Abstract
The perennial life style is a successful ecological strategy, and Arabis alpina is a recently developed model Brassicaceae species for studying it. One aspect, poorly investigated until today, concerns the differing patterns of allocation, storage, and metabolism of nutrients between perennials and annuals and the yet unknown signals that regulate this process. A. alpina has a complex lateral stem architecture with a proximal vegetative perennial (PZ) and a distal annual flowering zone (AZ) inside the same stems. Lipid bodies (LBs) with triacylglycerols (TAGs) accumulate in the PZ. To identify potential processes of lipid metabolism linked with the perennial lifestyle, we analyzed lipid species in the PZ versus AZ. Glycerolipid fractions, including neutral lipids with mainly TAGs, phospholipids, and glycolipids, were present at higher levels in the PZ as compared to AZ or roots. Concomitantly, contents of specific long‐chain and very long‐chain fatty acids increased during formation of the PZ. Corresponding gene expression data, gene ontology term enrichment, and correlation analysis with lipid species pinpoint glycerolipid‐related genes to be active during the development of the PZ. Possibilities that lipid metabolism genes may be targets of regulatory mechanisms specifying PZ differentiation in A. alpina are discussed.
Keywords: fatty acid, glycerolipid, lipid, perennial, secondary growth, triacylglycerol
Abbreviations
- AZ
annual zone
- FA
fatty acid
- GL
glycolipid
- LB
lipid body
- LCFA
long‐chain fatty acid
- NL
neutral lipid
- Paj
Pajares
- PL
phospholipid
- PZ
perennial zone
- TAG
triacylglycerol
- VLCFA
very long‐chain fatty acid
Highlights.
Glycerolipid amounts increase in the developing vegetative perennial stem zone.
Expression of corresponding lipid metabolism genes correlates with lipid species.
Lipid metabolism genes are potential targets for perennial–annual phase transition.
1. INTRODUCTION
Perennial plants survive and reproduce throughout many years, and in most natural settings the perennial life style is of advantage to secure a dominant position in the environment. Arabis alpina is a herbaceous hemicryptophyte with perennial vegetative branches above the soil (according to the Raunkiaer's system of plant life forms). This species has attracted attention as a model for studying its specific perennial characters. A. alpina is polycarpic and produces new flowers during every new growth season at the distal ends of the main shoot and lateral branches. In proximal vegetative parts of the stems, buds develop either into non‐flowering branches, remain as perennating buds, or develop into flowering lateral branches that maintain vegetative growth (Lazaro et al., 2018; Vayssiѐres et al., 2020). This complex architecture of A. alpina is established during and after vernalization, a required cold season that induces flowering. The Arabis clade belongs to the Brassicaceae family (Karl & Koch, 2013; Kiefer et al., 2017) and genome sequence information, techniques, and tools developed for Arabidopsis thaliana, and other Brassicaceae are employed to study Arabis perennial behavior. Flowering repressor PERPETUAL FLOWERING 1 (PEP1), the ortholog of the Arabidopsis FLOWERING LOCUS C (FLC), controls the flowering response and plant architecture (Albani et al., 2012; Hughes et al., 2019; Lazaro et al., 2018; Michaels & Amasino, 1999; Wang et al., 2009, 2011). pep1‐1 mutant plants continuously form new lateral branches irrespective of a cold season, each of them with proximal vegetative and distal reproductive character (Wang et al., 2009).
The vegetative part of the stem has secondary growth, coupled with high molecular weight carbon storage in a so‐called perennial zone (PZ). Carbon storage metabolites accumulate in the PZ in the form of starch and triacylglycerol (TAG) in cambium and cambium‐derived secondary phloem and xylem tissues (Sergeeva et al., 2020). These stored nutrients likely serve bud outgrowth in the following season. Starch accumulation and lipid body (LB) formation both occur in the secondary phloem and xylem parenchyma, however, there are also cambium‐derived tissues where these processes are spatially separated in the PZ, for example, LBs but not starch are present in secondary phloem and cambium (Sergeeva et al., 2020). In contrast to that, reproductive parts of the shoots show primary growth and after flowering and seed formation turn senescent, so‐called annual zone (AZ). Starch and LBs accumulate there in cortical parenchyma tissue (Sergeeva et al., 2020). Additionally, LBs but not starch accumulate in the AZ in fascicular cambium and phloem of the vascular bundles (Sergeeva et al., 2020). In the AZ, these high‐energy carbon‐rich nutrients are used during reproduction. Interestingly, the secondary growth and carbon storage property of A. alpina is found in the wild‐type Pajares (Paj) as well as the flowering mutant pep1‐1. This makes it very attractive to study this trait irrespective of vernalization requirement.
Lipids play key roles in plant metabolism and nutrient storage in plants. Lipid composition is a key for membrane identity. For example, the plasma membrane contains primarily phosphoglycerolipids (Simon, 1974; Wewer et al., 2011), while predominant constituents of the chloroplast thylakoid membranes are galactoglycerolipids (Hurlock et al., 2014; Karki et al., 2019; LaBrant et al., 2018). Besides that, lipids are carbon storage products in plants and animals (van Erp et al., 2014; Haslam et al., 2013; Li et al., 2015; Lu et al., 2011; Shi et al., 2012; Tan et al., 2011; Vanhercke et al., 2013). Plants store lipids mainly as TAG in LBs. TAG‐containing LBs are present in oilseeds of various plant species (Cai et al., 2017; Cao & Huang, 1986; Lacey et al., 1999; Lee et al., 1995; Loer & Herman, 1993; Shockey et al., 2006), but also in vegetative tissues, such as leaves (Brocard et al., 2017; Pyc et al., 2017), stems (Madey et al., 2002; Sauter & Cleve, 1994; Wang et al., 2007), and roots (Chinnasamy et al., 2003; Næsted et al., 2000). The biosynthesis pathways of different glycerolipid classes are interconnected in plants. Lipids are synthesized in the prokaryotic pathway in the chloroplast and in the eukaryotic pathway in the endoplasmic reticulum (ER) (Jayawardhane et al., 2018; Li‐Beisson et al., 2013). Biosynthesis of neutral lipid (NL) TAG takes place in the ER in the Kennedy or glycerol phosphate pathway (Li‐Beisson et al., 2013). Diacylglycerol functions as intermediate for TAG biosynthesis and as backbone for the biosynthesis of glycolipids (GLs) and phospholipids (PLs), phosphatidylcholine, and phosphatidylethanolamine. Diacylglycerol is therefore an important intermediate for the synthesis of storage and membrane lipids. Apart from the de novo synthesized diacylglycerol, characterized by saturated and monounsaturated acyl compositions, TAG can be synthesized from diacylglycerol deriving from phosphatidylcholine (Bates, 2016). Thus, additional desaturation of acyl chains esterified to phosphatidylcholine in the course of the acyl‐editing cycle results in a phosphatidylcholine‐derived diacylglycerol and, subsequently, TAG pool with an elevated degree of unsaturated fatty acids (FAs) (Bates & Browse, 2012; Bates et al., 2009; Griffiths et al., 1988). Phosphatidylcholine‐derived diacylglycerol represents the major source for TAG biosynthesis in diverse oilseed tissues (Bates & Browse, 2011; Bates et al., 2009; Yang et al., 2017).
GLs, representative for plastidic membranes, are produced in the prokaryotic and eukaryotic pathways in the plastid and ER, respectively. In addition, phosphatidylglycerol can be synthesized in the plastid in a series of sequential reactions. Phosphatidic acid is the intermediate for the biosynthesis of above‐mentioned GLs and also of PLs, which are produced in the ER. PLs mainly represent structural elements of extraplastidic membranes. The nature of FAs of glycerolipids is indicative for their function. De novo FA synthesis takes place in the plastid, and after their transport to the ER, FAs may be elongated to yield very long‐chain fatty acids (VLCFAs) (Joubès et al., 2008).
Here, we study a new aspect of A. alpina stem architecture related to lipid metabolism. The simultaneous presence of PZ and AZ poses the conflict of nutrient allocation and usage for either seed production or development of perennating storage tissues. This raises the question which genes and signals regulate this process. We compared glycerolipid profiles between PZ and AZ and found that they differed between PZ and AZ. Using transcriptome data, we identified differentially regulated lipid metabolism genes in the PZ and AZ, irrespective of flowering control by PEP1. Lipid metabolism gene ontology (GO) terms were enriched during development of the PZ. Expression of lipid metabolism‐related genes correlated with levels of lipid species in the PZ. These findings serve to investigate the regulation of TAG storage and lipid metabolism as a perennial trait in a Brassicaceae species and study the potential differentiation signals in the PZ.
2. MATERIALS AND METHODS
2.1. Lipid analysis of plant material
Freeze‐dried plant samples from different developmental stages of the PZ and AZ of A. alpina wild‐type Paj and its mutant‐derivative perpetual flowering 1‐1 (pep1‐1) were used for lipid analysis (Figure 1; Sergeeva et al., 2020). At first, total amounts of glycerolipids were extracted using an acidic modified chloroform‐methanol method (Hajra, 1974; https://www.imbio.uni‐bonn.de/molekulare‐biotechnologie/lipidomics‐platform/acidic‐chloroform‐methanol; Wewer et al., 2011), (Figure S1). 10 mg of lyophilized, and ground (fine powdered in a Mixer Mill MM 200, Retsch, Haan, Germany) plant material was vortexed with 0.4 ml chloroform/methanol/formic acid (1:1:0.1) and 0.2 ml solution containing 0.2 M phosphoric acid and 1 M potassium chloride. After 5‐min centrifugation at 2,380 g, the resulting lower phase with glycerolipids was transferred to a new glass tube. Chloroform extractions were repeated twice and bottom phases combined as glycerolipid fraction, which included nonpolar and polar lipids.
FIGURE 1.
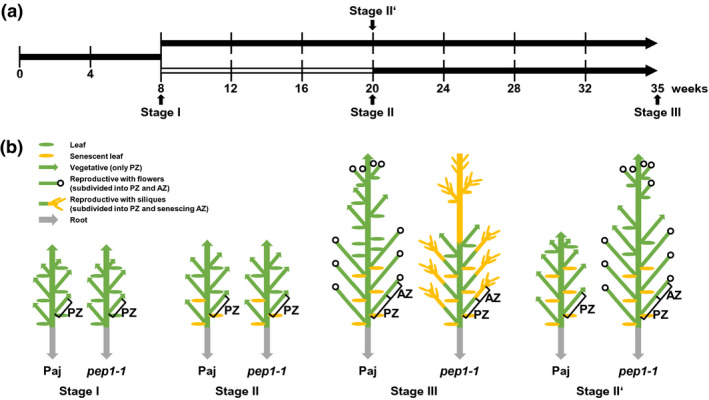
Schematic representation of plant growth stages for A. alpina Pajares (Paj, wild type) and its perpetual flowering 1‐1 (pep1‐1) mutant derivative used in lipid analysis. (a) Plant growth and harvesting scheme. Solid black lines, long‐day conditions at 20°C; open line, short‐day conditions at 4°C (vernalization); four developmental stages for harvesting, I, II, III, and II′. (b) Schematic representation of Paj and pep1‐1 architecture of lateral stems at stages I, II, III, and II′. Lateral stems were harvested, subdivided into perennial (PZ), and annual (AZ) zones, as indicated (see also Sergeeva et al., 2020). In addition to lateral stems, entire root systems of single plants were harvested as one root sample. The schematic plant representation was partially adopted from Wang et al. (2009), Lazaro et al. (2018), and Vayssiѐres et al. (2020)
Separation into nonpolar NLs and polar lipids containing GLs and PLs was performed using silica‐based solid‐phase extraction columns (55 µm, 70 Å) (Phenomenex) (Figure S1) (Wewer et al., 2011), equilibrated with 1 ml chloroform. Glycerolipid fractions were applied to the column after equilibration and the flow through collected in a new glass tube. The NL fraction was eluted into the same glass tube with 1 ml chloroform. One milliliter of acetone/isopropanol (1:1) was added to the same column to elute the GL fraction, which was collected in a second glass tube. The flow through containing the PL fraction eluted by addition of 1 ml methanol was retrieved in a third tube. Finally, organic solvents were evaporated from the tubes in a desiccator coupled to a vacuum pump.
Resulting NL, GL, and PL fractions were immediately applied to prepare fatty acid methylesters (FAMEs) according to Hielscher et al. (2017) (Figure S1), using heptadecanoic acid (C17) (1 mg/ml) as internal standard. One milliliter of 3 N methanolic hydrochloric acid and 20 µl of the C17 internal standard were added to each sample. The samples were incubated for 60 min at 90°C, cooled down at room temperature for 5 min. One ml n‐hexane as well as 1 ml 1% (w/v) sodium chloride were added. Tubes were vortexed for 10 s and centrifuged for 5 min at 753 g for phase separation. The upper hexane phase was transferred to a GC vial and stored at −20°C until further analysis.
Analysis of FAME extracts was performed by gas chromatography‐mass spectrometry (GC‐MS) using a 7200‐GC‐quadrupole time‐of‐flight mass spectrometer (GC‐QTOF) (Agilent Technologies) (Figure S1). Mass Hunter Software (Agilent Technologies) was used for integration of the resulting peaks. Absolute amounts of FAs in each sample were determined by relating the integrated peak area of detected FAs to the integrated peak area and known concentration of the C17 standard.
2.2. Gene expression analysis of lipid metabolism genes
The RNA‐seq dataset consisting of three biological replicates per sample that had been described (Sergeeva et al., 2020) was used to extract RNA‐seq gene expression data corresponding to lipid metabolism (Table S1; derived from the full transcriptome data set available under the GEO no. GSE152417). To identify enriched lipid metabolism GO terms (Tables S2–S7), GO term enrichment analysis was performed using topGO (Alexa & Rahnenfuhrer, 2010) with the closest A. thaliana ortholog Locus IDs (AGI code). The latest publicly available A. thaliana GO annotations (go_ensembl_arabidopsis_thaliana.gaf; downloaded from Gramene) were used. Fisher's exact test was applied to identify significantly enriched GO terms with p < .05. GO terms “NL metabolic process”, “galactolipid metabolic process”, “phosphatidylglycerol metabolic process”, “PL biosynthetic process”, “FA biosynthetic process”, and “FA catabolic process” (Tables S2 and S3) were screened for genes that were described to be involved in NL, GL, PL, and FA biosynthesis and catabolism using “GO Annotations” function of “The Arabidopsis Information Resource” web page (https://www.arabidopsis.org/index.jsp).
Validation of lipid metabolism gene expression was performed by reverse transcription‐qPCR using the cDNA samples and procedures as described (Sergeeva et al., 2020) and qPCR primers listed (Table S8). Absolute quantification of normalized gene expression was obtained using mass standard curve analysis (Ben Abdallah & Bauer, 2016) and normalization with reference genes CLATHRIN ADAPTOR COMPLEX MEDIUM SUBUNIT (CAC) and EUKARYOTIC TRANSLATION INITIATION FACTOR 4A1 (EIF4a) (Stephan et al., 2019).
2.3. Statistical and correlation analysis
One‐way analysis of variance (ANOVA) combined with Tukey's honest significant difference (HSD) test (α = 0.05) was performed using the R software. Different letters in the diagrams represent significant differences with p < .05.
Pearson correlation analysis was conducted via the R software, between individual gene expression and biochemical lipid analysis data, using stages III_PZ, IV_PZ, stage I and stage II’ for PZ, and stage IV_AZ_if and stage II’ for AZ, respectively. Only correlation coefficients with significant levels of correlation (p < .05) are displayed in the diagrams.
3. RESULTS
3.1. Levels of glycerolipids increase during development of the PZ
The PZ differentiates during development to form secondary growth tissues, in which LBs and TAGs accumulate, for example, for lipid storage (Sergeeva et al., 2020). The identity of lipids and TAGs had not been further elucidated previously. Besides TAGs, there are two further glycerolipid types, GLs and PLs, that are linked with lipid storage and metabolism in plants. These latter form cell and organellar membranes and membranous compartments for nutrient storage. Here, we present data for sub‐fractionated glycerolipid fractions corresponding to NLs, GLs, and PLs and their proportion as compared to total lipid amounts. Plant samples were lateral stem internodes divided into PZ and AZ and for comparison roots of A. alpina wild‐type Paj and mutant pep1‐1. These samples had been harvested along a developmental gradient at different growth stages during differentiation of PZ and AZ, namely, stages I, II, III, and II’ (for descriptions, see Figure 1). pep1‐1 does not require vernalization for triggering flowering, and Paj and pep1‐1 differ in the timing but not the outcome of PZ and AZ formation (Figure 1; Sergeeva et al., 2020).
NL, GL, and PL fractions were present at all stages in stems and roots of Paj and pep1‐1 (Figure 2) (for corresponding FA composition, see Figures S2 and S3). They were highest in the PZ at stages III and II’ in Paj and pep1‐1, and lower in PZ at stage I in Paj and stage II in pep1‐1 (Figure 2a,b). Together, the three glycerolipid fractions reached about 60% of total lipids in the PZ of Paj and even 80% or more in pep1‐1. On the other hand, the AZ of Paj and pep1‐1 contained lowest amounts of all three glycerolipid fractions. These findings point to a significant increase in storage‐related lipid forms in the PZ. In roots, on the other hand, NL, GL, and PL fractions remained constant and significant differences in their contents were not noted during the development at the same stages of Paj and pep1‐1 (Figure 2c,d). Glycerolipid fractions together did not exceed 30%–35% of all lipids in roots, which was lower than in most PZ stem samples.
FIGURE 2.
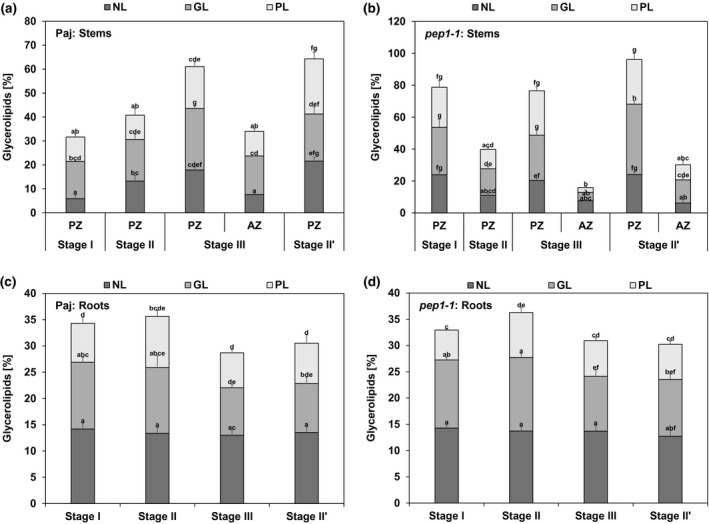
Glycerolipid accumulation in stems and roots. The three glycerolipid fractions are neutral lipids (NL), glycolipids (GL), and phospholipids (PL), represented as percentage of their fatty acids (FAs) in relation to total FA contents (as 100%) of dry matter of (a, b) perennial and annual internode stem zones (PZ, AZ) and (c, d) entire root systems of (a, c) A. alpina Pajares (Paj, wild type) and (b, d) perpetual flowering 1‐1 (pep1‐1) mutant derivative. Plant material was harvested at four different developmental stages, stage I, II, III, and II′, as indicated in Figure 1a. Data are represented as mean ± SD (n = 3–7). Different letters indicate statistically significant differences, determined by one‐way ANOVA‐Tukey‘s HSD test (p < .05)
Taken together, these data show the highest amounts of glycerolipids in developed PZ of lateral stems rather than roots or the AZ, suggesting the importance of lipid metabolism and lipid storage in the PZ. Moreover, Paj and pep1‐1 were similar in this respect showing that glycerolipid accumulation occurs irrespective of vernalization and flowering control by PEP1.
3.2. NLs with long‐chain and very long‐chain fatty acids increase during development of the PZ
The nature of FAs contained in glycerolipids is important to estimate their function. Storage lipids contained in glycerolipids often contain long‐chain FAs (LCFAs), comprising predominantly C16 and C18 FAs (palmitic (16:0), linoleic (18:2), and linolenic (18:3) acids), and VLCFAs, including C20, C22, and C24 FAs (arachidic (20:0), behenic (22:0), and lignoceric (24:0) acids).
At first, FAs present in the NL fraction of each investigated stage and line were therefore subdivided into the sum of total LCFAs (total of 14:0, 16:0, 16:1, 18:2, 18:3, and 18:1 FAs), and the sum of total VLCFAs (total of 20:0, 22:1, 22:0, 24:1, and 24:0 FAs) (Figure 3). Sums of total LCFAs were higher than those of total VLCFAs in all NL stem samples of Paj and pep1‐1. Generally, amounts of total LCFAs and total VLCFAs were higher in PZ than in AZ samples. Interestingly, contents of total VLCFAs increased in PZ with the progression of the development and reached highest levels at stage II’ (Figure 3a,b). Levels of total LCFAs and total VLCFAs in the NL fraction were not significantly differing from each other at every single investigated developmental stage in roots of Paj and pep1‐1 (Figure 3c,d). Contrary to stems, contents of total LCFAs and total VLCFAs in roots did not change during root development in both lines.
FIGURE 3.
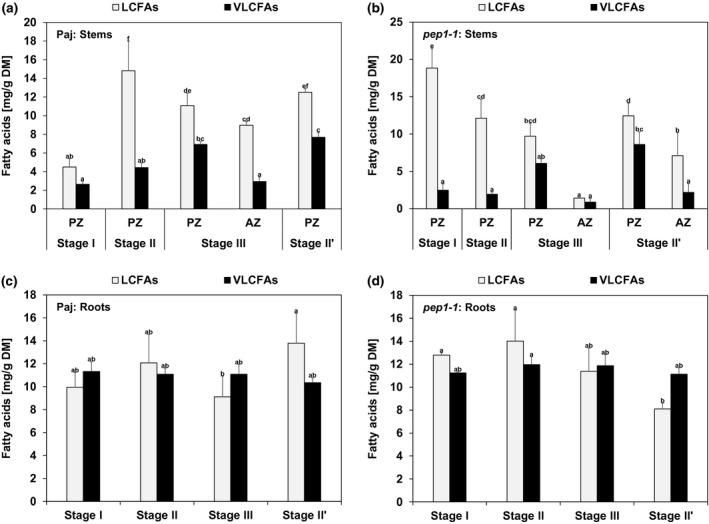
Long‐chain and very long‐chain fatty acid contents (LCFAs, VLCFAs) in neutral lipid (NL) fractions in stems and roots. Represented are the contents of NL fraction‐derived total LCFAs (total of 14:0, 16:0, 16:1, 18:2, 18:3, 18:1) and total VLCFAs (total of 20:0, 22:1, 22:0, 24:1, 24:0) per dry matter (DM) of (a, b) perennial (PZ) and annual (AZ) internode stem zones and (c, d) entire root systems of (a, c) A. alpina Pajares (Paj, wild type) and (b, d) perpetual flowering 1‐1 (pep1‐1) mutant derivative. Plant material was harvested at four different developmental stages, stage I, II, III, and II′, as indicated in Figure 1a. Data are represented as mean ± SD (n = 3–7). Different letters indicate statistically significant differences, determined by one‐way ANOVA‐Tukey's HSD test (p < .05)
Next, we investigated individual LCFAs and VLCFAs in the NL fractions. In NLs of both genotypes, 16:0 palmitic acid was the most abundant FA at examined stages, while in the same samples 14:0, 16:1, 18:1, 22:1, and 24:1 FAs were not abundant or varied significantly (Figure 4a,b). However, contents of 18:2 and 18:3 FAs along with VLCFAs 20:0, 22:0, and 24:0 increased markedly with progression of PZ development in both genotypes and only slightly during development of the AZ. Therefore, the increase in LCFAs 18:2 and 18:3 and in VLCFAs 20:0, 22:0, and 24:0 in PZ supports the storage character of the PZ and the relevance of NLs (Figure 4a,b). Interestingly, in pep1‐1 LCFAs 18:2 and 18:3 peaked at stage I and II in the PZ, but then declined (Figure 4b). Perhaps this decline is caused by catabolic activities during seed formation. GL and PL fractions of Paj and pep1‐1 stems, PZ and AZ, were mainly composed of LCFAs 16:0, 18:2, and 18:3 at every investigated developmental stage (Figure S4). Contrary to the NL fraction, amounts of VLCFAs, such as 20:0, 22:0, and 24:0, were low in the GL and PL fractions (Figure S4). Interestingly, the amounts of 18:2 and 18:3 FAs in the PL fraction of Paj increased slightly from stage I to stage III and II’ (Figure S4b), perhaps partially related to incorporation of these FAs in the PL monolayer of LBs that accumulate in the PZ. In roots, NLs of Paj and pep1‐1 were predominantly composed of LCFAs, 16:0, 18:2, and 18:3, and VLCFAs, 20:0, 22:0, and 24:0, that did not significantly vary between stages (Figure 4c,d). Differing from stems, 16:0 and 20:0 FAs represented the most abundant FAs at every investigated developmental stage in the roots of Paj and pep1‐1 (Figure 4c,d). The absence of a regulatory pattern of NL FAs across the root stages suggests again that roots do not develop as primary lipid storage organs. The GL fractions of the roots of Paj and pep1‐1 plants were marked by high contents of LCFAs 16:0, 18:2, and 18:3 and relatively large amounts of VLCFAs, 20:0, 22:0, and 24:0 (Figure S5a,c). The PL fractions of Paj and pep1‐1 roots had also high levels of LCFAs 16:0, 18:2, and 18:3, but low contents of all other investigated FAs (Figure S5b,d). Only slight variations in FA contents were noted for the PL and GL fractions in root samples across stages. Again, the high amount of LCFAs 16:0, 18:2, and 18:3 in the PL fraction may reflect their accumulation in the PL monolayer of LBs.
FIGURE 4.
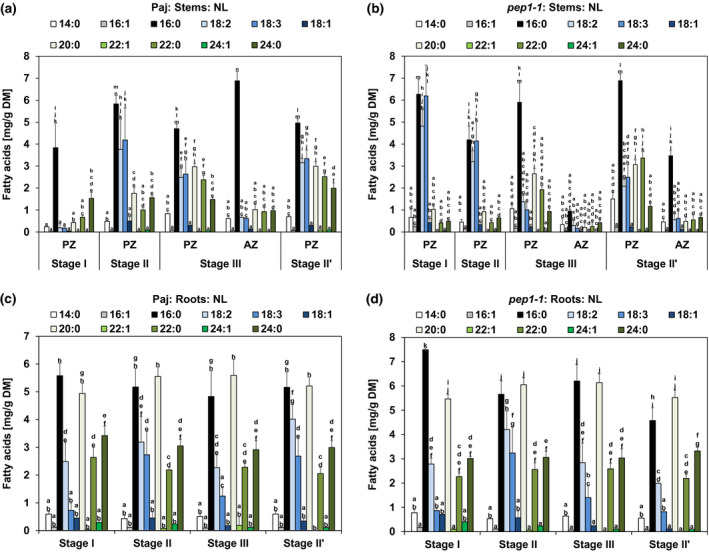
Long‐chain and very long‐chain fatty acid (FA) contents in neutral lipid (NL) fractions in stems and roots. Represented are the contents of NL fraction‐derived individual LCFAs (14:0, 16:1, 16:0, 18:2, 18:3, 18:1) and individual VLCFAs (20:0, 22:1, 22:0, 24:1, 24:0) per dry matter (DM) of (a, b) perennial (PZ) and annual (AZ) internode stem zones and (c, d) entire root systems of (a, c) A. alpina Pajares (Paj, wild type) and (b, d) perpetual flowering 1‐1 (pep1‐1) mutant derivative. Plant material was harvested at four different developmental stages, stage I, II, III, and II′, as indicated in Figure 1a. In addition, the PZ of pep1‐1 contained low contents of other FAs (compare with b), namely, 16:3 FA at stages I and II (0.03 mg/g DM) and of 20:1 FA at stages I and III (0.07 mg/g DM and 0.54 mg/g DM). For comparable representation, these FAs are not shown in the diagram, but were included in all calculations. Data are represented as mean ± SD (n = 3–7). Different letters indicate statistically significant differences, determined by one‐way ANOVA‐Tukey's HSD test (p < .05)
In summary, the FA profiles of glycerolipids show that the PZ is the primary place for lipid metabolic processes, while AZ and roots play minor roles in this. Similar patterns in Paj and pep1‐1 indicate that these processes occur irrespective of vernalization and flowering control by PEP1.
3.3. GO term enrichment and lipid metabolism gene expression confirm enhanced lipid metabolism during progression of the PZ
Enriched lipid metabolism GO terms in RNA‐seq data reflect importance of the respective pathways. To identify such, we examined GO term enrichment in three consecutive juvenile to adult pep1‐1 stages I_PZ (very early stage, prior to secondary growth), III_PZ, and IV_PZ in comparison with inflorescence stage IV_AZ_if, for which RNA‐seq‐based transcriptome data are available (Figure 5a, see also Sergeeva et al., 2020). We selected GO terms related to lipid metabolism (Figure 5b, Tables S1 and S2–S7). pep1‐1 was used for transcriptome analysis because PZ and AZ differentiation is present in lateral stems and the development of PZ is predictable in pep1‐1 irrespective of vernalization (Sergeeva et al., 2020).
FIGURE 5.
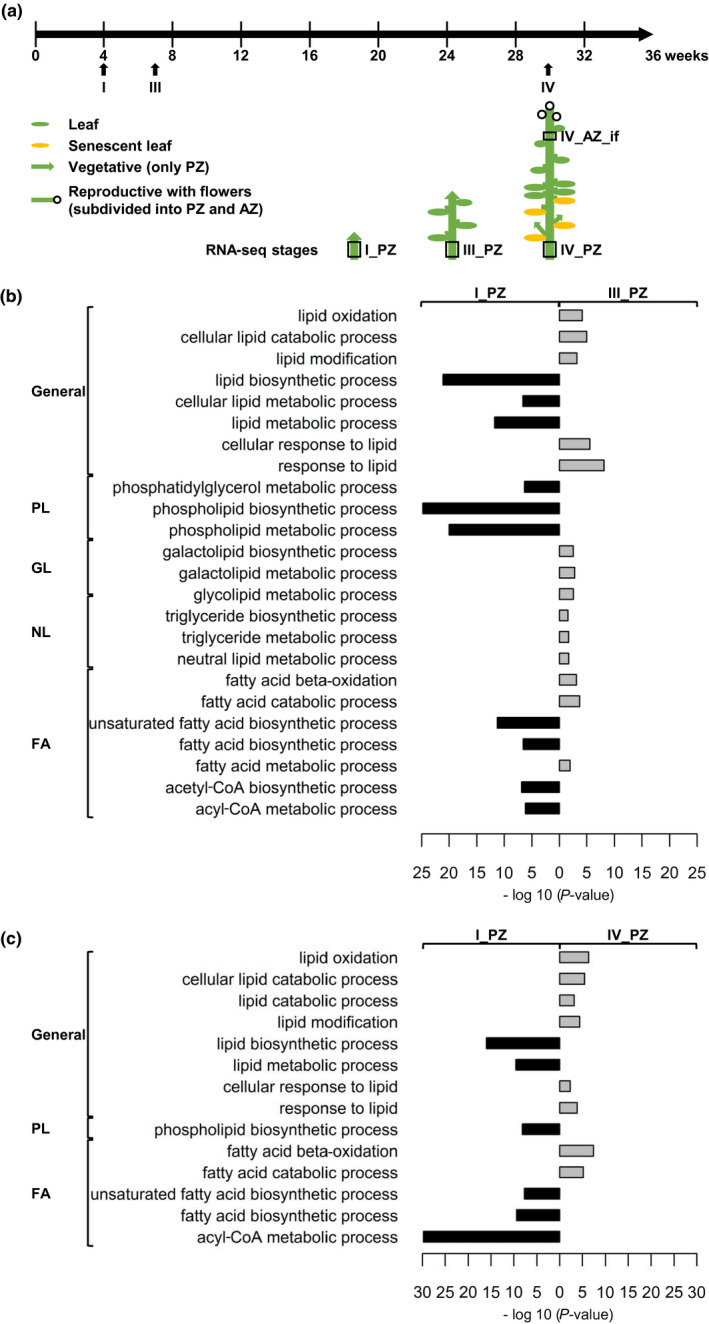
Lipid metabolism‐related GO term enrichment analysis of RNA‐seq data. (a) Plant growth and harvesting scheme for A. alpina perpetual flowering 1‐1 (pep1‐1) mutant with schematic representation of pep1‐1 architecture of the first and second lateral stems formed at lower internodes of the main stem at different stages. Solid black line, long‐day conditions at 20°C; three developmental RNA‐seq stages were considered, stages I_PZ, III_PZ, and IV_PZ as well as IV_AZ_if (see also Sergeeva et al., 2020). Lateral stem internodes had been subdivided into perennial (PZ) and annual (AZ) zones, as indicated. The schematic plant representation was partially adopted from Wang et al. (2009), Lazaro et al. (2018), and Vayssiѐres et al. (2020). (b, c) Lipid metabolism‐related GO terms enriched in (b) stage I_PZ (represented in black) versus stage III_PZ (represented in grey) and in (c) stage I_PZ (represented in black) versus stage IV_PZ (represented in grey). GO terms were assigned to the different categories, general, phospholipids (PL), glycolipids (GL), neutral lipids (NL), and fatty acids (FA). GO terms with a p‐value < .05 are enriched; P‐values, represented as −log 10 values. Further information about the represented GO term enrichment analysis is provided in Tables S2 and S3
When comparing the stages I_PZ with III_PZ, lipid biosynthesis GO term enrichment was pronounced at stage I_PZ, while this was the case for lipid catabolism at stage III_PZ (Figure 5b, Table S2). Interestingly, stage I_PZ showed an enrichment of PL and FA biosynthetic GO terms, while stage III_PZ was enriched in NL and GL biosynthetic GO terms. When comparing stages I_PZ and IV_PZ, general lipid metabolism, PL and FA‐related GO terms were similarly enriched but this was no longer the case for GL and NL categories (Figure 5c, Table S3). In general, all three single comparisons between IV_AZ_if and each PZ stage were marked by an enrichment of PL and FA biosynthetic GO terms in the AZ (Tables S2–S7). The enrichment of PL and FA biosynthetic GO terms in the AZ internodes indicates that lipid‐related metabolic processes are rather similar to those occurring in the early‐stage I_PZ. As we were seeking for differences between the PZ and AZ development, we concentrated therefore on the GO terms that were enriched during the PZ development.
We selected and compared the genes from NL, GL, PL, and FA‐related GO categories. These 50 genes have direct functions in lipid metabolism, as described in the corresponding literature available to date (Figure 6 and Table S1). Nineteen of them were first, up‐regulated during PZ progression from stage I_PZ to stages III_PZ and/or IV_PZ and, second, up‐regulated at either stage III_PZ or stage_IV_PZ or both versus stage IV_AZ_if (Figure 6, genes marked in red; Table S1; Figure S6). In the GO category, “NL metabolic process” (Table S2) LYSOPHOSPHATIDYLCHOLINE ACYLTRANSFERASE 1 (LPCAT1), WRINKLED 1 (WRI1), PHYTYL ESTER SYNTHASE 1 (PES1), DAD1‐LIKE SEEDLING ESTABLISHMENT‐RELATED LIPASE (DSEL), and SUGAR‐DEPENDENT 1 (SDP1) fulfilled these criteria (Figure 6a and Figure S6a‐e). All five genes were significantly up‐regulated from stage I_PZ to stage III_PZ. Expression of LPCAT1, PES1, DSEL, and SDP1 increased from stage I_PZ to stage IV_PZ (Figure 6a and Figure S6a,c,d,e). The five genes were less expressed at stage IV_AZ_if versus stages III and/or IV_PZ (Figure 6a and Figure S6a‐e). Among the examined GL genes, all assigned to GO term “GL metabolic process” (Table S2), PHOSPHOLIPASE D ζ 2 (PLDζ2), PATATIN‐RELATED PHOSPHOLIPASE A IIIβ (pPLAIIIβ), DIGALACTOSYL DIACYLGLYCEROL DEFICIENT 1 (DGD1), and PLASTID LIPASE 3 (PLIP3) fulfilled the above criteria (Figure 6a and Figure S6f‐I). PLDζ2, pPLAIIIβ, and PLIP3 were significantly up‐regulated from stage I_PZ to stage III_PZ (Figure 6a and Figure S6f,g,i). Expression of DGD1 increased from stage I_PZ to stage IV_PZ, where it was highest (Figure 6a and Figure S6h). For the four genes, expression was lower at stage IV_AZ_if versus stages III and/or IV_PZ (Figure 6a and Figure S6f‐i). Among the PL‐related genes, we examined closely genes of the GO terms “phosphatidylglycerol metabolic process” and “PL biosynthetic process” (Tables S2 and S3). Only a single gene, ACYLTRANSFERASE 1 (ACT1), fulfilled the criteria, as it was significantly up‐regulated from stage I_PZ to stage IV_PZ, but down‐regulated in stage IV_AZ_if (Figure 6a and Figure S6j). Among FA‐related genes of GO terms “FA biosynthetic process” (Tables S2 and S3) and “FA catabolic process” (Tables S2 and S3), nine genes fulfilled the criteria, 3‐KETOACYL‐COA SYNTHASE 4 (KCS4), ACYL‐COA OXIDASE 3 (ACX3), Δ3, Δ2‐ENOYL COA ISOMERASE 1 (ECI1), 3‐KETOACYL‐COA THIOLASE 2 (KAT2), PEROXISOMAL ADENINE NUCLEOTIDE CARRIER 1 (PNC1), MULTIFUNCTIONAL PROTEIN 2 (MFP2), ACYL‐COA OXIDASE 4 (ACX4), ENOYL‐COA HYDRATASE 2 (ECH2), and ABNORMAL INFLORESCENCE MERISTEM 1 (AIM1) (Figure 6b and Figure S6k‐s). Expression of KCS4 decreased from stage I_PZ to stages III_PZ and IV_PZ and was lowest at stage IV_AZ_if (Figure 6b and Figure S6k). ACX3, ECI1, KAT2, PNC1, MFP2, ACX4, ECH2, and AIM1 were significantly up‐regulated from stage I_PZ to stage III and/or IV_PZ and down‐regulated at stage IV_AZ_if (Figure 6b and Figure S6l‐s).
FIGURE 6.
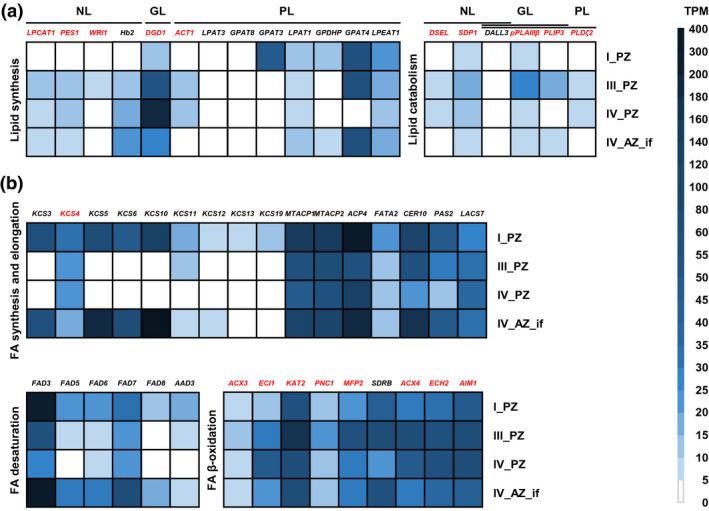
Gene expression profiles of 50 individual genes out of the NL, GL, PL, and FA‐related GO categories. Represented are RNA‐seq data (TPM, transcripts per million) of A. alpina perpetual flowering 1‐1 (pep1‐1) mutant lateral stem internodes of perennial (PZ) and annual (AZ) zones of RNA‐seq at stages I_PZ, III_PZ, IV_PZ, and IV_AZ_if (see Figure 5a). (a) NL, GL, and PL genes involved in lipid synthesis and catabolism: LPCAT1, LYSOPHOSPHATIDYLCHOLINE ACYLTRANSFERASE 1, PES1, PHYTYL ESTER SYNTHASE 1, WRI1, WRINKLED 1, Hb2, HAEMOGLOBIN 2, DGD1, DIGALACTOSYL DIACYLGLYCEROL DEFICIENT 1, ACT1, ACYLTRANSFERASE 1, LPAT3, LYSOPHOSPHATIDYL ACYLTRANSFERASE 3, GPAT8, GLYCEROL‐3‐PHOSPHATE SN‐2‐ACYLTRANSFERASE 8, GPAT3, GLYCEROL‐3‐PHOSPHATE SN‐2‐ACYLTRANSFERASE 3, LPAT1, LYSOPHOSPHATIDIC ACID ACYLTRANSFERASE 1, GPDHP, GLYCEROL‐3‐PHOSPHATE DEHYDROGENASE PLASTIDIC, GPAT4, GLYCEROL‐3‐PHOSPHATE SN‐2‐ACYLTRANSFERASE 4, LPEAT1, LYSOPHOSPHATIDYLETHANOLAMINE ACYLTRANSFERASE 1, DSEL, DAD1‐LIKE SEEDLING ESTABLISHMENT‐RELATED LIPASE, SDP1, SUGAR‐DEPENDENT1, DALL3, DAD1‐LIKE LIPASE 3, pPLAIIIβ, PATATIN‐RELATED PHOSPHOLIPASE A IIIβ, PLIP3, PLASTID LIPASE 3, PLDζ2, PHOSPHOLIPASE D ζ 2; (b) Genes involved in FA synthesis, elongation, desaturation, and β‐oxidation: KCS3, KCS4, KCS5, KCS6, KCS10, KCS11, KCS12, KCS13, KCS19, KETOACYL‐COA SYNTHASEs, MTACP1, MITOCHONDRIAL ACYL CARRIER PROTEIN 1, MTACP2, MITOCHONDRIAL ACYL CARRIER PROTEIN 2, ACP4, ACYL CARRIER PROTEIN 4, FATA2, OLEOYL‐ACYL CARRIER PROTEIN THIOESTERASE 2, CER10, ECERIFERUM 10, PAS2, PASTICCINO 2, LACS7, LONG‐CHAIN ACYL‐COA SYNTHETASE 7, FAD3, FAD5, FAD6, FAD7, FAD8, FATTY ACID DESATURASEs, AAD3, ACYL‐ACYL CARRIER PROTEIN DESATURASE 3, ACX3, ACYL‐COA OXIDASE 3, ECI1, Δ3, Δ2‐ENOYL COA ISOMERASE 1, KAT2, 3‐KETOACYL‐COA THIOLASE 2, PNC1, PEROXISOMAL ADENINE NUCLEOTIDE CARRIER 1, MFP2, MULTIFUNCTIONAL PROTEIN 2, SDRB, SHORT‐CHAIN DEHYDROGENASE‐REDUCTASE B, ACX4, ACYL‐COA OXIDASE 4, ECH2, ENOYL‐COA HYDRATASE 2, AIM1, ABNORMAL INFLORESCENCE MERISTEM 1. Candidate genes differentially regulated between PZ and AZ are represented in red. Data are represented as mean (n = 3)
Taken together, during progression of stage I_PZ to stage III_PZ, an enrichment of GL and NL‐related GO terms and analysis of individual genes support the biochemical data with increased presence of glycerolipids during progression of the PZ. The occurrence of catabolic processes related to lipid metabolism in the PZ at stages III_PZ and IV_PZ may indicate active turnover of lipids.
3.4. Biochemical lipid analysis and gene expression are correlated
We confirmed differentially regulated lipid metabolism by correlation analysis of the amounts of single FAs of NL, GL, and PL fractions and the expression levels of 50 genes in PZ and AZ samples (see Materials and Methods). We found clearly more correlations related to the PZ than AZ, supporting the coordinated presence of lipid species and involvement of corresponding genes in the PZ (Figures 7 and 8 and Figures S7–S9).
FIGURE 7.
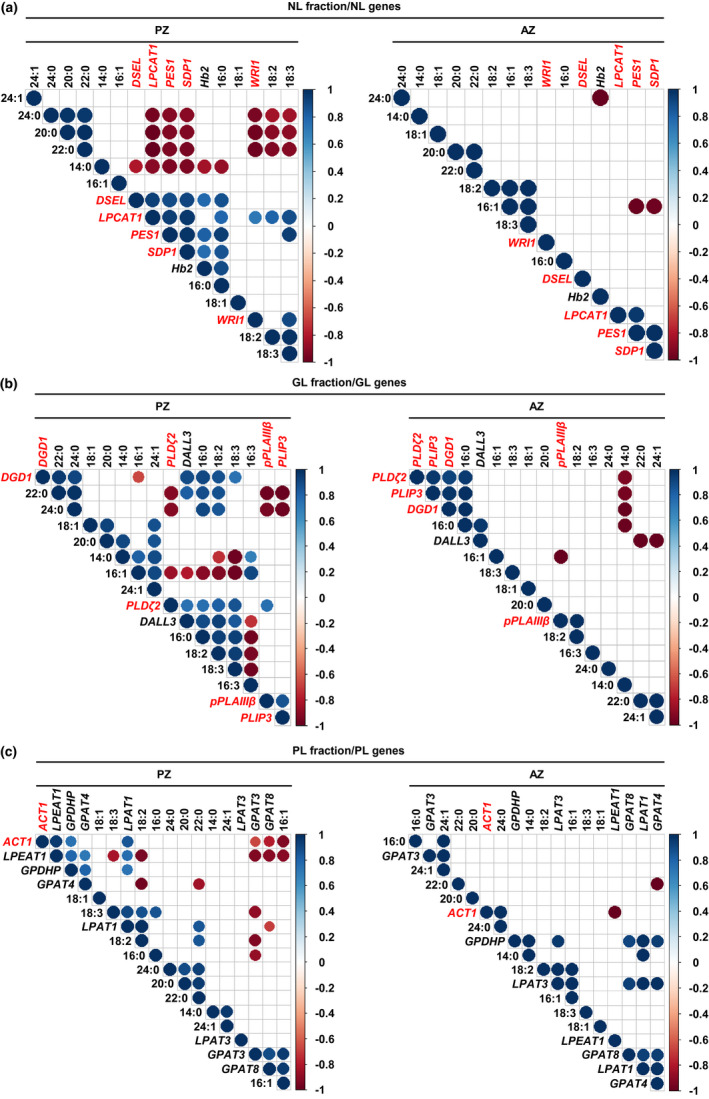
Pearson correlation analysis between gene expression levels of lipid metabolism genes and the amounts of single FAs of the NL, GL, and PL fraction of A. alpina perpetual flowering 1‐1 (pep1‐1) mutant lateral stem internodes of perennial (PZ) and annual (AZ) zones. The analysis was performed between stages III_PZ, IV_PZ, and I′, II′ for PZ and IV_AZ_if and II′ for AZ (see Figures 1 and 5a). (a) Correlation analysis between NL‐derived FAs and NL metabolism‐related genes. DSEL, DAD1‐LIKE SEEDLING ESTABLISHMENT‐RELATED LIPASE, Hb2, HAEMOGLOBIN 2, LPCAT1, LYSOPHOSPHATIDYLCHOLINE ACYLTRANSFERASE 1, PES1, PHYTYL ESTER SYNTHASE 1, SDP1, SUGAR‐DEPENDENT1, WRI1, WRINKLED 1. (b) Correlation analysis between GL‐derived FAs and GL metabolism‐related genes. DALL3, DAD1‐LIKE LIPASE 3, DGD1, DIGALACTOSYL DIACYLGLYCEROL DEFICIENT 1, PLDζ2, PHOSPHOLIPASE D ζ 2, PLIP3, PLASTID LIPASE 3, pPLAIIIβ, PATATIN‐RELATED PHOSPHOLIPASE A IIIβ. (c) Correlation analysis between PL‐derived FAs and PL metabolism‐related genes. ACT1, ACYLTRANSFERASE 1, GPAT3, GLYCEROL‐3‐PHOSPHATE SN‐2‐ACYLTRANSFERASE 3, GPAT4, GLYCEROL‐3‐PHOSPHATE SN‐2‐ACYLTRANSFERASE 4, GPAT8, GLYCEROL‐3‐PHOSPHATE SN‐2‐ACYLTRANSFERASE 8, GPDHP, GLYCEROL‐3‐PHOSPHATE DEHYDROGENASE PLASTIDIC, LPAT1, LYSOPHOSPHATIDIC ACID ACYLTRANSFERASE 1, LPAT3, LYSOPHOSPHATIDYL ACYLTRANSFERASE 3, LPEAT1, LYSOPHOSPHATIDYLETHANOLAMINE ACYLTRANSFERASE 1. Only correlation coefficients with significant levels of correlation (p < .05) are displayed in the diagrams. Candidate genes differentially regulated between PZ and AZ are represented in red
FIGURE 8.
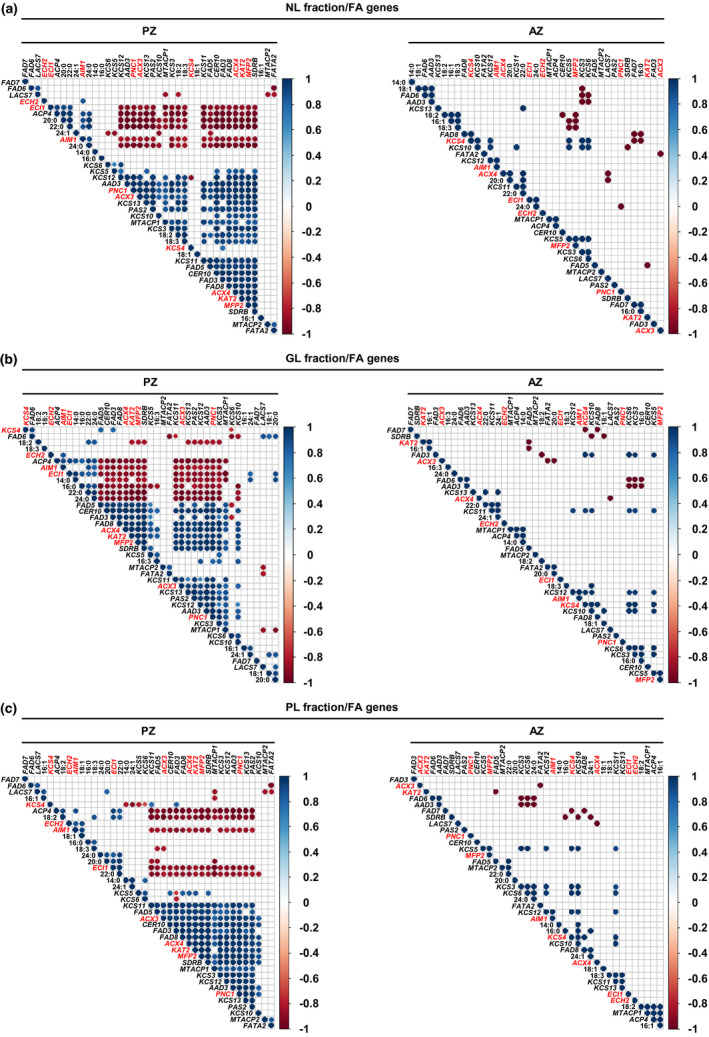
Pearson correlation analysis between gene expression levels of lipid metabolism genes and the amounts of single FAs of the NL, GL, and PL fraction of A. alpina perpetual flowering 1‐1 (pep1‐1) mutant lateral stem internodes of perennial (PZ) and annual (AZ) zones. The analysis was performed between stages III_PZ, IV_PZ, and I′, II′ for PZ and IV_AZ_if and II′ for AZ (see Figures 1 and 5a). Correlation analysis between (a) NL‐derived, (b) GL‐derived, (c) PL‐derived FAs and FA synthesis, elongation, desaturation, and β‐oxidation‐related genes. AAD3, ACYL‐ACYL CARRIER PROTEIN DESATURASE 3, ACP4, ACYL CARRIER PROTEIN 4, ACX3, ACYL‐COA OXIDASE 3, ACX4, ACYL‐COA OXIDASE 4, AIM1, ABNORMAL INFLORESCENCE MERISTEM 1, CER10, ECERIFERUM 10, ECH2, ENOYL‐COA HYDRATASE 2, ECI1, Δ3, Δ2‐ENOYL COA ISOMERASE 1, FAD3, FAD5, FAD6, FAD7, FAD8, FATTY ACID DESATURASEs, FATA2, OLEOYL‐ACYL CARRIER PROTEIN THIOESTERASE 2, KAT2, 3‐KETOACYL‐COA THIOLASE 2, KCS3, KCS4, KCS5, KCS6, KCS10, KCS11, KCS12, KCS13, KCS19, KETOACYL‐COA SYNTHASEs, LACS7, LONG‐CHAIN ACYL‐COA SYNTHETASE 7, MFP2, MULTIFUNCTIONAL PROTEIN 2, MTACP1, MITOCHONDRIAL ACYL CARRIER PROTEIN 1, MTACP2, MITOCHONDRIAL ACYL CARRIER PROTEIN 2, PAS2, PASTICCINO 2, PNC1, PEROXISOMAL ADENINE NUCLEOTIDE CARRIER 1, SDRB, SHORT‐CHAIN DEHYDROGENASE‐REDUCTASE B. Only correlation coefficients with significant levels of correlation (p < .05) are displayed in the diagrams. Candidate genes differentially regulated between PZ and AZ are represented in red
We investigated meaningful individual correlations in more detail. For example, NL gene expression levels of LPCAT1 positively correlated with amounts of 16:0, 18:2, and 18:3 FAs of the NL fraction of the PZ (Figure 7a, PZ). Gene expression levels of PES1 highly correlated with the amounts of 16:0 and 18:3 FAs. Positive correlation was also detected between WRI1 and the amounts of 18:3 FAs. Furthermore, 16:0 FAs positively correlated with gene expression levels of DSEL. Negative correlation was seen between SDP1 and VLCFAs, 24:0, 20:0, and 22:0 in PZ, as well as between 16:1 FAs, PES1 and SDP1 in AZ (Figure 7a). In addition, among the identified genes of the GO term “galactolipid metabolic process”, DGD1, pPLAIIIβ, PLIP3, and PLDζ2, up‐regulated at stage III_PZ and/or IV_PZ versus stage IV_AZ_if (Figure 6a), DGD1 positively correlated with 16:0, 18:2, 18:3, 22:0, and 24:0 FAs of the GL fraction of the PZ (Figure 7b). In the AZ, significant positive correlation for DGD1 was only detected with 16:0 FA. Negative correlation was detected for pPLAIIIβ and PLIP3 with 22:0 and 24:0 FAs in the PZ, while in the AZ pPLAIIIβ and PLIP3 negatively correlated with 16:1 and 14:0 Fas, respectively. Expression levels of PLDζ2 positively correlated with the amounts of 16:0, 18:2, and 18:3 FAs of the GL fraction of the PZ, but only with 16:0 FA in the AZ (Figure 7b).
Finally, among the PL‐related genes, ACT1 fulfilled the criteria of being expressed at higher level at stages III_PZ and IV_PZ than at stage IV_AZ_if (Figure 6a). ACT1 encodes a plastidic enzyme involved in the first step of phosphatidic acid biosynthesis (Li‐Beisson et al., 2013). No positive correlation was observed between ACT1 gene expression and amounts of single FAs of the PL fraction of the PZ (Figure 7c). Surprisingly, ACT1 positively correlated with 24:0 FAs in the AZ, indicating in fact a minor role in PL metabolism in the PZ and rather a prevalent function in the AZ. In addition to their involvement in GL metabolism, PLDζ2, pPLAIIIβ, and PLIP3 were reported to hydrolyze PLs (Li et al., 2011; Su et al., 2018; Wang et al., 2018). We therefore analyzed correlation of these genes with FAs present in the PL fraction, assuming a negative correlation (Figure S9b). Indeed, the three genes negatively correlated with 16:1 and 22:0 FAs in the PZ. In addition, the analysis resulted in negative correlation between PLDζ2 and 20:0 FAs.
Among the identified FA genes, KCS4, ACX3, ECI1, KAT2, PNC1, MFP2, ACX4, ECH2, and AIM1 were regulated according to the above‐described criteria (Figure 6b). While KCS4 is involved in the elongation of VLCFAs, the other FA genes are needed for peroxisomal FA β‐oxidation (Li‐Beisson et al., 2013). We therefore assumed that the importance of these genes for NL, GL, and PL metabolism in the PZ would be reflected by positive correlation between KCS4 and single FAs and by negative correlation between the genes of peroxisomal FA β‐oxidation and single FAs. Gene expression levels of KCS4 did not positively correlate with any of the identified FAs of the three lipid fractions present in the PZ and AZ (Figure 8). Supporting the assumption, significant negative correlation between the gene expression levels and the amounts of single FAs of each fraction of the PZ was detected for all of the mentioned FA β‐oxidation genes except for ECH2 (Figure 8a–c). In the NL fraction of the PZ, ECI1 and AIM1 negatively correlated with 18:2 and 18:3 FAs (Figure 8a). Gene expression levels of PNC1, ACX3, ACX4, KAT2, and MFP2 negatively correlated with the amounts of VLCFAs, 20:0, 22:0, and 24:0. Except for MFP2 and PNC1, no correlation was detected between the candidate genes and the amounts of NL‐derived FAs in the AZ (Figure 8a). Analysis between the FA genes and FAs of the GL fraction of the PZ resulted in high negative correlation between ACX4, KAT2, MFP2, ACX3, PNC1, and 16:0, 22:0, 24:0 FAs (Figure 8b). In addition, KAT2, MFP2, and PNC1 negatively correlated with 18:2 FAs. Except for ACX3 negatively correlating with 20:0 FAs, no correlation was detected between the FA β‐oxidation‐related genes and the amounts of GL‐derived FAs in the AZ (Figure 8b). Gene expression levels of ACX3, ACX4, KAT2, MFP2, and PNC1 negatively correlated with 18:2 and 22:0 FAs present in the PL fraction of the PZ (Figure 8c). In addition, the analysis resulted here in negative correlation between ACX3 and 18:3 FAs. Expression levels of the FA β‐oxidation‐related genes did not correlate with the amounts of PL‐derived FAs in the AZ (Figure 8c).
Taken together, a broad set of correlations is present between NL, GL, PL, and FA genes and the FA species in the various glycerolipid fractions in the PZ in contrast to the AZ. Correlation analysis therefore supported the involvement of candidate genes in lipid metabolism in the PZ.
4. DISCUSSION
Glycerolipids in A. alpina contain predominantly LCFAs and additionally VLCFAs in the NL fraction, more abundant in the PZ than AZ, pointing toward an active lipid metabolic function in the PZ. We have identified 19 lipid metabolism genes that have an expression pattern in favor of their function in storage and different steps of NL, GL, PL, and FA metabolism in the PZ, and all of them show correlation with certain FA species amounts. Positive and negative correlations of these 19 enzymatic functions support their involvement in biosynthesis or catabolism of NL, GL, and PL‐related lipid species in the PZ. These genes could be targets of signals that regulate PZ development and delimit PZ and AZ.
4.1. Biosynthesis and catabolism of NLs are differentially regulated during progression of PZ and between PZ and AZ
The high proportion of glycerolipids among total lipids is indicative of carbon storage. The diacylglycerol‐ and TAG‐containing NL fraction increased during development of PZ in Paj and pep1‐1. Significant amounts of VLCFAs are known to be present in TAGs. Millar and Kunst (1999) demonstrated that seed oil from 100 ecotypes of A. thaliana contained large proportions of VLCFAs including C20 and C22 FAs. These long‐chain FAs are more energy‐rich than short‐chain FAs. Interestingly, VLCFA contents indeed increased in the NL fractions of PZ, but were unchanged in corresponding PL and GL fractions during PZ progression. These findings support that PZs of lateral branches of A. alpina are high‐energy lipid storage sites.
Elongation of VLCFAs occurs in a sequence of reactions in the ER (Figure 9). The first enzyme involved in the process of FA elongation is KCS. KCS catalyzes condensation of a long‐chain acyl‐CoA with malonyl‐CoA. A. thaliana has 21 KCS genes (Joubès et al., 2008). Characterization of the members of the KCS family showed their diverse chain‐length specificities (Yang et al., 2018). We identified KCS4 to be differentially regulated during PZ progression. The exact chain‐length specificity for KCS4 has still to be elucidated. However, Blacklock and Jaworski (2006) hypothesized the possible activity of several KCS proteins including KCS4 toward acyl‐CoAs longer than C20. Beside 20:0 FAs, levels of 22:0 and 24:0 FAs increased in the NL fraction during PZ progression. Moreover, lowest levels of expression of KCS4 were detected in the AZ in comparison to the PZ tissues, indicating the possible involvement of KCS4 in the synthesis of TAG‐derived VLCFAs in PZ. However, gene expression levels of KCS4 neither correlated with any of the NL‐derived FAs nor with FAs from the GL and PL fraction. KCS4 may therefore also be involved in the elongation of FAs deriving from other lipid classes, for example, cuticular waxes. Thus, the exact role of KCS4 in lipid metabolism of A. alpina has to be elucidated in future studies.
FIGURE 9.
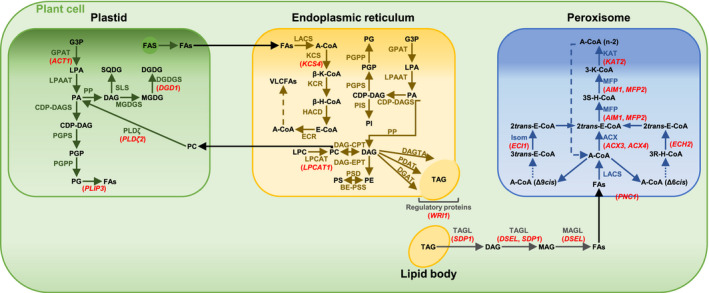
Candidate genes for lipid metabolism and their involvement in glycerolipid biosynthesis and fatty acid β‐oxidation. Overview of prokaryotic and eukaryotic glycerolipid biosynthesis pathways and fatty acid β‐oxidation in the plastid, in or at the endoplasmic reticulum (ER), and in the peroxisome, showing the interconversion of pathways. Candidate genes differentially regulated between PZ and AZ are represented in red, except PES1 and pPLAIIIβ. Abbreviations: A‐CoA: acyl‐CoA; ACX: acyl‐CoA thioesterase; β/3R/3S‐H‐CoA: β/3R/3S‐hydroxyacyl‐CoA; β/3‐K‐CoA: β/3‐ketoacyl‐CoA; BE‐PSS: base‐exchange‐type phosphatidylserine synthase; CDP‐DAGS: CDP‐DAG synthase; CoA: coenzyme A; DAG: diacylglycerol; DAGTA: diacylglycerol transacylase; DAG‐CPT: CDP‐choline: diacylglycerol cholinephosphotransferase; DAG‐EPT: CDP‐ethanolamine:diacylglycerol cholinephosphotransferase; DGAT: diacylglycerol acyltransferase; DGDG: digalactosyldiacylglycerol; DGDGS: digalactosyldiacylglycerol transferase; E‐CoA: enoyl‐CoA; ECR: enoyl‐CoA reductase; FAS: fatty acid synthase; FAs: fatty acids; G3P: glycerol 3‐phosphate; GPAT: glycerol‐3‐phosphate acyltransferase; HACD: hydroxyacyl‐CoA dehydratase; Isom: isomerase; KAT: 3‐ketothiolase; KCR: ketoacyl‐CoA reductase; KCS: 3‐ketoacyl‐CoA synthase; LACS: long‐chain acyl‐CoA synthetase; LPA: lysophosphatidic acid; LPAAT: lysophosphatidic acid acyltransferase; LPC: lysophosphatidylcholine; LPCAT: lysophosphatidylcholine acyltransferase; MAG: monoacylglycerol; MAGL: monoacylglycerol lipase; MFP: multifunctional protein; MGDG: monogalactosyldiacylglycerol; MGDGS: monogalactosyldiacylglycerol transferase; PA: phosphatidic acid; PC: phosphatidylcholine; PDAT: phospholipid:diacylglycerol acyltransferase; PE: phosphatidylethanolamine; PG: phosphatidylglycerol; PGP: phosphatidylglycerol phosphate; PGPP: phosphatidylglycerol phosphate phosphatase; PGPS: phosphatidylglycerol phosphate synthase; PI: phosphatidylinositol; PIS: PI synthase; PP: phosphatidate phosphatase; PS: phosphatidylserine; PSD: phosphatidylserine decarboxylae; SLS: sulfolipid synthase; SQDG: sulfoquinovosyldiacylglycerol; TAG: triacylglycerol; TAGL: triacylglycerol lipase; VLCFAs: very long‐chain fatty acids. Adopted from Li‐Beisson et al. (2013). Candidate genes: ACT1, ACYLTRANSFERASE 1, ACX3, ACYL‐COA OXIDASE 3, ACX4, ACYL‐COA OXIDASE 4, AIM1, ABNORMAL INFLORESCENCE MERISTEM 1, DGD1, DIGALACTOSYL DIACYLGLYCEROL DEFICIENT 1, DSEL, DAD1‐LIKE SEEDLING ESTABLISHMENT‐RELATED LIPASE, ECH2, ENOYL‐COA HYDRATASE 2, ECI1, Δ3, Δ2‐ENOYL COA ISOMERASE 1, KAT2, 3‐KETOACYL‐COA THIOLASE 2, KCS4, 3‐KETOACYL‐COA SYNTHASE 4, LPCAT1, LYSOPHOSPHATIDYLCHOLINE ACYLTRANSFERASE 1, MFP2, MULTIFUNCTIONAL PROTEIN 2, PES1, PHYTYL ESTER SYNTHASE 1, PLDζ2, PHOSPHOLIPASE D ζ 2, PLIP3, PLASTID LIPASE 3, PNC1, PEROXISOMAL ADENINE NUCLEOTIDE CARRIER 1, pPLAIIIβ, PATATIN‐RELATED PHOSPHOLIPASE A IIIβ, SDP1, SUGAR‐DEPENDENT1, WRI1, WRINKLED 1. Acyl‐CoA chains with a cis double bond on an even‐numbered carbon are exemplified by Δ6cis, while fatty acids with cis double bonds at odd‐numbered positions are exemplified by Δ9cis
LPCAT1, WRI1, PES1, DSEL, and SDP1 encode enzymes and regulators for TAG metabolism. These genes are expressed at higher level during development of the PZ and correlate with LCFAs and VLCFAs in comparison to the AZ, which supports their function in the accumulation of TAGs in the PZ, but not in the AZ. LPCAT1 is involved in acylation of lysophosphatidylcholine during acyl editing, the entry point of FAs into phosphatidylcholine (Bates et al., 2012) (Figure 9). In the ER, phosphatidylcholine can be further converted into diacylglycerol, a precursor of TAG biosynthesis. Moreover, Bates et al. (2012) demonstrated decreased polyunsaturated FA contents in TAG in lpcat1 lpcat2 mutant Arabidopsis plants. Polyunsaturated 18:2 and 18:3 FAs constitute a great proportion of total NL‐derived FAs in Paj and pep1‐1. The increased expression of LPCAT1 and the positive correlation with 18:2 and 18:3 FAs thus not only supports TAG biosynthesis and storage in the PZ, but also indicates that the pool of these polyunsaturated FAs in diacylglycerols and, respectively, in TAGs may derive from phosphatidylcholine produced via the activity of LPCAT1.
In accordance with the observed accumulation of NLs in the PZ, there was increased expression of PES1 and high positive correlation with 16:0 and 18:3 FAs in the same tissues. Involvement of PES1 in TAG biosynthesis was demonstrated by lower accumulation of TAGs, a phenotype of Arabidopsis pes1 pes2 mutants (Lippold et al., 2012). Regulation of TAG biosynthesis involves a complex of numerous transcription factors. One transcription factor, WRI1, known to regulate genes responsible for carbon allocation from starch to FAs of TAG (Baud et al., 2007; Cernac & Benning, 2004; Maeo et al., 2009), was most expressed at stage III_PZ. This indicates the enhanced rate of TAG biosynthesis at this stage with preferentially 18:3 FAs according to the correlation analysis. In addition, this points to lower rate of TAG biosynthesis in juvenile and developmentally more advanced PZ tissues, as compared to stage III_PZ. According to Sergeeva et al. (2020), starch rather accumulates in well‐developed PZ stages with pronounced secondary growth and not at stage III_PZ. WRI1 may therefore be involved in this process. Allocation of carbon from starch to FAs of TAG may be reduced with progression of the development of PZ due to the decreased activity of WRI1. Thus, starch can accumulate to higher levels in developmentally more advanced PZ tissues. Secondary growth and thus establishment of secondary phloem parenchyma tissue for TAG storage do not occur in the AZ, which is indirectly reflected by low expression and no FA‐related correlation of WRI1 in the AZ.
An intriguing finding was that NL genes did not correlate positively with VLCFAs. We explain this with the higher amounts of LCFAs in the PZ in comparison with VLCFAs at every investigated stage. For some genes, there could be catabolic VLCFA activities. SDP1 is a key enzyme of TAG hydrolysis (Fan et al., 2017; Kelly et al., 2013) and can hydrolyze also diacylglycerol (Eastmond, 2006) (Figure 9). The observed negative correlation between SDP1 and NL‐derived VLCFAs, 20:0, 22:0, and 24:0 may be indicative of catabolic processes related to VLCFA‐containing TAGs accumulated in the PZ at later stages of development.
The acylhydrolase DSEL catalyzes the hydrolysis of di‐ and monoacylglycerol (Kim et al., 2011) (Figure 9). Moreover, DSEL was suggested to negatively regulate storage oil mobilization by a still unknown mechanism. A. thaliana mutants overexpressing AtDSEL maintained numerous LBs in cotyledons in comparison to wild type. Moreover, FA β‐oxidation was inhibited in AtDSEL overexpressers. Likewise, DSEL may not only participate in the TAG hydrolysis pathway in A. alpina, but may also influence the levels of available energy by preventing TAG hydrolysis. Higher expression levels of DSEL in developmentally more progressed PZs fit with accumulation of TAG‐containing LBs in these tissues (Sergeeva et al., 2020). Contrary to that, DSEL expression in the AZ and early PZ was low, indirectly stressing the importance of oil storage in developmentally advanced PZ.
In summary, TAG storage at advanced stages of the PZ correlates with NL biosynthesis and catabolism gene expression, suggesting that these genes are involved in lipid storage in the PZ. WRI1 and DSEL showed the most striking difference of gene expression between PZ and AZ. These two candidates may thus represent important regulators of lipid metabolism, discriminating PZ‐AZ transition.
4.2. Catabolism of PLs is characteristic at late PZ stages and differentiates between PZ and AZ
GLs and PLs function primarily as structural elements of organellar membranes and membranous compartments. As the pathways of NL, GL, and PL biosynthesis are interconnected, GLs and PLs also contribute to storage in TAG‐containing LBs and energy storage in general. GLs and PLs were present in higher amounts at PZ stages than corresponding AZ stages and generally there was an increase from stages I and II to stage III and/or stage II’. Levels of DGD1 raised up to stage IV_PZ, positively correlated with GL‐derived 16:0, 18:2, 18:3, 22:0, and 24:0 FAs in the PZ, but were low at stage IV_AZ_if. DGD1 catalyzes synthesis of galactolipid digalactosyldiacylglycerol, constituent of plastid membranes (Benning & Ohta, 2005; Dörmann et al., 1995) (Figure 9). The gradual increase in DGD1 expression and enhanced digalactosyldiacylglycerol biosynthesis for incorporation into plastid membranes may be indicative of cell division, cell differentiation, and plastid differentiation during secondary growth. On the other side, DGD1 activity alone may also indicate a requirement for phosphate mobilization at the late PZ stage. Under phosphate deficiency, extraplastidial PLs are replaced by galactolipids (Essigmann et al., 1998; Härtel et al., 1998, 2000; Jouhet et al., 2004; Kelly & Dörmann, 2002; Kelly et al., 2003; Pant et al., 2015). A corresponding PL decrease indicative of remobilization of phosphate was not found and speaks against phosphate mobilization taking place in the PZ. Interestingly, turnover of PLs and GLs was supported by negative correlation of three catabolic lipase genes PLDζ2, PLIP3, and p PLAIIIβ with PL‐ and GL‐derived FAs of the PZ. PLDζ2 expression raised during progression of the development of PZ with highest levels of expression at stage III_PZ. PLDζ2 is related to inorganic phosphate deficiency (Li et al., 2006; Su et al., 2018). PLDζ2 hydrolyzes phosphatidylcholine (Figure 9) which finally leads to the synthesis of digalactosyldiacylglycerol and release of free phosphate (Cruz‐Ramírez et al., 2006; Su et al., 2018). Perhaps, there is an active turnover of PLs and supposedly also GLs during progression of the development of the PZ. We noted higher expression levels of PLIP3 and pPLAIIIβ at stage III_PZ. In addition to phosphatidylglycerol (Figure 9), both enzymes were reported to hydrolyze monogalactosyldiacylglycerol and digalactosyldiacylglycerol (Li et al., 2011; Wang et al., 2018). Moreover, higher expression of PLDζ2, PLIP3, and pPLAIIIβ in developmentally more progressed PZ may indicate catabolic processes related to TAG‐containing LBs. The TAG core of LBs is surrounded by a PL monolayer. Once hydrolysis of TAG occurs, PLs of the monolayer membrane are supposedly also subjected to catabolic processes. Expression levels of PLDζ2, PLIP3, and pPLAIIIβ correlate with the gene expression levels of TAG‐hydrolyzing lipase SDP1, thus supporting this idea.
In general, our data indicate that regarding PL metabolism differences during development of the PZ and between the PZ and the AZ occur for PL catabolic processes. In addition to TAGs, PLs thus may represent a source of acetyl‐CoA units as well as phosphate for developmental processes.
4.3. Enhanced FA β‐oxidation during progression of PZ indicates turnover of stored lipid resources
In peroxisomal β‐oxidation, FAs are broken down to yield finally C2 acetyl units (Figure 9). Especially FAs released from TAGs and PLs may be targets for peroxisomal FA β‐oxidation during progression of the PZ in comparison to the AZ. Eight genes, ACX3, ECI1, KAT2, PNC1, MFP2, ACX4, ECH2, and AIM1, involved in peroxisomal FA β‐oxidation, were up‐regulated during PZ progression and down‐regulated in the AZ (Figure 9). All of these genes except for ECH2 negatively correlated with LCFAs and VLCFAs present in the three investigated lipid fractions, supporting catabolic processes related to NL, GL, and PL metabolism. The first step of peroxisomal FA β‐oxidation, the conversion of acyl‐CoA to 2trans‐enoyl‐CoA, is catalyzed by ACXs (Li‐Beisson et al., 2013). AIM1 is an isoform of multifunctional protein (MFP) involved in the second and the third step of the core β‐oxidation pathway (Li‐Beisson et al., 2013; Richmond & Bleeker, 1999), leading to conversion of 2trans‐enoyl‐CoA into 3S‐hydroxyacyl‐CoA and production of 3‐ketoacyl‐CoA. KAT2 catalyzes cleavage of 3‐ketoacyl‐CoA into acyl‐CoA. Acyl‐CoA chains with a cis double bond on an even‐numbered carbon are converted in the final step into 2trans‐enoyl‐CoA by ECH2, while 3trans‐enoyl‐CoA is converted into 2trans‐enoyl‐CoA by the isomerase ECI1 in the case of acyl‐CoA chains with cis double bonds at odd‐numbered positions (Li‐Beisson et al., 2013) (Figure 9). Interestingly, the LCFAs 18:2 and 18:3, deriving from NLs, decreased with progression of the development of PZ, indicating that these FAs might be catabolized to generate acetyl‐CoA for developmental processes and seed setting. This decrease fits to increasing levels of ECI1 and ECH2 expression and is supported by the negative correlation between ECI1 and NL‐derived 18:2 and 18:3 FAs. Moreover, ECH2 was demonstrated to play an important role with regard to mobilization of storage lipids (Katano et al., 2016; Li et al., 2019; Strader et al., 2011). The peroxisomal adenine nucleotide carrier protein PNC1 catalyzes the import of adenosine triphosphate into peroxisomes, thus playing an important role with regard to energy supply for subsequent peroxisomal reactions (Linka et al., 2008) (Figure 9).
In summary, gene expression levels of enzymes involved in peroxisomal FA β‐oxidation correlate with degradation of certain VLCFAs and LCFAs indicating turnover of stored lipid resources.
5. CONCLUSIONS AND PERSPECTIVES
This study sheds light on lipid metabolism and corresponding genes catalyzing and regulating metabolic steps that potentially differentiate PZ and AZ in the perennial A. alpina. On one side, glycerolipids increase during PZ progression with a tendency for higher amounts of VLCFAs. On the other side, catabolic activities interconvert lipids or degrade them for energy consumption during specific developmental steps. In future studies, the exact role of these candidate genes in lipid metabolism and thereby the importance of metabolic steps themselves can be investigated. Detailed lipid analysis based on, for example, liquid chromatography (LC)‐MS can be used in the future to distinguish individual species of neutral and polar glycerolipids. Thereby, lipid metabolism gene expression will help dissecting the regulatory pathways for storage lipid deposition, potentially controlled by PZ‐AZ transition signals.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
AUTHOR CONTRIBUTIONS
AS, TMA, and PB designed the research; AS, TMA, and HL performed research; AS and HJM contributed new analytic/computational/etc. tools; AS, TMA, HL, HJM, and PB analyzed data; AS wrote the manuscript; TMA, HJM, and PB made corrections to the manuscript.
Supporting information
Fig S1‐S9
Table S1‐S8
ACKNOWLEDGEMENTS
This project was “funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany´s Excellence Strategy – EXC‐2048/1 – project ID 390686111”. We acknowledge the excellent technical assistance of M. Graf, E. Klemp, and K. Weber for GC‐MS measurements. We also want to thank Dr. Vera Wewer (University of Cologne) for her advice regarding the establishment of lipid extraction. The authors thank E. Wieneke for help with gene expression studies. H.L. was the recipient of a doctoral fellowship from Northwest A&F University, College of Horticulture, Yangling, China.
Sergeeva A, Mettler‐Altmann T, Liu H, Mai H‐J, Bauer P. Glycerolipid profile differences between perennial and annual stem zones in the perennial model plant Arabis alpina . Plant Direct. 2021;5:1–18. 10.1002/pld3.302
REFERENCES
- Albani, M. C. , Castaings, L. , Wotzel, S. , Mateos, J. L. , Wunder, J. , Wang, R. , Reymond, M. , & Coupland, G. (2012). PEP1 of Arabis alpina is encoded by two overlapping genes that contribute to natural genetic variation in perennial flowering. PLoS Genetics, 8, e1003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa, A. , & Rahnenfuhrer, J. (2010). topGO: enrichment analysis for gene ontology. R package version 2.
- Bates, P. D. (2016). Understanding the control of acyl flux through the lipid metabolic network of plant oil biosynthesis. Biochimica Et Biophysica Acta, 1861(9 Pt B), 1214–1225. [DOI] [PubMed] [Google Scholar]
- Bates, P. D. , & Browse, J. (2011). The pathway of triacylglycerol synthesis through phosphatidylcholine in Arabidopsis produces a bottleneck for the accumulation of unusual fatty acids in transgenic seeds. The Plant Journal, 68, 387–399. [DOI] [PubMed] [Google Scholar]
- Bates, P. D. , & Browse, J. (2012). The significance of different diacylglycerol synthesis pathways on plant oil composition and bioengineering. Frontiers in Plant Science, 3(147), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, P. D. , Durrett, T. P. , Ohlrogge, J. B. , & Pollard, M. (2009). Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiology, 150, 55–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, P. D. , Fatihi, A. , Snapp, A. R. , Carlsson, A. S. , Browse, J. , & Lu, C. (2012). Acyl editing and headgroup exchange are the major mechanisms that direct polyunsaturated fatty acid flux into triacylglycerols. Plant Physiology, 160, 1530–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud, S. , Santos Mendoza, M. , To, A. , Harscoët, E. , Lepiniec, L. , & Dubreucq, B. (2007). WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. The Plant Journal, 50, 825–838. [DOI] [PubMed] [Google Scholar]
- Ben Abdallah, H. , & Bauer, P. (2016). Quantitative reverse transcription‐qPCR‐based gene expression analysis in plants In Botella J., & Botella M. (Eds.), Plant signal transduction. Methods in molecular biology 1363. Humana Press. [DOI] [PubMed] [Google Scholar]
- Benning, C. , & Ohta, H. (2005). Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. Journal of Biological Chemistry, 280, 2397–2400. [DOI] [PubMed] [Google Scholar]
- Blacklock B. J., & Jaworski J. G. (2006). Substrate specificity of Arabidopsis 3‐ketoacyl‐CoA synthases. Biochemical and Biophysical Research Communications, 346(2), 583–590. 10.1016/j.bbrc.2006.05.162 [DOI] [PubMed] [Google Scholar]
- Brocard, L. , Immel, F. , Coulon, D. , Esnay, N. , Tuphile, K. , Pascal, S. , Claverol, S. , Fouillen, L. , Bessoule, J. J. , & Brehelin, C. (2017). Proteomic analysis of lipid droplets from Arabidopsis aging leaves brings new insight into their biogenesis and functions. Frontiers in Plant Science, 8, 894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Y. , McClinchie, E. , Price, A. , Nguyen, T. N. , Gidda, S. K. , Watt, S. C. , Yurchenko, O. , Park, S. , Sturtevant, D. , Mullen, R. T. , Dyer, J. M. , & Chapman, K. D. (2017). Mouse fat storage‐inducing transmembrane protein 2 (FIT2) promotes lipid droplet accumulation in plants. Plant Biotechnology Journal, 15, 824–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y. , & Huang, A. H. C. (1986). Diacylglycerol acyltransferase in maturing oil seeds of maize and other species. Plant Physiology, 82, 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac, A. , & Benning, C. (2004). WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. The Plant Journal, 40, 575–585. [DOI] [PubMed] [Google Scholar]
- Chinnasamy, G. , Davis, P. J. , & Bal, A. K. (2003). Seasonal changes in oleosomic lipids and fatty acids of perennial root nodules of beach pea. Journal of Plant Physiology, 160, 355–365. [DOI] [PubMed] [Google Scholar]
- Cruz‐Ramírez, A. , Oropeza‐Aburto, A. , Razo‐Hernández, F. , Ramírez‐Chávez, E. , & Herrera‐Estrella, L. (2006). Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proceedings of the National Academy of Sciences of the United States of America, 103, 6765–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann, P. , Hoffmann‐Benning, S. , Balbo, I. , & Benning, C. (1995). Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. The Plant Cell, 7, 1801–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond, P. J. (2006). SUGAR‐DEPENDENT1 encodes a Patatin Domain Triacylglycerol Lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. The Plant Cell, 18, 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essigmann, B. , Güler, S. , Narang, R. A. , Linke, D. , & Benning, C. (1998). Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 95, 1950–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J. , Yu, L. , & Xu, C. (2017). A central role for triacylglycerol in membrane lipid breakdown, fatty acid beta‐oxidation, and plant survival under extended darkness. Plant Physiology, 174, 1517–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, G. , Stymne, S. , & Stobart, A. K. (1988). Phosphatidylcholine and its relationship to triacylglycerol biosynthesis in oil‐tissues. Phytochemistry, 27, 2089–2093. [Google Scholar]
- Hajra, A. K. (1974). On extraction of acyl and alkyl dihydroxyacetone phosphate from incubation mixtures. Lipids, 9, 502–505. [DOI] [PubMed] [Google Scholar]
- Härtel, H. , Dörmann, P. , & Benning, C. (2000). DGD1‐independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 97, 10649–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtel, H. , Essigmann, B. , Lokstein, H. , Hoffmann‐Benning, S. , Peters‐Kottig, M. , & Benning, C. (1998). The phospholipid‐deficient pho1 mutant of Arabidopsis thaliana is affected in the organization, but not in the light acclimation, of the thylakoid membrane. Biochimica Et Biophysica Acta, 1415, 205–218. [DOI] [PubMed] [Google Scholar]
- Haslam, R. P. , Ruiz‐Lopez, N. , Eastmond, P. , Moloney, M. , Sayanova, O. , & Napier, J. A. (2013). The modification of plant oil composition via metabolic engineering–better nutrition by design. Plant Biotechnology Journal, 11, 157–168. [DOI] [PubMed] [Google Scholar]
- Hielscher, B. , Charton, L. , Mettler‐Altmann, T. , & Linka, N. (2017). Analysis of peroxisomal β‐oxidation during storage oil mobilization in Arabidopsis thaliana seedlings In Schrader M. (Eds.), Peroxisomes. Methods in molecular biology (Vol. 1595). New York, NY: Humana Press, Springer Nature. [DOI] [PubMed] [Google Scholar]
- Hughes, P. W. , Soppe, W. J. J. , & Albani, M. C. (2019). Seed traits are pleiotropically regulated by the flowering time gene PERPETUAL FLOWERING 1 (PEP1) in the perennial Arabis alpina. Molecular Ecology, 28, 1183–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlock, A. K. , Roston, R. L. , Wang, K. , & Benning, C. (2014). Lipid trafficking in plant cells. Traffic, 15, 915–932. [DOI] [PubMed] [Google Scholar]
- Jayawardhane, K. N. , Singer, S. D. , Weselake, R. J. , & Chen, G. (2018). Plant sn‐glycerol‐3‐phosphate acyltransferases: Biocatalysts involved in the biosynthesis of intracellular and extracellular lipids. Lipids, 53, 469–480. [DOI] [PubMed] [Google Scholar]
- Joubès, J. , Raffaele, S. , Bourdenx, B. , Garcia, C. , Laroche‐Traineau, J. , Moreau, P. , Domergue, F. , & Lessire, R. (2008). The VLCFA elongase gene family in Arabidopsis thaliana: Phylogenetic analysis, 3D modelling and expression profiling. Plant Molecular Biology, 67, 547–566. [DOI] [PubMed] [Google Scholar]
- Jouhet, J. , Marechal, E. , Baldan, B. , Bligny, R. , Joyard, J. , & Block, M. A. (2004). Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. Journal of Cell Biology, 167, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki, N. , Johnson, B. S. , & Bates, P. D. (2019). Metabolically distinct pools of phosphatidylcholine are involved in trafficking of fatty acids out of and into the chloroplast for membrane production. The Plant Cell, 31, 2768–2788. 10.1105/tpc.19.00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl, R. , & Koch, M. A. (2013). A world‐wide perspective on crucifer speciation and evolution: Phylogenetics, biogeography and trait evolution in tribe Arabideae. Annals of Botany, 112, 983–1001. 10.1093/aob/mct165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katano, M. , Takahashi, K. , Hirano, T. , Kazama, Y. , Abe, T. , Tsukaya, H. , & Ferjani, A. (2016). Suppressor screen and phenotype analyses revealed an emerging role of the monofunctional peroxisomal Enoyl‐CoA Hydratase 2 in compensated cell enlargement. Frontiers in Plant Science, 7, 132 10.3389/fpls.2016.00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, A. A. , & Dörmann, P. (2002). DGD2, an arabidopsis gene encoding a UDP‐galactose‐dependent digalactosyldiacylglycerol synthase is expressed during growth under phosphate‐limiting conditions. Journal of Biological Chemistry, 277, 1166–1173. [DOI] [PubMed] [Google Scholar]
- Kelly, A. A. , Froehlich, J. E. , & Dörmann, P. (2003). Disruption of the two digalactosyldiacylglycerol synthase genes DGD1 and DGD2 in Arabidopsis reveals the existence of an additional enzyme of galactolipid synthesis. The Plant Cell, 15, 2694–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, A. A. , van Erp, H. , Quettier, A. L. , Shaw, E. , Menard, G. , Kurup, S. , & Eastmond, P. J. (2013). The sugar‐dependent1 lipase limits triacylglycerol accumulation in vegetative tissues of Arabidopsis. Plant Physiology, 162, 1282–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer, C. , Severing, E. , Karl, R. , Bergonzi, S. , Koch, M. , Tresch, A. , & Coupland, G. (2017). Divergence of annual and perennial species in the Brassicaceae and the contribution of cis‐acting variation at FLC orthologues. Molecular Ecology, 26, 3437–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, E. Y. , Seo, Y. S. , & Kim, W. T. (2011). AtDSEL, an Arabidopsis cytosolic DAD1‐like acylhydrolase, is involved in negative regulation of storage oil mobilization during seedling establishment. Journal of Plant Physiology, 168, 1705–1709. 10.1016/j.jplph.2011.03.004 [DOI] [PubMed] [Google Scholar]
- LaBrant, E. , Barnes, A. C. , & Roston, R. L. (2018). Lipid transport required to make lipids of photosynthetic membranes. Photosynthesis Research, 138, 345–360. [DOI] [PubMed] [Google Scholar]
- Lacey, D. J. , Beaudoin, F. , Dempsey, C. E. , Shewry, P. R. , & Napier, J. A. (1999). The accumulation of triacylglycerols within the endoplasmic reticulum of developing seeds of Helianthus annuus. The Plant Journal, 17, 397–405. [Google Scholar]
- Lazaro, A. , Obeng‐Hinneh, E. , & Albani, M. C. (2018). Extended vernalization regulates inflorescence fate in Arabis alpina by stably silencing PERPETUAL FLOWERING1. Plant Physiology, 176, 2819–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. , Ratnayake, C. , & Huang, A. H. C. (1995). Genetic dissection of the co‐expression of genes encoding the two isoforms of oleosins in the oil bodies of maize kernel. The Plant Journal, 7, 603–611. 10.1046/j.1365-313X.1995.7040603.x [DOI] [PubMed] [Google Scholar]
- Li, M. , Bahn, S. C. , Guo, L. , Musgrave, W. , Berg, H. , Welti, R. , & Wang, X. (2011). Patatin‐related phospholipase pPLAIIIβ‐induced changes in lipid metabolism alter cellulose content and cell elongation in Arabidopsis. The Plant Cell, 23, 1107–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Qin, C. , Welti, R. , & Wang, X. (2006). Double knockouts of phospholipases Dzeta1 and Dzeta2 in Arabidopsis affect root elongation during phosphate‐limited growth but do not affect root hair patterning. Plant Physiology, 140, 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Wei, F. , Tawfall, A. , Tang, M. , Saettele, A. , & Wang, X. (2015). Overexpression of patatin‐related phospholipase AIIIdelta altered plant growth and increased seed oil content in camelina. Plant Biotechnology Journal, 13, 766–778. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Liu, Y. , & Zolman, B. K. (2019). Metabolic alterations in the enoyl‐CoA hydratase 2 mutant disrupt peroxisomal pathways in seedlings. Plant Physiology, 180, 1860–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li‐Beisson, Y. , Shorrosh, B. , Beisson, F. , Andersson, M. X. , Arondel, V. , Bates, P. D. , Baud, S. , Bird, D. , Debono, A. , Durrett, T. P. , Franke, R. B. , Graham, I. A. , Katayama, K. , Kelly, A. A. , Larson, T. , Markham, J. E. , Miquel, M. , Molina, I. , Nishida, I. , … Ohlrogge, J. (2013). Acyl‐lipid metabolism. Arabidopsis Book. Rockville, MD: American Society of Plant Biologists. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linka, N. , Theodoulou, F. L. , Haslam, R. P. , Linka, M. , Napier, J. A. , Neuhaus, H. E. , & Weber, A. P. M. (2008). Peroxisomal ATP import is essential for seedling development in Arabidopsis thaliana. The Plant Cell, 20, 3241–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippold, F. , vom Dorp, K. , Abraham, M. , Holzl, G. , Wewer, V. , Yilmaz, J. L. , Lager, I. , Montandon, C. , Besagni, C. , Kessler, F. , Stymne, S. , & Dörmann, P. (2012). Fatty acid phytyl ester synthesis in chloroplasts of Arabidopsis. The Plant Cell, 24, 2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loer, D. S. , & Herman, E. M. (1993). Cotranslational integration of soybean (Glycien max) oil body membrane protein oleosin into microsomal membranes. Plant Physiology, 101, 993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C. , Napier, J. A. , Clemente, T. E. , & Cahoon, E. B. (2011). New frontiers in oilseed biotechnology: Meeting the global demand for vegetable oils for food, feed, biofuel, and industrial applications. Current Opinion in Biotechnology, 22, 252–259. [DOI] [PubMed] [Google Scholar]
- Madey, E. , Nowack, L. M. , & Thompson, J. E. (2002). Isolation and characterization of lipid in phloem sap of canola. Planta, 214, 625–634. [DOI] [PubMed] [Google Scholar]
- Maeo, K. , Tokuda, T. , Ayame, A. , Mitsui, N. , Kawai, T. , Tsukagoshi, H. , Ishiguro, S. , & Nakamura, K. (2009). An AP2‐type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW‐box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. The Plant Journal, 60, 476–487. [DOI] [PubMed] [Google Scholar]
- Michaels, S. D. , & Amasino, R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. The Plant Cell, 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A. A. , & Kunst, L. (1999). The natural genetic variation of the fatty‐acyl composition of seed oild in different ecotypes of Arabidopsis thaliana. Phytochemistry, 52, 1029–1033. [DOI] [PubMed] [Google Scholar]
- Næsted, H. , Frandsen, G. I. , Jauh, G. Y. , Hernandez‐Pinzon, I. , Nielsen, H. B. , Murphy, D. J. , Rogers, J. C. , & Mundy, J. (2000). Caleosins: Ca2+‐binding proteins associated with lipid bodies. Plant Molecular Biology, 44, 463–476. [DOI] [PubMed] [Google Scholar]
- Pant, B. D. , Burgos, A. , Pant, P. , Cuadros‐Inostroza, A. , Willmitzer, L. , & Scheible, W. R. (2015). The transcription factor PHR1 regulates lipid remodeling and triacylglycerol accumulation in Arabidopsis thaliana during phosphorus starvation. Journal of Experimental Botany, 66, 1907–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyc, M. , Cai, Y. , Greer, M. S. , Yurchenko, O. , Chapman, K. D. , Dyer, J. M. , & Mullen, R. T. (2017). Turning over a new leaf in lipid droplet biology. Trends in Plant Science, 22, 596–609. [DOI] [PubMed] [Google Scholar]
- Richmond, T. A. , & Bleecker, A. B. (1999). A defect in beta‐oxidation causes abnormal inflorescence development in Arabidopsis. The Plant Cell, 11, 1911–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter, J. J. , & van Cleve, B. (1994). Storage, mobilization and interrelations of starch, sugars, protein and fat in the ray storage tissue of poplar trees. Trees, 8, 297–304. [Google Scholar]
- Sergeeva, A. , Liu, H. , Mai, H. J. , Mettler‐Altmann, T. , Kiefer, C. , Coupland, G. , & Bauer, P. (2020). Cytokinin‐promoted secondary growth and nutrient storage in the perennial stem zone of Arabis alpina. The Plant Journal. Advance online publication. 10.1101/2020.06.01.124362 [DOI] [PubMed] [Google Scholar]
- Shi, L. , Katavic, V. , Yu, Y. , Kunst, L. , & Haughn, G. (2012). Arabidopsis glabra2 mutant seeds deficient in mucilage biosynthesis produce more oil. The Plant Journal, 69, 37–46. [DOI] [PubMed] [Google Scholar]
- Shockey, J. M. , Gidda, S. K. , Chapital, D. C. , Kuan, J. C. , Dhanoa, P. K. , Bland, J. M. , Rothstein, S. J. , Mullen, R. T. , & Dyer, J. M. (2006). Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. The Plant Cell, 18, 2294–2313. 10.1105/tpc.106.043695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, E. W. (1974). Phospholipids and plant membrane permeability. New Phytologist, 73, 377–420. [Google Scholar]
- Stephan, L. , Tilmes, V. , & Hulskamp, M. (2019). Selection and validation of reference genes for quantitative Real‐Time PCR in Arabis alpina. PLoS One, 14, e0211172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader, L. C. , Wheeler, D. L. , Christensen, S. E. , Berens, J. C. , Cohen, J. D. , Rampey, R. A. , & Bartel, B. (2011). Multiple facets of Arabidopsis seedling development require indole‐3‐butyric acid‐derived auxin. The Plant Cell, 23, 984–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y. , Li, M. , Guo, L. , & Wang, X. (2018). Different effects of phospholipase Dζ2 and non‐specific phospholipase C4 on lipid remodeling and root hair growth in Arabidopsis response to phosphate deficiency. The Plant J, 94, 315–326. 10.1111/tpj.13858 [DOI] [PubMed] [Google Scholar]
- Tan, H. , Yang, X. , Zhang, F. , Zheng, X. , Qu, C. , Mu, J. , Fu, F. , Li, J. , Guan, R. , Zhang, H. , Wang, G. , & Zuo, J. (2011). Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1‐LIKE in developing seeds. Plant Physiology, 156, 1577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp, H. , Kelly, A. A. , Menard, G. , & Eastmond, P. J. (2014). Multigene engineering of triacylglycerol metabolism boosts seed oil content in Arabidopsis. Plant Physiology, 165, 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhercke, T. , Wood, C. C. , Stymne, S. , Singh, S. P. , & Green, A. G. (2013). Metabolic engineering of plant oils and waxes for use as industrial feedstocks. Plant Biotechnology Journal, 11, 197–210. [DOI] [PubMed] [Google Scholar]
- Vayssiѐres, A. , Mishra, P. , Roggen, A. , Neumann, U. , Ljung, K. , & Albani, M. C. (2020). Vernalization shapes shoot architecture and ensures the maintenance of dormant buds in the perennial Arabis alpina. New Phytologist, 227, 99–115. [DOI] [PubMed] [Google Scholar]
- Wang, G. , Lin, Q. , & Xu, Y. (2007). Tetraena mongolica Maxim can accumulate large amounts of triacylglycerol in phloem cells and xylem parenchyma of stems. Phytochemistry, 68, 2112–2117. [DOI] [PubMed] [Google Scholar]
- Wang, K. , Guo, Q. , Froehlich, J. E. , Hersh, H. L. , Zienkiewicz, A. , Howe, G. A. , & Benning, C. (2018). Two abscisic acid‐responsive plastid lipase genes involved in jasmonic acid biosynthesis in Arabidopsis thaliana. The Plant Cell, 30, 1006–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. , Albani, M. C. , Vincent, C. , Bergonzi, S. , Luan, M. , Bai, Y. , Kiefer, C. , Castillo, R. , & Coupland, G. (2011). Aa TFL1 confers an age‐dependent response to vernalization in perennial Arabis alpina. The Plant Cell, 23, 1307–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. , Farrona, S. , Vincent, C. , Joecker, A. , Schoof, H. , Turck, F. , Alonso‐Blanco, C. , Coupland, G. , & Albani, M. C. (2009). PEP1 regulates perennial flowering in Arabis alpina. Nature, 459, 423–427. [DOI] [PubMed] [Google Scholar]
- Wewer, V. , Dombrink, I. , vom Dorp, K. , & Dormann, P. (2011). Quantification of sterol lipids in plants by quadrupole time‐of‐flight mass spectrometry. Journal of Lipid Research, 52, 1039–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W. , Wang, G. , Li, J. , Bates, P. D. , Wang, X. , & Allen, D. K. (2017). Phospholipase Dζ enhances diacylglycerol flux into triacylglycerol. Plant Physiology, 174, 110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Wang, Z. , Feng, T. , Li, J. , Huang, L. , Yang, B. , Zhao, H. , Jenks, M. A. , Yang, P. , & Lü, S. (2018). Evolutionarily conserved function of the sacred lotus (Nelumbo nucifera Gaertn.) CER2‐LIKE family in very‐long‐chain fatty acid elongation. Planta, 248, 715–727. 10.1007/s00425-018-2934-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S9
Table S1‐S8


