Abstract
Macroautophagy (hereafter referred to as autophagy) is a evolutionarily conserved pathway in which proteins and organelles are delivered to the lysosome for degradation. In neurons, autophagy was originally described as associated with disease states and neuronal survival. Over the last decade, however, evidence has accumulated that autophagy controls synaptic function in both the axon and dendrite. Here, we review this literature, highlighting the role of autophagy in the pre- and postsynapse, synaptic plasticity, and behavior. We end by discussing open questions in the field of synaptic autophagy.
Keywords: Autophagy, synaptic transmission, synaptic plasticity, protein degradation, endocytosis
I. Introduction
Research over the past 40 years has defined roles for protein synthesis in neuronal plasticity and brain function [1]. Recently, however, the controlled degradation of proteins and organelles has also been appreciated to play critical roles in neurotransmission [2]. These steps occur via the degradation of specific cytosolic proteins through the ubiquitin / proteasome pathway and by multiple pathways that lead to degradation within the lysosome [3].
Christian De Duve, who with Alex Novikoff, co-discovered the lysosome, introduced the term autophagy to describe how “portions of a cell somehow find their way inside the cell’s own lysosomes and are broken down” [4]. De Duve also coined the terms lysosome, endocytosis / endosome, phagocytosis/ phagosome, / autophagosome and peroxisome [5].
De Duve’s definition of autophagy encompasses the breakdown of any of a cell’s own components within the lysosome, but even in 1963 it was clear that proteins, lipids, and organelles could use multiple mechanisms of entry into the lysosomal lumen.
In the endosomal-lysosomal degradation pathway, membrane, lumenal or formerly extracellular components that were endocytosed are delivered to lysosomes by fusion with endosomes. In this highly regulated pathway, extracellular proteins and intrinsic proteins on the cell surface are internalized into small vesicles and sorted through a series of intermediate vesicular structures. Some components may be recycled to other membranes, while others that reach a late endosome can fuse with a lysosome that is capable of proteolysis and can degrade proteins in both the endosomal membrane and lumen [6,7].
A relatively newly discovered autophagic pathway is known as chaperone-mediated autophagy (CMA) [8], in which cytosolic proteins are bound by the chaperone Hsc70 and delivered to the lysosomal membrane, where they are unfolded and transported into the lysosomal lumen by a membrane protein, Lamp2A [9]. Proteins within the lumen are then exposed to lysosomal proteases and degraded. While a great deal of research has identified cytosolic proteins that play roles in synapses to be degraded by CMA including alpha-synuclein [10] and tau protein [11], there has been little research on CMA at synapses in contrast to in cell bodies.
In endosomal microautophagy (eMI), cytosolic proteins are similarly bound by Hsc70, but are internalized into small vesicles within endosomes which subsequently fuse with lysosomes for degradation [12]. These structures may also participate in producing exosomes secreted from the neurons, and so may participate in signaling and disease spread. As multivesicular bodies are very common in synapses and axons, these are likely to be important for synaptic turnover, and early evidence suggests that eMI may play a role in presynaptic function [13,14].
At this writing, most research has been conducted on roles for macroautophagy (which by convention will be called autophagy in this review), a process that can degrade cytosolic proteins, lipids and organelles. This form of autophagy has been particularly tractable because it involves the participation of large double membrane vesicles, termed autophagosomes, that fuse with lysosomes for degradation that can be examined by microscopy and because it can be manipulated by modulating the activity of genes initially characterized in yeast that are responsible for steps in the pathway.
Here, we will first review the molecular machinery involved in autophagy in mammalian cells. We will then briefly describe the history of the discovery of autophagy in neurons and explore how the machinery that mediates autophagy in dividing cells has been adapted to ensure appropriate autophagic protein degradation in post-mitotic neurons.
Recent work has highlighted a potential role for autophagy in the regulation of synaptic transmission, plasticity and behavior. These studies indicate that autophagy plays some roles that can be revealed simply by blocking specific steps in the pathway, while others that require both blockade and a triggering additional factor, particularly via cellular stress responses [15–18]. Both classes of synaptic autophagic function may regulate pre- and postsynaptic functions during normal neuronal development and in disease. We conclude with three outstanding questions that we feel are critical for the field to address to deepen our understanding of the role of autophagy in controlling neurotransmission.
II. Molecular Machinery of Autophagy
The molecular machinery that controls autophagosome formation, maturation and fusion with lysosomes were originally described in yeast by Yoshinori Ohsumi and colleagues [19]. Autophagy in mammalian cells is controlled by both proteins that are homologous to the yeast autophagy (atg) proteins and additional mammalian-specific components [20].
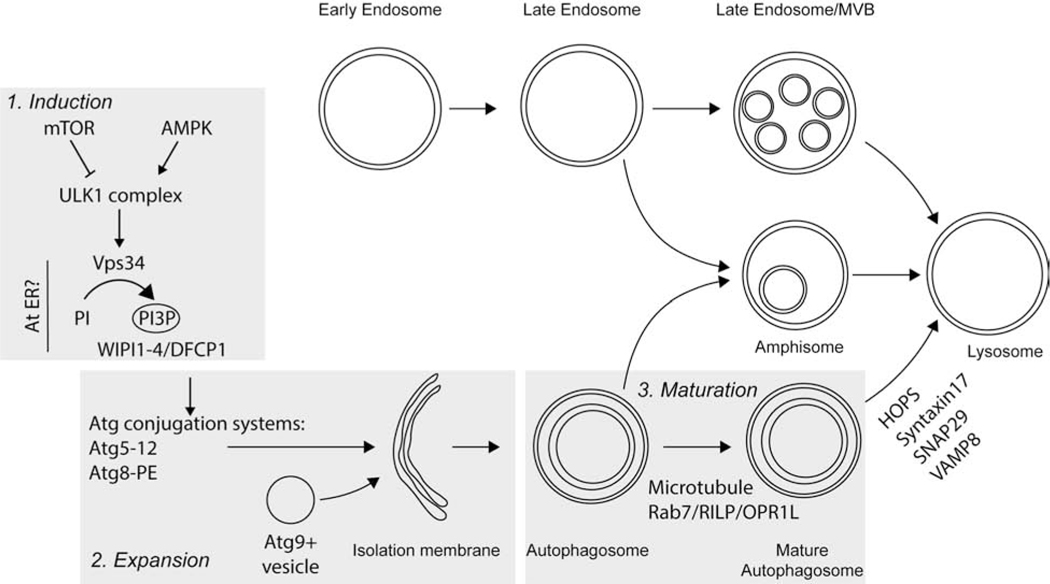
In yeast and dividing mammalian cells, autophagy is strongly activated by nutrient or serum starvation and can be divided into distinct stages: (1) induction, (2) expansion, (3) maturation (Figure 1).
Figure 1. Schematic representation of the mechanisms of autophagy.
Autophagy induction is driven by mTOR and AMPK. These metabolic kinases stimulate PI3P synthesis which recruits the PI3P binding proteins DFCP1 and WIPI1-4 to a membrane source. The activity of the Atg conjugations systems and Atg9+ vesicles expand the autophagic membrane. Once the membrane closes, the autophagosome matures, traffics to the perinuclear area and fuses with lysosomes. Late endosomes can fuse with closed autophagosomes to form amphisomes. Cargo is not depicted for simplicity.
Autophagy is induced following nutrient or serum deprivation via the activity of the metabolic kinases, mTOR [21] and AMPK [22]. mTOR inhibits and AMPK activates the kinase ULK1, in complex with Atg13 and FIP200 [23–27]. ULK2, a ULK1 homolog in mammalian cells, may also control autophagy but it is presently unclear whether these kinases are redundant [24,28]. ULK complex activation leads to the phosphorylation of Beclin-1 and activation of the class iii phosphoinositol-3-kinase (PI3K), Vps34 [29,30]. The PI3K activity of Vps34 is critical both for endocytic activity [31,32] and autophagy [30] and is directed to a particular pathway by its interacting proteins. For example, when partnered with Atg14, Vps34 activity stimulates autophagy [33], and ULK1-dependent phosphorylation of Beclin-1 specifically activates Vps34 activity when Vps34 is complexed with Atg14 and Ambra1 [29].
Vps34 activity induces autophagy via local synthesis of phosphoinositol-3-phosphate (PI3P). The precise sites of PI3P synthesis and initial formation of the autophagic membrane may be at the endoplasmic reticulum [34–36], but contributions from plasma membrane [37], mitochondria [38], or recycling endosomes [39–42] are also involved. Subsequent recruitment of PI3P binding proteins, such as DFCP1 [34] and WIPI1-4 [39,43–45], initiate an enzymatic cascade that leads to the assembly of the autophagosomal membrane.
Expansion of the autophagosome membrane is driven by two ubiquitin-like conjugation systems. In the first, Atg12 is conjugated to Atg5 by the E1-like enzyme, Atg7, and the E2-like enzyme, Atg10 [46]. The Atg5-12 conjugate subsequently interacts with Atg16L1 [47]. In the second pathway, Atg8 is processed by the protease, Atg4 [48], and subsequently conjugated to phophatidylethanolamine (PE) by Atg7 and the E2-like enzyme, Atg3 [46]. Atg8 with PE conjugation requires the E3-like activity of Atg5-12 complexed with Atg16L1 [49,50].
Mammals have numerous Atg8 homologs, including the LC3 and GABARAP families [51]. The conjugated form of LC3 is referred to below as LC3-II and the unmodified protein is referred to LC3-I. Vesicles containing Atg9, a transmembrane protein, then fuse with the growing autophagosome and provide an additional membrane source [52].
How are proteins and organelles selected for autophagic degradation [53]? Upon nutrient deprivation, bulk cytosolic proteins can be sequestered into autophagosomes and degraded to increase the availability of amino acids [54–56]. Alternatively, proteins and organelles can be selectively degraded by binding to autophagy cargo adapters such as p62 [57,58], NBR1 [59], Tax1BP1 [60], optineurin [61] and NDP52 [62–65]. Proteins may contain an LC3-interacting motif that promotes binding to LC3 and sequestration within the autophagosome, such as Dishevelled-2 [66–68]. Proteins and organelles that are targeted for selective degradation can also be modified by post-translational modifications such as ubiquitination or acetylation, which mediate interactions with autophagy cargo proteins such as p62 [69].
Following cargo sequestration, autophagosomes close and are trafficked retrogradely to the perinuclear region, where most late endosomes and lysosomes are localized [70–72]. Retrograde trafficking of autophagosomes occurs on microtubules in a dynein-dependent manner [70]. Autophagosomes are linked to dynein via a Rab7, RILP, and OPR1L complex [73,74] and in the absence of these factors, mature autophagosomes accumulate. Interestingly, actin-dependent autophagosome transport has also been demonstrated, although this does not occur during starvation-induced autophagy [75]. Following retrograde transport, autophagosome fusion with lysosomes depends on the Qa SNARE, synataxin 17; the Qbc SNARE, SNAP29; and the R SNARE, VAMP8 [76]. This process also depends on the HOPS complex [77–79] and the autophagosome adapter PLEKHM1 [78]. The lysosomal GTPase Arl8 also controls autophagosomal and endosomal fusion with lysosomes [80–84] via recruitment of the HOPS complex [82] and lysosomal positioning [80]. The fusion of autophagosomes with lysosomes leads to degradation of autophagic cargo.
III. Autophagy in Neurons and Neurites
The initial studies of autophagy by De Duve, Novikoff and others were in dividing cells. Neurons have long been known to possess lipofuscin, neuromelanin, and in ceroid lipofuscinosis disorders, ceroid pigments, that are accumulated within autophagosomes [85]. Nevertheless, to our knowledge, the initial reports in neurons of autophagosomes (also known as autophagic vacuoles) were in two mid-1970s electron microscopy reports of pathological specimens from the brain of Huntington’s disease patients [86,87]. The latter study stressed a highly increased abundance of lipofuscin in the neurons, astrocytes and microglia of these patients, and conjectured that this was due to insufficient autophagy, an impression supported by many subsequent studies in human tissue and animal models. This same study further noted enlarged mitochondria devoid of cristae and suggested that this could be due to deficient autophagy associated with Huntington’s disease, as borne out by later research by Marianne DiFiglia and colleagues [88]. Cultured striatal neurons with mutant huntingtin gene were found to form prominent autophagosomes when stressed by dopamine-mediated oxidative stress [89], predicting one theme of this review: in synapses, some effects of autophagy become apparent under conditions of cellular stress. Mechanistic studies by Ana Maria Cuervo and collaborators indicated that in the case of huntingtin mutations that this could result from a lack of appropriate autophagic cargo recognition [90].
In addition to the lipofuscin pigment, work by Luigi Zecca and collaborators has shown that the neuromelanin pigment of substantia nigra and locus coeruleus neurons is encased with autophagic organelles that apparently do not degrade these contents [85,91], leading to an initial focus in the field on neuronal autophagy as a product of disease and normal aging related stress [92]. Subsequent studies by Randy Nixon, Anne Cataldo and colleagues revealed the presence of neuronal autophagic vacuoles in tissue from Alzheimer’s patients and other disorders [93–95] and further work by Anne Tolkovsky, David Rubinsztein, Zhenyu Yue, Richard Youle and many others have extended the observation of aberrant autophagy to a wide variety of associated neuronal disorders [96–98].
The presence of autophagic organelles in neurites and synapses awaited identification by Peter Hollenbeck [99], who observed retrograde axonal transport and lysosomal delivery of fluorescent cargo by autophagic vacuoles in cultured sympathetic neurons. This study suggested, in contrast to the other studies of the era, that autophagy might be associated with synaptic activity and may occur independently from disease or aging-oriented stress.
It took a long time for Hollenbeck’s pioneering observation to be appreciated by the field at large. One reason is that in dividing cells, autophagy was typically initiated by nutrient deprivation [54–56]. While nutrient deprivation and starvation is a potent trigger for autophagic degradation in non-neuronal cells, it has little effect in most populations of central neurons [100]. An exception is in the hypothalamus [101,102], a brain region that is critical for the maintenance of energy homeostasis [103], where autophagy senses energy balance to control neuronal activity (see below). Another reason may be that the autophagosomes, particularly in neurites, may be relatively short-lived and transported rapidly.
An experimental tool that has vastly changed the study of autophagy in synaptic transmission is the use of fluorescently tagged LC3 to specifically label autophagosome membrane in living cells. A study from Erica Holzbaur and colleagues introduced an elegant model in which autophagosomes were primarily generated in the distal processes of neurons and then trafficked toward the cell body to fuse with lysosomes in proximal processes and the cell body [104–106]. Live imaging GFP-LC3 puncta in primary cultures of dorsal root ganglia (DRG) suggested that autophagosome formation occurs almost exclusively within the axon tip. This may not be surprising, as DRG neurons differ from most CNS axons in that they do not make en passant synapses but instead have a single release site from the distal axon [106]. Similar imaging studies in synaptically mature primary hippocampal neurons yielded a more complex picture, with autophagosome formation occurring predominantly in distal processes but also in the cell body and more proximally in dendrites and axons [106]. Shehata et al reported the activity-dependent formation of autophagosomes in dendrites of primary hippocampal neurons [15]. Similar findings suggesting distal formation of autophagosomes have been reported in C. elegans [107]. These data suggest that autophagosome formation occurs within pre- and post-synaptic compartments and provide a means for retrograde transport of autophagic cargo from distal regions of the neuron to the cell body for degradation.
What specifies the location of autophagosome formation in neurons? Holzbaur and Maday proposed that PI3P synthesis occurs distally within axons, as DFCP1 puncta, which mark sites of PI3P synthesis and nascent autophagosome formation, were located distally in neuronal processes [106]. In C. elegans, Colon-Ramos and colleagues identified Atg9 trafficking events within the distal axon as being required for spatial organization of autophagosome formation [107]. Whether additional (neuron-specific) regulators are involved in orchestrating autophagosome formation and maturation within these highly complex cells remains to be seen.
The identification of autophagosomes and their regulated formation and trafficking, in neurons without ongoing pathology suggests that autophagy may play a role in the regulation of neurotransmission. To address this, transgenic mice lacking required autophagy genes could be used to identify changes in behavior and neurophysiology associated with loss of autophagy. Unfortunately, whole-body, constitutive knockout of many autophagy genes yield perinatal lethality [108]. Furthermore, the consequences of autophagy loss in the CNS could not be disambiguated from the effect of loss of autophagy in other organs.
Fundamental contributions to elucidating roles for autophagy in neurons were made by Hara, Komatsu and colleagues in the mid-2000’s who addressed this issue by generating mice with conditional alleles of Atg5 or Atg7 that could be knocked out in genetically-defined cell types following expression of Cre recombinase [109,110]. CNS-specific knockout of Atg5 or Atg7 yielded similar age-dependent neurodegeneration, accumulation of cytoplasmic proteinaceous inclusions and motor dysfunction. These phenotypes were similar in both mouse lines, indicating that as in yeast, Atg5 and Atg7 function at similar steps in the mammalian CNS. These transgenic mouse lines have become critical tools that have enabled the identification of cell-type specific roles for autophagy in the CNS (see below).
IV. Autophagy and neurotransmission
Throughout the remainder of this review, we discuss the reciprocal roles of autophagic protein degradation in neurotransmission
1. Autophagy in synaptic development
As we and others have recently reviewed the role of autophagy in synaptic development, we limit our review of this topic to two particular studies specifically relevant to the role of autophagy in neurotransmission [111,112].
In C. elegans, clustering of postsynaptic GABAA receptors in muscle depends on presynaptic GABA release [113]. In the absence of presynaptic innervation by GABAergic terminals, GABAA receptors are diffusely localized on the plasma membrane of the muscle. Interestingly, however, the absence of both GABAergic and cholinergic inputs leads to endocytosis and degradation GABAA receptors via autophagy [16], while deletion of autophagy in non-innervated muscles rescues deficits in neurotransmission. These data provide initial evidence that transmembrane postsynaptic receptors are degraded via autophagy, in an endocytosis-dependent manner. Notably, one family of mammalian Atg8 homologs, the GABARAPs, are critical for anterograde trafficking of GABAA receptors [114,115], suggesting that autophagy-associated proteins are critical regulators of biosynthesis and degradation of these receptors.
In addition to its role in degradation of neurotransmitter receptors during development, autophagy is also critical for the maturation of synaptic morphology. In Drosophila, developmental loss of autophagy decreases neuromuscular junction (NMJ) size, while enhancing neuronal autophagy by overexpression of Atg1, a homolog of ULK1, leads to an increased number of synaptic sites [116]. In contrast, however, autophagy is required for dendritic spine pruning during mouse cortical development [117] and plays a critical role in axon pathfinding in the developing CNS [118–120], indicating that autophagy has been adapted for a range of synaptic mechanisms.
Finally, the lysosomal GTPase Arl8 [121,122] contributes to the delivery of active zone and synaptic vesicle precursor proteins within non-degradative lysosome-like vesicle in Drosophila, C. elegans and in mammalian neurons [123–125]. Whether delivery of these components involves lysosomal degradation or intersects with autophagy remains to be seen.
Little is known how neuronal autophagy regulates synaptic structure. Autophagic degradation of neurotransmitter receptors or neurotransmitter release (see below) may contribute to activity-dependent synaptic development. Similarly, autophagic degradation of mitochondria or endoplasmic reticulum, organelles that regulate synapse development in part due to their position in neurites [126,127] may be critical for the establishment and maturation of synaptic contacts. Experiments in which the degradation of specific autophagic cargo are disrupted during CNS development will be required to distinguish between these mechanisms.
2. Autophagy and neurotransmitter release
Autophagosome formation at presynaptic terminals suggests that autophagy shapes neurotransmitter release. Our lab provided early evidence of a cell-autonomous role for autophagy in neurotransmitter release [128]. We took advantage of the fact that dopamine (DA) release can be directly measured using cyclic voltammetry, in contrast to the release of other neurotransmitters which depends on postsynaptic recordings of receptor activation, to unambiguously demonstrate that DA neurons lacking autophagy release more DA following electrical stimulation. This was a clear result of the role of autophagy in axons, as opposed to a role in cell bodies, as DA somata were not present in the brain slice preparation we used. Ultrastructural analysis of DA axons in the absence of autophagy revealed enlarged presynaptic terminals, and changes in the morphology and number of synaptic vesicles. These data implicated autophagy in the homeostasis of synaptic vesicles and neurotransmitter release machinery.
Recent work from Ackermann, Garner and colleagues further implicates autophagy in the clearance of damaged synaptic vesicles. Synaptic vesicles undergo rapid cycles of release and reformation, and this has been hypothesized to lead to damage to synaptic vesicle associated proteins [129]. To address whether autophagy is involved in the clearance of synaptic vesicle proteins, Hoffman et al fused supernova, a protein which creates reactive oxygen species (ROS) in response to light [130], to synaptotagmin, synapsin or synaptophysin and demonstrated that ROS-induced damage leads to autophagosome formation within the activated presynaptic terminal and degradation of synaptic vesicle-associated proteins [131]. Activation of autophagy was required to counteract the damage caused by ROS on neurotransmitter release and synaptic vesicle endocytosis. These data suggest that autophagy contributes to synaptic homeostasis by degrading damaged synaptic vesicles on demand.
Does normal physiological activity also induce presynaptic autophagy? High-frequency stimulation of the Drosophila NMJ induces autophagosome formation within the presynaptic terminal, suggesting that autophagy may be required for the maintenance of presynaptic machinery during stimuli with elevated energy demands [132]. In combination, these data provide tantalizing evidence that autophagy is critical for the regulation of neurotransmitter release via a contribution to synaptic vesicle quality control.
Additional insights into the dynamics of presynaptic autophagy have arisen from experiments aimed at defining the role of presynaptic proteins in the control of autophagy. Synaptojanin is a lipid phosphatase that induces synaptic vesicle endocytosis via the dephosphorylation of PI(4,5)P2. The SAC1 domain of synaptojanin has recently been implicated in the maturation of autophagosomes at the presynaptic terminal in Drosophila [133]. EndophilinA, another protein involved in synaptic vesicle endocytosis, has recently been found to promote presynaptic autophagy by recruiting autophagy-associated proteins to highly curved membranes [132]. Finally, the presynaptic proteins Bassoon and Piccolo suppress presynaptic autophagy by sequestering Atg5 [134]. In the absence of Bassoon and Piccolo, synaptic vesicle pools are depleted and synaptic sites are lost via activation of autophagy. These data suggest not only that autophagy plays important roles in presynaptic function, but also that presynaptic proteins contribute to autophagic function within the axon.
3. Autophagy and postsynaptic functio
Autophagy further controls synaptic transmission via postsynaptic mechanisms. Loss of autophagy in cortical pyramidal neurons or using nestin-cre to knockout autophagy throughout the nervous system leads to elevated levels of components of the postsynaptic density such as PSD-95, SHANK3, and PICK1 [102,117]. Furthermore, in a mouse model of Fragile X syndrome, elevations in PSD95 and the immediate early gene, Arc, can be rescued by activation of autophagy [135]. Whether these proteins are sequestered into nascent autophagosomes formed into the dendrite or their levels are affected by autophagy through an alternative mechanism remains unknown. The ability of autophagy to degrade postsynaptic scaffolding proteins provides a possible mechanism through which autophagy could control synapse morphology and the efficacy of synaptic transmission.
Autophagy also contributes to the degradation of neurotransmitter receptors. In addition to the degradation of GABAA receptors during C. elegans development [16], chemically-induced long-term depression activates autophagic degradation of the AMPA receptor, GluR1 [15]. The mechanism by which autophagy contributes to GluR1 degradation and whether this is endocytosis-dependent, remains unknown.
4. Autophagy and synaptic plasticity
Changes in the strength of synaptic transmission, termed synaptic plasticity, are thought to represent a cellular correlate of learning and memory. Disrupted synaptic plasticity has been reported in mouse models lacking autophagy; however, much of the mechanism through which autophagy regulates synaptic plasticity remains unknown.
Long-term potentiation appears to be regulated by autophagy in some cases. Glatigny et al demonstrated that pharmacological inhibition of autophagy with Spautin-1 blocks theta burst stimulation-induced LTP in CA1 [136]. Nikoletopoulou and colleagues suggest that ongoing autophagy in the hippocampus is suppressed by brain-derived neurotrophic factor (BDNF) to permit LTP [102]. Autophagy may also play a role in long-term depression (LTD), as impaired autophagy has been implicated in exaggerated hippocampal mGluR-LTD in a mouse model of Fragile X syndrome [135].
Autophagy may contribute to synaptic plasticity in several ways. First, autophagy may actively degrade AMPA receptors to reduce synaptic strength during LTD [15]. Second, autophagy may degrade other synapse-associated proteins required for reorganization of the postsynaptic membrane during plasticity [102,117,135]. Autophagy may further regulate the levels of cytosolic calcium within the pre- or post-synaptic elements via degradation of mitochondria or endoplasmic reticulum [98,126]. It should also be noted that several kinases that regulate autophagic activity, including mTOR, Akt, and AMPK, are involved in synaptic plasticity [137,138], although whether these kinases act through autophagy to modulate synaptic plasticity remains a key question.
5. Autophagy contribution to behavior
The contribution of autophagy to brain function has also been explored through cell-type specific knockouts of autophagy genes.
a. Hypothalamus and energy homeostasis
The role of autophagy in hypothalamic control of food intake and energy homeostasis has been extensively studied. The arcuate nucleus of the hypothalamus (ARC) controls food intake in response to the status of an organism’s energy store [103]. Two neuronal populations in the ARC, Agouti-related peptide (AgRP)-expressing (ARCAgRP) and pre-opiomelanocortin (POMC)-expressing (ARCPOMC), oppositely control feeding [139–142]. ARCPOMC neurons release α-MSH, a cleavage product of POMC [143], which stimulates Melanocortin-4 Receptors (MC4R) on neurons in the paraventricular hypothalamus (PVH) [144–146], and other brain regions, to inhibit feeding. AgRP, which is also released in the PVH by ARCAgRP neurons, is an antagonist of the MC4R and stimulates feeding [140,147]. Circulating hormones such as leptin, insulin and ghrelin, and extracellular levels of glucose and lipids alter ARC neuron firing to elicit feeding behavior or satiety [103]. These extrinsic cues activate intracellular biochemical cascades. In particular, mTOR and AMPK activity within ARC neurons control firing rates and neuropeptide release to affect feeding in response to energy status [148–151]. Thus, neurons of the ARC represent a critical central node in energy homeostasis.
As described above, autophagy is tightly regulated by the coordinated activity of AMPK and mTOR. In contrast to most of the CNS, fasting induces autophagic activity in the hypothalamus [101,102]. To define the role of autophagy in ARC neurons and central control of energy homeostasis, Kaushik et al conditionally deleted the required autophagy protein Atg7 in ARCAgRP neurons using the Cre-LoxP system [101]. Mice lacking Atg7 in ARCAgRP neurons (which promote feeding) had lower body weight and fat mass relative to controls suggesting a deficit in ARCAgRP neuron function as these neurons promote feeding. One mechanism through which ARCAgRP neurons promote feeding in response to food deprivation is to increase the expression of AgRP mRNA. Kaushik et al reported that in mice lacking Atg7 in ARCAgRP neurons, fasting failed to induce an increase in AgRP expression. They concluded that intact autophagy is required for the fasting-induced mobilization of free fatty acids (FFA) from lipid droplets, a process known as lipophagy [152], and that FFA signalling normally activates AgRP mRNA expression by ARCAgRP neurons. These results elegantly demonstrate a cell-type specific role for autophagy in the central control of energy homeostasis.
In contrast to ARCAgRP neurons, conditional deletion of Atg7 in ARCPOMC neurons leads to increased body weight. Bouret and colleagues reported that this effect correlated with decreased axonal arborization of ARCPOMC neurons and innervation of target regions such as the PVH [153]. Both the Bouret and Lee groups demonstrated that autophagy was required in ARCPOMC neurons for a normal response to leptin: systemic or intracerebral administration of leptin did not suppress feeding in mice lacking autophagy in ARCPOMC neurons [153,154]. This lack of appetite suppression was associated with increased circulating leptin levels, further suggesting a state of relative leptin resistance. The precise mechanism, however, through which autophagy regulates ARCPOMC axonal outgrowth or biochemical response to leptin remains obscure. Furthermore, autophagy also contributes to the cellular response to neuropeptide Y, another key contributor of feeding within the hypothalamus [155].
AMPK also mediates fasting-induced synaptic plasticity within the ARC [156]. Whether changes in AMPK activity affect autophagic flux within the ARC and whether autophagy contributes to fasting-induced and AMPK-dependent synaptic plasticity in the ARC remains unknown. Although some AMPK targets, such as p21-associated kinase (PAK), are implicated in fasting-induced synaptic plasticity, it is not clear if PAK modulates autophagy in the ARC [156]. While lipophagy is implicated in feeding behavior, identification of additional potentially relevant autophagic targets required for fasting-induced plasticity, such as neurotransmitter receptors or mitochondria, requires further investigation.
b. Hippocampus and spatial memory
Recent reports highlight a role for autophagy in hippocampus-dependent behavioral tasks. Glatigny et al demonstrated that knockdown of the required autophagy proteins Beclin-1, FIP200, and Atg12 in the hippocampus reduced novel object recognition and contextual fear conditioning [136]. This effect was similar when the shRNA was expressed under the control of a neuron-specific promoter or a general promoter suggesting that loss of neuronal autophagy led to these behavioral phenotypes. This group then used pharmacological tools to acutely manipulate autophagy and found that autophagy inhibition in the hippocampus, using Spautin-1, during but not after, the training phase of the contextual fear conditioning task reduced freezing during a probe trial while stimulation of autophagy during, but not after the training phase, increased freezing during the probe trial. These results suggest that hippocampal autophagy is required for the formation of contextual and object recognition memories.
A decrease in hippocampal autophagy also mediates hippocampal dysfunction in a mouse model of Fragile X syndrome (FXS). Suzanne Zukin and colleagues found reduced autophagy in hippocampal neurons lacking Fmr1, a model for FXS [135]. Reduction of mTOR signaling in hippocampal region CA1 rescued novel object recognition in FXS mice and this was dependent on the required autophagy protein Atg7. Thus, at least a subset of behavioral phenotypes in FXS mice arise from a deficit in hippocampal autophagy.
V. Speculation and future directions
1. Basal vs induced autophagy
One key remaining question in the study of neuronal autophagy is the distinction between ongoing basal autophagy and autophagy induced by extrinsic factors including cellular stress or synaptic input. Deletion of the required autophagy proteins Atg5 and Atg7 throughout the CNS leads to widespread changes in behavior, formation of cellular inclusions and eventual neurodegeneration [17,18]. These reports, along with the observation of relatively high basal levels of autophagosome biogenesis in neuronal cultures, supports a model in which ongoing autophagic activity is required for normal neuronal homeostasis and survival in the CNS [99,106]. This hypothesis seems particularly attractive because neurons are post-mitotic and unable to dilute toxic or damaged proteins and organelles by cell division, and may contribute to why autophagic dysfunction is found in many neurodegenerative diseases [157]. In contrast, however, recent findings that autophagic degradation can be induced on rapid (minute) timescales [131] and by neuronal activity [15] suggest that autophagy may in some cases provide a permissive or downstream role in the control of neuronal plasticity.
Defining the relative importance of basal autophagy versus activity-dependent inducible autophagy for neuronal function remains an important open issue. Furthermore, whether the targets of autophagic degradation are altered during induced autophagy, or whether the rate of autophagy alone changes is unknown. These questions are presently challenging to address, as cell-type specific manipulation of autophagic activity generally necessitates genetic approaches that occur over long timescales and lead to compensatory responses in vesicle trafficking pathways that may obscure the responses to autophagy disruption. Furthermore, while rapid pharmacological manipulation of autophagic responses can be accomplished with newly developed compounds, many of these target kinases upstream of autophagy, such as Vps34 and ULK1, that have pleiotropic effects on cellular pathways. The advent of temporally specific, genetically encoded regulators of autophagic activity will more easily permit dissection of basal versus inducible autophagy in the CNS.
2. Substrate specificity
While early studies of autophagy were often interpreted to demonstrate that cytosolic macromolecules were non-selectively sequestered into autophagic vacuoles [54–56], more recent work has found that specific proteins or damaged organelles can be selectively degraded by autophagy [53]. In neurons, this specificity may be particularly important as the requirements for synaptic plasticity are distinct between brain regions and across developmental stage. It seems likely that autophagy may selectively degrade a subset of proteins within particular neuronal populations or during specific developmental stages. Such specificity could be achieved through: 1) selective expression of adapter proteins that only sequester a subset of possible autophagic cargo, 2) regulated post-translational modification of autophagic cargo which directs substrates for autophagy only under specific conditions, 3) a yet-to-be described recycling pathway that removes sequestered cargo from mature autophagosomes prior to fusion with the lysosome. Identifying the list of autophagic substrates in specific cell types or during specific developmental time points could be achieved using either genetic or biochemical approaches. Eliminating autophagy with temporal specificity in distinct cell types would lead to the accumulation of cell-type specific autophagy substrates. Alternatively, new proteomics tools that label autophagosome associated proteins in distinct cell-types could be used to define autophagic cargo biochemically [158]. These approaches will provide insight into the dynamics of autophagic degradation within neurons and how autophagy contributes to synaptic function.
3. Intersection with endolysosomal system
Finally, it is critical to elucidate the relationship between the endosomal system and autophagy within neurons. These two cellular pathways are deeply interconnected in terms of their molecular regulation [79,159,160] and because late endosomes and mature autophagosomes fuse prior to fusion with lysosomes and cargo degradation [161–167]. Interestingly, early steps in both autophagy and endocytosis are both regulated by Vps34 [159], endophilin [132] and synaptojanin [133], suggesting that these shared regulators may coordinate a balance between these two pathways. Second, Atg9, a transmembrane protein involved in early autophagosome biogenesis, is moreover present on endocytic vesicles that fuse with nascent autophagic membranes, suggesting that endosomes potentially act as a membrane source to growing autophagosomes [40,42,52]. Later steps in the maturation of both endosomes and autophagosomes depend on the same proteins including Rab7 [160,168,169] and the same proteins have been found to be degraded by the endolysosomal system and autophagy (see [15]).
The connection between autophagy and endocytosis presents experimental and conceptual challenges. First, many experimental perturbations that affect autophagy may either also directly disrupt endocytosis, such as PI3K inhibition [30–32], or cause compensatory changes in the endolysosomal pathway and be responsible for the observed phenotypes. Combined pharmacological and genetic manipulations, genetic complementation and convergent phenotypes from distinct manipulations of each pathway would strengthen conclusions that one pathway is directly involved in the process of interest.
Second, implicating autophagy per se in the degradation of membrane proteins is difficult. As a key class of autophagy substrates that can be degraded by autophagy to control neurotransmission [15,16], identifying the precise mechanism of their degradation by autophagy represents a key conceptual question. For example, while Shehata et al demonstrated that GluR1 degradation during chemical long-term depression (chem-LTD) depends on expression of the required autophagy protein Atg7 [15]. GluR1 degradation during chem-LTD depends on endocytosis and endolysosomal trafficking/degradation [15,170–172]. Whether GluR1 truly enters bona fide autophagosomes or whether Atg7 plays a distinct role in the endolysosomal degradation of GluR1 during chem-LTD remains unknown. This participation of both pathways is a strong possibility considering that endosomal fusion with autophagosomes to form amphisomes may provide a key final step in the degradation of endosomal contents. In contrast, GABAA receptor degradation at the NMJ in C. elegans depends on autophagy and these receptors are observed in LC3-positive vesicles [16]. Key experiments defining the localization putative autophagic substrates would enhance our understanding of the mechanisms of autophagy at synapses.
VI. Conclusion
Here, we have reviewed the accumulating evidence that autophagic protein degradation plays an important role in both disease-associated and non-pathogenic states within the CNS and contributes to synaptic transmission, plasticity and behavior. The reports that autophagy contributes to presynaptic homeostasis and postsynaptic function suggest that neurons have coopted an evolutionarily conserved stress response pathway to meet its needs in both synaptic compartments. We look forward to a deeper understanding of the mechanism through which autophagy acts at the synapse and contributes to synaptic function that we are sure will soon emerge.
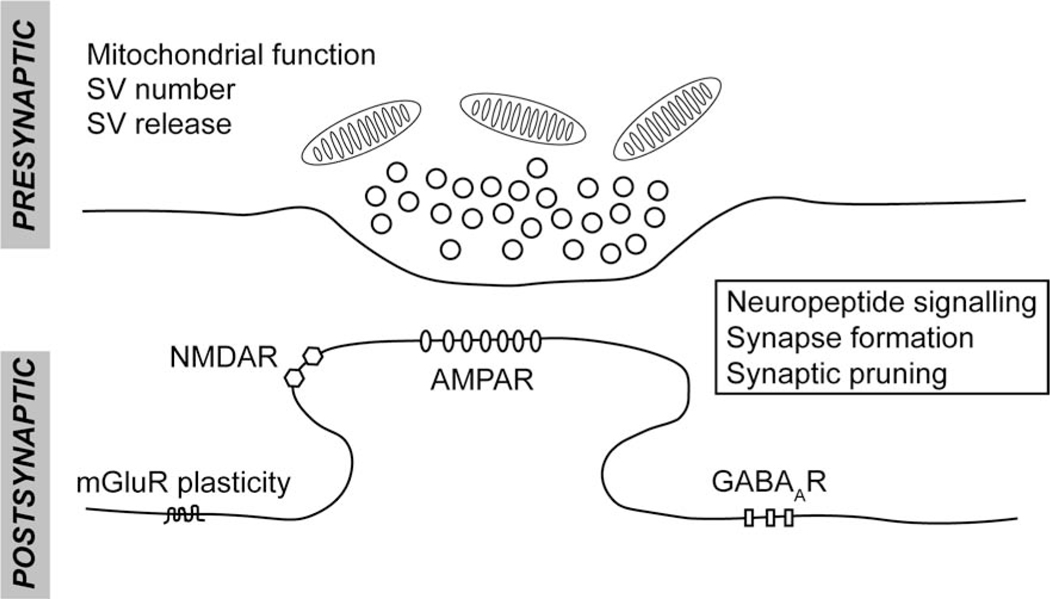
Figure 2. Aspect of neurotransmission that are regulated by autophagy.
Autophagy can control neurotransmission both pre- and postsynaptically. In the presynapse, autophagy controls synaptic vesicle (SV) homeostasis and release as well as mitochondrial function. Postsynaptically, autophagy controls excitatory neurotransmission by degrading AMPA receptors and GABAA receptors. The absence of autophagy disrupts synaptic plasticity that is dependent on metabotropic glutamate receptors. Finally, autophagy modulates neuropeptide signaling, synapse formation and synaptic pruning; the locus of action is unknown in these cases.
Highlights.
The molecular machinery of autophagy is evolutionarily conserved from yeast to mammals.
Neuronal autophagy occurs in the pre- and post-synaptic compartments.
Autophagy controls neurotransmitter release, receptor trafficking, and synaptic plasticity.
Neurons have coopted a ubiquitous cellular stress response pathway to regulate neurotransmission.
Acknowledgements
We would like to thank Dr. Ai Yamamoto for insightful discussions. Work on autophagy in the Sulzer lab is supported by the JPB and Simons Foundations (Grant number 514813). OJL is supported by NIMH F30 MH114390.
Footnotes
Declaration of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- [1].Davis HP, Squire LR. Protein synthesis and memory: a review. Psychol Bull 1984;96:518–59. doi: 10.1037/0033-2909.96.3.518. [DOI] [PubMed] [Google Scholar]
- [2].Alvarez-Castelao B, Schuman EM. The regulation of synaptic protein turnover. J Biol Chem 2015;290:28623–30. doi: 10.1074/jbc.R115.657130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dikic I. Proteasomal and autophagic degradation systems. Annu Rev Biochem 2017;86:193–224. doi: 10.1146/annurev-biochem-061516-044908. [DOI] [PubMed] [Google Scholar]
- [4].De Duve C. The lysosome. Sci Am 1963;208:64–72. doi: 10.1038/scientificamerican0563-64. [DOI] [PubMed] [Google Scholar]
- [5].Sabatini DD, Adesnik M. Christian de Duve: Explorer of the cell who discovered new organelles by using a centrifuge. Proc Natl Acad Sci USA 2013;110:13234–5. doi: 10.1073/pnas.1312084110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Klumperman J, Raposo G. The complex ultrastructure of the endolysosomal system. Cold Spring Harb Perspect Biol 2014;6:a016857. doi: 10.1101/cshperspect.a016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Huotari J, Helenius A. Endosome maturation. EMBO J 2011;30:3481–500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cuervo AM, Terlecky SR, Dice JF, Knecht E. Selective binding and uptake of ribonuclease A and glyceraldehyde-3-phosphate dehydrogenase by isolated rat liver lysosomes. J Biol Chem 1994;269:26374–80. [PubMed] [Google Scholar]
- [9].Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science 1996;273:501–3. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- [10].Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 2004;305:1292–5. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- [11].Caballero B, Wang Y, Diaz A, Tasset I, Juste YR, Stiller B, et al. Interplay of pathogenic forms of human tau with different autophagic pathways. Aging Cell 2018;17. doi: 10.1111/acel.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell 2011;20:131–9. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Uytterhoeven V, Lauwers E, Maes I, Miskiewicz K, Melo MN, Swerts J, et al. Hsc70–4 Deforms Membranes to Promote Synaptic Protein Turnover by Endosomal Microautophagy. Neuron 2015;88:735–48. doi: 10.1016/j.neuron.2015.10.012. [DOI] [PubMed] [Google Scholar]
- [14].Mukherjee A, Patel B, Koga H, Cuervo AM, Jenny A. Selective endosomal microautophagy is starvation-inducible in Drosophila. Autophagy 2016;12:1984–99. doi: 10.1080/15548627.2016.1208887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shehata M, Matsumura H, Okubo-Suzuki R, Ohkawa N, Inokuchi K. Neuronal stimulation induces autophagy in hippocampal neurons that is involved in AMPA receptor degradation after chemical long-term depression. J Neurosci 2012;32:10413–22. doi: 10.1523/JNEUROSCI.4533-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rowland AM, Richmond JE, Olsen JG, Hall DH, Bamber BA. Presynaptic terminals independently regulate synaptic clustering and autophagy of GABAA receptors in Caenorhabditis elegans. J Neurosci 2006;26:1711–20. doi: 10.1523/JNEUROSCI.2279-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- [18].Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- [19].Ohsumi Y. Historical landmarks of autophagy research. Cell Res 2014;24:9–23. doi: 10.1038/cr.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 2007;8:931–7. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- [21].Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem 1995;270:2320–6. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- [22].Meley D, Bauvy C, Houben-Weerts JHPM, Dubbelhuis PF, Helmond MTJ, Codogno P, et al. AMP-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem 2006;281:34870–9. doi: 10.1074/jbc.M605488200. [DOI] [PubMed] [Google Scholar]
- [23].Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011;13:132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chan EYW, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem 2007;282:25464–74. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- [25].Ganley IG, Lam DH, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 2009;284:12297–305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jung CH, Jun CB, Ro S-H, Kim Y-M, Otto NM, Cao J, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 2009;20:1981–91. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lee E-J, Tournier C. The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy. Autophagy 2011;7:689–95. doi: 10.4161/auto.7.7.15450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Russell RC, Tian Y, Yuan H, Park HW, Chang Y-Y, Kim J, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 2013;15:741–50. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3’-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem 2000;275:992–8. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- [31].Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, et al. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol 1999;1:249–52. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- [32].Vieira OV, Bucci C, Harrison RE, Trimble WS, Lanzetti L, Gruenberg J, et al. Modulation of Rab5 and Rab7 recruitment to phagosomes by phosphatidylinositol 3-kinase. Mol Cell Biol 2003;23:2501–14. doi: 10.1128/mcb.23.7.2501-2514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol 2010;190:511–21. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol 2009;11:1433–7. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- [36].Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen E-L. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 2009;5:1180–5. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- [37].Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol 2010;12:747–57. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 2010;141:656–67. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Puri C, Vicinanza M, Ashkenazi A, Gratian MJ, Zhang Q, Bento CF, et al. The RAB11A-Positive Compartment Is a Primary Platform for Autophagosome Assembly Mediated by WIPI2 Recognition of PI3P-RAB11A. Dev Cell 2018;45:114–131.e8. doi: 10.1016/j.devcel.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 2013;154:1285–99. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol 2012;197:659–75. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM, et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell 2012;23:1860–73. doi: 10.1091/mbc.E11-09-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Proikas-Cezanne T, Waddell S, Gaugel A, Frickey T, Lupas A, Nordheim A. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene 2004;23:9314–25. doi: 10.1038/sj.onc.1208331. [DOI] [PubMed] [Google Scholar]
- [44].Gaugel A, Bakula D, Hoffmann A, Proikas-Cezanne T. Defining regulatory and phosphoinositide-binding sites in the human WIPI-1 β-propeller responsible for autophagosomal membrane localization downstream of mTORC1 inhibition. J Mol Signal 2012;7:16. doi: 10.1186/1750-2187-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Dooley HC, Razi M, Polson HEJ, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell 2014;55:238–52. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, et al. A protein conjugation system essential for autophagy. Nature 1998;395:395–8. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- [47].Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci 2003;116:1679–88. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- [48].Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 2000;151:263–76. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 2007;282:37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- [50].Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 2008;19:2092–100. doi: 10.1091/mbc.e07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol 2011;12:226. doi: 10.1186/gb-2011-12-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol 2012;198:219–33. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zaffagnini G, Martens S. Mechanisms of selective autophagy. J Mol Biol 2016;428:1714–24. doi: 10.1016/j.jmb.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Mortimore GE, Schworer CM. Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature 1977;270:174–6. doi: 10.1038/270174a0. [DOI] [PubMed] [Google Scholar]
- [55].Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol 1992;119:301–11. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kopitz J, Kisen GO, Gordon PB, Bohley P, Seglen PO. Nonselective autophagy of cytosolic enzymes by isolated rat hepatocytes. J Cell Biol 1990;111:941–53. doi: 10.1083/jcb.111.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Pankiv S, Clausen TH, Lamark T, Brech A, Bruun J-A, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- [58].Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kirkin V, Lamark T, Sou Y-S, Bjørkøy G, Nunn JL, Bruun J-A, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 2009;33:505–16. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- [60].Tumbarello DA, Waxse BJ, Arden SD, Bright NA, Kendrick-Jones J, Buss F. Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nat Cell Biol 2012;14:1024–35. doi: 10.1038/ncb2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 2011;333:228–33. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015;524:309–14. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mostowy S, Sancho-Shimizu V, Hamon MA, Simeone R, Brosch R, Johansen T, et al. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J Biol Chem 2011;286:26987–95. doi: 10.1074/jbc.M111.223610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Thurston TLM, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 2009;10:1215–21. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- [65].Heo J-M, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Mol Cell 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Birgisdottir ÅB, Lamark T, Johansen T. The LIR motif - crucial for selective autophagy. J Cell Sci 2013;126:3237–47. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- [67].Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T, et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol 2010;12:781–90. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
- [68].Zhang Y, Wang F, Han L, Wu Y, Li S, Yang X, et al. GABARAPL1 negatively regulates Wnt/β-catenin signaling by mediating Dvl2 degradation through the autophagy pathway. Cell Physiol Biochem 2011;27:503–12. doi: 10.1159/000329952. [DOI] [PubMed] [Google Scholar]
- [69].Khaminets A, Behl C, Dikic I. Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol 2016;26:6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- [70].Kimura S, Noda T, Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct 2008;33:109–22. [DOI] [PubMed] [Google Scholar]
- [71].Aplin A, Jasionowski T, Tuttle DL, Lenk SE, Dunn WA. Cytoskeletal elements are required for the formation and maturation of autophagic vacuoles. J Cell Physiol 1992;152:458–66. doi: 10.1002/jcp.1041520304. [DOI] [PubMed] [Google Scholar]
- [72].Köchl R, Hu XW, Chan EYW, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic 2006;7:129–45. doi: 10.1111/j.1600-0854.2005.00368.x. [DOI] [PubMed] [Google Scholar]
- [73].Wijdeven RH, Janssen H, Nahidiazar L, Janssen L, Jalink K, Berlin I, et al. Cholesterol and ORP1L-mediated ER contact sites control autophagosome transport and fusion with the endocytic pathway. Nat Commun 2016;7:11808. doi: 10.1038/ncomms11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, et al. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol 2001;11:1680–5. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- [75].Lee J-Y, Koga H, Kawaguchi Y, Tang W, Wong E, Gao Y-S, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J 2010;29:969–80. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 2012;151:1256–69. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- [77].Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T, et al. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell 2014;25:1327–37. doi: 10.1091/mbc.E13-08-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell 2015;57:39–54. doi: 10.1016/j.molcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- [79].Liang C, Lee J, Inn K, Gack MU, Li Q, Roberts EA, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol 2008;10:776–87. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Farías GG, Guardia CM, De Pace R, Britt DJ, Bonifacino JS. BORC/kinesin-1 ensemble drives polarized transport of lysosomes into the axon. Proc Natl Acad Sci USA 2017;114:E2955–64. doi: 10.1073/pnas.1616363114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Rosa-Ferreira C, Sweeney ST, Munro S. The small G protein Arl8 contributes to lysosomal function and long-range axonal transport in Drosophila. Biol Open 2018;7. doi: 10.1242/bio.035964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Garg S, Sharma M, Ung C, Tuli A, Barral DC, Hava DL, et al. Lysosomal trafficking, antigen presentation, and microbial killing are controlled by the Arf-like GTPase Arl8b. Immunity 2011;35:182–93. doi: 10.1016/j.immuni.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Lund VK, Madsen KL, Kjaerulff O. Drosophila Rab2 controls endosome-lysosome fusion and LAMP delivery to late endosomes. Autophagy 2018;14:1520–42. doi: 10.1080/15548627.2018.1458170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Nakae I, Fujino T, Kobayashi T, Sasaki A, Kikko Y, Fukuyama M, et al. The arf-like GTPase Arl8 mediates delivery of endocytosed macromolecules to lysosomes in Caenorhabditis elegans. Mol Biol Cell 2010;21:2434–42. doi: 10.1091/mbc.E09-12-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Sulzer D, Mosharov E, Talloczy Z, Zucca FA, Simon JD, Zecca L. Neuronal pigmented autophagic vacuoles: lipofuscin, neuromelanin, and ceroid as macroautophagic responses during aging and disease. J Neurochem 2008;106:24–36. doi: 10.1111/j.1471-4159.2008.05385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Roizin L, Stellar S, Willson N, Whittier J, Liu JC. Electron microscope and enzyme studies in cerebral biopsies of Huntington’s chorea. Trans Am Neurol Assoc 1974;99:240–3. [PubMed] [Google Scholar]
- [87].Tellez-Nagel I, Johnson AB, Terry RD. Studies on brain biopsies of patients with Huntington’s chorea. J Neuropathol Exp Neurol 1974;33:308–32. doi: 10.1097/00005072-197404000-00008. [DOI] [PubMed] [Google Scholar]
- [88].Kegel KB, Kim M, Sapp E, McIntyre C, Castaño JG, Aronin N, et al. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci 2000;20:7268–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Petersén A, Larsen KE, Behr GG, Romero N, Przedborski S, Brundin P, et al. Expanded CAG repeats in exon 1 of the Huntington’s disease gene stimulate dopamine-mediated striatal neuron autophagy and degeneration. Hum Mol Genet 2001;10:1243–54. doi: 10.1093/hmg/10.12.1243. [DOI] [PubMed] [Google Scholar]
- [90].Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci 2010;13:567–76. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Sulzer D, Bogulavsky J, Larsen KE, Behr G, Karatekin E, Kleinman MH, et al. Neuromelanin biosynthesis is driven by excess cytosolic catecholamines not accumulated by synaptic vesicles. Proc Natl Acad Sci USA 2000;97:11869–74. doi: 10.1073/pnas.97.22.11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Larsen KE, Sulzer D. Autophagy in neurons: a review. Histol Histopathol 2002;17:897–908. doi: 10.14670/HH-17.897. [DOI] [PubMed] [Google Scholar]
- [93].Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, et al. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol 2005;64:113–22. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- [94].Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci 2008;28:6926–37. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Cataldo AM, Hamilton DJ, Barnett JL, Paskevich PA, Nixon RA. Properties of the endosomal-lysosomal system in the human central nervous system: disturbances mark most neurons in populations at risk to degenerate in Alzheimer’s disease. J Neurosci 1996;16:186–99. doi: 10.1523/JNEUROSCI.16-01-00186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Yue Z, Friedman L, Komatsu M, Tanaka K. The cellular pathways of neuronal autophagy and their implication in neurodegenerative diseases. Biochim Biophys Acta 2009;1793:1496–507. doi: 10.1016/j.bbamcr.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Rubinsztein DC, DiFiglia M, Heintz N, Nixon RA, Qin Z-H, Ravikumar B, et al. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy 2005;1:11–22. doi: 10.4161/auto.1.1.1513. [DOI] [PubMed] [Google Scholar]
- [98].Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Hollenbeck PJ. Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport. J Cell Biol 1993;121:305–15. doi: 10.1083/jcb.121.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 2004;15:1101–11. doi: 10.1091/mbc.e03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Kaushik S, Rodriguez-Navarro JA, Arias E, Kiffin R, Sahu S, Schwartz GJ, et al. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab 2011;14:173–83. doi: 10.1016/j.cmet.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Nikoletopoulou V, Sidiropoulou K, Kallergi E, Dalezios Y, Tavernarakis N. Modulation of autophagy by BDNF underlies synaptic plasticity. Cell Metab 2017;26:230–242.e5. doi: 10.1016/j.cmet.2017.06.005. [DOI] [PubMed] [Google Scholar]
- [103].Andermann ML, Lowell BB. Toward a wiring diagram understanding of appetite control. Neuron 2017;95:757–78. doi: 10.1016/j.neuron.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Maday S, Wallace KE, Holzbaur ELF. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol 2012;196:407–17. doi: 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Maday S, Holzbaur ELF. Compartment-Specific Regulation of Autophagy in Primary Neurons. J Neurosci 2016;36:5933–45. doi: 10.1523/JNEUROSCI.4401-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Maday S, Holzbaur ELF. Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev Cell 2014;30:71–85. doi: 10.1016/j.devcel.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Stavoe AKH, Hill SE, Hall DH, Colón-Ramos DA. KIF1A/UNC-104 Transports ATG-9 to Regulate Neurodevelopment and Autophagy at Synapses. Dev Cell 2016;38:171–85. doi: 10.1016/j.devcel.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Kuma A, Komatsu M, Mizushima N. Autophagy-monitoring and autophagy-deficient mice. Autophagy 2017;13:1619–28. doi: 10.1080/15548627.2017.1343770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- [111].Lieberman OJ, McGuirt AF, Tang G, Sulzer D. Roles for neuronal and glial autophagy in synaptic pruning during development. Neurobiol Dis 2019;122:49–63. doi: 10.1016/j.nbd.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Winckler B, Faundez V, Maday S, Cai Q, Guimas Almeida C, Zhang H. The endolysosomal system and proteostasis: from development to degeneration. J Neurosci 2018;38:9364–74. doi: 10.1523/JNEUROSCI.1665-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Gally C, Bessereau J-L. GABA is dispensable for the formation of junctional GABA receptor clusters in Caenorhabditis elegans. J Neurosci 2003;23:2591–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Kittler JT, Rostaing P, Schiavo G, Fritschy JM, Olsen R, Triller A, et al. The subcellular distribution of GABARAP and its ability to interact with NSF suggest a role for this protein in the intracellular transport of GABA(A) receptors. Mol Cell Neurosci 2001;18:13–25. doi: 10.1006/mcne.2001.1005. [DOI] [PubMed] [Google Scholar]
- [115].Wang H, Bedford FK, Brandon NJ, Moss SJ, Olsen RW. GABA(A)-receptor-associated protein links GABA(A) receptors and the cytoskeleton. Nature 1999;397:69–72. doi: 10.1038/16264. [DOI] [PubMed] [Google Scholar]
- [116].Shen W, Ganetzky B. Autophagy promotes synapse development in Drosophila. J Cell Biol 2009;187:71–9. doi: 10.1083/jcb.200907109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Tang G, Gudsnuk K, Kuo S-H, Cotrina ML, Rosoklija G, Sosunov A, et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 2014;83:1131–43. doi: 10.1016/j.neuron.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Dragich JM, Kuwajima T, Hirose-Ikeda M, Yoon MS, Eenjes E, Bosco JR, et al. Autophagy linked FYVE (Alfy/WDFY3) is required for establishing neuronal connectivity in the mammalian brain. Elife 2016;5. doi: 10.7554/eLife.14810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].De Pace R, Skirzewski M, Damme M, Mattera R, Mercurio J, Foster AM, et al. Altered distribution of ATG9A and accumulation of axonal aggregates in neurons from a mouse model of AP-4 deficiency syndrome. PLoS Genet 2018;14:e1007363. doi: 10.1371/journal.pgen.1007363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Yamaguchi J, Suzuki C, Nanao T, Kakuta S, Ozawa K, Tanida I, et al. Atg9a deficiency causes axon-specific lesions including neuronal circuit dysgenesis. Autophagy 2018;14:764–77. doi: 10.1080/15548627.2017.1314897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Hofmann I, Munro S. An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J Cell Sci 2006;119:1494–503. doi: 10.1242/jcs.02958. [DOI] [PubMed] [Google Scholar]
- [122].Bagshaw RD, Callahan JW, Mahuran DJ. The Arf-family protein, Arl8b, is involved in the spatial distribution of lysosomes. Biochem Biophys Res Commun 2006;344:1186–91. doi: 10.1016/j.bbrc.2006.03.221. [DOI] [PubMed] [Google Scholar]
- [123].Vukoja A, Rey U, Petzoldt AG, Ott C, Vollweiter D, Quentin C, et al. Presynaptic Biogenesis Requires Axonal Transport of Lysosome-Related Vesicles. Neuron 2018;99:1216–1232.e7. doi: 10.1016/j.neuron.2018.08.004. [DOI] [PubMed] [Google Scholar]
- [124].Niwa S, Tao L, Lu SY, Liew GM, Feng W, Nachury MV, et al. BORC Regulates the Axonal Transport of Synaptic Vesicle Precursors by Activating ARL-8. Curr Biol 2017;27:2569–2578.e4. doi: 10.1016/j.cub.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Klassen MP, Wu YE, Maeder CI, Nakae I, Cueva JG, Lehrman EK, et al. An Arf-like small G protein, ARL-8, promotes the axonal transport of presynaptic cargoes by suppressing vesicle aggregation. Neuron 2010;66:710–23. doi: 10.1016/j.neuron.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 2015;522:359–62. doi: 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- [127].Li Z, Okamoto K-I, Hayashi Y, Sheng M. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell 2004;119:873–87. doi: 10.1016/j.cell.2004.11.003. [DOI] [PubMed] [Google Scholar]
- [128].Hernandez D, Torres CA, Setlik W, Cebrián C, Mosharov EV, Tang G, et al. Regulation of presynaptic neurotransmission by macroautophagy. Neuron 2012;74:277–84. doi: 10.1016/j.neuron.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci 2004;27:509–47. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- [130].Takemoto K, Matsuda T, Sakai N, Fu D, Noda M, Uchiyama S, et al. SuperNova, a monomeric photosensitizing fluorescent protein for chromophore-assisted light inactivation. Sci Rep 2013;3:2629. doi: 10.1038/srep02629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Hoffmann S, Orlando M, Andrzejak E, Bruns C, Trimbuch T, Rosenmund C, et al. Light-Activated ROS Production Induces Synaptic Autophagy. J Neurosci 2019;39:2163–83. doi: 10.1523/JNEUROSCI.1317-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Soukup S-F, Kuenen S, Vanhauwaert R, Manetsberger J, Hernández-Díaz S, Swerts J, et al. A LRRK2-Dependent EndophilinA Phosphoswitch Is Critical for Macroautophagy at Presynaptic Terminals. Neuron 2016;92:829–44. doi: 10.1016/j.neuron.2016.09.037. [DOI] [PubMed] [Google Scholar]
- [133].Vanhauwaert R, Kuenen S, Masius R, Bademosi A, Manetsberger J, Schoovaerts N, et al. The SAC1 domain in synaptojanin is required for autophagosome maturation at presynaptic terminals. EMBO J 2017;36:1392–411. doi: 10.15252/embj.201695773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Okerlund ND, Schneider K, Leal-Ortiz S, Montenegro-Venegas C, Kim SA, Garner LC, et al. Bassoon Controls Presynaptic Autophagy through Atg5. Neuron 2018;97:727. doi: 10.1016/j.neuron.2018.01.010. [DOI] [PubMed] [Google Scholar]
- [135].Yan J, Porch MW, Court-Vazquez B, Bennett MVL, Zukin RS. Activation of autophagy rescues synaptic and cognitive deficits in fragile X mice. Proc Natl Acad Sci USA 2018;115:E9707–16. doi: 10.1073/pnas.1808247115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Glatigny M, Moriceau S, Rivagorda M, Ramos-Brossier M, Nascimbeni AC, Lante F, et al. Autophagy Is Required for Memory Formation and Reverses Age-Related Memory Decline. Curr Biol 2019;29:435–448.e8. doi: 10.1016/j.cub.2018.12.021. [DOI] [PubMed] [Google Scholar]
- [137].Horwood JM, Dufour F, Laroche S, Davis S. Signalling mechanisms mediated by the phosphoinositide 3-kinase/Akt cascade in synaptic plasticity and memory in the rat. Eur J Neurosci 2006;23:3375–84. doi: 10.1111/j.1460-9568.2006.04859.x. [DOI] [PubMed] [Google Scholar]
- [138].Jaworski J, Sheng M. The growing role of mTOR in neuronal development and plasticity. Mol Neurobiol 2006;34:205–19. doi: 10.1385/MN:34:3:205. [DOI] [PubMed] [Google Scholar]
- [139].Fenselau H, Campbell JN, Verstegen AMJ, Madara JC, Xu J, Shah BP, et al. A rapidly acting glutamatergic ARC→PVH satiety circuit postsynaptically regulated by α-MSH. Nat Neurosci 2017;20:42–51. doi: 10.1038/nn.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Aponte Y, Atasoy D, Sternson SM. AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nat Neurosci 2011;14:351–5. doi: 10.1038/nn.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Zhan C, Zhou J, Feng Q, Zhang J-E, Lin S, Bao J, et al. Acute and long-term suppression of feeding behavior by POMC neurons in the brainstem and hypothalamus, respectively. J Neurosci 2013;33:3624–32. doi: 10.1523/JNEUROSCI.2742-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Krashes MJ, Koda S, Ye C, Rogan SC, Adams AC, Cusher DS, et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J Clin Invest 2011;121:1424–8. doi: 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Dubé D, Lissitzky JC, Leclerc R, Pelletier G. Localization of alpha-melanocyte-stimulating hormone in rat brain and pituitary. Endocrinology 1978;102:1283–91. doi: 10.1210/endo-102-4-1283. [DOI] [PubMed] [Google Scholar]
- [144].Atasoy D, Betley JN, Su HH, Sternson SM. Deconstruction of a neural circuit for hunger. Nature 2012;488:172–7. doi: 10.1038/nature11270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Ghamari-Langroudi M, Digby GJ, Sebag JA, Millhauser GL, Palomino R, Matthews R, et al. G-protein-independent coupling of MC4R to Kir7.1 in hypothalamic neurons. Nature 2015;520:94–8. doi: 10.1038/nature14051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Liu H, Kishi T, Roseberry AG, Cai X, Lee CE, Montez JM, et al. Transgenic mice expressing green fluorescent protein under the control of the melanocortin-4 receptor promoter. J Neurosci 2003;23:7143–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Krashes MJ, Shah BP, Koda S, Lowell BB. Rapid versus delayed stimulation of feeding by the endogenously released AgRP neuron mediators GABA, NPY, and AgRP. Cell Metab 2013;18:588–95. doi: 10.1016/j.cmet.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Minokoshi Y, Alquier T, Furukawa N, Kim Y-B, Lee A, Xue B, et al. AMP-kinase regulates food intake by responding to hormonal and nutrient signals in the hypothalamus. Nature 2004;428:569–74. doi: 10.1038/nature02440. [DOI] [PubMed] [Google Scholar]
- [149].Cota D, Proulx K, Smith KAB, Kozma SC, Thomas G, Woods SC, et al. Hypothalamic mTOR signaling regulates food intake. Science 2006;312:927–30. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- [150].Kohno D, Sone H, Minokoshi Y, Yada T. Ghrelin raises [Ca2+]i via AMPK in hypothalamic arcuate nucleus NPY neurons. Biochem Biophys Res Commun 2008;366:388–92. doi: 10.1016/j.bbrc.2007.11.166. [DOI] [PubMed] [Google Scholar]
- [151].Kohno D, Sone H, Tanaka S, Kurita H, Gantulga D, Yada T. AMP-activated protein kinase activates neuropeptide Y neurons in the hypothalamic arcuate nucleus to increase food intake in rats. Neurosci Lett 2011;499:194–8. doi: 10.1016/j.neulet.2011.05.060. [DOI] [PubMed] [Google Scholar]
- [152].Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature 2009;458:1131–5. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Coupé B, Ishii Y, Dietrich MO, Komatsu M, Horvath TL, Bouret SG. Loss of autophagy in pro-opiomelanocortin neurons perturbs axon growth and causes metabolic dysregulation. Cell Metab 2012;15:247–55. doi: 10.1016/j.cmet.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Quan W, Kim H-K, Moon E-Y, Kim SS, Choi CS, Komatsu M, et al. Role of hypothalamic proopiomelanocortin neuron autophagy in the control of appetite and leptin response. Endocrinology 2012;153:1817–26. doi: 10.1210/en.2011-1882. [DOI] [PubMed] [Google Scholar]
- [155].Aveleira CA, Botelho M, Carmo-Silva S, Pascoal JF, Ferreira-Marques M, Nóbrega C, et al. Neuropeptide Y stimulates autophagy in hypothalamic neurons. Proc Natl Acad Sci USA 2015;112:E1642–51. doi: 10.1073/pnas.1416609112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Kong D, Dagon Y, Campbell JN, Guo Y, Yang Z, Yi X, et al. A Postsynaptic AMPK→p21-Activated Kinase Pathway Drives Fasting-Induced Synaptic Plasticity in AgRP Neurons. Neuron 2016;91:25–33. doi: 10.1016/j.neuron.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Yamamoto A, Yue Z. Autophagy and its normal and pathogenic states in the brain. Annu Rev Neurosci 2014;37:55–78. doi: 10.1146/annurev-neuro-071013-014149. [DOI] [PubMed] [Google Scholar]
- [158].Le Guerroué F, Eck F, Jung J, Starzetz T, Mittelbronn M, Kaulich M, et al. Autophagosomal Content Profiling Reveals an LC3C-Dependent Piecemeal Mitophagy Pathway. Mol Cell 2017;68:786–796.e6. doi: 10.1016/j.molcel.2017.10.029. [DOI] [PubMed] [Google Scholar]
- [159].Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 2008;19:5360–72. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Feng Y, Press B, Wandinger-Ness A. Rab 7: an important regulator of late endocytic membrane traffic. J Cell Biol 1995;131:1435–52. doi: 10.1083/jcb.131.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Eskelinen E-L. Maturation of autophagic vacuoles in Mammalian cells. Autophagy 2005;1:1–10. doi: 10.4161/auto.1.1.1270. [DOI] [PubMed] [Google Scholar]
- [162].Sanchez-Wandelmer J, Reggiori F. Amphisomes: out of the autophagosome shadow? EMBO J 2013;32:3116–8. doi: 10.1038/emboj.2013.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Berg TO, Fengsrud M, Strømhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem 1998;273:21883–92. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- [164].Liou W, Geuze HJ, Geelen MJ, Slot JW. The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J Cell Biol 1997;136:61–70. doi: 10.1083/jcb.136.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Punnonen EL, Autio S, Kaija H, Reunanen H. Autophagic vacuoles fuse with the prelysosomal compartment in cultured rat fibroblasts. Eur J Cell Biol 1993;61:54–66. [PubMed] [Google Scholar]
- [166].Lucocq J, Walker D. Evidence for fusion between multilamellar endosomes and autophagosomes in HeLa cells. Eur J Cell Biol 1997;72:307–13. [PubMed] [Google Scholar]
- [167].Tooze J, Hollinshead M, Ludwig T, Howell K, Hoflack B, Kern H. In exocrine pancreas, the basolateral endocytic pathway converges with the autophagic pathway immediately after the early endosome. J Cell Biol 1990;111:329–45. doi: 10.1083/jcb.111.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci 2004;117:4837–48. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- [169].Gutierrez MG, Munafó DB, Berón W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci 2004;117:2687–97. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- [170].Man HY, Lin JW, Ju WH, Ahmadian G, Liu L, Becker LE, et al. Regulation of AMPA receptor-mediated synaptic transmission by clathrin-dependent receptor internalization. Neuron 2000;25:649–62. doi: 10.1016/S0896-6273(00)81067-3. [DOI] [PubMed] [Google Scholar]
- [171].Widagdo J, Chai YJ, Ridder MC, Chau YQ, Johnson RC, Sah P, et al. Activity-Dependent Ubiquitination of GluA1 and GluA2 Regulates AMPA Receptor Intracellular Sorting and Degradation. Cell Rep 2015;10:783–95. doi: 10.1016/j.celrep.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Ehlers MD. Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron 2000;28:511–25. doi: 10.1016/s0896-6273(00)00129-x. [DOI] [PubMed] [Google Scholar]