SUMMARY:
Artificial intelligence technology is a rapidly expanding field with many applications in acute stroke imaging, including ischemic and hemorrhage subtypes. Early identification of acute stroke is critical for initiating prompt intervention to reduce morbidity and mortality. Artificial intelligence can help with various aspects of the stroke treatment paradigm, including infarct or hemorrhage detection, segmentation, classification, large vessel occlusion detection, Alberta Stroke Program Early CT Score grading, and prognostication. In particular, emerging artificial intelligence techniques such as convolutional neural networks show promise in performing these imaging-based tasks efficiently and accurately. The purpose of this review is twofold: first, to describe AI methods and available public and commercial platforms in stroke imaging, and second, to summarize the literature of current artificial intelligence–driven applications for acute stroke triage, surveillance, and prediction.
Stroke is the second leading cause of death worldwide with an annual mortality of about 5.5 million.1,2 In the United States, nearly 800,000 people have a stroke annually, and the economic burden of stroke is estimated at $34 billion per year.3 Morbidity is high, with more than half of patients with stroke left chronically disabled.2 Neuroimaging is an important tool for the detection, characterization, and prognostication of acute strokes, including ischemic and hemorrhagic subtypes. Artificial intelligence (AI) technology is a rapidly burgeoning field, providing a promising avenue for fast and efficient imaging analysis.4 AI applications for imaging of acute cerebrovascular disease have been implemented, including tools for triage, quantification, surveillance, and prediction. This review aims to summarize the current landscape of AI-driven applications for acute cerebrovascular disease assessment focusing primarily on deep learning (DL) methods.
Overview of AI
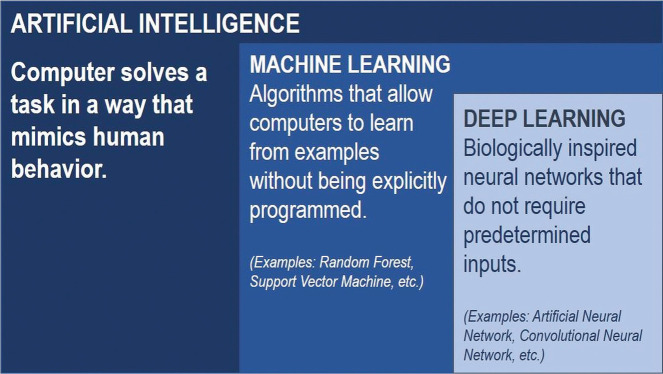
Although AI, machine learning (ML), and DL are used interchangeably, these in fact represent subdisciplines. Specifically, DL is a subset of ML, and ML is a subset of AI (Fig 1). Broadly, AI uses computers to perform tasks that typically require human knowledge. ML, a subset of AI, uses statistical approaches to enable machines to optimize outcome prediction as they are exposed to data and train computers for pattern recognition, a task generally requiring human intelligence.5 ML offers several potential advantages over visual inspection by human experts, including objective and quantitative evaluation, the ability to detect subtle voxel-level patterns, speed, and large-scale implementation. Feature selection, classifier type, and DL are key considerations for the application of ML techniques to imaging.
FIG 1.
AI uses computers to mimic human intelligence. ML is a subset of AI, and DL is a subset of ML.
Feature Selection
Just as a radiologist summarizes an image with a few key descriptors (eg, hemorrhage volume), ML algorithms attempt to do the same with a matrix of voxels. Different feature selection methods can identify a subset of variables to develop a predictive model. Selecting relevant features is important for the explainability, speed, and cost efficiency of a model and to avoid overfitting.6
Classifier Type
After each image is converted into numeric descriptors, a method is chosen to leverage this information to predict 1 of multiple potential classes. For certain cases, even very simple models such as basic logistic or linear regression could be effective.7 However, if nonindependent, nonlinear relationships are expected between the various chosen features, a more complex model is required. Many such ML classifiers exist, and the most popular include random forest (RF), support vector machine (SVM), k-nearest neighbor clustering, and neural networks.8 In general, these techniques are modeled by an underlying finite number of adjustable parameters. As a given set of features is passed through the model, these adjustable parameters act to convert the input descriptors into a predicted output class. Starting with randomly initialized parameters, a series of iterative updates is performed until an accurate mapping between numeric features and correct class is achieved, thus “training” the ML model.9
Deep Learning
DL through neural networks is distinguished by the ability to independently learn abstract, high-order features from data without requiring feature selection. Artificial neural networks (ANNs) are a subtype of DL that mimic biologic neurons and are composed of an input, 1 or more hidden layers, and an output. Generally, in computer vision, convolutional neural networks (CNNs) are most successful and popular for image classification in medical imaging. CNNs represent all recent winning entries within the annual ImageNet Classification challenge, consisting of more than 1 million photographs in 1000 object categories, with a 3.6% classification error rate.10,11 CNNs are distinguished from traditional ML approaches by automatically identifying patterns in complex imaging datasets, thus combining both feature selection and classification into 1 algorithm and removing the need for direct human interaction during the training process. Recent advances in CNNs have achieved human accuracy in identification of everyday objects such as cats and dogs, which had previously been impossible to model using rigid mathematical formulas.12 CNNs have already shown promise in the detection of pulmonary nodules,13 colon cancer,14 and cerebral microbleeds.15
Evaluation of AI Performance
Table 1 details performance metrics and limitations of AI methods.
Table 1:
Machine learning performance metrics and limitations
| Performance metrics | Classification | Sensitivity (recall): TP/(TP + FN) Specificity (true-negative rate): TN/(TN + FP) Accuracy: number of correct predictions/total predictions AUC: plot of true positive rate (sensitivity) against false positive rate (1 – specificity) |
| Segmentation | Dice similarity coefficient: overlap of 2 samples Pearson correlation coefficient: strength of linear relationship between 2 variables |
|
| Limitations and ways to address them | Requires large datasets: multisite collaboration, open-source datasets Interpretability: saliency maps Overfitting: more training data, regularization, and batch normalization |
|
Note:—FP indicates false positive; FN, false-negative; ROC, receiver operating characteristic; TN, true-negative; TP, true positive.
Accuracy
It is imperative that evaluation of ML models assess the accuracy of algorithms. Often, when testing large numbers of potential features, a few numeric descriptors meet the threshold for statistical significance between 2 target classes. However, P values are more often a reflection of the underlying power (sample size) of an experiment and may or may not relate to the clinical significance of the identified difference in features. As a result, it is critical not only to prove that a difference in features exists but also to assess the sensitivity, specificity, and accuracy of the feature(s) to predict a given end point. For classification, receiver operating characteristic curves can evaluate a model’s performance, with the area under the curve (AUC) representing an aggregate measure for performance across all possible classification thresholds of a receiver operating characteristic curve. For segmentation analysis, Dice similarity coefficients and Pearson correlation coefficients are typically used. The Dice score measures the spatial overlap between the manually segmented and neural network-derived segmentations. Dice scores range from 0 (no overlap) to 1 (perfect overlap) and are commonly used to evaluate segmentation performance.16
Limitations
ML and DL approaches have limitations that should be considered. First, the development of algorithms requires data sets that are large, organized, well-classified, and accurate. Interpretability is challenging, especially for DL algorithms. To mitigate this “black box” effect, explainable AI models incorporate tools such as saliency maps. Overfitting is a limitation for ML, when a model mistakenly learns the “noise” instead of the “signal” in a training dataset and thus does poorly with unseen data and is limited in generalizability.17 More training data, regularization, and batch normalization are ways to mitigate overfitting. Differences in image acquisition and data storage among institutions and difficulties in sharing data are obstacles to collecting enough data to obtain useful models. Standardization of imaging methods and open-source data collection can address this issue. Additionally, several proprietary ML software platforms have recently been introduced in the market that incorporate various aspects of the stroke pathway into their algorithms; however, comparison and validation of their performance are still necessary to ensure their robustness in routine use.18 Despite limitations, ML remains a powerful tool for detection and management of stroke and hemorrhage.
AI Platforms in Stroke and Hemorrhage
Open-Source Datasets
Large datasets are required for ML algorithms to perform optimally. However, the availability of high-quality large-scale data remains a challenge given barriers in data sharing across institutions, the complexity of building imaging processing pipelines, and the time and cost of data annotation. To address these challenges, many publicly available imaging datasets are now available for ML in stroke (Table 2).19-24 These datasets are valuable because they are already anonymized, postprocessed, and annotated, and they can be used for testing and comparing algorithms in diagnosing ischemic stroke and hemorrhage. Many of these datasets are initiated as AI challenges such as the RSNA (Radiology Society of North America) Head CT Challenge for Hemorrhage, ASFNR (American Society of Functional Neuroradiology) Head CT Challenge for Ischemic and Hemorrhagic Stroke, and ISLES (Ischemic Stroke Lesion Segmentation) Challenge for Ischemic Stroke, supporting worldwide collaboration and new algorithm development.
Table 2:
Open-source datasets for stroke and hemorrhage
| Dataset | Cerebrovascular Disease | Annotated Data | Number of Scans | Imaging Technique |
|---|---|---|---|---|
| Anatomical Tracings of Lesions After Stroke (ATLAS)19 | Subacute or chronic ischemic strokes | Manually segmented stroke lesions | 304 | T1-weighted MR imaging |
| CQ50020 | Hemorrhage | Hemorrhage, subtype, location, and associated hemorrhage findings | 491 | CT |
| RSNA Brain Hemorrhage CT Dataset21 | Hemorrhage | Hemorrhage, subtypes | 874,035 | CT |
| Ischemic Stroke Lesion Segmentation (ISLES) 2016–201722 | Ischemic stroke | Perfusion and diffusion MR imaging of patients with stroke and clinical outcomes | 35 training and 19 testing (2016); 43 training and 32 testing (2017) | MR imaging, MRP |
| ISLES 201823 | Ischemic stroke | CT and perfusion of patients with stroke | 94 labeled training images and 62 unlabeled testing images | CT, CTP |
Note:—MRP indicates MR perfusion.
Commercially Available Software Platforms
Increasingly, commercially available platforms providing automated information about various components of the acute stroke triage pathway are being integrated into routine clinical practice and clinical trials.25-28 These tools offer fast and efficient analyses that seek to optimize the delivery of stroke care at spoke and hub hospitals and reduce turnaround times in the clinical workflow.29 Table 3 lists some of the most popular commercially available stroke platforms and highlights their capabilities and AI-based algorithms. Figs 2–6 show the various web and mobile interfaces of these software platforms.
Table 3:
Commercially available software platforms and their applicationsa
| Software | Applications | Machine Learning Algorithm | Imaging Technique |
|---|---|---|---|
| Aidoc | ICH: identifies ICH, triage, and notification | DL | CT |
| LVO: identifies LVO, triage, and notification | DL | CTA | |
| CTP: orchestration of third-party perfusion results | Other | CTP | |
| Avicenna.AI | CINA ICH: identifies ICH, triage, and notification | DL | CT |
| CINA LVO: identifies LVO, triage, and notification | DL | CTA | |
| CINA ASPECTS: ASPECTS scoring; provides heat map | DL | CT | |
| Brainomix | e-Blood: identifies and quantifies ICH volume with mask overlay | DL | CT |
| e-ASPECTS: identifies ASPECTS, voxelwise map of early ischemic change, and core infarct volume | Predominantly ML | CT | |
| e-CTA: identifies and notifies LVO, collateral score, and collateral vessel attenuation; voxelwise map of collateral deficit | Combination of DL and traditional ML | CTA | |
| e-ASPECTS HDVS: identifies and measures hyperattenuated vessel | DL | CT | |
| e-Mismatch: identifies mismatch on CTP and MR imaging | Deconvolution | CTP, MR imaging, MRP | |
| RapidAI | Rapid ICH: identifies and classifies ICH | DL | CT |
| Rapid ASPECTS: identifies ASPECTS, measurement, and scoring | RF | CT | |
| Rapid CTA: identifies and notifies LVO and collateral vessel attenuation | Other | CTA | |
| Rapid CTP: identifies mismatch on CTP, collateral maps, and scoring | Other | CTP | |
| Rapid MR: identifies mismatch on MR, collateral maps, and scoring | Other | MR imaging, MRP | |
| Viz.ai | Viz ICH: identifies and triages ICH | DL | CT |
| Viz LVO: identifies and triages LVO | DL | CTA | |
| Viz CTP: automated perfusion color maps and calculations | DL | CTP |
Note:—HDVS indicates hyperattenuated vessel sign.
Some, but not all, of these products have FDA, European, and/or worldwide regulatory clearance at the time of publication.
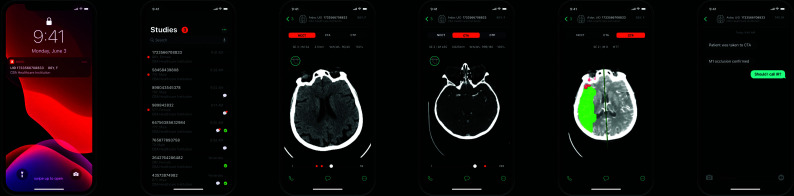
FIG 2.
Aidoc stroke triage mobile interface. From left to right, a notification alert, a study list of cases, NCCT of an acute stroke, CTA of an LVO of the right MCA, CTP mean transit time in the right MCA territory, and a text messaging system. Images courtesy of Aidoc.
FIG 3.
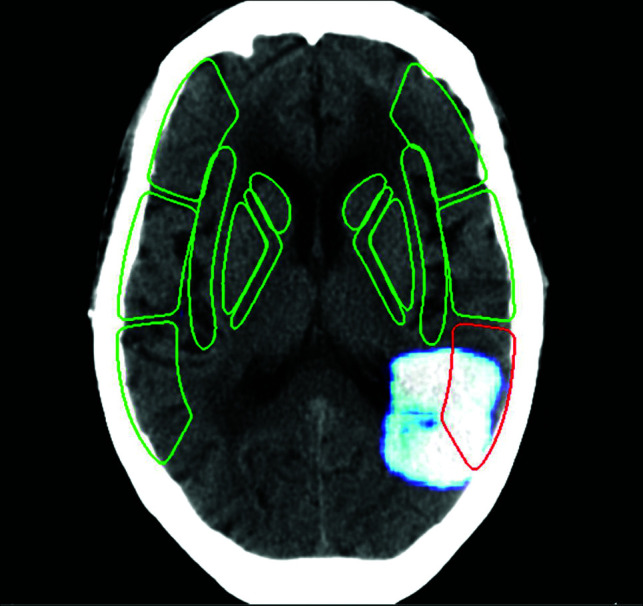
Avicenna.AI DL-based ASPECTS tool demonstrating identification of ASPECTS and a heat map overlay (white). Image courtesy of Avicenna.AI.
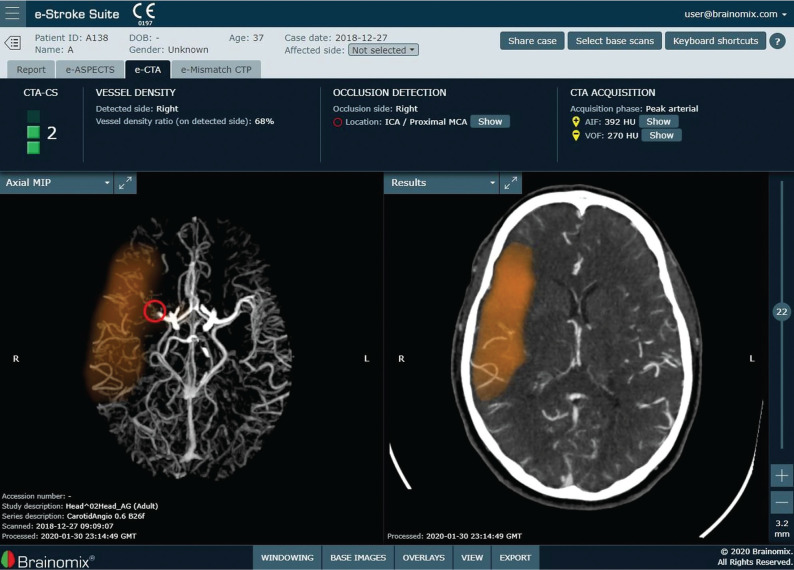
FIG 4.
Brainomix e-CTA tool demonstrating identification and localization of an LVO of the right MCA, collateral score and collateral vessel attenuation, and a heat map of the collateral deficit (orange). Images courtesy of Brainomix.
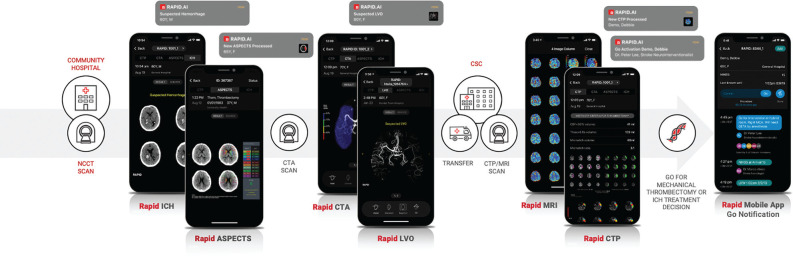
FIG 5.
The RapidAI stroke triage or transfer mobile interface, which integrates the hub and spoke model. From left to right, ICH and ASPECTS scoring and alerts on NCCT, LVO detection on CTA, perfusion mismatch on MR imaging or CTP with FDA mechanical thrombectomy indication, and a mobile communication platform with “GO” notification system for rapid treatment decision making. Images courtesy of RapidAI.
FIG 6.
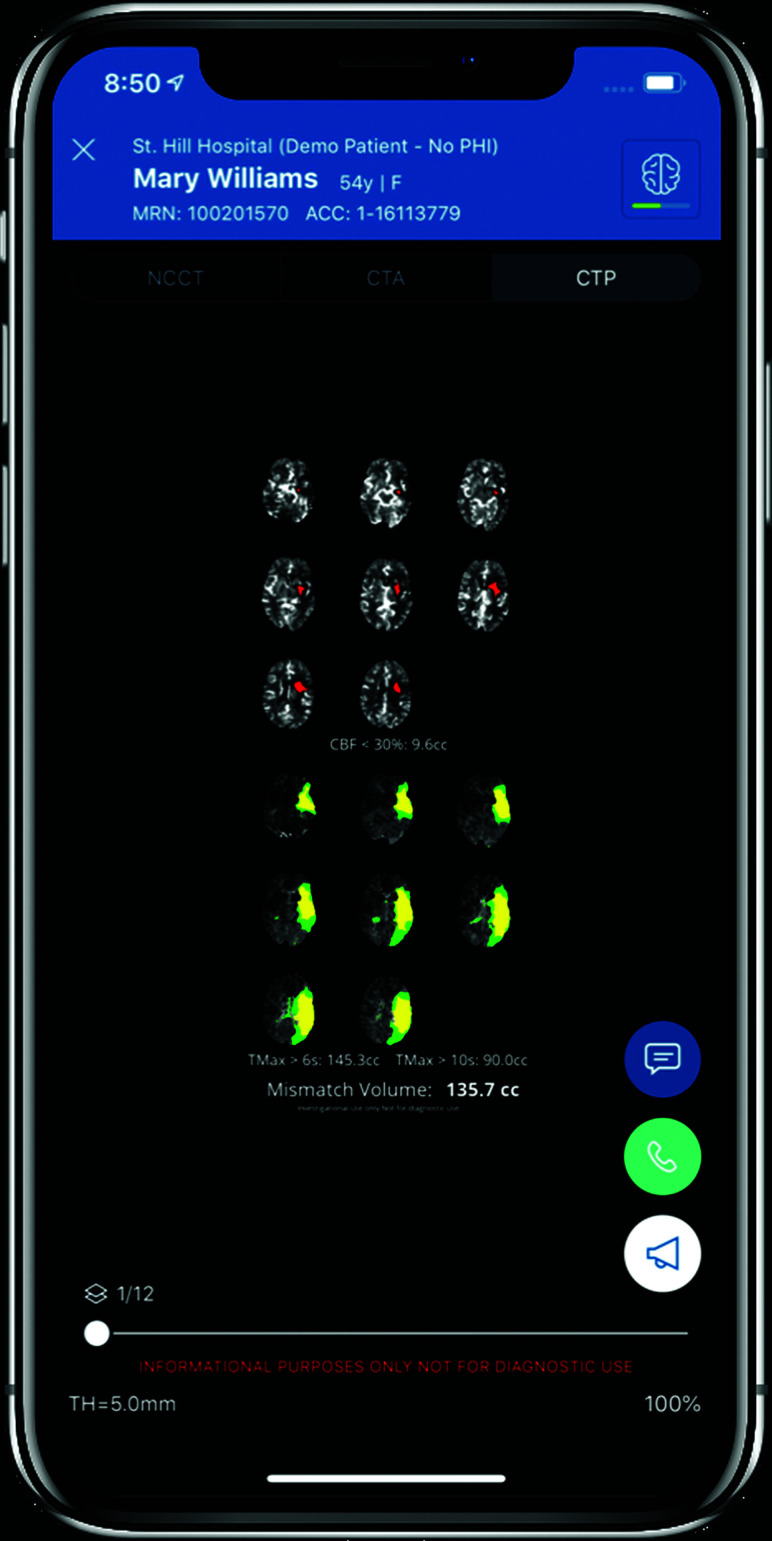
Viz.ai mobile interface showing a left MCA territory infarction with mismatch on CTP. Image courtesy of Viz.ai.
AI Evaluation of Ischemic Stroke
Online Tables 1–4 provide an overview of the AI-based models of evaluating ischemic stroke discussed in this section, including detection and core infarct segmentation, identification of large-vessel occlusion (LVO), Alberta Stroke Program Early CT Score (ASPECTS) grading and additional factors in treatment selection, and prognostication.
Detection Methods
Rapid detection of ischemic infarction is important for triaging patients as potential candidates for thrombolysis because of the narrow window of therapeutic efficacy. Several studies have used ML algorithms for identification of ischemic infarction on CT or MR imaging.
Tang et al30 developed a computer-automated detection (CAD) scheme using a circular adaptive region of interest (CAROI) method on noncontrast head CT to detect subtle changes in attenuation in patients with ischemic stroke. They found that CAD improved detection of stroke for emergency physicians and radiology residents (AUC of 0.879 improved to 0.942 for emergency physicians and AUC of 0.965 improved to 0.990 for radiology residents) but did not improve significantly detection for experienced radiologists who already had high stroke detection rates.30 Another study showed that an ANN was able to distinguish acute stroke from stroke mimics within 4.5 hours of onset (which was verified by clinical and CT and MR imaging data), with a mean sensitivity of 80.0% and specificity of 86.2%.31
Core Infarct Volume Segmentation
Establishing infarct volumes is important to triage patients for appropriate therapy. AI has been able to establish core infarct volumes on DWI through automatic lesion segmentation. For example, 1 study used an ensemble of 2 CNNs to segment DWI lesions of any size and remove false positives.32 This combined CNN approach had a Dice score of 0.61 for small lesions (<37 pixel size) and 0.83 for large lesions and outperformed other CNNs.32 Guerrero et al33 developed a CNN (uResNet) that segmented and differentiated white matter hyperintensities (WMHs) caused by chronic small-vessel disease from cortically or subcortically based strokes. The uResNet CNN mean Dice scores were 0.7 for white matter hyperintensities and 0.4 for strokes.33 The uResNet slightly outperformed the DeepMedic CNN in distinguishing white matter hyperintensities and strokes compared with expert analysis (R2 values 0.951 and 0.791 for white matter hyperintensities and strokes, respectively, using uResNet and 0.942 and 0.688 using DeepMedic).33 One limitation of the study was the reliance on FLAIR and T1 images that do not fully account for timing of stroke occurrence, and the value of uResNet in detection of acute strokes needs evaluation. The first study to use a DL approach on CTA source images to detect acute middle cerebral artery ischemic stroke, a 3D CNN (DeepMedic), performed with a sensitivity of 0.93, specificity of 0.82, AUC of 0.93, and Dice score of 0.61.34 Specificity was maximized when the contralateral cerebral hemisphere on CTA was included, and a marginal reduction in false positives was seen when NCCT was included in the algorithm.34 Limitations of this CNN were its tendency to overestimate the volume of small infarcts and underestimate large infarcts compared with manual segmentation by expert radiologists and difficulty in distinguishing old versus new strokes.34
The largest cohort using CTP for core infarct determination based on an ANN was able to accurately identify core infarct volume (AUC = 0.85; sensitivity = 0.9; specificity = 0.62) and was not significantly different from a model incorporating clinical data (AUC = 0.87; sensitivity = 0.91; specificity = 0.65).35 Although the study minimized the time between CTP and MR imaging DWI reference standard acquisition, any time delay between the CTP and MR imaging may have limited accurate core infarct determination because of core expansion or reversal. A model incorporating a U-net architecture CNN and RF classifier segmented acute ischemic stroke on NCCT with high concordance with manually segmented DWI core volumes (r = 0.76, P < .001) and manually segmented DWI ASPECTS scores (r = −0.65, P < .001). Furthermore, the agreement approached significance when dichotomizing infarcts using a volume threshold of 70 mL (McNemar test, P = .11). Discrepancies in volumes were attributed to nondetectable early ischemic findings, partial volume averaging, and stroke mimics on CT.36
Large Vessel Occlusion
Diagnosing LVO is essential for identifying candidates who could potentially benefit from mechanical thrombectomy. On NCCT, an SVM algorithm detected the MCA dot sign in patients with acute stroke with high sensitivity (97.5%).37 A neural network that incorporated various demographic, imaging, and clinical variables in predicting LVO outperformed or equaled most other prehospital prediction scales with an accuracy of 0.820.38 A CNN-based commercial software, Viz-AI-Algorithm v3.04, detected proximal LVO with an accuracy of 86%, sensitivity of 90.1%, specificity of 82.5, AUC of 86.3% (95% CI, 0.83–0.90; P ≤ .001), and intraclass correlation coefficient (ICC) of 84.1% (95% CI, 0.81–0.86; P ≤ .001), and Viz-AI-Algorithm v4.1.2 was able to detect LVO with high sensitivity and specificity (82% and 94%, respectively).39,40 No study has yet shown whether AI methods can accurately identify other potentially treatable lesions such as M2, intracranial ICA, and posterior circulation occlusions.
ASPECTS Grading
ASPECTS is a widely used clinical grading system for assessing extent of early ischemic stroke on NCCT and has been used in randomized clinical trials to select thrombectomy candidates.26,41,42 However, grading can be challenging, and interobserver agreement is variable. One commercial software platform with automated ASPECTS scoring (e-ASPECTS, Brainomix) performed as well as neuroradiologists when scoring ASPECTS on NCCT in patients with acute stroke (P < .003).43 However, e-ASPECTS did not perform as well as neuroradiologists when scoring ASPECTS in patients with acute stroke with baseline non–normal-appearing CT (eg, leukoencephalopathy, old infarcts, or other parenchymal defects), demonstrating a correlation coefficient of 0.59 versus 0.71–0.80 for experts.44 One study found that an automated ASPECTS detection algorithm on NCCT using texture feature extraction to train a RF classifier generated ASPECTS values that had high agreement with expert-generated DWI ASPECTS scores (ICC = 0.76 and κ = 0.6 when used for all 10 ASPECTS regions).45
Another commercial software platform with automated ASPECTS scoring (Rapid ASPECTS, version 4.9; iSchemaView) showed higher agreement with a consensus ASPECTS grade that takes into account follow-up DWI (κ = 0.9) compared with neuroradiologists’ moderate agreement (κ = 0.56–0.57), and the software performed well in the immediate time interval 1 hour after stroke onset (κ = 0.78) and even better 4 hours after stroke onset (κ = 0.92).46 This platform had better agreement of ASPECTS grading with DWI infarct volume in patients with large hemispheric infarct compared with experienced readers (median DWI ASPECTS, 3 [IQR, 2–4]; Rapid ASPECTS, 3 [1–6]; and CT ASPECTS for the clinicians, 5 [4–7].47
Additional Factors in Treatment Selection
Various factors, including collaterals, penumbra, and stroke onset time, are important for evaluating potentially salvageable tissue and determining treatment eligibility. An automated commercial software program (e-CTA; Brainomix) combining deep and traditional ML techniques for CTA collateral status determination improved consensus scoring among expert neuroradiologists compared with visual inspection alone, with an ICC of 0.58 (0.46–0.67) improving to 0.77 (0.66–0.85; P = .003).48 Penumbra prediction on a noncontrast MR imaging pseudocontinuous arterial spin labeling technique using a DL model performed well (AUC = 0.958).49 This algorithm outperformed traditional ML algorithms and was able to predict endovascular treatment eligibility based on DEFUSE 3 (Endovascular Therapy Following Imaging Evaluation for Ischemic Stroke) trial criteria. Another study evaluating various traditional ML models in predicting stroke onset time demonstrated that incorporation of DL features to the models improved AUC compared with the ground truth (ie, a DWI–FLAIR mismatch), with the optimal AUC of 0.765 incorporating logistic regression and DL features of MR imaging and MR perfusion (MRP) images.50 Lee et al51 used DWI–FLAIR mismatch to predict stroke onset time <4.5 hours and found that traditional ML models were more sensitive than stroke neurologists (sensitivity = 48.5% for stroke neurologists vs 75.8% for logistic regression; P = .020; 72.7% for SVM, P = .033; 75.8% for RF, P = .013).
Prognostication
Various ML algorithms have been used to predict imaging and clinical outcomes after ischemic stroke. An early classical ML study found that a generalized linear model combining DWI and perfusion-weighted imaging MR images was better than DWI (P = .02) or PWI (P = .04) alone at predicting voxelwise tissue outcomes.52 A CNN-based patch sampling of the Tmax feature on MRP outperformed a single voxel-based regression model in predicting final infarct volume, with a mean accuracy of 85.3 ± 9.1% compared with 78.3 ± 5.5%, respectively.53 Another CNN performed better than other ML methods in predicting final infarct volume by incorporating MR imaging DWI, MRP, and FLAIR data, with an AUC of 0.88 ± 0.12.54 This CNN could predict tissue fate based on whether intravenous tissue plasminogen activator was administered, showing significantly different final infarct volumes (P = .048).54 A CNN based on MRP source images was able to predict final infarct volume with an AUC of 0.871 ± 0.024.55 A multicenter study showed that an attention-gated U-Net DL algorithm with DWI and MRP as inputs could predict final infarct volume regardless of reperfusion status, with a median AUC of 0.92 (IQR, 0.87–0.96) and significant overlap with the ground truth of a FLAIR sequence obtained 3–7 days after baseline presentation (Dice score, 0.53; IQR, 0.31–0.68).56
The e-ASPECTS software was able to predict poor clinical outcomes after thrombectomy (Spearman correlation = −0.15; P = .027) and was an independent predictor of poor outcome in a multivariate analysis (OR, 0.79; 95% CI, 0.63–0.99) while also demonstrating high consensus with 3 expert ASPECTS readers (ICC = 0.72, 0.74, and 0.76).57 Traditional ML techniques combining clinical data and core-penumbra mismatch ratio derived from MR imaging and MRP to determine postthrombolysis clinical outcomes performed with an AUC of 0.863 (95% CI, 0.774–0.951) for short-term (day 7) outcomes and 0.778 (95% CI, 0.668–0.888) for long-term (day 90) outcomes.58 Decision tree–based algorithms including extreme gradient boosting and gradient boosting machine were able to predict 90-day modified Rankin scale (mRS) > 2 using imaging and clinical data with AUC of 0.746 (extreme gradient boosting) and 0.748 (gradient boosting machine), and performance improved when incorporating NIHSS at 24 hours and recanalization outcomes.59 ML techniques, including regularized logistic regression, linear SVM, and RF, outperformed existing pretreatment scoring methods in predicting good clinical outcomes (mRS ≤2 at 90 days) of patients with LVO who will undergo thrombectomy, with AUC 0.85–0.86 for ML models compared with 0.71–0.77 for pretreatment scores.60 A combination CNN and ANN approach incorporating clinical and NCCT data predicted functional thrombolysis outcomes with accuracy 0.71 for 24-hour NIHSS improvement of ≥4 and accuracy 0.74 for 90-day mRS of 0–1.61 Finally, traditional ML techniques and neural networks were used to predict hemorrhagic transformation of acute ischemic stroke before treatment from MRP source images and DWI, with the highest AUC of 0.837 ± 2.6% using a kernel spectral regression ML technique.62 One limitation of this study was the variable recanalization of the participants, which may have confounded results.
AI Evaluation of Hemorrhage
This section focuses primarily on DL methods that have been used for intracranial hemorrhage (ICH) detection and classification, quantification, and prognostication (Online Table 5).
Detection and Classification
A study using two 2D convolutional neural networks, GoogLeNet and AlexNet, to detect basal ganglia hemorrhages on NCCT found that GoogLeNet with augmented data in a pretrained network was the most accurate (AUC = 1.0; sensitivity and specificity = 100%) compared with the highest performing augmented, untrained AlexNet (AUC = 0.95; sensitivity = 100%; and specificity = 80%).63 False positive results from basal ganglia calcification were seen in some of the methods, and sensitivity of detection of small basal ganglia hemorrhages remains to be investigated.
One of the largest cohorts for detection and classification of ICH examined more than 30,0000 NCCTs from different hospitals in India using DL algorithms.64 The algorithm performed well on 2 different validation datasets, Qure25k and CQ500, achieving AUCs of 0.92 (95% CI, 0.91–0.93) and 0.94 (CI, 0.92–0.97), respectively, for detecting ICH. The algorithm was also able to classify subtypes of hemorrhage (parenchymal, intraventricular, subdural, extradural/epidural, and subarachnoid) with AUCs ranging from 0.90 to 0.96 for the Qure25K dataset and 0.93 to 0.97 for the CQ500 dataset. An additional feature of the algorithm was its ability to recognize associated pertinent CT findings, such as calvarial fracture, midline shift, and mass effect.
Another study using a fully 3D CNN with a large patient cohort was able to detect ICH and reprioritize studies as “stat” (defined as a positive ICH study) versus “routine.”65 The AUC was 0.846 (95% CI, 0.837–0.856), specificity was 0.80 (0.790–0.809), and sensitivity was 0.73 (0.713–0.748). The algorithm was integrated into the radiologist’s workflow, and time to detection was reduced from 512 to 19 minutes.
An explainable pretrained 2D convolutional neural networks system performed at a similar level to expert neuroradiologists on a relatively small cohort of cases when detecting acute ICH and classifying the 5 ICH subtypes on NCCT.66 The algorithm incorporated techniques such as attention maps and prediction based modules to help mitigate the “black box” of the DL system. The system displayed a robust performance when detecting ICH on a retrospective dataset of 200 cases (AUC = 0.99; sensitivity = 98%; and specificity = 95%) and prospective dataset of 196 cases (AUC = 0.96; sensitivity = 92%; and specificity = 95%). Furthermore, the overall localization accuracy of the attention maps was 78.1% compared with bleeding points annotated by expert neuroradiologists.
Quantification
A custom DL-trained hybrid 3D–2D CNN was able to detect and quantify ICHs on NCCT in a retrospective training cohort and a prospective testing cohort from the emergency department.67 Accuracy, AUC, sensitivity, specificity, positive predictive value, and negative predictive value for ICH detection for the training cohort were 0.975, 0.983, 0.971, 0.975, 0.793, and 0.997, respectively, and for the prospective cohort were 0.970, 0.981, 0.951, 0.973, 0.829, and 0.993. For ICH quantification, Dice scores were 0.931, 0.863, and 0.772, and Pearson correlation coefficients were 0.999, 0.987, and 0.953 for intraparenchymal hemorrhage, epidural or subdural hemorrhage, and SAH, respectively, compared with semiautomated segmentation by a radiologist. This study used real-life prospective testing of the algorithm and quantified hemorrhage volume during segmentation. The study also addresses the black box critique with the use of a custom mask ROI-based CNN architecture.
A patch-based fully DL CNN simultaneously classified and quantified hemorrhages at a level equal to or above that of expert radiologists (AUC = 0.991 ± 0.006).68 The algorithm was able to identify some small hemorrhages that were missed by radiologists and performed well on a relatively small dataset. The strongly supervised approach took into account the heterogeneous morphology of hemorrhages and showed perfect sensitivity (1.00) while maintaining high specificity (0.87).
Prognostication
Identifying patients at risk for ICH expansion is important for prognostication. One study showed good performance when applying a SVM that incorporated various clinical and imaging variables to predict hematoma expansion on NCCT (AUC = 0.89; mean sensitivity = 81.3%; and mean specificity = 84.8%).69 Rapid and accurate identification of ICH by AI methods could aid with triaging of positive studies.
Conclusions
Prompt detection and treatment of acute cerebrovascular disease is critical to reduce morbidity and mortality. The current application of AI in this field has allowed for vast opportunities to improve treatment selection and clinical outcomes by aiding in all parts of the diagnostic and treatment pathway, including detection, triage, and outcome prediction. Future studies validating AI techniques are needed to allow for more widespread use in various practice environments.
Acknowledgments
The authors thank Aidoc, Avicenna, Brainomix, RapidAI, and Viz.ai for providing information regarding commercially available products and sample images of their applications for publication.
ABBREVIATIONS:
- AI
artificial intelligence
- ANN
artificial neural network
- AUC
area under the curve
- CNN
convolutional neural network
- DL
deep learning
- ICC
intraclass correlation coefficient
- ICH
intracranial hemorrhage
- LVO
large vessel occlusion
- ML
machine learning
- MRP
MR perfusion
- RF
random forest
- SVM
support vector machine
Footnotes
Disclosures: Daniel Chow—RELATED: Grant: Avicenna.ai*; UNRELATED: Consultancy: Canon Medical; Expert Testimony: Cullins & Grandy; Grants/Grants Pending: Canon Medical, Novocure; Stock/Stock Options: Avicenna.ai. Christopher Filippi—UNRELATED: Consultancy: Guerbet, Syntactx, Comments: Advisor on AI (Guerbet) and interpret brain MR for clinical trials (Syntactx); Grants/Grants Pending: FASNR grant and National MS Society grant; Stock/Stock Options: Minority stakeholder in start-up Avicenna. Wengui Yu—UNRELATED: Employment: University of California Irvine. Peter Chang—UNRELATED: Consultancy: Canon Medical, Comments: Consulting, travel expenses, honorarium for invited keynote speaker (RSNA); Stock/Stock Options: Avicenna.ai, Comments: Co-founder, board member. *Money paid to institution.
References
- 1.Ovbiagele B, Nguyen-Huynh MN. Stroke epidemiology: advancing our understanding of disease mechanism and therapy. Neurotherapeutics 2011;8:319–29 10.1007/s13311-011-0053-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donkor ES. Stroke in the 21(st) century: a snapshot of the burden, epidemiology, and quality of life. Stroke Res Treat 2018;2018:3238165 10.1155/2018/3238165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boehme AK, Esenwa C, Elkind MSV. Stroke risk factors, genetics, and prevention. Circ Res 2017;120:472–95 10.1161/CIRCRESAHA.116.308398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee EJ, Kim YH, Kim N, et al. . Deep into the brain: artificial intelligence in stroke imaging. J Stroke 2017;19:277–85 10.5853/jos.2017.02054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodfellow I, Bengio Y, Courville A. Deep Learning. The MIT Press; 2016 [Google Scholar]
- 6.Saeys Y, Inza I, Larrañaga P. A review of feature selection techniques in bioinformatics. Bioinformatics 2007;23:2507–17 10.1093/bioinformatics/btm344 [DOI] [PubMed] [Google Scholar]
- 7.Dreiseitl S, Ohno-Machado L. Logistic regression and artificial neural network classification models: a methodology review. J Biomed Inform 2002;35:352–59 10.1016/S1532-0464(03)00034-0 [DOI] [PubMed] [Google Scholar]
- 8.Wang S, Summers RM. Machine learning and radiology. Med Image Anal 2012;16:933–51 10.1016/j.media.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jordan MI, Mitchell TM. Machine learning: trends, perspectives, and prospects. Science 2015;349:255–60 10.1126/science.aaa8415 [DOI] [PubMed] [Google Scholar]
- 10.He K, Zhang X, Ren S, et al. . Deep residual learning for image recognition. arXiv:1512:03385;10 Dec 2015;1–12 [Google Scholar]
- 11.Krizhevsky A, Sutskever I, Hinton G. Imagenet classification with deep convolutional neural networks. Proc Advances in Neural Information Processing Systems 2012;1090–98 [Google Scholar]
- 12.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Neural information processing systems. Stateline 2012;1097–1105 [Google Scholar]
- 13.Setio AA, Ciompi F, Litjens G, et al. . Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans Med Imaging 2016;35:1160–69 10.1109/TMI.2016.2536809 [DOI] [PubMed] [Google Scholar]
- 14.Roth HR, Lu L, Liu J, et al. . Improving computer-aided detection using convolutional neural networks and random view aggregation. IEEE Trans Med Imaging 2016;35:1170–81 10.1109/TMI.2015.2482920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi D, Hao C, Lequan Y, et al. . Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Trans Med Imaging 2016;35:1182–95 10.1109/TMI.2016.2528129 [DOI] [PubMed] [Google Scholar]
- 16.Zou KH, Warfield SK, Bharatha A, et al. . Statistical validation of image segmentation quality based on a spatial overlap index1: scientific reports. Acad Radiol 2004;11:178–89 10.1016/S1076-6332(03)00671-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamashita R, Nishio M, Do RKG, et al. . Convolutional neural networks: an overview and application in radiology. Insights Imaging 2018;9:611–29 10.1007/s13244-018-0639-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray NM, Unberath M, Hager GD, et al. . Artificial intelligence to diagnose ischemic stroke and identify large vessel occlusions: a systematic review. J Neurointerv Surg 2020;12:156–64 10.1136/neurintsurg-2019-015135 [DOI] [PubMed] [Google Scholar]
- 19.Liew S-L, Anglin JM, Banks NW, et al. . A large, open source dataset of stroke anatomical brain images and manual lesion segmentations. Sci Data 2018;5:180011 10.1038/sdata.2018.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chilamkurthy S, Ghosh R, Tanamala S, et al. . Development and validation of deep learning algorithms for detection of critical findings in head CT scans. arXiv :1803:05854;30 March 2018; 1–18 [DOI] [PubMed] [Google Scholar]
- 21.Flanders AE, Prevedello LM, Shih G, et al. . Construction of a machine learning dataset through collaboration: the RSNA 2019 brain CT hemorrhage challenge. Radiol Art Int 2020;2:e190211 10.1148/ryai.2020190211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winzeck S, Hakim A, McKinley R, et al. . ISLES 2016 and 2017—benchmarking ischemic stroke lesion outcome prediction based on multispectral MRI. Front Neurol 2018;9:679 10.3389/fneur.2018.00679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clèrigues A, Valverde S, Bernal J, et al. . Acute ischemic stroke lesion core segmentation in CT perfusion images using fully convolutional neural networks. Comput Biol Med 2019;115:103487 10.1016/j.compbiomed.2019.103487 [DOI] [PubMed] [Google Scholar]
- 24.American Society of Functional Neuroradiology. ASFNR Ai challenge. https://aichallenge.asfnr.org. Accessed July 5, 2020
- 25.Nogueira RG, Jadhav AP, Haussen DC, et al. . Thrombectomy 6 to 24 hours after stroke with a mismatch between deficit and infarct. N Engl J Med 2018;378:11–21 10.1056/NEJMoa1706442 [DOI] [PubMed] [Google Scholar]
- 26.Saver JL, Goyal M, Bonafe A, et al. . Stent-retriever thrombectomy after intravenous t-PA vs. t-PA alone in stroke. N Engl J Med 2015;372:2285–95 10.1056/NEJMoa1415061 [DOI] [PubMed] [Google Scholar]
- 27.Albers GW, Marks MP, Kemp S, et al. . Thrombectomy for stroke at 6 to 16 hours with selection by perfusion imaging. N Engl J Med 2018;378:708–18 10.1056/NEJMoa1713973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell BCV, Mitchell PJ, Kleinig TJ, et al. . Endovascular therapy for ischemic stroke with perfusion-imaging selection. N Engl J Med 2015;372:1009–18 10.1056/NEJMoa1414792 [DOI] [PubMed] [Google Scholar]
- 29.Wismüller A, Stockmaster L. A prospective randomized clinical trial for measuring radiology study reporting time on artificial Intelligence-based detection of intracranial hemorrhage in emergent care head CT. arXiv :2002:12515; 28 Feb 2020; 1–7 [Google Scholar]
- 30.Tang FH, Ng DK, Chow DH. An image feature approach for computer-aided detection of ischemic stroke. Comput Biol Med 2011;41:529–36 10.1016/j.compbiomed.2011.05.001 [DOI] [PubMed] [Google Scholar]
- 31.Abedi V, Goyal N, Tsivgoulis G, et al. . Novel screening tool for stroke using artificial neural network. Stroke 2017;48:1678–81 10.1161/STROKEAHA.117.017033 [DOI] [PubMed] [Google Scholar]
- 32.Chen L, Bentley P, Rueckert D. Fully automatic acute ischemic lesion segmentation in DWI using convolutional neural networks. NeuroImage Clin 2017;15:633–43 10.1016/j.nicl.2017.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guerrero R, Qin C, Oktay O, et al. . White matter hyperintensity and stroke lesion segmentation and differentiation using convolutional neural networks. NeuroImage Clin 2018;17:918–34 10.1016/j.nicl.2017.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oman O, Makela T, Salli E, et al. . 3D convolutional neural networks applied to CT angiography in the detection of acute ischemic stroke. Eur Radiol Exp 2019;3:8 10.1186/s41747-019-0085-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kasasbeh AS, Christensen S, Parsons MW, et al. . Artificial neural network computer tomography perfusion prediction of ischemic core. Stroke 2019;50:1578–81 10.1161/STROKEAHA.118.022649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu W, Kuang H, Teleg E, et al. . Machine learning for detecting early infarction in acute stroke with non–contrast-enhanced CT. Radiology 2020;294:638–44 10.1148/radiol.2020191193 [DOI] [PubMed] [Google Scholar]
- 37.Takahashi N, Lee Y, Tsai D-Y, et al. . An automated detection method for the MCA dot sign of acute stroke in unenhanced CT. Radiol Phys Technol 2014;7:79–88 10.1007/s12194-013-0234-1 [DOI] [PubMed] [Google Scholar]
- 38.Chen Z, Zhang R, Xu F, et al. . Novel prehospital prediction model of large vessel occlusion using artificial neural network. Front Aging Neurosci 2018;10:181 10.3389/fnagi.2018.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chatterjee A, Somayaji NR, Kabakis IM. Abstract WMP16: artificial intelligence detection of cerebrovascular large vessel occlusion—nine month, 650 patient evaluation of the diagnostic accuracy and performance of the Viz.ai LVO algorithm. Stroke 2019;50:AWMP16 10.1161/str.50.suppl_1.WMP16 [DOI] [Google Scholar]
- 40.Barreira C, Bouslama M, Lim J, et al. . E-108 Aladin study: automated large artery occlusion detection in stroke imaging study—a multicenter analysis. J Neurointerv Surg 2018;10:A101–02 10.1136/neurointsurg-2018-SNIS.184 [DOI] [Google Scholar]
- 41.Goyal M, Demchuk AM, Menon BK, et al. . Randomized assessment of rapid endovascular treatment of ischemic stroke. N Engl J Med 2015;372:1019–30 10.1056/NEJMoa1414905 [DOI] [PubMed] [Google Scholar]
- 42.Jovin TG, Chamorro A, Cobo E, et al. . Thrombectomy within 8 hours after symptom onset in ischemic stroke. N Engl J Med 2015;372:2296–2306 10.1056/NEJMoa1503780 [DOI] [PubMed] [Google Scholar]
- 43.Nagel S, Sinha D, Day D, et al. . e-ASPECTS software is non-inferior to neuroradiologists in applying the ASPECT score to computed tomography scans of acute ischemic stroke patients. Int J Stroke 2017;12:615–22 10.1177/1747493016681020 [DOI] [PubMed] [Google Scholar]
- 44.Guberina N, Dietrich U, Radbruch A, et al. . Detection of early infarction signs with machine learning-based diagnosis by means of the Alberta Stroke Program Early CT score (ASPECTS) in the clinical routine. Neuroradiology 2018;60:889–901 10.1007/s00234-018-2066-5 [DOI] [PubMed] [Google Scholar]
- 45.Kuang H, Najm M, Chakraborty D, et al. . Automated ASPECTS on noncontrast CT scans in patients with acute ischemic stroke using machine learning . AJNR Am J Neuroradiol 2019;40:33–38 10.3174/ajnr.A5889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maegerlein C, Fischer J, Monch S, et al. . Automated calculation of the Alberta Stroke Program Early CT score: feasibility and reliability. Radiology 2019;291:141–48 10.1148/radiol.2019181228 [DOI] [PubMed] [Google Scholar]
- 47.Albers GW, Wald MJ, Mlynash M, et al. . Automated calculation of Alberta Stroke Program Early CT score: validation in patients with large hemispheric infarct. Stroke 2019;50:3277–79 10.1161/STROKEAHA.119.026430 [DOI] [PubMed] [Google Scholar]
- 48.Grunwald IQ, Kulikovski J, Reith W, et al. . Collateral automation for triage in stroke: evaluating automated scoring of collaterals in acute stroke on computed tomography scans. Cerebrovasc Dis 2019;47:217–22 10.1159/000500076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang K, Shou Q, Ma SJ, et al. . Deep learning detection of penumbral tissue on arterial spin labeling in stroke. Stroke 2020;51:489–97 10.1161/STROKEAHA.119.027457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ho KC, Speier W, Zhang H, et al. . A machine learning approach for classifying ischemic stroke onset time from imaging. IEEE Trans Med Imaging 2019;38:1666–76 10.1109/TMI.2019.2901445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee H, Lee E-J, Ham S, et al. . Machine learning approach to identify stroke within 4.5 hours. Stroke 2020;51:860–66 10.1161/STROKEAHA.119.027611 [DOI] [PubMed] [Google Scholar]
- 52.Wu O, Koroshetz WJ, Østergaard L, et al. . Predicting tissue outcome in acute human cerebral ischemia using combined diffusion- and perfusion-weighted MR imaging. Stroke 2001;32:933–42 10.1161/01.str.32.4.933 [DOI] [PubMed] [Google Scholar]
- 53.Stier N, Vincent N, Liebeskind D, et al. . Deep learning of tissue fate features in acute ischemic stroke. Proceedings (IEEE Int Conf Bioinformatics Biomed) 2015;2015:1316–21 10.1109/BIBM.2015.7359869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nielsen A, Hansen MB, Tietze A, et al. . Prediction of tissue outcome and assessment of treatment effect in acute ischemic stroke using deep learning. Stroke 2018;49:1394–1401 10.1161/STROKEAHA.117.019740 [DOI] [PubMed] [Google Scholar]
- 55.Ho KC, Scalzo F, Sarma K, et al. . Predicting ischemic stroke tissue fate using a deep convolutional neural network on source magnetic resonance perfusion images. J Med Imag 2019;6:1 10.1117/1.JMI.6.2.026001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu Y, Xie Y, Thamm T, et al. . Use of deep learning to predict final ischemic stroke lesions from initial magnetic resonance imaging. JAMA Netw Open 2020;3:e200772 10.1001/jamanetworkopen.2020.0772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfaff J, Herweh C, Schieber S, et al. . e-ASPECTS correlates with and is predictive of outcome after mechanical thrombectomy. AJNR Am J Neuroradiol 2017;38:1594–99 10.3174/ajnr.A5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang T-Y, Jiao Y, Cui Y, et al. . Development and validation of a penumbra-based predictive model for thrombolysis outcome in acute ischemic stroke patients. EBioMedicine 2018;35:251–59 10.1016/j.ebiom.2018.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie Y, Jiang B, Gong E, et al. . Use of gradient boosting machine learning to predict patient outcome in acute ischemic stroke on the basis of imaging, demographic, and clinical information. AJR Am J Roentgenol 2019;212:44–51 10.2214/AJR.18.20260 [DOI] [PubMed] [Google Scholar]
- 60.Nishi H, Oishi N, Ishii A, et al. . Predicting clinical outcomes of large vessel occlusion before mechanical thrombectomy using machine learning. Stroke 2019;50:2379–88 10.1161/STROKEAHA.119.025411 [DOI] [PubMed] [Google Scholar]
- 61.Bacchi S, Zerner T, Oakden-Rayner L, et al. . Deep learning in the prediction of ischaemic stroke thrombolysis functional outcomes: a pilot study. Acad Radiol 2020;27:e19–23 10.1016/j.acra.2019.03.015 [DOI] [PubMed] [Google Scholar]
- 62.Yu Y, Guo D, Lou M, et al. . Prediction of hemorrhagic transformation severity in acute stroke from source perfusion MRI. IEEE Trans Biomed Eng 2018;65:2058–65 10.1109/TBME.2017.2783241 [DOI] [PubMed] [Google Scholar]
- 63.Desai V, Flanders A, Lakhani P. Application of deep learning in neuroradiology: automated detection of basal ganglia hemorrhage using 2D-convolutional neural networks. arXiv :1710:03823;10 Oct 2017;1–7
- 64.Chilamkurthy S, Ghosh R, Tanamala S, et al. . Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. Lancet 2018;392:2388–96 10.1016/S0140-6736(18)31645-3 [DOI] [PubMed] [Google Scholar]
- 65.Arbabshirani MR, Fornwalt BK, Mongelluzzo GJ, et al. . Advanced machine learning in action: identification of intracranial hemorrhage on computed tomography scans of the head with clinical workflow integration. NPJ Digital Med 2018;1:9 10.1038/s41746-017-0015-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee H, Yune S, Mansouri M, et al. . An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nat Biomed Eng 2019;3:173–82 10.1038/s41551-018-0324-9 [DOI] [PubMed] [Google Scholar]
- 67.Chang PD, Kuoy E, Grinband J, et al. . Hybrid 3D/2D convolutional neural network for hemorrhage evaluation on head CT. AJNR Am J Neuroradiol 2018;39:1609–16 10.3174/ajnr.A5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuo W, Häne C, Mukherjee P, et al. . Expert-level detection of acute intracranial hemorrhage on head computed tomography using deep learning. Proc Natl Acad Sci USA 2019;116:22737–45 10.1073/pnas.1908021116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu J, Xu H, Chen Q, et al. . Prediction of hematoma expansion in spontaneous intracerebral hemorrhage using support vector machine. EBioMedicine 2019;43:454–59 10.1016/j.ebiom.2019.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]