Abstract
Background
It has been well established that the TMEM106B gene rs1990622 variant was a frontotemporal dementia (FTD) risk factor. Until recently, growing evidence highlights the role of TMEM106B in Alzheimer’s disease (AD). However, it remains largely unclear about the role of rs1990622 variant in AD.
Methods
Here, we conducted comprehensive analyses including genetic association study, gene expression analysis, eQTLs analysis, and colocalization analysis. In stage 1, we conducted a genetic association analysis of rs1990622 using large-scale genome-wide association study (GWAS) datasets from International Genomics of Alzheimer’s Project (21,982 AD and 41,944 cognitively normal controls) and UK Biobank (314,278 participants). In stage 2, we performed a gene expression analysis of TMEM106B in 49 different human tissues using the gene expression data in GTEx. In stage 3, we performed an expression quantitative trait loci (eQTLs) analysis using multiple datasets from UKBEC, GTEx, and Mayo RNAseq Study. In stage 4, we performed a colocalization analysis to provide evidence of the AD GWAS and eQTLs pair influencing both AD and the TMEM106B expression at a particular region.
Results
We found (1) rs1990622 variant T allele contributed to AD risk. A sex-specific analysis in UK Biobank further indicated that rs1990622 T allele only contributed to increased AD risk in females, but not in males; (2) TMEM106B showed different expression in different human brain tissues especially high expression in cerebellum; (3) rs1990622 variant could regulate the expression of TMEM106B in human brain tissues, which vary considerably in different disease statuses, the mean ages at death, the percents of females, and the different descents of the selected donors; (4) colocalization analysis provided suggestive evidence that the same variant contributed to AD risk and TMEM106B expression in cerebellum.
Conclusion
Our comprehensive analyses highlighted the role of FTD rs1990622 variant in AD risk. This cross-disease approach may delineate disease-specific and common features, which will be important for both diagnostic and therapeutic development purposes. Meanwhile, these findings highlight the importance to better understand TMEM106B function and dysfunction in the context of normal aging and neurodegenerative diseases.
Keywords: TMEM106B, Alzheimer’s disease, Genome-wide association study, Neurological diseases, eQTLs
Background
TMEM106B is a lysosomal protein and belongs to the TMEM106 family of proteins with relatively unknown function [1, 2]. In 2010, a common TMEM106B genetic variant rs1990622 (T>C) was first identified to be associated with frontotemporal dementia (FTD) risk (OR = 1.64 for T allele, allele frequency = 0.679, and P = 1.08E−11) [3]. Since the identification of rs1990622 as an FTD risk variant, kinds of studies have been conducted to understand the role of this non-coding mutation, which is located downstream 6.9 kb 3′ of TMEM106B [4–6].
Li and colleagues conducted a cell type quantitative trait loci (cQTL) analysis using 2008 brain samples derived from 1536 unique individuals including 640 AD, 488 cognitively healthy controls, 11 FTD, 75 progressive supranuclear palsy, 28 pathological aging, 189 schizophrenia, 30 bipolar disorders, and 75 individuals with other unknown dementia or no diagnosis information [4]. Interestingly, Li and colleagues identified TMEM106B variant rs1990621 C allele, which is in high linkage disequilibrium with rs1990622 variant T allele, to be significantly associated with the reduced neuronal proportion [4].
Until recently, Yang and colleagues conducted a module quantitative trait loci (modQTL) analysis to identify genetic variants regulating the average expression of the genes found in the gene co-expression modules [5]. Interestingly, they found rs1990622 variant to show a significant modQTL effect, and highlighted TMEM106B as key aging human brain transcriptome regulator [5]. Meanwhile, Yang and colleagues identified that myelination and lysosomal genes regulated by TMEM106B could connect amyloid-β (Aβ) and TAR DNA-binding protein 43 kDa (TDP-43) [5]. It is known that increased Aβ is a key Alzheimer’s disease (AD) neuropathology. Hence, Yang and colleagues provided important findings about the key pathogenic link between AD and TDP-43 proteinopathy [5]. However, Yang and colleagues did not directly evaluate the association between rs1990622 variant and AD risk. Until now, it remains unclear whether rs1990622 variant is associated with AD risk, although a lack of significant association between rs1990622 variant and AD risk [7]. We think that this may be caused by inadequate sample sizes (300 AD cases and 137 controls) [7], and large-scale samples are needed.
Meanwhile, Yang and colleagues conducted an expression quantitative trait loci (eQTLs) analysis of rs1990622 variant using 494 human prefrontal cortex samples from the Religious Orders Study and Memory and Aging Project (ROSMAP) [8]. They found that rs1990622 variant T allele could significantly increase TMEM106B expression (β = 0.067, and P = 5.90E−05) [5]. However, gene expression analysis did not support the increased TMEM106B expression in human brain tissues. Satoh et al. evaluated the expression levels of TMEM106B in AD and control frontal cortex and the hippocampus tissues [9]. They selected 6 sporadic AD patients and 13 controls including 4 normal subjects without neurological disease, 3 patients with sporadic Parkinson’s disease, 4 patients with sporadic amyotrophic lateral sclerosis, and 2 patients with sporadic multiple system atrophy [9]. They demonstrated that both the mRNA and protein levels of TMEM106B were significantly reduced in AD brains compared control brains [9].
In discussion, Yang and colleagues concluded that the pre-existing neurodegenerative proteinopathies were not necessary to drive the association between rs1990622 variant and TMEM106B transcriptome dysregulation [5]. However, recent findings from other TMEM106B variants did not support this conclusion. Ren and colleagues conducted a stratification analysis and highlighted more pronounced effects of TMEM106B rs3173615 variant on the transcriptome in neurodegenerative diseases than in healthy controls [6]. Li and colleagues conducted a stratification analysis and found that TMEM106B rs1990621 variant could regulate the neuronal proportion in AD cases, other neurodegenerative diseases, elderly cognitively healthy controls, but not young controls [4]. All these findings indicated that the link between TMEM106B haplotype and transcriptome dysregulation is context dependent [4, 6]. Importantly, rs1990622 variant is in high linkage disequilibrium with rs3173615 (r2 = 0.98 and D′ = 1) and rs1990621 (r2 = 0.99 and D′ = 1). Hence, we consider that the association between rs1990622 variant and TMEM106B transcriptome dysregulation may also be context dependent.
Until now, large-scale AD genome-wide association study (GWAS) datasets and large-scale eQTLs datasets in both the neuropathologically normal individuals and neurological disease individuals have provided strong support to answer these concerns [10, 11]. Here, we conducted comprehensive analyses using publicly available datasets. In stage 1, we conducted a genetic association analysis to investigate the effect of rs1990622 variant on AD risk using multiple large-scale GWAS datasets. In stage 2, we performed a gene expression analysis of TMEM106B in 49 different human tissues. In stage 3, we performed an eQTLs analysis to evaluate the effect of rs1990622 variant on TMEM106B expression in multiple human brain tissues with different disease statuses. In stage 4, we performed a colocalization analysis to provide evidence of the AD GWAS and eQTLs pair influencing both AD and the TMEM106B expression at a particular region.
Methods
AD GWAS datasets
We selected two independent large-scale AD GWAS dataset resources from International Genomics of Alzheimer’s Project (IGAP) stage 1 [10] and UK Biobank [11]. The IGAP stage 1 consisted of 21,982 AD and 41,944 cognitively normal controls of European descent [10]. These individuals are from four consortia including Alzheimer Disease Genetics Consortium (ADGC), Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE), the European Alzheimer’s Disease Initiative (EADI), and Genetic and Environmental Risk in AD/Defining Genetic, Polygenic and Environmental Risk for Alzheimer’s Disease Consortium (GERAD/PERADES) [10]. AD patients are diagnosed using the NINCDS-ADRDA criteria or DSM-IV guidelines [10]. In UK Biobank, AD GWAS was conducted in 314,278 participants including 27,696 maternal cases and 14,338 paternal cases [11]. Meanwhile, UK Biobank also included two sex-specific AD GWAS datasets including one AD GWAS dataset in males diagnosed by paternal history of AD (14,338 cases and 245,941 controls) and one AD GWAS dataset in females diagnosed by maternal history of AD (27,696 cases and 260,980 controls) [11].
eQTLs datasets from neuropathologically normal and disease individuals
We selected two independent eQTLs dataset resources from the neuropathologically normal individuals. The first resource is from the UK Brain Expression Consortium (UKBEC), which included 134 neuropathologically normal individuals of European descent [12]. The UKBEC consisted of 10 eQTLs datasets in 10 brain regions including cerebellar cortex, frontal cortex, hippocampus medulla, occipital cortex, putamen, substantia nigra, temporal cortex, thalamus, and intralobular white matter [12]. The second resource is from the Genotype-Tissue Expression (GTEx, version 8) [13]. The GTEx included 13 eQTLs datasets in 13 brain tissues (amygdala/amygdalae, anterior cingulate cortex, caudate basal ganglia, cerebellar hemisphere, cerebellum, cortex, frontal cortex, hippocampus, hypothalamus, nucleus accumbens, putamen, spinal cord, and substantia nigra) [13]. About 99% of the donors of these brain tissues are neuropathologically normal individuals, and 1% of the donors of these brain tissues died of neurological diseases (1.3% in age 20–39 and 1.2% in age 60–71) [13]. Recent studies have provided more detailed information about these datasets [14–19].
The eQTLs dataset resource in neurological disease individuals is from the Mayo RNAseq Study [20]. Mayo eQTLs datasets included 773 brain samples, which could be further divided into 197 AD cerebellar samples, 202 AD temporal cortex samples, 177 non-AD cerebellar samples, and 197 non-AD temporal cortex samples [20]. The non-AD samples have several brain pathologies including PSP, LBD, corticobasal degeneration, FTD, multiple system atrophy, and vascular dementia [20].
The main demographic profiles of the selected eQTLs datasets are provided in Table 1. In brief, the selected donors in UKBEC are of European descent with mean age at death 59 and 26% of these donors were female. The selected donors in GTEx are of multiple descents including European (85.3%), African (12.3%), Asian (1.4%), and Hispanic or Latino (1.9%), with mean age at death 55, and 33% of these donors were female. The selected donors in Mayo are of European descent with mean age at death 74 in AD and 72 in non-AD, and 51–53% of these donors were female in AD and 36–40% of these donors were female in non-AD. In order to further compare the findings from ROSMAP, we also included the demographic profiles of the ROSMAP eQTLs datasets, as described in Table 1. In ROSMAP, the selected donors are of European descent with mean age at death 88 and 62% of these donors were female. Thirty-nine percent of these donors were diagnosed with clinical AD, and 58% were pathological AD.
Table 1.
Demographic profiles of the selected eQTLs datasets
| Dataset | Diagnosis | Donors # | Mean age at death | Descent | Females % |
|---|---|---|---|---|---|
| UKBEC | Neuropathologically normal | 134 | 59 | European | 26 |
| GTEx | Neuropathologically normal | 838 | 55 | European (85.3%) | 33 |
| African (12.3%) | |||||
| Asian (1.4%) | |||||
| Hispanic or Latino (1.9%) | |||||
| Mayo | AD (cerebellar) | 197 | 74 | European | 51 |
| Mayo | Non-AD (cerebellar) | 177 | 72 | European | 36 |
| Mayo | AD (temporal cortex) | 202 | 74 | European | 53 |
| Mayo | Non-AD (temporal cortex) | 197 | 72 | European | 40 |
| ROSMAP | 39% clinical AD | 494 | 88 | European | 62 |
| 58% pathological AD |
Genetic association analysis
For the genetic association analysis, we first used the AD GWAS summary statistics to directly evaluate the association of rs1990622 variant with AD in IGAP and UK Biobank. We extract the corresponding summary statistics of rs1990622 variant including the beta coefficient (effect size) and standard error in these datasets, respectively. We then determine the heterogeneity of rs1990622 variant in both datasets using Cochran’s Q test [21–26]. Finally, we conducted a meta-analysis to evaluate the association between rs1990622 variant and AD using R package (meta: General Package for Meta-Analysis) [21–26]. The overall OR is calculated by the fixed effect model (Mantel-Haenszel) or random-effect model (DerSimonian-Laird), which is determined by the heterogeneity [21–26]. Meanwhile, we further perform additional sex stratification analysis only using the UK Biobank GWAS summary datasets. The statistically significant association for heterogeneity test and meta-analysis is defined to be P < 0.05.
Gene expression analysis of TMEM106B
In order to evaluate the expression of TMEM106B in different human tissues, we conduct a gene expression analysis using the gene expression data in GTEx (version 8, dbGaP Accession phs000424.v8.p2) [27]. GTEx (version 8) consists of 49 tissues, 838 donors, and 15,201 samples with the number of samples with genotype ≥ 70 [27]. Illumina TruSeq RNA sequencing and Affymetrix Human Gene 1.1 ST Expression Array were selected to measure the levels of gene expression, which was quantified by transcripts per million (TPM) based on the GENCODE 26 annotation [27]. Here, T test or analysis of variance (ANOVA) was selected to evaluate the potential difference of TMEM106B expression in different human tissues. The statistical significance is P < 0.05.
eQTLs analysis of rs1990622 variant
In all these selected eQTLs datasets, an additive model was used to indicate the rs1990622 genotype dosages including CC = 0, CT = 1, and TT = 2, and a linear regression analysis was used to conduct the eQTLs analysis. For eQTLs analysis in UKBEC, we first downloaded the TMEM106B gene expression data and the rs1990622 genotype data [12]. We then evaluated the association between rs1990622 variant and TMEM106B gene expression using a linear regression analysis [12]. For eQTLs analysis in GTEx (version 8), we used the online GTEx eQTL Calculator with the linear regression method to evaluate the association rs1990622 variant and TMEM106B expression [13]. In Mayo RNAseq Study, a linear regression was used to perform the eQTLs analysis by correcting for APOE ε4 dosage, age at death, gender, and multiple technical variables [20]. Here, we downloaded the summary results from the Mayo RNAseq Study to directly evaluate the association rs1990622 variant and TMEM106B expression [20]. The statistically significant association is defined to be P < 0.05/27 = 1.85E−03. The suggestive association is defined to be P < 0.05.
Colocalization analysis
In order to provide evidence of the AD GWAS and eQTLs pair influencing both AD and the TMEM106B expression at a particular region, we performed a colocalization analysis using Coloc: a package for colocalization analyses [28, 29]. Coloc could test five hypotheses based on the posterior probability (PP) for colocalization: H0—neither trait has a genetic association in the region; H1/H2—only trait 1/trait 2 has a genetic association in the region; H3—both traits are associated, but with different causal variants; and H4—both traits are associated and share a single causal variant [28, 29].
Results
Genetic association analysis of rs1990622 with AD
In IGAP, the results showed that rs1990622 T allele was significantly associated with increased AD risk (P = 5.42E−03). Interestingly, this finding was successfully replicated in UK Biobank (P = 1.20E−02). Importantly, the sex-specific analysis indicated that rs1990622 T allele was only significantly associated with increased AD risk in females (P = 5.74E−04), but not in males (P = 6.48E−01), as provided in Table 2. In both IGAP and UK Biobank, we found no significant heterogeneity with Cochran’s Q test P = 0.3833. A meta-analysis using the fixed effect model showed significant association between rs1990622 variant T allele and AD risk (OR = 1.03, 95% CI 1.01–1.05, P = 2.00E−04).
Table 2.
Association between rs1990622 variant T allele and AD
| Dataset | Disease | Beta | SE | P value |
|---|---|---|---|---|
| IGAP | AD | 0.040 | 0.014 | 5.42E−03 |
| UK Biobank | AD | 0.025 | 0.010 | 1.20E−02 |
| UK Biobank | AD in females | 0.043 | 0.012 | 5.74E−04 |
| UK Biobank | AD in males | 0.008 | 0.017 | 6.48E−01 |
SE standard error. Beta is the regression coefficient based on the effect allele. Beta > 0 and beta < 0 mean that this effect allele could increase and reduce AD risk, respectively. The statistically significant association is defined to be P < 0.05
Gene expression analysis of TMEM106B
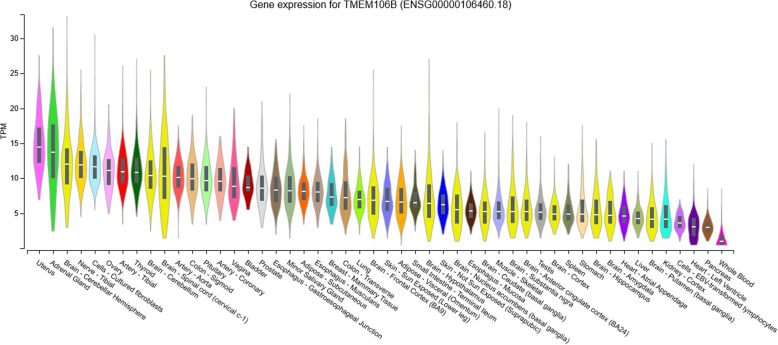
In these selected 49 human tissues from GTEx, the top 10 human tissues with high TMEM106B expression are uterus (n = 142, TPM median = 14.48), adrenal gland (n = 258, TPM median = 13.72), cerebellar hemisphere (n = 215, TPM median = 12.02), tibial nerve (n = 619, TPM median = 11.90), cultured fibroblast cells (n = 504, TPM median = 11.66), ovary (n = 180, TPM median = 11.14), tibial artery (n = 663, TPM median = 10.94), thyroid (n = 653, TPM median = 10.90), cerebellum (n = 241, TPM median = 10.36), and spinal cord (n = 159, TPM median = 10.32). Meanwhile, TMEM106B shows low expression in other 10 human brain tissues including frontal cortex (n = 209, TPM median = 6.904), hypothalamus (n = 202, TPM median = 6.468), nucleus accumbens (n = 246, TPM median = 5.533), caudate (n = 246, TPM median = 5.286), substantia nigra (n = 139, TPM median = 5.242), anterior cingulate cortex (n = 176, TPM median = 5.237), cortex (n = 255, TPM median = 4.939), hippocampus (n = 197, TPM median = 4.848), amygdala/amygdalae (n = 197, TPM median = 4.848), and putamen (n = 205, TPM median = 4.213). The box plots about TMEM106B gene expression in different tissues are provided in Fig. 1.
Fig. 1.
The box plots for the expression of TMEM106B in different tissues in GTEx. The gene expression values are shown in transcripts per million (TPM). The gene expression level was quantified by TPM based on the GENCODE 26 annotation, collapsed to a single transcript model for each gene using a custom isoform collapsing procedure [27]
eQTLs analysis
In UKBEC, eQTLs analysis showed that rs1990622 T allele was not significantly associated with TMEM106B expression in all these 10 brain regions, as provided in Table 3. In GTEx, eQTLs analysis indicated that rs1990622 T allele was significantly associated with TMEM106B expression in cerebellum (P = 1.90E−06), cortex (P = 2.20E−05), and cerebellar hemisphere (P = 1.50E−03). Meanwhile, rs1990622 T allele also showed suggestive association with TMEM106B expression in frontal cortex (P = 2.20E−02). Importantly, the rs1990622 T allele could only significantly reduce TMEM106B expression in these brain tissues, as provided in Table 3. Hence, the eQTLs findings in UKBEC and GTEx from neuropathologically normal individuals did not support the association between rs1990622 T allele and increased TMEM106B expression even in the same brain tissue, as reported in ROSMAP by Yang and colleagues.
Table 3.
Association between rs1990622 variant T allele and TMEM106B expression
| Dataset | Beta | SE | P value | Brain tissue | Number |
|---|---|---|---|---|---|
| UKBEC | 0.024 | 0.046 | 5.96E−01 | Cerebellar cortex | 134 |
| UKBEC | 0.019 | 0.053 | 7.20E−01 | Frontal cortex | 134 |
| UKBEC | 0.037 | 0.054 | 4.92E−01 | Hippocampus | 134 |
| UKBEC | 6.57E−05 | 0.044 | 9.99E−01 | Medulla | 134 |
| UKBEC | 0.015 | 0.065 | 8.19E−01 | Occipital cortex | 134 |
| UKBEC | − 0.003 | 0.063 | 9.63E−01 | Putamen | 134 |
| UKBEC | − 0.025 | 0.058 | 6.69E−01 | Substantia nigra | 134 |
| UKBEC | − 0.030 | 0.049 | 5.38E−01 | Temporal cortex | 134 |
| UKBEC | − 0.019 | 0.058 | 7.48E−01 | Thalamus | 134 |
| UKBEC | 0.046 | 0.052 | 3.78E−01 | Intralobular white matter | 134 |
| GTEx | − 0.084 | 0.065 | 1.90E−01 | Amygdala | 129 |
| GTEx | − 0.100 | 0.059 | 9.60E−02 | Anterior cingulate cortex | 147 |
| GTEx | − 0.025 | 0.033 | 4.50E−01 | Caudate | 194 |
| GTEx | − 0.130 | 0.041 | 1.50E−03 | Cerebellar hemisphere | 175 |
| GTEx | − 0.150 | 0.031 | 1.90E−06 | Cerebellum | 209 |
| GTEx | − 0.140 | 0.032 | 2.20E−05 | Cortex | 205 |
| GTEx | − 0.089 | 0.039 | 2.20E−02 | Frontal cortex | 175 |
| GTEx | − 0.009 | 0.037 | 8.20E−01 | Hippocampus | 165 |
| GTEx | − 0.048 | 0.044 | 2.80E−01 | Hypothalamus | 170 |
| GTEx | 0.048 | 0.034 | 1.60E−01 | Nucleus accumbens | 202 |
| GTEx | − 0.022 | 0.032 | 5.00E−01 | Putamen | 170 |
| GTEx | − 0.047 | 0.062 | 4.50E−01 | Spinal cord | 126 |
| GTEx | 0.100 | 0.067 | 1.40E−01 | Substantia nigra | 114 |
| Mayo | − 0.036 | – | 2.11E−01 | Cerebellum in AD | 186 |
| Mayo | 0.052 | – | 1.18E−01 | Cerebellum in non-AD | 170 |
| Mayo | − 0.001 | – | 9.61E−01 | Temporal cortex in AD | 191 |
| Mayo | 0.031 | – | 1.75E−01 | Temporal cortex in non-AD | 181 |
SE standard error. Beta is the regression coefficient based on the effect allele. Beta > 0 and beta < 0 mean that this effect allele could increase and reduce gene expression, respectively. The statistically significant association is defined to be P < 0.05/27 = 1.85E−03. The suggestive association is defined to be P < 0.05
In Mayo datasets, eQTLs analysis indicated no significant association of rs1990622 T allele with TMEM106B expression in cerebellum and temporal cortex (Table 3). Hence, the most significant association between rs1990622 T allele and reduced TMEM106B expression identified in neuropathologically normal individuals was not successfully replicated in neurological disease individuals. Here, we provided more detailed results about eQTLs analysis in Table 3.
Colocalization analysis
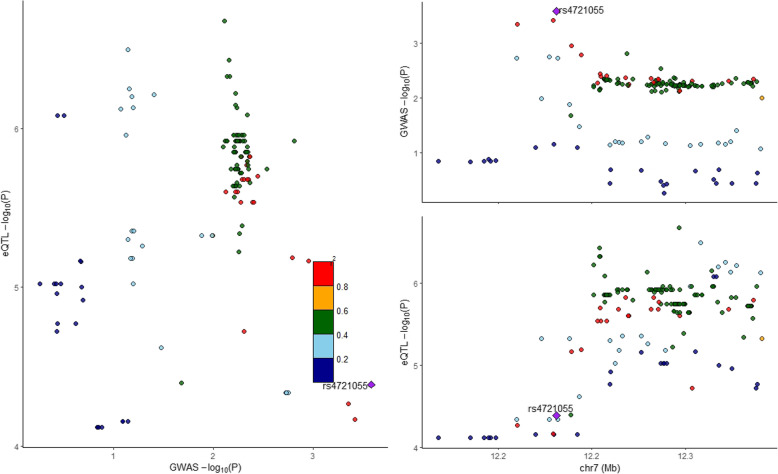
In GTEx cerebellum eQTLs dataset, we got 181 genetic variants, which could regulate TMEM106B expression with the genome-wide significance level P < 1.00E−04. We then integrated these 181 genetic variants with AD GWAS dataset from IGAP using Coloc. The results showed that TMEM106B had suggestive evidence (PP4 = 20%) of sharing the same variant with AD risk and TMEM106B expression in cerebellum (PP0 = 0.031, PP1 = 0.002, PP2 = 0.735, PP3 = 0.037), as described in Fig. 2.
Fig. 2.
Colocalization analysis of genetic variants associated with TMEM106B expression in GTEx cerebellum and AD risk. Created using locuscomparer R package. Coloc PP0 = 0.031, PP1 = 0.002, PP2 = 0.735, PP3 = 0.037, and PP4 = 0.196. The eQTLs dataset is from GTEx cerebellum (n = 209). The AD GWAS dataset is from the IGAP including 21,982 AD and 41,944 cognitively normal controls
Discussion
It has been well established that the TMEM106B rs1990622 variant was a FTD risk factor [30, 31]. Until recently, growing evidence highlights the role of TMEM106B in other neurological processes including hippocampal sclerosis of aging [32], neuronal loss [31], cognitive deficits [31], better residual cognition [30], AD [9, 33], Parkinson’s disease [34], and amyotrophic lateral sclerosis [34]. Importantly, Yang and colleagues identified the TMEM106B rs1990622 variant to show a significant modQTL effect, and highlighted the converging effects of APOE-Aβ and TMEM106B [5]. However, it remains largely unclear about the role of rs1990622 variant in AD. Here, we conducted comprehensive analyses including genetic association study, gene expression analysis, eQTLs analysis, and colocalization analysis.
Using the genetic association analysis, we evaluated the association of rs1990622 variant with AD using two independent large-scale GWAS datasets from IGAP and UK Biobank, and then conducted a meta-analysis [10, 11]. Interestingly, the results are consistent in both IGAP and UK Biobank, which indicated that rs1990622 was significantly associated with AD risk in both datasets (Table 2). A sex-specific analysis in UK Biobank further indicated that rs1990622 T allele only contributed to increased AD risk in females, but not in males (Table 2). Tropea and colleagues have also evaluated the association of rs1990622 variant with AD using 300 AD cases and 137 neurologically normal control subjects [7]. However, Tropea and colleagues did not identify any significant association of rs1990622 with AD [7]. We think that this may be caused by inadequate sample sizes.
It is known that rs1990622 variant is a non-coding mutation. Hence, it remains unclear how rs1990622 variant affects AD risk. eQTLs analysis is an important method to evaluate the roles of non-coding genetic variants especially the GTEx project, which established a data resource and tissue bank to study the relationship between genetic variation and gene expression in multiple human tissues [35]. To explore the effect of rs1990622 variant in AD risk by regulating TMEM106B expression, eQTLs analysis should be conducted in neuropathologically normal individuals or in a general population based on the three considerations. First, eQTLs analysis in AD patients could not be interpreted in terms of AD risk or susceptibility as lack of healthy controls or general individuals [36]. Second, it is well known that disease statuses could change the expression of a specific gene. Hence, gene expression analysis often indicated dysregulated genes in cases compared with controls [37]. Take TREM2 for example, its expression is upregulated in multiple pathological conditions such as Parkinson’s disease, amyotrophic lateral sclerosis, stroke, traumatic brain injury, and AD, compared with normal controls [37]. Until now, most eQTLs studies focusing genetic variants associated with neurological diseases were conducted in neuropathologically normal individuals, such as AD (UKBEC [38], and GSE15745 [38], and 128 normal subjects [39]), progressive supranuclear palsy (387 normal subjects) [40], schizophrenia (UKBEC [41], GTEx [41], 128 normal subjects [39], and 120 normal subjects [42]), Parkinson’s disease (128 normal subjects [39], GTEx [43]), and bipolar disorder (120 normal subjects [42], GTEx [44], and UKBEC [44]). Meanwhile, other eQTLs studies using both AD and other controls were also reported by adjusting for disease status and some critical covariates [45, 46]. Third, Nicholson and colleagues have reviewed recent findings and found that the significant association between rs1990622 and TMEM106B mRNA expression identified in lymphoblast cells could not be successfully replicated in postmortem brain tissues [2]. It is possible that the variable levels of neuronal loss and cell type composition may have masked the association between rs1990622 and TMEM106B mRNA expression [2]. Hence, Nicholson and Rademakers suggested that eQTLs studies might be best conducted in non-diseased tissues [2].
Considering these above findings, we then performed an eQTLs analysis to evaluate the effect of rs1990622 variant on TMEM106B expression in multiple human brain tissues from neuropathologically normal individuals (UKBEC and GTEx), and further compared the findings from neurological disease individuals (Mayo and ROSMAP). In UKBEC, we found no significant association between rs1990622 variant and TMEM106B expression in 10 brain regions, as provided in Table 3. In GTEx, we found that rs1990622 T allele could significantly reduce TMEM106B expression in cerebellum (P = 1.90E−06), cortex (P = 2.20E−05), and cerebellar hemisphere (P = 1.50E−03). In Mayo, we found no significant association of rs1990622 with TMEM106B expression in cerebellum and temporal cortex. In ROSMAP, Yang and colleagues found that the rs1990622 T allele could increase TMEM106B expression in human prefrontal cortex [5]. In summary, rs1990622 variant showed different association with TMEM106B expression in neuropathologically normal individuals and neurological disease individuals (Mayo and ROSMAP). The differences were even observed across the neuropathologically normal individuals, such as UKBEC and GTEx, and across the neurological disease individuals, such as Mayo and ROSMAP.
We consider that four reasons may contribute to explain these differences. First, disease statuses may have caused these differences. Satoh and colleagues found that both the mRNA and protein levels of TMEM106B were significantly reduced in AD brains compared control brains [9]. Hence, the different expression of TMEM106B may further cause different eQTLs findings in AD and controls. Importantly, eQTLs or cQTL analysis using other TMEM106B variants including rs3173615 and rs1990621 further supported our findings [4, 6]. Meanwhile, our and other studies have clearly indicated that eQTLs could vary considerably in different disease statuses [16–19, 47–52]. Second, the mean ages at death in different eQTLs datasets may have driven these differences. The mean ages at death were 55 (UKBEC), 59 (GTEx), 72 or 74 (Mayo), and 88 (ROSMAP), respectively, as provided in Table 1. Nicholson and colleagues explained that the variable levels of neuronal loss and cell type composition may have masked the association between rs1990622 and TMEM106B mRNA expression in the older population [2]. Third, the percents of females in different eQTLs datasets may have driven these differences. The percents of females were 26% (UKBEC), 33% (donors), 36–53% (Mayo), and 62% (ROSMAP), respectively, as provided in Table 1. This explanation was supported by our genetic association findings that rs1990622 T allele only contributed to increased AD risk in females, but not in males (Table 2). Importantly, recent findings from GTEx also highlighted the impact of sex on gene expression across human tissues [53]. Fourth, the different descents of the selected donors may also have contributed to these differences. The donors in UKBEC, Mayo, and ROSMAP were of European descent. However, about 85.3% donors were of European descent, and others 14.7% were of African, Asian, and Hispanic or Latino descents, as provided in Table 1.
Using the gene expression analysis, we showed that TMEM106B had high expression in cerebellar hemisphere, tibial nerve, cerebellum, and spinal cord, but low expression in other 10 human brain tissues including frontal cortex, hypothalamus, nucleus accumbens, caudate, substantia nigra, anterior cingulate cortex, cortex, hippocampus, amygdala/amygdalae, and putamen. Hence, these findings may explain the significant eQTLs results in cerebellum. Importantly, the colocalization analysis further provided suggestive evidence of sharing the same variant with AD risk and TMEM106B expression in cerebellum.
We also realized some limitations in our study, although these above findings. First, we only conducted a sex stratification genetic association analysis using the UK Biobank GWAS summary datasets. The sex stratification datasets in IGAP are not publicly available. Meanwhile, the original GWAS genotype datasets from IGAP and UK Biobank are not publicly available, or a long time is needed to request. Hence, we could not determine the interaction between the sex and rs1990622 genotypes using the raw data. Second, our genetic association analysis identified the female-specific role of rs1990622 in AD risk, but female- or male-specific eQTLs datasets are not publicly available. Third, we performed the eQTLs analysis to investigate the role of rs1990622 variant. In fact, the modQTL analysis may also be important, as did by Yang and colleagues. However, the original gene expression datasets limit our further modQTL analysis. Hence, we will further conduct additional sex stratification analysis, female-specific eQTLs analysis, and modQTL analysis when all these datasets are publicly available.
Conclusions
Here, we performed comprehensive analyses and found (1) FTD risk variant rs1990622 contributed to AD risk. This cross-disease approach may delineate disease-specific and common features, which will be important for both diagnostic and therapeutic development purposes; (2) TMEM106B showed different expression in different human brain tissues especially high expression in cerebellum; (3) rs1990622 variant could regulate the expression of TMEM106B in human brain tissues, which vary considerably in different disease statuses, the mean ages at death, the percents of females, and the different descents of the selected donors; (4) colocalization analysis provided suggestive evidence that the same variant contributed to AD risk and TMEM106B expression in cerebellum. These findings highlight the importance to better understand TMEM106B function and dysfunction in the context of normal aging and neurodegenerative diseases.
Acknowledgements
We thank the International Genomics of Alzheimer’s Project (IGAP) and UK Biobank for the GWAS summary statistics. We thank the Braineac, Mayo Clinic, and GTEx for the eQTLs dataset resources. We also thank Marioni et al. for the GWAS datasets about the family history of AD. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in the analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients, and their families. The i-Select chips was funded by the French National Foundation on AD and related disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2, and the Lille University Hospital. GERAD was supported by the Medical Research Council (grant no. 503480), Alzheimer’s Research UK (grant no. 503176), the Wellcome Trust (grant no. 082604/2/07/Z), and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant nos. 01GI0102, 01GI0711, 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01-AG-12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer’s Association grant ADGC-10-196728. This work was partially supported by funding from the Science and Technology Beijing One Hundred Leading Talent Training Project (Z141107001514006), Beijing Municipal Administration of Hospitals’ Mission Plan (SML20150802), and the National Natural Science Foundation of China (81620108011).
Abbreviations
- AD
Alzheimer’s disease
- cQTL
Cell type quantitative trait loci
- eQTLs
Expression quantitative trait loci
- GWAS
Genome-wide association study
- modQTL
Module quantitative trait loci
Authors’ contributions
GYL conceived and initiated the project. YH, JYS, and GYL analyzed the data, and YH, JYS, and GYL wrote the first draft of the manuscript. All authors contributed to the interpretation of the results and critical revision of the manuscript for important intellectual content and approved the final version of the manuscript.
Funding
This work was supported by funding from the National Natural Science Foundation of China (grant nos. 82071212, 61801147 and 81901181), the Mathematical Tianyuan Fund of the National Natural Science Foundation of China (grant no. 12026414), and Beijing Ten Thousand Talents Project (grant no. 2020A15). This work was also partially supported by funding from the Science and Technology Beijing One Hundred Leading Talent Training Project (Z141107001514006), the Beijing Municipal Administration of Hospitals’ Mission Plan (SML20150802), and the Funds of Academic Promotion Programme of Shandong First Medical University & Shandong Academy of Medical Sciences (no. 2019QL016, no. 2019PT007).
Availability of data and materials
All relevant data are within the paper. The authors confirm that all data underlying the findings are either fully available without restriction through consortia websites, or may be made available from consortia upon request. UK Biobank: http://www.ukbiobank.ac.uk/scientists-3/genetic-data/; https://www.ccace.ed.ac.uk/node/335; IGAP consortium data are available at http://web.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php; https://www.niagads.org/datasets/ng00075; eQTLs in UKBEC: http://www.braineac.org/; eQTLs in GTEx: http://www.braineac.org/; eQTLs in Mayo: https://www.niagads.org/datasets/ng00025.
Ethics approval and consent to participate
This article contains human participants collected by several studies performed by previous studies. All participants gave informed consent in all the corresponding original studies, as described in the “Methods” section. Here, our study is based on the publicly available, large-scale datasets, and not the individual-level data. Hence, ethical approval was not sought.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhou X, Nicholson AM, Ren Y, Brooks M, Jiang P, Zuberi A, Phuoc HN, Perkerson RB, Matchett B, Parsons TM, et al. Loss of TMEM106B leads to myelination deficits: implications for frontotemporal dementia treatment strategies. Brain. 2020;143(6):1905–1919. doi: 10.1093/brain/awaa141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicholson AM, Rademakers R. What we know about TMEM106B in neurodegeneration. Acta Neuropathol. 2016;132(5):639–651. doi: 10.1007/s00401-016-1610-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Deerlin VM, Sleiman PM, Martinez-Lage M, Chen-Plotkin A, Wang LS, Graff-Radford NR, Dickson DW, Rademakers R, Boeve BF, Grossman M, et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet. 2010;42(3):234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Z, Farias FHG, Dube U, Del-Aguila JL, Mihindukulasuriya KA, Fernandez MV, Ibanez L, Budde JP, Wang F, Lake AM, et al. The TMEM106B FTLD-protective variant, rs1990621, is also associated with increased neuronal proportion. Acta Neuropathol. 2020;139(1):45–61. doi: 10.1007/s00401-019-02066-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang HS, White CC, Klein HU, Yu L, Gaiteri C, Ma Y, Felsky D, Mostafavi S, Petyuk VA, Sperling RA, et al. Genetics of gene expression in the aging human brain reveal TDP-43 proteinopathy pathophysiology. Neuron. 2020;107(3):496–508. doi: 10.1016/j.neuron.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren Y, van Blitterswijk M, Allen M, Carrasquillo MM, Reddy JS, Wang X, Beach TG, Dickson DW, Ertekin-Taner N, Asmann YW, et al. TMEM106B haplotypes have distinct gene expression patterns in aged brain. Mol Neurodegener. 2018;13(1):35. doi: 10.1186/s13024-018-0268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tropea TF, Mak J, Guo MH, Xie SX, Suh E, Rick J, Siderowf A, Weintraub D, Grossman M, Irwin D, et al. TMEM106B effect on cognition in Parkinson disease and frontotemporal dementia. Ann Neurol. 2019;85(6):801–811. doi: 10.1002/ana.25486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng B, White CC, Klein HU, Sieberts SK, McCabe C, Patrick E, Xu J, Yu L, Gaiteri C, Bennett DA, et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat Neurosci. 2017;20(10):1418–1426. doi: 10.1038/nn.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satoh J, Kino Y, Kawana N, Yamamoto Y, Ishida T, Saito Y, Arima K. TMEM106B expression is reduced in Alzheimer’s disease brains. Alzheimers Res Ther. 2014;6(2):17. doi: 10.1186/alzrt247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, Boland A, Vronskaya M, van der Lee SJ, Amlie-Wolf A, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet. 2019;51(3):414–430. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marioni RE, Harris SE, Zhang Q, McRae AF, Hagenaars SP, Hill WD, Davies G, Ritchie CW, Gale CR, Starr JM, et al. GWAS on family history of Alzheimer’s disease. Transl Psychiatry. 2018;8(1):99. doi: 10.1038/s41398-018-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, De T, Coin L, de Silva R, Cookson MR, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17(10):1418–1428. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Human genomics The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu G, Wang T, Tian R, Hu Y, Han Z, Wang P, Zhou W, Ren P, Zong J, Jin S, et al. Alzheimer’s disease risk variant rs2373115 regulates GAB2 and NARS2 expression in human brain tissues. J Mol Neurosci. 2018;66(1):37–43. doi: 10.1007/s12031-018-1144-9. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Wang L, Jia H, Liao M, Chen X, Xu J, Bao Y, Liu G. Genetic variants regulate NR1H3 expression and contribute to multiple sclerosis risk. J Neurol Sci. 2018;390:162–165. doi: 10.1016/j.jns.2018.04.037. [DOI] [PubMed] [Google Scholar]
- 16.Liu G, Zhang H, Liu B, Wang T, Han Z, Ji X. rs4147929 variant minor allele increases ABCA7 gene expression and ABCA7 shows increased gene expression in Alzheimer’s disease patients compared with controls. Acta Neuropathol. 2020;139(5):937–940. doi: 10.1007/s00401-020-02135-9. [DOI] [PubMed] [Google Scholar]
- 17.Liu G, Zhang H, Liu B, Ji X. Rs2293871 regulates HTRA1 expression and affects cerebral small vessel stroke and Alzheimer’s disease. Brain. 2019;142(11):e61. doi: 10.1093/brain/awz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu G, Hu Y, Han Z, Jin S, Jiang Q. Genetic variant rs17185536 regulates SIM1 gene expression in human brain hypothalamus. Proc Natl Acad Sci U S A. 2019;116(9):3347–3348. doi: 10.1073/pnas.1821550116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu G, Jin S, Hu Y, Jiang Q. Disease status affects the association between rs4813620 and the expression of Alzheimer’s disease susceptibility gene TRIB3. Proc Natl Acad Sci U S A. 2018;115(45):E10519–E10520. doi: 10.1073/pnas.1812975115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou F, Chai HS, Younkin CS, Allen M, Crook J, Pankratz VS, Carrasquillo MM, Rowley CN, Nair AA, Middha S, et al. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet. 2012;8(6):e1002707. doi: 10.1371/journal.pgen.1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu G, Bao X, Jiang Y, Liao M, Jiang Q, Feng R, Zhang L, Ma G, Chen Z, Wang G, et al. Identifying the association between Alzheimer’s disease and Parkinson’s disease using genome-wide association studies and protein-protein interaction network. Mol Neurobiol. 2015;52(3):1629–1636. doi: 10.1007/s12035-014-8946-8. [DOI] [PubMed] [Google Scholar]
- 22.Liu G, Jiang Y, Chen X, Zhang R, Ma G, Feng R, Zhang L, Liao M, Miao Y, Chen Z, et al. Measles contributes to rheumatoid arthritis: evidence from pathway and network analyses of genome-wide association studies. Plos One. 2013;8(10):e75951. doi: 10.1371/journal.pone.0075951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y, Zheng L, Cheng L, Zhang Y, Bai W, Zhou W, Wang T, Han Z, Zong J, Jin S, et al. GAB2 rs2373115 variant contributes to Alzheimer’s disease risk specifically in European population. J Neurol Sci. 2017;375:18–22. doi: 10.1016/j.jns.2017.01.030. [DOI] [PubMed] [Google Scholar]
- 24.Liu G, Sun JY, Xu M, Yang XY, Sun BL. SORL1 variants show different association with early-onset and late-onset Alzheimer’s disease risk. J Alzheimers Dis. 2017;58(4):1121–1128. doi: 10.3233/JAD-170005. [DOI] [PubMed] [Google Scholar]
- 25.Liao M, Wang G, Quan B, Qi X, Yu Z, Feng R, Zhang L, Jiang Y, Zhang Y, Liu G. Analyzing large-scale samples confirms the association between rs16892766 polymorphism and colorectal cancer susceptibility. Sci Rep. 2015;5:7957. doi: 10.1038/srep07957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Y, Cheng L, Zhang Y, Bai W, Zhou W, Wang T, Han Z, Zong J, Jin S, Zhang J, et al. Rs4878104 contributes to Alzheimer’s disease risk and regulates DAPK1 gene expression. Neurol Sci. 2017;38(7):1255–1262. doi: 10.1007/s10072-017-2959-9. [DOI] [PubMed] [Google Scholar]
- 27.Battle A, Brown CD, Engelhardt BE, Montgomery SB. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. Plos Genet. 2014;10(5):e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace C. Eliciting priors and relaxing the single causal variant assumption in colocalisation analyses. PLoS Genet. 2020;16(4):e1008720. doi: 10.1371/journal.pgen.1008720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White CC, Yang HS, Yu L, Chibnik LB, Dawe RJ, Yang J, Klein HU, Felsky D, Ramos-Miguel A, Arfanakis K, et al. Identification of genes associated with dissociation of cognitive performance and neuropathological burden: multistep analysis of genetic, epigenetic, and transcriptional data. Plos Med. 2017;14(4):e1002287. doi: 10.1371/journal.pmed.1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhinn H, Abeliovich A. Differential aging analysis in human cerebral cortex identifies variants in TMEM106B and GRN that regulate aging phenotypes. Cell Syst. 2017;4(4):404–415. doi: 10.1016/j.cels.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Katsumata Y, Nelson PT, Ellingson SR, Fardo DW. Gene-based association study of genes linked to hippocampal sclerosis of aging neuropathology: GRN, TMEM106B, ABCC9, and KCNMB2. Neurobiol Aging. 2017;53:193 e117–193 e125. doi: 10.1016/j.neurobiolaging.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jun G, Ibrahim-Verbaas CA, Vronskaya M, Lambert JC, Chung J, Naj AC, Kunkle BW, Wang LS, Bis JC, Bellenguez C, et al. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry. 2016;21(1):108–117. doi: 10.1038/mp.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu T, Chen Y, Ou R, Wei Q, Cao B, Zhao B, Wu Y, Song W, Chen X, Shang HF. Association analysis of polymorphisms in VMAT2 and TMEM106B genes for Parkinson’s disease, amyotrophic lateral sclerosis and multiple system atrophy. J Neurol Sci. 2017;377:65–71. doi: 10.1016/j.jns.2017.03.028. [DOI] [PubMed] [Google Scholar]
- 35.GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369(6509):1318–1330. [DOI] [PMC free article] [PubMed]
- 36.Yau AC, Houtman M, Padyukov L, Holmdahl R. Reply to Liu et al.: translation of rat congenic data to humans on a conserved MHC-III haplotype associated with rheumatoid arthritis. Proc Natl Acad Sci U S A. 2016;113(42):E6323–E6324. doi: 10.1073/pnas.1614662113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gratuze M, Leyns CEG, Holtzman DM. New insights into the role of TREM2 in Alzheimer’s disease. Mol Neurodegener. 2018;13(1):66. doi: 10.1186/s13024-018-0298-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karch CM, Ezerskiy LA, Bertelsen S, Goate AM. Alzheimer’s disease risk polymorphisms regulate gene expression in the ZCWPW1 and the CELF1 loci. Plos One. 2016;11(2):e0148717. doi: 10.1371/journal.pone.0148717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blauwendraat C, Francescatto M, Gibbs JR, Jansen IE, Simon-Sanchez J, Hernandez DG, Dillman AA, Singleton AB, Cookson MR, Rizzu P, et al. Comprehensive promoter level expression quantitative trait loci analysis of the human frontal lobe. Genome Med. 2016;8(1):65. doi: 10.1186/s13073-016-0320-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoglinger GU, Melhem NM, Dickson DW, Sleiman PM, Wang LS, Klei L, Rademakers R, de Silva R, Litvan I, Riley DE, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. 2011;43(7):699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhalala OG, Nath AP, Inouye M, Sibley CR. Identification of expression quantitative trait loci associated with schizophrenia and affective disorders in normal brain tissue. Plos Genet. 2018;14(8):e1007607. doi: 10.1371/journal.pgen.1007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Brien HE, Hannon E, Hill MJ, Toste CC, Robertson MJ, Morgan JE, McLaughlin G, Lewis CM, Schalkwyk LC, Hall LS, et al. Expression quantitative trait loci in the developing human brain and their enrichment in neuropsychiatric disorders. Genome Biol. 2018;19(1):194. doi: 10.1186/s13059-018-1567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pierce S, Coetzee GA. Parkinson’s disease-associated genetic variation is linked to quantitative expression of inflammatory genes. Plos One. 2017;12(4):e0175882. doi: 10.1371/journal.pone.0175882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akkouh IA, Skrede S, Holmgren A, Ersland KM, Hansson L, Bahrami S, Andreassen OA, Steen VM, Djurovic S, Hughes T. Exploring lithium’s transcriptional mechanisms of action in bipolar disorder: a multi-step study. Neuropsychopharmacology. 2020;45(6):947–955. doi: 10.1038/s41386-019-0556-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chung J, Zhang X, Allen M, Wang X, Ma Y, Beecham G, Montine TJ, Younkin SG, Dickson DW, Golde TE, et al. Genome-wide pleiotropy analysis of neuropathological traits related to Alzheimer’s disease. Alzheimers Res Ther. 2018;10(1):22. doi: 10.1186/s13195-018-0349-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sieberts SK, Perumal TM, Carrasquillo MM, Allen M, Reddy JS, Hoffman GE, Dang KK, Calley J, Ebert PJ, Eddy J, et al. Large eQTL meta-analysis reveals differing patterns between cerebral cortical and cerebellar brain regions. Sci Data. 2020;7(1):340. doi: 10.1038/s41597-020-00642-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu Y, Jin S, Cheng L, Liu G, Jiang Q. Autoimmune disease variants regulate GSDMB gene expression in human immune cells and whole blood. Proc Natl Acad Sci U S A. 2017;114(38):E7860–E7862. doi: 10.1073/pnas.1712127114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soldner F, Stelzer Y, Shivalila CS, Abraham BJ, Latourelle JC, Barrasa MI, Goldmann J, Myers RH, Young RA, Jaenisch R. Parkinson-associated risk variant in distal enhancer of alpha-synuclein modulates target gene expression. Nature. 2016;533(7601):95–99. doi: 10.1038/nature17939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu G, Hu Y, Jin S, Jiang Q. Genetic variant rs763361 regulates multiple sclerosis CD226 gene expression. Proc Natl Acad Sci U S A. 2017;114(6):E906–E907. doi: 10.1073/pnas.1618520114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu G, Hu Y, Jin S, Zhang F, Jiang Q, Hao J. Cis-eQTLs regulate reduced LST1 gene and NCR3 gene expression and contribute to increased autoimmune disease risk. Proc Natl Acad Sci U S A. 2016;113(42):E6321–E6322. doi: 10.1073/pnas.1614369113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peters JE, Lyons PA, Lee JC, Richard AC, Fortune MD, Newcombe PJ, Richardson S, Smith KG. Insight into genotype-phenotype associations through eQTL mapping in multiple cell types in health and immune-mediated disease. Plos Genet. 2016;12(3):e1005908. doi: 10.1371/journal.pgen.1005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu G, Hu Y, Jiang Q. Population difference and disease status affect the association between genetic variants and gene expression. Gastroenterology. 2019;157(3):894–896. doi: 10.1053/j.gastro.2019.01.278. [DOI] [PubMed] [Google Scholar]
- 53.Oliva M, Munoz-Aguirre M, Kim-Hellmuth S, Wucher V, Gewirtz ADH, Cotter DJ, Parsana P, Kasela S, Balliu B, Vinuela A et al. The impact of sex ongene expression across human tissues. Science. 2020;369(6509):eaba3066. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper. The authors confirm that all data underlying the findings are either fully available without restriction through consortia websites, or may be made available from consortia upon request. UK Biobank: http://www.ukbiobank.ac.uk/scientists-3/genetic-data/; https://www.ccace.ed.ac.uk/node/335; IGAP consortium data are available at http://web.pasteur-lille.fr/en/recherche/u744/igap/igap_download.php; https://www.niagads.org/datasets/ng00075; eQTLs in UKBEC: http://www.braineac.org/; eQTLs in GTEx: http://www.braineac.org/; eQTLs in Mayo: https://www.niagads.org/datasets/ng00025.