Abstract
This paper describes the chemical synthesis, X-ray crystallographic structure, and antibiotic activity assay of lactam analogues of teixobactin and explores ring-expanded analogues of teixobactin with β3-homo amino acids. Lactam analogues of teixobactin containing all four stereoisomers of azathreonine at position 8 were synthesized on a solid support from commercially available stereoisomeric threonine derivatives. The threonine stereoisomers are converted to the diastereomeric aza-threonines by mesylation, azide displacement, and reduction during the synthesis. d-aza-Thr8,Arg10-teixobactin exhibits 2–8-fold greater antibiotic activity than the corresponding macrolactone Arg10-teixobactin. Aza-teixobactin analogues containing other stereoisomers of aza-threonine are inactive. A dramatic 16–128-fold increase in the activity of teixobactin and teixobactin analogues is observed with the inclusion of 0.002% of the mild detergent polysorbate 80 in the MIC assay. The X-ray crystallographic structure of N-Me-d-Gln4,d-aza-Thr8,Arg10-teixobactin reveals an amphipathic hydrogen-bonded antiparallel β-sheet dimer that binds chloride anions. In the binding site, the macrolactam amide NH groups of residues 8, 10, and 11, as well as the extra amide NH group of the lactam ring, hydrogen bond to the chloride anion. The teixobactin pharmacophore tolerates ring expansion of the 13-membered ring to 14-,15-, and 16-membered rings containing β3-homo amino acids with retention of partial or full antibiotic activity.
Graphical Abstract
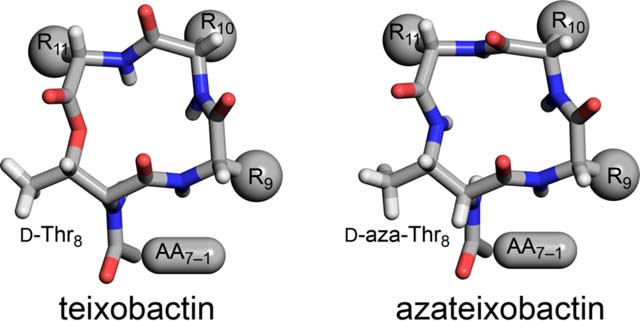
INTRODUCTION
Teixobactin is a new class of peptide antibiotic against Gram-positive bacteria that inhibits cell-wall formation, interrupting both the synthesis of peptidoglycan and the synthesis of teichoic acid, and ultimately causing bacterial cell lysis.1 ,2 Teixobactin is thought to bind the highly conserved prenyl-pyrophosphate-saccharide regions of lipid II and related membrane-bound cell-wall precursors, and thus precludes the development of antibiotic resistance.3 Furthermore, these targets are extracellular and represent the bottleneck of peptidoglycan synthesis. Teixobactin exhibits remarkable antibiotic activity against all important Gram-positive pathogens, including methicillin-resistant Staphylococcus aureus (MRSA), drug-resistant Streptococcus pneumonia, and vancomycin-resistant Enterococci (VRE).
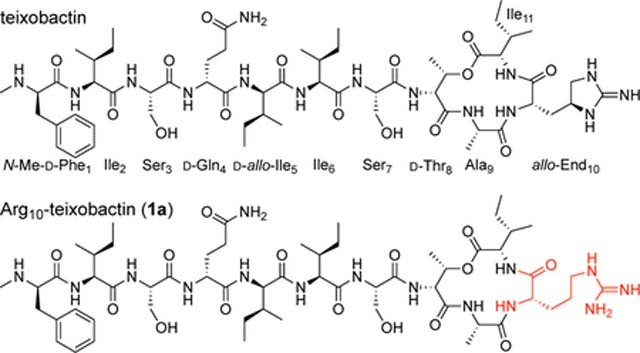
Teixobactin is a non-ribosomal undecapeptide containing a linear tail (residues 1–7) and a macrocyclic ring (residues 8–11). It contains four d-amino acids at positions 1, 4, 5, and 8, namely N-Me-d-Phe1, d-Gln4, d-allo-Ile5, and d-Thr8, as well as seven l-amino acids at positions 2, 3, 6, 7, 9, 10, and 11. The tail contains a unique pattern of d-l-l-d-d-l-l stereochemistry with a hydrophobic–hydrophobic–hydrophilic–hydrophilic–hydrophobic–hydrophobic–hydrophilic pattern of sidechains. The macrocyclic ring is composed of d-Thr8–Ala9–allo-End10–Ile11, in which the C-terminal Ile11 and the hydroxy group of d-Thr8 form an ester bond to close the 13-membered lactone ring. Residue 10 is the rare amino acid allo-enduracididine (allo-End10), a cyclic arginine analogue.
We recently reported the X-ray crystallographic structure of a teixobactin analogue.4 The analogue forms hydrogen-bonded antiparallel β-sheet dimers that bind sulfate anions (Figure 1A). In the X-ray crystallographic structure, the three NH groups of the macrolactone ring form three hydrogen bonds to each sulfate anion (Figure 1B). The macrolactone ring oxygen points toward the bound anion but is unable to form a hydrogen bond. The N-terminal ammonium group of the second monomer subunit of the dimer also hydrogen bonds to the sulfate anion. The dimers further assemble to form a double-helix of β-sheet fibrils. The binding of the sulfate anions suggests how teixobactin might bind to the anionic pyrophosphate group of lipid II and related cell-wall precursors, and thus inhibit cell-wall biosynthesis.4,5
Figure 1.
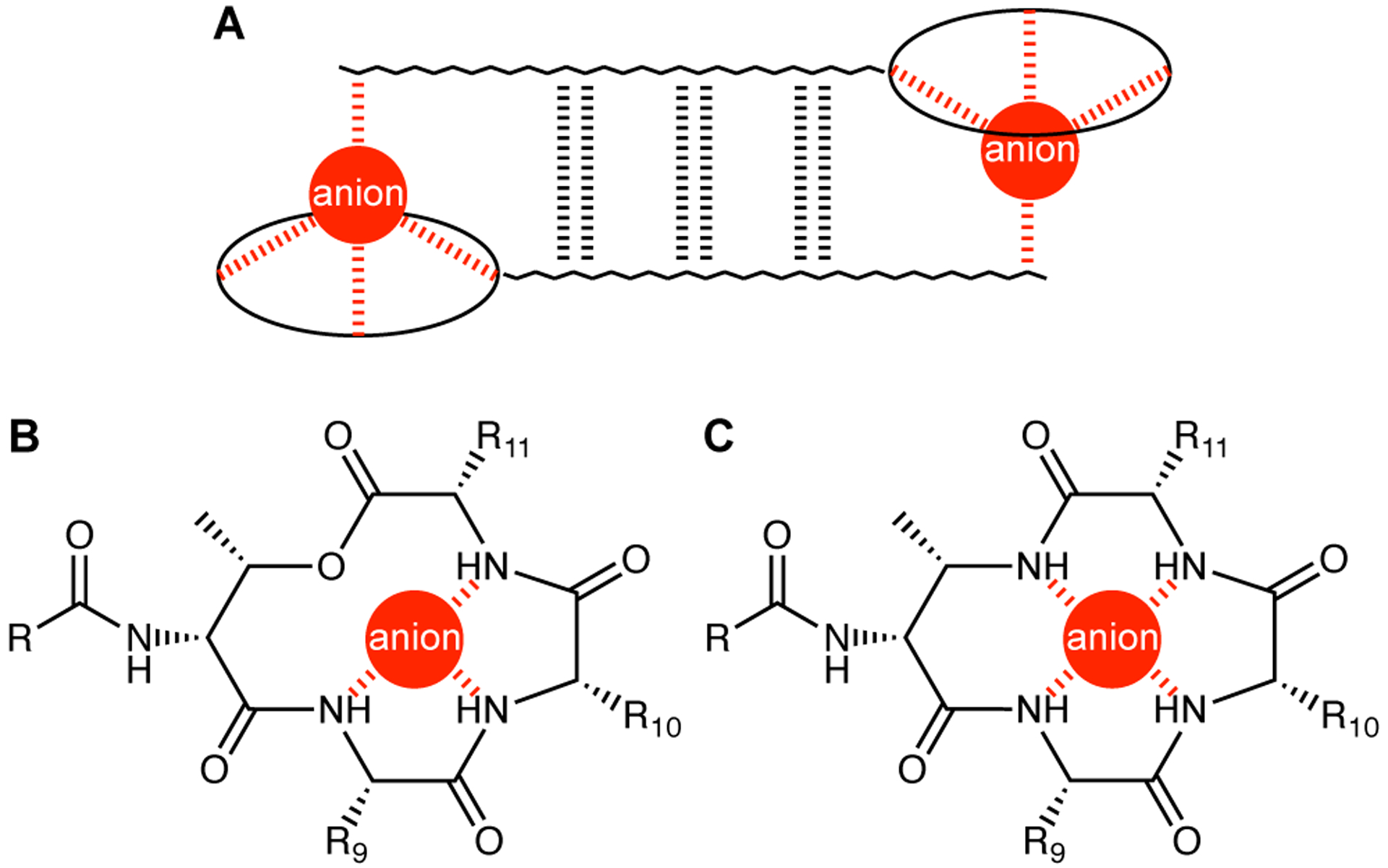
(A) Antiparallel β-sheet dimer formed by teixobactin. (B) Coordination of an anion by the teixobactin macrolactone ring. (C) Hypothesized coordination of an anion by an azateixobactin macrolactam ring.
The current paper begins by exploring the hypothesis that replacement of the lactone ring oxygen with an NH group will allow the resulting macrolactam ring to better bind anions by forming an additional hydrogen bond to the bound anion and thus lead to enhanced antibiotic activity (Figure 1C).6 We report a synthesis of macrolactam derivatives of teixobactin that contain d-aza-threonine at position 8 and find that a lactam derivative of teixobactin is two to eight times more active than the corresponding lactone. We then explore whether the 13-membered macrocyclic ring composed of residues 8–11 is optimal for binding the pyrophosphate group of lipid II and related cell-wall precursors by expanding the ring with β3-homo amino acids and find that the teixobactin pharmacophore tolerates ring expansion to 14-, 15-, and 16-membered rings with retention or partial retention of activity. Through these studies, we further elucidate the role of the macrocyclic ring in the teixobactin pharmacophore.
RESULTS AND DISCUSSION
Solid-Phase Syntheses of Lactam Analogues of Teixobactin Containing Aza-Threonine at Position 8.
We had previously reported a synthetic route to teixobactin analogues through solid-phase peptide synthesis (SPPS) on 2-chlorotrityl chloride resin followed by solution-phase macrolactamization.7 In this route, d-Thr8 is introduced without a protecting group at the hydroxy position. Because d-aza-threonine is not commercially available in a suitably protected form, we envisioned adapting this route by incorporating d-allo-Thr at position 8 and converting it to d-aza-threonine on the solid support by an SN2 displacement reaction with a nitrogen-containing nucleophile.
We tested this approach using the tripeptide Boc-Ala-d-allo-Thr-Gln(Trt) on 2-chlorotrityl chloride resin and found that attempted conversion of the d-allo-Thr residue to the mesylate and then to the azide instead gave elimination to form the corresponding dehydropeptide (Figure S1). We hypothesized that the elimination reaction could be avoided by introducing the azide group before elongating the peptide chain (Figure S2). Because the Fmoc protecting group proved labile to sodium azide, we first replaced the Fmoc group on d-allo-Thr with an Alloc group, to give the dipeptide Alloc-d-allo-Thr-Gln(Trt) on resin. This dipeptide was then converted to the mesylate and then to Alloc-d-azido-Thr-Gln(Trt) on resin by treating with triethylamine and mesyl chloride in dichloromethane, followed by sodium azide in a mixture of 15-crown-5 and DMF. Alloc deprotection with (Ph3P)4Pd and phenylsilane liberated the α-amino group of d-azido-threonine for subsequent SPPS. This sequence of steps proceeded cleanly and afforded the tripeptide Boc-Ala-d-azido-Thr-Gln(Trt)-OH in greater than 90% conversion by HPLC analysis (Figures S3–S7).
Reduction of the azide group of the resin-bound Boc-Ala-d-azido-Thr-Gln(Trt) to an amino group proved challenging. Treatment with stannous chloride (SnCl2, PhSH, DIPEA)8 resulted in approximately 60% conversion after four treatments with the reducing cocktail; Staudinger reduction (Ph3P and H2O/THF)9 stalled at the imino-phosphorane intermediate, and no hydrolysis to the amine was observed, even at high temperatures (50 °C) and in various mixtures of solvents. Saneyoshi et al. reported a derivative of triphenylphosphine, triphenylphosphine-2-carboxamide (Ph2P-o-C6H4CONH2), which contains an ortho-carboxamide group to facilitate the hydrolysis of the imino-phosphorane intermediate by providing anchimeric assistance.10 When we used triphenylphosphine-2-carboxamide in our model system, we observed good conversion from the azido group to the amino group (Figures S8–S9).
We applied the conditions that we developed to the synthesis of d-aza-Thr8,Arg10-teixobactin (2a). The synthesis began by attaching Fmoc-Ala-OH to 2-chlorotrityl resin (Figure 2).11 Fmoc-d-allo-Thr-OH was then introduced by standard Fmoc-based SPPS using HCTU as the coupling reagent. The Fmoc group was removed by treatment with 20% piperidine in DMF, and the N-terminus was protected with allyl chloroformate. The hydroxy group of d-allo-Thr8 was then converted to a mesyl group with methanesulfonyl chloride and to an azide group with sodium azide. The Alloc protecting group was removed with (Ph3P)4Pd and phenylsilane, and residues 7 through 1 were introduced by SPPS. The azide group of d-azido-Thr8 was reduced to the corresponding amine with triphenylphosphine-2-carboxamide and H2O. Residues 11 and 10 were then introduced by SPPS.11 Fmoc deprotection followed by cleavage from the resin with 20% hexafluoroisopropanol (HFIP) in CH2Cl2 afforded the protected acyclic precursor. Macrolactamization with HBTU and HOBt, followed by global deprotection with trifluoroacetic acid (TFA), RP-HPLC purification, and lyophilization, afforded 13.5 mg of d-aza-Thr8,Arg10-teixobactin (2a) as the trifluoroacetate salt with ≥95% purity, from a 0.15 mmol scale synthesis.
Figure 2.
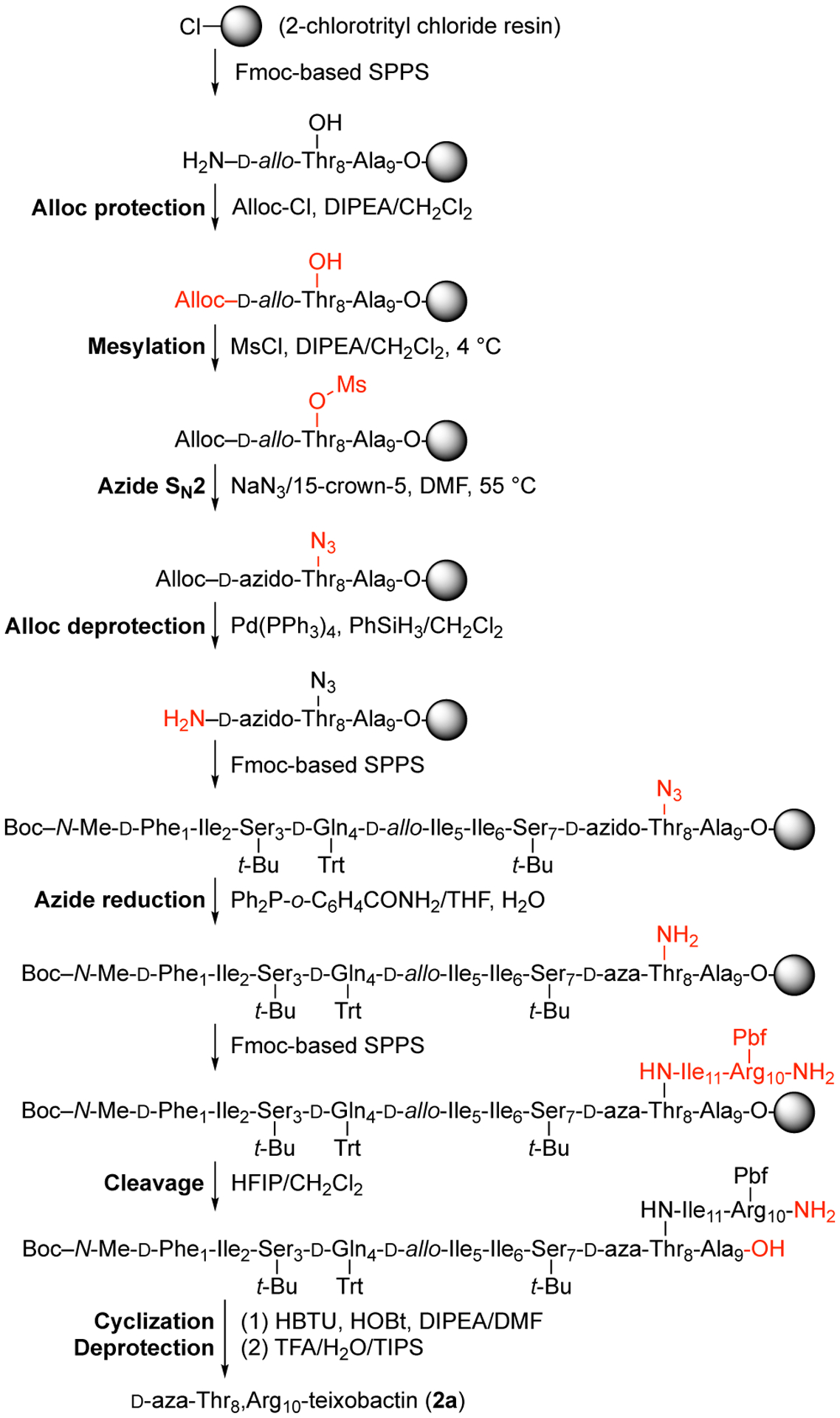
Synthesis of d-aza-Thr8,Arg10-teixobactin (2a).
This synthetic route proved versatile and also allowed us to prepare the three diastereomeric analogues, d-allo-aza-Thr8,Arg10-teixobactin (2b), l-aza-Thr8,Arg10-teixobactin (2c), and l-allo-aza-Thr8,Arg10-teixobactin (2d) (Figure 3). Replacing d-allo-threonine with d-threonine in the synthesis afforded d-allo-aza-threonine and gave diastereomer 2b. Replacing d-allo-threonine with l-allo-threonine in the synthesis afforded l-aza-threonine and gave diastereomer 2c. Replacing d-allo-threonine with l-threonine in the synthesis afforded l-allo-aza-threonine and gave diastereomer 2d.12 The stereoisomers containing l-aza-threonine and l-allo-aza-threonine (2c and 2d) exhibited an exceptionally strong propensity to form gels, making purification difficult and affording only low yields of these diastereomeric teixobactin analogues after purification by RP-HPLC.
Figure 3.
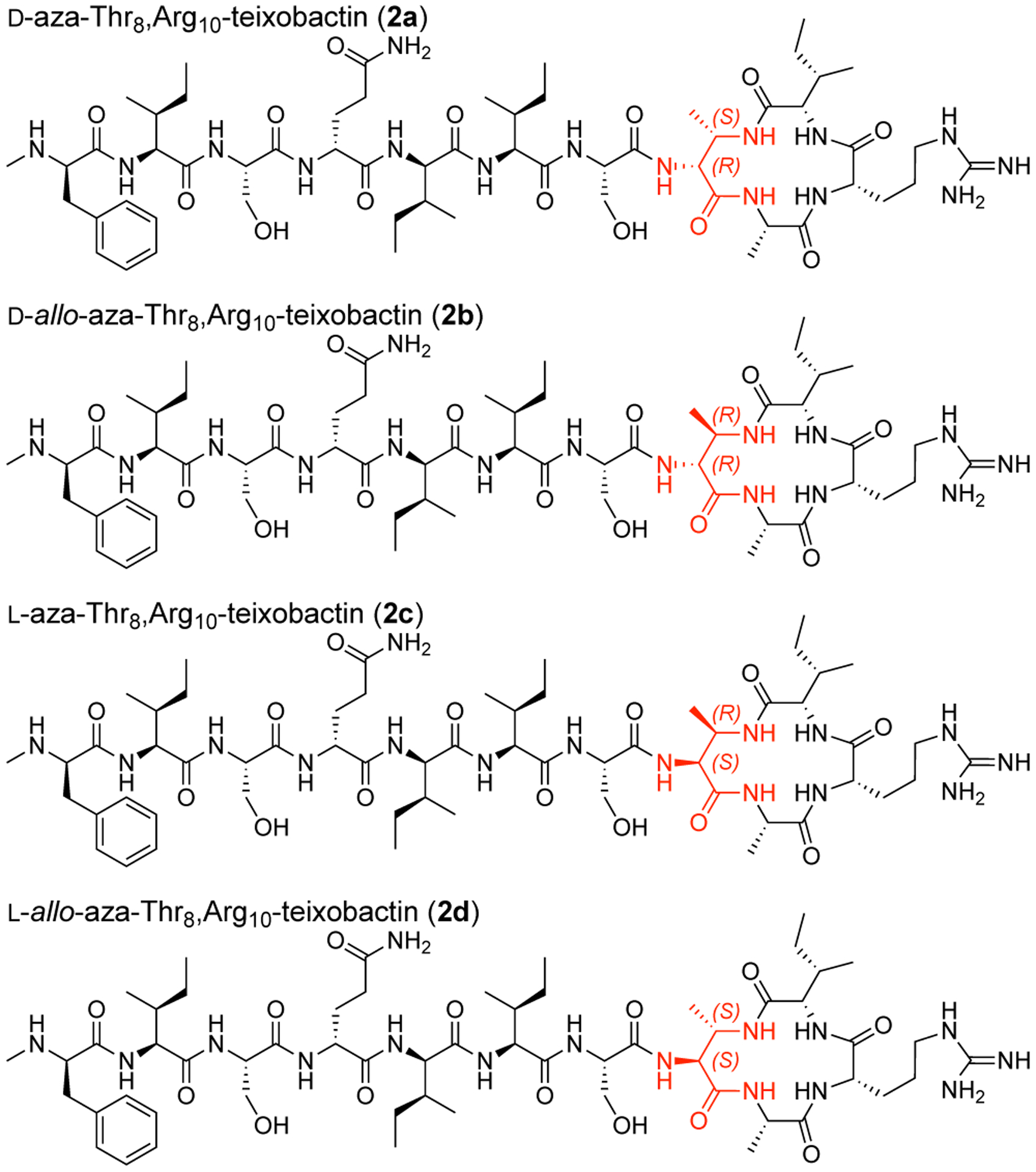
Diastereomeric azateixobactin analogues 2a–2d.
Minimum Inhibitory Concentration (MIC) Assays of Lactam Analogues of Teixobactin.
We assessed the antibiotic activity of d-aza-Thr8,Arg10-teixobactin (2a) and stereoisomeric analogues (2b–2d) in minimum inhibitory concentration (MIC) assays against three Gram-positive bacteria (S. aureus, S. epidermidis, and B. subtilis, Table 1). We used teixobactin and vancomycin as positive controls and the Gram-negative bacterium E. coli as a negative control. d-aza-Thr8,Arg10-teixobactin (2a) exhibits MIC values of 0.5–1 μg/mL across the Gram-positive bacteria and is twice as potent as Arg10-teixobactin (1a).13,14 The teixobactin analogues containing other stereoisomers of aza-threonine (2b–2d) exhibited no antibiotic activity (MIC >32 μg/mL).
Table 1.
MIC values of teixobactin and teixobactin analogues (μg/mL).
| Staphylococcus aureus ATCC 29213 | Staphylococcus epidermidis ATCC 14990 | Bacillus subtilis ATCC 6051 | Escherichia coli ATCC 10798 | |
|---|---|---|---|---|
| Arg10-teixobactin (1a) | 2 | 1 | 2 | >32 |
| d-aza-Thr8,Arg10-teixobactin (2a) | 1 | 0.5 | 1 | >32 |
| d-allo-aza-Thr8,Arg10-teixobactin (2b) | >32 | >32 | >32 | >32 |
| l-aza-Thr8,Arg10-teixobactin (2c) | >32 | >32 | >32 | >32 |
| l-allo-aza-Thr8,Arg10-teixobactin (2d) | >32 | >32 | >32 | >32 |
| teixobactin | 0.25 | 0.25 | 0.25–0.5 | >32 |
| vancomycin | 0.25 | 0.25 | 0.5 | >32 |
Effect of Polysorbate 80 on Antibiotic Activity.
In the original report of teixobactin, the authors described the use of the mild detergent polysorbate 80 at 0.002% concentration in their MIC assays.1 Having performed our previous MIC assays without polysorbate 80, we decided to investigate the effect of polysorbate 80 on the MIC values of azateixobactin analogue 2a. When we performed the MIC assay with broth containing 0.002% polysorbate 80, the MIC values of azateixobactin analogue 2a decreased from 0.5–1 μg/mL to 0.008–0.03 μg/mL, a dramatic 16–128-fold increase in antibiotic activity (Table 2). We also observe a similar decrease in the MIC of teixobactin, from 0.25–0.5 μg/mL to <0.008 μg/mL, the lowest concentration we tested. In contrast, we observe only a modest two-fold decrease in the MIC of vancomycin.15 We observe a similar but smaller increase in the activity of teixobactin analogue 1a with 0.002% polysorbate 80. Thus, azateixobactin analogue 2a proved 4–8 times more active than teixobactin analogue 1a in the presence of 0.002% polysorbate 80. Although the enhanced antibiotic activity of azateixobactin analogue 2a is consistent with a model in which the additional NH group better binds the anionic pyrophosphate group of lipid II and related cell-wall precursors, other mechanisms of enhanced antibiotic activity cannot be precluded.
Table 2.
MIC values of teixobactin and teixobactin analogues (μg/mL) with 0.002% polysorbate 80.
| Staphylococcus aureus ATCC 29213 | Staphylococcus epidermidis ATCC 14990 | Bacillus subtilis ATCC 6051 | Escherichia coli ATCC 10798 | |
|---|---|---|---|---|
| Arg10-teixobactin (1a) | 0.06 | 0.13 | 0.06 | >8 |
| d-aza-Thr8,Arg10-teixobactin (2a) | 0.008 | 0.03 | 0.016 | >8 |
| teixobactin | <0.008 | <0.008 | <0.008 | >8 |
| vancomycin | 0.125 | 0.25 | 0.25 | >8 |
The authors of the original teixobactin report suggest that polysorbate 80 prevents the binding of the antibiotic to plastic surfaces.1,16 We believe that other effects may also be involved. Having observed that teixobactin and its analogues form gels and amyloid-like fibrils upon addition to buffer or culture media,4,17 we believe that inclusion of polysorbate 80 in the broth may help solubilize the gels or fibrils, and thus increases the bioavailability and activity of teixobactin and its analogues.
X-ray Crystallographic Structure of N-Me-d-Gln4, d-aza-Thr8,Arg10-teixobactin (3a).
To assess the effect of the additional amide NH group on the structure of azateixobactin analogues, we turned to X-ray crystallography. We had previously found that N-methylation of the peptide backbone of d-Gln4 facilitated crystallization of a teixobactin analogue by attenuating its propensity to form a gel.4 In the current study we utilized this finding and synthesized N-Me-d-Gln4, d-aza-Thr8,Arg10-teixobactin (3a).18
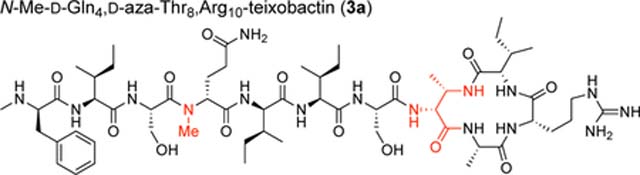
We screened N-Me-d-Gln4, d-aza-Thr8,Arg10-teixobactin (3a) for crystallization in 864 conditions in a 96-well plate format using crystallization kits from Hampton Research (PEG/Ion, Index, and Crystal Screen). Hexagonal prism -shaped crystals grew in conditions containing polyethylene glycol (PEG) and chloride salts. With further optimization in 24-well plates, a crystallization buffer of 0.16 M CaCl2, 0.1 M HEPES sodium at pH 7.00, and 24% PEG 400 afforded crystals of azateixobactin analogue 3a suitable for X-ray diffraction. Three X-ray diffraction data sets were acquired at the Advanced Light Source (ALS) synchrotron at a wavelength of 1.77 Å (7000 eV). The data sets were processed using XDS19 and merged using BLEND.20 The structure was solved by single-wavelength anomalous diffraction (SAD) phasing using the anomalous signal from the chloride anion. The structure was refined with PHENIX21 in the P3221 space group at 2.10 Å resolution. The asymmetric unit contains one N-Me-d-Gln4, d-aza-Thr8,Arg10-teixobactin (3a) molecule, as well as one chloride anion and four ordered water molecules (Figure 4, PDB 6PSL). We refined the N-methyl terminus (N-Me-d-Phe1) as the free amine, rather than as the ammonium ion, because there is only a single chloride anion in the asymmetric unit, which balances the positive charge of the arginine sidechain. We found no electron density or voids in the lattice that could account for an additional anion.
Figure 4.
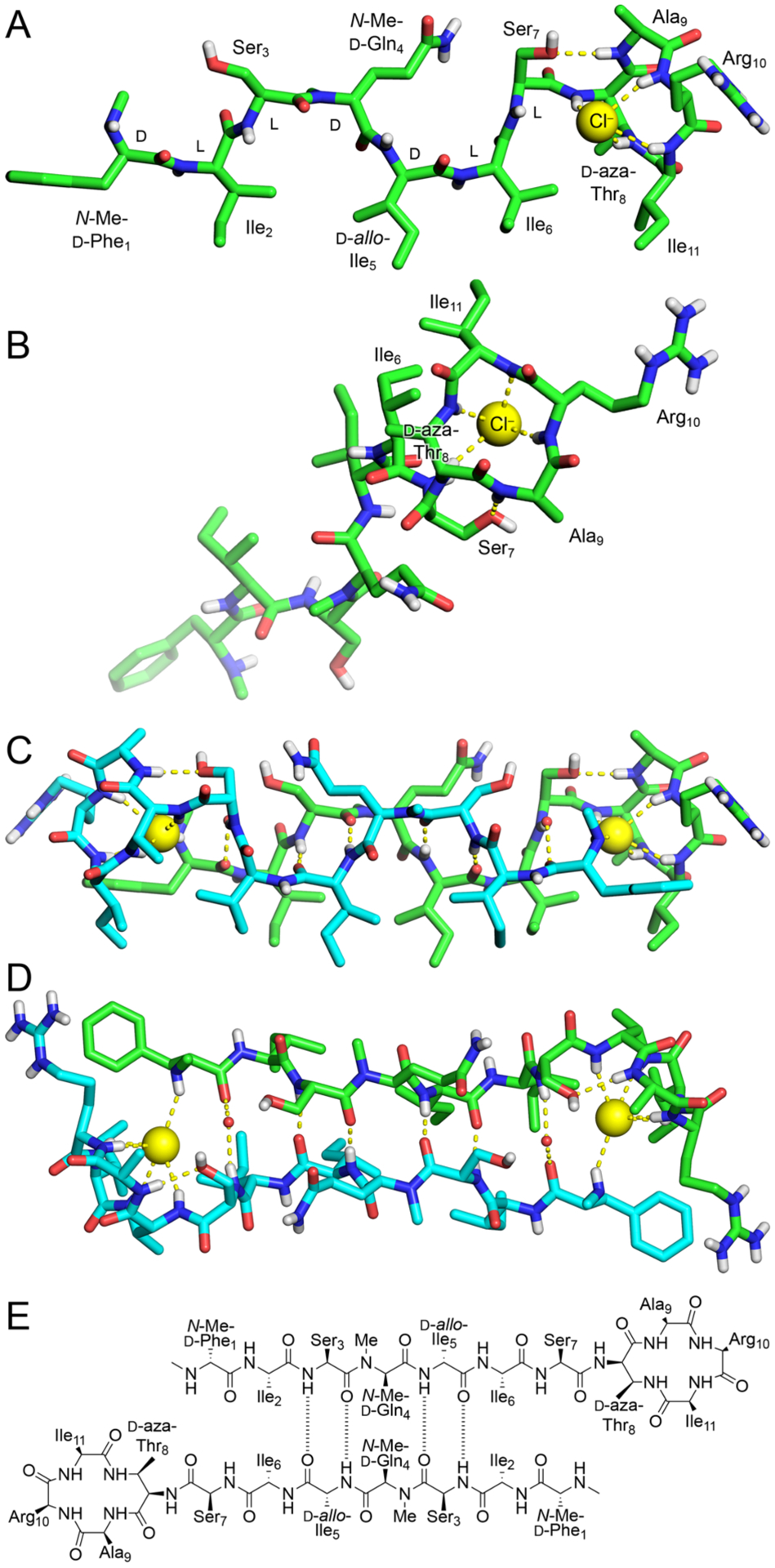
X-ray crystallographic structure of N-Me-d-Gln4, d-aza-Thr8,Arg10-teixobactin (3a) binding chloride anions (PDB 6PSL). (A and B) Monomer side and top views. (C and D) Dimer side and top views with two bridging water molecules shown as non-bonded spheres. (E) Alignment of the dimer assembly.
The X-ray crystallographic structure reveals an amphipathic hydrogen-bonded antiparallel β-sheet dimer that binds chloride anions (Figure 4). Residues 1–7 form the dimerization interface and create an amphipathic antiparallel β-sheet containing both D and L residues. The macrolactam amide NH groups of residues 8, 10, and 11, as well as the extra amide NH group of the lactam ring, hydrogen bond to the chloride anion. The amide NH group of Ala9 hydrogen bonds to the hydroxy group of Ser7.22
In the antiparallel β-sheet assembly, the sidechains of hydrophobic residues N-Me-d-Phe1, Ile2, d-allo-Ile5, Ile6, and Ile11, as well as the β-methyl group of d-aza-Thr8, create a hydrophobic face, and the sidechains of hydrophilic residues Ser3, N-Me-d-Gln4, Ser7, and Arg10, as well as the N-methylamine terminus, create a hydrophilic face. The N-methyl group of N-Me-d-Gln4 points outward from the dimer, blocking further assembly. The antiparallel β-sheet dimer brings the N-methylamine terminus of each monomer subunit into proximity with the macrolactam ring of the other monomer subunit and allows each methylamine group to hydrogen bond to a chloride anion.
The X-ray crystallographic structure of azateixobactin analogue 3a reveals a mode of antiparallel β-sheet assembly distinct from that which we had previously reported a related teixobactin analogue (N-Me-d-PheI1,N-Me-d-Gln4,Arg10-teixobactin, PDB 6E00).4 In our previous X-ray crystallographic antiparallel β-sheet dimer, residues 1–7 also create an amphipathic dimerization interface (Figure S32). The antiparallel β-sheet dimer assembly also brings each N-terminus into the proximity with the anion binding site and allows each N-terminus to hydrogen bond to the bound anion. However, there are three significant differences between the current antiparallel β-sheet dimer (3a) and our previously reported antiparallel β-sheet dimer: (1) The dimerization interfaces involve opposite edges of the β-sheets. In the current structure, residues 3 and 5 form hydrogen-bonded pairs; in our previously reported structure, residues 2 and 6 form hydrogen-bonded pairs (Figures 4E and S32). (2) The psi and phi angles of Ser7 differ dramatically between the two structures, thus rotating the macrocycle toward the dimerization interface of each structure. (3) The dimer in the current structure binds two chloride anions, while the dimer in our previously reported structure binds two sulfate anions. Although the details of the crystallographic structures of the previous and current dimers differ, both structures are consistent with the model shown in Figure 1A.
Ring-Expanded Teixobactin Analogues.
The 13-membered macrolactone ring of teixobactin is substantially smaller than that of many other cyclic depsipeptide antibiotics that bind lipid II,23 including ramoplanin24 (49-membered macrolactone), lysobactin/katanosin B25 (28-membered macrolactone), and plusbacin A326 (28-membered macrolactone). Our X-ray crystallographic structures of teixobactin analogues bound to chloride and sulfate anions suggest that a larger ring might allow teixobactin analogues to better accommodate the larger pyrophosphate group of lipid II and related cell-wall precursors. Inspired by our X-ray crystallographic structures and the larger macrolactone rings of other antibiotics, we set out to explore the effect of the teixobactin macrolactone ring size upon antibiotic activity. In this section, we expand the 13-membered macrolactone ring to 14-, 15-, and 16-membered macrolactone rings with β3-homo amino acids.
We synthesized seven ring-expanded teixobactin analogues containing one, two, or three β3-homo amino acids at positions 9, 10, and 11 (Figure 5) and assessed their activities in MIC assays (Table 3 and Table S1). Six of the seven ring-expanded teixobactin analogues were active against Gram-positive bacteria, indicating that the teixobactin pharmacophore tolerates ring expansion. β3h-Arg10-teixobactin (5) and β3h-Arg10,β3h-Ile11-teixobactin (9) exhibit comparable activity to Arg10-teixobactin, while the other ring-expanded analogues exhibit diminished activity. Molecular modeling studies suggest that the ring expanded analogues are more flexible than teixobactin or Arg10-teixobactin and that the NH groups of the rings are less well aligned to bind anions (Figures S33–S34). Collectively, the MIC and molecular modeling studies suggest that the 13-membered ring of teixobactin may provide an optimal balance of size and preorganization for binding the pyrophosphate group of lipid II and related cell-wall precursors.
Figure 5.
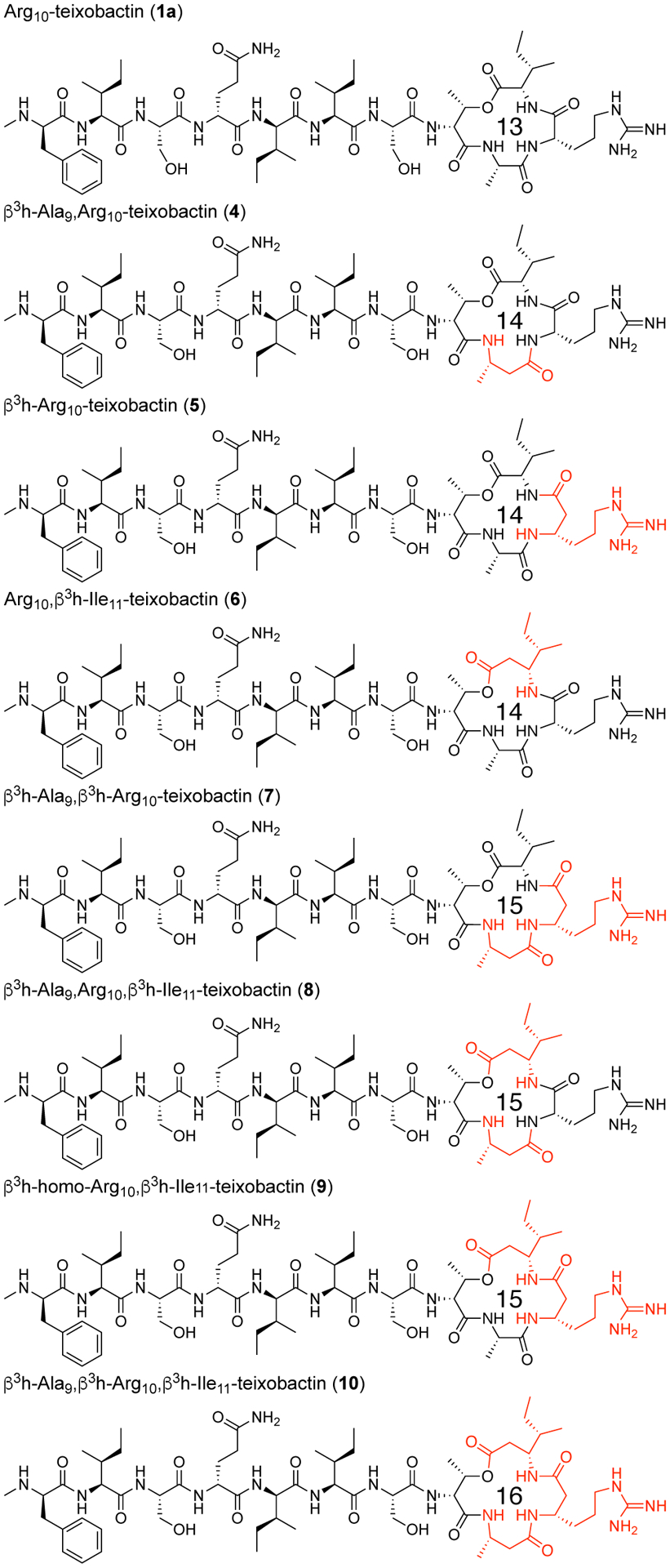
Arg10-teixobactin and ring-expanded Arg10-teixobactin analogues containing β3-homo amino acids at positions 9, 10, and 11. The ring-expanded analogues contain 14-, 15-, and 16-membered macrolactone rings.
Table 3.
MIC values of ring-expanded teixobactin analogues (μg/mL).
| Staphylococcus aureus ATCC 29213 | Staphylococcus epidermidis ATCC 14990 | Bacillus subtilis ATCC 6051 | Escherichia coli ATCC 10798 | |
|---|---|---|---|---|
| Arg10-teixobactin (1a) | 2 | 1 | 2 | >32 |
| β3h-Ala9,Arg10-teixobactin (4) | 32 | 4 | 8 | >32 |
| β3h-Arg10-teixobactin (5) | 8 | 1 | 2 | >32 |
| Arg10,β3h-Ile11-teixobactin (6) | 4 | 2 | 4 | >32 |
| β3h-Ala9, β3h-Arg 10-teixobactin (7) | >32 | >32 | >32 | >32 |
| β3h-Ala9,Arg10,β3h-Ile11-teixobactin (8) | 32 | 16 | 16 | >32 |
| β3h-Arg10,β3h-Ile11-teixobactin (9) | 4 | 1 | 4 | >32 |
| β3h-Ala9,β3h-Arg10,β3h-Ile11-teixobactin (10) | 16 | 2 | 4 | >32 |
| teixobactin | 0.25 | 0.25 | 0.25–0.5 | >32 |
| vancomycin | 0.25 | 0.25 | 0.5 | >32 |
CONCLUSION
Lactam analogues of teixobactin containing aza-threonine at position 8 are readily prepared by solid-phase synthesis, with conversion of the corresponding diastereomeric threonine analogue to the aza-threonine analogue by mesylation, azide SN2 displacement, and Staudinger reduction with triphenylphosphine-2-carboxamide. Teixobactin analogues containing all four diastereomers of aza-threonine can be prepared by this route. Replacement of the lactone ring oxygen with an NH group increases antibiotic activity, with d-aza-Thr8,Arg10-teixobactin (2a) exhibiting 2–8-fold greater antibiotic activity than Arg10-teixobactin (1a). Polysorbate 80 exhibits a dramatic effect on the antibiotic activity of teixobactin and teixobactin analogues; d-aza-Thr8,Arg10-teixobactin exhibits an MIC of 0.008 μg/mL against S. aureus in the presence of 0.002% polysorbate 80.
X-ray crystallography reveals that the additional NH group of azateixobactin analogue 3a hydrogen bonds to a bound chloride anion and supports a model in which azateixobactin analogues achieve enhanced antibiotic activity by better binding to the anionic pyrophosphate group of lipid II and related cell-wall precursors. The X-ray crystallographic structure further supports a model in which the formation of hydrogen-bonded dimers or amyloid-like higher order assemblies is central to antibiotic activity,4 with two molecules of teixobactin or a teixobactin analogue coordinating to the bound anion. Although ring-expanded teixobactin analogues were hypothesized to better accommodate the pyrophosphate group of lipid II and related cell-wall precursors, teixobactin analogues containing β3-homo amino acids exhibited no greater antibiotic activity. Collectively, these studies illustrate how chemical synthesis, X-ray crystallography, and antibiotic activity assays may be used together to provide insights into the unique activity of teixobactin. We anticipate that these studies will facilitate the design of teixobactin analogues with improved properties that are useful in the clinic.
EXPERIMENTAL SECTION
General information
Methylene chloride (CH2Cl2) was passed through alumina under argon prior to use. Amine-free N,N-dimethylformamide (DMF) was purchased from Alfa Aesar. Fmoc-d-allo-Ile-OH was purchased from Santa Cruz Biotechnology. Fmoc-N-Me-d-Gln(Trt)-OH was purchased from ChemPep. Other protected amino acids were purchased from CHEM-IMPEX. 2-(Diphenylphosphino)benzoic acid was purchased from Arctom chemicals. Preparative reverse-phase HPLC was performed on a Rainin Dynamax instrument equipped with an Agilent Zorbax SB-C18 column. Analytical reverse-phase HPLC was performed on an Agilent 1260 Infinity II instrument equipped with a Phenomonex Aeris PEPTIDE 2.6μ XB-C18 column. HPLC grade acetonitrile (MeCN) and deionized water (18 MΩ) containing 0.1% trifluoroacetic acid (TFA) were used as solvents for both preparative and analytical reverse-phase HPLC. Deionized water (18 MΩ) was obtained from a Barnstead NANOpure Diamond purification system or a ThermoScientific Barnstead GenPure Pro water purification system. Glass solid-phase peptide synthesis vessels with fritted disks and BioRad Polyprep columns were used for solid-phase peptide synthesis. Teixobactin analogues 2a–10 were prepared and studied as the trifluoroacetate salts.
Synthesis of d-aza-Thr8,Arg10-teixobactin (2a)
Resin Loading.
2-Chlorotrityl chloride resin (300 mg, 1.2 mmol/g) was added to a 10-mL Bio-Rad Poly-Prep chromatography column. The resin was suspended in dry CH2Cl2 (5 mL) and allowed to swell for 15 min. The CH2Cl2 was drained with a flow of nitrogen. The resin was loaded with a solution of Fmoc-Ala-OH (90 mg, 0.29 mmol) and 2,4,6-collidine (300 μL) in dry CH2Cl2 (7 mL) and rocked for 4 h.
Resin Capping.
The solution was drained with a flow of nitrogen, and the resin was washed with dry CH2Cl2 (3x). A mixture of CH2Cl2 (5 mL), MeOH (0.8 mL), and DIPEA (0.4 mL) was made and poured into the Poly-Prep column containing the resin, and the column was rocked for 1 h to cap any unreacted sites in the resin. The solution was drained with a flow of nitrogen, and the resin was washed with dry CH2Cl2 (3x). The resin was washed with MeOH and blown with nitrogen until dry resin was observed.
Loading check.
Approximately 1 mg of the dry resin was weighed out into a scintillation vial and 20% piperidine in DMF (3 mL) was added. The vial was rocked for 30 min. The mixture was filtered using a Pasteur pipet plugged with glass wool. The UV/Vis spectrometer was blanked at 290 nm with a cuvette filled with 20% piperidine in DMF. The absorbance of the filtered mixture was measured [1.4 mg of resin weighed; A290 = 1.1234; 0.15 mmol loading].
Fmoc deprotection.
The loaded resin was transferred to a solid-phase peptide hand coupling vessel. The resin was washed with dry CH2Cl (3x) and then dry DMF (3x). To the reaction vessel, 20% piperidine in dry DMF (5 mL) was added. Using a nitrogen flow to bubble the hand coupling vessel, the reaction was mixed for 20 min. The resin was washed with dry DMF (3x).
Coupling Fmoc-d-allo-Thr-OH with HCTU.
Based on loading, Fmoc-d-allo-Thr-OH (105 mg, 0.30 mmol, 2 equiv) and HCTU (121 mg, 0.30 mmol, 2 equiv) were weighed out and dissolved in 20% collidine in dry DMF. This solution was added to the reaction vessel containing the deprotected peptide on resin. Using a nitrogen flow to bubble the hand coupling vessel, the reaction was mixed for 4 h. The resin was washed with dry DMF (3x).
Fmoc deprotection.
To the reaction vessel, 20% piperidine in dry DMF (5 mL) was added. Using a nitrogen flow to bubble the hand coupling vessel, the reaction was mixed for 20 min. The resin was washed with dry DMF (3x).
Alloc protection.
The resin was transferred to a Poly-Prep column with dry DMF and the solution was drained with a flow of nitrogen. The resin was washed with dry CH2Cl2 (3x). To the resin in the Poly-Prep column, dry CH2Cl2 (5 mL), DIPEA (38 μL, 0.23 mmol, 1.5 equiv) and allyl chloroformate (23 μL, 0.23 mmol, 1.5 equiv) were added sequentially. The column was then capped and rocked for 1 h. The resin was washed with dry CH2Cl (3x).
Mesylation.
Dry CH2Cl2 (6 mL) was added to the resin in the Poly-Prep column. The column was then capped and rocked in a cold room (4 °C) for 15 min. DIPEA (254 μL, 1.5 mmol, 10 equiv) was added, and the mixture was rocked in a cold room (4) for an additional 15 min. Methanesulfonyl chloride (113 μL, 1.5 mmol, 10 equiv) was added, and mixture was rocked in a cold room (4 °C) for an additional 15 min. The resin was then washed with dry CH2Cl2 (3x) and then with dry DMF (3x). The resin was transferred to the hand coupling vessel.
SN2 with NaN3.
NaN3 (474 mg, 7.5 mmol, 50 equiv) was added to the resin in the hand coupling vessel. Dry DMF (1 mL) and 15-crown-5 (1 mL) were added to the hand coupling vessel. [Not all of the NaN3 dissolves in the solvent mixture.] Plastic tubing with a continuous water flow at 55 °C was wrapped around the hand coupling vessel to provide heating. Using a gentle nitrogen flow, the mixture was bubbled for 12 h at 55 °C. The resin was washed with 10 mL of 20% H2O in THF (5x) to remove excess NaN3. The resin was transferred to a Poly-Prep column with dry DMF and then washed with dry CH2Cl2 (3x).
Alloc deprotection.
A mixture of CH2Cl2 (5 mL), (Ph3P)4Pd (16.9 mg, 0.015 mmol, 0.1 equiv) and PhSiH3 (360 μL, 3 mmol, 20 equiv) was added to the resin in the Poly-Prep column, and the column was rocked for 30 min. The resin was washed with dry CH2Cl2 (3x) and then with dry DMF (3x) and transferred to a hand coupling vessel.
Peptide coupling.
The linear peptide was synthesized through the following cycles: i. coupling of amino acid (0.60 mmol, 4 equiv) with HCTU (241 mg, 0.60 mmol, 4 equiv) in 20% (v/v) 2,4,6-collidine in dry DMF (3 mL) for 30 min, ii. resin washing with dry DMF (3x), iii. Fmoc deprotection with 20% (v/v) piperidine in dry DMF (3 mL) for 20 min, and iv. resin washing with dry DMF (3x). For d-to-l and l-to-d amino acid couplings, the reaction time in step i was increased to 1 h. After completing the linear synthesis, the resin was transferred to a 10-mL Bio-Rad Poly-Prep chromatography column. The resin was then washed with dry DMF (3x) and then dry THF (3x).
Azide reduction.
Triphenylphosphine-2-carboxamide27 (223 mg, 0.45 mmol, 5 equiv) in THF (5 mL) was added to the resin in a Poly-Prep column, and the column was rocked for 12 h. The solution was drained with a flow of nitrogen and 20% H2O in THF (5 mL) was added and rocked for 4 h. The resin was washed with dry DMF (3x) and transferred to a hand coupling vessel using DMF.
Peptide coupling.
The coupling and Fmoc deprotection of Ile11 and Arg10 was performed as described above. After Fmoc deprotection of Arg10, the resin containing branched linear peptide was transferred to 10-mL Bio-Rad Poly-Prep chromatography column using DMF. The resin was washed with dry DMF (3x) and then with dry CH2Cl2 (3x).
Cleavage of the branched linear peptide from the resin.
The protected peptide was cleaved from the resin by adding 20% hexafluoroisopropanol in dry CH2Cl2 (7 mL) to the column and rocking for 30 min. The filtrate was collected in a round-bottom flask. The resin was washed with a second aliquot of 20% hexafluoroisopropanol (7 mL). The filtrates were combined and concentrated under reduced pressure to afford a clear oil. The oil was placed under vacuum (≤ 100 mTorr) to remove any residual solvents.
Macrolactamization.
To the round-bottom flask containing cleaved peptide, a mixture of HBTU (332 mg, 0.9 mmol, 6 equiv) and HOBT (118 mg, 0.9 mmol, 6 equiv) in dry DMF (50 mL) was added and stirred for 15 min under nitrogen. Diisopropylethylamine (153 μL, 0.9 mmol, 6 equiv) was added to the stirring solution and then stirred for 12 h under nitrogen. The solution was evaporated by rotary evaporator and the residue was dried under vacuum (≤ 100 mTorr) to give a pale-yellow pellet.
Global deprotection.
A solution of TFA (9 mL), H2O (0.5 mL), and TIPS (0.5 mL) was added to the round-bottom flask containing cyclized peptide and stirred for 1 h under nitrogen. The solution was evaporated by rotary evaporator and the residue was dried under vacuum (≤ 100 mTorr).
Purification.
The globally deprotected peptide was dissolved in approximately 35% CH3CN in H2O (10 mL), and the solution was centrifuged at 14,000 rpm for 5 min. The solution was filtered through a 0.20-μm nylon filter. The peptide was purified by reverse-phase HPLC with H2O/CH3CN (gradient elution of 20–95% CH3CN with 0.1% TFA). Pure fractions were identified by analytical HPLC and electrospray ionization (ESI) mass spectrometry and were combined and lyophilized. d-aza-Thr8,Arg10-teixobactin (2a) was isolated as trifluoroacetic acid (TFA) salt of a 13.5 mg white powder with ≥95% purity.
Synthesis of diastereomeric azateixobactin analogues (2b–2d)
Other diastereomeric azateixobactin analogues (2b–2d) were prepared using the procedures described above. Replacing d-allo-threonine with d-threonine in the synthesis afforded d-allo-aza-threonine and gave diastereomer 2b. Replacing d-allo-threonine with l-allo-threonine in the synthesis afforded l-aza-threonine and gave diastereomer 2c. Replacing d-allo-threonine with l-threonine in the synthesis afforded l-allo-aza-threonine and gave diastereomer 2d. For synthesis of l-allo-aza-Thr8,Arg10-teixobactin (2d), the azide reduction with triphenylphosphine-2-carboxamide was performed for 24 h instead of 12 h. The stereoisomers containing l-aza-threonine and l-allo-aza-threonine (2c and 2d) exhibited an exceptionally strong propensity to form gels, making purification difficult and affording only low yields of these diastereomeric teixobactin analogues after purification by RP-HPLC.
Synthesis of N-Me-d-Gln4, d-aza-Thr8,Arg10-teixobactin (3a)
N-Methylated azateixobactin analogue (3a) was prepared using the procedures described above. Coupling of Fmoc-Ser3(tBu)-OH after N-Me-d-Gln4 was performed using 4 equiv Fmoc-Ser3(tBu)-OH with coupling reagent HATU (4 equiv), HOAt (4 equiv) in 20% (v/v) collidine in dry DMF (5 mL) for 12 h.
Synthesis of ring-expanded teixobactin analogues (4–10)
Ring-expanded teixobactin analogues containing β3-homo amino acids (4–10) were synthesized using the procedures we have previously reported.7 Dry DMF was used instead of a mixture of CH3CN/THF/CH2Cl2 for the cyclization step.
HPLC conditions and MS results
Analytical RP-HPLC was performed on a C18 column with an elution gradient of 5–67% CH3CN over 15 min for teixobactin analogues 5, 8, 9, and 10, and with an elution gradient of 5–100% CH3CN over 20 min for other analogues.
d-aza-Thr8,Arg10-teixobactin (2a). MS (ESI-MS): m/z: [M+H]+ Calcd for C58H99N16O14 1243.8; found 1243.3. d-allo-aza-Thr8,Arg10-teixobactin (2b). MS (ESI-MS): m/z: [M+H]+ Calcd for C58H99N16O14 1243.8; found 1243.4. l-aza-Thr8,Arg10-teixobactin (2c). MS (ESI-MS): m/z: [M+H]+ Calcd for C58H99N16O14 1243.8; found 1243.9. l-allo-aza-Thr8,Arg10-teixobactin (2d). MS (MALDI-MS): m/z: [M+H]+ Calcd for C58H99N16O14 1243.8; found 1243.6. N-Me-d-Gln4, d-aza-Thr8,Arg10-teixobactin (3a). MS (ESI-MS): m/z: [M+H]+ Calcd for C59H101N16O14 1257.8; found 1257.8. β3h-Ala9,Arg10-teixobactin (4). MS (ESI-MS): m/z: [M+H]+ Calcd for C59H100N15O15 1258.8; found 1258.6. β3h-Arg10-teixobactin (5). MS (ESI-MS): m/z: [M+H]+ Calcd for C59H100N15O15 1258.8; found 1258.5. Arg10,β3h-Ile11-teixobactin (6). MS (ESI-MS): m/z: [M+H]+ Calcd for C59H100N15O15 1258.8; found 1258.6. β3h-Ala9,β3h-Arg10-teixobactin (7). MS (ESI-MS): m/z: [M+H]+ Calcd for C60H102N15O15 1272.8; found 1272.6. β3h-Ala9,Arg10,β3h-Ile11-teixobactin (8). MS (ESI-MS): m/z: [M+H]+ Calcd for C60H102N15O15 1272.8; found 1272.5. β3h-Arg10,β3h-Ile11-teixobactin (9). MS (ESI-MS): m/z: [M+H]+ Calcd for C60H102N15O15 1272.8; found 1272.6. β3h-Ala9,β3h-Arg10,β3h-Ile11-teixobactin (10). MS (ESI-MS): m/z: [M+H]+ Calcd for C61H104N15O15 1286.8; found 1286.6.
Minimum inhibitory concentration (MIC) assay of teixobactin analogue
MIC assays of teixobactin and teixobactin analogues (2a–10) were performed using the procedure we have previously reported.7,17
Preparing the peptide analogue stock.
Solutions of d-aza-Thr8,Arg10-teixobactin (2a), other teixobactin analogues (2b–10), teixobactin and vancomycin were prepared gravimetrically by dissolving an appropriate amount of peptide in an appropriate volume of sterile DMSO to make 20 mg/mL stock solutions. The stock solutions were stored at −20 °C for subsequent experiments.
Prior to transferring the peptide solution in broth to 96-well plates, the peptide solution was vigorously vortexed with a vortex mixer for ca. 30 seconds to ensure thorough dispersion and/or dissolution of teixobactin and teixobactin analogues, which have a propensity to form gels.
Serial dilution of peptides in a 96-well plate.
In the serial dilution, the first well containing 200-uL peptide solution was mixed with pipette up and down at least ten times before moving to the next well. In the following serial dilutions, each well was also mixed with pipette up and down at least ten times.
Performing the minimum inhibitory concentration (MIC) assays with Mueller-Hinton broth containing 0.002% polysorbate 80.
Mueller-Hinton broth containing 0.002% (v/v) polysorbate 80 was autoclaved and used in the MIC assay. The broth was used to dilute the 20 mg/mL DMSO solution of teixobactin and teixobactin analogues to 16 μg/mL and then to serially dilute the peptide solutions in the 96-well plate to the following concentrations: 16, 8, 4, 2, 1, 0.5, 0.25, 0.13, 0.063, 0.031, and 0.016 μg/mL. Each well contained 100 μL of solution. The broth was used to dilute bacterial suspension OD600 of 0.075 and to further dilute the bacterial suspension to 1 × 106 CFU/mL. 100-μL aliquots of the diluted bacteria were added to each well to give final concentration of 5 × 105 CFU/mL and final peptide concentrations of 8, 4, 2, 1, 0.5, 0.25, 0.13, 0.062, 0.031, 0.016, and 0.008 μg/mL.
Crystallization of N-Me-d-Gln4, d-aza-Thr8,Arg10-teixobactin (3a)28
N-Me-d-Gln4,d-aza-Thr8,Arg10-teixobactin (3a) was dissolved in 0.2 micron syringe filtered NANOpure H2O (10 mg/mL). Crystallization conditions were screened by screening in a 96-well plate format using three crystallization kits from Hampton Research (PEG/Ion, Index, and Crystal Screen). Each well was loaded with 100 μL of a different mother liquor solution from the kits. The hanging drops were set up using a TTP Labtech Mosquito® liquid handling instrument. Hanging drops were made by combining an appropriate volume of N-Me-d-Gln4,d-aza-Thr8,Arg10-teixobactin (3a) with an appropriate volume of well solution to create three 150-nL hanging drops with 1:1, 1:2, and 2:1 peptide:well solution. Hexagonal prism-shaped crystals grew in all conditions that contained polyethylene glycol (PEG) and chloride salts.
Crystal growth was optimized using conditions containing HEPES Na, PEG 400 and CaCl2. In the optimization, the HEPES Na (pH 5.5–8.0), CaCl2, and PEG 400 concentrations were varied across the 4×6 matrix of a Hampton VDX 24-well plate to afford crystals suitable for X-ray diffraction. The hanging drops for these optimizations were prepared on glass slides by combining 1 or 2 μL of teixobactin solution with 1 or 2 μL of well solution in ratios of 1:1, 2:1, and 1:2. Crystals that formed were checked for diffraction using a Rigaku Micromax-007 HF diffractometer with a Cu anode at 1.54 Å. As a result of the optimization, 0.16 M CaCl2, 0.1 M HEPES Na pH 7.00, and 24% PEG 400 afforded crystals suitable for X-ray diffraction.
X-ray crystallographic data collection, data processing, and structure determination
Data collection was performed with the BOS/B3 software at Advanced Light Source (ALS) using beamline 8.2.2 at a wavelength of 1.771190 Å (7000 eV). The rotation method was employed and three sets of 360 images each were collected at a 1.0° rotation interval (a total of three complete 360° rotations). The three sets were processed with XDS,29 and the resulting datasets were merged with BLEND.30 The structure was solved with SAD phasing implemented in the Hybrid Substructure Search (HySS)31 module of the Phenix suite32 using the anomalous signal from the chloride ion. The initial electron density maps were generated using the substructure coordinates as initial positions in Autosol.33 The structure was then refined with Phenix.refine34 under Phenix using Coot35 for model building. All B-factors were refined isotropically and riding hydrogen atoms coordinates were generated geometrically. The bond length, angles, and torsions restraints for unnatural amino acids (N-Me-d-Gln, d-aza-Thr, and d-allo-Ile) were generated with eLBOW36 under Phenix. We refined the N-methyl terminus (N-Me-d-Phe1) as the free amine, rather than as the ammonium ion, because there is only a single chloride anion in the asymmetric unit, which balances the positive charge of the arginine sidechain. We found no electron density or voids in the lattice that could account for an additional anion.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (grants R21AI121548 and R56AI137258). H.Y. acknowledges Allergan for fellowship support. We thank NovoBiotic Pharmaceuticals, LLC for generously providing teixobactin, which we have used as a positive control in MIC assays. We thank the Berkeley Center for Structural Biology (BCSB) of the Advanced Light Source (ALS) for synchrotron data collection. The BCSB is supported in part by the Howard Hughes Medical Institute. The ALS is supported by DOE Office of Science User Facility under contract no. DE-AC02-05CH11231.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.joc.9b02631.
Supplementary text, figures and table: HPLC and MS characterization data for teixobactin analogues; X-ray crystallographic statistics for N-Me-d-Gln4,d-aza-Thr8,Arg10-teixobactin (3a). Crystallographic coordinates of N-Me-d-Gln4, d-aza-Thr8,Arg10-teixobactin (3a) deposited into the Protein Data Bank (PDB) with code 6PSL.
The authors declare no competing financial interest.
References and Notes
- 1.Ling LL; Schneider T; Peoples AJ; Spoering AL; Engels I; Conlon BP; Mueller A; Schäberle TF; Hughes DE; Epstein S; Jones M; Lazarides L; Steadman VA; Cohen DR; Felix CR; Fetterman KA; Millett WP; Nitti AG; Zullo AM; Chen C; Lewis K A New Antibiotic Kills Pathogens Without Detectable Resistance. Nature 2015, 517, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Homma T; Nuxoll A; Gandt AB; Ebner P; Engels I; Schneider T; Götz F; Lewis K; Conlon BP Dual Targeting of Cell Wall Precursors by Teixobactin Leads to Cell Lysis. Antimicrob Agents Chemother. 2016, 60, 6510–6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breukink E; de Kruijff B Lipid II as a Target for Antibiotics. Nat. Rev. Drug Discov 2006, 5, 321–323. [DOI] [PubMed] [Google Scholar]
- 4.Yang H; Wierzbicki M; Du Bois DR; Nowick JS X-ray Crystallographic Structure of a Teixobactin Derivative Reveals Amyloid-like Assembly. J. Am. Chem. Soc 2018, 140, 14028–14032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zong Y; Fang F; Meyer KJ; Wang L; Ni Z; Gao H; Lewis K; Zhang J; Rao Y Gram-Scale Total Synthesis of Teixobactin Promoting Binding Mode Study and Discovery of More Potent Antibiotics. Nat. Commun 2019, 10, 3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(1) For related studies, see: Nam J; Shin D; Rew Y; Boger DL Alanine Scan of [L-Dap2]Ramoplanin A2 Aglycon: Assessment of the Importance of Each Residue. J. Am. Chem. Soc 2007, 129, 8747–8755. [DOI] [PMC free article] [PubMed] [Google Scholar]; (2) Xie J; Okano A; Pierce JG; James RC; Stamm S; Crane CM; Boger DL Total Synthesis of [Ψ[C(=S)NH]Tpg4]Vancomycin Aglycon, [Ψ[C(=NH)NH]Tpg4]Vancomycin Aglycon, and Related Key Compounds: Reengineering Vancomycin for Dual d-Ala-d-Ala and d-Ala-d-Lac Binding. J. Am. Chem. Soc 2012, 134, 1284–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang H; Chen KH; Nowick JS Elucidation of the Teixobactin Pharmacophore. ACS Chem. Biol 2016, 11, 1823–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Izzo I; Acosta GA; Tulla-Puche J; Cupido T; Martin-Lopez MJ; Cuevas C; Albericio F Solid-Phase Synthesis of Aza-Kahalalide F Analogues: (2R,3R)-2-Amino-3-azidobutanoic Acid as Precursor of the Aza-Threonine. Eur. J. Org. Chem 2010, 2536–2543. [Google Scholar]
- 9.Bartra M; Romea P Urpí F; Vilarrasa J A Fast Procedure for the Reduction of Azides and Nitro Compounds Based on the Reducing Ability of Sn(SR)3-Species. Tetrahedron 1990, 46, 587–594. [Google Scholar]
- 10.Saneyoshi H; Ochikubo T; Mashimo T; Hatano K; Ito Y; Abe H Triphenylphosphinecarboxamide: An Effective Reagent for the Reduction of Azides and Its Application to Nucleic Acid Detection. Org. Lett 2014, 16, 30–33. [DOI] [PubMed] [Google Scholar]
- 11.We prefer to load Ala9 on the resin and cyclize onto Arg10, rather than loading Arg10 and cyclizing onto Ile11, because loading of Fmoc-Arg(Pbf)-OH onto 2-chlorotriyl resin is less reproducible than loading Fmoc-Ala-OH onto 2-chlorotrityl resin.
- 12.The reduction of l-allo-azido-threonine with triphenylphosphine-2-carboxamide proceeds more slowly than that of the other diastereomers in our Boc-Ala-l-allo-azido-Thr-Gln(Trt) model system, but the reaction could be driven to completion by prolonged reaction time.
- 13.Yang H; Du Bois DR; Ziller JW; Nowick JS X-ray Crystallographic Structure of a Teixobactin Analogue Reveals Key Interactions of the Teixobactin Pharmacophore. Chem. Commun 2017, 53, 2772–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zong et al. reported that a lactam derivative of Lys10-teixobactin bearing d-aza-threonine at position 8 and a biphenyl derivative of phenylalanine at position 1 exhibits antibiotic activity comparable to natural teixobactin.; [Zong Y; Sun X; Gao H; Meyer KJ; Lewis K; Rao Y Developing Equipotent Teixobactin Analogues Against Drug-Resistant Bacteria and Discovering a Hydrophobic Interaction Between Lipid II and Teixobactin. J. Med. Chem 2018, 61, 3409–3421. [DOI] [PubMed] [Google Scholar]
- 15.Similar enhancements in the activity of teixobactin analogues by polysorbate 80 have been observed by Parmer et al.:; Parmar A; Iyer A; Prior SH; Lloyd DG; Leng Goh ET; Vincent CS; Palmai-Pallag T; Bachrati CZ; Breukink E; Madder A; Lakshminarayanan R; Taylor EJ; Singh I Teixobactin Analogues Reveal Enduracididine to be Non-Essential for Highly Potent Antibacterial Activity and Lipid II Binding. Chem. Sci 2017, 8, 8183–8192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arhin FF; Sarmiento I; Belley A; McKay GA; Draghi DC; Grober P; Sahm DF; Parr TR Jr.; Moeck G Effect of Polysorbate 80 on Oritavancin Binding to Plastic Surfaces: Implications for Susceptibility Testing. Antimicrob. Agents Chemother 2008, 52, 1597–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen KH; Le SP; Han X; Frias JM; Nowick JS Alanine Scan Reveals Modifiable Residues in Teixobactin. Chem. Commun 2017, 53, 11357–11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.N-Methylation substantially decreases the antibiotic activity of teixobactin analogues. The MIC of azateixobactin analogue 3a was determined to be 32 μg/mL against S. epidermidis and B. subtilis in the absence of polysorbate 80. We have previously observed similar diminished antibiotic activity of N-Me-d-PheI1,N-Me-d-Gln4,Arg10-teixobactin [ref 4]
- 19.Kabsch W XDS. Acta Crystallogr., Sect. D: Biol. Crystallogr 2010, 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foadi J; Aller P; Alguel Y; Cameron A; Axford D; Owen RL; Armour W; Waterman DG; Iwata S; Evans G Clustering Procedures for the Optimal Selection of Data Sets from Multiple Crystals in Macromolecular Crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr 2013, 69, 1617–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams PD; Afonine PV; Bunkoczi G; Chen VB; Davis IW; Echols N; Headd JJ; Hung LW; Kapral GJ; Grosse-Kunstleve RW; McCoy AJ; Moriarty NW; Oeffner R; Read RJ; Richardson DC; Richardson JS; Terwilliger TC; Zwart PH PHENIX: a Comprehensive Python-Based system for Macromolecular Structure Solution. Acta Crystallogr., Sect. D: Biol. Crystallogr 2010, 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The macrolactam-chloride anion complex formed by azateixobactin analogue 3a is similar in conformation to the macrolactone-chloride anion complex formed by a truncated teixobactin analogue, which we have previously reported [ref 13].
- 23.Breukink E; de Kruijff B Lipid II as a Target for Antibiotics. Nat. Rev. Drug Discov 2006, 5, 321–323. [DOI] [PubMed] [Google Scholar]
- 24.Cavalleri B; Pagani H; Volpe G; Selva E; Parenti F Ramoplanin (A-16686), a New Glycolipodepsipeptide Antibiotic. J. Antibiot 1984, 37, 309–317. [DOI] [PubMed] [Google Scholar]
- 25.(a) Bonner DP; O’Sullivan J; Tanaka SK; Clark JM; Whitney RR Lysobactin, a Novel Antibacterial Agent Produced by Lysobacter sp. II. Biological Properties. J. Antibiot 1988, 41, 1745–1751. [DOI] [PubMed] [Google Scholar]; (b) Kato T; Hinoo H; Terui Y; Kikuchi J; Shoji J The Structures of Katanosins A and B. J. Antibiot 1988, 41, 719–725. [DOI] [PubMed] [Google Scholar]; (c) Shoji J; Hinoo H; Matsumoto K; Hattori T; Yoshida T; Matsuura S; Kondo E Isolation and Characterization of Katanosins A and B. J. Antibiot 1988, 41, 713–718. [DOI] [PubMed] [Google Scholar]
- 26.Shoji J; Hinoo H; Katayama T; Nakagawa Y; Ikenishi Y; Iwatani K; Yoshida T Structures of New Peptide Antibiotics, Plusbacins A1~A4 and B1~B4. J. Antibiot 1992, 45, 824–831. [DOI] [PubMed] [Google Scholar]
- 27.Triphenylphosphine-2-carboxamide was synthesized using the procedure reported by Saneyoshi et al. MS (positive ion mode) calcd for C19H17NOP+ [M+H]+ m/z 306.10, found 306.12.; [Saneyoshi H; Ochikubo T; Mashimo T; Hatano K; Ito Y; Abe H Triphenylphosphinecarboxamide: An Effective Reagent for the Reduction of Azides and Its Application to Nucleic Acid Detection. Org. Lett 2014, 16, 30–33. [DOI] [PubMed] [Google Scholar]
- 28.The procedure in this section is adapted from and in some cases taken verbatim from ref 4.
- 29.Kabsch W XDS. Acta Crystallogr., Sect. D: Biol. Crystallogr 2010, 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foadi J; Aller P; Alguel Y; Cameron A; Axford D; Owen RL; Armour W; Waterman DG; Iwata S; Evans G Clustering Procedures for the Optimal Selection of Data Sets from Multiple Crystals in Macromolecular Crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr 2013, 69, 1617–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grosse-Kunstleve RW; Adams PD Substructure Search Procedures for Macromolecular Structures. Acta Crystallogr., Sect. D: Biol. Crystallogr 2003, 59, 1966–1973. [DOI] [PubMed] [Google Scholar]
- 32.Adams PD; Afonine PV; Bunkoczi G; Chen VB; Davis IW; Echols N; Headd JJ; Hung LW; Kapral GJ; Grosse-Kunstleve RW; McCoy AJ; Moriarty NW; Oeffner R; Read RJ; Richardson DC; Richardson JS; Terwilliger TC; Zwart PH PHENIX: A Comprehensive Python-Based System for Macromolecular Structure Solution. Acta Crystallogr., Sect. D: Biol. Crystallogr 2010, 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terwilliger TC; Adams PD; Read RJ; McCoy AJ; Moriarty NW; Grosse-Kunstleve RW; Afonine PV; Zwart PH; Hung LW Decision-Making in Structure Solution using Bayesian Estimates of Map Quality: the PHENIX AutoSol Wizard. Acta Crystallogr., Sect. D: Biol. Crystallogr 2009, 65, 582–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afonine PV; Grosse-Kunstleve RW; Echols N; Headd JJ; Moriarty NW; Mustyakimov M; Terwilliger TC; Urzhumtsev A; Zwart PH; Adams. P. Towards Automated Crystallographic Structure Refinement with phenix.refine. Acta Crystallogr., Sect. D: Biol. Crystallogr 2012, 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P; Lohkamp B; Scott WG; Cowtan K Features and Development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr 2010, 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moriarty NW; Grosse-Kunstleve RW; Adams PD Electronic Ligand Builder and Optimization Workbench (eLBOW): A Tool for Ligand Coordinate and Restraint Generation. Acta Crystallogr., Sect. D: Biol. Crystallogr 2009, 65, 1074–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


