Abstract
Regarding the urgency of therapeutic measures for coronavirus disease 2019 (COVID-19) pandemic, the use of available drugs with FDA approval is preferred because of the less time and cost required for their development. In silico drug repurposing is an accurate way to speed up the screening of the existing FDA-approved drugs to find a therapeutic option for COVID-19. The similarity in SARS-CoV-2 and HIV-1 fusion mechanism to host cells can be a key point for Inhibit SARS-CoV-2 entry into host cells by HIV fusion inhibitors. Accordingly, in this study, an HIV-1 fusion inhibitor called Enfuvirtide (Enf) was selected. The affinity and essential residues involving in the Enf binding to the S2 protein of SARS-CoV-2, HIV-1 gp41 protein and angiotensin-converting enzyme 2 (ACE-2) as a negative control, was evaluated using molecular docking. Eventually, Enf-S2 and Enf-gp41 protein complexes were simulated by molecular dynamics (MD) in terms of binding affinity and stability. Based on the most important criteria such as docking score, cluster size, energy and dissociation constant, the strongest interaction was observed between Enf with the S2 protein. In addition, MD results confirmed that Enf-S2 protein interaction was remarkably stable and caused the S2 protein residues to undergo the fewest fluctuations. In conclusion, it can be stated that Enf can act as a strong SARS-CoV-2 fusion inhibitor and demonstrates the potential to enter the clinical trial phase of COVID-19.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2, COVID-19, Enfuvirtide, molecular docking, molecular dynamics, in silico drug repurposing
1. Introduction
The outbreak of coronavirus disease 2019 (COVID-19) which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was started from the Wuhan (China) (Arshad Ali et al., 2020; Li & Liu, 2020). Afterward, the terrible disease rapidly spread all over the world so that by December 25, 2020, more than 79.4 million people have been confirmed to be infected with SARS-CoV-2 and more than 1,740,000 deaths due to COVID-19 have been recorded worldwide (Chen, 2020; Li & Liu, 2020). On 11th March 2020, WHO declared COVID-19 as a pandemic due to its high contagion and mortality rate (Cucinotta & Vanelli, 2020). No approved vaccine or effective drug has been reported for COVID-19 yet (Bhagavathula et al., 2020). Regarding the urgency of therapeutic measures for COVID-19, the use of available drugs with FDA approval is preferred because they need to the less time and cost for the evaluation and development (Durojaiye et al., 2020). Computer-aided drug repurposing represents an accurate strategy to speed up the screening of existing drugs with FDA approval to find the suitable treatment options for COVID-19 (Ciliberto & Cardone, 2020; Wang, 2020). Similar to SARS-CoV, SARS-CoV-2 is an enveloped positive-strand RNA virus (Romano et al., 2020). The cell surface receptor of both viruses is angiotensin-converting enzyme 2 (ACE-2), however the apparent affinity of SARS-CoV-2 to ACE-2 is 10 to 20 times higher than that of SARS-COV (Fani et al., 2020). The genetics alignments showed that the SARS-CoV-2 genome had 88–89% similarity to two other bats-derived SARS-like coronaviruses namely bat-SL-CoVZC45 and bat-SL-CoVXC21. Also, 82 and 50% of the SARS-CoV-2 genome is similar to SARS-CoV and MERS-CoV genomes, respectively (Lai et al., 2020; Morse et al., 2020). Studies revealed that one-third of SARS-CoV-2 genome encoded structural proteins including the glycosylated spike (S), the envelope protein (E), the membrane protein (M), and the nucleoprotein (N) (Li et al., 2020). The virus typically enters the cell by binding of the S protein to its receptor, ACE-2 (Hoffmann et al., 2020). The S protein contains two subunits namely S1 and S2. The S1 subunit contains a receptor-binding domain (RBD) which is responsible for binding to ACE-2, while the S2 subunit forms a fusion core and causes fusion of the viral membrane and the host cell membrane (Hoffmann et al., 2020; Song et al., 2019). Therefore, the S2 subunit represents a proper target for drug design to inhibit virus entry into the host cells (Prajapat et al., 2020; Shang et al., 2020; Wu et al., 2020; Yi et al., 2004). As the RBD of the S1 subunit binds to the ACE-2 receptor on the host cell surface, the heptad repeats 1 and 2 (HR1 and HR2) trimeric domains of the S2 subunit interact with each other and form a six-helix bundle (6-HB) fusion core that is responsible for the virus fusion to the host cells membrane (Shang et al., 2020; Xia et al., 2020a, 2020b). A similar mechanism happens for human immunodeficiency virus-1 (HIV-1) entrance to the host cells (Belouzard et al., 2012; Guillén et al., 2005). The glycoprotein gp120 in HIV-1 has an equivalent role with the S1 subunit in SARS-CoV-2 and causes the binding of HIV-1 to its main receptor, CD4, and its specific co-receptors namely CXCR4 and CCR5. Following this binding, a structural change occurs in the HIV-1 virus gp41 protein, which is equivalent to the S2 subunit of the SARS-CoV-2 virus. This structural change allows the fusion peptide (FP) of the gp41 protein inserts to the target cell membrane and forms the pre-hairpin fusion intermediate N-terminal heptad repeat (NHR)-trimer (Belouzard et al., 2012; Guillén et al., 2005; Pan et al., 2010). The NHR-trimer structure contains a pocket-forming domain (PFD), a heptad repeat (HR) sequence, and a glycineisoleucine-valine (GIV) motif (Pan et al., 2010). In the other groove of this trimer structure, a deep envelope with highly conserved and hydrophobic residues is formed by PFD, which plays a significant role in the stability of the helix bundle (6-HB) structure and the entrance of the virus into the cells (Lu et al., 2015; Pan et al., 2010). Subsequently, three specific molecules of the C-terminal-heptad region (CHR) of the gp41 protein are placed in anti-parallel and diagonally configuration inside the conserved hydrophobic pocket on the surface of NHR, leading to the formation of the 6H core of gp41 protein and occurrence of the successful fusion of the virus and the host cells (Lu et al., 2016; Nelson et al., 2008; Pan et al., 2010). The 6H core structure of SARS-CoV-2, which is formed by the interaction of HR1 and HR2 trimers of the S2 subunit, is similar to that of the HIV-1 virus (Belouzard et al., 2012; Guillén et al., 2005; Kawase et al., 2019; Xia et al., 2020a). Therefore, interfering drugs with each of the 6H core components can inhibit the entry of SARS-CoV-2 into the host cell (Xia et al., 2020a).
Enfuvirtide (Enf) or T20 (Fuzeon) is a fusion inhibitor peptide with 36 residues that inhibits the fusion of HIV-1 and the host cell membrane (Eggink et al., 2019; Zhang et al., 2018). This drug inhibits gp41 structural change and consequently fusion of the virus to the host cell membrane by binding to HR1 from the pre-hairpin fusion intermediate NHR-trimer (Eggink et al., 2019; He, 2013; Pang et al., 2009; A Yi et al., 2016; Zhang et al., 2018). Given the remarkable similarity of the cell fusion mechanism of SARS-CoV-2 and HIV-1 and also the similarities between structures and even the amino acid contents of the 6H core of these two viruses, the question arose as to whether the approved Enf drug could be used to inhibit the SARS-CoV-2 virus fusion into the host cell membrane? To answer this question, regarding the structural similarity of S2 protein of SARS-CoV-2 and gp41 of HIV-1, as well as the mechanism of action of approved drugs, Enf was selected to be studied. The affinity and the state of binding as well as the essential residues involving in the binding of Enf and the S2 protein of SARS-CoV-2 was analyzed using molecular docking. In order to compare the affinity of Enf-S2 protein in the molecular docking step, HIV-1 gp41 and ACE-2 were employed as positive and negative controls, respectively. Finally, Enf-S2 and Enf-gp41 protein complexes were simulated by molecular dynamics (MD) in terms of binding affinity and stability of the drug.
2. Material and methods
2.1. Receptor and ligand preparation and identifying the binding sites
The three-dimensional structures of S2 protein of SARS-CoV-2 (PDB ID: 6lxt), ACE2 receptor (PDB ID: 1R4L), and gp41 of HIV (PDB ID: 1F23) were obtained from the protein data bank (PDB, https://www.rcsb.org/) (Berman et al., 2000). Enfuvirtide (DB00109) sequence was obtained from Drug bank (https://www.drugbank.ca/). The structures were cleaned so that crystallographic water molecules and other co-crystallized molecules were removed and only the monomer forms of each protein were kept. Subsequently, the structures of the four receptors and the drug candidate were prepared by the Dock prep tool in UCSF Chimera software (Pettersen et al., 2004). As identifying the binding site of receptors is a crucial step in the drug target finding and functional studies, the binding sites of all three receptors were determined based on the previous studies (Chan et al., 1997; Towler et al., 2004; Xia et al., 2020a). Multiple sequence alignments of the HR1 of S2 protein and HR2 domain of gp41 protein was performed using CLC Genomics Workbench software (CLC bio; Qiagen) with default parameters (https://digitalinsights.qiagen.com). In the following, the three-dimensional structure of Enf as the ligand was predicted by PEP-FOLD3 server (https://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD3/) (Lamiable et al., 2016).
2.2. Molecular docking
The HADDOCK (High Ambiguity Driven protein-protein DOCKing) and ClusPro web servers were used to validate the peptide-protein interaction modes. HADDOCK server is a flexible docking approach and provides complete structural flexibility for both side chains and backbone (de Vries et al., 2010). The binding affinity and the dissociation constant (Kd) of the peptide-protein complexes were predicted using PRODIGY (PROtein binDIng enerGY prediction) web server (Kurkcuoglu et al., 2018). The docking results were screened based on the Kd scores and the top cluster with the lowest intermolecular energy was selected. Next, the peptide-protein complexes were analyzed by Ligplot software (Wallace et al., 1995) which identifies the hydrophobic and hydrogen interactions within the complex.
2.3. MD simulation
To perform the MD simulation for Enf-S2 and Enf-gp41 complexes, GROMOS 53a6 (Gromacs 5.1 package) was applied (Van Der Spoel et al., 2005). The complexes were solvated by a water model of transferable intermolecular potential with 3 points (TIP3P) in a cubic box with a distance of 10 angstrom from the furthermost atom of the protein (Jorgensen et al., 1983). Subsequent to the solvation, Cl− and Na+ ions were added to neutralize the system. Next, NaCl at a concentration of 150 mM was added to the systems (Batoulis et al., 2016; Reis et al., 2014) and the energy minimization was done by applying the steepest descent method. Each system was equilibrated by 1 ns simulation in the both isothermal–isobaric (NPT) and canonical (NVT) ensembles applying position restraints on the protein heavy atoms to allow the solvent equilibration. In order to fix the system temperature at 310 K, the Nose–Hoover thermostat was used. The Parrinello–Rahman pressure coupling method was applied for maintaining the pressure of system at a fixed 1 bar pressure (Akya et al., 2019). The electrostatic interactions were calculated using the Particle Mesh Ewald (PME) approach with 1.0 nm short-range van der Waals and electrostatic cutoffs (Farasat et al., 2017; Ochoa et al., 2018). Finally, for each peptide-protein complex, the process of 100 ns simulation was carried out with time steps of 2 fs on the equilibrated systems.
2.4. PMF (potential of mean force)
The free energy pattern which is frequently cited as PMF along a special physical reaction coordinate can be obtained by the umbrella sampling (US) method. Further structural insights can be achieved by using this physical reaction coordinate (Naughton et al., 2018). In the present study, the binding energy of Enf-S2 and Enf-gp41 complexes were estimated from PMF through the US method. At first, the simulations were performed to drive the peptide far away from S2 and gp41 proteins which were stable during the process of MD simulation. Next, 50 configurations were created on the z axis coordinate. In each configuration, the z coordinates of center of mass (COM) interval among Enf and the proteins differed by 0.5 Å with the force constant of 10 kcal/mol Å. For each window, the process of equilibration was performed in a period of 10 ns. Furthermore, a production run of 10 ns was extended for sampling (Gheibi et al., 2019; Lemkul & Bevan, 2010). Finally, the PMF patterns were supplied through the method of Weighted Histogram Analysis (WHAM) which was done by GROMACS as the ‘g_wham’ command (Zeng et al., 2016). Moreover, for the better recognition of the MD process, the root mean square deviation (RMSD), the root mean square fluctuation (RMSF), the electrostatic and van der Waals energies (Mudedla et al., 2015), and the SASA (solvent accessible surface area) of each system were analyzed by GROMACS available tools during the MD simulation. The final PDB files of the MD simulations were plotted using Pymol software.
2.5. Molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) method
The molecular model affinities such as ligand-protein and protein-protein interactions are widely evaluated by the MMPBSA method (Wu et al., 2016). The MM-PBSA calculation was carried out on the last 10 ns trajectories via g-mmpbsa tool in the GROMACS software (Kumari et al., 2014). The binding-free energy calculation was done by the MM-PBSA as follows:
3. Results
3.1. Comparison of key residues of binding sites of S2 and gp41 proteins
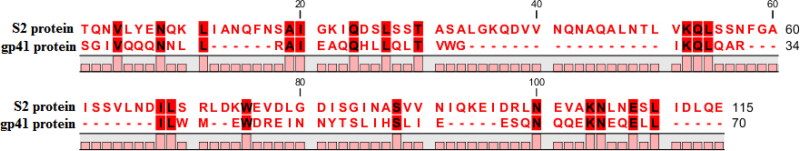
Multiple sequence alignments of the HR1 and HR2 domains of S2 protein (PDB ID: 6lxt) using the CLC Workbench showed 28.57% identity to gp41 protein (PDB ID: 1F23). The results also showed that most of the gp41 binding site-located essential residues responsible for interact with the Enf were also conserved in the S2 binding site (Figure 1) (Chan et al., 1997; Xia et al., 2020a).
Figure 1.
The multiple sequence alignments of the HR1 and HR2 domains of S2 and gp41 proteins using CLC Workbench. The conserved residues between the binding site of S2 and gp41 proteins are marked in red highlight.
3.2. Analysis of interactions between drug and different protein receptors and evaluation of their binding affinity
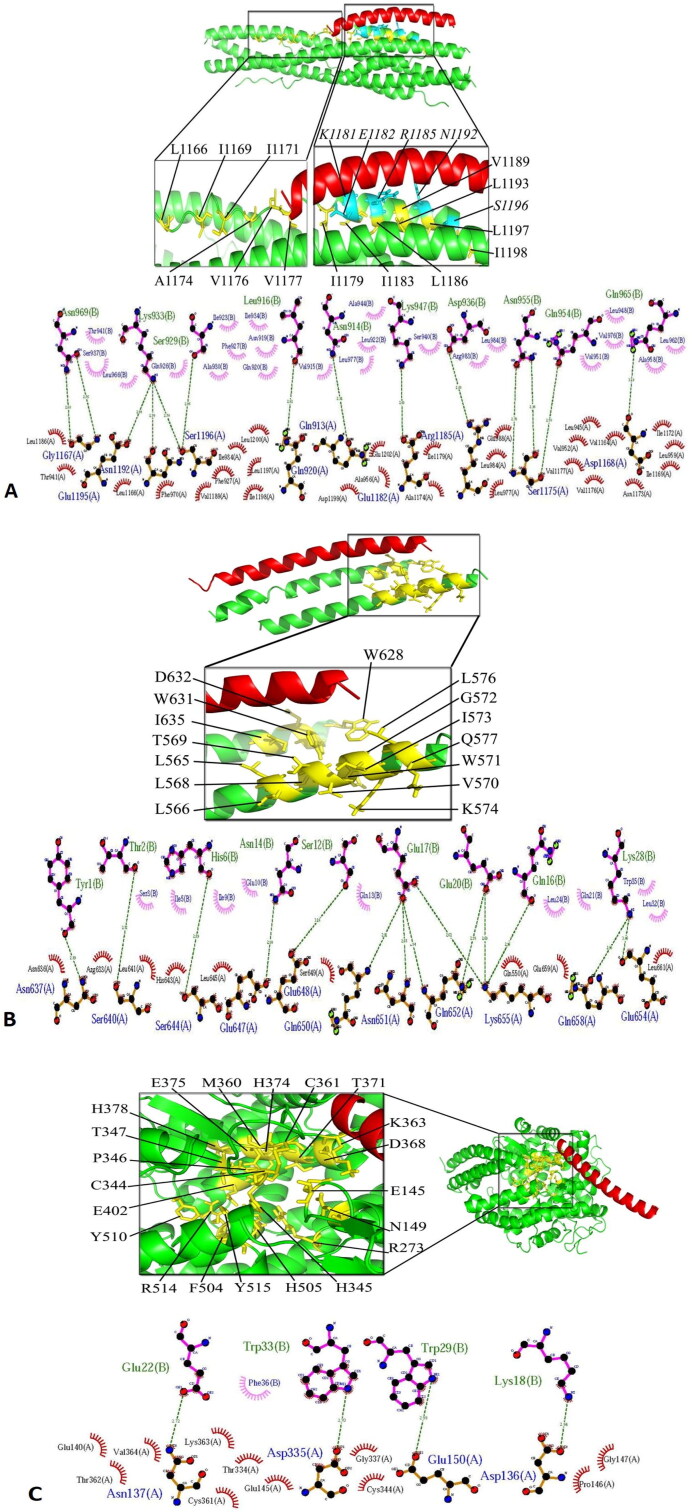
The highest cluster size, the most negative global energy (kcal/mol), the lowest PRODIGY (Kd) score, and the highest number of residues involved in the hydrogen and hydrophobic intermolecular bindings were considered as the key criteria for choosing the most potent complexes. The most significant interactions between Enf and key residues of the binding sites of HIV-1 gp41, S2 protein of SARS-CoV-2 and the ACE-2 receptor are shown in Figure 2 and Table 1. The best docking score belonged to the Enf-S2 protein complex which formed the most interaction bonds between Enf and the critical amino acids of the S2 protein (Figure 2 and Table 1).
Figure 2.
(A) The molecular docking of Enf-S2 protein complex. In the close-up view, the residues involved in the binding site of the S2 protein are demonstrated in yellow while the residues essential for HR1-HR2 interaction are shown in blue. (B) The molecular docking of Enf-gp41 protein complex. The binding site of gp41 protein is presented in yellow. (C) The molecular docking of Enf-ACE2 receptor complex. The binding site of the ACE2 receptor is indicated in yellow. In all parts, the S2, gp41 and ACE2 proteins are shown in green while Enf is shown in red.
Table 1.
Ligplot analyses of the docking result of Enf interaction with the selected proteins.
| Proteins | Cluster size | Energy (kcal/mol) | Hydrophobic interaction | Hydrogen interaction | PRODIGY (Kd) |
|---|---|---|---|---|---|
| gp41 | 36 | −93 | Asn636, Arg633, Leu641, His643, Leu645, Ser649, Gln550, Glu659, Leu661 | Asn637, Ser640, Ser644, Glu647, Glu648, Gln650, Asn651, Gln652, Lys655, Gln658, Glu654 | 3.7E − 06 |
| S2 | 48 | −95 | Leu1166a, Phe970, Thr941, Leu 1186, Val 1189, Leu 1197, Ile 1198, Phe 927, Ile 934, Leu 1200, Glu1202, Asp1199, Leu 977, Ile1179, Glu988, Leu 984, Ala 956, Ala1174, Val1176, Val 1177, Leu 945, Val 1164, Ile 1172, Asn 1173, Ile 1169, Leu 959 | Gly1167, Glu1195, Asn1192, Ser1196, Gln 920, Gln 913, Glu1182, Arg 1185, Ser1175, Asp1168 | 1.6E − 10 |
| ACE2 | 13 | −64 | Val298, Val364, Gly377, Pro336, Asp335, Thr362, Pro146, Glu145, Thr334, Cys344, Lys363, Thr365 | Asn137, Asp335, Glu150 and Asp136 | 1.5E − 6 |
3.3. MD simulation and PMF analysis
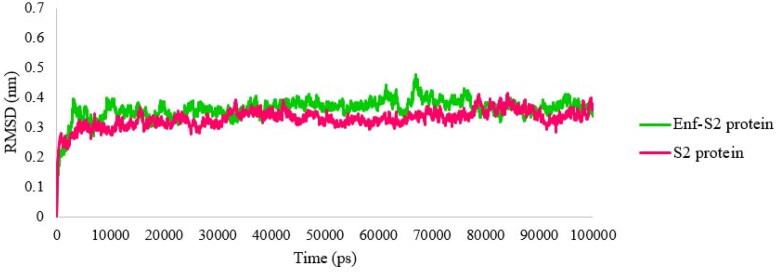
RMSD is an essential parameter that predicts system equilibration during the MD simulation (Kufareva & Abagyan, 2011; Kumar et al., 2019; Shen et al., 2012). The RMSD profiles were evaluated during 100 ns of the MD simulation to assess the stability of the complexes. As shown in Figure 3, both Enf-S2 protein complex and S2 protein alone reached stability after 3 ns. The average values of the last 5 ns of the simulation were 0.36 nm and 0.34 nm for the Enf-S2 protein complex and the S2 protein alone, respectively.
Figure 3.
The root mean square deviation (RMSD) values of Cα of the S2 protein-Enf complex and the S2 protein alone.
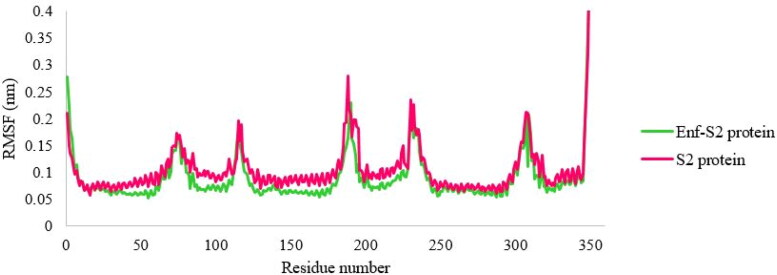
RMSF comprises a valuable parameter which allows for a more proper understanding of protein flexibility and structural fluctuations (Chen et al., 2016; Mahapatra et al., 2018). RMSF was calculated to evaluate the flexibility of the S2 protein in the presence or absence of the peptide (Enf). As shown in Figure 4, the least flexibility and fluctuation occurred in most residues of S2 protein in the presence of the peptide.
Figure 4.
The RMSF fluctuation values of the S2 protein-Enf complex and the S2 protein alone.
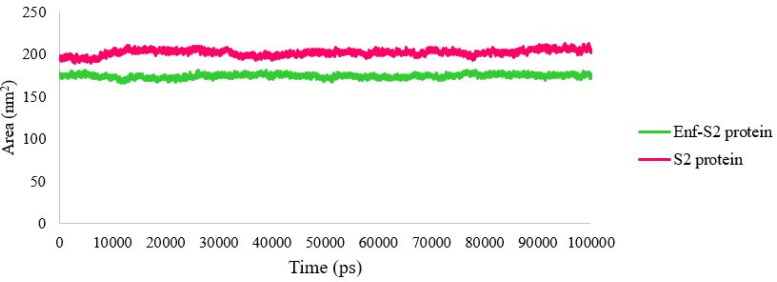
SASA represents a parameter that shows the accessibility level of molecules against solvents, hence any change in this parameter is an indicator of the interaction level (Mitternacht, 2016). The SASA profile of S2 protein in the presence and absence of Enf is shown in Figure 5. The accessible level of the S2 protein was decreased in the presence of Enf indicating that Enf interacts with the S2 protein and undergoes a structural change of the protein (Figure 5).
Figure 5.
The SASA values of the S2 protein-Enf complex and the S2 protein alone.
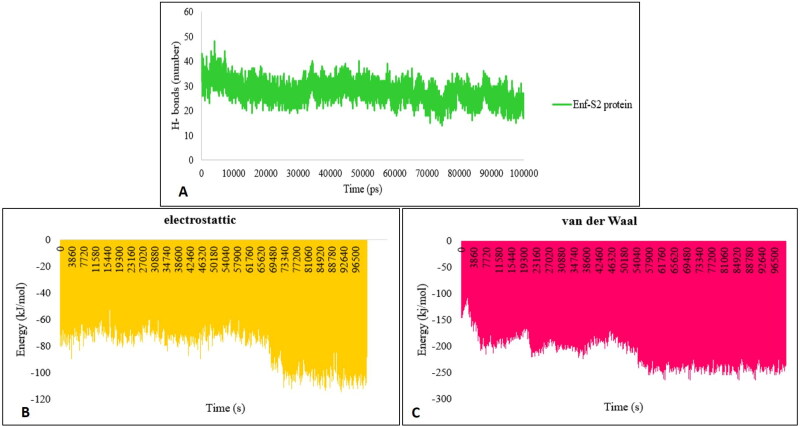
The formation of the hydrogen bonds (H-bonds) in the protein complexes plays an important role in their stability (Mallamace et al., 2018). The formation of H-bonds in the Enf-S2 protein complex was evaluated during the MD simulation. The average number of H-bonds between S2 protein and Enf in the last 5 ns of the MD simulation was 22 (Figure 6A). The electrostatic and van der Waals energies were estimated to evaluate the major interactions involved in Enf-S2 formation by the MD simulation method. The results indicated that the electrostatic and van der Waals energies were strong enough to keep the S2 protein and Enf peptide in contact with each other (Figure 6B and 6C).
Figure 6.
(A) The number of H-bonds between S2 protein and Enf. (B) Analysis of the electrostatic and (C) van der Waals energies.
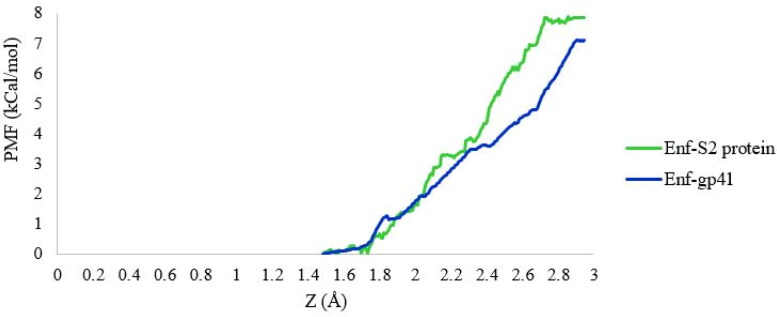
Umbrella sampling (US) is a method that is widely used to calculate binding free energy and explore the dissociation process of ligand-receptor systems (Bowman & Lindert, 2018; You et al., 2019). In the present study, to determine the binding free energy between S2 and gp41 proteins with Enf peptide, the PMF were calculated by applying the US method. As shown in Figure 7, the binding free energies between S2 and gp41 proteins with Enf peptide were 7.9 kcal/mol and 7.1 kcal/mol, respectively. Also, MMPBSA binding energy calculations of Enf and S2/gp41 complexes showed the affinity of −182.497 and −179.259 kJ/mol (−43.617 and −42.843 kcal/mol), respectively.
Figure 7.
The binding free energy profile of the S2/gp41 proteins dissociated from the S2 protein-Enf and the gp41-Enf complexes.
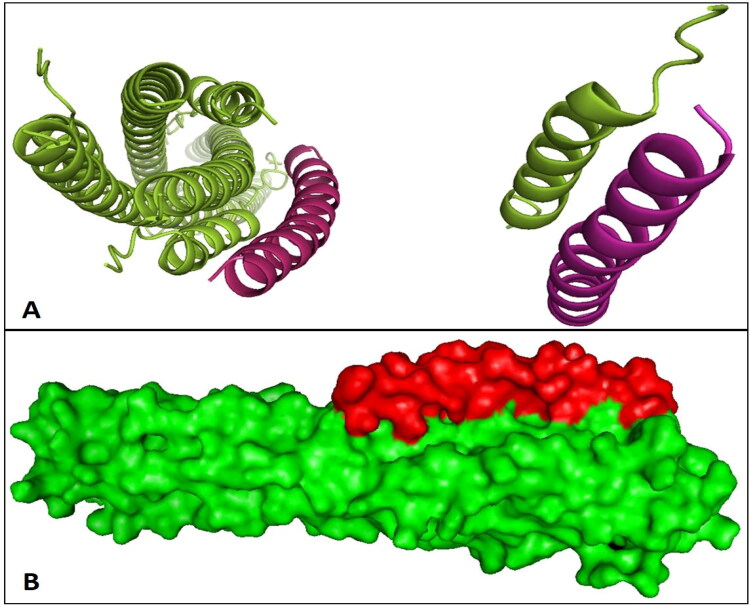
The three-dimensional (3D) structure of the S2 protein-Enf complex after 100 ns of MD simulation and the location of Enf in the complex is shown in Figure 8.
Figure 8.
The interactions of the S2 protein and Enf created by Pymol software. (A) The cartoon 3D image of S2 protein-Enf complex. (B) The surface structure of S2 protein-Enf complex. The 3D images indicated that Enf binds tightly to the S2 protein and can fill the cavities, as proved by the binding energies. The S2 protein and Enf are shown in green and red, respectively.
4. Discussion
In this study, because of the similarity in fusion mechanisms of HIV-1 and SARS-CoV-2 and the structure similarity of gp41 and S2 proteins, Enf, an HIV-1 fusion inhibitor, was selected. ACE2 was selected as a negative control to determine whether Enf could specifically bind to the S2 protein or not. HADDOCK-2 server and ClusPro web servers were used for this purpose. HADDOCK-2 server is a powerful docking tool to analyze interactions of protein-protein, protein-peptide, protein-nucleic acid and protein-ligand complexes based on the information-driven approach (Kurkcuoglu et al., 2018). ClusPro web server is used for protein-protein and protein-peptide docking that including DOT or ZDOCK algorithms for performing rigid-body docking based on the fast Fourier transform (FFT) correlation method (Kozakov et al., 2017; Vajda et al., 2017).
HR1 domain of the S2 protein contains residues 912 to 984 while its HR2 domain is composed of residues 1163 to 1213 (Xia et al., 2020a). The binding site of the HR2 domain includes Val1164, Leu1166, Ile1169, Ile1172, Ala1174, Val1176, Val1177, Ile1179, Ile1183, Leu1186, Val1189, Leu1193, Leu1197, Ile1198, Ser1196, Glu1182, Arg1185, and Asn1192 residues. Also, residues Ser1196, Glu1182, Arg1185, Asn1192, which are essential for the interaction between HR1 and HR2, are present in the S2 binding site (Xia et al., 2020a). The binding site of gp41 protein is composed of Ile635, Trp631, Trp628, Asp632, Thr569, Ile573, Leu576, Leu566, Leu565, Val570, Lys574, Gln577, Leu568, Trp571, and Gly572 (Chan et al., 1997). Active site of ACE2 receptor includes Glu145, Cys344, His345, Cys361, Asp368, His374, Glu375, His378, Glu402, Phe504, His505, Arg514, Tyr515, Asn149, Arg273, Pro346, Thr347, Met360, Lys363, Thr371, and Tyr510 (Towler et al., 2004). The multiple sequence alignment results showed that the essential residues of the gp41 binding site for interaction with Enf peptide are also conserved in the S2 protein binding site. It should be noted that these two proteins (S2 and gp41) are not the same in terms of length, so the total percentage of identity or similarity is low. However, in the same similarity percentage, the binding site-located essential residues responsible for Enf binding are very similar in the gp41 and S2 proteins. According to the Ligplot analyzes and above data on the amino acid contents of the binding sites, Enf had the most interaction with essential residues in the binding site of S2 protein of SARS-CoV-2. Enf binds to the HR1 domain of gp41 and inhibits the HIV-1 fusion to the target cell membrane but our result predicted that Enf interacts to both HR1 (residues Gln 913, Gln 920, Phe 927, Ile 934, Thr941, Leu 945, Ala 956, Leu 959, Phe970, Leu 977, and Leu 984) and HR2 domain (residues Val 1164, Leu1166, Gly1167, Asp1168, Ile 1169, Ile 1172, Asn 1173, Ala1174, Ser1175, Val1176, Val 1177, Ile1179, Glu1182, Arg 1185, Leu 1186, Val 1189, Asn 1192, Glu1195, Ser1196, Leu 1197, Ile 1198, Asp1199, Leu 1200, and Glu1202) of S2 protein.
Kd value for Enf-S2 and Enf-gp41 complexes was 1.6E − 10 and 3.7E − 06, respectively. The less Kd value indicates higher binding affinity. Kd value of Enf-S2 was significantly lower than that of Enf-gp41 complex. However, the binding energy difference between the two complexes was negligible that indicated the stability of both Enf-S2 protein and Enf-gp41 complexes. In addition, PRODIGY (Kd) also correlates with the number of interfacial contacts at the interface of a protein-protein complex and the high effective predictive model based on intermolecular contacts and properties derived from the non-interface surface (Xue et al., 2016). In general, it can be stated that the S2 protein has the best complex in interaction with Enf peptide because it has the highest cluster size (43), the lowest energy (−95), the highest number of residues involved in hydrogen and hydrophobic bonds (26 hydrophobic bonds and 10 hydrogen bonds), and the best PRODIGY (Kd) score (1.6E − 10). In this study, according to the number of hydrophobic and hydrogen interactions of the complexes, the number of interfacial contacts at the Enf-S2 protein complex is more than the Enf-gp41 complex. The Enf-ACE2 showed the weakest docking result and binding affinity. This could be due to poor connection of ACE2 active site in the cavity-like position and the large peptide size.
The results of MD simulation through RMSD, RMSF and SASA parameters showed that the simulated systems reached stability. These parameters sufficiently pointed out that most residues were involved in the S2 protein-Enf interaction, demonstrated the greatest flexibility, and had the principal role in establishing the interaction. In addition, the SASA results indicated that the S2 protein-Enf interaction makes a reduction in the level of surface accessibility of the S2 protein. Furthermore, during the MD simulation, the number of hydrogen bonds was constant which shows the system stability during the simulation and supports the aforementioned results.
The PMF, which shows the thermodynamic properties of the system during the simulation, exhibited suitable binding free energies for Enf-S2 and Enf-gp41 complexes. The MM-PBSA is a method that is currently used to estimate the binding free energy (Verma et al., 2016). In addition, MM-PBSA binding energy calculations showed that S2 protein had the lowest binding free energy (−182.497 kJ/mol) and higher binding affinity in interaction with Enf peptide compared with Enf-gp41 complex (−179.259 kJ/mol), suggesting a more stable ligand conformation. In both the PMF and MMPBSA analyzes, the Enf-S2 complex showed a more stable interaction than the Enf-gp41 complex. The RMSF values indicated that the structures were remarkably stable in their natural function, and the structural analyses showed that the peptide could fill the cavities and bind tightly to the S2 protein.
Similar to our results, the results of Kliger et al. study in 2003 suggested that Enf can bind to the C-terminal HR of the S2 protein may serve as inhibitors for SARS-CoV entry (Kliger & Levanon, 2003). In 2005, Veiga et al by investigating of biophysical features showed a significant interaction between a SARS-CoV HR1-derived peptide and Enf (Veiga et al., 2006). In their study, Enf (T20) established a stronger binding to a SARS-CoV S protein-derived peptide called Pep1D (ENQKQIANQFNKAISQIQESLT) than T-1249, another HIV-1 fusion inhibitor peptide (Veiga et al., 2006). The study showed that Enf could inhibit the fusion of SARS-CoV with the target cells but its effect is not strong enough for the therapeutic application (Veiga et al., 2006). However, with a few limited in vitro tests, the effectiveness of this drug cannot be ruled out with certainty and requires further studies in more advanced areas such as inhibiting virus entry into target cells and other in vitro and in vivo studies and clinical trials.
In another study by Calligari et al., Enf was studied as an anti-SARS-CoV-2 drug and their results showed that this drug could be a suitable candidate for the treatment of SARS-CoV-2. They used Autodock Vina to identify the best binding situation between Umifenovir, Enfuvirtide, and Pleconaril drugs with spike protein of SARS-CoV-2. Their study showed that Enf can interact with the spike protein (Vina score: −5.9 kcal/mol) (Calligari et al., 2020). Autodock Vina performs properly for short peptides (up to four residues) because it is a standard docking tool for small-molecule ligands (Ciemny et al., 2018; Li et al., 2014; Rentzsch & Renard, 2015). Here, we used HADDOCK and Cluspro web servers because they are more suitable and accurate for peptide docking (Dominguez et al., 2003; Kozakov et al., 2017; Kurkcuoglu et al., 2018). Although the results of our study confirm their data, the study of Calligari et al. has been limited to perform proper molecular docking.
5. Conclusion
Altogether, in the present in silico study, we examined the amino acids involved in the Enf-S2 protein interaction and compared the binding strength with gp41 HIV and ACE2 receptor. Also, the stability of this interaction was evaluated by MD simulation with various analyzes. The results showed that Enf has a good interaction with the most important residues of the HR2 domain of SARS-CoV-2 S2 protein and can act as a fusion inhibitor of this virus. Therefore, based on our results and present information about the pharmacological properties of Enfuvirtide, we recommend that this FDA approved antiviral drug has the potential to enter the clinical trial phase for the treatment of the COVID-19.
Acknowledgements
We also thank the staff on the Department and Center for Biotechnology Research from Semnan University of Medical Sciences.
Glossary
Abbreviations
- ACE-2
angiotensin-converting enzyme 2
- Enf
Enfuvirtide
- FDA
Food and Drug Administration
- HIV
human immunodeficiency virus
- MD
molecular dynamics
- RBD
receptor binding domain
Funding Statement
This project was approved and supported by the Semnan University of Medical Sciences (grant No. A-10-500-8).
Disclosure statement
The authors confirm that this article content has no conflict of interest.
References
- Akya, A., Farasat, A., Ghadiri, K., & Rostamian, M. (2019). Identification of HLA-I restricted epitopes in six vaccine candidates of Leishmania tropica using immunoinformatics and molecular dynamics simulation approaches. Infection, Genetics and Evolution, 75, 103953. 10.1016/j.meegid.2019.103953 [DOI] [PubMed] [Google Scholar]
- Arshad Ali, S., Baloch, M., Ahmed, N., Arshad Ali, A., & Iqbal, A. (2020). The outbreak of Coronavirus Disease 2019 (COVID-19)-An emerging global health threat. Journal of Infection and Public Health, 13(4), 644–646. 10.1016/j.jiph.2020.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoulis, H., Schmidt, T. H., Weber, P., Schloetel, J.-G., Kandt, C., & Lang, T. (2016). Concentration dependent ion-protein interaction patterns underlying protein oligomerization behaviours. Scientific Reports, 6(1), 24131. 10.1038/srep24131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard, S., Millet, J. K., Licitra, B. N., & Whittaker, G. R. (2012). Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses, 4(6), 1011–1033. 10.3390/v4061011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., & Bourne, P. E. (2000). The Protein Data Bank. Nucleic Acids Research, 28(1), 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagavathula, A. S., Aldhaleei, W. A., Rovetta, A., & Rahmani, J. (2020). Vaccines and drug therapeutics to lock down novel coronavirus disease 2019 (COVID-19): A systematic review of clinical trials. Cureus, 12(5), e8342. 10.7759/cureus.8342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J. D., & Lindert, S. (2018). Molecular dynamics and umbrella sampling simulations elucidate differences in troponin C isoform and mutant hydrophobic patch exposure. The Journal of Physical Chemistry B, 122(32), 7874–7883. 10.1021/acs.jpcb.8b05435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calligari, P., Bobone, S., Ricci, G., & Bocedi, A. (2020). Molecular investigation of SARS–CoV-2 proteins and their interactions with antiviral drugs. Viruses, 12(4), 445. 10.3390/v12040445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, D. C., Fass, D., Berger, J. M., & Kim, P. S. (1997). Core structure of gp41 from the HIV envelope glycoprotein. Cell, 89(2), 263–273. 10.1016/S0092-8674(00)80205-6 [DOI] [PubMed] [Google Scholar]
- Chen, J. (2020). Pathogenicity and transmissibility of 2019-nCoV-A quick overview and comparison with other emerging viruses. Microbes and Infection, 22(2), 69–71. 10.1016/j.micinf.2020.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Wang, J., & Zhu, W. (2016). Molecular mechanism and energy basis of conformational diversity of antibody SPE7 revealed by molecular dynamics simulation and principal component analysis. Scientific Reports, 6(1), 36900. 10.1038/srep36900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciemny, M., Kurcinski, M., Kamel, K., Kolinski, A., Alam, N., Schueler-Furman, O., & Kmiecik, S. (2018). Protein-peptide docking: Opportunities and challenges. Drug Discovery Today, 23(8), 1530–1537. 10.1016/j.drudis.2018.05.006 [DOI] [PubMed] [Google Scholar]
- Ciliberto, G., & Cardone, L. (2020). Boosting the arsenal against COVID-19 through computational drug repurposing. Drug Discovery Today, 25(6), 946–948. 10.1016/j.drudis.2020.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta, D., & Vanelli, M. (2020). WHO declares COVID-19 a pandemic. Acta Bio-Medica: Atenei Parmensis, 91(1), 157–160. 10.23750/abm.v91i1.9397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries, S. J., van Dijk, M., & Bonvin, A. M. J. J. (2010). The HADDOCK web server for data-driven biomolecular docking. Nature Protocols, 5(5), 883–897. 10.1038/nprot.2010.32 [DOI] [PubMed] [Google Scholar]
- Dominguez, C., Boelens, R., & Bonvin, A. M. J. J. (2003). HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. Journal of the American Chemical Society, 125(7), 1731–1737. 10.1021/ja026939x [DOI] [PubMed] [Google Scholar]
- Durojaiye, A. B., Clarke, J.-R. D., Stamatiades, G. A., & Wang, C. (2020). Repurposing cefuroxime for treatment of COVID-19: A scoping review of in silico studies. Journal of Biomolecular Structure & Dynamics, 1–8. 10.1080/07391102.2020.1777904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggink, D., Bontjer, I., de Taeye, S. W., Langedijk, J. P. M., Berkhout, B., & Sanders, R. W. (2019). HIV-1 anchor inhibitors and membrane fusion inhibitors target distinct but overlapping steps in virus entry. The Journal of Biological Chemistry, 294(15), 5736–5746. 10.1074/jbc.RA119.007360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fani, M., Teimoori, A., & Ghafari, S. (2020). Comparison of the COVID-2019 (SARS-CoV-2) pathogenesis with SARS-CoV and MERS-CoV infections. Future Virology, 15(5), 317–323. 10.2217/fvl-2020-0050 [DOI] [Google Scholar]
- Farasat, A., Rahbarizadeh, F., Hosseinzadeh, G., Sajjadi, S., Kamali, M., & Keihan, A. H. (2017). Affinity enhancement of nanobody binding to EGFR: In silico site-directed mutagenesis and molecular dynamics simulation approaches. Journal of Biomolecular Structure & Dynamics, 35(8), 1710–1728. 10.1080/07391102.2016.1192065 [DOI] [PubMed] [Google Scholar]
- Gheibi, N., Ghorbani, M., Shariatifar, H., & Farasat, A. (2019). In silico assessment of human Calprotectin subunits (S100A8/A9) in presence of sodium and calcium ions using Molecular Dynamics simulation approach. PLOS One, 14(10), e0224095. 10.1371/journal.pone.0224095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén, J., Pérez-Berná, A. J., Moreno, M. R., & Villalaín, J. (2005). Identification of the membrane-active regions of the severe acute respiratory syndrome coronavirus spike membrane glycoprotein using a 16/18-mer peptide scan: Implications for the viral fusion mechanism. Journal of Virology, 79(3), 1743–1752. 10.1128/JVI.79.3.1743-1752.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y. (2013). Synthesized peptide inhibitors of HIV-1 gp41-dependent membrane fusion. Current Pharmaceutical Design, 19(10), 1800–1809. 10.2174/1381612811319100004 [DOI] [PubMed] [Google Scholar]
- Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., Schiergens, T. S., Herrler, G., Wu, N.-H., Nitsche, A., Müller, M. A., Drosten, C., & Pöhlmann, S. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(2), 271–280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., & Klein, M. L. (1983). Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics, 79(2), 926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- Kawase, M., Kataoka, M., Shirato, K., & Matsuyama, S. (2019). Biochemical analysis of coronavirus spike glycoprotein conformational intermediates during membrane fusion. Journal of Virology, 93(19), 1-19. 10.1128/JVI.00785-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliger, Y., & Levanon, E. Y. (2003). Cloaked similarity between HIV-1 and SARS-CoV suggests an anti-SARS strategy. BMC Microbiology, 3, 20. 10.1186/1471-2180-3-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov, D., Hall, D. R., Xia, B., Porter, K. A., Padhorny, D., Yueh, C., Beglov, D., & Vajda, S. (2017). The ClusPro web server for protein–protein docking. Nature Protocols, 12(2), 255–278. 10.1038/nprot.2016.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufareva, I., & Abagyan, R. (2011). Methods of protein structure comparison in homology modeling. Methods in Molecular Biology (Methods and Protocols). Humana Press, 857, 231–257. 10.1007/978-1-61779-588-6_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A., Srivastava, G., Negi, A. S., & Sharma, A. (2019). Docking, molecular dynamics, binding energy-MM-PBSA studies of naphthofuran derivatives to identify potential dual inhibitors against BACE-1 and GSK-3β. Journal of Biomolecular Structure & Dynamics, 37(2), 275–290. 10.1080/07391102.2018.1426043 [DOI] [PubMed] [Google Scholar]
- Kumari, R., Kumar, R., & Lynn, A. (2014). g_mmpbsa-a GROMACS tool for high-throughput MM-PBSA calculations. Journal of Chemical Information and Modeling, 54(7), 1951–1962. 10.1021/ci500020m [DOI] [PubMed] [Google Scholar]
- Kurkcuoglu, Z., Koukos, P. I., Citro, N., Trellet, M. E., Rodrigues, J. P. G. L. M., Moreira, I. S., Roel-Touris, J., Melquiond, A. S. J., Geng, C., Schaarschmidt, J., Xue, L. C., Vangone, A., & Bonvin, A. M. J. J. (2018). Performance of HADDOCK and a simple contact-based protein-ligand binding affinity predictor in the D3R Grand Challenge 2. Journal of Computer-Aided Molecular Design, 32(1), 175–185. 10.1007/s10822-017-0049-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, C.-C., Shih, T.-P., Ko, W.-C., Tang, H.-J., & Hsueh, P.-R. (2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. International Journal of Antimicrobial Agents, 55(3), 105924. 10.1016/j.ijantimicag.2020.105924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamiable, A., Thévenet, P., Rey, J., Vavrusa, M., Derreumaux, P., & Tufféry, P. (2016). PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Research, 44(W1), W449–W454. 10.1093/nar/gkw329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemkul, J. A., & Bevan, D. R. (2010). Assessing the stability of Alzheimer's amyloid protofibrils using molecular dynamics. The Journal of Physical Chemistry B, 114(4), 1652–1660. 10.1021/jp9110794 [DOI] [PubMed] [Google Scholar]
- Li, X., Geng, M., Peng, Y., Meng, L., & Lu, S. (2020). Molecular immune pathogenesis and diagnosis of COVID-19. Journal of Pharmaceutical Analysis, 10(2), 102–108. 10.1016/j.jpha.2020.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., & Liu, W. (2020). Puzzle of highly pathogenic human coronaviruses (2019-nCoV). Protein & Cell, 11(4), 235–238. 10.1007/s13238-020-00693-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Liu, X., Dong, X., Zhang, L., & Sun, Y. (2014). Biomimetic design of affinity peptide ligand for capsomere of virus-like particle. Langmuir, 30(28), 8500–8508. 10.1021/la5017438 [DOI] [PubMed] [Google Scholar]
- Lu, L., Yu, F., Cai, L ., Debnath, AK., & Jiang, S. (2016). Development of small-molecule HIV entry inhibitors specifically targeting gp120 or gp41. Current Topics in Medicinal Chemistry, 16(10), 1074–1090. 10.2174/1568026615666150901114527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra, M. K., Bera, K., Singh, D. V., Kumar, R., & Kumar, M. (2018). In silico modelling and molecular dynamics simulation studies of thiazolidine based PTP1B inhibitors. Journal of Biomolecular Structure & Dynamics, 36(5), 1195–1211. 10.1080/07391102.2017.1317026 [DOI] [PubMed] [Google Scholar]
- Mallamace, D., Fazio, E., Mallamace, F., & Corsaro, C. (2018). The role of hydrogen bonding in the folding/unfolding process of hydrated lysozyme: A review of recent NMR and FTIR results. International Journal of Molecular Sciences, 19(12), 3825. 10.3390/ijms19123825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitternacht, S. (2016). FreeSASA: An open source C library for solvent accessible surface area calculations. F1000Research, 5, 189. 10.12688/f1000research.7931.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse, J. S., Lalonde, T., Xu, S., & Liu, W. R. (2020). Learning from the past: Possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem, 21(5), 730–738. 10.1002/cbic.202000047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudedla, S. K., Azhagiya Singam, E. R., Balamurugan, K., & Subramanian, V. (2015). Influence of the size and charge of gold nanoclusters on complexation with siRNA: A molecular dynamics simulation study. Physical Chemistry Chemical Physics, 17(45), 30307–30317. 10.1039/c5cp05034k [DOI] [PubMed] [Google Scholar]
- Naughton, F. B., Kalli, A. C., & Sansom, M. S. P. (2018). Modes of interaction of pleckstrin homology domains with membranes: Toward a computational biochemistry of membrane recognition. Journal of Molecular Biology, 430(3), 372–388. 10.1016/j.jmb.2017.12.011 [DOI] [PubMed] [Google Scholar]
- Nelson, J. D., Kinkead, H., Brunel, F. M., Leaman, D., Jensen, R., Louis, J. M., Maruyama, T., Bewley, C. A., Bowdish, K., Clore, G. M., Dawson, P. E., Frederickson, S., Mage, R. G., Richman, D. D., Burton, D. R., & Zwick, M. B. (2008). Antibody elicited against the gp41 N-heptad repeat (NHR) coiled-coil can neutralize HIV-1 with modest potency but non-neutralizing antibodies also bind to NHR mimetics. Virology, 377(1), 170–183. 10.1016/j.virol.2008.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa, R., Soler, M. A., Laio, A., & Cossio, P. (2018). Assessing the capability of in silico mutation protocols for predicting the finite temperature conformation of amino acids. Physical Chemistry Chemical Physics, 20(40), 25901–25909. 10.1039/c8cp03826k [DOI] [PubMed] [Google Scholar]
- Pan, C., Liu, S., & Jiang, S. (2010). HIV-1 gp41 fusion intermediate: A target for HIV therapeutics. Journal of the Formosan Medical Association = Taiwan yi Zhi, 109(2), 94–105. 10.1016/S0929-6646(10)60029. [DOI] [PubMed] [Google Scholar]
- Pang, W., Tam, S.-C., & Zheng, Y.-T. (2009). Current peptide HIV Type-1 fusion inhibitors. Antiviral Chemistry & Chemotherapy, 20(1), 1–18. 10.3851/IMP1369 [DOI] [PubMed] [Google Scholar]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera-a visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Prajapat, M., Sarma, P., Shekhar, N., Avti, P., Sinha, S., Kaur, H., Kumar, S., Bhattacharyya, A., Kumar, H., Bansal, S., & Medhi, B. (2020). Drug targets for corona virus: A systematic review. Indian Journal of Pharmacology, 52(1), 56–65. 10.4103/ijp.IJP_115_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis, R. A. G., Bortot, L. O., & Caliri, A. (2014). In silico assessment of S100A12 monomer and dimer structural dynamics: Implications for the understanding of its metal-induced conformational changes. Journal of Biological Inorganic Chemistry, 19(7), 1113–1120. 10.1007/s00775-014-1149-y [DOI] [PubMed] [Google Scholar]
- Rentzsch, R., & Renard, B. Y. (2015). Docking small peptides remains a great challenge: An assessment using AutoDock Vina. Briefings in Bioinformatics, 16(6), 1045–1056. 10.1093/bib/bbv008 [DOI] [PubMed] [Google Scholar]
- Romano, M., Ruggiero, A., Squeglia, F., Maga, G., & Berisio, R. (2020). A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells, 9(5), 1267. 10.3390/cells9051267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, J., Wan, Y., Luo, C., Ye, G., Geng, Q., Auerbach, A., & Li, F. (2020). Cell entry mechanisms of SARS-CoV-2. Proceedings of the National Academy of Sciences of the United States of America, 117(21), 11727–11734. 10.1073/pnas.2003138117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, M., Guan, J., Xu, L., Yu, Y., He, J., Jones, G. W., & Song, Y. (2012). Steered molecular dynamics simulations on the binding of the appendant structure and helix-β2 in domain-swapped human cystatin C dimer. Journal of Biomolecular Structure & Dynamics, 30(6), 652–661. 10.1080/07391102.2012.689698 [DOI] [PubMed] [Google Scholar]
- Song, Z., Xu, Y., Bao, L., Zhang, L., Yu, P., Qu, Y., Zhu, H., Zhao, W., Han, Y., & Qin, C. (2019). From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses, 11(1), 59. 10.3390/v11010059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler, P., Staker, B., Prasad, S. G., Menon, S., Tang, J., Parsons, T., Ryan, D., Fisher, M., Williams, D., Dales, N. A., Patane, M. A., & Pantoliano, M. W. (2004). ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. The Journal of Biological Chemistry, 279(17), 17996–18007. 10.1074/jbc.M311191200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajda, S., Yueh, C., Beglov, D., Bohnuud, T., Mottarella, S. E., Xia, B., Hall, D. R., & Kozakov, D. (2017). New additions to the ClusPro server motivated by CAPRI. Proteins, 85(3), 435–444. 10.1002/prot.25219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E., & Berendsen, H. J. C. (2005). GROMACS: Fast, flexible, and free. Journal of Computational Chemistry, 26(16), 1701–1718. 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
- Veiga, S., Yuan, Y., Li, X., Santos, N. C., Liu, G., & Castanho, M. A. R. (2006). Why are HIV-1 fusion inhibitors not effective against SARS-CoV? Biophysical evaluation of molecular interactions. Biochimica et Biophysica Acta, 1760(1), 55–61. 10.1016/j.bbagen.2005.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, S., Grover, S., Tyagi, C., Goyal, S., Jamal, S., Singh, A., & Grover, A. (2016). Hydrophobic interactions are a key to MDM2 inhibition by polyphenols as revealed by molecular dynamics simulations and MM/PBSA free energy calculations. PLOS One, 11(2), e0149014. 10.1371/journal.pone.0149014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, A. C., Laskowski, R. A., & Thornton, J. M. (1995). LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Engineering, 8(2), 127–134. http://www.ncbi.nlm.nih.gov/pubmed/7630882 10.1093/protein/8.2.127 [DOI] [PubMed] [Google Scholar]
- Wang, J. (2020). Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. Journal of Chemical Information and Modeling, 60(6), 3277–3286. 10.1021/acs.jcim.0c00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S., Duan, N., Gu, H., Hao, L., Ye, H., Gong, W., & Wang, Z. (2016). A review of the methods for detection of staphylococcus aureus enterotoxins. Toxins, 8(7), 176. 10.3390/toxins8070176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., Wang, Q., Xu, Y., Li, M., Li, X., Zheng, M., Chen, L., & Li, H. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B, 10(5), 766–788. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, S., Liu, M., Wang, C., Xu, W., Lan, Q., Feng, S., Qi, F., Bao, L., Du, L., Liu, S., Qin, C., Sun, F., Shi, Z., Zhu, Y., Jiang, S., & Lu, L. (2020a). Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Research, 30(4), 343–355. 10.1038/s41422-020-0305-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, S., Zhu, Y., Liu, M., Lan, Q., Xu, W., Wu, Y., Ying, T., Liu, S., Shi, Z., Jiang, S., & Lu, L. (2020b). Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cellular & Molecular Immunology, 17(7), 765–767. 10.1038/s41423-020-0374-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, L. C., Rodrigues, J. P., Kastritis, P. L., Bonvin, A. M., & Vangone, A. (2016). PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics (Oxford, England), 32(23), 3676–3678. 10.1093/bioinformatics/btw514 27503228 [DOI] [PubMed] [Google Scholar]
- A Yi, H, C Fochtman, B. , C Rizzo, R., & Jacobs, A. (2016). Inhibition of HIV entry by targeting the envelope transmembrane subunit gp41. Current HIV Research, 14(3), 283–294. 10.2174/1570162x14999160224103908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, L., Li, Z., Yuan, K., Qu, X., Chen, J., Wang, G., Zhang, H., Luo, H., Zhu, L., Jiang, P., Chen, L., Shen, Y., Luo, M., Zuo, G., Hu, J., Duan, D., Nie, Y., Shi, X., Wang, W., … Xu, X. (2004). Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. Journal of Virology, 78(20), 11334–11339. 10.1128/JVI.78.20.11334-11339.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, W., Tang, Z., & Chang, C. A. (2019). Potential mean force from umbrella sampling simulations: What can we learn and what is missed? Journal of Chemical Theory and Computation, 15(4), 2433–2443. 10.1021/acs.jctc.8b01142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, S., Zhou, G., Guo, J., Zhou, F., & Chen, J. (2016). Molecular simulations of conformation change and aggregation of HIV-1 Vpr13-33 on graphene oxide. Scientific Reports, 6(1), 24906. 10.1038/srep24906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Zhu, Y., Hu, H., Zhang, S., Wang, P., Chong, H., He, J., Wang, X., & He, Y. (2018). Structural insights into the mechanisms of action of short-peptide HIV-1 fusion inhibitors targeting the Gp41 pocket. Frontiers in Cellular and Infection Microbiology, 8, 51. 10.3389/fcimb.2018.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]