Abstract
Pannexin 1 (PANX1) is an ATP-permeable channel crucially involved in a variety of physiological functions such as blood pressure regulation1, apoptotic cell clearance2, and human oocyte development3. We present several human PANX1 structures in a heptameric assembly at resolutions up to 2.8 Å, including an apo state, a caspase 7-cleaved state, a CBX-bound state. We elaborate a novel gating mechanism that involves the use of two ion-conducting pathways. Under normal cellular conditions, the intracellular entry of the wide main pore is physically plugged by the C-terminal tail. Small anions are conducted through narrow tunnels in the intracellular domain. These tunnels connect to the main pore and are gated by a long linker between the N-terminal helix and the first transmembrane helix. During apoptosis, the C-terminal tail is cleaved by caspase, allowing the release of ATP through the main pore. We identified a carbenoxolone (CBX) binding site embraced by W74 in the extracellular entrance and also a role for CBX as a channel blocker. We captured a gap junction-like structure using a glycosylation-deficient mutant, N255A. Our studies provided a solid foundation for understanding the molecular mechanisms underlying the channel gating and inhibition of PANX1 and related large-pore channels.
Introduction
Purinergic signaling is crucially involved in activities of the nervous system as diverse as neuroprotection, synaptic transmission, nociception, and inflammation4–7. This signaling is initiated by releasing ATP across the membrane through exocytosis8, or through ATP-permeable channels9–11. The pannexin family is one of the ATP-permeable channels and consists of three members in human12, PANX1, 2 and 3. Widely expressed throughout the body, PANX1 plays crucial roles in physiological processes such as blood pressure regulation, glucose uptake, inflammation, and cell death1,13–17. Gain-of-function mutations in PANX1 are implicated in defects of oocyte development3. Thus, PANX1 has been considered a promising therapeutic target for a diverse array of diseases. Compounds such as CBX and trovafloxacin have been reported to inhibit the activity of PANX118,19. However, little is known about where they bind or the inhibition mechanisms.
PANXs are nonselective large-pore channels that share a 4-transmembrane domain arrangement with connexins, innexins, volume-regulated anion channels (VRACs), and calcium homeostasis modulators (CALHMs)20–28. Unlike connexins and innexins, PANXs have been thought to not form gap junctions due to N-glycosylation29–31, although one study suggested that PANX1 may form gap junction in a cell type specific manner32.
PANX1 has been found to be activated under various physiological/pathological conditions. Under normal cellular conditions, PANX1 is autoinhibited by its C-terminal tail (CTT)33,34. However, the channel can still be activated by membrane depolarization, extracellular potassium, intracellular calcium, tyrosine phosphorylation, and mechanical stretching through unknown mechanisms35–40. Cleavage of the CTT either during apoptosis or experimentally by caspase 3 or 7 results in channel activation and ATP release2,33.
To gain insight into the channel assembly, gating mechanism, ion conducting pathway and inhibitor binding site, we studied hsPANX1 using a combination of single-particle cryo-electron microscopy (cryo-EM) and patch-clamp electrophysiology.
Overall structure
We determined structures of the wild type hsPANX1 (wt-hsPANX1) in the presence of EDTA, Ca2+ or K+. These structures are indistinguishable, suggesting that likely neither Ca2+ nor K+ directly activates hsPANX1. To study the location and role of the CTT and the N-terminal helix (NTH), we cleaved the CTT from the wt-hsPANX1 and from an NTH truncation mutant using caspase 7, and determined their structures (ΔCTT-hsPANX1, ΔNTH/ΔCTT-hsPANX1). We also determined structures of hsPANX1 bound to CBX (CBX-ΔCTT-hsPANX1, CBX-ΔNTH/ΔCTT-hsPANX1). To understand the role of N-glycosylation of PANX1, we studied the structure of a glycosylation-deficient mutant N255A, which yielded both gap junction (N255A-hsPANX1Gap) and hemichannel (N255A-hsPANX1Hemi). The procedure of structural determination is detailed in the Methods, and the results are summarized in Extended Data Tables 1–3. The structure of ΔCTT-hsPANX1 has the highest overall quality, and is thus used for the discussion throughout the text unless otherwise noted.
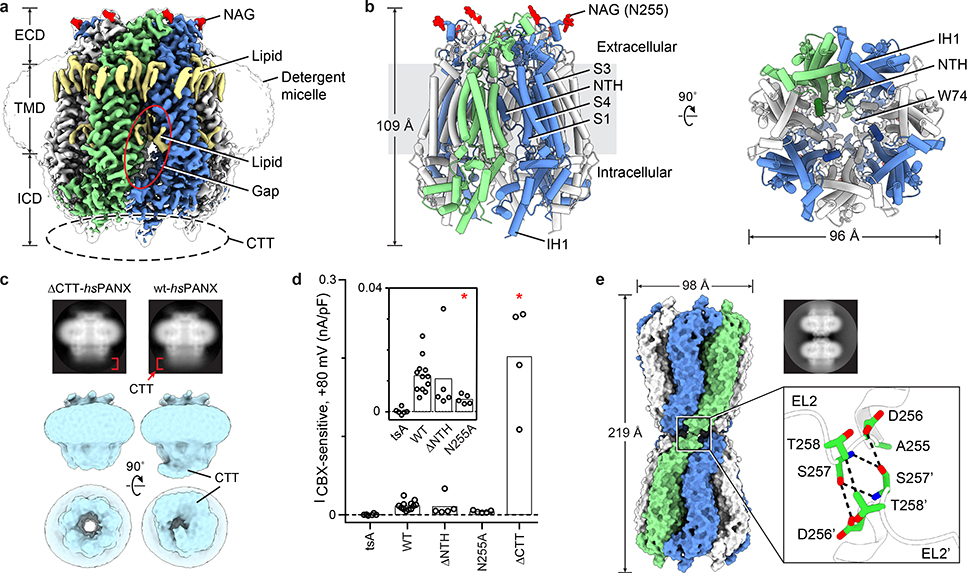
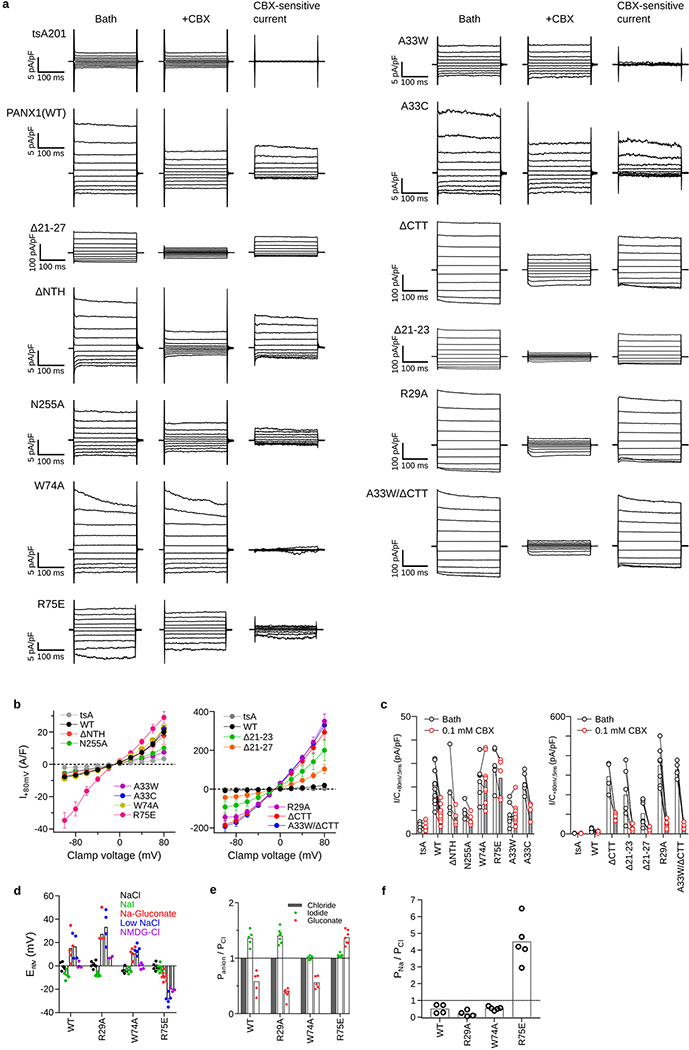
The hsPANX1 channel forms a heptamer that has, from top to bottom, a narrow extracellular domain (ECD), a cone-shaped transmembrane domain (TMD), and an intracellular domain (ICD), with the unstructured CTT blocking the ICD entrance to the ion pathway along the symmetry axis (referred to as the main pore) (Fig. 1a, b). Caspase 7 treatment released the blockage by cleaving the CTT, opening the main pore (Fig. 1c). Removal of the CTT resulted in larger currents compared to the wild type (Fig. 1d, Extended Data Fig. 7a–c). The main pore narrows along the pore axis toward the extracellular side, with a small opening at the ECD entrance restricted by seven W74 residues.
Figure 1: PANX1 overall architecture.
Odd- and even-numbered subunits are in blue and white, respectively; the 7th subunit is in green. a, The cryo-EM map of wt-hsPANX1 viewed parallel to the membrane. The unsharpened map is shown as a transparent envelope. Lipid-like densities and N-acetylglucosamine (NAG) densities are in yellow and red, respectively. The position of the disordered CTT is outlined by a dashed ellipse. b, The structure of wt-hsPANX1, side (left) and bottom (right) views. c, The 2D class averages in side view, and the 3D classes obtained without imposing symmetry in both side and bottom views for ΔCTT-hsPANX1 and apo-wt-hsPANX. The CTT density in the 2D averages and 3D maps are labeled. d, Whole-cell CBX-sensitive current density (+80 mV, 5 ms) from patch-clamped tsA201 cells (n=6 cells), and tsA201 cells overexpressing wt-hsPANX1(n=12) and mutants (Δ2–20, n=5; N255A, n=5; ΔCTT, n=4). Each point represents one cell and the bar represents the mean. For statistical comparisons to WT, a two-tailed unpaired Mann-Whitney test with Bonferroni correction was applied (The p values are 0.748, 0.011, and 0.013, respectively, for Δ2–20, N255A, and ΔCTT). Asterisk indicates p<0.05. e, The structure and 2D class average (side view) of N255A-hsPANX1 gap junction shown in surface representation. The docking of two hemichannels is mediated through the EL2 linker, where D256, S257, and T258 in the paired subunits form hydrophilic and hydrophobic interactions.
In the TMD, four transmembrane helices (S1-S4) frame the exterior wall, while the NTH lines the interior wall in a domain-swapped manner—the NTH of one subunit interacts with the TMD of the adjacent subunit (Fig. 1b). Truncating the NTH yielded a TMD in a truncated elliptic cone shape (Extended Data Fig. 2b, 6a, b), yet generated CBX-sensitive, voltage-dependent currents similar to that of the wild type (Fig. 1d, Extended Data Fig. 7a–c). We thus suggest a role for the NTH in maintaining a rigid TMD while keeping a large pore.
While the ECD and the lower part of the ICD form close contacts between neighboring subunits, there are gaps in the TMD and upper part of the ICD (Fig. 1a). The gap in the TMD is filled with lipid-like densities, similar to LRRC8A25,26. However, the gap in the upper ICD is solvent-exposed and connects to the main pore, implying a novel ion pathway. We also observed several strong lipid-like densities surrounding the upper part of the TMD, forming a lipid belt (Fig. 1a).
The gap junction N255A-hsPANX1Gap is formed by two hemichannels docked in a head-to-head manner solely through the extracellular linker 2 (EL2) in the ECD (Fig. 1e; Extended Data Fig. 4c, d). This results in a substantially smaller contact area (122 Å2 per paired subunit) than that of connexin-46 (942 Å2) or innexin-6 (1550 Å2), in which both the EL1 and EL2 linkers participate in the docking. The N255A mutant also forms a hemichannel and produces CBX-sensitive current (Fig. 1d; Extended Data Fig. 7a–c). Our data provide direct evidence that glycosylation is a key determinant for PANX1 gap junction formation, though it is worthwhile noting that the N255A gap junction structure may not represent a normal physiological conformation of the channel41.
CBX blocks the extracellular entrance
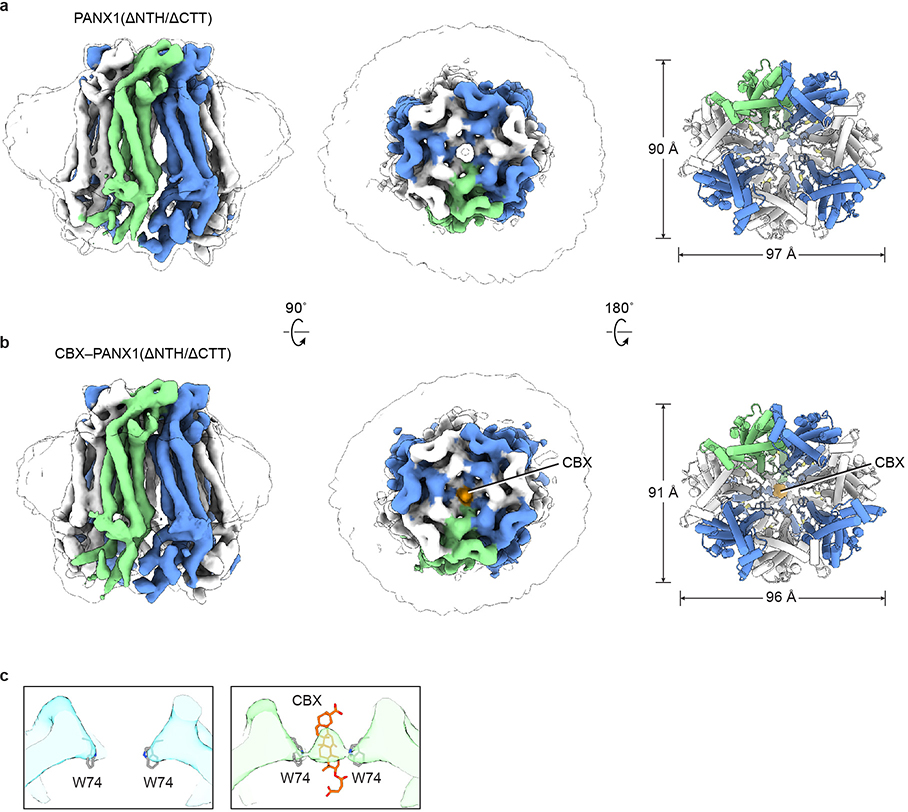
In the CBX-bound structures, there was a strong density at the extracellular entrance, which is surrounded by seven W74 residues (Fig. 2a–c). We assigned this density to CBX based on several pieces of evidence. First, this density does not exist in the structures without CBX (Fig. 2c; Extended Data Fig. 2, 6). Second, the extracellular entrance provides an ideal chemical environment for the binding of CBX: the aromatic side chain of W74 interacts with the steroid-like core of CBX; the N-terminus of helix EH1 and the side-chain of R75 provide favorable electrostatic potential to accommodate the two terminal carboxyl groups of CBX; and the molecular size of CBX fits well in the extracellular entrance. Third, the W74A and R75E mutations abolished and reduced CBX-dependent PANX1 inhibition18,42, respectively (Fig. 2d, Extended Data Fig. 7a, c). A recent study also showed that the CBX inhibition is much reduced for the R75A mutant of frog PANX143. Finally, this binding site is supported by the loss of symmetry of the structure—the asymmetric CBX overlaps with the symmetry axis, which breaks the C7 symmetry (Fig. 2a, Extended Data Fig. 2b). Taken together, our data support CBX being a pore blocker for PANX1.
Figure 2: CBX binding site of hsPANX1.
a, The structure of CBX-ΔCTT-hsPANX1 viewed from the intracellular side. b, The CBX binding site in the extracellular entrance. Subunits are shown alternately in blue and white. The three subunits in the front are hidden for clarity. c, The slice view of the extracellular entrance of ΔCTT-hsPANX1 and CBX-ΔCTT-hsPANX1 maps, respectively. The CBX and the side chain of W74 are shown as stick. d, Whole-cell current density (+80 mV, 5 ms) from patch-clamped tsA201 cells overexpressing wt-hsPANX1 (n=12 cells), W74A (n=8), and R75E (n=5) before and after superfusion of extracellular solution containing 0.1 mM CBX. Each paired point represents an individual cell and the bar represents the mean. CBX inhibition for each construct was evaluated using a two-tailed paired Wilcoxon test with Bonferroni correction. The p values are 0.0015, 1.00, and 0.18, respectively for WT, W74A, and R75E.
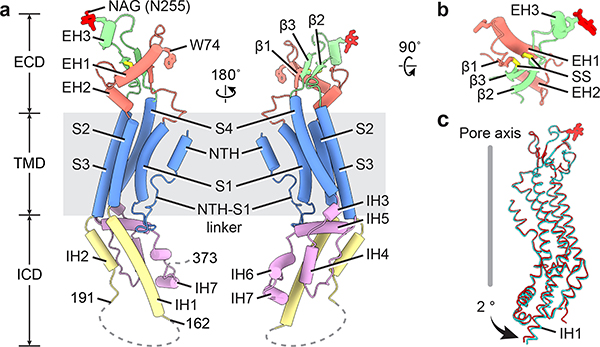
Single subunit
Each hsPANX1 protomer consists of a TMD connecting the ECD and ICD (Fig. 3a). The ECD is formed by two extracellular linkers, EL1 and EL2, mainly through two types of interaction. The β1 strand in EL1 forms an intact sheet with the β2 and β3 in EL2, and this interaction is strengthened by two disulfide bonds, one connecting β1 and β3 and the other connecting helix EH1 and β2 (Fig. 3b). The EH1 helix has an angle of approximately 45° relative to the membrane and is the major component that forms the extracellular entrance through its W74 residue. At the top of the ECD in helix EH3, N255A is glycosylated, which prevents two hemichannels from docking into a gap junction (Fig. 3a, b, Extended Data Fig. 1g). The N255A mutation caused a subtle change of the EH3, and a conformational rearrangement in the ICD characterized by an outward shift in the intracellular helix IH1, which results in an expansion of the ICD (Fig. 3c).
Figure 3: A single hsPANX1 subunit.
a, The disordered IH1-IH2 linker (residues 163–190) and the CTT (residues after 373) are indicated by dashed lines. b, The ECD of single subunit viewed from the extracellular side. SS stands for disulfide bond. c, Superimposition of the subunits of wt-hsPANX1 (red) and N255A-hsPANX1 (cyan) aligned by ECD.
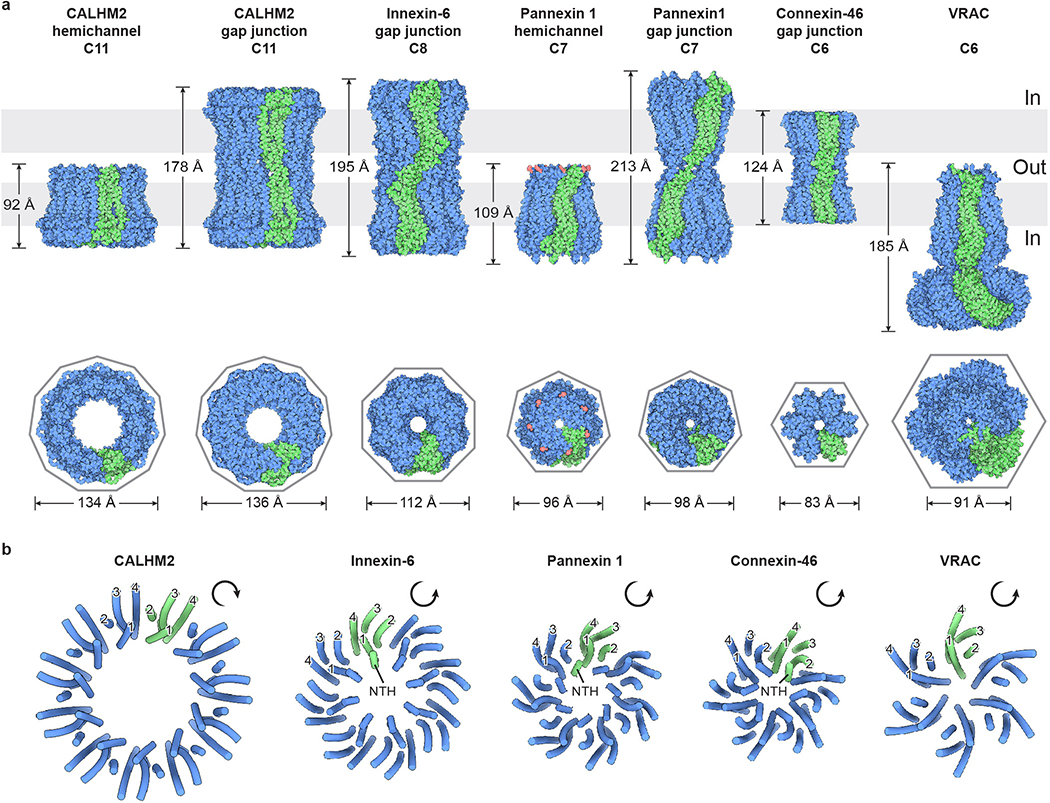
The TMD consists of a short NTH helix and a tight helix bundle formed by S1–4 (Fig. 3a). The NTH of hsPANX1 is unique among the large-pore channels because of its location. It spans the extracellular half of the TMD and, as a result, connects to S1 through a long and flexible linker (NTH-S1 linker); both the NTH and the linker line the main pore. In contrast, the NTH in connexins and innexins spans the intracellular half and is connected to S1 by a short linker. The NTH in VRAC is only partially visible in the structure and runs along the intracellular face of the membrane23–26. Notably, in the recently published frog PANX1 structures, the NTH and NTH-S1 linker are positioned in the intracellular side but not within the TMD42,43. The helices S1–4 of PANX1 are arranged in a counterclockwise manner as viewed from intracellular side, with S1 tightly clamped between S2 and S4. Such an arrangement is the same as in connexins, innexins, and VRAC, but it is distinct from that of CALHM2, in which the S1 to S4 are arranged clockwise and S1 is loosely attached to S3 (Extended Data Fig. 8).
The ICD is a helix-rich structure formed by the linker connecting S2 and S3, and the residues after S4 (Fig. 3a). The IH1 helix is long, extending deeply into the intracellular space as an extension of S2; it constitutes the main body of the ICD and is surrounded by six short helices. Such a long IH1 is reminiscent of the long cytoplasmic helix CH1 of CALHM2, yet they have different structural roles. The IH1 in hsPANX1 has an angle of approximately 60° relative to the membrane and is not involved in intersubunit interactions, whereas the CH1 in CALHM2 is nearly parallel to the membrane and participates extensively in intersubunit interactions27,28. The CTT is located downstream of the last intracellular helix, IH7, right after the caspase cleavage site. Although we were unable to model the CTT due to its intrinsically disordered nature, we have defined its key role in caspase-dependent channel gating (see Ion pathways and channel gating).
Channel assembly
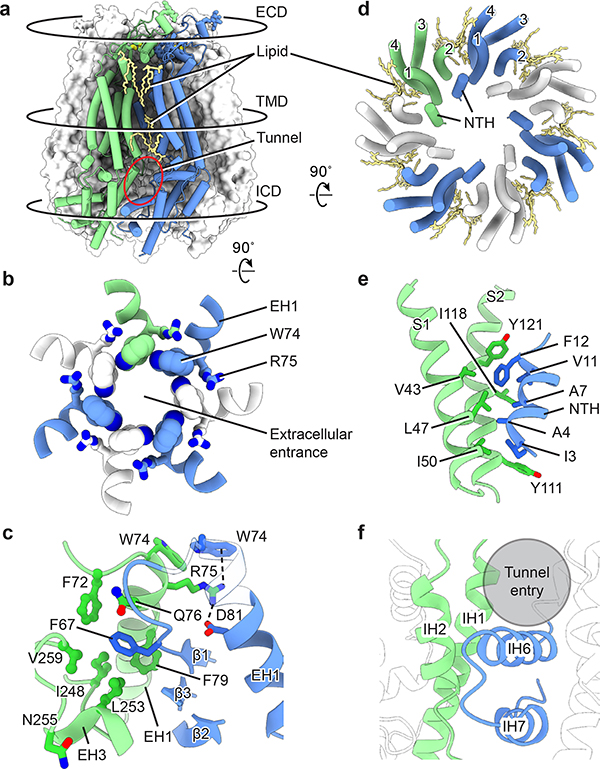
hsPANX1 is assembled through extensive interactions between the adjacent ECDs and between the adjacent lower ICDs, whereas the TMDs and upper ICD barely form contacts, creating a shallow crevice (Fig. 4a). While the crevice in the TMD is occupied by lipids, the one in the ICD is solvent-exposed, resulting in a tunnel toward the main pore.
Figure 4: Channel assembly of hsPANX1.
a, An overview of the intersubunit interfaces at the ECD, TMD and ICD using wt-hsPANX1 model. b, The intersubunit interface at the ECD, viewed from the intracellular side. The extracellular entrance is formed by W74 on the EH1 helix. c, The ECD intersubunit interface viewed parallel to the membrane. R75 forms a cation-π interaction and a salt bridge with adjacent W74 and D81, respectively; F67 is inserted in a hydrophobic pocket in the adjacent subunit. Part of the EH1 in is transparent. d, The intersubunit interface of the TMD between NTH and the adjacent S1 and S2, viewed from the intracellular side. Only the TMD helices are shown. The gap between adjacent TMDs is filled with lipids. e, The TMD interface viewed parallel to the membrane. f, The intersubunit interface at the lower part of the ICD between the IH1 and IH2 helices and the adjacent IH6 and IH7 helices. A crevice in the upper part of the ICD forms a tunnel that connects to the main pore.
The ECD of hsPANX1 has a contact area of 1069 Å2 between adjacent subunits, which is larger than that of VRAC (936 Å2), innexin (889 Å2), connexin (342 Å2), and CALHM2 (57 Å2). The most remarkable feature are seven W74 residues ringing the pore center and forming an extracellular entrance (Fig. 4b), with the Trp pyrrole ring’s Nε1 atom pointing to the benzene ring in the adjacent residue. This entrance is encircled by a second layer of interaction where R75 is sandwiched between W74 and D81 of the adjacent helix EH1, forming a cation-π interaction and a salt bridge, respectively (Fig. 4c). These two layers of interaction form a rigid extracellular entrance containing positive charges. Interestingly, W74R or R75D/E mutations decreased the stability of hsPANX1 (Extended Data Fig. 1h–j), suggesting an important role of the cation-π interaction and salt bridge in channel assembly. The radius of the extracellular entrance is 4.4 Å, which is about the size of an ATP molecule. In addition to EH1, a β sheet tightly organizes the close contact between adjacent ECDs (Fig. 4c). Specifically, in the center of the ECD, F67 is inserted into a hydrophobic pocket formed by helices EH1 and EH3 of the neighboring subunit. On the peripheral side of the ECD, strands β2 and β3 form multiple hydrophilic interactions with the EH3 of the adjacent subunit.
The TMD of hsPANX1 has a two-layer architecture—the helix bundle S1–4 forms the exterior, while the NTH lines the ion-conducting pore (Fig. 4d). The NTH is clamped between the adjacent S1 and S2 in a domain-swapped manner, creating the only interactions between neighboring TMDs (Fig. 4e). Such a domain-swapped interaction in the TMD is not present in connexin, innexin, or VRAC (Extended Data Fig. 8).
The interface in the ICD is restricted to its lower part, where helices IH1 and IH2 interact with helices IH6 and IH7 from the adjacent subunit through hydrophobic and hydrophilic interactions (Fig. 4f). In contrast, the upper part of the ICD lacks interfaces and has several positively charged residues, resulting in a tunnel between adjacent ICDs that is favorable for anions such as chloride but is not large enough for small molecules such as ATP.
Ion pathways and channel gating
To understand how hsPANX1 gates under different physiological conditions, we investigated potential ion-conducting pathways in the structures other than the main pore along the symmetry axis. We have strong evidence that the seven side tunnels in the upper ICD (Fig. 5a, c) serve as an alternative pathway for conducting ions, and we have thus established a gating mechanism involving the two pathways by combining data from cryo-EM, patch-clamp electrophysiology, and molecular dynamics simulation.
Figure 5: Ion conducting pathways and channel gating.
a, An overview of the main pore (gray body) and the side tunnels (green) in wt-hsPANX1. The pore lining structural elements of are shown as cartoons. The position of the disease-causing, gain-of-function mutation in the NTH-S1 linker (Δ21–23) is highlighted by a red box. b, The interior surface of wt-hsPANX1 colored according to the electrostatic surface potential from –3 to 3 kT/e (red to blue). c, The key elements that constitute the side tunnel. IH5 is shown as a transparent tube for clarity. d, The size and electrostatic potential (EP) profiles of the main pore and side tunnel. In the upper panel, the y-axis is numbered according to PDB coordinates along z dimension; the location of the side tunnel and W74 is marked. e, MD simulation of hsPANX1. Left, density of water molecules projected using a range of 127.6–145.6 Å along the z dimension (PDB coordinates) covering the side tunnel. Right, average root mean square fluctuation (RMSF) of single hsPANX1 subunit Cα atoms from 100-ns MD trajectory. The NTH-S1 linker (red), IH1 and IH2 (blue), and IH6 and IH7 (green) showed the highest flexibility; the NTH-S1 linker gates the tunnel, whereas the IH1, IH2, IH6, and IH7 constitute the periphery of the tunnel. f, Whole-cell CBX-sensitive current density (+80 mV, 5 ms) from patch-clamped tsA201 cells overexpressing wt-hsPANX1 (n=12) and mutants A33W (n=10), A33C (n=5), Δ21–27 (n=7), Δ21–23 (n=6), R29A (n=9), ΔCTT (n=4), A33W/ΔCTT (n=4). For statistical comparisons to WT, a two-tailed Mann-Whitney test with Bonferroni correction was applied. The p values are 0.0006, 1.00, 0.011, 0.006, 0.003, 0.031, and 0.031, respectively, for A33W, A33C, Δ21–27, Δ21–23, R29A, ΔCTT, and A33W/ΔCTT. Asterisk indicates p<0.05. g, A cartoon showing the two ion pathways, and the mechanism of CBX blocking.
The main pore is in the shape of a truncated cone. Both the extracellular and intracellular entrances are positively charged (Fig. 5b, d), making it favorable for negatively charged cargos such as chloride and ATP. Its extracellular entrance is constricted by seven W74/R75 residue pairs in a rigid conformation. We investigated the functional importance of these residues by performing electrophysiological experiments. While the W74A mutant remained voltage-dependent similar to the wild type, its anion selectivity was altered so that iodide and chloride showed equal permeability, contrasting to wild type which prefers iodide over chloride44 (Extended Data Fig. 7a, b d–f). Moreover, a charge reversing mutant, R75E, showed not only a loss of voltage-dependence and anion selectivity, but also a striking increase in permeability of sodium over chloride (Extended Data Fig. 7a, b, d–f). We thus suggest that the extracellular constriction site plays a role in discriminating cargo based on the charge and size, but it is unlikely to act as a gate that alters its conformation during channel opening and closing. This is in agreement with a recent study where mutants of the extracellular entrance in frog PANX1 also affect ion selectivity42.
Although the NTH lines the main pore within the TMD, it likely does not act as a gate either, because it is clamped by the adjacent S1 and S2 thus lacks the flexibility to close the pore. Indeed, an NTH-deletion construct produced CBX-sensitive conductance similar to that of the wild type (Fig. 1d, Extended Data Fig. 7a–c). Instead, we observed that the intracellular entry is blocked by the unstructured CTT (Fig. 1c). Although it has been proposed that the autoinhibitory CTT is cleaved by caspase 3 or 7 to open the channel during apoptosis2,33,34, our study provided a first visualization of such a gating mechanism, supporting the concept that the main pore is gated by CTT in a caspase-dependent manner.
This leads to the question of how hsPANX1 is gated under normal (nonapoptotic) conditions, given that it produces a small but measurable ionic conductance without caspase treatment33,35 (Fig. 5f; Extended Data Fig. 7a, b, c). We believe the seven tunnels at the ICD inter-subunit interface play a role. These tunnels contain several positively charged residues along with other polar residues, which make them accessible to solvent and anions (Fig. 5a–d). The peripheral part of the tunnel, surrounded by the S1 and IH5 and the S2 and IH1 of adjacent subunits, is wide enough to accommodate fully hydrated chloride ions (Fig. 5d). The tunnel gradually narrows toward the interior part, which is formed by the adjacent NTH-S1 linkers. Notably, deletions (Δ21–23 disease mutant3 and Δ21–27) on this linker result in a gain-of-function phenotype (Fig. 5a, f; Extended Data Fig. 7a–c). We thus propose that the NTH-S1 linker may act as a gate, and its shortening may result in less blockage of the tunnel. Within the tunnel, we identified two constriction sites formed by R29 and A33 on the NTH-S1 linker, having a radius of 2.9 and 2.0 Å, respectively, suggesting that partial dehydration is likely required for chloride ions to pass through the tunnel.
To provide evidence for our hypothesis that the side tunnel is an alternative ion-conducting pathway and that the flexible NTH-S1 linker functions as a gate, we performed molecular dynamics simulations and electrophysiology experiments. The simulation showed that this tunnel was hydrated and that the NTH-S1 linker had high flexibility (Fig. 5e). Next, we generated mutants on the two constriction residues, R29A and A33W, aiming to respectively enlarge or shrink the tunnel. As expected, the R29A mutant yielded ~30-fold larger CBX-sensitive current relative to the wild type, but the A33W mutant completely abolished CBX-sensitive current (Fig 5f; Extended Data Fig. 7a–c). In contrast, truncating the CTT from the A33W mutant generated large currents comparable to the ΔCTT-hsPANX1, indicating that the side tunnel and the main pore are gated independently (Fig. 5f). As a control, mutating A33 to a residue of similar size, i.e., Cys, did not alter the channel function (Fig. 5f). Together, our structural and functional data strongly support the idea that the tunnels constitute an alternative ion pathway that is gated by the NTH-S1 linker and operates independently from the CTT. We found that such a side tunnel also exists in innexin and VRAC; thus, such tunnels might be common feature of these large-pore channels.
Discussion
Our studies on hsPANX1 addressed long-sought questions regarding its channel architecture, gap junction formation, ligand-binding site, ion pathways, and mechanisms of channel gating and inhibition. Our structures support a previous study that suggests the TM1-β1 linker lines the channel pore, and the NTH is involved in channel assembly45. We have identified two ion-conducting pathways—the main pore along the symmetry axis, and seven side tunnels in the ICD (Fig. 5g). Under normal physiological conditions, the intracellular entrance of the main pore is blocked by the unstructured CTT, so anions—but not ATP—can move solely through the side tunnels, which explains why PANX1 currents can be evoked with voltage clamp during which no ATP release was observed1,46. During apoptosis, the CTT is cleaved by caspase, which opens the main pore for the release of ATP, triggering signals that attract phagocytes to engulf the dying cells. Both pathways can be blocked by CBX because it occupies the entryway at the extracellular side.
Notably, PANX1 has been reported to release ATP or open into large conductance state through tyrosine phosphorylation38,39. In our structures, these tyrosine residues are located in the lower portion of the ICD and their phosphorylation might regulate the conformation of the CTT nearby. Further studies are required to fully understand the phosphorylation-dependent activation mechanism.
Methods
Constructs and cloning
Full-length hsPANX1 (UniprotID: Q96RD7) was synthesized from GenScript and subcloned into a pEG Bacmam vector between XbaI and NotI restriction sites47. The C-terminus of the hsPANX1 gene contains a thrombin digestion site (Leu-Val-Pro-Arg-ll-Gly-Ser), an enhanced green fluorescent protein (eGFP), and StrepII and 8xHis tags. Caspase 3 and 7 plasmids were obtained from addgene (plasmid 11821 and 11825)48. Primers for site-directed mutagenesis were designed using NEBaseChanger (https://nebasechanger.neb.com/) and synthesized from Eurofins Genomics LLC. Q5 site-directed mutagenesis protocol was used to generate all the mutants (ΔNTH, W74A, N255A, Δ21–23, R29A, A33W, A33W/ΔCTT, ΔCTT, and A33C). The ΔNTH represents a hsPANX1 mutant in which residues 2–20 were deleted. The ΔCTT corresponds to a G380* (stop codon) mutation to mimic the caspase cleaved hsPANX1. Positive clones were confirmed via Sanger sequencing through Eurofins Genomics LLC.
Transfection and solubilization test
Adherent tsA201 cells were grown at 37°C in DMEM media with 10% FBS. Cells were transfected using lipofectamine-2000 by following the manufacturer’s protocol. The next day, sodium butyrate was added to 10 mM and the cells were incubated at 30°C. After 24 hours, cells were harvested and stored at −80°C. For fluorescence size-exclusion chromatography (FSEC) experiment, cells were solubilized using 500 μl of 150 mM NaCl, 20 mM Tris-HCl pH 8.0 (TBS) buffer with 1% GDN or SMA30010 for 1 hour at 4°C. The solubilized sample was clarified by ultracentrifugation at 87,000 g and analyzed by high-performance liquid chromatography (HPLC). The supernatant (20μl) was injected onto a Superose 6 Increase 5/150 GL column (GE Health) at a flow rate of 0.3 ml/min. GFP (480 nm) and tryptophan (280 nm) absorbance were monitored to determine protein solubility and retention volume.
Deglycosylation test
Adherent tsA201 cells infected with eGFP tagged wt-hsPANX1 or N255A-hsPANX1 were solubilized using 100 μL TBS buffer with 1% GDN at 4°C for 1h. The samples were then centrifuged at 20,000 g for 30 minutes. The supernatant (16 μL) was mixed with 2 μL of GlycoBuffer 2 and 2 μL PNGase F enzyme. The control reaction was made by adding 2 μL of water instead of PNGase F enzyme. After overnight incubation at room temperature, the sample was mixed with 10 μL 2x SDS loading buffer (with 5% βME) and resolved by SDS-PAGE. GFP signal (UV488) of the gel was detected using the ChemiDoc imaging system.
Protein expression and purification
Caspase 3 and 7 protein was expressed in BL21 E. coli. strain (NEB) and purified using a protocol established previously49. For hsPANX1 protein, a mammalian cell expression system optimized for membrane ion channel protein was used50. Specifically, bacmid was produced by transforming pEG-hsPANX1 plasmid into DH10α cells. Positive colonies were selected from a LB plate with 50 μg/mL kanamycin, 10 μg/mL tetracycline, 7 μg/mL gentamicin, 40 μg/mL IPTG, and 100 μg/mL Bluo-gal. Bacmid from positive clones was extracted and transfected into adherent Sf9 cells using Cellfectin II reagent using the manufacturer’s protocol. Five days after transfection, P1 virus was filtered and harvested. P2 virus was produced by infecting suspension Sf9 cells at 0.9×10^6 cells/ml in a 5000:1 v/v ratio. Five days after infection, P2 virus was harvested and supplemented with 2% FBS. P2 virus was stored at 4°C and light protected.
Suspension tsA201 cells were grown at 37°C in FreeStyle 293 media supplemented with 1% FBS. The cells were infected with 5% v/v P2 virus at a density of 3.5×10^5 cells/ml and grown at 37°C. Twelve hours after infection, sodium butyrate was added to a final concentration of 5 mM and cells were grown at 30°C. Cells were harvested 56 hours post-infection and stored at −80°C until further use.
Infected tsA201 cells expressing hsPANX1(200 mL culture) were thawed on ice and suspended with TBS buffer. Cells were then solubilized with 1% GDN and a protease inhibitor cocktail (1 mM PMSF, 0.8 μM aprotinin, 2 μg/ml leupeptin, and 2 mM pepstatin A) for 1 hour at 4°C. The sample was clarified by centrifugation at 5000 g for 15 minutes and ultracentrifugation at 186,000 g for 1 hour. The supernatant was mixed with 2.5 mL TALON resin using a batch protocol. Imidazole (10mM) was added to the sample to prevent nonspecific binding. After 1 hour, the resin was washed using 2×10mL TBS buffer with 0.01% GDN with 20 mM imidazole. Protein was eluted with 4x TBS buffer with 0.01% GDN with 250 mM imidazole. The eluent was concentrated to 500 μL using a 100 kDa concentrator (Millipore). The C-terminal GFP was cleaved by thrombin (0.005 mg/ml) or caspase 7 (0.02 mg/mL). GFP-cleaved protein was further purified by size-exclusion chromatography (SEC) using a Superose 6 Increase 10/300 GL column (GE Healthcare) with TBS buffer containing 0.01% GDN as the mobile phase. Peak fractions were concentrated to 4–8 mg/mL for grid freezing.
For hsPANX1 protein purification using styrene-maleic acid (SMA) copolymer, 200 mL of thawed tsA201 cells were initially solubilized in 20 mL TBS buffer with 1% SMA30010 at 4°C. After 2 hours, the sample was diluted to 50 mL using TBS buffer, followed by 2 rounds of ultracentrifugation at 186,000 g for 1 hour each. The supernatant was then diluted to 150 mL final using TBS buffer and applied to 7.5 mL talon resin to allow batch binding at 4°C overnight. The resin was washed with 60 mL TBS buffer containing 10 mM imidazole. Protein was eluted with 22.5 mL TBS buffer containing 250 mM imidazole and concentrated to 500 μL using a 100 kDa concentrator. The GFP tag was cleaved with thrombin (0.05 mg/mL) at room temperature for 4 hours. GFP-free protein was further purified by SEC using TBS buffer. Peak fractions were concentrated to 7 mg/mL for grid freezing.
Cryo-EM sample preparation and data collection
Purified full-length hsPANX1 showed well-behaved biochemical properties in detergent or styrene maleic acid polymers (SMAs) (Extended Data Fig. 1a–f). The hsPANX1 protein sample was first mixed with designated additives/ligand to the desired concentration prior to the grid freezing. Quantifoil 300 mesh 2/1 Au or Cu grids were glow discharged for 30 s. The protein (3 μL) was added to the grid in Vitrobot Mark III at 100% humidity. Grids were then blotted for 2 s, flash-frozen in liquid ethane, and transferred/stored in liquid nitrogen.
Preliminary grid screening was conducted using a 200 kV Talos Arctica microscope equipped with a Falcon III direct electron detection detector (FEI). High-resolution data collection was conducted using a 300 kV Talos Krios microscope equipped with a K2 Summit or K3 direct electron detector (Gatan). SerialEM was used to automate the data collection using a super-resolution counting mode51. The calibrated pixel size for the K2 and K3 cameras are 1.026 Å and 0.812 Å, respectively. For the K2 camera, the total exposure time is 8 s for 40 frames with a dose rate of 6.2 e-/Å2/s for each micrograph. For the K3 camera, the exposure time is 1.5 s for 75 frames with a dose rate of 31.5 e-/Å2/s for each micrograph. The nominal defocus range was specified from −1.2 μm to −1.9 μm with a 0.1 μm step. The raw movies were saved in LZW compressed tif format for further processing.
Cryo-EM single particle analysis
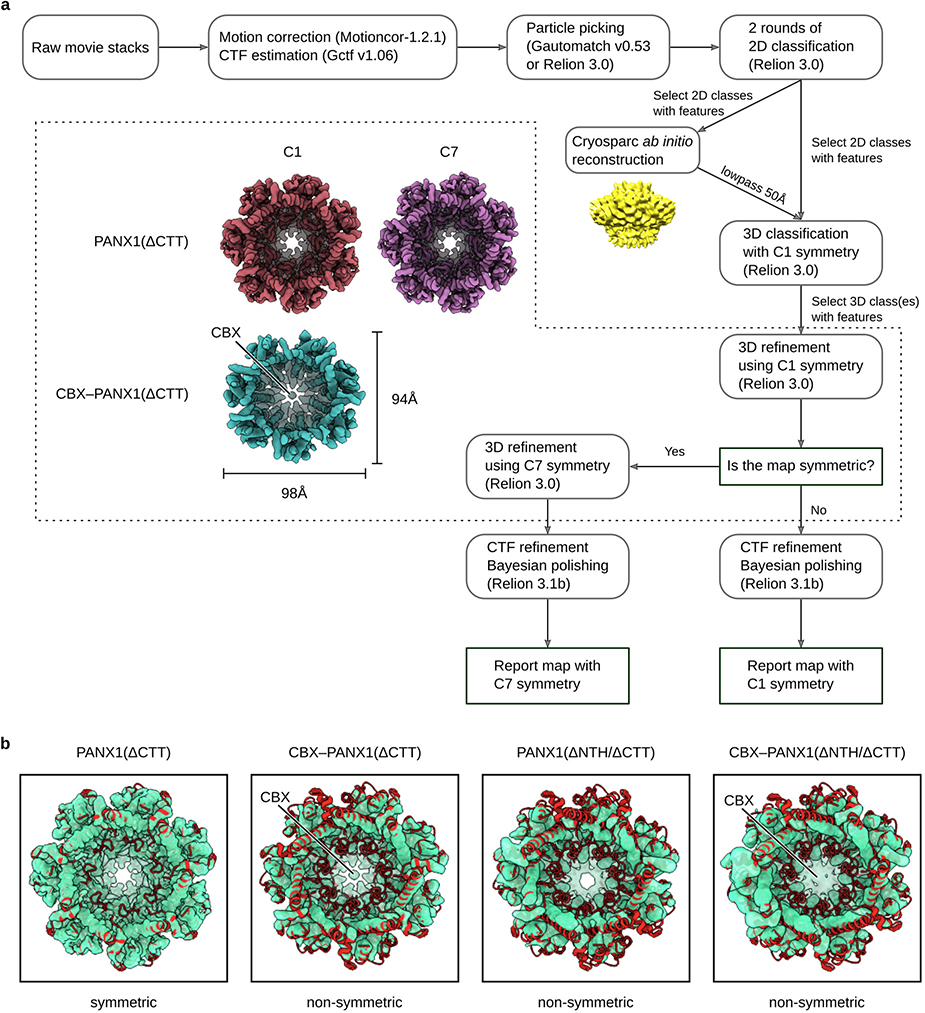
The data processing workflow and statistics is summarized in Extended Data Fig. 2 and 5, and Extended Data Tables 1–3. Specifically, raw movies were first motion-corrected using motioncor2 version 1.2.1 (ref 52). The contrast transfer function (CTF) of each micrograph was estimated using gctf version 1.06 or ctffind version 4.1.10 (ref 53,54). For K2 data, particle picking was conducted using gautomatch (http://www.mrc-lmb.cam.ac.uk/kzhang/). For K3 data, relion 3.0 was used for particle picking55. Junk particles were removed by 2 rounds of reference-free 2D classifications in relion 3.0. The initial 3D model was generated using Cryosparc56. Particles were further cleaned by 3D classification. High-resolution classes with clear secondary structure features were used for 3D refinement. The initial refinement was conducted using C1 symmetry without a solvent mask. A solvent mask was then generated and used for subsequent 3D refinement jobs. The map is then checked manually to identify if the structure adheres to C7 symmetry. CTF refinement and Bayesian polishing were also performed in relion3.1b to further improve the refinement result57,58.
For the N255A-hsPANX1 dataset, side and tilted views of the hemichannel particles and gap junction particles were separated during 2D classification. The top/down views of hemichannel and gap junction were difficult to separate from 2D class averages. We combined all top/down views with the hemichannel and gap junction dataset, and classified the particles using 3D classification (Extended Data Fig. 5). After a homogeneous particle set for hemichannel or gap junction was obtained, the map was refined using the same procedure as the other non-gap junction datasets.
Structural determination
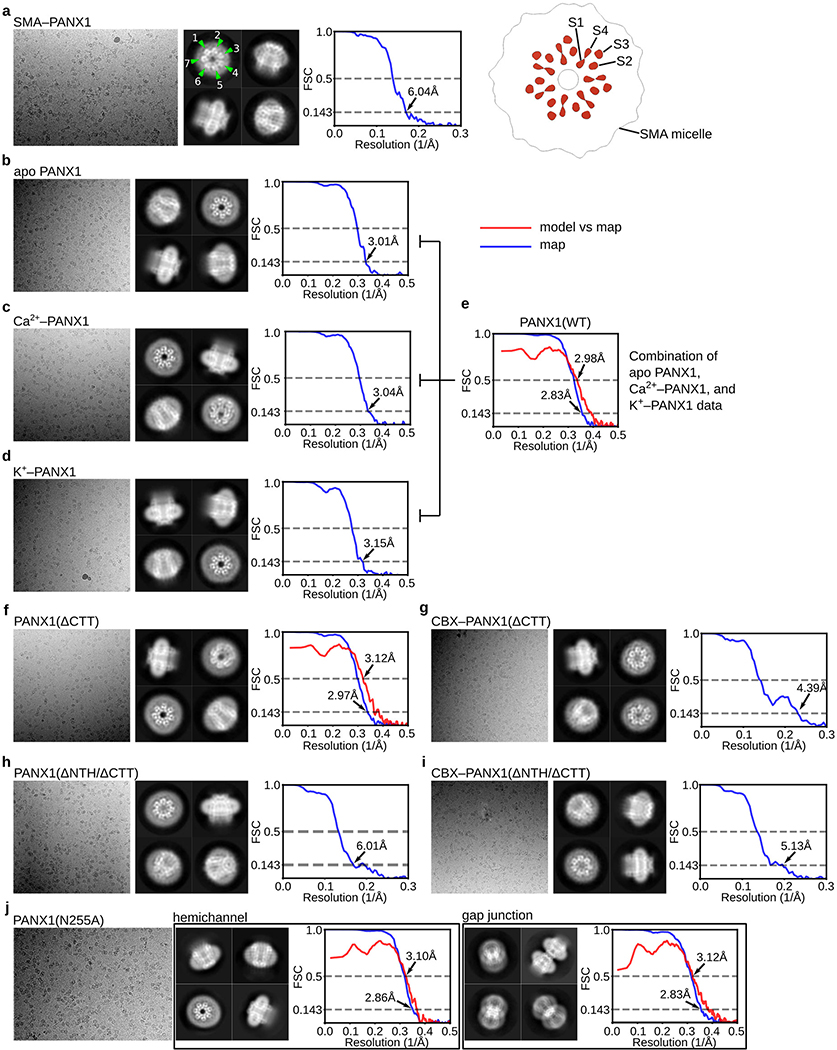
We focused on hsPANX1 in detergent because it yielded cryo-EM maps at substantially higher resolutions than hsPANX1 in SMA (Extended Data Tables 1–3). We first determined the hsPANX1 structures (Extended Data Fig. 2a, 3b–d) in the presence of 1 mM EDTA, 5 mM Ca2+, or 150 mM KCl, aiming to capture the apo, Ca2+-, and K+-activated states, respectively. We also wanted to address the question of whether Ca2+ or K+ directly activates hsPANX1 given the contradictory evidence in the literature16,34,36,59–61. All three structures formed only hemichannels having 7-fold symmetry. The heptameric stoichiometry was confirmed by the structure of SMA-extracted hsPANX1 (Extended Data Fig. 3a). Because no noticeable difference was observed among these three structures, we combined the data, which yielded a structure with improved quality at 2.8 Å (wt-hsPANX1; Fig. 1a, b; Extended Data Fig. 3e; Extended Data Fig. 4a, b; Extended Data Table 1). Our structures suggest that likely neither Ca2+ nor K+ directly activates hsPANX1.
The CTT after the caspase cleavage site is predicted to be disordered62; it is invisible in the cryo-EM maps refined with C7 symmetry (Fig. 1a). However, we observed densities covering the intracellular entrance in the 3D class obtained without imposing symmetry (Fig. 1c). Because the location matches where the CTT was predicted to be by functional studies34, we determined the structure of hsPANX1 after cleaving the CTT by using caspase 7 (ΔCTT-hsPANX1; Extended Data Table 2). Although the C7 refined structure of ΔCTT-hsPANX1 is identical to the full-length hsPANX1 structure, the densities blocking the intracellular entrance disappeared in the 3D class without imposing symmetry (Fig. 1c). We thus concluded that these densities are indeed derived from the flexible CTT.
We observed extra density associated with the side chain of N255, indicating that this asparagine is glycosylated, which was confirmed by a deglycosylation assay (Extended Data Fig. 1g). PANX1 was thought to not form gap junction due to the N-glycosylation in its extracellular loop30,31,63. To provide a structural basis for this idea, we studied the structure of a glycosylation-deficient mutant N255A (Extended Data Fig. 5), which yielded both gap junctions (N255A-hsPANX1Gap) and hemichannels (N255A-hsPANX1Hemi) at an approximate ratio of 1:5 (grid concentration, about 23 μM) at resolutions of 2.83 and 2.86 Å, respectively (Extended Data Fig. 3j, 4c, 4d; Extended Data Table 3).
To study PANX1 inhibition, we determined the structure of the complex with CBX (CBX-ΔCTT-hsPANX1). Because the structure showed obvious asymmetry and the CBX site overlapped with the symmetry axis, we refined the structure without imposing any symmetry to avoid symmetry axis–associated noise or artifacts; the final resolution was 4.4 Å (Extended Data Fig. 3g; Extended Data Table 2). In addition to the change in overall structure, we noticed two differences between the CBX-ΔCTT-hsPANX1 and the ΔCTT-hsPANX1 map. First, the center of the extracellular restriction site (W74) is filled with strong density in the CBX-ΔCTT-hsPANX1 map, but not in the ΔCTT-hsPANX1 map. Second, the NTH density is weaker and seems to have been shifted in the CBX-ΔCTT-hsPANX1 map when compared to the ΔCTT-hsPANX1 map. We were initially not confident to assign NTH to this density in the cryo-EM map of CBX-ΔCTT-hsPANX1. To resolve this uncertainty, we studied the structure of a hsPANX1 mutant in which residues 2–20 were deleted (ΔNTH mutant). Both ΔNTH/ΔCTT-hsPANX1 and CBX-ΔNTH/ΔCTT-hsPANX1 structures are non-symmetric and adopt an elliptical shape similar to CBX-ΔCTT-hsPANX1 (Extended Data Fig. 6a, b; Extended Data Table 2). In addition, the “shifted” NTH density is not observed in both ΔNTH/ΔCTT-hsPANX1 and CBX-ΔNTH/ΔCTT-hsPANX1 structure, suggesting that the “shifted” density in CBX-ΔCTT-hsPANX1 is indeed NTH. Additionally, we observed strong density located in the center of the extracellular restriction site for CBX-ΔNTH/ΔCTT-hsPANX1, but not for the ΔNTH/ΔCTT-hsPANX1 dataset (Extended Data Fig. 6c), strongly suggesting that CBX binds to the location. Therefore, our structural analysis has ruled out the possibility that CBX can compete with NTH for hsPANX1 binding, but instead supports the idea that CBX functions as a pore blocker and binds to the extracellular restriction site (W74). We were unable to unambiguously define the exact positioning of CBX for two reasons. First, the two carboxyl groups of CBX lack direct interaction with the protein and are thus flexible. Second, the density is an average of the many different poses which CBX may adopt within the extracellular entrance.
Model building, validation, and structural analysis
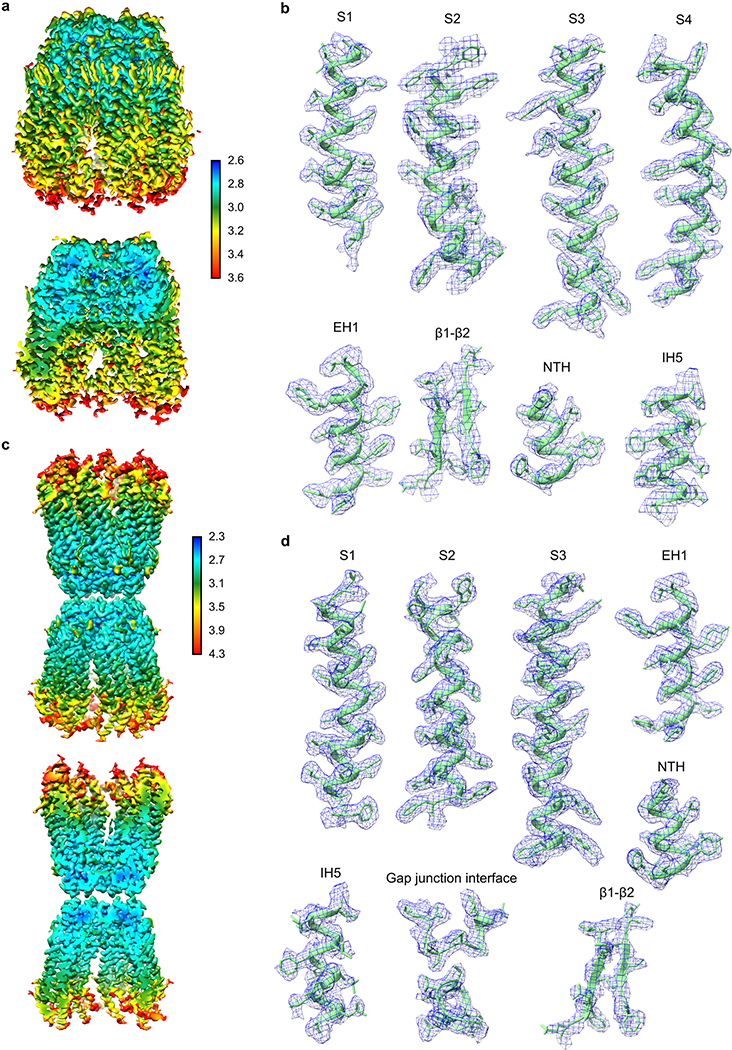
The cryo-EM map of wt-hsPANX1 and ΔCTT-hsPANX1 are of excellent quality, allowing us to model almost the entire protein. De novo model building was initially conducted using Coot64. Registers were guided by secondary structure prediction (Extended Data Fig. 9) using JPred65 webserver and the bulky residues of hsPANX1. We were able to model most of the wt-hsPANX1 except for the linker connecting the intracellular helices IH1 and IH2 (residues 163–190), and the CTT (residues 374–426). For regions of the map with limited side-chain information (residue 19–26, 302–305, 316–327, and 356–373), we only modeled the residue backbone. Extra density in N255 is modeled as N-acetyl-d-glucosamine (NAG) based on biochemical analysis on WT and N255A mutant. Putative lipid density in the map is modeled as cholesterol (CLR), diacylglycerol (DGA), phosphatidylethanolamine (PTY), distearoyl-3-sn-phosphatidylethanolamine (PEH) based on the shape of the lipid density. However, we don’t have experimental evidence to claim the exact identity of these lipid molecules. The atomic model was refined using phenix.real_space_refine and evaluated using phenix.mtriage66.
CBX-ΔCTT-hsPANX1, ΔNTH/ΔCTT-hsPANX1, and CBX-ΔNTH/ΔCTT-hsPANX1 mutant models were generated using molecular dynamics based flexible fitting (MDFF) with the wt-hsPAXN1 model as the input67. The resulting model was manually adjusted and refined using phenix real-space refinement66. CBX molecules were manually fitted into the center of the extracellular entrance. We provided strong structural and functional evidence to support the proposed CBX binding site. However, the two terminal carboxyl groups of CBX do not make specific interactions with the protein, and are therefore not well defined in the map. Models were validated using phenix.molprobity68.
The N255A-hsPANX1Gap and N255A-hsPANX1Hemi channel models were built by docking the wt-hsPANX1 model into the gap junction and hemichannel map. We manually adjusted the position of IH1 and IH7 helices and mutate N255 to alanine in Coot64. The resulting model was subjected to phenix real-space refinement and validation66,68.
Solvent accessible contact interface is measured using the get_area function in PyMOL69. Electrostatic potential was calculated using the APBS Plugin embedded in PyMOL70. Tunnel and size analysis were done using Caver 3.071. Figures were generated using PyMOL and UCSF Chimera/ChimeraX72,73.
Electrophysiology
Using patch-clamp analysis, whole-cell currents were measured in tsA201 cells plated on poly-l-lysine-coated (Sigma) coverslips following 1–2 days of over-expression. C-terminal GFP tagged hsPANX1 constructs were used for visual identification. The only untagged mutants were ΔCTT and A33C/ΔCTT which were co-expressed with a plasmid encoding GFP. The ΔCTT-hsPANX1 used in Fig. 1f and Extended Data 7 is a G380 stop codon mutation to mimic the CTT-cleaved hsPANX1. Both viral infection (cells infected at 5% v/v of P2 virus, incubated at 37°C for 10–12 hours and then at 30°C with media containing 5 mM sodium butyrate) and Lipofectamine 2000 transfection were used for overexpression. Since no differences in current amplitudes were observed between methods for wt-hsPANX1 and various mutants, the data were pooled. Patch pipettes contained (in mM) 145 NaCl, 10 Hepes, 10 EGTA, pH 7.3 and the bath solution contained 160 NaCl, 10 Hepes, 3 KCl, 2 CaCl2, and 1 MgCl2, pH 7.4. Voltage steps of 0.25 s were imposed from −100 mV to +80 mV and membrane currents were recorded (digitized at 10 kHz and filtered at 2 kHz) using a Multiclamp 700B microamplifier (Axon Instruments) and Clampex software (11.0.3). CBX (Sigma) was added to the bath solution at a final concentration of 0.1 mM. Currents were analyzed off-line using ClampFit (11.0.3). Current amplitudes were measured 5 ms at a clamp of −80mV. CBX-sensitive currents were calculated from the difference in current amplitude in a cell measured with and without CBX. For measuring ion selectivity, whole cell currents were measured from patch-clamped cells imposed with 2 s voltage ramps (−100 mV to +100 mV) using a AgCl reference electrode and a 3M KCl-agar bridge. Cells were first measured in symmetrical NaCl concentrations where patch pipettes contained (in mM): 145 NaCl, 10 HEPES, 10 EGTA (pH 7.35) and the bath solution contained: 145 NaCl, 10 HEPES, 2 Ca-gluconate, 1 Mg-gluconate, pH 7.4. Sucrose was added to all bath solutions for a final osmolarity of about 325 mOsm / L. Sequential measurements were performed following superfusion with various bath solutions where NaCl was replaced with equimolar amounts of NaI, Na-gluconate, and NMDG-Cl. Anion permeability ratios were calculated using shifts (relative to NaCl) in the zero-current reversal potential using a modified Goldman-Hodgkin-Katz (GHK) equation74. Liquid junction potentials were minimal (< 2 mV) and uncorrected. For Na/Cl permeability ratios, zero-current reversal potentials were measured using a reduced salt bath solution containing (in mM): 14.5 NaCl, 10 Hepes, 2 Ca-gluconate, 1 Mg-gluconate, 245 sucrose and calculated using the GHK equation. Recordings were performed at room temperature. Statistical analysis of carbenoxolone-sensitive currents was performed with Mann-Whitney test (GraphPad Prism). P-values of <0.05 were deemed significant.
Molecular dynamics (MD) simulation
Lipid bilayer MD system with wt-hsPANX1 was prepared using CHARMM-GUI75. Specifically, the wt-hsPANX1 model was first oriented with respect to the membrane normal using the OPM server76. CHARMM-GUI was then used to build the lipid bilayer system with POPC on both sides of the membrane. The wt-hsPANX1 is protonated at neutral pH. The system was then solvated with 603 TIP3P water, and neutralized with approximately 150 mM NaCl (144 NA, and 165 CL ions). The final system has a dimension of 130Åx130Åx154Å.
CHARMM36m force field parameters are used for the simulation77. Initial energy minimization was performed using the steepest-descent algorithm with a Fmax tolerance of 500 kJ mol−1 nm-1. Position restraint was enabled for non-hydrogen atoms of the protein and phosphorus atom of POPC. The system then underwent 6 rounds of equilibration with the strength of the position restraint gradually decreased (from 4000 kJ mol−1 nm−2 to 200 kJ mol−1 nm−2 for backbone atoms, and 2000 kJ mol−1 nm−2 to 200 kJ mol−1 nm−2 for side-chain atoms). The temperature of the system was brought to 310 K using a Berendsen thermostat by coupling protein, POPC membrane, and solvent in NVT ensemble for the first 3 rounds of equilibration. Subsequently, NPT ensemble was launched using a Berendsen semiisotropic pressure coupling at 1bar. The linear constraint solver (LINCS) algorithm was used to constrain the hydrogen atoms78. NVT and NPT equilibration simulations were carried out using 1 fs and 2 fs timestep, respectively. The un-restraint production simulation continued from the NPT ensemble using Nose-Hoover thermostat and Parrinello-Rahman barostat. The trajectory was accumulated for 100 ns using a timestep of 2 fs. The Verlet cutoff scheme was used to generate the neighbor list (12 Å) for short-range electrostatic and van der Waals interactions. Gromacs version 2019.2 was used as the MD engine79. Trajectory data was analyzed using the utilities of gromacs and visualized using PyMOL69.
Extended Data
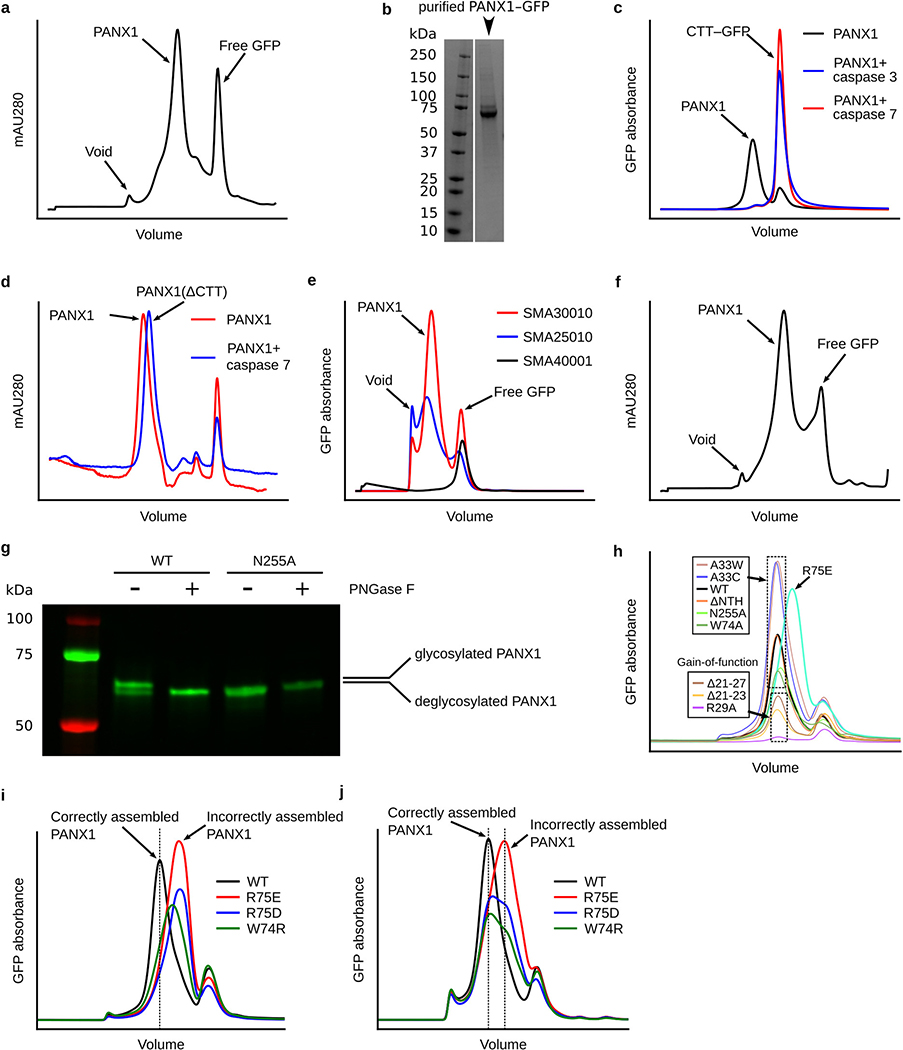
Extended Data Figure 1: Purification and biochemical analysis of hsPANX1.
a, Size-exclusion chromatography (SEC) profile of wt-hsPANX1 purification using glyco-diosgenin (GDN). b, SDS gel of purified wt-hsPANX1-GFP (For gel source data, see Supplementary Figure 1a for gel source data). c, Fluorescence size-exclusion chromatography (FSEC) experiment on caspase 3/7 cleavage of wt-hsPANX1-GFP. GFP absorbance (480 nm) is shown in y-axis. d, Caspase 7 cleavage of purified wt-hsPANX1-GFP. The cleavage of CTT results in a peak shift. Tryptophan absorbance (280 nm) is shown in y-axis. e, Styrene maleic acid (SMA) solubilization screening of wt-hsPANX1 using FSEC. Three SMA polymers (SMA25010, SMA30010, and SMA40001) were tested. GFP absorbance (480 nm) is shown in y-axis. f, SEC profile of hsPANX1 purification using SMA30010. Tryptophan absorbance (280 nm) is shown in y-axis. g, Deglycosylation test of wt-hsPANX1-GFP and N255A-hsPANX1-GFP using PNGase F. Bands correspond to the glycosylated and non-glycosylated hsPANX1 are marked. See Supplementary Figure 1b for gel source data. h, FSEC analysis on the hsPANX1 mutations with electrophysiology data. Cells expressing hsPANX1 WT or mutants are solubilized using GDN. Gain-of-function mutations with less expression level are labeled. R75E mutant contains a peak position shifted to the right. i and j, FSEC analysis on extracellular gate mutations of hsPANX1 solubilized using GDN (i) or SMA30010 (j). Peak positions of correctly assembled hsPANX1 and incorrectly assembled hsPANX1 are indicated by arrows and vertical bars. The W74R, R75D, and R75E showed decreased stability relative to wild-type because, when extracted using detergent, they mostly ran at positions representing incorrect assemblies (i). Nevertheless, the SMA-extracted W74R, R75D, and R75E all showed peaks at positions representing correct assemblies (j), indicating that they are able to form correctly assembled channel complex in a native lipid environment.
Extended Data Figure 2: The workflow of cryo-EM data processing of hsPANX1 constructs that do not form gap junctions.
a, The data analysis pipeline for hsPANX1 cryo-EM analysis with no gap junction. Special attention was paid to determine whether the dataset indeed adheres to C7 symmetry. Two examples of hsPANX1 adopting a symmetric conformation (ΔCTT-hsPANX1) or a non-symmetric conformation (CBX-ΔCTT-hsPANX1) are shown. A detailed description of the data analysis procedure can be found in the Method section. b, The overlay of the C1 refined maps of ΔCTT-hsPANX1, CBX-ΔCTT-hsPANX1, ΔNTH/ΔCTT-hsPANX1 and CBX-ΔNTH/ΔCTT-hsPANX1 with the symmetric model of wt-hsPANX1. The CBX-ΔCTT-hsPANX1, ΔNTH/ΔCTT-hsPANX1 and CBX-ΔNTH/ΔCTT-hsPANX1 maps adopt a non-symmetric shape.
Extended Data Figure 3: Representative micrographs, 2D class averages, and Fourier shell correlation (FSC) curves for all datasets in this study.
For each dataset, a representative micrograph, four 2D class averages and FSC curve plot are shown, except for N255A-hsPANX1 dataset, in which two structures are shown. The map resolution is determined based on the “gold-standard” 0.143 criterion. If an atomic model is available for the dataset, a model vs map FSC curve is also provided. The model vs map resolution is determined based on the 0.5 FSC criterion. Panel (a) contains a slice view of the SMA-wt-hsPANX1 map showing the organization of TMD.
Extended Data Figure 4: Local resolution estimation and representative densities.
a, b, wt-hsPANX1 map. c, d, The N255A-hsPANX1Gap map. To provide better visualization on the exterior and interior map quality, a non-sliced and a sliced view of the maps are shown. The unit for the color bar is in Å. The representative densities were shown for a few selected secondary structure elements of hsPANX1.
Extended Data Figure 5: The workflow of cryo-EM data processing of N255A-hsPANX1.
To separate gap junction particles from hemichannel particles, we relied on 2D classification to distinguish tilted and side views. The top/down views were separated during 3D classification. A more detailed description of the data analysis pipeline can be found in the Methods section.
Extended Data Figure 6: The structures of ΔNTH/ΔCTT-hsPANX1.
a, The apo state. b, In complex with CBX. The CBX is shown in orange. For both panels, odd- or even-numbered subunits are in blue or white, respectively; the 7th subunit is in green. The left and middle panels are cryo-EM maps viewed parallel to the membrane and from the extracellular side, respectively; the unsharpened map is shown as a transparent envelope. The right panels are structural models viewed from the intracellular side. c, The slice view of the extracellular entrance of ΔNTH/ΔCTT-hsPANX1 (left) and CBX-ΔNTH/ΔCTT-hsPANX1 (right) maps. The CBX molecule and the side chain of W74 are shown as stick.
Extended Data Figure 7: Patch-clamp analysis of the wild type hsPANX1 and its mutants.
a, Representative traces of whole-cell current density from patch-clamped tsA201 control cells (N=6) and tsA201 cells overexpressing: wt-hsPANX1 (n=12), ΔNTH (n=5), W74A (n=8), R75E (n=5), N255A (n=5), Δ21–23 (n=6), Δ21–27 (n=7), R29A (n=9), A33W (n=10), A33W/ΔCTT (n=4), ΔCTT (n=4), and A33C (n=5). Voltage steps (0.25 s) of 20 mV were imposed from −100 mV to +80 mV from a holding potential of −10 mV. Cells were first measured in standard bath solution and then re-measured following the superfusion of a bath solution containing 0.1 mM carbenoxolone. b, Mean current measured at 5 ms of experiments in (a) plotted as a function of clamp voltage. c, Current amplitudes of experiments in (a) with and without carbenoxolone. Each paired point represents an individual cell and the bar represents the mean. d, Plot of zero-current reversal potentials using a 2 s voltage ramp for various bath solutions containing either (in mM): 145 NaCl, 145 Na-Iodide, 145 Na-Gluconate, 14.5 NaCl, or 145 NMDG-Cl (see Methods for complete solutions). The number of cells patched (indicated in parenthesis) for the various bath solutions were as follows, 145 mM NaCl: WT (5), R29A (7), W74A (5), R75E (7); 145 mM NaI: WT (5), R29A (7), W74A (5), R75E (7); 145 mM Na-gluconate: WT (5), R29A (7), W74A (5), R75E (7); 14.5 NaCl mM WT (4), R29A (4), W74A (5), R75E (5); 145 mM NMDG-Cl WT (3), R29A (2), W74A (5), R75E (3). e, and f, Plots of the calculated (see methods) permeability of iodide relative to chloride, PI/PCl, the permeability of Gluconate relative to chloride, PGluconate/PCl, and permeability of sodium relative to chloride, PNa/PCl for WT and mutant channels. For statistical comparisons to WT, one-way ANOVA with Bonferroni correction was performed. The p values for the PI/PCl ratios are 0.99, 3.7×10−5, 5.9×10−5 for R29A, W74A, and R75E, respectively. The p values for the PGluconate/PCl ratios are 0.046, 0.99, 6.8×10−9 for R29A, W74A, and R75E, respectively. The p values for the PNa/PCl ratios are 0.99, 0.99, 2.2×10−6 for R29A, W74A, and R75E, respectively. Each point represents an individual cell and the bar represents the mean value. All error bars are SEM.
Extended Data Figure 8: Comparison of large-pore channels.
a, The structures of large-pore channels, viewed parallel (upper) or perpendicular (lower) to the membrane. One subunit (or one pair of subunits) is in green. The diameter of VRAC is calculated without the cytoplasmic leucine-rich repeat domain. b, Organization of the transmembrane domain, viewed from the intracellular side. The NTH and transmembrane helices S1, S2, S3, and S4 are labeled for two subunits. Only CALHM2 has its transmembrane helices arranged in a clockwise manner. The contact between adjacent TMDs in PANX1 is made by the NTH with the S1and S2 helices in the neighboring subunit; the same contact in CALHM2 is made by the S2 and S4 of adjacent subunits, and in connexin by the S1 and S2 of adjacent subunits. There is no major contact in innexin and VRAC.
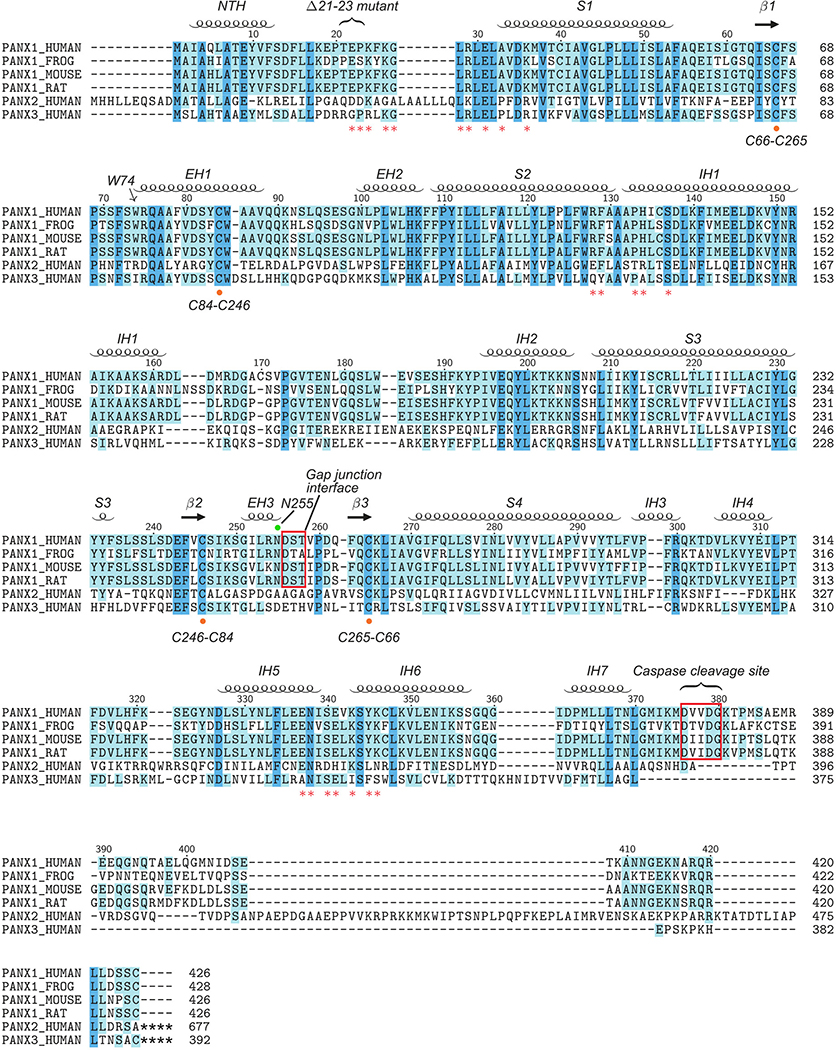
Extended Data Figure 9: Secondary structure arrangement and sequence alignment.
Secondary structures based on the hsPANX1 structure model are labeled. The W74 forming the extracellular entry is marked with an arrow. Key residues forming the side tunnel are labeled with a red asterisk. The cysteine residues forming the extracellular disulfide bonds are highlighted by an orange dot. The N255 glycosylation site is marked with a green dot. The gap junction interface and caspase 3/7 cleavage site are indicated with a red frame. A gain-of-function disease mutation (Δ21–23) of hsPANX1 is also marked.
Extended Data Table 1:
Cryo-EM data collection, refinement and validation statistics
| PANX1(WT) (EMDB-21588) (PDB 6WBF) |
apo PANX1 (EMDB-21595) |
Ca2+–PANX1 (EMDB-21596) |
K+–PANX1 (EMDB-21597) |
|
|---|---|---|---|---|
| Data collection and processing | ||||
| Magnification | 130,000 | 130,000 | 130,000 | 130,000 |
| Voltage (kV) | 300 | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 49.6 | 49.6 | 49.6 | 49.6 |
| Defocus range (μm) | −1.2 – −1.9 | −1.2 – −1.9 | −1.2 – −1.9 | −1.2 – −1.9 |
| Pixel size (Å) | 1.026 | 1.026 | 1.026 | 1.026 |
| Symmetry imposed | C7 | C7 | C7 | C7 |
| Initial particle images (no.) | 1,741,195 | 687,650 | 724,686 | 328,859 |
| Final particle images (no.) | 179,855 | 87,000 | 74,625 | 45,156 |
| Map resolution (Å) | 2.83 | 3.01 | 3.04 | 3.15 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 2.83 – 246.2 | 3.01 – 246.2 | 3.04 – 246.2 | 3.15 – 246.2 |
| Refinement | ||||
| Initial model used (PDB code) | De novo | |||
| Model resolution (Å) | 2.98 | |||
| FSC threshold | 0.5 | |||
| Model resolution range (Å) | −103.879 | |||
| Map sharpening B factor (Å2) | ||||
| Model composition | ||||
| Non-hydrogen atoms | 19712 | |||
| Protein residues | 2408 | |||
| Ligands | 35 | |||
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.005 | |||
| Bond angles (°) | 0.608 | |||
| Validation | ||||
| MolProbity score | 1.98 | |||
| Clashscore | 13.27 | |||
| Poor rotamers (%) | 0.05 | |||
| Ramachandran plot | ||||
| Favored (%) | 95.65 | |||
| Allowed (%) | 4.35 | |||
| Disallowed (%) | 0.00 | |||
Extended Data Table 2:
Cryo-EM data collection, refinement and validation statistics for ΔCTT-hsPANX1, CBX-ΔCTT-hsPANX1, ΔNTH/ΔCTT-hsPANX1, CBX-ΔNTH/ΔCTT-hsPANX1.
| PANX1(ΔCTT) (EMDB-21589) (PDB 6WBG) |
CBX–PANX1(ΔCTT) (EMDB-21590) (PDB 6WBI) |
PANX1(ΔNTH/ΔCTT) (EMDB-21591) (PDB 6WBK) |
CBX–PANX1(ΔNTH/ΔCTT) (EMDB-21592) (PDB 6WBL) |
|
|---|---|---|---|---|
| Data collection and processing | ||||
| Magnification | 130,000 | 130,000 | 130,000 | 130,000 |
| Voltage (kV) | 300 | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 49.6 | 49.6 | 49.6 | 49.6 |
| Defocus range (μm) | −1.2 – −1.9 | −1.2 – −1.9 | −1.2 – −1.9 | −1.2 – −1.9 |
| Pixel size (Å) | 1.026 | 1.026 | 1.026 | 1.026 |
| Symmetry imposed | C7 | C1 | C1 | C1 |
| Initial particle images (no.) | 745,206 | 712,835 | 573,968 | 779,920 |
| Final particle images (no.) | 88,601 | 59,332 | 85,884 | 92,116 |
| Map resolution (Å) | 2.97 | 4.39 | 6.01 | 5.13 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 2.97 – 246.2 | 4.39 – 246.2 | 6.01 – 246.2 | 5.13 – 246.2 |
| Refinement | ||||
| Initial model used (PDB code) | De novo | MDFF | MDFF | MDFF |
| Model resolution (Å) | 3.12 | 7.04 | 7.17 | 7.17 |
| FSC threshold | 0.5 | 0.5 | 0.5 | 0.5 |
| Model resolution range (Å) | ||||
| Map sharpening B factor (Å2) | −97.296 | |||
| Model composition | ||||
| Non-hydrogen atoms | 19712 | 13018 | 11135 | 11170 |
| Protein residues | 2408 | 2051 | 1948 | 1948 |
| Ligands | 35 | 1 | 0 | 1 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.006 | 0.01 | 0.02 | 0.02 |
| Bond angles (°) | 0.74 | 1.25 | 1.28 | 1.30 |
| Validation | ||||
| MolProbity score | 1.98 | 2.07 | 1.74 | 1.75 |
| Clashscore | 13.29 | 11.46 | 8.23 | 8.61 |
| Poor rotamers (%) | 0.38 | 1.2 | 0.00 | 0.00 |
| Ramachandran plot | ||||
| Favored (%) | 95.00 | 94.64 | 95.82 | 95.82 |
| Allowed (%) | 5.00 | 5.32 | 4.13 | 4.14 |
| Disallowed (%) | 0.00 | 0.13 | 0.05 | 0.05 |
Extended Data Table 3:
Cryo-EM data collection, refinement and validation statistics for N255A-hsPANX1Hemi, N255A-hsPANX1Gap, SMA-wt-hsPANX1.
| PANX1(N255A)Hemi (EMDB-21593) (PDB 6WBM) |
PANX1 (N255A)Gap (EMDB-21594) (PDB 6WBN) |
SMA–PANX1 (EMDB-21598) |
|
|---|---|---|---|
| Data collection and processing | |||
| Magnification | 105,000 | 105,000 | 105,000 |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 47.25 | 47.25 | 47.25 |
| Defocus range (μm) | −1.2 – −1.8 | −1.2 – −1.8 | −1.2 – −1.8 |
| Pixel size (Å) | 0.812 | 0.812 | 0.812 |
| Symmetry imposed | C7 | C7 | C7 |
| Initial particle images (no.) | 3,945,051 | 3,945,051 | 1,098,021 |
| Final particle images (no.) | 407,341 | 78,983 | 125,025 |
| Map resolution (Å) | 2.86 | 2.83 | 6.04 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 2.86 – 246.2 | 2.83 – 246.2 | 6.04 – 246.2 |
| Refinement | |||
| Initial model used (PDB code) | De novo | De novo | |
| Model resolution (Å) | 3.10 | 3.12 | |
| FSC threshold | 0.5 | 0.5 | |
| Model resolution range (Å) | |||
| Map sharpening B factor (Å2) | −119.822 | −70.501 | |
| Model composition | |||
| Non-hydrogen atoms | 19201 | 38402 | |
| Protein residues | 2387 | 4774 | |
| Ligands | 42 | 84 | |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.006 | 0.01 | |
| Bond angles (°) | 0.71 | 1.15 | |
| Validation | |||
| MolProbity score | 2.03 | 1.88 | |
| Clashscore | 13.68 | 11.23 | |
| Poor rotamers (%) | 0.8 | 0.8 | |
| Ramachandran plot | |||
| Favored (%) | 94.40 | 95.51 | |
| Allowed (%) | 5.60 | 4.49 | |
| Disallowed (%) | 0.00 | 0.00 | |
Supplementary Material
Acknowledgements
We thank Becca Roth for the initial construct screening, and G. Zhao and X. Meng for the support with data collection at the David Van Andel Advanced Cryo-Electron Microscopy Suite. We appreciate the HPC team of VARI for computational support. We thank David Nadziejka for technical editing. W.L. is supported by the National Institute of Health (NIH) (grant R56HL144929). J.D. is supported by a McKnight Scholar Award, a Klingenstein-Simon Scholar Award, a Sloan Research Fellowship in neuroscience and the NIH (grant R01NS111031). Z.R. is supported by an American Heart Association postdoctoral fellowship (grant 20POST35120556).
Data availability
The cryo-EM density maps of wt-hsPANX1, ΔCTT-hsPANX1, CBX-ΔCTT-hsPANX, ΔNTH/ΔCTT-hsPANX1, CBX-ΔNTH/ΔCTT-hsPANX1, N255A-hsPANX1Hemi, N255A-hsPANX1Gap, Apo-wt-hsPANX1, Ca2+-wt-hsPANX1, K+-wt-hsPANX1, and SMA-wt-hsPANX1 have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-21588, EMD-21589, EMD-21590, EMD-21591, EMD-21592, EMD-21593, EMD-21594, EMD-21595, EMD-21596, EMD-21597, and EMD-21598, respectively. The structure models of wt-hsPANX1, ΔCTT-hsPANX1, CBX-ΔCTT-hsPANX, ΔNTH/ΔCTT-hsPANX1, CBX-ΔNTH/ΔCTT-hsPANX1, N255A-hsPANX1Hemi, and N255A-hsPANX1Gap have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank under accession codes 6WBF, 6WBG, 6WBI, 6WBK, 6WBL, 6WBM, and 6WBN, respectively.
Main text references
- 1.Billaud M et al. A molecular signature in the pannexin1 intracellular loop confers channel activation by the α1 adrenoreceptor in smooth muscle cells. Sci. Signal. 8, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chekeni FB et al. Pannexin 1 channels mediate ‘find-me’ signal release and membrane permeability during apoptosis. Nature 467, 863–867 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sang Q et al. A pannexin 1 channelopathy causes human oocyte death. Sci. Transl. Med 11, (2019). [DOI] [PubMed] [Google Scholar]
- 4.Tozaki-Saitoh H, Tsuda M & Inoue K Role of purinergic receptors in CNS function and neuroprotection. Adv. Pharmacol 61, 495–528 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Ren J & Bertrand PP Purinergic receptors and synaptic transmission in enteric neurons. Purinergic Signal. 4, 255–266 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsuda M, Tozaki-Saitoh H & Inoue K Pain and purinergic signaling. Brain Res. Rev 63, 222–232 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Le T-TT et al. Purinergic Signaling in Pulmonary Inflammation. Front. Immunol 10, 1633 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z et al. Regulated ATP release from astrocytes through lysosome exocytosis. Nat. Cell Biol 9, 945–953 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Dahl G ATP release through pannexon channels. Philos. Trans. R. Soc. Lond. B. Biol. Sci 370, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taruno A et al. CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes. Nature 495, 223–226 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anselmi F et al. ATP release through connexin hemichannels and gap junction transfer of second messengers propagate Ca2+ signals across the inner ear. Proc. Natl. Acad. Sci. U. S. A 105, 18770–18775 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penuela S, Gehi R & Laird DW The biochemistry and function of pannexin channels. Biochim. Biophys. Acta 1828, 15–22 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Adamson SE et al. Pannexin 1 is required for full activation of insulin-stimulated glucose uptake in adipocytes. Mol. Metab 4, 610–618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makarenkova HP, Shah SB & Shestopalov VI The two faces of pannexins: new roles in inflammation and repair. J. Inflamm. Res 11, 273–288 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson RJ et al. Activation of pannexin-1 hemichannels augments aberrant bursting in the hippocampus. Science 322, 1555–1559 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Silverman WR et al. The pannexin 1 channel activates the inflammasome in neurons and astrocytes. J. Biol. Chem 284, 18143–18151 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crespo Yanguas S et al. Pannexin1 as mediator of inflammation and cell death. Biochim. Biophys. acta. Mol. cell Res 1864, 51–61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michalski K & Kawate T Carbenoxolone inhibits Pannexin1 channels through interactions in the first extracellular loop. J. Gen. Physiol 147, 165–174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poon IKH et al. Unexpected link between an antibiotic, pannexin channels and apoptosis. Nature 507, 329–334 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myers JB et al. Structure of native lens connexin 46/50 intercellular channels by cryo-EM. Nature 564, 372–377 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maeda S et al. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature 458, 597–602 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Oshima A, Tani K & Fujiyoshi Y Atomic structure of the innexin-6 gap junction channel determined by cryo-EM. Nat. Commun 7, 13681 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kefauver JM et al. Structure of the human volume regulated anion channel. Elife 7, e38461 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasuya G et al. Cryo-EM structures of the human volume-regulated anion channel LRRC8. Nat. Struct. Mol. Biol 25, 797–804 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Deneka D, Sawicka M, Lam AKM, Paulino C & Dutzler R Structure of a volume-regulated anion channel of the LRRC8 family. Nature 558, 254–259 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Kern DM, Oh S, Hite RK & Brohawn SG Cryo-EM structures of the DCPIB-inhibited volume-regulated anion channel LRRC8A in lipid nanodiscs. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi W, Clemente N, Sun W, Du J & Lu W The structures and gating mechanism of human calcium homeostasis modulator 2. Nature 576, 163–167 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Syrjanen JL et al. Structure and assembly of calcium homeostasis modulator proteins. Nat. Struct. Mol. Biol 27, 150–159 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sosinsky GE et al. Pannexin channels are not gap junction hemichannels. Channels (Austin). 5, 193–197 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boassa D et al. Pannexin1 channels contain a glycosylation site that targets the hexamer to the plasma membrane. J. Biol. Chem 282, 31733–31743 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Beckmann A, Grissmer A, Krause E, Tschernig T & Meier C Pannexin-1 channels show distinct morphology and no gap junction characteristics in mammalian cells. Cell Tissue Res. 363, 751–763 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Sahu G, Sukumaran S & Bera AK Pannexins form gap junctions with electrophysiological and pharmacological properties distinct from connexins. Sci. Rep 4, 4955 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandilos JK et al. Pannexin 1, an ATP release channel, is activated by caspase cleavage of its pore-associated C-terminal autoinhibitory region. J. Biol. Chem 287, 11303–11311 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiu Y-H et al. A quantized mechanism for activation of pannexin channels. Nat. Commun 8, 14324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michalski K, Henze E, Nguyen P, Lynch P & Kawate T The weak voltage dependence of pannexin 1 channels can be tuned by N-terminal modifications. J. Gen. Physiol 150, 1758–1768 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J et al. The membrane protein Pannexin1 forms two open-channel conformations depending on the mode of activation. Sci. Signal. 7, ra69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Locovei S, Wang J & Dahl G Activation of pannexin 1 channels by ATP through P2Y receptors and by cytoplasmic calcium. FEBS Lett 580, 239–244 (2006). [DOI] [PubMed] [Google Scholar]
- 38.DeLalio LJ et al. Constitutive SRC-mediated phosphorylation of pannexin 1 at tyrosine 198 occurs at the plasma membrane. J. Biol. Chem 294, 6940–6956 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weilinger NL et al. Metabotropic NMDA receptor signaling couples Src family kinases to pannexin-1 during excitotoxicity. Nat. Neurosci 19, 432–442 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Furlow PW et al. Mechanosensitive pannexin-1 channels mediate microvascular metastatic cell survival. Nat. Cell Biol 17, 943–952 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Penuela S, Celetti SJ, Bhalla R, Shao Q & Laird DW Diverse subcellular distribution profiles of pannexin1 and pannexin3. Cell Commun. Adhes 15, 133–142 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Michalski K et al. The cryo-EM structure of a pannexin 1 reveals unique motifs for ion selection and inhibition. Elife 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng Z et al. Cryo-EM structures of the ATP release channel pannexin 1. bioRxiv 2020.01.05.895235 (2020) doi: 10.1101/2020.01.05.895235. [DOI] [PubMed] [Google Scholar]
- 44.Ma W et al. Pannexin 1 forms an anion-selective channel. Pflugers Arch 463, 585–592 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Wang J & Dahl G SCAM analysis of Panx1 suggests a peculiar pore structure. J. Gen. Physiol 136, 515–527 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romanov RA et al. The ATP permeability of pannexin 1 channels in a heterologous system and in mammalian taste cells is dispensable. J. Cell Sci 125, 5514–5523 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional References
- 47.Goehring A et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc 9, 2574–2585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Q et al. Target protease specificity of the viral serpin CrmA. Analysis of five caspases. J. Biol. Chem 272, 7797–7800 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Denault J-B & Salvesen GS Expression, purification, and characterization of caspases. Curr. Protoc. protein Sci Chapter 21, Unit 21.13 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Haley E et al. Expression and Purification of the Human Lipid-sensitive Cation Channel TRPC3 for Structural Determination by Single-particle Cryo-electron Microscopy. J. Vis. Exp (2019) doi: 10.3791/58754. [DOI] [PubMed] [Google Scholar]
- 51.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang K Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohou A & Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheres SHW RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Zivanov J, Nakane T & Scheres SHW A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zivanov J, Nakane T & Scheres SHW Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seminario-Vidal L et al. Thrombin promotes release of ATP from lung epithelial cells through coordinated activation of rho- and Ca2+-dependent signaling pathways. J. Biol. Chem 284, 20638–48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chiu Y-H, Schappe MS, Desai BN & Bayliss DA Revisiting multimodal activation and channel properties of Pannexin 1. J. Gen. Physiol 150, 19–39 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nielsen BS et al. Pannexin 1 activation and inhibition is permeant-selective. J. Physiol 598, 361–379 (2020). [DOI] [PubMed] [Google Scholar]
- 62.Dourado M, Wong E & Hackos DH Pannexin-1 is blocked by its C-terminus through a delocalized non-specific interaction surface. PLoS One 9, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Penuela S, Bhalla R, Nag K & Laird DW Glycosylation regulates pannexin intermixing and cellular localization. Mol. Biol. Cell 20, 4313–4323 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Drozdetskiy A, Cole C, Procter J & Barton GJ JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43, W389–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Afonine PV et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. Sect. D, Struct. Biol 74, 814–840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trabuco LG, Villa E, Schreiner E, Harrison CB & Schulten K Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods 49, 174–180 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Williams CJ et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schrödinger LLC. The {PyMOL} Molecular Graphics System, Version~2.1. (2020). [Google Scholar]
- 70.Baker NA, Sept D, Joseph S, Holst MJ & McCammon JA Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A 98, 10037–10041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chovancova E et al. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput. Biol 8, e1002708 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Goddard TD et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shen MR et al. Differential expression of volume-regulated anion channels during cell cycle progression of human cervical cancer cells. J. Physiol 529 Pt 2, 385–394 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jo S, Kim T, Iyer VG & Im W CHARMM-GUI: a web-based graphical user interface for CHARMM. Journal of computational chemistry vol. 29 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 76.Lomize MA, Pogozheva ID, Joo H, Mosberg HI & Lomize AL OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res 40, D370–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang J et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hess B P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation. J. Chem. Theory Comput 4, 116–122 (2008). [DOI] [PubMed] [Google Scholar]
- 79.Abraham MJ et al. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density maps of wt-hsPANX1, ΔCTT-hsPANX1, CBX-ΔCTT-hsPANX, ΔNTH/ΔCTT-hsPANX1, CBX-ΔNTH/ΔCTT-hsPANX1, N255A-hsPANX1Hemi, N255A-hsPANX1Gap, Apo-wt-hsPANX1, Ca2+-wt-hsPANX1, K+-wt-hsPANX1, and SMA-wt-hsPANX1 have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-21588, EMD-21589, EMD-21590, EMD-21591, EMD-21592, EMD-21593, EMD-21594, EMD-21595, EMD-21596, EMD-21597, and EMD-21598, respectively. The structure models of wt-hsPANX1, ΔCTT-hsPANX1, CBX-ΔCTT-hsPANX, ΔNTH/ΔCTT-hsPANX1, CBX-ΔNTH/ΔCTT-hsPANX1, N255A-hsPANX1Hemi, and N255A-hsPANX1Gap have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank under accession codes 6WBF, 6WBG, 6WBI, 6WBK, 6WBL, 6WBM, and 6WBN, respectively.