Abstract
BACKGROUND AND PURPOSE:
Increased cellular density is a hallmark of gliomas, both in the bulk of the tumor and in areas of tumor infiltration into surrounding brain. Altered cellular density causes altered imaging findings, but the degree to which cellular density can be quantitatively estimated from imaging is unknown. The purpose of this study was to discover the best MR imaging and processing techniques to make quantitative and spatially specific estimates of cellular density.
MATERIALS AND METHODS:
We collected stereotactic biopsies in a prospective imaging clinical trial targeting untreated patients with gliomas at our institution undergoing their first resection. The data included preoperative MR imaging with conventional anatomic, diffusion, perfusion, and permeability sequences and quantitative histopathology on biopsy samples. We then used multiple machine learning methodologies to estimate cellular density using local intensity information from the MR images and quantitative cellular density measurements at the biopsy coordinates as the criterion standard.
RESULTS:
The random forest methodology estimated cellular density with R2 = 0.59 between predicted and observed values using 4 input imaging sequences chosen from our full set of imaging data (T2, fractional anisotropy, CBF, and area under the curve from permeability imaging). Limiting input to conventional MR images (T1 pre- and postcontrast, T2, and FLAIR) yielded slightly degraded performance (R2 = 0.52). Outputs were also reported as graphic maps.
CONCLUSIONS:
Cellular density can be estimated with moderate-to-strong correlations using MR imaging inputs. The random forest machine learning model provided the best estimates. These spatially specific estimates of cellular density will likely be useful in guiding both diagnosis and treatment.
Increased cellular density (CD) is a hallmark of cancer and a key feature in histologic glioma analysis.1 Mapping cellular density throughout a tumor would be a valuable tool to probe how tumors infiltrate and analyze the transition between diseased and healthy brain. However, measuring CD requires tissue, which entails additional risks and is expensive to obtain. There is no currently accepted clinical algorithm to translate imaging data into quantitative assessments of CD.
There is great need for a method to estimate CD noninvasively in human patients with gliomas. In this article, we describe the development of such a method using MR imaging data inputs by correlating with multiple biopsy specimens acquired during a prospective human clinical trial. We obtained comprehensive MR imaging, including conventional, diffusion, perfusion, and permeability imaging sequences. We used machine learning approaches to correlate imaging findings with CD measurements from pathology, devised an algorithm to estimate CD from MR imaging inputs, and generated CD maps for the visual display of the predictions. We identified the most informative imaging data subset. This work has multiple applications in the diagnosis and treatment of patients with gliomas: For example, the method can be used to guide biopsy, resection, and surgery and delineate tumor borderzones both pre- and postoperatively.2
MATERIALS AND METHODS
Data were collected as part of a Health Insurance Portability and Accountability Act–compliant institutional review board–approved imaging clinical trial protocol (NCT03458676) for adult, treatment-naïve patients with gliomas at MD Anderson Cancer Center. See Table 1 for demographics. We have previously reported results in the estimation of Ki-67 and tumor grade.3,4 Up to 5 separate biopsies were collected per patient, which were targeted by 2 alternative methods:
The conventional biopsy site was defined as enhancing tumor if present or T2 bright signal closest to an accessible brain surface
The advanced biopsy site was defined, in order, by high relative CBV, high volume transfer constant, and/or restricted diffusion.
Table 1:
Patient demographics
| Demographics | |
|---|---|
| No. (Sex) | 23 Patients (14 women, 9 men) |
| Age (mean) (range) (yr) | 43.9 [SD, 16.9] range, 21–80 |
| WHO grade (I/II/III/IV) | 0/7/9/7 |
| Biopsy samples/patient (mean) | 2.26 [SD, 0.54] |
| No. of biopsy samples in final analysis | 52 real + 52 virtual |
Note:—WHO indicates World Health Organization.
Additional biopsies could be obtained en route to the sites defined above, with preference given to sampling normal brain and the brain-tumor interface when possible. Biopsy locations were sampled under stereotactic guidance using iPlan cranial neuronavigation software (Brainlab), and the final sampling coordinate locations were recorded. Tissue samples were sectioned at 4-μm thickness and stained with H&E. Some slides were also immunohistochemically stained with proliferation and vascularity markers, but these data were not used in the cell density measurement. The H&E-stained tissue was analyzed by a board-certified neuropathologist, and the cell density was measured semiautomatically in nuclei/square millimeter using Aperio ImageScope software (Leica Biosystems).
Image Acquisition
Four clinical categories of imaging were obtained for every patient on a Signa HDxt 3T or Discovery MR750 3T clinical MR imaging scanner (GE Healthcare): anatomic/conventional, diffusion, perfusion, and permeability. Detailed acquisition parameters are given in the Online Appendix and Online Tables 1 and 2. A total of 23 imaging parameters were acquired or generated.
Anatomic images were T1-weighted, T1 postgadolinium, T2-weighted, T2*/susceptibility, FLAIR, and T2*-weighted angiography (SWAN).
DWI and DTI were both acquired for each patient. They were then processed to generate maps of ADC, exponential ADC (eADC), and fractional anisotropy.5,6
Finally, we acquired 2 image series using a dynamic acquisition after the injection of contrast. The dynamic contrast-enhanced (DCE) image series was acquired using a 0.1-mmol/kg contrast bolus, and this same bolus was used for the conventional T1 contrast-enhanced image. The raw data were processed using nordicICE (NordicNeuroLab) using the full extended Tofts model, including leakage correction7-9 and arterial input deconvolution. This process yielded maps of forward and backwards transfer constants (Ktrans and kep respectively), plasma and extravascular extracellular contrast fractions (vp and ve), time to peak enhancement (TTP), area under enhancement curve (AUC), and peak enhancement.7-9 Later in the same study, a second bolus of contrast (again 0.1 mmol/kg) was injected to acquire dynamic susceptibility contrast image data. This time-series was similarly processed to yield maps of relative CBV, CBF, MTT, delay time, and leakage correction K2.10-12
Image Normalization and Measurement
Anatomic images were normalized on the basis of patient-specific average tissue intensities. The T1-weighted, FLAIR, and T2*-weighted angiography images were linearly scaled so that white matter (WM) and CSF had average intensities of 1 and 0, respectively. Similarly, T2-weighted and T2*-weighted images were scaled with WM and CSF having average intensities of 0 and 1. T1 postcontrast was scaled using CSF and gray matter. Parametric maps from DWI, DSC, and DCE are quantitative and were used as provided. Although all images were acquired in the same study, we mutually coregistered them using a 6-df and 12-df affine registration in ANTs13 to correct for patient motion and geometric distortion.14,15 For each biopsy, a 5-mm spheric VOI was placed at the sampling coordinates, and the average intensity in this VOI was recorded for each image parameter.
Cell Density in Control Regions
To record imaging characteristic in normal brain, a neuroradiologist placed an ROI in normal-appearing white matter contralateral to the location of each tissue biopsy, and the average image intensity was recorded. These “virtual biopsies” were assumed to have cell density equal to that of normal white matter. We used values provided by Roetzer et al16 and corrected the CD estimates for histologic section thickness using the Abercrombie method (see On-line Appendix).17,18 We obtained a best estimate for normal white matter cell density of 2912 [SD, 673] nuclei/mm2. Table 2 lists values reproduced from Roetzer et al,16 and the corrected values were used in our study.
Table 2:
Normal brain tissue cell density estimates (1/mm2)
Model-Building Procedure
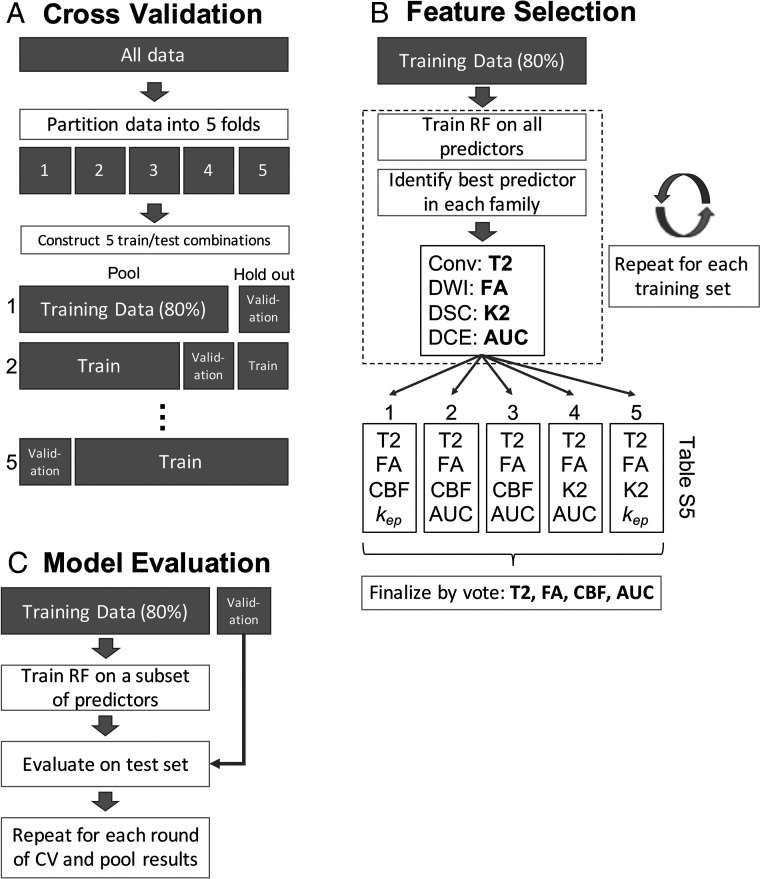
To avoid overfitting, we performed 5-fold cross-validation using disjoint training and testing subsets of the biopsy data within each round (Fig 1A). We partitioned our data into 5 groups, each containing about 20% of our data. To further preserve independence, we placed each sample in the same fold as its paired virtual biopsy.
FIG 1.
Schematic of each of the 3 main stages of the model-building procedure. A, The data are partitioned into 5 homogeneous folds, each comprising 80% of the data for training and 20% for validation, repeated 5 times. Within each round of cross-validation (CV) 1 fold is held out for validation while the other 4 (80%) are pooled to form a training set. B, A random forest (RF) model was fit to the training data, and the best predictor variable from each imaging family was selected. Because there are 5 rounds of CV, this procedure was repeated 5 times and the selections (listed in Online Tables 4 and 5) were combined by voting. C, Using the consensus subset of predictor variables after feature selection, we trained another random forest on each training set and made predictions on the corresponding holdout set. The average correlations between predicted and observed values across all rounds of CV are given in Table 4. Kep indicates reverse transfer constant from DCE imaging.
Variable selection was performed to reduce the number of model inputs from the 23 available parameters to something more parsimonious and to reduce information redundancy among related imaging variables. Within each round of cross-validation, we fit a random forest model to the training subset (4 of 5 groups) using all input variables and computed a variable importance ranking on the basis of the permuted out-of-bag data.19 After the rankings were computed, the most important predictor from each imaging family was used to select 4 final predictors, 1 each from anatomic, diffusion, perfusion, and permeability.3,20 Because this was repeated for each round of cross-validation, 5 variable selections resulted. When the 5 rankings gave different results regarding the most important imaging variable, simple voting was used to determine a consensus. This process is illustrated in Fig 1B and yielded a final set of imaging variables to include in the final model. To provide a fair comparison, we used the final 4-variable set in all model classes tested.
The cross-validation procedure was performed using both the selected variables within each round and the final 4 variables based on consensus among all rounds. The predicted estimates of CD were compared with the actual values known in the 20% validation set and assessed using the Pearson correlation (R2) between predicted and observed CD. This process is explained in Fig 1C. Figure 1 illustrates the full process with the random forest model (our eventual winner), but several models were tested (single decision tree, single-layer neural network, and linear regression). Model descriptions are given in Online Table 3. The model building process was implemented in R, Version 3.4.2 (http://www.r-project.org/).
RESULTS
Patient Data
Thirty-one patients were enrolled in the trial between 2013 and 2016 (mean age, 46 ± [SD, 16] years). Patient demographics are summarized in Table 1. Tissue could not be harvested from 5 patients due to surgical complexity or technical difficulties. For another 3 patients, missing DCE imaging data (1 patient) or unanalyzable tissue samples (2 patients) excluded them from analysis. After exclusions, 23 evaluable patients with 52 image-guided biopsies remained in the final analysis. Of these patients, 7 had clinical grade II gliomas, 9 had grade III, and 7 had grade IV. Among the tissue samples themselves, 9 were collected from contrast-enhancing regions, 2 were collected from just outside the visible T2 hyperintensity, none were collected from necrotic regions, and the remainder were collected from the visible T2-hyperintense tumor. The cell density among all tissue samples increased with increasing sample grades as listed in Table 3.
Table 3:
Number of samples of each WHO gradea
| Sample Grade | No. of Samples | Cell Density (Mean) (SD) |
|---|---|---|
| Normalb | 3 | 1777 [SD, 371] |
| II | 39 | 5790 [SD, 2667] |
| II/III | 3 | 6085 [SD, 2022] |
| IIIc | 2 | 2584, 14,634 |
| IV | 5 | 11,547 [SD, 4252] |
The grade of a sample is not necessarily the same as the patient’s clinical grade. As expected, the cell density of the samples increases with increasing sample grade.
These samples appeared histologically tumor-free but showed Ki-67-positive endothelial cells.
Because only 2 samples were grade III, the values are listed.
Three samples were histologically graded as normal cortex and provide some comparison with the normal cell densities used for virtual biopsies. The recorded CD values were 2169, 1729, and 1432 nuclei/mm2, comparing well with the CD values of Roetzer et al16 for normal cortex at 2011 [SD, 582] nuclei/mm2. See Table 2 for the original estimates and corrected values comparable with our measurements.
Variable Selection and Predictive Modeling
The best predictor from each imaging sequence family for the random forest model was determined by the most votes from each fold of cross-validation and is summarized in Online Table 4. Combining the selections produced a 4-variable set: T2, fractional anisotropy, CBF, and AUC. Of these 4 predictors in the final model, the T2-weighted image intensity had the greatest importance as measured by the random forest with a 27% increase in mean-square error when variables are permuted. Fractional anisotropy and AUC have a smaller importance at 18.0% and 16.1%, respectively. Finally, CBF, despite being the best predictor from the DSC family in 3 of 5 folds of cross-validation, had only an importance of 11.3% increased mean-square error in the final model.
Similar magnitude variable importance was measured for the conventional-only model, ranging from 26.7% for the T2-weighted image down to 9.4% for the FLAIR image, shown in Online Table 5. While the importance measures provide some information about how each predictor relates to CD, they do not measure the combined nonlinear relationships modeled by the random forest.
We compared the reduced variable set selected with modeling done with no selection to show the effects of variable selection. We found comparable modeling performance using the random forest trained on all 23 variables and using the much smaller 4-variable set chosen by random forest importance (R2 = 0.572 versus 0.586). The predicted-versus-observed R2 values are given in Table 4 and Fig 2. Additionally, root-mean-square error values are listed in Online Table 6. Overall, the high average R2 = 0.586 with 4 imaging variables and the random forest model suggest a strong ability to predict cell density with imaging data.
Table 4:
Average R2 for predicted-versus-observed cell density for cross-validationa
| All Variables (23 Inputs) | Variables Selected by RF Importance (4 Inputs) | All Conventional Variables (6 Inputs) | Variables Selected by RF Importance: Conventional Only (4 Inputs) | |
|---|---|---|---|---|
| Random forest | 0.572 | 0.586 | 0.513 | 0.523 |
| Linear | 0.542 | 0.572 | 0.444 | 0.475 |
| Neural network | 0.265 | 0.460 | 0.382 | 0.379 |
| Decision tree | 0.301 | 0.325 | 0.376 | 0.376 |
The columns list variables used to train the predictive model. “All Variables” is simply using all 23 imaging parameters of all 6 conventional sequences, whereas “RF Importance” and “RF Importance, Conventional” use the final 4 variable sets shown in Online Tables 4 and 5. A larger average R2 indicated better performance.
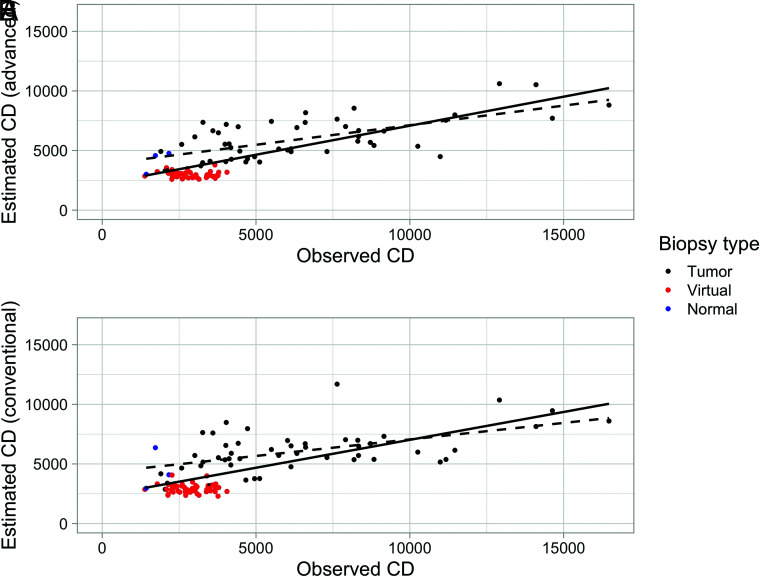
FIG 2.
Predicted and observed cell density (nuclei/square millimeter) for the random forest model using 4 fixed inputs from conventional-plus-advanced imaging. A, The solid line is the best fit for all predictions (R2 = 0.56), and the dashed black line is the best fit for real-sample predictions only (R2 = 0.39), that is, excluding virtual samples (red points). B, The predictions using conventional imaging only. The model performance decreased slightly with R2 = 0.50 for all observations and R2 = 0.30 for real observations only. Blue points are real tissue biopsies that were graded as histologically normal-appearing. The cell density for these points is generally lower than the CD for tumor samples and falls within the range of the virtual biopsies.
Using only conventional imaging variables yielded marginally lesser predictive performance. For conventional imaging only, the highest performance was also with all 6 conventional images as variables in a random forest model, and there was a small improvement made by reducing the number of input variables from 6 to 4 (R2 = 0.513 versus 0.523). The 4-variable set consisted of T2, T1 postcontrast (T1C), FLAIR, and T1 (Online Table 5). These predictions also are strong enough to still be clinically meaningful. No other model tested using only conventional imaging performed better than the random forest.
Maps of cell density generated using the random forest model and the selected conventional and advanced imaging variables are shown in Figs 3 and 4. As expected, the estimated cell density is heightened in regions of increased blood flow, permeability, and contrast enhancement. The estimated CD is also increased in regions of T2 hyperintensity relative to the normal white matter, which may represent infiltrative tumor growth. By looking at a line profile in Fig 3, we observe how the estimates based on the final predictive model change with the input images across the tumor. The importance of AUC and CBF in predicting CD within the visible tumor can be visually appreciated by studying these plotted transects. Additionally, these maps demonstrate the increased CD and heterogeneity of high-grade gliomas (Fig 4), showing the potential value of making local predictions of cell density because such a map may be useful for guiding therapeutic interventions for a range of glioma grades.
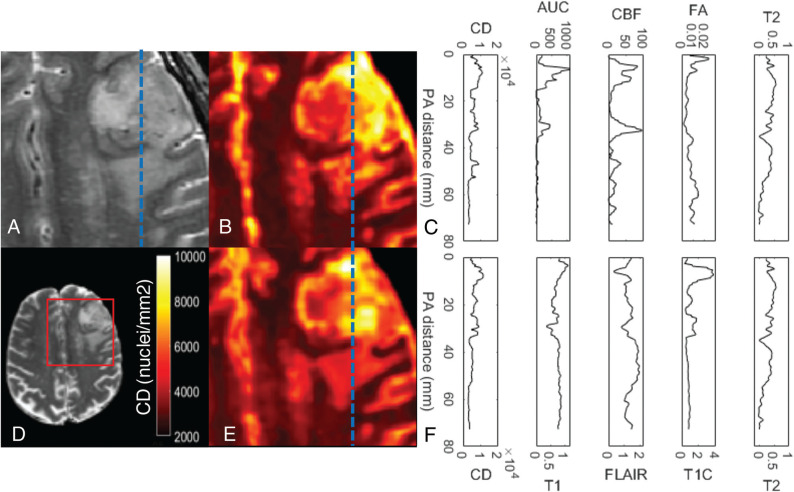
FIG 3.
Estimated map of cell density using machine learning models. A, The T2-weighted image of a glioblastoma. B, The estimated cell density map using the 4 inputs to the final model (T2, fractional anisotropy, CBF, and AUC) selected from all the available imaging data (conventional and advanced) and smoothed by a 1-mm radius filter. The profiles shown in C correspond to the dashed blue line in A, B, and E. We can see in C that the predicted CD is strongly related to the AUC and CBF within the tumor volume. E, The estimated cell density map using conventional imaging only with the analogous profiles shown in F. The model predicts less extreme values than seen with the model using advanced imaging. D, The whole axial section of the T2-weighted image for reference with the cropped area for A, B, and E is outlined in red. PA indicates posteroanterior distance.
FIG 4.
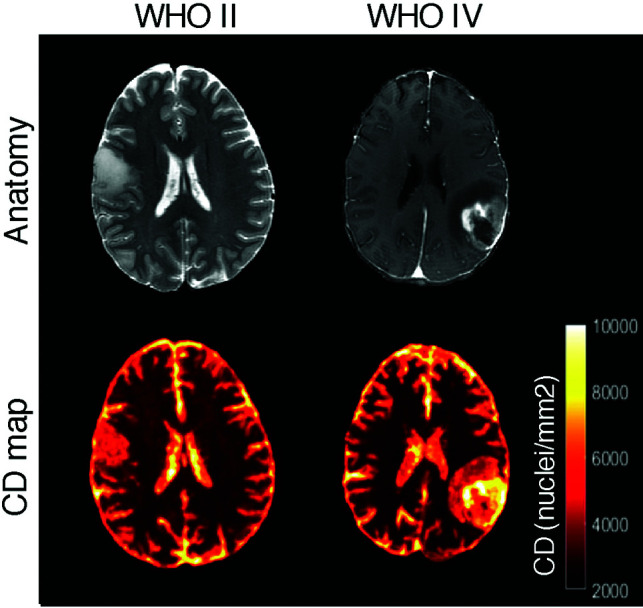
Sample cell density maps for a low-grade glioma (World Health Organization II, left images) and a high-grade glioblastoma (World Health Organization IV, images). The T2-weighted or T1-weighted postcontrast images are shown for reference. As expected, the low-grade tumor shows lower and more homogeneous cell density estimates.
DISCUSSION
We have derived an algorithm for the point-wise local estimation of CD in gliomas using MR imaging data. Using only conventional anatomic imaging sequences, T1, FLAIR, T1C, and T2, CD can be estimated to a R2 of 0.523. When advanced imaging from diffusion tensor imaging, perfusion, and permeability sequences is also used, CD can be estimated to an R2 of 0.586. The final inputs to these predictions are areas under the curve from DCE, cerebral blood flow, fractional anisotropy, and T2. This algorithm allows the construction of CD maps of the brain, translating imaging information into quantitative pathologic estimates that may be useful to guide biopsy and treatment.
Our work has avoided global-level image analysis, including “radiomics,”20-22 due to biologic limitations imposed by histologic heterogeneity exhibited by gliomas.23 Instead, we performed point-wise spatially specific analysis using tissue samples as the criterion standard, which allows local correlation with MR image characteristics and, in turn, point-wise estimates of CD. CD is 1 characteristic that varies considerably between tumor and healthy brain tissue16,24 and also correlates with tumor aggressiveness, making it a useful target for predictive modeling.
In previous work, similar methods have shown effective correlation of imaging with proliferation, grade, and genetic heterogeneity.3,4,25 We used a similar and effective methodology here to develop predictive models for CD as reported in our previous work.3,4 However, estimating cell density represents a very different biologic target with different applications and stands separately from estimating proliferation or tumor grade. Our final model also takes advantage of different derivates of advanced imaging sequences.
Other previous studies have also estimated glioma cell density with spatially specific tissue samples. These studies are characterized by similar imaging protocols, including DTI or DSC, and generally find correlations with CD.24,26 However, the exact image features like CBV versus CBF, ADC versus fractional anisotropy, or mean intensity versus 90th percentile intensity vary. Possible explanations for these discrepancies are differences in sample grades, such as only including samples from glioblastoma,27 or subtle differences in measurement methods for imaging or cell density, cellularity, or related quantities. Overall, our work finds similar results and includes the addition of imaging features from DCE, a greater number of samples, advanced machine learning modeling via random forest, and a smaller number of variables included in the final model.
Our study confirmed the relationship between nuclear density and fractional anisotropy,24 which is a natural expectation, considering that increased nuclei mean increased cell packing. This reduces water movement and affects diffusion parameters. While not used in the final predictive model, we also observed the well-established correlation between ADC and cell density.27 The selection of fractional anisotropy over ADC within the 5 folds of cross-validation does not exclude ADC as a strong predictor of CD; it means simply that FA was a stronger predictor and we chose a priori to include only 1 diffusion parameter in the final model to reduce redundancy. There are many strongly correlated quantities derived from similar source data that are equally useful for predictive modeling and could be substituted with similar performance.
These correlations among sequences may also explain our success in predicting CD (R2 = 0.523) using only conventional imaging sequences: T1, T1 postcontrast, T2, and FLAIR. The DCE parameter AUC is a rough measure of total blood-brain-barrier disruption;28 thus, we would expect similar information to be contained in the T1 postcontrast image data. Correlating imaging with pathology (like cell density) is a rich way to help understand the degree to which these advanced sequences and their similar conventional sequences may probe the same underlying biologic processes. Such sequences have evolved in clinical practice because they are informative of biology and highlight pathology (ie, heightened CD).
The technical demands of collecting spatially specific tissue samples and comprehensive preoperative imaging with diffusion-weighted, DSC, and DCE imaging mean that our study is somewhat limited by the small sample size. However, the 52 samples used in the final analysis and the additional 52 corresponding virtual controls were sufficient to make useful estimates of cell density. Increased sample size would help improve model confidence and would likely help stabilize some of the variable selections that changed between cross-validation folds. An additional limitation is the assumption of cell density for normal white matter. Ethical constraints prohibit sampling healthy brain, so we must impute values for normal tissue on the basis of literature values.16,17 The values we used from Roetzer et al16 do appear to agree with our values in both normal samples and tumor.
CONCLUSIONS
Our methodology allows noninvasive estimation of CD at points in the gliomatous brain with clinically useful accuracy using a combination of MR imaging sequences that are already in wide clinical use. CD estimates can be used to generate a map of estimated cell density for the whole brain. Our work is consistent with previous studies3,24 and with clinical intuition. Maps of CD could be a useful clinical tool to guide biopsies and resections, measure the extent of resection, or plan radiation treatments. Additional trials to prospectively validate our estimating algorithms are justified.
ABBREVIATIONS:
- AUC
area under the curve
- CD
cellular density
- DCE
dynamic contrast-enhanced
- RF
random forest
- T1C
T1 postcontrast
Footnotes
E.D.H. Gates is supported by a training fellowship from the Gulf Coast Consortia and the National Library of Medicine Training Program in Biomedical Informatics & Data Science (T15LM007093) and acknowledges support from the American Legion Auxiliary. Partial support for this study is provided by the Dunn Chair funds (to Dr Bill Murphy), the MD Anderson Cancer Center Internal Research Grant and Clinical Research Support mechanisms for physician-sponsored clinical trials, and the Greenspun neurosurgical research fund. Funding was also provided by the National Cancer Institute (P30 CA016672).
Disclosures: Veera Baladandayuthapani—UNRELATED: Employment: University of Michigan.
References
- 1.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016;131:803–20 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 2.Li YM, Suki D, Hess K, et al. The influence of maximum safe resection of glioblastoma on survival in 1229 patients: can we do better than gross-total resection? J Neurosurg 2016;124:977–88 10.3171/2015.5.JNS142087 [DOI] [PubMed] [Google Scholar]
- 3.Gates EDH, Lin JS, Weinberg JS, et al. Guiding the first biopsy in patients with gliomas using estimated Ki-67 maps derived from MRI: conventional versus advanced imaging. Neuro Oncol 2019;21:527–36 10.1093/neuonc/noz004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gates ED, Lin JS, Weinberg JS, et al. Imaging-based algorithm for the local grading of glioma. AJNR Am J Neuroradiol 2020;41:400–07 10.3174/ajnr.A6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arlinghaus L, Yankeelov TE. Diffusion-Weighted MRI In: Yankeelov TE, Pickens DR, Price RR, eds. Quantitative MRI in Cancer. Taylor & Francis; 2011:81–98 [Google Scholar]
- 6.Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. J Magn Reson B 1996;111:209–19 10.1006/jmrb.1996.0086 [DOI] [PubMed] [Google Scholar]
- 7.Tofts PS, Brix G, Buckley DL, et al. Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging 1999;10:223–32 [DOI] [PubMed] [Google Scholar]
- 8.Sourbron SP, Buckley DL. Classic models for dynamic contrast-enhanced MRI. NMR Biomed 2013;26:1004–27 10.1002/nbm.2940 [DOI] [PubMed] [Google Scholar]
- 9.Sung YS, Park B, Choi Y, et al. Dynamic contrast-enhanced MRI for oncology drug development. J Magn Reson Imaging 2016;44:251–64 10.1002/jmri.25173 [DOI] [PubMed] [Google Scholar]
- 10.Ostergaard L, Sorensen AG, Kwong KK, et al. High-resolution measurement of cerebral blood flow using intravascular tracer bolus passages, Part II: experimental comparison and preliminary results. Magn Reson Med 1996;36:726–36 10.1002/mrm.1910360511 [DOI] [PubMed] [Google Scholar]
- 11.Østergaard L, Weisskoff RM, Chesler DA, et al. High-resolution measurement of cerebral blood flow using intravascular tracer bolus passages, Part I: mathematical approach and statistical analysis. Magn Reson Med 1996;36:715–25 10.1002/mrm.1910360510 [DOI] [PubMed] [Google Scholar]
- 12.Boxerman JL, Schmainda KM, Weiskoff RM. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am J Neuroradiol 2006;27:859–67 [PMC free article] [PubMed] [Google Scholar]
- 13.Avants BB, Tustison NJ, Stauffer M, et al. The Insight ToolKit image registration framework. Front Neuroinf 2014;8:44 10.3389/fninf.2014.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin JS, Fuentes D, Chandler A, et al. Performance assessment for brain MR imaging registration methods. AJNR Am J Neuroradiol 2017;38:973–80 10.3174/ajnr.A5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avants BB, Tustison NJ, Song G, et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 2011;54:2033–44 10.1016/j.neuroimage.2010.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roetzer T, Leskovar K, Peter N, et al. Evaluating cellularity and structural connectivity on whole brain slides using a custom-made digital pathology pipeline. J Neurosci Methods 2019;311:215–21 10.1016/j.jneumeth.2018.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abercrombie M. Estimation of nuclear population from microtome sections. Anat Rec 1946;94:239–47 10.1002/ar.1090940210 [DOI] [PubMed] [Google Scholar]
- 18.Hedreen JC. What was wrong with the Abercrombie and empirical cell counting methods? A review. Anat Rec 1998;250:373–80 [DOI] [PubMed] [Google Scholar]
- 19.Breiman L. Random Forests. Mach Learn 2001;45:5–32 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 20.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parmar C, Grossmann P, Bussink J, et al. Machine learning methods for quantitative radiomic biomarkers. Sci Rep 2015;5:13087 10.1038/srep13087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magn Reson Imaging 2012;30:1234–48 10.1016/j.mri.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Autry A, Phillips JJ, Maleschlijski S, et al. Characterization of metabolic, diffusion, and perfusion properties in GBM: contrast-enhancing versus non-enhancing tumor. Transl Oncol 2017;10:895–903 10.1016/j.tranon.2017.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durst CR, Raghavan P, Shaffrey ME, et al. Multimodal MR imaging model to predict tumor infiltration in patients with gliomas. Neuroradiology 2014;56:107–15 10.1007/s00234-013-1308-9 [DOI] [PubMed] [Google Scholar]
- 25.Gates ED, Yang J, Fukumura K, et al. Spatial distance correlates with genetic distance in diffuse glioma. Front Oncol 2019;9:676 10.3389/fonc.2019.00676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadeghi N, D’Haene N, Decaestecker C, et al. Apparent diffusion coefficient and cerebral blood volume in brain gliomas: relation to tumor cell density and tumor microvessel density based on stereotactic biopsies. AJNR Am J Neuroradiol 2008;29:476–82 10.3174/ajnr.A0851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barajas RF Jr, Phillips JJ, Parvataneni R, et al. Regional variation in histopathologic features of tumor specimens from treatment-naive glioblastoma correlates with anatomic and physiologic MR imaging. Neuro Oncol 2012;14:942–54 10.1093/neuonc/nos128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heye AK, Culling RD, Valdés Hernández M del C, et al. Assessment of blood–brain barrier disruption using dynamic contrast-enhanced MRI: a systematic review. Neuroimage Clin 2014;6:262–74 10.1016/j.nicl.2014.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]