Abstract
Background
The nucleotide-binding site–leucine-rich repeat (NBS-LRR) genes are important for plant development and disease resistance. Although genome-wide studies of NBS-encoding genes have been performed in several species, the evolution, structure, expression, and function of these genes remain unknown in radish (Raphanus sativus L.). A recently released draft R. sativus L. reference genome has facilitated the genome-wide identification and characterization of NBS-encoding genes in radish.
Results
A total of 225 NBS-encoding genes were identified in the radish genome based on the essential NB-ARC domain through HMM search and Pfam database, with 202 mapped onto nine chromosomes and the remaining 23 localized on different scaffolds. According to a gene structure analysis, we identified 99 NBS-LRR-type genes and 126 partial NBS-encoding genes. Additionally, 80 and 19 genes respectively encoded an N-terminal Toll/interleukin-like domain and a coiled-coil domain. Furthermore, 72% of the 202 NBS-encoding genes were grouped in 48 clusters distributed in 24 crucifer blocks on chromosomes. The U block on chromosomes R02, R04, and R08 had the most NBS-encoding genes (48), followed by the R (24), D (23), E (23), and F (17) blocks. These clusters were mostly homogeneous, containing NBS-encoding genes derived from a recent common ancestor. Tandem (15 events) and segmental (20 events) duplications were revealed in the NBS family. Comparative evolutionary analyses of orthologous genes among Arabidopsis thaliana, Brassica rapa, and Brassica oleracea reflected the importance of the NBS-LRR gene family during evolution. Moreover, examinations of cis-elements identified 70 major elements involved in responses to methyl jasmonate, abscisic acid, auxin, and salicylic acid. According to RNA-seq expression analyses, 75 NBS-encoding genes contributed to the resistance of radish to Fusarium wilt. A quantitative real-time PCR analysis revealed that RsTNL03 (Rs093020) and RsTNL09 (Rs042580) expression positively regulates radish resistance to Fusarium oxysporum, in contrast to the negative regulatory role for RsTNL06 (Rs053740).
Conclusions
The NBS-encoding gene structures, tandem and segmental duplications, synteny, and expression profiles in radish were elucidated for the first time and compared with those of other Brassicaceae family members (A. thaliana, B. oleracea, and B. rapa) to clarify the evolution of the NBS gene family. These results may be useful for functionally characterizing NBS-encoding genes in radish.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-020-02803-8.
Keywords: Raphanus sativus L., NBS-encoding gene, Gene duplication, Evolution, Synteny, Fusarium oxysporum
Background
Plants contain numerous resistance (R) genes that are vital for immunity against viral, fungal, and bacterial pathogens [1–3]. Specifically, R gene-mediated disease resistance is one of the most important plant mechanisms related to defense against pathogens [1]. The R genes are grouped in the subsequent five functionally diverse category based on respective domains: first, nucleotide-binding site–leucine-rich repeat (NBS-LRR) genes, sub-grouped as Toll/interleukin 1 receptor (TIR)-NBS-LRR (TNL) and coiled-coil (CC)-NBS-LRR (CNL) genes; second, receptor-like transmembrane proteins; third, serine–threonine kinases; fourth, receptor-like kinases (RLKs); and fifth, atypical R genes [4]. The predominant class of R genes includes genes with NBS and LRR domains [1, 5, 6]. To date, in all plant species over 300 R genes have been detected and cloned, of which more than 60% encode NBS and LRR domains [7]. The three NBS-LRR subclasses are TNL, CNL, and resistance to powdery mildew 8 (RPW8)-NBS-LRR (RNL), which are distinguished based on the differences in the N-terminal domains in angiosperms [8]. The TNL and CNL proteins are mainly responsible for the recognition of specific pathogens, whereas RNL proteins participate in downstream defense signal transduction pathways [9]. Therefore, Bonardi et al. described RNL proteins as helper NBS-LRRs [10].
The LRR domain, which is located at the C-terminal of plant NBS-LRR proteins, comprises tandem LRRs involved in the detection of invading pathogens [11]. The NBS domain is a functional ATPase domain, with its nucleotide-binding state believed to regulate R protein activities and function as a molecular switch [12, 13]. Although the TIR and CC domains are implicated in signaling and resistance specificity, their associated pathways differ. The TIR domain is mostly involved in self-association and homotypic interactions with other TIR domains [14, 15], whereas the CC domain may be related to protein–protein interactions and signaling [16]. To protect against diverse and rapidly evolving pathogens, a single plant genome usually encodes hundreds of NBS-LRR genes. The data generated in recent whole-genome sequencing analyses have enabled researchers to comprehensively analyze NBS-LRR genes in economically important plants [17–20].
Radish (Raphanus sativus L.), which is one of the more prominent members of the family Brassicaceae, is an economically valuable root vegetable crop grown worldwide [21]. Radish quality and yield are influenced by biotic stresses, including fungal and bacterial diseases as well as infestations by insect pests [22]. Specifically, Fusarium wilt caused by the soil-borne fungal pathogen Fusarium oxysporum, can severely damage radish and numerous other types of vegetables [23]. Because of its broad host range, F. oxysporum can survive at relatively high soil temperatures (> 24 °C) and remain viable even in the absence of any host plant, making it difficult to control [24, 25]. In addition to the TNL-type resistance genes FOC1 and FocBo1, which are responsible for the resistance of Brassica oleracea to F. oxysporum, many NBS-LRR genes related to Fusarium wilt resistance have been recently identified in diverse plant species [26–28].
Other genes belonging to the NBS-LRR family have been detected in various plants, including Arabidopsis thaliana [17, 29], Brassica rapa [30], chickpea [31], and Gossypium species [32]. Unfortunately, to the best of our knowledge, the potential roles of radish NBS-LRR genes related to disease resistance have not been investigated. The available radish genome sequence is a useful resource for the whole-genome identification of transcription factor families [33, 34]. However, the effects of F. oxysporum infections on radish NBS-LRR genes and their families remain unexplored. Therefore, combining bioinformatics and gene expression analyses to systematically study the evolution, expression, and potential functions of NBS-LRR genes may help to improve our understanding of the regulatory networks involved in radish plant growth and in response to F. oxysporum.
In this study, a genome-wide analysis of the radish genome identified 225 NBS-encoding genes (99 full NBS-LRR and 126 partial NBS genes) divided into two subclasses (CNL and TNL). These genes were further characterized regarding chromosomal locations, structures, and duplications. Additionally, with a focus on the NBS-LRR genes, we examined the encoded conserved domains as well as phylogenetic relationships and synteny with genes from A. thaliana, B. rapa, and B. oleracea. Furthermore, the pathogen-induced NBS-LRR gene expression profiles indicated that some R genes are differentially expressed in genetically different germplasm of radish (resistant and susceptible). Our results provide crucial insights into the evolution of this gene family in the radish genome. Moreover, the extensive R gene data presented herein may be useful for accelerating future breeding efforts aimed at improving the disease resistance of radish and other Brassicaceae crops.
Results
Identification and classification of NBS-encoding genes in R. sativus
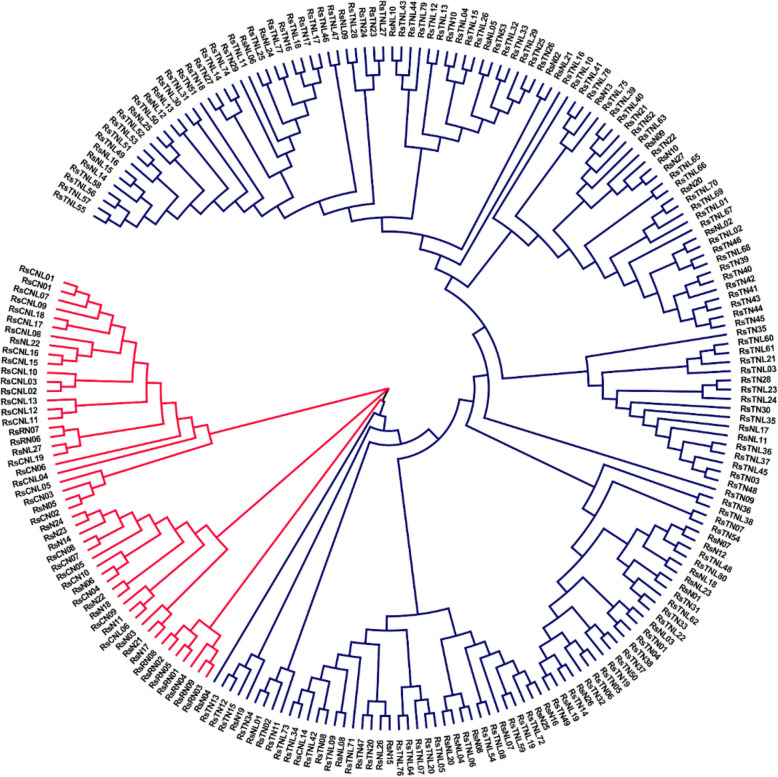
To comprehensively identify potential NBS-encoding genes in radish, the hidden Markov model (HMM) profile NB-ARC (Pfam: PF00931) from the Pfam database was used to screen the protein sequences encoded in the radish genome [34]. A total of 488 gene candidates with an NBS-LRR domain were identified. These candidate NBS-encoding genes were manually screened and functionally annotated according to the closest A. thaliana homolog. Finally, 225 non-redundant NBS-encoding R gene candidates were identified in the Rs1.0 genome (Table 1, Additional file 1: Table S1). The NBS-containing candidate proteins were classified into the TNL or CNL subfamilies based on their HMM profiles and the NCBI Conserved Domain Database, with relationships visualized in a phylogenetic tree (Fig. 1, Table 1). The phylogenetic tree clearly distinguished the TNL and CNL genes in two separate clades (Fig. 1). Gene names were assigned based on their domain type and chromosomal position. The TNL subfamily included 80 genes with full-length domains (TIR, NBS, and LRR) as well as 54 TN (TIR and NBS, but no LRR), 25 NLTNL (NBS and LRR, but no TIR), and 15 NTNL (no TIR or LRR) genes. The remaining 51 genes belonging to the CNL subfamily included 19 with full-length domains (CC, NBS, and LRR) as well as 10 CN (CC and NBS, but no LRR), 9 RN (RPW8 and NBS, but no LRR), 2 NLCNL (NBS and LRR, but no CC), and 11 NCNL (no CC or LRR) genes. Genes encoding RPW8-NBS-LRR proteins were not detected in the radish genome. We also analyzed the well-characterized NBS-encoding genes from the genetically closely related plant species A. thaliana, B. rapa, and B. oleracea (Table 1) using the same method. A total of 164 (A. thaliana), 212 (B. rapa), and 244 (B. oleracea) NBS-encoding genes were identified.
Table 1.
Summary of NBS gene in R. sativus, A. thaliana, B. rapa and B. oleracea.
| Predicted Type | Letter code | R.sativus | A.thaliana | B.rapa | B.oleracea |
|---|---|---|---|---|---|
| TNL type | |||||
| TIR NB LRR | TNL | 80 | 75 | 81 | 93 |
| NB LRR | NLTNL | 25 | 4 | 7 | 11 |
| TIR NB | TN | 54 | 26 | 38 | 50 |
| NB | NTNL | 15 | 2 | 5 | 23 |
| CNL type | |||||
| CC NB LRR | CNL | 19 | 18 | 19 | 15 |
| RW8 NB LRR | RNL | 0 | 1 | 1 | 1 |
| NB LRR | NLCNL | 2 | 3 | 15 | 3 |
| CC NB | CN | 10 | 11 | 15 | 16 |
| RW8 NB | RN | 9 | 4 | 6 | 12 |
| NB | NCNL | 11 | 20 | 25 | 20 |
| Total | 225 | 164 | 212 | 244 | |
Fig. 1.
Phylogenetic analysis of the radish NBS-LRR proteins. A maximum likelihood phylogenetic tree was constructed based on 225 NBS-encoding genes. The bootstrap values (1000 iterations) are provided on each branch. The proteins are designated as follows: Rs + Domain (TNL, TN, NL, CNL, CN, RN, N, and NL). Red and blue correspond to CNL and TNL clades, respectively. The radish NBS-encoding gene ID and the protein sequence information are provided in Additional file 1: Table S1
Genomic distribution among radish chromosomes
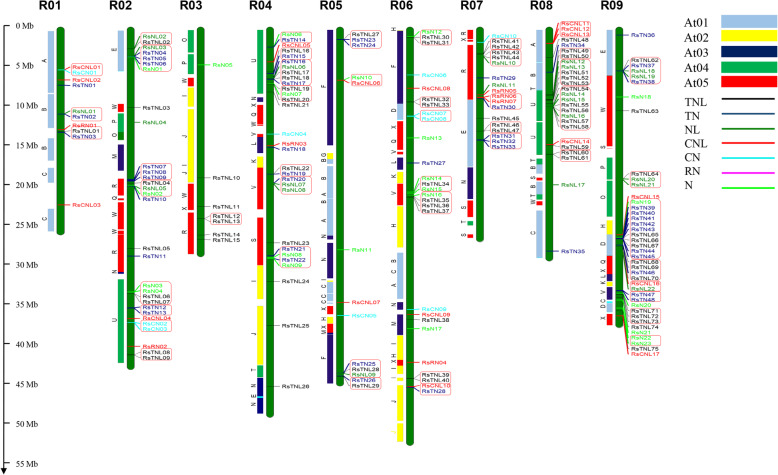
Of the 225 NBS-encoding genes, 202 were mapped onto nine radish chromosomes, whereas the other 23 genes were located on several scaffolds which were not mapped on the chromosome of R. sativus (Fig. 2, Additional file 1: Table S1). We determined the distribution of CNL, TNL, and partial NBS-encoding genes on different chromosomes (Additional file 2: Fig. S1). Moreover, the TNL genes were almost uniformly distributed on chromosomes R01 to R09, whereas the CNL genes were not detected on chromosomes R03 and R06. Furthermore, chromosome R09 had the most NBS-encoding genes (41), whereas chromosome R03 had the fewest (7). The ratio of radish TNL:CNL genes was almost 4:1 (80:19), which was consistent with the corresponding ratios in A. thaliana (78:18), B. rapa (74:20), and B. oleracea (91,20) [17].
Fig. 2.
Chromosomal localization and clustering of NBS-encoding genes in the R. sativus genome. The distribution of 202 genes on nine chromosomes (R01–R09) is presented. Different NBS types are marked by different colors. Gene clusters are indicated by red rectangles. The left side of chromosomes represents the syntenic regions in A. thaliana. The A. thaliana chromosomes are designated as At01–05, which are presented in different colors. The conserved chromosomal blocks in crucifer genomes are indicated by A–X [35]
There is no known pattern in the chromosomal distribution of NBS-encoding genes, with most of them detected in clusters. This distribution may facilitate sequence exchanges via recombination mispairing. We identified NBS-encoding gene clusters based on previously established criteria that an NBS-encoding cluster should have two or more genes separated by fewer than 200 kb, with no more than eight non-NBS-encoding genes in between [36]. On radish chromosomes, 146 (72%) NBS genes were mapped in 48 clusters, whereas the remaining 55 genes were detected as singletons (Fig. 2 and Additional file S2). Our analysis revealed that chromosome R09 has the most NBS genes (41; 20.30% of the mapped genes) distributed in eight clusters, in addition to nine singletons. Cluster sizes varied across the genome (2–11 genes). Cluster 44 was the largest, with 11 genes belonging to the TNL subfamily.
To clarify the evolutionary relationships among genes, we analyzed the distribution of NBS-LRR genes among the crucifer blocks in the radish genome. Of the 24 identified blocks (Fig. 2) [35], the U block on chromosomes R02, R04, and R08 was the largest (48 NBS-encoding genes), followed by the R block (24 genes), D block (23 genes), E block (23 genes), and F block (17 genes). The U block may be one of the most important in terms of NBS-encoding genes.
Gene characteristics and structure
The lengths of the genomic and coding sequences of the 225 NBS-encoding genes as well as the length, molecular weight (MW), and isoelectric point (pI) of the corresponding proteins were comprehensively analyzed (Additional file 1: Table S1). The genomic and coding sequence lengths ranged from 336 bp (RsN02) to 11,267 bp (RsTNL06) and from 1149 bp (RsN02) to 4899 bp (RsTNL15), respectively. The protein lengths ranged from 111 amino acids (RsN02) to 1632 amino acids (RsTNL15). There were also significant variations in the MW and pI, which ranged from 12.72 kDa (RsN11) to 182.40 kDa (RsTNL15) and from 4.77 to 9.60, respectively. Additionally, the average MW of the TNL (122.27 kDa) and CNL (99.84 kDa) proteins were markedly different.
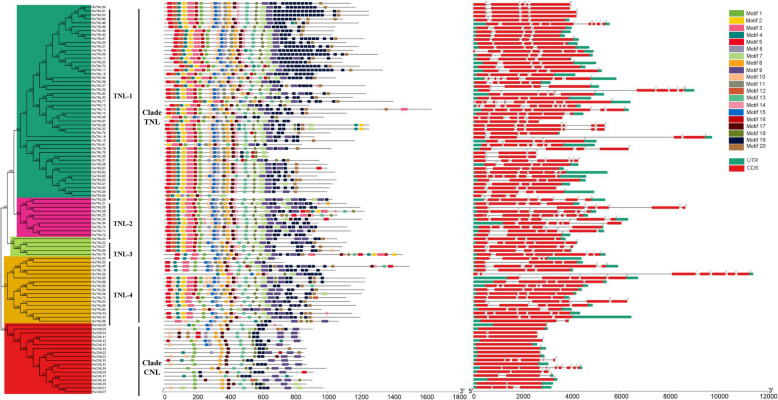
To assess the structural diversity of the R. sativus NBS-encoding genes, we compared the number of exons. The full-length CNL and TNL genes in the radish genome had an average of 2.42 and 5.26 exons, respectively (Fig. 3), which is consistent with the analyses of A. thaliana, B. rapa, and B. oleracea, in which the CNL and TNL genes have an average of 2.2, 2.3, and 3.4, and 5.3, 5.2, and 6.4 exons, respectively. Moreover, 47.37% of the CNL genes were encoded by a single exon. These results indicate that the number of introns may have increased and decreased during the structural evolution of the two types of NBS-LRR resistance genes in radish.
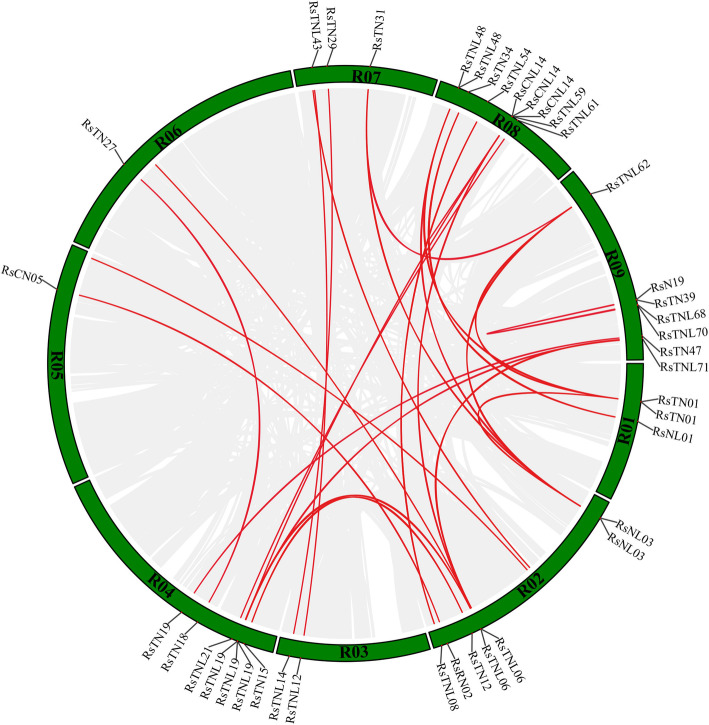
Fig. 3.
Circos diagram of segmentally duplicated NBS-encoding genes in the radish genome. Gray lines indicate the syntenic blocks in the radish genome. Red lines indicate the duplicated pairs of NBS-encoding genes. Chromosome numbers are provided on each chromosome
Cis-element analysis
Cis-elements in promoters are usually involved in gene regulation. Thus, to further investigate the potential regulatory networks of the NBS-encoding genes, the cis-elements in the sequences 2 kb upstream of the start codon of 225 NBS-encoding genes were analyzed with PlantCARE (Additional file 1: Table S3). A total of 70 cis-elements were detected, including 10 hormone-responsive elements, 33 light-responsive elements, 4 elements related to abiotic stress, 8 elements associated with tissue-specific expression, and 15 other elements. The cis-element DRE, which is a common cis-acting element in promoter and enhancer regions, was detected in the promoter region of 218 NBS-encoding genes. Additionally, an AT-rich fragment, which is a core promoter element located approximately 30 bp upstream of the transcription start site, was detected in 216 NBS-encoding genes. This sequence was considered to be an essential element in the promoter of the NBS-encoding genes. The promoter region of 159 NBS-encoding genes included the CGTCA-motif, which is a cis-acting regulatory element associated with methyl jasmonate-responsiveness. The abscisic acid-responsive element influencing abscisic acid responsiveness was detected in the promoter region of 165 genes, suggesting that NBS-encoding genes are involved in plant responses to pathogen infections. Moreover, a TCA-element, which is involved in salicylic acid responses, was detected in 97 gene promoters, whereas the TGA-element related to auxin responses was identified in 82 gene promoters. Furthermore, the P-box and GARE-motif associated with gibberellin-responsiveness were present in 55 and 40 NBS-encoding gene promoters, respectively. These results may be relevant for developing a method for identifying candidate genes related to disease resistance.
Conserved motifs and phylogenetic relationships among TNLs and CNLs
To investigate the TNL and CNL gene and protein structures, we built a phylogenetic tree based on the full-length amino acid sequences encoded by the R. sativus TNL and CNL genes. The phylogenetic analysis indicated that the radish NBS-LRRs can be divided into two large groups, namely CNL and TNL (Fig. 4 and Additional file 1: Table S4). The RsTNL group comprised four subgroups. Of the 80 RsTIR proteins, 49, 10, 5, and 16 belonged to subgroups RsTNL-1, RsTNL-2, RsTNL-3, and RsTNL-4, respectively. These results were identical to those of phylogenetic analyses of R. sativus and A. thaliana (Additional file Fig. 2).
Fig. 4.
Phylogenetic relationships, structure, and the encoded conserved motifs of the CNL and TNL genes. a Phylogenetic tree constructed based on the full-length radish protein sequences with the MEGA-X program. Different clades are presented in different colors. b Motif compositions of radish CNL and TNL proteins. Motifs 1–20 are displayed in different colored boxes. The sequence information for each motif is provided in Additional file S5. c Exon–intron structures of radish CNL and TNL genes. Green boxes represent untranslated 5′ and 3′ regions. Red boxes and gray lines indicate exons and introns, respectively. Gene and protein lengths can be estimated with the scale at the bottom
To further elucidate the potential functions and diversification of the TNL and CNL genes in R. sativus, 20 encoded conserved motifs were identified and numbered 1–20 based on the MEME program (Additional file 1: Tables S4 and S5). The TIR domain was detected in all RsTNLs. Additionally, the RsTNL-1 subgroup members had most of the motifs. The RsTNL-2 and RsTNL-3 proteins lacked motifs 20 and 11, respectively. The proteins in subgroup RsTNL-4 were missing motifs 11, 12, and 16 (Fig. 3). The RsCNL group members mostly had only six motifs, including the CC domain. Common motif compositions were revealed within subgroups. However, regarding the motif types and numbers, there was considerable diversity among subgroups. This suggests the proteins within subgroups are functionally similar.
Tandem duplication and synteny analyses of NBS-encoding genes
Whole-genome and tandem duplications are critical events for enhancing genome complexity and evolutionary novelty. In the R. sativus genome, 34 of 225 NBS-encoding genes (15.11%) were associated with tandem duplications and were distributed in 15 tandem arrays of 2–5 genes (Additional file 1: Table S6). Our data also revealed variability in the number of duplicated genes per tandem duplication event and an uneven distribution of these duplications on five of nine chromosomes. Genes encoding domains (e.g., CNL, TNL, and TN) were present in the tandem arrays. The 14 tandem duplication events detected for chromosome R09 involved five RsTN genes. In contrast, single tandem duplication events occurred on chromosomes R02, R06, R07, R08, and R09, each involving two genes. Chromosome R08 had the most tandem arrays (six tandem groups containing 13 genes), reflecting a hot spot for the distribution of NBS-encoding genes. An analysis of our data according to BLASTP and MCScanX methods identified 20 segmental duplication events involving 32 NBS-encoding genes (Fig. 4 and Additional file 1: Table S7).
The Ka/Ks values (Additional file 1: Table S8) of the pairs of segmentally duplicated genes were less than 1, indicating these genes evolved under negative selection. These results suggest that both tandem and segmental duplication events were a major driving force for the evolutionary expansion of the NBS-encoding genes in the radish genome.
Comparative synteny analyses of orthologous pairs of NBS-encoding genes
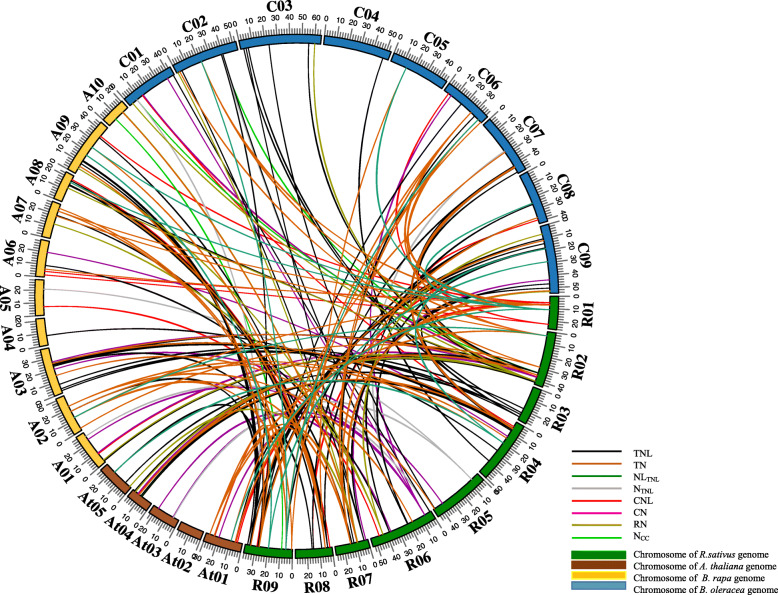
To further investigate the phylogenetic relationships among the radish NBS-encoding genes, we constructed three synteny maps comparing radish with A. thaliana, B. rapa, and B. oleracea. A total of 209 pairs of NBS-encoding genes (Additional file 1: Table S9) had syntenic relationships between R. sativus and A. thaliana, B. rapa, and B. oleracea. Specifically, 39 orthologous gene pairs were detected between R. sativus and A. thaliana, whereas there were 73 pairs between R. sativus and B. rapa as well as 97 pairs between R. sativus and B. oleracea (Fig. 5). Of the 39 pairs between R. sativus and A. thaliana, all were single-copy genes, except for RsCN06, which had two copies. Four partial NBS genes (RsNTIR19, RsNLTIR07, RsTN31, and RsTN39) in the radish genome were detected as homologous to A. thaliana TNL genes (AT1G63730, AT5G45210, and AT1G63860). The A. thaliana partial NBS gene AT5G18350 corresponded to the complete NBS gene RsTNL12. Of the 73 gene pairs between R. sativus and B. rapa, 46 radish NBS-LRR genes were present as a single copy, whereas there were two copies of 12 genes and three copies of one gene (RsTN01). The Bra027122 (no TIR domain) and Bra030779 (no CC domain) B. rapa genes corresponded to RsTNL75 and RsCNL11, respectively. Moreover, four RsTN genes (RsTN26, 34, 39, and 44) and RsNL01 were syntenic with TNL genes (Bra001160, Bra024652, Bra027779, and Bra027772), whereas RsCN02 was syntenic with a CNL gene (Bra019063) in the B. rapa genome. Overall, we detected 59 radish NBS-encoding genes syntenic with 64 NBS-encoding genes in the B. rapa genome (Additional file 1: Table S9). There were about 97 homologous gene pairs between R. sativus and B. oleracea, among which 47, 19, and 4 NBS-encoding genes in radish were retained as one, two, and three copies, respectively, in B. oleracea, with the remaining genes lacking syntenic relationships. The partial NBS-encoding genes RsTN11, RsTN31, RsNL07, and RsN19 were syntenic with complete TNL genes (Bo2g010720, Bo8g104700, Bo9g061200, and Bo9g029350) in the B. rapa genome. Additionally, two RsCNL genes (RsCNL03 and RsCNL05) as well as four RsTNL genes (RsTNL09, RsTNL14, RsTNL62, and RsTNL72) corresponded to partial NBS genes (Bo6g020950, Bo1g048080, Bo3g154220, Bo6g089230, Bo2g126980, and Bo3g006960) in the B. rapa genome. We detected a total of 70 radish NBS-encoding genes syntenic with 67 NBS-encoding genes in the B. oleracea genome (Additional file 1: Table S9). An analysis of the synteny among the radish, A. thaliana, B. rapa, and B. oleracea genomes revealed the complete NBS-LRR genes were more syntenic than the partial genes.
Fig. 5.
Syntenic relationships between radish NBS-encoding genes and A. thaliana, B. rapa, and B. oleracea genes. Green bars represent R. sativus chromosomes (R01–R09). Brown, yellow, and blue bars represent the chromosomes of A. thaliana (At01–At05), B. rapa (A01–A10), and B. oleracea (C01–C09), respectively
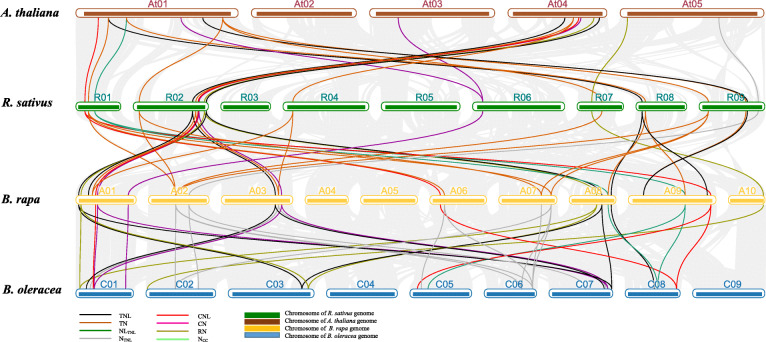
Finally, a comparative analysis of the orthologous pairs of NBS-LRR genes among four species (R. sativus, A. thaliana, B. rapa, and B. oleracea) revealed 22 R. sativus NBS-encoding genes with corresponding copies in the A. thaliana, B. rapa, and B. oleracea genomes (Fig. 6 and Additional file 1: Table S10). There was a one-to-one relationship among the NBS-encoding genes between the R. sativus and A. thaliana genomes. Additionally, half of the radish genes had single-copy syntenic genes in the B. rapa and B. oleracea genomes. Furthermore, the B. rapa and B. oleracea genomes had two and three copies of RsTN38, respectively, as well as three copies of RsTN01. Accordingly, these genes may have been important for the evolution of the NBS-LRR gene family.
Fig. 6.
Synteny of 22 NBS-encoding genes between radish and A. thaliana, B. rapa, and B. oleracea. Gray lines in the background indicate the collinear blocks within the radish and other genomes. Different colored lines represent the relationships among orthologous gene pairs among the plant species
The Ka/Ks ratio indicates the selective pressure on genes during evolution. We examined the Ka/Ks ratio for the orthologous gene pairs to determine the evolutionary selection patterns of NBS-LRR genes among R. sativus, A. thaliana, B. rapa, and B. oleracea. The Ka/Ks ratio reflects the number of non-synonymous substitutions per non-synonymous site (Ka) and the number of synonymous substitutions per synonymous site (Ks). The ratios for the orthologous gene pairs were estimated for each branch of the phylogenetic tree using the KaKs Calculator. The segmentally and tandemly duplicated NBS-LRR gene pairs as well as all orthologous NBS-LRR gene pairs had a Ka/Ks ratio < 1 (Additional file 1: Table S11), suggesting that the radish NBS-LRR gene family might have experienced strong purifying selection pressure during evolution.
Radish NBS-LRR gene expression profiles in response to F. oxysporum f. sp. raphanin 59 (FOR59)
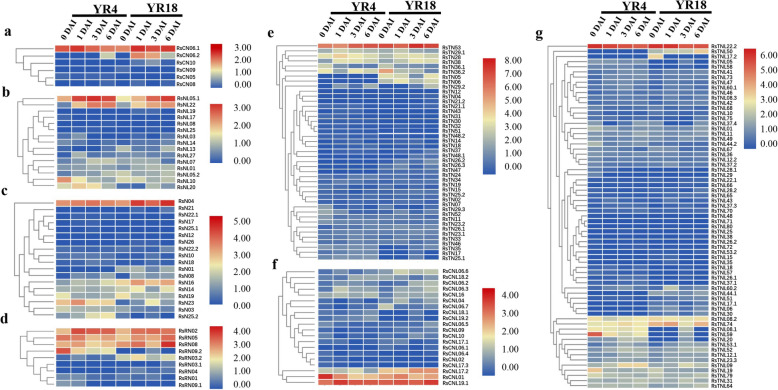
To identify NBS-LRR genes responsive to a FOR59 infection and determine their spatiotemporal expression patterns, we analyzed the transcriptome data for all NBS-LRR genes. The transcriptome data were generated for whole plant seedlings of the ‘YR4’ and ‘YR18’ genotypes infected with FOR59; the whole seeding of ‘YR4’ and ‘YR18’ were collected on days 0, 1, 3, and 6 after the inoculation of F. oxysporum for the subsequent sequencing on the Illumina platform. Furthermore, we extracted the NBS-LRR genes from the generated RNA-seq data. Transcriptome data were obtained for 171 of 225 NBS-encoding genes based on the conserved domains and gene IDs. The remaining unidentified genes may be unexpressed in response to a FOR59 infection. Among these 171 NBS-encoding genes, 29 were not differentially expressed between ‘YR4’ and ‘YR18’, and were minimally expressed at all time intervals. Thus, these genes were excluded from the gene expression analysis. The fragments per kilobase of exon per million fragments mapped (FPKM) values obtained for the 142 remaining NBS-LRR genes varied from 0 to 278.884. A total of 80 NBS-encoding genes had an FPKM value less than 1 FPKM (low expression), whereas there were 84 genes with more than 1 FPKM value, and less than 10 (high expression). Extremely high expression represented in 13 genes with more than 10, and less than 50; 1 gene with more than 50, less than 100; and 1 gene with more than 100, less than 300. The differentially expressed genes following the FOR59 inoculation were analyzed with the R package edgeR [37], which revealed 36 genes that were more highly expressed in the resistant ‘YR4’ line than in the susceptible ‘YR18’ line at various time-points (Fig. 7). Seven genes (RsCN10, RsN21, RsN26, RsNL19, RsTN04, RsTNL15, and RsTNL38) were up-regulated only in ‘YR18’, whereas six genes (RsCN05, RsCNL17.3, RsN22.1, RsTN31, RsTN32, and RsTN43) were up-regulated in ‘YR4’. Most importantly, the expression of five genes (RsN25.2, RsNL07, RsTN18, RsTNL09, and RsTN17) gradually increased over time in the ‘YR4’ plants, but remained relatively stable in the ‘YR18’ plants, implying they may be crucial for the resistance to Fusarium wilt. However, the RsTNL51 expression level was higher in ‘YR18’ than in ‘YR4’, and gradually increased during the infection period (Fig. 7g). These findings suggest a total of 75 NBS-LRR genes contribute to the resistance of radish to Fusarium wilt, with six genes (RsN25.2, RsNL07, RsTN18, RsTNL09, RsTN17, and RsTNL51) potentially crucial for the resistance.
Fig. 7.
Expression profiles of NBS-LRR genes in whole seeding plant of radish. The ‘YR4’ and ‘YR18’ plants are resistant and susceptible plant lines, respectively. a Expression of RsCN genes. b Expression of RsNL genes. c Expression of RsN genes. d Expression of RsRN genes. e Expression of RsTN genes. f Expression of RsCNL genes. g Expression of RsTNL genes. DAI: days after inoculation
Responses to FOR59 infections
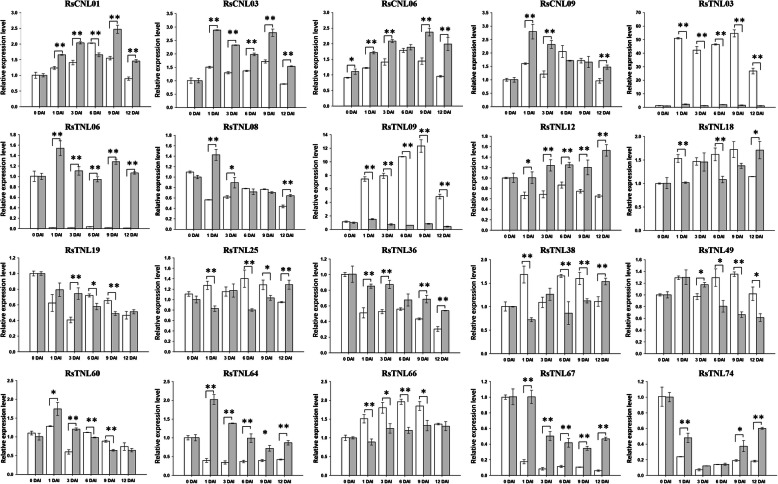
The expression patterns of NBS-LRR-encoding genes in ‘YR4’ and ‘YR18’ plants were confirmed by a quantitative real-time (qRT)-PCR assay. Specifically, we validated the expression of only the CNL and TNL genes. Of the 19 RsCNL and 80 RsTNL genes in the radish genome, approximately 40 genes encoded proteins with amino acid differences between ‘YR4’ and ‘YR18’. The expression patterns of these genes in ‘YR4’ and ‘YR18’ at 0, 1, 3, 6, 9, and 12 days after inoculation (DAI) were determined, with the 18S rRNA gene used as an internal control. The RsTNL03 (Rs093020) and RsTNL09 (Rs042580) genes on chromosome R02 were significantly more highly expressed in the resistant ‘YR4’ plants than in the susceptible ‘YR18’ plants (Fig. 8), indicating these two genes are related to the Fusarium wilt resistance of radish. Interestingly, both genes were similarly expressed in ‘YR4’ and ‘YR18’ on day 0, but the infection rapidly up-regulated the expression levels in ‘YR4’ plants. The expression levels continued to increase until 9 DAI, further suggesting their potential role in Fusarium wilt resistance. Homologs of RsTNL09 (Rs042580) were identified in A. thaliana (AT4G36150), B. rapa (Bra010552 and Bra011666), and B. oleracea (Bo3g154220 and Bo7g117810).
Fig. 8.
Relative expression levels of 4 RsCNL and 16 RsTNL radish genes. The ‘YR4’ (resistant) and ‘YR18’ (susceptible) lines are represented by white and gray bars, respectively. The y-axis represents the relative gene expression levels, whereas the time-points (0, 1, 3, 6, 9, and 12 DAI) are presented on the x-axis. The error bars indicate the standard deviation of three replicates; Asterisks indicate significant differences between the ‘YR4’ and ‘YR18’ lines based on a t-test (independent), ‘*’ indicates P value < 0.05 and > 0.01, and ‘**’ indicates P value < 0.01
Discussion
The NBS-LRR genes form a large family of stress resistance genes that are ubiquitous in all plant species. Genome-wide analyses of NBS-LRR gene families have been conducted for numerous species with sequenced genomes [17, 20, 38]. In the current study, we identified candidate NBS-LRR genes and studied their distribution, structure, clustering, duplication, synteny, and conservation. We identified 225 non-redundant NBS-encoding R gene candidates in the radish genome (Table 1, Additional file 1: Table S1) using highly stringent HMM and Pfam approaches. To confirm the accuracy of our method, a similar approach was used to identify NBS-encoding genes in B. rapa, B. oleracea, and A. thaliana. We identified 164, 212, and 244 NBS-encoding genes in A. thaliana, B. rapa, and B. oleracea, respectively, which is consistent with the results of an earlier investigation by Zhang et al. (2016), in which 165 and 204 NBS-encoding genes were detected in A. thaliana and B. rapa, respectively. Similar to our study, Yu et al. (2014) also identified 239 NBS-encoding genes in B. oleracea. However, the number of NBS-LRR genes identified in A. thaliana, B. oleracea, and B. rapa genomes differed in other studies [17, 18, 30]. These discrepancies are likely due to our stringent HMMER E-value as well as the manual checking for genes with partial NBS domains.
On the basis of a phylogenetic analysis of NBS-encoding candidate genes, 225 NBS-encoding genes were classified into the TNL and CNL subfamilies. The TNL subfamily included 80 TNL, 54 TN, 25 NLTNL, and 15 NTIR genes, with the remaining 51 genes in the CNL subfamily comprising 19 CNL, 10 C, 9 RN, 2 NLCC, and 11 NCC genes. Genes encoding RPW8-NBS-LRR proteins were not detected in radish. The CNL:TNL gene ratio was approximately 1:4, which is similar to that in A. thaliana, B. oleracea, and B. rapa [17]. This distinct pattern of TNL gene abundance suggests that the TIR domain is more functionally active than the CC domain in Brassicaceae species [39].
The NBS-LRR genes were unevenly distributed across the radish genome, with chromosomes R09 and R03 having the most and fewest genes, respectively. Additionally, 23 of the genes were located on diverse scaffolds (Fig. 2). Similar to other species, the NBS-LRR genes in radish mainly exist in clusters because of their rapid evolution [40, 41]. About 72% of the radish NBS-LRR genes were detected in 48 clusters, which is higher than the corresponding percentages in A. thaliana (61.7%), B. rapa (59.4%), and B. oleracea (60.3%) [17]. Moreover, 11 genes were included in cluster 44 of the radish genome. Gene families may expand because of polyploidizations, and tandem and local duplications are the most commonly evaluated mechanisms underlying gene family expansions [42]. We identified 66 NBS-encoding genes (29.33%) in radish that underwent tandem and segmental duplications, which is lower than the duplication rates for A. thaliana (55.7%), B. rapa (47.1%), and B. oleracea (43.3%) [17]. The TN genes were predominantly present in tandem arrays, and only three pairs of CN genes were detected in tandem arrays 2 (RsCn02 and RsCN03), 4 (RsCN07 and RsCN08), and 8 (RsCNL11 and RsCNL12). Interestingly, RsCNL14 and RsTNL59, which belong to two distinct subgroups (CNL and TNL), had undergone segmental duplications. These two genes were located in cluster 40 of the radish genome. We speculate that the NBS-LRR genes (especially TNL subfamily genes) in the radish genome had undergone inter- and intraspecific replications. Because radish and A. thaliana are Brassicaceae species, we investigated the crucifer blocks in the radish genome containing the identified NBS-LRR genes. A total of 45 NBS-LRR genes were located in the U block distributed on different chromosomes (R02, R04, and R08). The genes were also relatively abundant in the R (24 genes), D (23 genes), E (23 genes), and F (17 genes) blocks (Fig. 2). These observations suggest that the R. sativus genome structure arose following the rearrangement and divergence from a common ancestor with A. thaliana [43, 44]. All tandemly duplicated genes were found in the same clusters (i.e., highly similar), whereas most of the segmentally duplicated genes did not form clusters and were located on different chromosomes. The relatively few tandem and segmental duplications of NBS-encoding genes may help to explain why radish has evolved more slowly than other Brassicaceae species [45].
Gene duplication is considered as a major force for evolution [46]. The frequent sequence variation that can occur in the duplication process, can lead to domain’s structural variation, directly involved in protein functional role [47, 48]. In addition to this expansion, variation leads to neofunctionalization in family members. The neofunctionalization has been reported in KCS gene family and MADS box gene of Arabidopsis [49, 50]. According to the genome comparison in Brassica family members, we identified that the structural variation happened in many NBS-encoding genes which influenced the domain structure (Additional file 1: Table S12). For example, RsNL01 has the homolog gene in A. thaliana, B. rapa, and B. oleracea, but the number of exons was reduced, additionally LRR domain was added and TIR domain was lost in the R. sativus. Taken together of this results, we possibly suggest that these variation in the NBS-encoding genes might result in neofunctionalization or loss of function during the radish evolution.
The conserved structural domains of the radish TNL and CNL proteins were examined in this study. The NBS-LRR genes encoding the TIR domain were more common than the genes encoding the CC domain in the analyzed species (R. sativus, A. thaliana, B. oleracea, and B. rapa). However, the functions of several partial NBS-encoding genes lacking one or two domains (TIR, CC, and LRR) remain unknown, but their presence in various plant species imply they are important [17]. In the radish genome, the average number of exons was higher for TNL genes than for CNL genes, with half of the CNL genes containing only one exon (Fig. 4). This difference between gene types may be due to the conservative nature of CNL gene replications, which involve many regulatory components. This result is also consistent with those reported for A. thaliana, B. rapa, and B. oleracea [17, 20, 38]. Although the TNL and CNL genes are related to signaling and resistance specificity during pathogen recognition, the genes in these two subfamilies vary regarding sequences and associated signaling pathways and they cluster separately according to phylogenetic analyses [51]. Previous studies proved that the TNL group forms four phylogenetic clades in B. rapa [18] and B. oleracea [38]. Our motif analysis with the MEME program uncovered diverse motif compositions in the different RsTNL subfamilies. For example, the RsTNL-4 subgroup members had lost motifs 11, 12, and 16, whereas these motifs are present in the RsTNL-1 subgroup members. These differences may contribute to the functional divergence among the TNL proteins in radish.
Radish is an agronomically important root vegetable crop. Its genome, which contains triplicated segments, has intermediate characteristics between the Brassica A/C and B genomes, suggesting radish originated from a Brassica species [34]. According to a comparison with the Brassica A (Br), B (Bn), and C (Bo) genomes, the radish genome has been positioned between the Brassica A/C and B genomes [34]. Thus, an analysis of the synteny among the NBS-LRR genes of radish, A. thaliana, B. rapa, and B. oleracea may lead to novel insights into the evolutionary characteristics of RsNBS-encoding genes as well as the phylogenetic relationships with the genes in the other three species. In the current study, 39, 73, and 97 orthologous pairs were respectively identified between radish and A. thaliana, B. rapa, and B. oleracea (Fig. 5), implying that R. sativus is more closely related to B. oleracea than to A. thaliana or B. rapa.
Moreover, we identified genes in the A. thaliana, B. rapa, and B. oleracea genomes that correspond to 22 R. sativus NBS-encoding genes (Fig. 6). Additionally, the orthologous gene pairs may have existed before the ancestral divergence, with important roles related to the evolution of the NBS-LRR gene family [52]. Furthermore, we compared the Ka/Ks values of the orthologous gene pairs between R. sativus and A. thaliana, B. rapa, and B. oleracea lineages. For the CNL and TNL gene types, there were no significant differences in the orthologous gene pairs among the three species (Additional file 1: Table S11). We speculate that the NBS-LRR genes in R. sativus, A. thaliana, B. rapa, and B. oleracea may have been exposed to diverse selection pressures to develop genetic resistance to the same pathogen. Additionally, some pathogens may be specific to certain Brassica species.
In a previous study regarding radish resistance to Fusarium wilt, we isolated 68 NBS-RGAs and 46 SRLK-RGAs from two Fusarium wilt-resistant radish inbred lines, and the genetic diversity was analyzed with RGA-specific primers [53]. We also identified a major QTL (Fwr1) containing the ORF4 gene, encoding a serine/arginine-rich protein kinase that may mediate Fusarium wilt resistance [54]. To further explore the mechanism underlying Fusarium wilt resistance, we profiled the expression of NBS-LRR genes by screening transcriptome data for the resistant ‘YR4’ and susceptible ‘YR18’ lines at different time-points during an infection (Fig. 7). A total of 75 NBS-LRR genes were identified as likely involved in the Fusarium wilt resistance of the ‘YR4’ line, including RsNL07, RsTNL09 (chromosome R02), RsTN17 and RsTN18 (chromosome R04), and RsN25 (scaffold), which exhibited gradually up-regulated expression in infected ‘YR4’ plants. Accordingly, the genes belonging to the same cluster were similarly expressed. The identified genes and expression profiles based on transcriptome data were verified by a qRT-PCR analysis of the ‘YR4’ and ‘YR18’ lines at several time-points after plants were inoculated (Fig. 8). Of the analyzed genes, RsTNL03 and RsTNL09 were more highly expressed in ‘YR4’ (resistant) than in ‘YR18’ (susceptible) at different time-points. The RsTNL09 (Rs042580) gene is homologous to AT4G36150 in A. thaliana, Bra010552 and Bra011666 in B. rapa, and Bo3g154220 and Bo7g117810 in B. oleracea. The Bra010552 gene encodes a TNL protein, and is one of the candidate genes for clubroot resistance in B. rapa [55]. The RsTNL03 gene, which is a homolog of AT4G16890 in A. thaliana, is likely involved in salicylic acid-dependent early plant defense responses [38–40]. Our RNA-seq and qRT-PCR data also indicated that RsTNL06 (Rs053740) was up regulated in ‘YR18’ than in ‘YR4’ after the FOR59 infection, taken together with the morphological characteristics (Additional file 1: Table S13) of ‘YR18’ (severe infected) suggests that RsTNL06 might be potential negative regulators of Fusarium wilt resistance in radish. Interestingly, RsTNL06 was identified as an ortholog of the B. oleracea gene Bo7g106630, and may contribute to plant responses to Fusarium species [38]. The potential positive and negative regulators of Fusarium wilt resistance that were identified following the comprehensive analyses of RNA-seq and qRT-PCR data will need to be thoroughly functionally characterized to assess their utility for enhancing the Fusarium wilt resistance of radish.
Conclusions
The NBS-LRR genes are important for the resistance of radish plants to various pathogens. A comprehensive analysis of the radish genome revealed 225 NBS-encoding genes, including 99 NBS-LRR genes and 126 partial NBS genes. Additionally, of the 202 NBS-encoding genes mapped onto nine chromosomes, 72% were located in clusters. Another 23 genes were located on different scaffolds. An analysis of syntenic and phylogenetic relationships among the NBS-LRR genes from A. thaliana, B. rapa, and B. oleracea provided valuable insights into the evolutionary characteristics of radish NBS-LRR genes. Moreover, 99 complete NBS-LRR genes were grouped in two main families (TNL and CNL), with the TNL genes further divided into four subgroups with highly similar exon-intron structures and motif compositions. In this study, we identified 75 NBS-LRR genes that protect radish plants from Fusarium wilt based on the transcriptome data. We also determined that RsTNL03 (Rs093020) and RsTNL09 (Rs042580) positively associated with the resistance to F. oxysporum, whereas RsTNL06 (Rs053740) negatively associated with Fusarium wilt resistance in radish. The phylogenetic and gene expression data presented herein may help to clarify NBS-LRR gene functions.
Methods
Identification of NBS-encoding genes
To identify the NBS-encoding genes, we downloaded the whole-genome sequence from the radish database (http://radish-genome.org/). We applied hmmsearch of the HMMER (version 3.3) program [56] based on the HMM corresponding to the Pfam NBS (NB-ARC) domain (PF00931) to screen and identify the NBS-encoding genes in the radish genome. We also selected proteins with an E value <1e− 20 for sequence alignments with ClustalW [57]. We constructed the radish-specific NBS HMM with the hmmbuild module of HMMER (version 3.3) to rescan the radish protein database, and proteins with an E-value less than 0.01 were selected for further analyses [17, 58].
The N-terminal of NBS-containing proteins usually include the TIR and CC domains, whereas the C-terminal contains the RPW8 and LRR domains. We used the CLC main workbench (version 7.9) [59] and the Pfam (version 32) program [60] to detect domains in the NBS-containing proteins. These results were confirmed with the NCBI Conserved Domains Tool [61] and MEME (Multiple Expression motifs for Motif Elicitation) [62]. The CC domain in the protein sequences was identified with Paircoil2 [63], with a P score cut-off of 0.025. The NBS-encoding genes in the genomes of A. thaliana (Araport11), B. rapa (version 1.5), and B. oleracea (version 1.1) were also identified using the same method. The A. thaliana genome was download from the TAIR database (www.arabidopsis.org), whereas the B. rapa and B. oleracea genomes were downloaded from the database on the BRAD website (http://brassicadb.org/brad/).
Multiple sequence alignment and phylogenetic analysis of NBS-encoding genes
These analyses confirmed the segregation between the two major NBS-encoding subpopulations (TNL and CNL) in the radish genome and clarified the phylogenetic relationships among the genes on the major branches. Multiple full NBS-containing protein sequences were aligned with the MUSCLE [64] program. The MEGA-X [65] program was used for the phylogenetic analysis, which was completed with the maximum likelihood method based on the Whelan and Goldman model [66]. Finally, the maximum likelihood method involving the LG model with 500 bootstrap replicates was used to construct the phylogenetic tree. Two additional phylogenetic trees were constructed with the same method. One tree was used for analyzing the relationships between the CNL and TNL genes in radish, whereas the other trees were used to verify that the radish CNL and TNL genes are consistent with the corresponding A. thaliana genes.
Chromosomal locations, structures, and duplication events of NBS-encoding genes
The Mapchart (version 2.2) software was used to map all identified NBS-encoding genes onto R. sativus chromosomes based on their physical positions indicated in the R. sativus genome database. Several genes were organized in diverse NBS-LRR clusters, in which at least two NBS-encoding genes were localized in a 200-kb region and were separated by a maximum of eight non-NBS-LRR genes [35].
The Pepstats program (http://www.ebi.ac.uk/Tools/seqstats/emboss_pepstats) was used to calculate the pI and MW of the NBS-containing proteins. The promoter sequence (2000 bp upstream of the start codon) of each gene was examined with PlantCARE (http://bioinformatics.psb.ugen.be/webtools/plantcare/html/) to identify cis-elements [67].
The tandem and segmental duplications of the NBS-encoding genes in the radish genome were analyzed with the default parameters of MCScanX [68] and TBtools [69]. Gene duplications were confirmed based on the following two criteria: (a) the length of the shorter aligned sequence covered > 70% of the longer sequence; (b) the similarity of the two aligned sequences was > 70% [70]. The non-synonymous (Ka) and synonymous (Ks) substitutions for each duplication event were calculated with the KaKs Calculator (version 2.0) [71].
The predicted TNL and CNL proteins were analyzed with MEME [72], with default iterative cycles and the maximum number of motifs set to 20. Additionally, TBtools was used to visualize the structures of the TNL and CNL genes according to the genomic and coding sequences.
Analysis of the orthologous gene pairs between R. sativus and three Brassicaceae species
Orthologous genes are important for investigating the evolutionary associations among diverse species. In this study, we identified the orthologous gene pairs between the R. sativus and A. thaliana, B. rapa, and B. oleracea genomes with the MCScanX program. Specifically, the following parameters were used: e = 1e− 20, u = 1, and s = 5 [73]. The orthologous pairs of NBS-encoding genes were extracted and the maps for analyzing synteny were prepared with TBtools. The 24 genome building blocks from the ancestral karyotype were assigned to the radish genome as previously described [34].
Plant growth and Fusarium oxysporum inoculation
The two radish plant materials, inbred lines ‘YR4’ and ‘YR18’, were kindly provided by Seungho Kim (Neo Seed Co., Anseong, Republic of Korea). ‘YR4’ and ‘YR18’ are highly resistant and susceptible to F. oxysporum, respectively. To do the sampling, pathogen inoculation was used the modified root-dipping methods following the method by Baik et al. [74]. F. oxysporum f. sp. raphanin 59 was supported from the Korea Research Institute of Chemical Technology (Daejeon, Korea). Ten-day-old seedling of germplasm were grown at control condition at 25 °C and further were subjected to water rinse to remove the soil from the root, and inoculated in spore suspension of F. oxysporum with concentration of conidia. 3 × 106 mL− 1 for 30 min. Later, all individual seedlings were transplanted into 50-well seedling tray containing autoclaved soil. After inoculation, all plants were incubated in the controlled chamber, 80% humidity and 25 °C for 1 day without light. Further, these plants were grown in the controlled chamber with 60% humidity and 25 °C with 12 h/12 h condition. Individual seedlings of ‘YR4’ and ‘YR18’ were collected at 0, 1, 3, 6, 9, and 12 days after inoculation (DAI). Three individuals were used for subsequent RNA isolation and qRT-PCR analysis.
Total RNA extraction and gene expression analysis
Total RNA was isolated from the ‘YR4’ and ‘YR18’ plants with the RNeasy Mini Kit (QIAGEN, Germany). The RNA quality and quantity were investigated through agarose gel electrophoresis and with the Nanodrop 2000 spectrophotometer (Thermo Scientific, USA). The purified total RNA (2 μg) was reverse transcribed to cDNA with the oligo-(dT) primer and TOPscript™ reverse transcriptase (Enzynomics Company, Korea). A qRT-PCR assay was performed with the SYBR Green Supermix and the CFX96™ Real-Time System (Bio-Rad Company, USA) using subsequent conditions: 95 °C for 3 min; 39 cycles of 95 °C for 15 s and 58 °C for 20 s. Relative gene expression levels were calculated as per the 2−∆∆Ct method [75].
Previously generated transcriptome data for the FOR59-inoculated ‘YR4’ and ‘YR18’ lines at four time-points (0, 1, 3, and 6 DAI) were analyzed (Accession number PRJNA643982) (Additional file 1: Table S14). The growth conditions of plant materials were mentioned earlier and two biological replications were used for RNA-Seq. The brief summary of RNA sequencing and library preparation is as follows. Firstly rRNA was removed from total RNA using RIBO COP rRNA depletion kit (LEXOGEN, Inc., Austria). Total RNA was used to prepare cDNA libraries using the NEBNext Ultra II Directional RNA-Seq Kit (NEW ENGLAND BioLabs, Inc., UK) as per manufacture’s instruction. Then, libraries were examined with the Agilent 2100 bioanalyzer (DNA High Sensitivity Kit) to estimate the fragment size and quantified with the library quantification kit via a StepOne Real-Time PCR System (Life Technologies, USA). High-throughput paired-end sequencing was carried out using Hiseq X10 (Illumina, Inc., USA). Total RNA-Seq reads were mapped using TopHat software tool [76] to get bam file (alignment file). These alignment files were used to assemble transcripts, estimate their abundances and detects differential expression of genes using cufflinks. We used the FPKM (fragments per kilobase of exon per million fragments) method for determining the expression level of the gene regions. The FPKM data were normalized based on Quantile normalization method using EdgeR within R [37]. Additionally, heat maps were produced with TBtools.
Supplementary Information
Additional file 1: Table S1. Information regarding the radish NBS-LRR genes. Table S2. Clusters of radish NBS-LRR genes. Table S3 Predicted promoter elements of NBS-encoding genes in the radish genome. Table S4.Analysis and distribution of conserved motifs in radish CNL and TNL proteins. Table S5. MEME analysis of the CNL and TNL proteins. Table S6. Tandem arrays of the radish NBS-LRR genes. Table S7. Segmentally duplicated radish NBS-LRR genes. Table S8. Ka/Ks values for the segmentally duplicated genes in the radish genome. Table S9. Segmentally duplicated NBS-LRR genes in the radish/A. thaliana, radish/B. oleracea, and radish/B. rapa genomes. Table S10. Common segmentally duplicated NBS-LRR genes in the radish/A. thaliana, radish/B. oleracea, and radish/B. rapa genomes. Table S11. Ka/Ks values between radish and three related plant species. Table S12. The information for gene structure and domain change of radish/A. thaliana, radish/B. oleracea, and radish/B. rapa genomes. Table S13. The resistance of radish lines to F. oxysporum f. sp. Raphani 59. Table S14. The summary of the RNA-Seq database. Table S15. Details regarding the qRT-PCR primers. Table S16. The transcriptome data of the radish lines after inoculated with F. oxysporum f. sp. Raphani 59 (XLSX 15325 kb)
Additional file 2: Figure S1. Distribution of TNL, CNL, and partial genes. Distributions for each class are presented across the radish chromosomes. RUS, scaffold summary. Bars are divided into CNL genes (orange), TNL genes (blue), and partial genes (green)
Additional file 3: Figure S2. Phylogenetic tree representing the relationships of NBS-encoding genes between radish and arabidopsis. Different colored arcs represent the different groups (or subgroups) of NBS-encoding genes.
Additional file 4: Figure S3. Relative expression levels of RsCNL and RsTNL radish genes. The ‘YR4’ (resistant) and ‘YR18’ (susceptible) lines are represented by white and gray bars, respectively. The y-axis represents the relative gene expression levels, whereas the time-points (0, 1, 3, 6, 9, and 12 DAI) are presented on the x-axis.
Acknowledgements
The authors appreciate all editors and reviewers for their comments and suggestion to improve on this manuscript.
Abbreviations
- CC
Coiled-coil
- CNL
Coiled-coil nucleotide-binding site–leucine-rich repeat
- FOR
Fusarium oxysporum f. sp. raphanin 59
- FPKM
Fragments per kilobase of exon per million fragments mapped
- HMM
Hidden Markov model
- NBS-LRR
Nucleotide-binding site–leucine-rich repeat
- RPW8
Resistance to powdery mildew 8
- TNL
Toll/interleukin 1 receptor nucleotide-binding site–leucine-rich repeat
Authors’ contributions
SSC, SRC and YPL conceived and designed the study. YM carried out experiments, generated data and wrote manuscript. SSC executed data analysis and wrote draft of the manuscript. SO, LL, SK, GJC were participated in phenotype evaluations and performing the experiments. SS and CSK provided comments in the writing of manuscript. YPL and SRC participated as a director, supervised this study, corrected and modified the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, and Forestry (IPET) through Golden Seed Project (213006–05-5-SBO 20, 213006–05-5-SB110), funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development Administration (RDA) and Korea Forest Services (KFS). These funding bodies had no role in the designing of this study, collection, analysis, interpretation of the data, and in the writing of the manuscript.
Availability of data and materials
All datasets generated and analyzed during this study are included in this article and its additional files. The RNA-seq data generated during the current study have been deposited and publicly available at the National Center of Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/) repository under the accession number PRJNA643982.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yinbo Ma and Sushil Satish Chhapekar contributed equally to this work.
Contributor Information
Yinbo Ma, Email: mayinbo@126.com.
Sushil Satish Chhapekar, Email: sushilchhapekar@gmail.com.
Lu Lu, Email: dlLL220186@126.com.
Sangheon Oh, Email: rederaser64@gmail.com.
Sonam Singh, Email: sonamsingh688@gmail.com.
Chang Soo Kim, Email: changsookim@cnu.ac.kr.
Seungho Kim, Email: raskkim@hanmail.net.
Gyung Ja Choi, Email: kjchoi@krict.re.kr.
Yong Pyo Lim, Email: yplim@cnu.ac.kr.
Su Ryun Choi, Email: srchoi@cnu.ac.kr.
References
- 1.Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411(6839):826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 2.Maekawa T, Kufer TA, Schulze-Lefert P. NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol. 2011;12(9):817. doi: 10.1038/ni.2083. [DOI] [PubMed] [Google Scholar]
- 3.Dangl JL, Horvath DM, Staskawicz BJ. Pivoting the plant immune system from dissection to deployment. Science. 2013;341(6147):746–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong Y, Cheng Z-MM. A unique RPW8-encoding class of genes that originated in early land plants and evolved through domain fission, fusion, and duplication. Sci Rep. 2016;6:32923. doi: 10.1038/srep32923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yue JX, Meyers BC, Chen JQ, Tian D, Yang S. Tracing the origin and evolutionary history of plant nucleotide-binding site–leucine-rich repeat (NBS-LRR) genes. New Phytol. 2012;193(4):1049–1063. doi: 10.1111/j.1469-8137.2011.04006.x. [DOI] [PubMed] [Google Scholar]
- 6.Marone D, Russo MA, Laidò G, De Leonardis AM, Mastrangelo AM. Plant nucleotide binding site–leucine-rich repeat (NBS-LRR) genes: active guardians in host defense responses. Int J Mol Sci. 2013;14(4):7302–7326. doi: 10.3390/ijms14047302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kourelis J, van der Hoorn RA. Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell. 2018;30(2):285–299. doi: 10.1105/tpc.17.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao Z-Q, Xue J-Y, Wu P, Zhang Y-M, Wu Y, Hang Y-Y, Wang B, Chen J-Q. Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol. 2016;170(4):2095–2109. doi: 10.1104/pp.15.01487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collier SM, Hamel L-P, Moffett P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol Plant-Microbe Interact. 2011;24(8):918–931. doi: 10.1094/MPMI-03-11-0050. [DOI] [PubMed] [Google Scholar]
- 10.Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL. Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci. 2011;108(39):16463–16468. doi: 10.1073/pnas.1113726108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeYoung BJ, Innes RW. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat Immunol. 2006;7(12):1243–1249. doi: 10.1038/ni1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersen EJ, Ali S, Byamukama E, Yen Y, Nepal MP. Disease resistance mechanisms in plants. Genes. 2018;9(7):339. doi: 10.3390/genes9070339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Ooijen G, Mayr G, Kasiem MM, Albrecht M, Cornelissen BJ, Takken FL. Structure–function analysis of the NB-ARC domain of plant disease resistance proteins. J Exp Bot. 2008;59(6):1383–1397. doi: 10.1093/jxb/ern045. [DOI] [PubMed] [Google Scholar]
- 14.Bernoux M, Ve T, Williams S, Warren C, Hatters D, Valkov E, Zhang X, Ellis JG, Kobe B, Dodds PN. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe. 2011;9(3):200–211. doi: 10.1016/j.chom.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ve T, Williams SJ, Kobe B. Structure and function of toll/interleukin-1 receptor/resistance protein (TIR) domains. Apoptosis. 2015;20(2):250–261. doi: 10.1007/s10495-014-1064-2. [DOI] [PubMed] [Google Scholar]
- 16.van Ooijen G, van den Burg HA, Cornelissen BJ, Takken FL. Structure and function of resistance proteins in solanaceous plants. Annu Rev Phytopathol. 2007;45:43–72. doi: 10.1146/annurev.phyto.45.062806.094430. [DOI] [PubMed] [Google Scholar]
- 17.Yu J, Tehrim S, Zhang F, Tong C, Huang J, Cheng X, Dong C, Zhou Y, Qin R, Hua W. Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genomics. 2014;15(1):3. doi: 10.1186/1471-2164-15-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lv S, Changwei Z, Tang J, Li Y, Wang Z, Jiang D, Hou X. Genome-wide analysis and identification of TIR-NBS-LRR genes in Chinese cabbage (Brassica rapa ssp. pekinensis) reveal expression patterns to TuMV infection. Physiol Mol Plant Pathol. 2015;90:89–97. doi: 10.1016/j.pmpp.2015.04.001. [DOI] [Google Scholar]
- 19.Jupe F, Pritchard L, Etherington GJ, MacKenzie K, Cock PJ, Wright F, Sharma SK, Bolser D, Bryan GJ, Jones JD. Identification and localisation of the NB-LRR gene family within the potato genome. BMC Genomics. 2012;13(1):75. doi: 10.1186/1471-2164-13-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alamery S, Tirnaz S, Bayer P, Tollenaere R, Chaloub B, Edwards D, Batley J. Genome-wide identification and comparative analysis of NBS-LRR resistance genes in Brassica napus. Crop and Pasture Science. 2018;69(1):72–93. doi: 10.1071/CP17214. [DOI] [Google Scholar]
- 21.Rakow G. Species origin and economic importance of Brassica. Brassica: Springer; 2004. pp. 3–11. [Google Scholar]
- 22.Reddy PP. Recent advances in crop protection: Springer Science & Business Media. 2012. [Google Scholar]
- 23.Patil S, Sriram S. Biological control of Fusarium wilt in crop plants using non-pathogenic isolates of Fusarium species. Indian Phytopathology. p. 1–9.
- 24.Z-j P. Shimizu M, Zhang Y-j, Nagaoka T, Hayashi T, Hori H, Matsumoto S, Fujimoto R, Okazaki K: genetic mapping of a fusarium wilt resistance gene in Brassica oleracea. Mol Breed. 2012;30(2):809–818. doi: 10.1007/s11032-011-9665-8. [DOI] [Google Scholar]
- 25.Okungbowa F, Shittu H. Fusarium wilts: an overview. Environ Res J. 2012;6(2):122–134. [Google Scholar]
- 26.Lv H, Fang Z, Yang L, Zhang Y, Wang Q, Liu Y, Zhuang M, Yang Y, Xie B, Liu B. Mapping and analysis of a novel candidate Fusarium wilt resistance gene FOC1 in Brassica oleracea. BMC Genomics. 2014;15(1):1094. doi: 10.1186/1471-2164-15-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimizu M. Pu Z-j, Kawanabe T, Kitashiba H, Matsumoto S, Ebe Y, Sano M, Funaki T, Fukai E, Fujimoto R: map-based cloning of a candidate gene conferring Fusarium yellows resistance in Brassica oleracea. Theor Appl Genet. 2015;128(1):119–130. doi: 10.1007/s00122-014-2416-6. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Chitwood J, Menda N, Mueller L, Hutton SF. Linkage between the I-3 gene for resistance to Fusarium wilt race 3 and increased sensitivity to bacterial spot in tomato. Theor Appl Genet. 2018;131(1):145–155. doi: 10.1007/s00122-017-2991-4. [DOI] [PubMed] [Google Scholar]
- 29.Meyers BC, Vu TH, Tej SS, Ghazal H, Matvienko M, Agrawal V, Ning J, Haudenschild CD. Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat Biotechnol. 2004;22(8):1006–1011. doi: 10.1038/nbt992. [DOI] [PubMed] [Google Scholar]
- 30.Mun J-H, Yu H-J, Park S, Park B-S. Genome-wide identification of NBS-encoding resistance genes in Brassica rapa. Mol Gen Genomics. 2009;282(6):617. doi: 10.1007/s00438-009-0492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sagi MS, Deokar AA, Tar’an B. Genetic analysis of NBS-LRR gene family in chickpea and their expression profiles in response to Ascochyta blight infection. Front Plant Sci. 2017;8:838. doi: 10.3389/fpls.2017.00838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang L, Liu J, Wu C, Deng Y, Cai C, Zhang X, Cai Y. Genome-wide comparative analysis of NBS-encoding genes in four Gossypium species. BMC Genomics. 2017;18(1):292. doi: 10.1186/s12864-017-3682-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitsui Y, Shimomura M, Komatsu K, Namiki N, Shibata-Hatta M, Imai M, Katayose Y, Mukai Y, Kanamori H, Kurita K. The radish genome and comprehensive gene expression profile of tuberous root formation and development. Sci Rep. 2015;5(1):1–14. doi: 10.1038/srep10835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeong Y-M, Kim N, Ahn BO, Oh M, Chung W-H, Chung H, Jeong S, Lim K-B, Hwang Y-J, Kim G-B. Elucidating the triplicated ancestral genome structure of radish based on chromosome-level comparison with the Brassica genomes. Theor Appl Genet. 2016;129(7):1357–1372. doi: 10.1007/s00122-016-2708-0. [DOI] [PubMed] [Google Scholar]
- 35.Richly E, Kurth J, Leister D. Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol Biol Evol. 2002;19(1):76–84. doi: 10.1093/oxfordjournals.molbev.a003984. [DOI] [PubMed] [Google Scholar]
- 36.Schranz ME, Lysak MA, Mitchell-Olds T. The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 2006;11(11):535–542. doi: 10.1016/j.tplants.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 37.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z, Xie J, Wang H, Zhong X, Li H, Yu J, Kang J. Identification and expression profiling analysis of NBS–LRR genes involved in Fusarium oxysporum f. sp. conglutinans resistance in cabbage. 3 Biotech. 2019;9(5):202. doi: 10.1007/s13205-019-1714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng F, Wu H, Zhang R, Li S, He W, Wong F-L, Li G, Zhao S, Lam H-M. Molecular phylogeny and dynamic evolution of disease resistance genes in the legume family. BMC Genomics. 2016;17(1):402. doi: 10.1186/s12864-016-2736-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDowell JM, Woffenden BJ. Plant disease resistance genes: recent insights and potential applications. Trends Biotechnol. 2003;21(4):178–183. doi: 10.1016/S0167-7799(03)00053-2. [DOI] [PubMed] [Google Scholar]
- 41.Friedman AR, Baker BJ. The evolution of resistance genes in multi-protein plant resistance systems. Curr Opin Genet Dev. 2007;17(6):493–499. doi: 10.1016/j.gde.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 42.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4(1):10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y-W, Lai K-N, Tai P-Y, Li W-H. Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J Mol Evol. 1999;48(5):597–604. doi: 10.1007/PL00006502. [DOI] [PubMed] [Google Scholar]
- 44.Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol Biol Evol. 2000;17(10):1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- 45.Panchy N, Lehti-Shiu M, Shiu S-H. Evolution of gene duplication in plants. Plant Physiol. 2016;171(4):2294–2316. doi: 10.1104/pp.16.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Magadum S, Banerjee U, Murugan P, Gangapur D, Ravikesavan R. Gene duplication as a major force in evolution. J Genet. 2013;92(1):155–161. doi: 10.1007/s12041-013-0212-8. [DOI] [PubMed] [Google Scholar]
- 47.Ponting CP, Russell RR. The natural history of protein domains. Annu Rev Biophys Biomol Struct. 2002;31(1):45–71. doi: 10.1146/annurev.biophys.31.082901.134314. [DOI] [PubMed] [Google Scholar]
- 48.Sun Q-L, Zhou H-J, Lin K. The evolutionary relationship of the domain architectures in the RhoGEF-containing proteins. Genomics, proteomics & bioinformatics. 2005;3(2):94–106. doi: 10.1016/S1672-0229(05)03014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo H-S, Zhang Y-M, Sun X-Q, Li M-M, Hang Y-Y, Xue J-Y. Evolution of the KCS gene family in plants: the history of gene duplication, sub/neofunctionalization and redundancy. Mol Gen Genomics. 2016;291(2):739–752. doi: 10.1007/s00438-015-1142-3. [DOI] [PubMed] [Google Scholar]
- 50.Erdmann R, Gramzow L, Melzer R, Theißen G, Becker A. GORDITA (AGL63) is a young paralog of the Arabidopsis thaliana Bsister MADS box gene ABS (TT16) that has undergone neofunctionalization. Plant J. 2010;63(6):914–924. doi: 10.1111/j.1365-313X.2010.04290.x. [DOI] [PubMed] [Google Scholar]
- 51.McHale L, Tan X, Koehl P, Michelmore RW. Plant NBS-LRR proteins: adaptable guards. Genome Biol. 2006;7(4):212. doi: 10.1186/gb-2006-7-4-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR–encoding genes in Arabidopsis. Plant Cell. 2003;15(4):809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu X, Kang DH, Choi SR, Ma Y, Lu L, Oh SH, Chhapekar SS, Lim YP. Isolation and characterization of fusarium wilt resistance gene analogs in radish. 3 Biotech. 2018;8(5):255. doi: 10.1007/s13205-018-1279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu X, Lu L, Ma Y, Chhapekar SS, Yi SY, Lim YP, Choi SR. Fine-mapping of a major QTL (Fwr1) for fusarium wilt resistance in radish. Theor Appl Genet. 2019:1–12. [DOI] [PubMed]
- 55.Chang A, Lamara M, Wei Y, Hu H, Parkin IA, Gossen BD, Peng G, Yu F. Clubroot resistance gene Rcr6 in Brassica nigra resides in a genomic region homologous to chromosome A08 in B. rapa. BMC plant biology. 2019;19(1):224. doi: 10.1186/s12870-019-1844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic acids research. 2011;39(suppl_2):W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. Clustal W and Clustal X version 2.0. bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 58.Lozano R, Hamblin MT, Prochnik S, Jannink J-L. Identification and distribution of the NBS-LRR gene family in the cassava genome. BMC Genomics. 2015;16(1):360. doi: 10.1186/s12864-015-1554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bioinformatics Q: CLC Main workbench. In: Release; 2014.
- 60.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47(D1):D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015;43(D1):D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bailey TL, Johnson J, Grant CE, Noble WS. The MEME suite. Nucleic Acids Res. 2015;43(W1):W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McDonnell AV, Jiang T, Keating AE, Berger B. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics. 2006;22(3):356–358. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- 64.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18(5):691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 67.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X. Lee T-h, Jin H, Marler B, Guo H: MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49–9. [DOI] [PMC free article] [PubMed]
- 69.Chen C, Chen H, He Y, Xia R. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv. 2018;289660.
- 70.Gu Z, Cavalcanti A, Chen F-C, Bouman P, Li W-H. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol Biol Evol. 2002;19(3):256–262. doi: 10.1093/oxfordjournals.molbev.a004079. [DOI] [PubMed] [Google Scholar]
- 71.Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics, proteomics & bioinformatics. 2010;8(1):77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic acids research. 2009;37(suppl_2):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu J, Zhao M, Wang X, Tong C, Huang S, Tehrim S, Liu Y, Hua W, Liu S. Bolbase: a comprehensive genomics database for Brassica oleracea. BMC Genomics. 2013;14(1):664. doi: 10.1186/1471-2164-14-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baik S-Y, Jang K-S, Choi Y-H, Kim J-C, Choi G-J. Resistance degree of radish cultivars to Fusarium oxysporum f. sp. raphani according to several conditions. Horticultural Science & Technology. 2011;29(1):48–52. [Google Scholar]
- 75.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 76.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Information regarding the radish NBS-LRR genes. Table S2. Clusters of radish NBS-LRR genes. Table S3 Predicted promoter elements of NBS-encoding genes in the radish genome. Table S4.Analysis and distribution of conserved motifs in radish CNL and TNL proteins. Table S5. MEME analysis of the CNL and TNL proteins. Table S6. Tandem arrays of the radish NBS-LRR genes. Table S7. Segmentally duplicated radish NBS-LRR genes. Table S8. Ka/Ks values for the segmentally duplicated genes in the radish genome. Table S9. Segmentally duplicated NBS-LRR genes in the radish/A. thaliana, radish/B. oleracea, and radish/B. rapa genomes. Table S10. Common segmentally duplicated NBS-LRR genes in the radish/A. thaliana, radish/B. oleracea, and radish/B. rapa genomes. Table S11. Ka/Ks values between radish and three related plant species. Table S12. The information for gene structure and domain change of radish/A. thaliana, radish/B. oleracea, and radish/B. rapa genomes. Table S13. The resistance of radish lines to F. oxysporum f. sp. Raphani 59. Table S14. The summary of the RNA-Seq database. Table S15. Details regarding the qRT-PCR primers. Table S16. The transcriptome data of the radish lines after inoculated with F. oxysporum f. sp. Raphani 59 (XLSX 15325 kb)
Additional file 2: Figure S1. Distribution of TNL, CNL, and partial genes. Distributions for each class are presented across the radish chromosomes. RUS, scaffold summary. Bars are divided into CNL genes (orange), TNL genes (blue), and partial genes (green)
Additional file 3: Figure S2. Phylogenetic tree representing the relationships of NBS-encoding genes between radish and arabidopsis. Different colored arcs represent the different groups (or subgroups) of NBS-encoding genes.
Additional file 4: Figure S3. Relative expression levels of RsCNL and RsTNL radish genes. The ‘YR4’ (resistant) and ‘YR18’ (susceptible) lines are represented by white and gray bars, respectively. The y-axis represents the relative gene expression levels, whereas the time-points (0, 1, 3, 6, 9, and 12 DAI) are presented on the x-axis.
Data Availability Statement
All datasets generated and analyzed during this study are included in this article and its additional files. The RNA-seq data generated during the current study have been deposited and publicly available at the National Center of Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/) repository under the accession number PRJNA643982.