DENV-1 | 14 (42.4) |
| DENV-2 | 2 (6.0) |
| DENV-3 | 14 (42.4) |
| DENV-4 | 1 (3.0) |
| Co-infection with DENV-1 & DENV-3 | 2 (6.0) |
Abbreviations: DENV, dengue virus; PCR, polymerase chain reaction.
Concerning CHIKV, only 3 patients (1%) out of 294 were CHIKV RT-PCR positive. Among the 3 positives, 2 were males. The 3 patients were a 22-year-old student, a 27-year-old farmer, and a 70-year-old farmer. Two CHIKV-positive cases were detected during the rainy season and 1 in the dry season.
DISCUSSION
This study reveals the presence of DENV and CHIKV among febrile patients seen at a district hospital in rural Tanzania in 2018 at the same time as a DENV outbreak in the coastal area of the country. Our findings demonstrate the occurrence of all 4 DENV serotypes as well as a co-infection with DENV-1 and DENV-3.
In the present study, the 9.9% prevalence of DENV was lower than previously reported in Dar es Salaam in 2019 (17 out of 20) [25]. This difference could be due to the study population, as the study from Dar es Salaam only involved dengue-suspected cases while the current study involved outpatient febrile patients from a health center. In addition, the earlier study was conducted in an urban setting with diverse breeding habitats for Aedes mosquitoes, which may contribute to urban transmission of the arboviral diseases, whereas the later study was conducted in a rural setting [30]. Also, the lower prevalence of DENV in our study could be a result of viral RNA detection sensitivity by RT-PCR declining with duration of fever, and therefore we might have missed patients with >5 days’ fever duration.
The 1% prevalence of CHIKV observed in this study is lower than that of Northern and Central Tanzania, where previous epidemiological studies reported a prevalence of 4%–8% [16, 17]. Furthermore, the low prevalence of CHIKV in Tanzania is in contrast to other countries in Africa (including East Africa), where outbreaks of CHIKV have been frequently reported since 1952 with a prevalence of up to 53% (n = 100/189) [31–34]. Our findings document the persistent circulation of CHIKV, supporting seroprevalence studies in other districts of Tanzania where anti-CHIKV antibodies (both immunoglobulin M and immunoglobulin G) have been detected; that is, 4.7% (n = 364) and 14% (n = 105), respectively, in the population, thus suggesting the endemicity of the disease [10, 35].
Unlike in the previous DENV outbreaks in Tanzania, which were mainly caused by DENV-2 [10, 20, 23], findings from the current study have shown that all DENV serotypes were circulating, with a predominance of DENV-1 and DENV-3 in the 2018 outbreak. Our results are in line with the findings from the study done in Dar es Salaam in 2019, which reported co-circulation of DENV-1 and -3 [25]. Furthermore, DENV-1 was detected in a traveler returning from Tanzania to Japan in 2019 [26], and DENV-3 had been reported in Japan from a traveler who had visited Tanzania in 2010 [36], supporting its existence in the country. During the repeated DENV outbreaks in Tanzania, the reported disease severity was low to moderate with few deaths, that is, 4 and 13 deaths in the 2014 and 2018–2019 outbreaks, respectively [22, 24]. Out of the 4 deceased cases in 2014, 1 had presented with DHF and 1 with multiple organ failure. Unfortunately, no data on the DENV serotype are available from the individuals who died [37]. With the increasing number of cases reported recently and the coexistence of all DENV serotypes, more severe disease or fatal outcomes might be observed in the future [14, 38].
The present study was conducted at a health care facility serving urban and semi-urban communities. Most DENV-positive cases seen in this study (26 samples) were from ITC and occurred in both the rainy and dry seasons. This is in line with other studies, which have demonstrated the occurrence of DENV in urban and semi-urban areas, correlating to the distribution of the vector A. aegypti mosquitoes [39, 40]. The surrounding wetlands in our study settings provide suitable breeding habitats for mosquitoes, and in addition the presence of short rains could further favor breeding of A. aegypti throughout the year. Thus, this increases the rate of CHIKV and DENV transmission.
Strengths and Limitations of the Study
To the best of our understanding, this study is the first to show the occurrence of DENV and CHIKV in Southwestern Tanzania. The study was conducted at a primary health care facility where most patients living in the area seek treatment and thus gives a good representation of the general population. Limitations of the study are that we tested the samples by real-time RT-PCR only, which might lead to underdetection of cases with a fever duration of >5 days. Our findings could be complemented by employing serological tests coupled with acute and convalescent serum and isolating RNA from whole blood and urine, as viral RNA persist longer in the urine. Also, we did not include very sick and hospitalized patients, which might have led to an underreporting of cases. Furthermore, due to limited resources, we excluded mRDT-positive individuals and hence missed chances to detect co-infections with malaria.
CONCLUSIONS
This study demonstrated the occurrence of all 4 DENV serotypes and CHIKV in Kilombero Valley, with DENV-4 being reported for the first time in Tanzania. In addition, we revealed the presence of co-infection with DENV-1 and -3 in 1 patient, possibly corresponding to a changing pattern of DENV serotypes from DENV-2 in the previous outbreaks to DENV-1 and DENV-3 in the 2018 outbreak. These findings are important in planning for future epidemics. We recommend further studies, particularly longitudinal ones that integrate febrile patients and vector surveillance to understand local transmission dynamics.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We would like to thank the Town Council Office of Ifakara and Kibaoni Health Center (KHC) for allowing us to use their facility to conduct this study. We are also grateful to participants for their willingness to participate in this study. We sincerely acknowledge the medical officer in charge of KHC for his cooperation throughout the study period. Special thanks go to the clinicians Pilly Azizi Ally and Joyce Karlo Mbata of KHC and the laboratory technician Sebastian Cobero of Ifakara Health Institute. Finally, we acknowledge the Infectious Diseases Horizontal Technology Centre (ID HTC) and Singapore Immunology Network (SIgN), Laboratory of Microbial Immunity, for providing us multiplex real-time RT-PCR kits for detection and differentiation of CHIKV and DENV serotypes 1–4.
Financial support. This research was funded by Ifakara Health Institute, Tanzania, through the Director’s Research and Innovation Fund award, the USAID-Zika Grand Challenges grant, and a core research grant provided to Singapore Immunology Network by Biomedical Research Council, A*STAR. The funders had no role in the study design, data collection or analysis, the decision to publish, or the preparation of the manuscript.
Potential conflicts of interest. L.F.P.N. has filed a technology disclosure for the RT-PCR multiplex (SIgN/P/11138/00/PCT). All other authors declare no competing interests. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author contributions. B.C., M.W., F.O., and L.F.P.N. conceived the study; B.C., R.D.S., and W.G. participated in study implementation; B.C., N.K.W.Y., and S.N.A. assisted in laboratory analysis; R.D.S. analyzed the data; B.C. and R.D.S. prepared the first draft; L.F.P.N. contributed reagents. All authors have read and approved the final manuscript.
Consent for publication. All participants gave consent for publication.
Availability of data and materials. The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- 1. Liang G, Gao X, Gould EA. Factors responsible for the emergence of arboviruses; strategies, challenges and limitations for their control. Emerg Microbes Infect 2015; 4:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Charrel RN, de Lamballerie X, Raoult D. Chikungunya outbreaks—the globalization of vectorborne diseases. N Engl J Med 2007; 356:769 –71. [DOI] [PubMed] [Google Scholar]
- 3. Mohan A, Kiran DH, Manohar IC, Kumar DP. Epidemiology, clinical manifestations, and diagnosis of Chikungunya fever: lessons learned from the re-emerging epidemic. Indian J Dermatol 2010; 55:54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bhatt S, Gething PW, Brady OJ, et al. The global distribution and burden of dengue. Nature 2013; 496:504–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sanyaolu A, Okorie C, Badaru O, et al. Chikungunya epidemiology: a global perspective. SM J Public Health Epidemiol 2016; 2:1028. [Google Scholar]
- 6. Petersen LR, Powers AM. Chikungunya: epidemiology. F1000Research 2016; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. ECDC. Chikungunya Worldwide Overview Stockholm, Sweden: European Centre; for Disease Prevention and Control; 2020. Available at: https://wwwecdceuropaeu/en/chikungunya-monthly. Accessed 7 March 2020. [Google Scholar]
- 8. Chipwaza B, Mugasa JP, Mayumana I, et al. Self-medication with anti-malarials is a common practice in rural communities of Kilosa district in Tanzania despite the reported decline of malaria. Malar J 2014; 13:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gautret P, Simon F, Hervius Askling H, et al. ; EuroTravNet Dengue type 3 virus infections in European travellers returning from the Comoros and Zanzibar, February-April 2010. Euro Surveill 2010; 15:19541. [PubMed] [Google Scholar]
- 10. Chipwaza B, Mugasa JP, Selemani M, et al. Dengue and Chikungunya fever among viral diseases in outpatient febrile children in Kilosa district hospital, Tanzania. PLoS Negl Trop Dis 2014; 8:e3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parreira R, Centeno-Lima S, Lopes A, et al. Dengue virus serotype 4 and Chikungunya virus coinfection in a traveller returning from Luanda, Angola, January 2014. Eurosurveillance 2014; 19:20730. [DOI] [PubMed] [Google Scholar]
- 12. Gubler DJ, Clark GG. Dengue/dengue hemorrhagic fever: the emergence of a global health problem. Emerg Infect Dis 1995; 1:55–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rodenhuis-Zybert IA, Wilschut J, Smit JM. Dengue virus life cycle: viral and host factors modulating infectivity. Cell Mol Life Sci 2010; 67:2773–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guzman MG, Alvarez M, Halstead SB. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: an historical perspective and role of antibody-dependent enhancement of infection. Arch Virol 2013; 158:1445–59. [DOI] [PubMed] [Google Scholar]
- 15. Afreen N, Deeba F, Khan WH, et al. Molecular characterization of dengue and Chikungunya virus strains circulating in New Delhi, India. Microbiol Immunol 2014; 58:688–96. [DOI] [PubMed] [Google Scholar]
- 16. Hertz JT, Munishi OM, Ooi EE, et al. Chikungunya and dengue fever among hospitalized febrile patients in northern Tanzania. Am J Trop Med Hyg 2012; 86:171–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kajeguka DC, Kaaya RD, Mwakalinga S, et al. Prevalence of dengue and Chikungunya virus infections in north-eastern Tanzania: a cross sectional study among participants presenting with malaria-like symptoms. BMC Infect Dis 2016; 16:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vairo F, Nicastri E, Meschi S, et al. Seroprevalence of dengue infection: a cross-sectional survey in mainland Tanzania and on Pemba Island, Zanzibar. Int J Infect Dis 2012; 16:e44–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hertz JT, Munishi OM, Ooi EE, et al. Chikungunya and dengue fever among hospitalized febrile patients in northern Tanzania. Am J Trop Med Hyg 2012; 86:171–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mboera LEG, Mweya CN, Rumisha SF, et al. The risk of Dengue virus transmission in Dar es Salaam, Tanzania during an epidemic period of 2014. PLoS Negl Trop Dis 2016; 10:e0004313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. SACIDS. Dengue Outbreaks in Tanzania: Recent Trends and Importance of Research Data in Disease Surveillance. Morogoro, Tanzania: Southern African Centre for Infectious Disease Surveillance; 2019. Available at: http://wwwsacidsorg/news/dengue-outbreaks-tanzania-recent-trends-importance-research-data-disease-surveillance/. Accessed 9 March 2020. [Google Scholar]
- 22. Msellemu D, Gavana T, Ngonyani H, et al. Description and lessons learned from the 2014 dengue outbreak in Dar es Salaam, Tanzania. Knowledge, attitudes and bite prevention practices among those with confirmed Dengue. bioRxiv 567396 [Preprint]. 4 March 2019. Available at: 10.1101/567396. Accessed 20 June 2020. [DOI]
- 23. Vairo F, Mboera LE, De Nardo P, et al. Clinical, virologic, and epidemiologic characteristics of dengue outbreak, Dar es Salaam, Tanzania, 2014. Emerg Infect Dis 2016; 22:895 –9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boillat-Blanco N, Klaassen B, Mbarack Z, et al. Dengue fever in Dar es Salaam, Tanzania: clinical features and outcome in populations of black and non-black racial category. BMC Infect Dis 2018; 18:644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mwanyika G, Mboera LE, Rugarabamu S, et al. Co-circulation of dengue virus serotypes 1 and 3 during the 2019 epidemic in Dar es Salaam, Tanzania. bioRxiv 763003 [Preprint]. 9 September 2019. Available at: 10.1101/763003. Accessed 10 July 2020. [DOI]
- 26. Okada K, Morita R, Egawa K, et al. Dengue virus type 1 infection in traveler returning from Tanzania to Japan, 2019. Emerg Infect Dis 2019; 25:1782–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moi ML, Takasaki T, Kotaki A, et al. Importation of dengue virus type 3 to Japan from Tanzania and Cote d’Ivoire. Emerg Infect Dis 2010; 16:1770–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Crump JA, Morrissey AB, Nicholson WL, et al. Etiology of severe non-malaria febrile illness in Northern Tanzania: a prospective cohort study. PLoS Negl Trop Dis 2013; 7:e2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Naing L, Winn T, Rusli B. Practical issues in calculating the sample size for prevalence studies. Arch Orofacial Sci 2006; 1:9–14. [Google Scholar]
- 30. Wilke ABB, Chase C, Vasquez C, et al. Urbanization creates diverse aquatic habitats for immature mosquitoes in urban areas. Sci Rep 2019; 9:15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.World Health Organization. Chikungunya – Mombasa, Kenya. Disease Outbreak News Geneva: World Health Organization; 2018. Available at: https://wwwwhoint/csr/don/27-february-2018-chikungunya-kenya/en/. Accessed 12 March 2020. [Google Scholar]
- 32. Konongoi SL, Nyunja A, Ofula V, et al. Human and entomologic investigations of Chikungunya outbreak in Mandera, Northeastern Kenya, 2016. PloS One 2018; 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zeller H, Van Bortel W, Sudre B. Chikungunya: its history in Africa and Asia and its spread to new regions in 2013–2014. J infect Dis 2016; 214:S436–40. [DOI] [PubMed] [Google Scholar]
- 34.World Health Organization. Chikungunya – Congo. Disease Outbreak News Geneva: World Health Organization; 2019. Available at: https://www.who.int/csr/don/01-may-2019-chikungunya-congo/en/. Accessed 12 March 2020. [Google Scholar]
- 35. Kinimi E, Shayo MJ, Patrick BN, et al. Evidence of Chikungunya virus infection among febrile patients seeking healthcare in selected districts of Tanzania. Infect Ecol Epidemiol 2018; 8:1553460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moi ML, Takasaki T, Kotaki A, et al. Importation of dengue virus type 3 to Japan from Tanzania and Cote d’Ivoire. Emerg Infect Dis 2010; 16:1770–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. ReliefWeb. Dengue outbreak in the United Republic of Tanzania (situation as of 30 May 2014) 2014. Available at: https://reliefwebint/report/united-republic-tanzania/dengue-outbreak-united-republic-tanzania-situation-30-may-2014. Accessed 12 March 2020.
- 38. Van Ta T, Tran HT, Ha QNT, et al. The correlation of clinical and subclinical presentations with dengue serotypes and plasma viral load: the case of children with dengue hemorrhagic fever in Vietnam. Int J Res Pharma Sci 2019; 10:2578–85. [Google Scholar]
- 39. Noordeen F, Raza M, Pitchai F, et al. Distribution of dengue vectors, Aedes aegypti and Aedes albopictus, in a few selected semi-urban areas of the Central Province of Sri Lanka. Sri Lankan J Infect Dis 2018; 8:36–9. [Google Scholar]
- 40. Dalpadado C, Amarasinghe L. Abundance and distribution pattern of Aedes aegypti and Aedes albopictus in selected urban, sub-urban and rural areas of Gampaha District, Sri Lanka. Paper presented at: Research Symposium on Pure and Applied Sciences, 26 October 2018, University of Kelaniya, Kelaniya, Sri Lanka; 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.