Abstract
Pectobacterium carotovorum has an incomplete Entner–Doudoroff (ED) pathway, including enzyme 2‐keto‐3‐deoxy‐6‐phosphogluconate aldolase (Eda) but lacking phosphogluconate dehydratase (Edd), while P. atrosepticum (Pba) has a complete pathway. To understand the role of the ED pathway in Pectobacterium infection, mutants of these two key enzymes, Δeda and Δedd, were constructed in Pba SCRI1039. Δeda exhibited significant decreased virulence on potato tubers and colonization in planta and was greatly attenuated in pectinase activity and the ability to use pectin breakdown products, including polygalacturonic acid (PGA) and galacturonic acid. These reduced phenotypes were restored following complementation with an external vector expressing eda. Quantitative reverse transcription PCR analysis revealed that expression of the pectinase genes pelA, pelC, pehN, pelW, and pmeB in Δeda cultured in pyruvate, with or without PGA, was significantly reduced compared to the wild type, while genes for virulence regulators (kdgR, hexR, hexA, and rsmA) remained unchanged. However, Δedd showed similar phenotypes to the wild type. To our knowledge, this is the first demonstration that disruption of eda has a feedback effect on inhibiting pectin degradation and that Eda is involved in building the arsenal of pectinases needed during infection by Pectobacterium.
Keywords: Eda, Entner–Doudoroff pathway, pathogenicity, Pectobacterium
A component of the Entner–Doudoroff pathway, 2‐keto‐3‐deoxy‐6‐phosphogluconate aldolase, plays a vital role in the virulence of Pectobacterium atrosepticum on potato by controlling expression of genes involved in pectin degradation.
1. INTRODUCTION
The Entner–Doudoroff (ED) pathway is one of the central glycolytic pathways in gram‐negative bacteria (Figure 1; Conway, 1992). It was first described in Pseudomonas saccharophila (Entner & Doudoroff, 1952) and was later discovered in a wide range of organisms from Archaea to plants (Flamholz et al., 2013). Although the ED pathway produces only one adenosine triphosphate (ATP) per glucose molecule, which is half that of the Embden–Meyerhof–Parnas (EMP) pathway, it has the advantage of requiring several‐fold less enzymatic protein to achieve the same glucose conversion rate. Furthermore, genomic analysis has revealed that the EMP pathway is the major choice for energy‐deprived anaerobes as it has a higher ATP yield, whereas the ED pathway is preferred by facultative anaerobes and aerobes (Flamholz et al., 2013).
FIGURE 1.
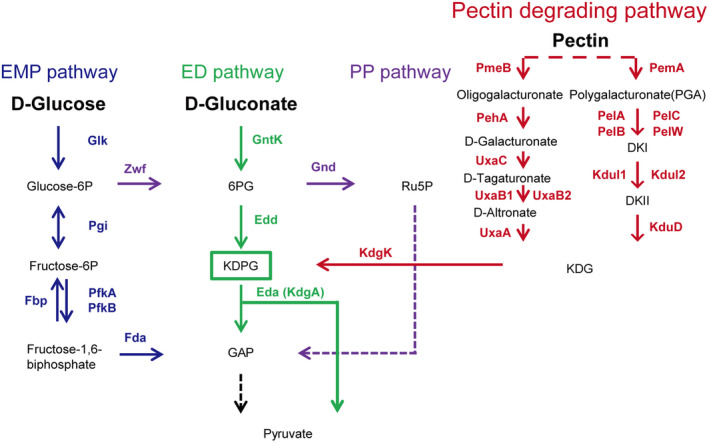
Major glycolytic pathways in Pectobacterium atrosepticum. Different metabolic pathways are displayed in different colours. Solid arrows represent reactions that are catalysed by a single enzyme, while dashed arrows indicate multiple reactions. The key products are boxed. The pectin degrading pathway is presented in red; EMP, Embden–Meyerhof–Parnas pathway (blue); ED, Entner–Doudoroff pathway (green); PP, pentose phosphate pathway (purple); 6PG, 6‐gluconate‐phosphate; Ru5P, ribulose 5‐phosphate; DKI, 5‐keto‐4‐deoxyuronate; DKII, 2,5‐diketo‐3‐deoxygluconate; KDG, 2‐keto‐3‐deoxygluconate; KDPG, 2‐keto‐3‐deoxy‐6‐phosphate‐gluconate; GAP, glyceraldehyde‐3‐phosphate
The ED pathway has two unique enzymes, 6‐phosphogluconate dehydratase (Edd, EC 4.2.1.12) and 2‐keto‐3‐deoxy‐6‐phosphogluconate (KDPG) aldolase (Eda, EC 4.1.2.14). 6‐phosphogluconate is dehydrated by Edd to form KDPG, which is then cleaved by Eda to form pyruvate and glyceraldehyde 3‐phosphate (GAP). The primary function of the ED pathway is the breakdown of sugar acids such as gluconate, which cannot be metabolized through the EMP pathway (glucose metabolism) (Peekhaus & Conway, 1998). Gluconate is a major carbon source used by Escherichia coli MG1655 in the colonization of the mouse intestine. It was found that a deletion within edd negatively affects both disease initiation and pathogen maintenance within the host (Chang et al., 2004). Transcriptional activation analyses and gene silencing revealed that in Vibrio cholerae the ED pathway is obligatory for gluconate utilization and regulating virulence (Patra et al., 2012). A recent study also suggested that nicotinamide adenine dinucleotide phosphate, generated by the ED pathway, is required for counteracting oxidative stress in Pseudomonas putida KT2440, which consumes glucose almost exclusively through the ED pathway (Chavarria et al., 2013). In addition to these virulence‐related roles, the ED pathway is also critical to the survival of the pathogenic organisms Legionella pneumophila and Shigella flexneri in their host systems (Harada et al., 2010; Waligora et al., 2014). However, the role of the ED pathway in virulence is not conserved in all pathogenic organisms as it has a limited impact on Helicobacter pylori colonization of mice (Wanken et al., 2003).
Gram‐negative bacterial species within the Pectobacterium genus are listed in the top 10 most important plant‐pathogenic bacteria, causing blackleg and soft rot on potato and diseases on many other crops and ornamental plants (Mansfield et al., 2012). Pectobacterium spp. employ a series of plant cell wall‐degrading enzymes (PCWDEs), which are mainly secreted through the Type II secretion system (T2SS), to break down the host plant cell wall (Toth et al., 2003). These are the main virulence determinants in Pectobacterium spp. but, in addition to the PCWDEs, small virulence proteins such as Nip and Svx, flagella and the Type III (T3SS) and Type VI (T6SS) secretion systems are also involved (Corbett et al., 2005; Holeva et al., 2004; Laasik et al., 2014; Liu et al., 2008; Mattinen et al., 2004). The regulation of these factors is controlled by a complex network of regulators, including the quorum‐sensing system (e.g., ExpI/ExpR), two‐component systems (e.g., GacA/GacR), LysR family regulators (e.g., KdgR, RexZ), and a small RNA system (RsmA/RsmB/RsmC) (Babujee et al., 2012; Barnard & Salmond, 2007; Faure & Dessaux, 2007; Liu et al., 2008; Lyon, 1989; Toth et al., 2006, 2015). Environmental factors such as oxygen, host plant extracts, and divalent cations have also been reported to affect virulence in Pectobacterium (Babujee et al., 2012; Flego et al., 1997; Mattinen et al., 2007). In Dickeya spp. (formerly Erwinia chrysanthemi) Eda (referred to as KdgA) was shown to be induced by pectin‐degrading products in vivo through the action of the negative regulator KdgR. However, no role in virulence was established (Hugouvieux‐Cotte‐Pattat & Robert‐Baudouy, 1994). Recently, the vgu operon involved in gluconate metabolism was reported to be required for correct expression of virulence through regulators KdgR, FlhD, HexA, and RsmA in P. carotovorum WPP14 (Mole et al., 2010). However, as results were based on a multigene deletion, and the WPP14 strain lacks the edd gene, the role of the ED pathway (including genes edd and eda) in these plant‐pathogenic organisms remains unclear.
In our previous work, we reported that the KDPG adolase (Eda) in the ED pathway is required for full virulence of P. carotovorum strain PccS1 (Wang et al., 2018). However, details of the role of Eda or the ED pathway in virulence were not determined. In this study, we conducted a full functional analysis of the ED pathway key enzymes (Edd and Eda) in the virulence of P. atrosepticum. We identified that, while the ED pathway is not directly involved in virulence of Pectobacterium, Eda is through a role in pectin degradation controlled through the expression of PCWDEs.
2. RESULTS
2.1. A complete ED pathway exists in P. atrosepticum, P. parmentieri, and P. wasabiae but not in P. carotovorum or other Pectobacterium species and is critical for gluconate utilization
In a previous study, we reported that a ∆eda mutant of P. carotovorum PccS1 was significantly attenuated in virulence on Zantedeschia elliottiana. However, bacterial growth in both Luria‐Bertani (LB) and minimal medium (MM) supplemented with 0.2% glucose (MM + 0.2% glucose) was not affected by the deletion (Wang et al., 2018). To explore the role of eda in the metabolism pathways, we performed a genomic comparison between P. carotovorum PccS1 and related pathogenic bacteria P. atrosepticum and E. coli. The results (Figure 2a) show that the genome of PccS1 carries the eda but not the edd gene, while P. atrosepticum (Pba) SCRI1043 and E. coli K‐12 carry both eda and edd genes, which are jointly necessary for the complete ED pathway (Conway, 1992).
FIGURE 2.
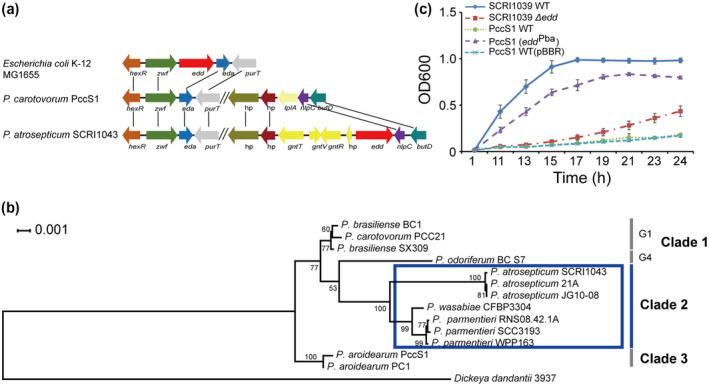
Gene edd is essential for Pectobacterium in utilization of gluconate. (a) Operon structure of edd and eda in different bacterial species. (b) Phylogenetic analysis of 13 Pectobacterium strains revealed a possible role in host adaption for the Entner–Doudoroff pathway. Protein sequences of 31 housekeeping genes from 14 bacterial genomes were extracted. Dickeya dadantii3937 was used as an outgroup for the analysis. A RAxML tree was developed using the IQ‐TREE web server with partitions. The blue rectangle indicates that the edd gene is present in these genomes. The bootstrap value was inferred from 1,000 replicates. (c) Growth curve over 24 hr in minimal media using gluconate as the sole carbon source of P. carotovorum PccS1 (which lacks gene edd) and following complementation with the edd gene (edd Pba) from P. atrosepticum SCRI1039 together with the SCRI1039 wild type and ∆edd mutant strain. Values are the means ± SE of three independent experiments
We conducted a phylogenetic analysis to further demonstrate the potential role of the ED pathway in Pectobacterium species. Based on the protein sequences of 31 housekeeping genes, a total of 13 Pectobacterium strains were used to conduct a phylogeny inference with Dickeya dadantii 3937 set as an outgroup. A rectangular phylogram shows that these Pectobacterium strains are generally subdivided into three clades (Figure 2b). Clade 1 includes P. carotovorum PCC21, P. brasiliense strains BC1 and BX309, and P. odoriferum BC S7. When compared with the phylogenetic tree obtained by Zhang et al. (2016), which showed similar groupings for these strains, Clade 1 can be subdivided into two groups: BC1, PCC21, and SX309 were classified into group 1 (G1), and BC S7 into group 4 (G4). The species in Clade 1 lack the edd gene and therefore do not have a complete ED pathway. Clade 2 includes three species, P. atrosepticum (three strains), P. wasabiae (one strain), and P. parmentieri (Ppa, three strains), all of which possess all components of the ED pathway, and Clade 2 forms an evolutionary group distinct from other Pectobacterium species (Clade 1 and Clade 3), which all lack the edd gene. Clade 3 includes two strains: PccS1, isolated from rotted Zantedeschia elliottiana (calla lily) in China, shared a high similarity with PC1 isolated from Ornithogalum dubium in Israel.
Although PccS1 could not utilize d‐gluconate, it could do so when the gene edd Pba was introduced into the strain (Figure 2c). The results therefore indicate that edd Pba was successfully expressed heterogeneously and confirms that PccS1 has an incomplete ED pathway. Moreover, a mutant of Pba SCRI1039 lacking edd showed attenuated growth in MM supplemented with gluconate at 0.2% as the sole carbon source compared with the wild‐type SCRI1039 (Figure 2c). The results further demonstrate that a completed ED pathway is necessary for the utilization of gluconate, which only Clade 2 isolates are able to undertake.
2.2. Mutant ∆eda shows significantly reduced pathogenicity by P. atrosepticum SCRI1039 on potato tubers, while mutant Δedd has no observable effect
To better understand the role of the key components of the ED pathway during infection of P. atrosepticum, two mutants, ∆eda and ∆edd, were constructed in Pba SCRI1039. The mutants were confirmed by PCR and sequencing of eda and edd fragments. To determine whether eda and edd are involved in virulence, we first compared tuber maceration by ∆eda and ∆edd mutants with that of the wild type. Each potato tuber slice was inoculated with a single strain using 10 mM MgSO4 buffer as a negative control. The wild type, wild type with empty pGemT vector, and Δedd mutant all macerated the tuber slices to a similar extent over a 3‐day period, which indicates that the edd gene had no observable effect on virulence. However, mutant Δeda and Δeda with the empty vector showed a lower maceration level on the potato tuber slices than the wild type (Figure 3a). ∆eda(pEda), which carried the complementing fragment of eda, restored the mutant's ability to macerate the tuber slices, although it did not reach wild‐type levels (Figure 3a).
FIGURE 3.
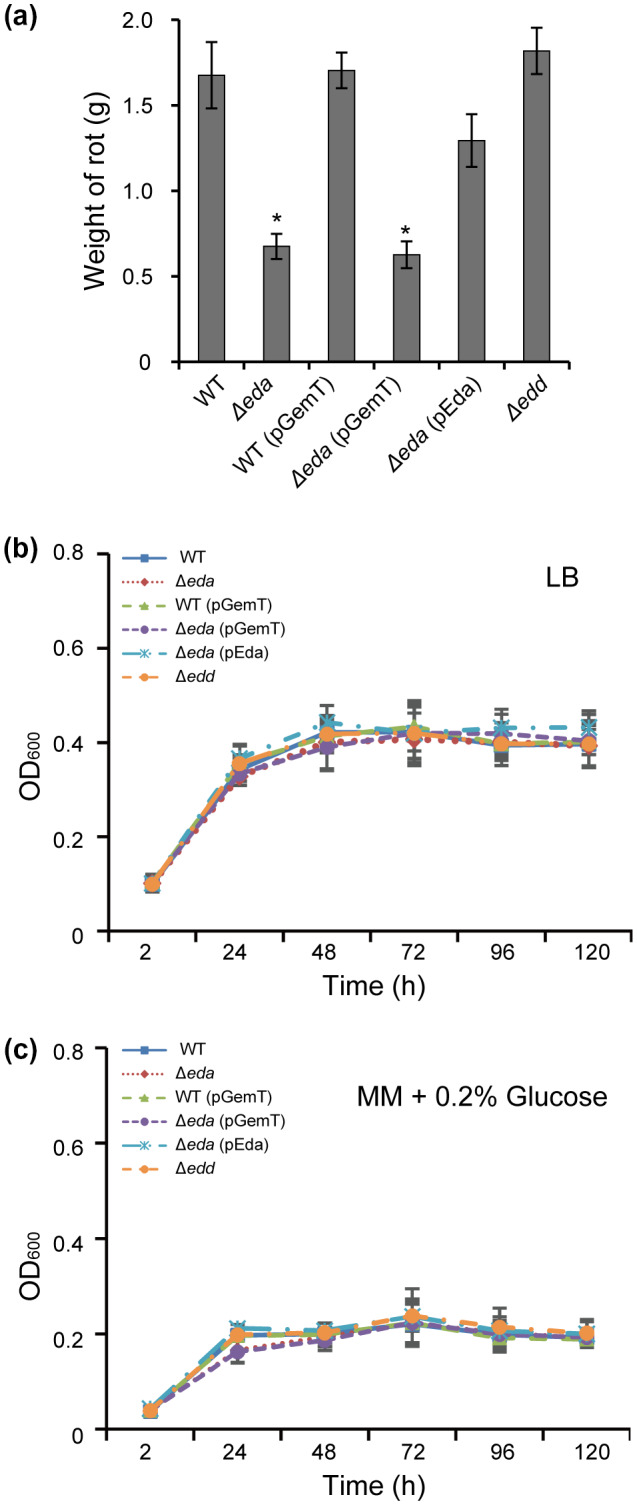
Mutant Δeda is defective in virulence on potato tubers. (a) Tuber slides were inoculated with Δeda and Δedd strains derived from Pectobacterium atrosepticum SCRI1039 (WT). The y axis shows grams of macerated tissues measured 72 hr after inoculation. The experiment had six internal replicates and was conducted on at least three occasions. Values are the means ± SE. *Significant difference between wild type and mutant (p < .05; Duncan's multiple range test). (b) and (c) Mutants Δeda and Δedd had no effect on growth in media. Growth curves of P. atrosepticum SCRI1039 in Luria‐Bertani (LB) (b) and minimal medium (MM) supplemented with 0.2% glucose (c) over 120 hr. Values are the means ± SE of three independent experiments
When cultured in LB medium and MM plus 0.2% glucose, Δeda grew at an equivalent level to the wild‐type strain over a 5‐day period (Figure 3b,c). This suggested that the attenuated virulence in Δeda was not due to its ability to grow using simple sugars.
2.3. P. atrosepticum SCRI1039 ∆eda shows significantly attenuated virulence and colonization on potato stems
To determine whether ∆eda was reduced in virulence on potato plants as well as tubers, the ability of the wild type and Δeda to colonize and cause disease on potato stems was assayed (Figure 4). Unlike the wild‐type strain, Δeda was unable to develop blackleg symptoms 3 days postinfiltration (Figure 4a). The population of the wild‐type strain in the stems increased from c.103 to 108 cfu/g during this period, showing a slight decline by day 7 (Figure 4b). Δeda, on the other hand, was 2 logs lower at c.106 cfu/g by day 3 and the population remained at that level up to day 7 (Figure 4b). Following complementation, both blackleg symptoms and a population level equivalent to that of the wild type were observed in Δeda (Figure 4a,b).
FIGURE 4.
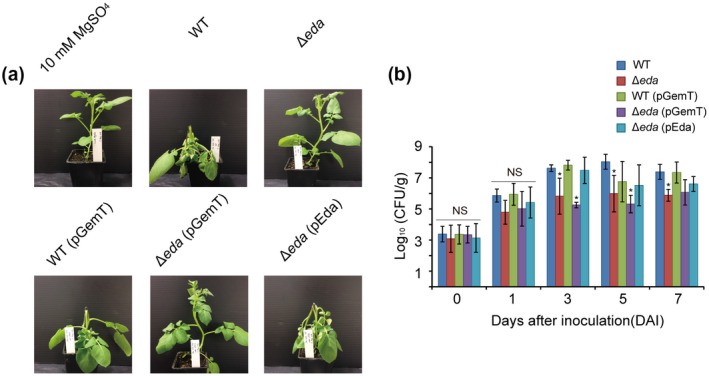
Mutant Δeda did not cause typical blackleg symptoms on potato plants and was reduced in its ability to colonize in planta. (a) Plants were inoculated with 105 cfu/ml Δeda derived from Pectobacterium atrosepticum SCRI1039 (WT) using 10 mM MgSO4 as a negative control. Images were collected at 3 days after inoculation when typical blackleg symptoms on potato (cv. Estima) appeared. (b) Infected plants were ground and bacteria extracted on the day of inoculation and 1, 3, 5, and 7 days later. Colonies were counted on crystal violet pectate medium. Each strain was inoculated into three plants and the experiment was repeated twice. Values are the means ± SE of log10cfu per gram plant tissue from three independent experiments
2.4. Mutant ∆eda is attenuated in pectin degradation
It has been demonstrated that PCWDEs are the main virulence factors of Pectobacterium spp. and are a crucial prerequisite during plant infection and subsequent disease development (Toth et al., 2003). Recently, we reported that the ∆eda mutant of Pectobacterium carotovorum PccS1 was significantly reduced in polygalacturonase activity on pectin agar plates (Wang et al., 2018). A similar result was observed when the Pba SCRI1039 ∆eda mutant was patched onto the assay plates (Figure 5a,b), while cellulase activity was unaffected compared with the wild type (Figure S1a,b) and neither of these two assays showed reduced activity in the ∆edd mutant.
FIGURE 5.
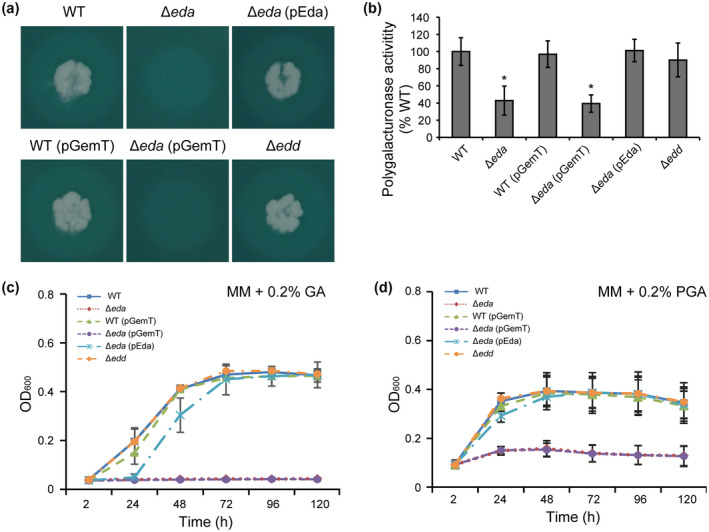
Mutant Δeda of Pectobacterium atrosepticum SCRI1039 is deficient in pectin degradation. The activities of pectinase (a) in P. atrosepticum SCRI1039 wild type (WT), ∆eda, and ∆edd were carried out on agar plates. Diameters of haloes around the colonies were calculated and statistically analysed (b). The y axis represents means of percentage compare to the WT (%) ± SE from three independent experiments. *Significant difference between the WT and mutant strains (p < .05; Duncan's multiple range test); NS, no significant difference (p < .05; Duncan's multiple range test). Bacterial growth in minimal medium (MM) supplemented with pectin degradation products (c) galacturonic acid (GA) and (d) polygalacturonic acid (PGA) were evaluated over 120 hr. Values are the means ± SE of three independent experiments
We further analysed the abilities of SCRI1039 ∆eda and ∆edd to utilize pectin breakdown products by determining their growth ability in MM supplemented with polygalacturonic acid (PGA) or galacturonic acid (GA). When these were used as a sole carbon source supplemented in MM, ∆eda did not grow or grew to a very low level on PGA and GA, respectively (Figure 5c,d). Expression of the eda gene in trans in ∆eda(pEda) fully restored polygalacturonase activity and the ability to grow (Figure 5a–d), which suggests that the decreased ability to grow in media and attenuated virulence on the host was due to the reduced ability of ∆eda to metabolize the breakdown products of pectin. The ∆edd mutant, however, showed no observable changes in enzyme activities or pectin utilization compared to the wild type (Figure 5a–d). The results indicate that Edd does not participate in pectin utilization in P. atrosepticum SCRI1039, whereas Eda is crucial in this role.
2.5. Mutant ∆eda exhibits changes in expression of genes encoding enzymes for pectin degradation
To understand the role of the ED pathway key enzyme KDPG adolase (Eda) in virulence and PCWDE activity in P. atrosepticum, a quantitative reverse transcription PCR (RT‐qPCR) analysis was conducted to determine whether the expression levels of genes encoding regulators of virulence and carbon metabolism (kdgR, hexA, hexR, and rsmA) were affected in ∆eda compared to that of the wild type. When pyruvate (the ultimate breakdown product of both the ED pathway and pectin degradation) was used as the sole carbon source, ∆eda exhibited similar RNA levels to that of the wild type for most of the regulators tested, and comparable levels were also seen for the wild type and ∆eda in the presence of PGA (Figure 6a–d). However, gene expression for both strains was significantly down‐regulated for all four regulators when grown in PGA compared to pyruvate (Figure 6a–d). This suggests that reduced growth on pectin agar plates was not due to the influence of the eda gene on the expression of these virulence regulators.
FIGURE 6.
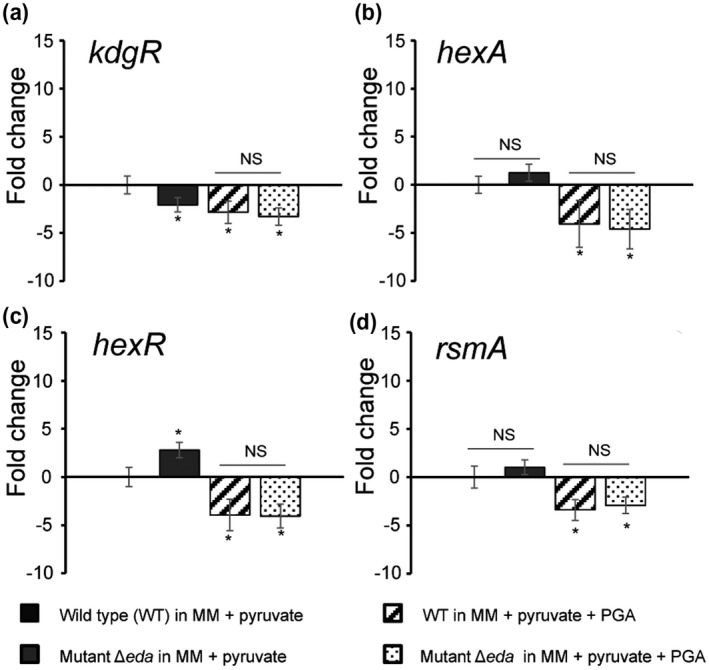
Relative transcript levels of regulators were measured in Pectobacterium atrosepticum strains wild type (WT) and ∆eda. P. atrosepticum SCRI1039 WT and ∆eda mutant were grown at 28 °C under different culture conditions (minimal medium [MM] plus 0.2% pyruvate and supplemented with or without polygalacturonic acid [PGA]) to OD600 0.4–0.6. The recA gene was used as a control housekeeping gene. Transcript levels of wild type in pyruvate were set as zero and the other three treatments were normalized to the wild type. The expression levels of up‐regulated genes were positive while those of down‐regulated genes were negative (a)–(f). Error bars represent standard errors for triplicate assays. *p < .05, **p < .01, NS, no significant difference, Duncan's multiple range test
To further determine whether reduced pectinase activity observed on pectin agar plates (Figure 5a,b) was due to changes in the expression of genes encoding PCWDEs, the expression profiles of eight such genes (pelA, pelB, pelC, pelZ, pehA, pehN, pelW, and pmeB) were analysed in the wild type or ∆eda mutant strains grown in pyruvate or in pyruvate supplemented with PGA. The results revealed that the expression levels of all eight genes in ∆eda were similar to that of the wild type when grown in pyruvate (Figure 7a–h). However, in the presence of pyruvate plus PGA, compared to pyruvate alone, the expression levels of four enzymes (pelA, pelC, pehN, pelW) were significantly up‐regulated in the wild type (>5‐fold) (Figure 7a,b,d,e), while six enzymes (pelA, pelC, pelZ, pehN [5–10‐fold], and pelW, pmeB [>10 fold]) were significantly down‐regulated in ∆eda (Figure 7a–f). In particular, pelW and pmeB were 40‐ and 180‐fold, respectively, down‐regulated in ∆eda. Overall, this shows that in the presence of PGA, increased expression of the genes encoding PCWDEs occurs through the action of eda but not via control of or through the above regulators, suggesting either direct control by Eda or control through regulators untested in this study.
FIGURE 7.
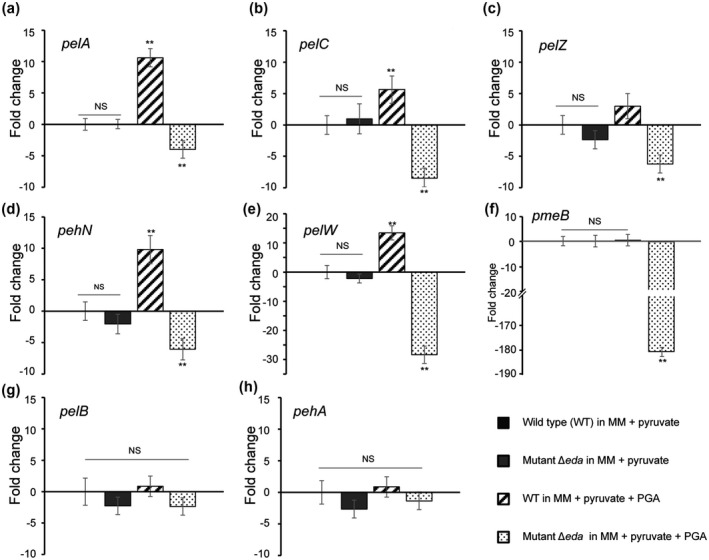
Relative transcript levels of selected genes were measured in Pectobacterium atrosepticum strains wild type (WT) and ∆eda. P. atrosepticum SCRI1039 WT and ∆eda mutant were grown at 28 °C under different culture conditions (minimal medium [MM] plus 0.2% pyruvate and supplemented with or without polygalacturonic acid [PGA]) to OD600 0.4–0.6. The recA gene was used as a control housekeeping gene. Transcript levels of WT in pyruvate were set as zero and the other three treatments were normalized to the WT. The expression levels of up‐regulated genes were positive while those of down‐regulated genes were negative (a)–(l). Error bars represent SE for triplicate assays. Difference between the treatment and WT in pyruvate condition, Duncan's multiple range test, *p < .05; **p < .01; NS, no significant difference p > .05
3. DISCUSSION
The genus Pectobacterium contains many species, some of which are found on a wide range of host plants. Our previous work showed that host plant extracts can induce expression of the Eda protein, which was found to be required for full virulence of P. carotovorum PccS1 (Wang et al., 2018). Eda is part of the ED metabolic pathway that also includes the Edd protein (Figure 1). The ED pathway was shown to be required for full virulence of the animal pathogenic bacteria E. coli MG1655 and V. cholerae (Chang et al., 2004; Patra et al., 2012). However, in our current study we observed that the edd gene is not present in the PccS1 genome (Figure 2a), raising questions about the role of the ED pathway, Eda and Edd in the virulence of Pectobacterium spp. We chose to further investigate this pathway using P. atrosepticum strains SCRI1043 and SCRI1039, in which we found both genes to be present.
To ensure that PccS1, SCRI1043, and SCRI1039 are not atypical in eda/edd gene content within the genus, we undertook a phylogenetic study of 14 strains from seven Pectobacterium species, using Dickeya dadantii 3937 (a close relative of Pectobacterium) as an outlier. Our phylogenetic analysis, based on 31 housekeeping genes, indicated that all the P. atrosepticum, P. wasabiae, and P. parmentieri strains tested (Clade 2) possess a complete ED pathway (Figure 2b). In our study, P. parmentieri SCC3193, WPP163, and RNS08.42.1A were clustered together in line with the results of the complete‐genome‐based phylogeny of Zhang et al. (2016), although in their paper they were referred to as P. wasabiae before being renamed P. parmentieri (Khayi et al., 2016). Thus, this supports our phylogenetic analysis based on 31 housekeeping genes in Pectobacterium species.
We then searched a further 65 Pectobacterium genomes and it was confirmed that of all Pectobacterium strains tested, the edd gene was only found in these three species (data not shown). Of the three clades identified, species in Clades 1 and 3 are associated with a wide host range (dicots and monocots) and monocots, respectively, while species in Clade 2 have narrow host ranges on dicot plants only (P. atrosepticum and P. parmentieri on potato and P. wasabiae on Japanese horseradish) (Khayi et al., 2016). Although not confirmed, it is interesting to speculate that the presence/absence of the edd gene, and therefore the complete ED pathway, may have a role to play in this host range differential.
Sugar metabolism provides various intermediates and energy for bacterial growth with the EMP, PP, and ED pathways involved in metabolism in gram‐negative bacteria (Conway, 1992). Pectobacterium produces large quantities of enzymes that utilize pectin from plant cell walls, with breakdown products that ultimately lead into these pathways (Toth et al., 2006). KDPG is one of the breakdown products in pectin metabolism, which feeds into the ED pathway (Figure 1). Because P. atrosepticum contains both the eda and edd genes, we used strains SCRI1043 and SCRI1039 to elucidate the function of the ED pathway. First, we showed that the pathway was functional in Pba but not in Pcc by showing a reduction in the growth of Δedd on gluconate (the primary carbon source of the ED pathway) compared to the wild‐type strain (Figure 2c). Also, by transferring the edd gene from Pba to Pcc we could restore this phenotype in PccS1, that is, gluconate utilization occurs specifically via this pathway and the pathway is dysfunctional in PccS1 due to the absence of the edd gene.
To demonstrate whether the ED pathway is required for full virulence in P. atrosepticum, after previously finding that the key gene (eda) of the ED pathway is important in PccS1 virulence (Wang et al., 2018), we undertook tuber assays and showed that the ∆eda exhibited reduced virulence compared to the wild‐type strain, while ∆edd was unaffected (Figure 3a). To then ensure that the lack of virulence was not due to a general growth defect, we confirmed that both mutants were able to grow in LB and MM containing glucose (Figure 3b,c). The eda result was confirmed in whole plant tests where ∆eda exhibited a clear reduction in bacterial numbers during infection compared to the wild‐type strain (Figure 4a,b).
Overall, this suggests that there is a conserved role for Eda in pathogenesis within the Pectobacterium genus, but this role does not require the ED pathway, instead pointing to a different mechanism(s) through which Eda operates. We showed previously that the impaired eda in PccS1 was affected in its ability to grow on pectin agar medium (Wang et al., 2018). While this was also the case in Pba, we went further by showing that the lack of growth was due to an inability of ∆eda to utilize GA and PGA, and that ∆edd was unaffected in growth on these substrates (Figure 5a–d). Neither mutant was affected in its ability to grow on cellulase plates (Figure S1a,b). This then prompted an examination of the effect of ∆eda on the expression of several transcriptional regulators of pectin degradation enzymes and virulence (kdgR, hexA, hexR, and rsmA), all of which affect PCWDE production (Charkowski et al., 2012). kdgR is a repressor that binds to 2‐keto‐3‐deoxygluconate (KDG), an intermediate upstream of KDPG in the pectin degradation pathway, thereby negatively regulating the expression of genes involved in pectin degradation (pelA, pelB, pelC, and pelE) and catabolism (kdgT, ogl, and kdul‐kdgF), and in pectinase secretion (outT) in Dickeya dadantii (previously Erwinia chrysanthemi) (Nasser et al., 1994). In human enteric pathogens, KdgR regulates gluconate metabolism (E. coli) and contributes to fitness (Salmonella enterica) (George et al., 2016; Pouyssegur & Stoeber, 1974). The hexA gene represses the production of PCWDEs, the quorum‐sensing signal molecule OHHL, and virulence in soft rot bacteria but also negatively regulates transcription of a second regulator, RsmB (Mukherjee et al., 2000). Analyses using comparative genomics and experimental approaches showed that HexR is a global regulator and serves as a repressor/activator to control carbon metabolism (Leyn et al., 2011). hexR is situated adjacent to the zwf‐eda operon in Pectobacterium strains PccS1 and SCRI1043 (Figure 1a), which is concordant with the findings in Pseudomonas putida (Kim et al., 2008). HexR controls the ED pathway in Pseudomonas fluorescens (Campilongo et al., 2017).
In the present work, the mRNA levels of all these four regulators were unchanged in the mutant ∆eda compared with the wild‐type SCRI1039 in MM supplemented with PGA (Figure 6), suggesting that Eda may influence virulence without involvement of these transcriptional regulators. Whether or not KDPG accumulates in the eda mutant still needs further investigation in the presence of PGA, as rapid accumulation of intercellular KDPG was reported to be bacteriostatic in an eda mutant of E. coli (Fuhrman et al., 1998). However, genes encoding enzymes involved in pectin degradation (pelA, pelC, pelZ, pehN, and pelW) were significantly induced in the wild type and repressed in the Δeda mutant in the presence of PGA but not pyruvate (Figure 7a–h). Therefore, Eda controls expression of genes involved in pectin degradation at the transcriptional level and not through the main regulators of these enzymes, with a lack of Eda resulting in defective utilization of products (GA and PGA) from pectin degradation (Figure 5c,d) and reduced pectinase (but not cellulase) activity on agar plates (Figures 5a,b and S1a,b).
In summary, our results reveal that the aldolase Eda plays a vital role in pectin degradation, and therefore virulence, with a mutation in eda preventing full breakdown of pectin and causing a feedback inhibition of pectinases in P. atrosepticum (Chang et al., 2004; Patra et al., 2012). However, the ED pathway appears not to be involved in Pectobacterium virulence, unlike in some animal pathogens where gluconate breakdown is necessary for virulence. The presence of a complete ED pathway in some species of Pectobacterium requires further investigation but may play an important role(s) in other environments, in host range or in the later stages of infection.
4. EXPERIMENTAL PREOCEDURES
4.1. Strains, media, and growth conditions
Bacterial strains and plasmids are listed in Table 1. P. carotovorum PccS1 and P. atrosepticum SCRI1039 were used as wild‐type strains for Pcc and Pba, respectively. Pectobacterium strains were maintained in LB medium (Wang et al., 2018) or MM supplemented with appropriate carbon sources (Jiang et al., 2017). Kanamycin (50 μg/ml), streptomycin (50 μg/ml), gentamycin (25 μg/ml), and ampicillin (100 μg/ml) were added as required. Prior to all experiments the bacteria were grown overnight in LB at 28 °C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strains or plasmid | Relative genotype or characteristic(s) a | Reference or source |
|---|---|---|
| Pectobacterium carotovorum strains | ||
| PccS1 | Rifr, wild‐type P. carotovorum | Wang et al. (2018) |
| WT(pBBR) | Rifr, Gmr, PccS1 wild type with empty pBBR | This study |
| PccS1(edd Pba) | Rifr, Gmr, PccS1 wild type with pBBR‐edd Pba | This study |
| P. atrosepticum strains | ||
| SCRI1039 | Wild‐type P. atrosepticum | JHI collection |
| ∆eda | SCRI1039 eda deletion mutant | This study |
| WT(pGemT) | Ampr, SCRI1039 wild type with empty pGemT | This study |
| ∆eda(pGemT) | Ampr, SCRI1039 ∆eda with empty pGemT | This study |
| ∆eda(pEda) | Ampr, SCRI1039 ∆eda complemented with pGemT‐eda | This study |
| ∆edd | SCRI1039 edd deletion mutant | This study |
| Escherichia coli strains | ||
| DH10B | F–, mcrA Δ(mrr‐hsdRMS‐mcrBC) φ80 lacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7,697 galU galK λ– rpsL nupG tonA | Life Technologies, Inc. |
| S17‐1 | Strepr, λ pir lysogen of S17‐1 | TakaRa |
| CC118 | Host for pKNG101‐based plasmids | JHI collection |
| HH26 | Mobilizing strain for conjugal transfer | JHI collection |
| Plasmids | ||
| pKNG101 | Strepr, allelic exchange vector, sacB, mobRK2, oriR6K | JHI collection |
| pBluscript‐II KS+ | Ampr, high copy cloning vector, multiple cloning site in lacZ’ | JHI collection |
| pBBR1‐MCS5 | Gmr, broad host range vector | Kovach et al. (1995) |
| pGem‐T Easy | Ampr, cloning vector | Promega |
| pBBR‐edd a | Gmr, pBBR with edd operon from Pba SCRI1039 | This study |
| pBlu‐eda | Ampr, pBluscript with two eda flanking fragments | This study |
| pKNG‐eda | Strepr, pKNG101 with two eda flanking fragments | This study |
| pBlu‐edd | Ampr, pBluscript with two edd flanking fragments | This study |
| pKNG‐edd | Strepr, pKNG101 with two edd flanking fragments | This study |
| pGem‐T‐eda | Ampr, pGem‐T with eda operon fragment | This study |
Rifr, rifampicin resistance; Gmr, gentamycin resistance; Strepr, streptomycin resistance; Ampr, ampicillin resistance. JHI, The James Hutton Institute, Dundee, UK.
4.2. Construction of plasmids and strains
To introduce the edd gene from P. atrosepticum into P. carotovorum, primers (Pba_edd‐F/Pba_edd‐R) were used to construct plasmid pBBR‐edd Pba containing an edd fragment from P. atrosepticum. The plasmid pBBR‐edd Pba was then introduced into P. carotovorum PccS1 by conjugation. Deletion mutants of genes (edd and eda) in P. atrosepticum SCRI1039 were constructed by homologous recombination as described previously (Coulthurst et al., 2008). The plasmids used in this study are detailed in Table 1. All mutants were checked by PCR and sequencing (data not shown). The primers used to construct and verify mutants are listed in Table S1.
4.3. Bacterial growth curves in different media
Bacteria were cultured at 28 °C overnight in LB medium. Cultures were collected by centrifugation at 4,000 × g for 15 min at room temperature and resuspended at 108 cfu/ml in 10 mM MgSO4. LB and MM plus 0.2% carbon source (glucose, pyruvate, gluconate, GA, and/or PGA) were used to assess growth conditions. For growth in 96‐well plates, 20 µl of culture and 180 µl of medium were added to each well. The plates were covered with lids, shaken lightly to mix, and incubated at 28 °C. Plates were assessed after incubating for 2, 24, 36, 48, 72, 96, and 120 hr to measure the OD600 values using a plate reader (Promega). Each strain had four replicates and three experiments were carried out.
4.4. RNA extraction
An overnight culture of bacteria was inoculated into fresh medium (1:50) and grown to exponential phase (OD600 = 0.4), after which a 5 ml sample was centrifuged at 10,000 × g for 5 min at 4 °C. A total of 20 µl lysozyme (100 mg/ml) was added to the centrifuged samples and vortexed vigorously. RNA was extracted by following the Qiagen mini Plant RNA extraction handbook and genomic DNA was removed using a DNase I kit (Qiagen). RNA was then quantified at 260 nm using a NanoDrop DE‐ND‐100 spectrophotometer (Thermo Fisher Scientific). The purity and integrity of RNA were monitored by loading a sample onto a 1% agarose gel.
4.5. Real‐time PCR analysis
The cDNA was synthesized using the GoScript Reverse Transcription System (Promega). Briefly, a total of 1 µg RNA was primed with random decamers and reverse transcribed with the qScript RT enzyme. To analyse the expression differences of the genes, RT‐qPCR analysis was performed using a PerfeCTa SYBR Green Fast Mix Kit (Quanta) on a StepOne Real‐Time PCR System (Applied Biosystems). The primers listed in Table S1 were designed by the online tool Primer3 web v. 4.0.0 based on the P. atrosepticum SCRI1043 genome sequence in NCBI. The ratio of gene expression was normalized to the level of expression of the housekeeping gene recA (Takle et al., 2007). Three independent experiments were performed.
4.6. Plant cell wall‐degrading enzyme activity assays
Polygalacturonase and cellulase activity was measured as previous described (Andro et al., 1984; Gilkes et al., 1984). Overnight bacterial cultures were grown to 104 cfu/ml. Samples of 10 μl aliquots were applied to the testing plates and incubated at 28 °C for 72 hr. For the polygalacturonase assay, plates were developed using 7.5% copper acetate for 1–2 hr. Cellulase activity was indicated using 0.2% (wt/vol) Congo red for 15–20 min and then washed using 1 M NaCl and 1 M HCl. The haloes around the colonies were measured. At least three plates were used for each assay, and the experiments were repeated at least three times with at least three replicates.
4.7. Potato virulence assay
Potato tubers (cv. Maris Piper) were used to perform the virulence assays of P. atrosepticum SCRI1039 wild type and mutants. Briefly, tubers were surface sterilized using 5% bleach and chopped with a sterile knife into 7 mm thick slices. A sterilized cork borer (5 mm diameter) was used to make a 5 mm deep well in the centre of each slice. A total of 50 µl of bacterial suspension (5 × 105 cfu) was added into each well and 10 mM MgSO4 was inoculated instead of bacteria as a negative control. At least six potato slices obtained from six different potato tubers were tested for each bacterial sample. The tuber slices were placed onto a sterilized Petri dish and incubated at 22 °C in a moist chamber for 3 days and then the rotted tissues were weighed. The entire experiment was repeated on at least three separate occasions.
4.8. Colonization in planta
The colonization in planta assay was performed on the stems of potato (cv. Estima) plants. The stems were stab‐inoculated with 105 cfu bacterial cells in 10 μl of 10 mM MgSO4 and the inoculation sites were covered with Vaseline. The plants were maintained at 22 °C. Infected stems were cut at 0, 1, 3, 5, and 7 days after inoculation (dai), weighed and ground in the presence of 10 ml Ringer's buffer (per litre 1.2 g NaCl; 0.62 g sodium lactate; 60 mg KCl, 40 mg CaCl2; pH 6.5). The cfu per gram of plant tissue was quantified by dilution plating on crystal violet pectate (CVP) agar plates. Photographs of the plants were taken 3 dai.
4.9. Phylogenetic analysis
To analyse the evolution of the ED pathway, phylogeny inference was conducted on 13 strains of Pectobacterium spp. The data set was limited to the 12 complete Pectobacterium genomes available in the PATRIC database (as listed in Table S2) at the time of study (Gillespie et al., 2011), together with one Pectobacterium (PccS1) genome sequenced by our group (unpublished data). The P. atrosepticum SCRI1043 (NC_004547) genome was used as a reference and the genome of D. dadantii 3937 was set as an outgroup.
The protein sequences of 31 housekeeping genes (listed in Table S3) were extracted and saved in FASTA format. The alignments were conducted with the MAFFT online software and quality checked in TOPALi v. 2.5 (Milne et al., 2004). The alignment columns where an amino acid only appeared in a single sequence were deleted manually. The data sets were concatenated using Sequence Matrix (Vaidya et al., 2011) and the concatenated sequences (7,960 amino acids) were then exported for further analysis.
Phylogenetic trees were estimated with the maximum‐likelihood (ML) method IQ‐TREE using the W‐IQ‐TREE web service (Nguyen et al., 2015). To simplify the analysis, we used Model Finder to find the best‐fit model to conduct the following analysis (models for each partition are listed in Table S3). An ultrafast bootstrap analysis was performed with 1,000 replicates. Bootstrap values from the ML analyses were used for adding statistical support onto congruent nodes of the trees drawn with Dendroscope 3 (Huson & Scornavacca, 2012).
4.10. Statistical analysis
All the phenotypic data were analysed using the IBM SPSS Statistics v. 20 program and p values were calculated using Duncan's multiple range test.
Supporting information
FIGURE S1 Mutants Δeda and Δedd of Pba SCRI1039 have no effect on cellulase activity. (a) The activities of cellulase in Pba SCRI1039 wild type, ∆eda, and ∆edd were carried out on agar plates. (b) Diameters of haloes around the colonies were calculated and statistically analysed. The y axis represents means of percentage compare to the wild type (%) ± SE from three independent experiments. NS, no significant difference (p < .05; Duncan’s multiple range test)
TABLE S1 Primers for gene modification and real‐time PCR used in this study
TABLE S2 Characteristics of Pectobacterium and Dickeya (outgroup) genomes used in this study
TABLE S3 Details of 31 housekeeping genes used in this study for the development of a phylogenetic tree of Pectobacterium strains
ACKNOWLEDGEMENTS
This work was supported by the Special Fund for Agro‐Scientific Research in the Public Interest of China (201303015) and the China Scholarship Council (201406850042). It was supported by a grant from the Scottish Government's Rural and Environmental Sciences and Analytical Services (RESAS) Division. The authors declare no conflicts of interest.
Wang H, Wang Y, Humphris S, et al. Pectobacterium atrosepticum KDPG aldolase, Eda, participates in the Entner–Doudoroff pathway and independently inhibits expression of virulence determinants. Mol Plant Pathol. 2021;22:271–283. 10.1111/mpp.13025
[Correction added on 19 December 2020, after first online publication: Jiaqin Fan has been included as the co‐corresponding author.]
Contributor Information
Jiaqin Fan, Email: fanjq@njau.edu.cn.
Ian Toth, Email: ian.toth@hutton.ac.uk.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Andro, T. , Chambost, J.P. , Kotoujansky, A. , Cattaneo, J. , Bertheau, Y. , Barras, F. et al. (1984) Mutants of Erwinia chrysanthemi defective in secretion of pectinase and cellulase. Journal of Bacteriology, 160, 1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babujee, L. , Apodaca, J. , Balakrishnan, V. , Liss, P. , Kiley, P.J. , Charkowski, A.O. et al. (2012) Evolution of the metabolic and regulatory networks associated with oxygen availability in two phytopathogenic enterobacteria. BMC Genomics, 13, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard, A.M.L. & Salmond, G.P.C. (2007) Quorum sensing in Erwinia species. Analytical and Bioanalytical Chemistry, 387, 415–423. [DOI] [PubMed] [Google Scholar]
- Campilongo, R. , Fung, R.K.Y. , Little, R.H. , Grenga, L. , Trampari, E. , Pepe, S. et al. (2017) One ligand, two regulators and three binding sites: How KDPG controls primary carbon metabolism in Pseudomonas . PLoS Genetics, 13, e1006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, D.E. , Smalley, D.J. , Tucker, D.L. , Leatham, M.P. , Norris, W.E. , Stevenson, S.J. et al. (2004) Carbon nutrition of Escherichia coli in the mouse intestine. Proceedings of the National Academy of Sciences of the United States of America, 101, 7427–7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charkowski, A. , Blanco, C. , Condemine, G. , Expert, D. , Franza, T. , Hayes, C. et al. (2012) The role of secretion systems and small molecules in soft‐rot Enterobacteriaceae pathogenicity. Annual Review of Phytopathology, 50, 425–449. [DOI] [PubMed] [Google Scholar]
- Chavarria, M. , Nikel, P.I. , Perez‐Pantoja, D. & de Lorenzo, V. (2013) The Entner‐Doudoroff pathway empowers Pseudomonas putida KT2440 with a high tolerance to oxidative stress. Environmental Microbiology, 15, 1772–1785. [DOI] [PubMed] [Google Scholar]
- Conway, T. (1992) The Entner‐Doudoroff pathway: history, physiology and molecular biology. FEMS Microbiology Reviews, 9, 1–27. [DOI] [PubMed] [Google Scholar]
- Corbett, M. , Virtue, S. , Bell, K. , Birch, P. , Burr, T. , Hyman, L. et al. (2005) Identification of a new quorum‐sensing‐controlled virulence factor in Erwinia carotovora subsp. atroseptica secreted via the type II targeting pathway. Molecular Plant‐Microbe Interactions, 18, 334–342. [DOI] [PubMed] [Google Scholar]
- Coulthurst, S.J. , Lilley, K.S. , Hedley, P.E. , Liu, H. , Toth, I.K. & Salmond, G.P.C. (2008) DsbA plays a critical and multifaceted role in the production of secreted virulence factors by the phytopathogen Erwinia carotovora subsp. atroseptica . Journal of Biological Chemistry, 283, 23739–23753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entner, N. & Doudoroff, M. (1952) Glucose and gluconic acid oxidation of Pseudomonas saccharophila . Journal of Biological Chemistry, 196, 853–862. [PubMed] [Google Scholar]
- Faure, D. & Dessaux, Y. (2007) Quorum sensing as a target for developing control strategies for the plant pathogen Pectobacterium . European Journal of Plant Pathology, 119, 353–365. [Google Scholar]
- Flamholz, A. , Noor, E. , Bar‐Even, A. , Liebermeister, W. & Milo, R. (2013) Glycolytic strategy as a trade‐off between energy yield and protein cost. Proceedings of the National Academy of Sciences of the United States of America, 110, 10039–10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flego, D. , Pirhonen, M. , Saarilahti, H. , Palva, T.K. & Palva, E.T. (1997) Control of virulence gene expression by plant calcium in the phytopathogen Erwinia carotovora . Molecular Microbiology, 25, 831–838. [DOI] [PubMed] [Google Scholar]
- Fuhrman, L.K. , Wanken, A. , Nickerson, K.W. & Conway, T. (1998) Rapid accumulation of intracellular 2‐keto‐3‐deoxy‐6‐phosphogluconate in an Entner‐Doudoroff aldolase mutant results in bacteriostasis. FEMS Microbiology Letters, 159, 261–266. [DOI] [PubMed] [Google Scholar]
- George, A.S. , Salas González, I. , Lorca, G.L. & Teplitski, M. (2016) Contribution of the Salmonella enteric KdgR regulon to persistence of the pathogen in vegetable soft rots. Applied and Environmental Microbiology, 82, 1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes, N.R. , Langsford, M.L. , Kilburn, D.G. , Miller, R.C. & Warren, R.A. (1984) Mode of action and substrate specificities of cellulases from cloned bacterial genes. Journal of Biological Chemistry, 259, 10455–10459. [PubMed] [Google Scholar]
- Gillespie, J.J. , Wattam, A.R. , Cammer, S.A. , Gabbard, J.L. , Shukla, M.P. , Dalay, O. et al. (2011) PATRIC: the comprehensive bacterial bioinformatics resource with a focus on human pathogenic species. Infection and Immunity, 79, 4286–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada, E. , Iida, K. , Shiota, S. , Nakayama, H. & Yoshida, S. (2010) Glucose metabolism in Legionella pneumophila: dependence on the Entner‐Doudoroff pathway and connection with intracellular bacterial growth. Journal of Bacteriology, 192, 2892–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holeva, M.C. , Bell, K.S. , Hyman, L.J. , Avrova, A.O. , Whisson, S.C. , Birch, P.R.J. et al. (2004) Use of a pooled transposon mutation grid to demonstrate roles in disease development for Erwinia carotovora subsp. atroseptica putative type III secreted effector (DspE/A) and helper (HrpN) proteins. Molecular Plant‐Microbe Interactions, 17, 943–950. [DOI] [PubMed] [Google Scholar]
- Hugouvieux‐Cotte‐Pattat, N. & Robert‐Baudouy, J. (1994) Molecular analysis of the Erwinia chrysanthemi region containing the kdgA and zwf genes. Molecular Microbiology, 11, 67–75. [DOI] [PubMed] [Google Scholar]
- Huson, D.H. & Scornavacca, C. (2012) Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Systematic Biology, 61, 1061–1067. [DOI] [PubMed] [Google Scholar]
- Jiang, H. , Jiang, M. , Yang, L. , Yao, P. , Ma, L. , Wang, C. et al. (2017) The ribosomal protein RplY is required for Pectobacterium carotovorum virulence and is induced by Zantedeschia elliottiana extract. Phytopathology, 107, 1322–1330. [DOI] [PubMed] [Google Scholar]
- Khayi, S. , Cigna, J. , Chong, T.M. , Quêtu‐Laurent, A. , Chan, K.‐G. , Hélias, V. et al. (2016) Transfer of the potato plant isolates of Pectobacterium wasabiae to Pectobacterium parmentieri sp. nov. International Journal of Systematic and Evolutionary Microbiology, 66, 5379–5383. [DOI] [PubMed] [Google Scholar]
- Kim, J. , Jeon, C.O. & Park, W. (2008) Dual regulation of zwf‐1 by both 2‐keto‐3‐deoxy‐6‐phosphogluconate and oxidative stress in Pseudomonas putida . Microbiology, 154, 3905–3916. [DOI] [PubMed] [Google Scholar]
- Kovach, M.E. , Elzer, P.H. , Hill, D.S. , Robertson, G.T. , Farris, M.A. , Roop, R.M. et al. (1995) Four new derivatives of the broad‐host‐range cloning vector pBBR1MCS, carrying different antibiotic‐resistance cassettes. Gene, 166, 175–176. [DOI] [PubMed] [Google Scholar]
- Laasik, E. , Pollumaa, L. , Pasanen, M. , Mattinen, L. , Pirhonen, M. & Mae, A. (2014) Expression of nip P.w of Pectobacterium wasabiae is dependent on functional flgKL flagellar genes. Microbiology, 160, 179–186. [DOI] [PubMed] [Google Scholar]
- Leyn, S.A. , Li, X. , Zheng, Q. , Novichkov, P.S. , Reed, S. , Romine, M.F. et al. (2011) Control of proteobacterial central carbon metabolism by the HexR transcriptional regulator: a case study in Shewanella oneidensis . Journal of Biological Chemistry, 286, 35782–35794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Coulthurst, S.J. , Pritchard, L. , Hedley, P.E. , Ravensdale, M. , Humphris, S. et al. (2008) Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum . PLoS Pathogens, 4, e1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon, G.D. (1989) The biochemical basis of resistance of potatoes to soft rot Erwinia spp.—a review. Plant Pathology, 38, 313–339. [Google Scholar]
- Mansfield, J. , Genin, S. , Magori, S. , Citovsky, V. , Sriariyanum, M. , Ronald, P. et al. (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Molecular Plant Pathology, 13, 614–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattinen, L. , Nissinen, R. , Riipi, T. , Kalkkinen, N. & Pirhonen, M. (2007) Host‐extract induced changes in the secretome of the plant pathogenic bacterium Pectobacterium atrosepticum . Proteomics, 7, 3527–3537. [DOI] [PubMed] [Google Scholar]
- Mattinen, L. , Tshuikina, M. , Mae, A. & Pirhonen, M. (2004) Identification and characterization of Nip, necrosis‐inducing virulence protein of Erwinia carotovora subsp. carotovora . Molecular Plant‐Microbe Interactions, 17, 1366–1375. [DOI] [PubMed] [Google Scholar]
- Milne, I. , Wright, F. , Rowe, G. , Marshall, D.F. , Husmeier, D. & McGuire, G. (2004) TOPALi: software for automatic identification of recombinant sequences within DNA multiple alignments. Bioinformatics, 20, 1806–1807. [DOI] [PubMed] [Google Scholar]
- Mole, B. , Habibi, S. , Dangl, J.L. & Grant, S.R. (2010) Gluconate metabolism is required for virulence of the soft‐rot pathogen Pectobacterium carotovorum . Molecular Plant‐Microbe Interactions, 23, 1335–1344. [DOI] [PubMed] [Google Scholar]
- Mukherjee, A. , Cui, Y. , Ma, W. , Liu, Y. & Chatterjee, A.K. (2000) hexA of Erwinia carotovora ssp. carotovora strain Ecc71 negatively regulates production of RpoS and rsmB RNA, a global regulator of extracellular proteins, plant virulence and the quorum‐sensing signal, N‐(3‐oxohexanoyl)‐l‐homoserine lactone. Environmental Microbiology, 2, 203–215. [DOI] [PubMed] [Google Scholar]
- Nasser, W. , Reverchon, S. , Condemine, G. & Robert‐Baudouy, J. (1994) Specific interactions of Erwinia chrysanthemi KdgR repressor with different operators of genes involved in pectinolysis. Journal of Molecular Biology, 236, 427–440. [DOI] [PubMed] [Google Scholar]
- Nguyen, L.‐T. , Schmidt, H.A. , von Haeseler, A. & Minh, B.Q. (2015) IQ‐TREE: A fast and effective stochastic algorithm for estimating maximum‐likelihood phylogenies. Molecular Biology and Evolution, 32, 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra, T. , Koley, H. , Ramamurthy, T. , Ghose, A.C. & Nandy, R.K. (2012) The Entner‐Doudoroff pathway is obligatory for gluconate utilization and contributes to the pathogenicity of Vibrio cholerae . Journal of Bacteriology, 194, 3377–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peekhaus, N. & Conway, T. (1998) What’s for dinner?: Entner‐Doudoroff metabolism in Escherichia coli . Journal of Bacteriology, 180, 3495–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouyssegur, J. & Stoeber, F. (1974) Genetic control of the 2‐keto‐3‐deoxy‐d‐gluconate metabolism in Escherichia coli K‐12: kdg regulon. Journal of Bacteriology, 117, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takle, G.W. , Toth, I.K. & Brurberg, M.B. (2007) Evaluation of reference genes for real‐time RT‐PCR expression studies in the plant pathogen Pectobacterium atrosepticum . BMC Plant Biology, 7, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth, I.K. , Bell, K.S. , Holeva, M.C. & Birch, P.R.J. (2003) Soft rot erwiniae: from genes to genomes. Molecular Plant Pathology, 4, 17–30. [DOI] [PubMed] [Google Scholar]
- Toth, I.K. , Humphris, S. , Campbell, E. & Pritchard, L. (2015) Why genomics research on Pectobacterium and Dickeya makes a difference. American Journal of Potato Research, 92, 218–222. [Google Scholar]
- Toth, I.K. , Pritchard, L. & Birch, P.R.J. (2006) Comparative genomics reveals what makes an enterobacterial plant pathogen. Annual Review of Phytopathology, 44, 305–336. [DOI] [PubMed] [Google Scholar]
- Vaidya, G. , Lohman, D.J. & Meier, R. (2011) SequenceMatrix: concatenation software for the fast assembly of multi‐gene datasets with character set and codon information. Cladistics, 27, 171–180. [DOI] [PubMed] [Google Scholar]
- Waligora, E.A. , Fisher, C.R. , Hanovice, N.J. , Rodou, A. , Wyckoff, E.E. & Payne, S.M. (2014) Role of intracellular carbon metabolism pathways in Shigella flexneri virulence. Infection and Immunity, 82, 2746–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Yang, Z. , Du, S. , Ma, L. , Liao, Y. , Wang, Y. et al. (2018) Characterisation of Pectobacterium carotovorum proteins differentially expressed during infection of Zantedeschia elliottiana in vivo and in vitro which are essential for virulence. Molecular Plant Pathology, 19, 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanken, A.E. , Conway, T. & Eaton, K.A. (2003) The Entner‐Doudoroff pathway has little effect on Helicobacter pylori colonization of mice. Infection and Immunity, 71, 2920–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Fan, Q. & Loria, R. (2016) A re‐evaluation of the taxonomy of phytopathogenic genera Dickeya and Pectobacterium using whole‐genome sequencing data. Systematic and Applied Microbiology, 39, 252–259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Mutants Δeda and Δedd of Pba SCRI1039 have no effect on cellulase activity. (a) The activities of cellulase in Pba SCRI1039 wild type, ∆eda, and ∆edd were carried out on agar plates. (b) Diameters of haloes around the colonies were calculated and statistically analysed. The y axis represents means of percentage compare to the wild type (%) ± SE from three independent experiments. NS, no significant difference (p < .05; Duncan’s multiple range test)
TABLE S1 Primers for gene modification and real‐time PCR used in this study
TABLE S2 Characteristics of Pectobacterium and Dickeya (outgroup) genomes used in this study
TABLE S3 Details of 31 housekeeping genes used in this study for the development of a phylogenetic tree of Pectobacterium strains
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


