Abstract
Erwinia amylovora is the causative agent of the devastating disease fire blight of pome fruit trees. After infection of host plant leaves at apple shoot tips, E. amylovora cells form biofilms in xylem vessels, restrict water flow, and cause wilting symptoms. Although E. amylovora is well known to be able to cause systemic infection, how biofilm cells of E. amylovora transit from the sessile mode of growth in xylem to the planktonic mode of growth in cortical parenchyma remains unknown. Increasing evidence has suggested the important modulatory roles of Hfq‐dependent small RNAs (sRNAs) in the pathogenesis of E. amylovora. Here, we demonstrate that the sRNA RprA acts as a positive regulator of amylovoran exopolysaccharide production, the type III secretion system (T3SS), and flagellar‐dependent motility, and as a negative regulator of levansucrase activity and cellulose production. We also show that RprA affects the promoter activity of multiple virulence factor genes and regulates hrpS, a critical T3SS regulator, at the posttranscriptional level. We determined that rprA expression can be activated by the Rcs phosphorelay, and that expression is active during T3SS‐mediated host infection in an immature pear fruit infection model. We further showed that overexpression of rprA activated the in vitro dispersal of E. amylovora cells from biofilms. Thus, our investigation of the varied role of RprA in affecting E. amylovora virulence provides important insights into the functions of this sRNA in biofilm control and systemic infection.
Keywords: biofilm dispersal, fire blight, posttranscriptional regulation, small RNA, systemic infection
The small RNA RprA is a positive regulator of pathogenicity factors in Erwinia amylovora, and overexpression of rprA activates the in vitro dispersal of E. amylovora cells from biofilms.
![]()
1. INTRODUCTION
Fire blight, caused by Erwinia amylovora, is a devastating bacterial disease threatening the worldwide production of many rosaceous fruits, such as apple and pear (Malnoy et al., 2012). The primary infection of E. amylovora is manifested by several distinct stages: flower infection, infection of leaves at shoot tips, xylem colonization, and systemic infection. During flower infection, large populations of E. amylovora cells (c.106–7 cfu/flower) are established on stigmas; these bacteria are then further disseminated down to the hypanthium, and ultimately initiate infection via natural openings in the flower nectaries (Thomson, 2000; Vanneste, 1995). Within host xylem vessels in leaves, E. amylovora cells block water transport and cause wilting symptoms by formation of biofilms, which are static microcolonies of bacterial cells enmeshed in a matrix of exopolysaccharides (EPSs) (Castiblanco & Sundin, 2016; Kharadi & Sundin, 2020; Koczan et al., 2009, 2011; Mina et al., 2019). E. amylovora cells can migrate further within the vascular tissue to cause systemic infection, but more commonly exit from xylem vessels and move further systemically through the host in cortical parenchyma tissue. The systemic spread of E. amylovora through host apple trees follows a downward path, ultimately ending at the crown–rootstock junction, where cankers can develop that girdle and kill the host (Norelli et al., 2003). Frequently, E. amylovora cells emerge from flower pedicel, leaf petiole, or stem tissues as ooze, which serves as the inoculum for secondary infection (Slack et al., 2017).
The pathogenesis of E. amylovora is mediated by several important virulence factors, including the type III secretion system (T3SS) (Oh et al., 2005; Zhao et al., 2009a), motility (Bayot & Ries, 1986), and several EPSs including amylovoran (Goodman et al., 1974; Sjulin & Beer, 1978), levan (Geier & Geider, 1993), and cellulose (Castiblanco & Sundin, 2018). The primary infection of E. amylovora through flowers requires motility to facilitate the migration of E. amylovora cells from stigmas to nectarthodes, and the T3SS to defeat host defence mechanisms and initiate pathogenesis (Bayot & Ries, 1986; Bogdanove et al., 1998; Kim et al., 1997; Oh et al., 2005). The T3SS is major pathogenicity factor of E. amylovora that has been well characterized to be regulated in a hierarchical manner. HrpL, an alternate sigma factor, activates transcription of the hrp genes, including the structural gene hrpA encoding the T3SS pilus, and the major effector gene dspE, through recognition of the “hrp box” motif (McNally et al., 2012; Triplett et al., 2009; Wei & Beer, 1995). hrpL expression is tightly regulated by the enhancer‐binding protein HrpS and the two‐component system HrpX/HrpY (Lee et al., 2016; Wei et al., 2000). Amylovoran, an acidic exopolysaccharide composed of repeating units of galactose molecules and a glucuronic acid residue, is another main pathogenicity factor of E. amylovora (Bellemann & Geider, 1992; Nimtz et al., 1996). Amylovoran biosynthesis is mediated by the 12‐gene ams operon (Bugert & Geider, 1995). Levan is a homopolymer of fructose molecules synthesized through hydrolysis of sucrose via the levansucrase enzyme that is encoded by the lsc gene (Geier & Geider, 1993; Gross et al., 1992). Amylovoran, levan, and cellulose together constitute the matrix of E. amylovora biofilms (Castiblanco & Sundin, 2018; Koczan et al., 2009).
The chaperone protein Hfq functions to stabilize its dependent small RNAs (sRNAs) and to facilitate the interactions of these sRNAs with their mRNA targets (Brennan & Link, 2007). Hfq‐dependent sRNA–mRNA interactions result in posttranscriptional regulation through either activation or inhibition of translation (Vogel & Luisi, 2011). Hfq and its associated sRNAs play critical roles in the regulation of pathogenesis in gram‐negative plant pathogenic bacteria such as Agrobacterium tumefaciens, Burkholderia glumae, Dickeya dadantii, E. amylovora, Pantoea ananatis, Pectobacterium carotovorum, and Xanthomonas campestris (Kim et al., 2018; Lai et al., 2018; Wang et al., 2018; Wilms et al., 2012; Yuan et al., 2019; Zeng et al., 2013; Zeng & Sundin, 2014). In E. amylovora, the Hfq‐dependent sRNA ArcZ was shown to positively affect amylovoran biosynthesis, the T3SS, levansucrase activity, and flagellar swimming motility, partially through posttranscriptional regulation of the leucine‐responsive regulatory protein Lrp (Schachterle & Sundin, 2019; Schachterle et al., 2019; Zeng & Sundin, 2014). However, the functions and regulatory mechanisms of most Hfq‐dependent sRNAs identified in E. amylovora are still enigmatic. A deletion mutant of the Hfq‐dependent sRNA rprA (Ea1189ΔrprA) was previously shown to cause decreased levels of virulence in an immature pear infection model, suggesting the positive involvement of this sRNA in the pathogenesis of E. amylovora through unknown mechanisms (Zeng et al., 2013). RprA was initially identified in Escherichia coli, and was shown to stimulate the translation of the stationary‐phase sigma factor RpoS in this bacterium (Majdalani et al., 2001). Through imperfect reverse complementarity with RprA, the inhibitory structure of the 5′ untranslated region (5′ UTR) of the rpoS mRNA is disengaged and the translation of rpoS is consequently activated (Majdalani et al., 2001; McCullen et al., 2010; Urban & Vogel, 2007). E. coli RprA has also been shown to down‐regulate csgD, encoding a stationary phase‐induced biofilm regulator, and ydaM, encoding a diguanylate cyclase (Andreassen et al., 2018; Mika et al., 2012). Expression of E. coli RprA has been demonstrated to be induced by the RcsCDB phosphorelay (Majdalani et al., 2002). On stimulation, the hybrid sensor kinase RcsC transfers a phosphoryl group to RcsD and then to the response regulator RcsB (Huang et al., 2006). The phosphorylated RcsB further modulates the expression of the downstream genes through binding to the “Rcs box” in promoter regions (Ancona et al., 2015; Huang et al., 2006; Wang et al., 2009). RprA expression in E. coli or Salmonella enterica serovar Typhimurium can also be activated by high population density and by environmental stressors, including osmolarity shock and low pH (Madhugiri et al., 2010; Srikumar et al., 2015).
We hypothesized that RprA affects the virulence of E. amylovora through modulation of its virulence factor(s), and that expression of rprA is activated or deactivated during E. amylovora pathogenesis upon perception of certain environmental cues. In this study, we demonstrate that RprA acts as a positive regulator of amylovoran, T3SS, and flagellar‐dependent motility, and as a negative regulator of levansucrase activity and cellulose production. We also identified the in vitro and in vivo conditions that activate RprA and demonstrated that activation of RprA decreased biofilm formation and promoted biofilm dispersal. This study provides important evidence for the involvement of RprA in the systemic movement of E. amylovora during pathogenesis.
2. RESULTS
2.1. RprA positively regulates amylovoran, T3SS, and motility, and negatively regulates levansucrase activity and cellulose production
RprA was previously identified as a 111‐nucleotide Hfq‐dependent sRNA that contributed to the full virulence of E. amylovora, as the deletion mutant Ea1189ΔrprA showed compromised virulence in an immature pear infection model (Zeng et al., 2013). To validate the positive involvement of RprA in the virulence of E. amylovora, we first examined the virulence of Ea1189ΔrprA (pJP‐rprA), in which rprA was expressed in trans. We showed that Ea1189ΔrprA (pJP‐rprA) exhibited strong virulence on the inoculated immature pears that was comparable to the E. amylovora wild‐type (WT) strain (Figure 1a), confirming the importance of RprA on the virulence of E. amylovora. Secondary structure modelling of RprA showed that RprA had four stem‐loops with a characteristic rho‐independent terminator in the 3′ end (Figure 1b). Sequence alignment of the RprA homologs in the Enterobacteriaceae family suggested that nucleotide sequences of RprA are highly conserved towards the 3′ end but much less conserved towards the 5′ end (Figure 1c).
FIGURE 1.

The effect of RprA on virulence of Erwinia amylovora and sequence characteristics of RprA. (a) E. amylovora strains at approximately 2 × 104 cfu in 2 µl were stab‐inoculated into immature pears. Images were captured 4 days postinoculation. The amylovoran‐null mutant Ea1189Δams was used as a negative control for the assays. (b) Secondary structure of RprA was predicted with the minimum free energy model of RNAfold (http://rna.tbi.univie.ac.at/cgi‐bin/RNAWebSuite/RNAfold.cgi). The positional entropy of each nucleotide is colour‐coded. (c) Sequence alignment of RprA homologs in representative species of Enterobacteriaceae. Shared nucleotide sequences are highlighted in green
To understand the molecular mechanisms of RprA in affecting the full virulence of E. amylovora, we examined how mutagenesis or overexpression of rprA affected virulence factors of this bacterium, including amylovoran production, hypersensitive response (HR), flagellar swimming motility, levansucrase activity, and cellulose production. We showed that amylovoran production was significantly reduced in the Ea1189ΔrprA mutant, and that production was restored to the WT level in the complementation strain (Figure 2a). In contrast, overproduction of RprA rendered a significantly increased level of amylovoran production in E. amylovora (Figure 2a). Our results therefore suggested that RprA is a positive regulator of amylovoran production. To investigate if RprA affected the function of T3SS, we examined whether deletion of rprA or overproduction affected the ability of E. amylovora to elicit the HR. Compared with the other strains examined, the Ea1189ΔrprA mutant exhibited a minimal HR on Nicotiana benthamiana leaves, suggesting a positive effect of RprA on the function of T3SS (Figure 2b). The rprA mutation had a small effect on the swimming motility of E. amylovora (Figure 2c). In contrast, the rprA overexpression strain Ea1189(pOE‐rprA) was hypermotile (Figure 2c). These results suggested that the basal level of RprA contributes little to the swimming motility of E. amylovora, but its effect on this phenotype was greater when rprA was overexpressed. Deletion mutation of rprA had negligible effects on the activity of levansucrase and the production of cellulose, but these EPSs were greatly compromised when rprA was overexpressed (Figure 2d,e). Therefore, a basal level of RprA had minimal effects on levansucrase activity and cellulose production; a high level of RprA, nevertheless, negatively correlated with the production of these extracellular polymeric substances. Taken together, RprA exhibited complex regulatory roles on the virulence factors of E. amylovora, and these impacts were greater when RprA was overproduced.
FIGURE 2.

Multifaced regulatory roles of RprA on virulence factors of Erwinia amylovora. (a) RprA positively regulates amylovoran production. Amylovoran was determined in cultures grown for 24 hr in MBMA medium with 1% galactose using a cetylpyridinium chloride‐binding assay. (b) Positive involvement of RprA in affecting the hypersensitive response (HR) elicited by E. amylovora. Approximately 100 μl cell suspension at OD600 = 0.05 was infiltrated into leaves of 10‐week‐old Nicotiana benthamiana plants. The HR was observed 16 hr postinoculation. (c) RprA increases the swimming motility of E. amylovora. Overnight cultures (2 µl) were inoculated into 0.3% agar LB plates, and the radius of the motility area was measured after 48 hr. (d) RprA inhibits levansucrase (Lsc) activity of E. amylovora; Lsc activity was quantified as previously described (Schachterle & Sundin, 2019). (e) RprA inhibits cellulose biosynthesis in E. amylovora determined through a Congo red‐binding assay. A greater amount of Congo red absorbance into the colony indicates an increased amount of cellulose production. For all of the in vitro assays, cultures were grown in the test media amended with 1 mM IPTG. Results represent the means of three biological replications and error bars represent the standard deviations. Different letters indicate significant differences (p < .05) using Tukey's HSD test. The assays were done at least three times with similar results
2.2. RprA regulates the promoter activity of virulence factor genes
To examine how RprA affected the transcriptional activity of virulence factor genes, we generated green fluorescent protein (gfp) transcriptional fusion reporter constructs of amsG, the first gene of the amylovoran biosynthetic gene operon, lsc, the levansucrase gene, and representative T3SS genes including hrpL, hrpA, and dspE. Compared with the empty vector strain, the rprA overexpression strain exhibited significantly higher promoter activity of amsG and of all of the T3SS genes examined (Figure 3). However, the promoter activity of lsc in the rprA overexpression strain was significantly lower; this was consistent with observation of reduced levansucrase activity in this strain compared with the control (Figure 3).
FIGURE 3.
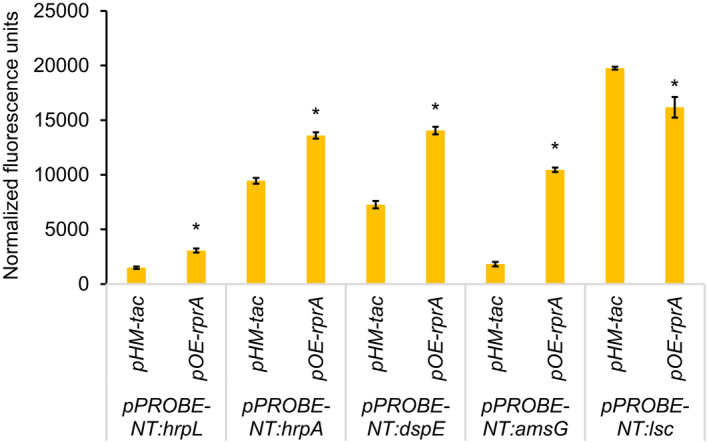
RprA regulates the promoter activity of Erwinia amylovora virulence factor genes. Relative fluorescence units of the indicated transcriptional fusion construct in Ea1189(pHM‐tac) and Ea1189(pOE‐rprA) were measured using a Tecan spectrophotometer followed by normalization with the corresponding OD600 values. IPTG at 1 mM was amended into the medium to induce rprA overexpression. Results represent the means of three biological replications and error bars represent the standard deviation of the means. Asterisks indicate significant difference (p < .05) using Student's t test. The assays were done three times with similar results
2.3. RprA regulates hrpS at a posttranscriptional level
To identify possible direct targets of RprA, we conducted a genome‐wide prediction of the targets of RprA using TargetRNA2 (Kery et al., 2014), which employs structural accessibility and sequence conservation for sRNA target screening. This analysis yielded prediction of 31 putative targets of RprA (Table S1). Of interest, the enhancer‐binding protein gene hrpS was predicted as a target of RprA. RprA was predicted to interact with the region from −20 to −7 bp relative to the translational start site of hrpS (Figure 4a). Two transcriptional start sites of hrpS were previously identified at 129 and 227 nucleotides (nt) upstream from its start codon, respectively (Lee & Zhao, 2018). To determine whether RprA affected hrpS mRNA posttranscriptionally, we constructed pxg‐20:hrpS129 and pxg‐20:hrpS227, which fused the 5′ untranslated region in 129 nt or 227 nt and the first 30 codons of hrpS in‐frame with gfp in pXG‐20. We found that overexpression of rprA resulted in significantly higher fluorescence in E. amylovora cells carrying pxg‐20:hrpS129, but no significant difference was observed in cells carrying the pxg‐20:hrpS227 construct (Figure 4b).
FIGURE 4.

RprA regulates hrpS at the posttranscriptional level. (a) Proposed interaction region between RprA and hrpS mRNA. (b) Relative fluorescence units of the indicated translational fusion in Ea1189(pHM‐tac) and Ea1189(pOE‐rprA) were measured using a Tecan spectrophotometer followed by normalization of their corresponding OD600 values. Results represent the means of three biological replications and error bars represent the standard deviations. Asterisks indicate significant difference (p < .05), whereas n.s. indicates no significant difference using Student's t test. The assays were done three times with similar results
2.4. rprA expression is activated by native and environmental cues
A previous study in E. coli showed that rprA was transcriptionally regulated by the Rcs phosphorelay, which is composed of the histidine kinase RcsC and the response regulator RcsB (Majdalani et al., 2002). We identified an “Rcs box,” the consensus binding site of RcsB, immediately upstream of the −35 region of the promoter of rprA (Figure S1), suggesting that rprA in E. amylovora may also be subject to the regulation by the Rcs phosphorelay. As expected, compared with that in the WT strain, the expression of rprA was reduced approximately two‐fold in the rcsB deletion mutant, Ea1189ΔrcsB (Figure 5a). Introduction of the complementation construct pJP‐rcsB completely restored the rprA expression defect in Ea1189ΔrcsB, which was four‐fold higher compared with the WT strain (Figure 5a). We also measured expression of rprA in Ea1189ΔrcsB(prcsB‐D56E) and Ea1189ΔrcsB(prcsB‐K154Q), which encoded RcsB with a D56E (aspartate to glutamic acid) substitution or a K154Q (lysine to glutamine) substitution to mimic the constitutively phosphorylated or the acetylated state of RcsB, respectively (Ancona et al., 2015; Hu et al., 2013). We showed that rprA expression was further elevated in Ea1189ΔrcsB(prcsB‐D56E) compared with Ea1189ΔrcsB(prcsB), whereas its expression in Ea1189ΔrcsB(prcsB‐K154Q) was reduced to a level comparable to that of Ea1189ΔrcsB (Figure 5a). Together, these results indicate that rprA expression is positively and negatively modulated by the phosphorylation and the acetylation of RcsB, respectively.
FIGURE 5.
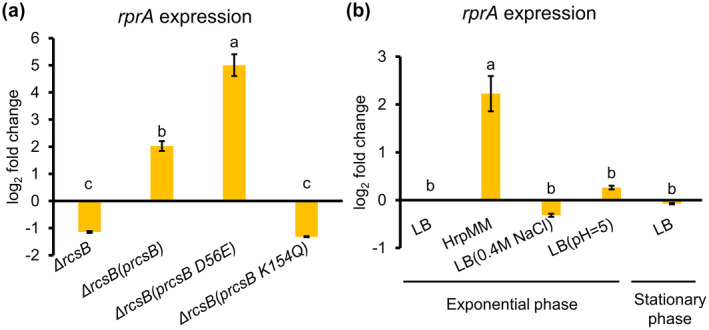
Activation of RprA by native and environmental cues. (a) Regulation of RprA expression by the Rcs phosphorelay system. (b) Effect of environmental stressors on rprA expression. Expression levels of rprA were quantified using quantitative reverse transcription PCR (RT‐qPCR), and fold changes were calculated using the 2−ΔΔ C t formula. The housekeeping gene recA was used as an endogenous control. Error bars indicate standard deviations of the means. Different letters indicate significant differences (p < .05) using Tukey's HSD test
To examine whether expression of rprA could be affected by external stressors, E. amylovora Ea1189 WT cultures were grown in Luria Bertani (LB) broth to exponential phase (OD600 = 0.5) and then challenged with either low pH (pH 5) or osmotic shock (0.4 M NaCl), or immersed in HrpMM medium, a low pH and low nutrient medium that mimics the conditions of the plant apoplast (Huynh et al., 1989). We also examined how the population density of E. amylovora (exponential phase versus stationary phase) may affect rprA expression. We did not observe any significant change of rprA expression in E. amylovora cells undergoing low pH stress or osmotic shock or in high population density (Figure 5b). However, rprA expression was significantly induced in E. amylovora cultures grown in HrpMM (Figure 5b), suggesting that rprA may be induced during host infection.
To enable in vivo examination of rprA transcriptional activity, we generated the dual reporter construct pNptII‐gfp‐rprA‐mCherry, which allowed cellular gfp expression in a constitutive manner and mCherry under the control of the native promoter of rprA. As observed using a confocal microscope, the transcriptional activity of rprA was very low in Ea1189(pNptII‐gfp‐rprA‐mCherry) cultures grown in LB broth (Figure 6a), as indicated by the very dim fluorescence from mCherry. In immature pear flesh tissue that was inoculated with Ea1189(pNptII‐gfp‐rprA‐mCherry), E. amylovora cells were shown to colonize the apoplast region of the flesh tissue (green fluorescence) and also exhibited strong levels of red fluorescence (Figure 6a). In pear ooze that contained a large population of E. amylovora cells emerging from the infection site of the inoculated pears, E. amylovora cells also exhibited very high levels of red fluorescence compared with that in LB broth. Through quantitative reverse transcription PCR (RT‐qPCR), we confirmed that rprA expression increased by c.30‐fold in E. amylovora cells from pear ooze compared with that grown in LB broth (Figure 6b). We also showed up‐regulation of the T3SS genes and the amylovoran biosynthetic gene amsG, and down‐regulation of the levansucrase gene lsc in E. amylovora cells from pear ooze (Figure 6b). Taken together, our results showed that rprA expression was activated in vitro by conditions mimicking the plant apoplast environment or in vivo during host infection.
FIGURE 6.

In vivo activation of RprA during host infection. (a) Confocal observation of rprA promoter activity in Erwinia amylovora Ea1189(pNptII‐gfp‐rprA‐mCherry) in LB medium and in flesh and ooze of inoculated immature pears. Gfp (ex/em = 488 nm/510 nm) is expressed constitutively, whereas mCherry (ex/em = 587 nm/610 nm) is expressed under the control of the promoter of rprA in Ea1189(pNptII‐gfp‐rprA‐mCherry) cultures. Images were captured through sequential scanning using a FluoView 1000 (Olympus) laser scanning confocal microscope. (b) Expression of rprA and several virulence factor genes in E. amylovora Ea1189 cells emerged in ooze from the inoculated immature pears and from cells that were grown overnight in LB medium. To ensure representativity of gene expression levels in E. amylovora cells from pear ooze, ooze from groups of six of the 18 inoculated pears were pooled together as one biological replication (labelled as “Ooze 1”, “Ooze 2”, and “Ooze 3”). Gene expression levels were quantified through quantitative reverse transcriptionPCR and fold changes were calculated using the 2−ΔΔ C t formula. The housekeeping gene recA was used as an endogenous control. Error bars indicate standard deviations of the mean within each biological replication
2.5. rprA inhibits biofilm formation and activates dispersal of biofilm cells
RprA activation induced amylovoran production, which promotes formation of biofilms; activation of RprA also induced flagellar‐dependent motility and inhibited production of levan and cellulose, which promotes dispersal of biofilm cells. To resolve this paradox, we examined the total effects of RprA on formation and dispersal of biofilm cells. We quantified biofilm formation in isopropyl‐β‐D‐1‐thiogalactopyranoside (IPTG)‐supplemented cultures of Ea1189(pHM‐tac) and Ea1189(pOE‐rprA) through a microtitre plate assay. Compared with the empty vector control, Ea1189(pOE‐rprA) cultures formed significantly less biofilm, suggesting an overall negative effect of RprA on biofilm formation (Figure 7a). We then questioned how induction of rprA affects biofilm dispersal. To investigate this, we first let strains of Ea1189(pHM‐tac) or Ea1189(pOE‐rprA) form biofilms on polystyrene beads without addition of any IPTG into the medium. Washed beads covered with biofilms were then transferred to fresh medium containing 1 mM IPTG. Cells that dispersed into the medium were periodically quantified through dilution plating. We showed that overexpression of rprA did not affect the number of dispersed cells in the first hour after induction (Figure 7b). However, significantly more cells of Ea1189(pOE‐rprA) were dispersed in the IPTG induction condition compared with Ea1189(pHM‐tac) from 2 to 5 hr after IPTG addition, and the differences were greater as the experiments continued (Figure 7b). These results indicated that induction of rprA negatively impacts biofilm formation and positively impacts dispersal of E. amylovora cells from biofilms. A working model for the functions of RprA is proposed (Figure 8).
FIGURE 7.

RprA negatively affects biofilm formation and activates biofilm dispersal in vitro. (a) Biofilm formation of Erwinia amylovora Ea1189(pHM‐tac) and Ea1189(pOE‐rprA) cultures. Cultures at OD600 = 1.0 were resuspended in 0.5 × LBmedium containing 1 mM IPTG and were inoculated into acetone‐etched microtitre plates for 48 hr. Biofilms were quantified through a crystal violet (CV) staining assay at the absorbance of 594 nm (A594). (b) Temporal dispersal of biofilm cells. Polystyrene beads (7 mm) were immersed into E. amylovora cultures in 0.5 × LB medium without any IPTG for 48 hr. Beads covered by biofilm were washed and dipped into fresh 0.5 × LB medium with 1 mM IPTG. Planktonic cultures were periodically withdrawn, and cfus were determined using dilution plating (line graph). To count the number of cells covered on the beads before and after dispersal, biofilm‐covered beads were immersed into 0.5 × phosphate‐buffered saline and were sonicated for 5 min to release the attached cells for cell count through dilution plating (bar graph). The assays were done three times with similar results
FIGURE 8.

Proposed model of the functions of the Hfq‐dependent sRNA RprA in modulating virulence factors and systemic infection of Erwinia amylovora. After infection of leaves at shoot tips, E. amylovora cells form biofilms within xylem vessels of the host plants. Expression of rprA is activated in E. amylovora cells upon perception of environmental cues, including RcsB phosphorylation and the host apoplast environment. Through regulation of virulence factor genes at the transcriptional (plain font) or posttranscriptional (bold font) level, RprA positively regulates amylovoran exopolysaccharide and the type III secretion system (T3SS), the two major pathogenicity factors of E. amylovora, and flagellar‐dependent motility, which has a known negative correlation with biofilm formation in E. amylovora; RprA also negatively affects production of levan and cellulose, which are constituents of the matrix of E. amylovora biofilms. The total effects of RprA promote the transition of biofilm cells of E. amylovora in the sessile mode of growth within host xylem tissue to the planktonic mode of growth; this consequently facilitates the further systemic infection of E. amylovora cells within the vascular or the cortical parenchyma tissue of host plants
3. DISCUSSION
Our results demonstrate that the Hfq‐dependent sRNA RprA regulates a varied group of virulence factors of E. amylovora that impact pathogenesis and systemic movement through the apple host. The regulatory impact of RprA on virulence would seem to mostly occur via direct interactions with mRNAs of transcriptional regulators of the associated virulence factor. For example, we demonstrated that RprA activates hrpS mRNA at a posttranscriptional level, probably via a direct interaction with hrpS mRNA. As the enhancer‐binding protein HrpS functions to activate the transcription of hrpL, encoding the alternative sigma factor HrpL, which is required for the transcription of all other genes within the Hrp regulon, the translational stimulation of hrpS due to rprA overexpression therefore explained its positive effects on the promoter activity of the downstream T3SS genes, including hrpL, hrpA, and dspE. Of note, RprA appeared to only affect the 129‐nt 5′ UTR but not the 227‐nt 5′ UTR of hrpS, suggesting that the longer 5′ UTR of hrpS forms a distinct structure that is inaccessible to RprA and may or may not require an additional but yet unidentified factor(s) to regulate its translation.
We also found that RprA is a positive regulator of amylovoran production through a positive effect on the promoter of amsG, the first gene of the amylovoran biosynthetic gene operon, and also has a negative effect on the expression of the levansucrase gene lsc. Transcriptional regulation of the 12‐gene ams operon, encoding amylovoran biosynthesis, is highly complex. This probably occurs for many reasons, including that the amylovoran EPS is required for pathogenicity (Norelli et al., 2003) and is the most important EPS component of E. amylovora biofilms (Koczan et al., 2009), and also reflects the inverse regulation of biofilm formation and expression of the T3SS (Edmunds et al., 2013). Positive transcriptional regulators of the ams operon include the Rcs phosphorelay via direct interaction with an Rcs box (Wang et al., 2009, 2012b; Zhao et al., 2009b); ams operon expression is also positively regulated by the second messenger molecule cyclic di‐GMP via an as yet unknown transcriptional regulator (Edmunds et al., 2013). Negative transcriptional regulators of the ams operon include the EnvZ/OmpR and GrrA/GrrS two‐component systems (Zhao et al., 2009b), and AmyR, a member of the enterobacterial YbjN family (Wang et al., 2012a). Thus, there are many potential targets that RprA might interact with to positively impact transcription of the ams operon.
It is noteworthy that we determined most functions of RprA from the overexpression studies but not from the mutagenesis studies. Compared with the WT strain, the Ea1189ΔrprA mutant had negligible effects on swimming motility, levansucrase activity, and cellulose production. This suggests that the basal level of RprA in E. amylovora cells grown in a rich medium is too low to have strong effects on the measured phenotypes. Indeed, the promoter activity of rprA was very low in E. amylovora cells grown in LB medium, as indicated by the dim red fluorescence using the dual reporter system. These observations are reminiscent of previous studies of RprA in E. coli. Through assays of an rprA‐lacZ fusion or northern blot, several studies have shown that rprA is nearly undetectable in WT E. coli cells grown in a rich medium (Madhugiri et al., 2010; Majdalani et al., 2001, 2005). In line with this, known targets of RprA in E. coli, including rpoS, csgD, and ydaM, were consistently identified through overexpression of this sRNA, whereas a rprA knockout mutation hardly affected any of these targets (Majdalani et al., 2002; Mika et al., 2012). Therefore, a basal level of RprA in the bacteria grown in a rich medium functions minimally in affecting its targets, which are nevertheless strongly perturbed by overproduction of this sRNA.
We demonstrated that rprA expression was under the tight regulation of the Rcs phosphorelay, as rprA expression was reduced by about one half in the Ea1189ΔrcsB mutant compared with the WT Ea1189 strain. A similar level of decrease of the transcriptional activity of rprA in E. coli was previously reported in a corresponding ΔrcsB mutant (Majdalani et al., 2002). In E. coli, rprA was shown to be positively regulated by RcsB when the protein was phosphorylated, and negatively regulated by RcsB when the protein was acetylated (Hu et al., 2013; Majdalani et al., 2002; Szczesny et al., 2018), which is consistent with our observation of the full activation of rprA expression in Ea1189ΔrcsB(prcsB‐D56E) but loss of rprA expression in Ea1189ΔrcsB(prcsB‐K154Q). We also examined rprA expression levels in E. amylovora cultures under conditions that are known to strongly activate rprA expression in S. enterica serovar Typhimurium or E. coli, including low pH stress, osmotic shock, and high population density (Madhugiri et al., 2010; Srikumar et al., 2015). None of these conditions significantly altered rprA expression in E. amylovora. Nevertheless, we found that rprA expression was strongly stimulated in E. amylovora cultures grown in HrpMM medium, a low nutrient and low pH medium that mimics the plant apoplast (Huynh et al., 1989), and was induced to a greater extent during infection of immature pears. Of note, we also observed differential expression of several virulence factor genes in E. amylovora cells from ooze that emerged from the inoculated immature pears in the same direction as in E. amylovora cells overexpressing rprA in vitro, suggesting the critical roles of RprA during E. amylovora pathogenesis on immature pears. Thus, although E. amylovora, E. coli, and S. enterica serovar Typhimurium are all phylogenetically closely related members of the Enterobacteriaceae family, rprA regulation responds to different environmental cues and is probably optimized for the regulation of specific virulence traits in each of these organisms.
The final stage of biofilm development is dispersal, which allows subpopulations of cells to be detached from the biofilm and resume the planktonic mode of growth (Koczan et al., 2011; Rumbaugh & Sauer, 2020). Although biofilm dispersal in E. amylovora, to the best of our knowledge, has not been previously genetically or phenotypically characterized, dispersal is evidently an important step in E. amylovora pathogenesis from several lines of evidence: First, E. amylovora is capable of migrating systemically within host xylem (Thomson, 2000), which clearly requires cells in the sessile mode of growth to switch back to the planktonic mode of growth to move to new infection sites. Using scanning electron microscopy analyses of longitudinal sections of the central vein of infected apple leaves, we previously visualized the discontinuous aggregation of E. amylovora microcolonies (Koczan et al., 2011), suggesting that a cycle of dispersal and re‐establishment of biofilms contributes to the systemic movement of E. amylovora in apple leaf xylem. In addition, masses of E. amylovora cells have been demonstrated to break out of xylem vessels to reach the surrounding intercellular spaces of the cortical parenchyma cells (Bogs et al., 1998), which is a further example of dispersal from the biofilm and transitioning back to T3SS‐mediated pathogenesis.
Our evidence suggests that RprA activates biofilm dispersal in E. amylovora and regulates several virulence traits associated with a transition from biofilm development to T3SS‐mediated pathogenesis. The induction of motility has been observed during biofilm dispersal in bacteria such as E. coli and Pseudomonas aeruginosa (Jackson et al., 2002; Purevdorj‐Gage et al., 2005; Sauer et al., 2002), and we found that motility is also positively regulated by RprA in E. amylovora. Cellulose and levan are important EPS constituents of the biofilm matrix of E. amylovora (Castiblanco & Sundin, 2018; Koczan et al., 2009), thus RprA‐mediated down‐regulation of both cellulose and levan production also suggests a cellular transition away from biofilm development. In contrast to the reduction in cellulose and levan we observed when rprA was overexpressed in E. amylovora Ea1189, amylovoran production was significantly increased (Figure 2a). This seemingly paradoxical observation can be resolved by the knowledge that amylovoran is a pathogenicity factor in E. amylovora (Bellemann & Geider, 1992), and this further indicates that amylovoran production of peripheral cells that are dispersing from biofilms may not be at the level necessary for continuing planktonic stage T3SS‐mediated pathogenesis.
In summary, we demonstrated that the Hfq‐dependent sRNA RprA exhibits important regulatory roles in orchestrating virulence factors of E. amylovora and affects transcriptional or posttranscriptional activity of the virulence factor genes. We showed that rprA is maintained at a very low basal level in a rich medium and can be activated by RcsB phosphorylation and by the host apoplast environment. Finally, we provided evidence that RprA plays an important role in the dispersal of E. amylovora cells from biofilms. This study sheds light on future mechanistic research into this important biological process during pathogenesis that has not yet been characterized in most phytopathogenic bacteria.
4. EXPERIMENTAL PROCEDURES
4.1. Bacterial strains, plasmids, and media
The E. amylovora strains, plasmids, and oligonucleotide primers used in this study are listed in Table 1. All strains were routinely maintained in 15% glycerol at −80 °C. Single colonies were grown overnight in LB broth at 28 °C with shaking at 200 rpm for 20 hr. The following antibiotics were amended to media as needed: ampicillin (100 μg/ml), chloramphenicol (30 μg/ml), gentamicin (15 μg/ml), and kanamycin (50 μg/ml).
TABLE 1.
Bacteria strains or plasmids used in this study
| Strain or plasmid | Genotype | Reference | |
|---|---|---|---|
| Strains | Ea1189 | Wild type | Yu et al. (2020) |
| Ea1189ΔrprA | rprA deletion mutant, CmR | Zeng et al. (2013) | |
| Ea1189ΔrcsB | rcsB deletion mutant, CmR | Wang et al. (2009) | |
| Plasmids | pBBR1MCS5 | Broad‐host‐range cloning vector, GmR | Kovach et al. (1995) |
| pJP‐rprA | A region spanning the rprA gene and its corresponding native promoter in pBBR1MCS‐5; GmR | This study | |
| prcsB | A region spanning the rcsBD operon along with its native promoter region in pBBR1MCS5; GmR | This study | |
| prcsB‐D56E | prcsB with an allele change resulting in the amino acid substitution of aspartic acid to glutamic acid at codon 56 in RcsB | This study | |
| prcsB‐K154Q | prcsB with an allele change resulting in the amino acid substitution of lysine to glutamine at codon 154 in RcsB | This study | |
| pHM‐tac | IPTG‐inducible sRNA overexpression vector, ApR | Park et al. (2013) | |
| pOE‐rprA | pHM‐tac::rprA; overexpression vector; ApR | This study | |
| pPROBE‐NT | Broad‐host‐range promoter‐probe vector; KmR | Miller et al. (2000) | |
| pPROBE‐hrpS | pPROBE‐NT::hrpS; native promoter of hrpS in pPROBE‐NT; KmR | This study | |
| pPROBE‐hrpL | pPROBE‐NT::hrpL; native promoter of hrpL in pPROBE‐NT; KmR | This study | |
| pPROBE‐hrpA | pPROBE‐NT::hrpA; native promoter of hrpA in pPROBE‐NT; KmR | This study | |
| pPROBE‐dspE | pPROBE‐NT::dspE; native promoter of dspE in pPROBE‐NT; KmR | This study | |
| pPROBE‐amsG | pPROBE‐NT::amsG; native promoter of amsG in pPROBE‐NT; KmR | This study | |
| pPROBE‐lsc | pPROBE‐NT::lsc; native promoter of lsc in pPROBE‐NT; KmR | This study | |
| pxg‐20 | Broad‐host‐range translational fusion vector; CmR | Urban & Vogel (2007) | |
| pxg‐20:hrpS129 | 5′ UTR (129 nt) of hrpS and 90 nt into the coding region of hrpS in pxg‐20; CmR | This study | |
| pxg‐20:hrpS227 | 5′ UTR (227 nt) of hrpS and 90 nt into the coding region of hrpS in pxg‐20; CmR | This study |
4.2. DNA manipulations
To generate the pJP‐rprA construct for rprA complementation, the rprA gene along with its native promoter region was cloned into the low‐copy plasmid pBBR1MCS5. To generate the rprA overexpression construct, the rprA full‐length gene sequence was cloned into pHM‐tac immediate downstream of the isopropyl β‐d‐1‐thiogalactopyranoside (IPTG)‐inducible tac promoter. To generate the construct prcsBD for rcsB complementation, the rcsBD operon along with its native promoter region was cloned into pBBR1MCS5. The prcsB‐D56E and prcsB‐K154Q constructs, which allow expression of rcsB with a D56E substitution or a K154Q substitution, respectively, were generated through site‐directed mutagenesis using the QuikChange Lightning kit. To generate the transcriptional fusion constructs, including pPROBE‐NT:hrpL, pPROBE‐NT:hrpA, pPROBE‐NT:dspE, pPROBE‐NT:amsG, and pPROBE‐NT:lsc, the promoter region of the corresponding genes (c.500‐bp amplicons upstream of the start codon) were cloned immediately upstream of the promoterless gfp gene in pPROBE‐NT (Miller et al., 2000). To generate the translation fusion constructs pxg‐20:hrpS129 and pxg‐20:hrpS227, the 5′ UTR regions of hrpS were amplified from the two transcriptional start sites to 90 nt into the coding region and were cloned in‐frame with gfp in pXG‐20 (Urban & Vogel, 2007). The transcription start sites of hrpS were identified previously (Lee & Zhao, 2018). To generate pNptII‐gfp‐rprA‐mCherry, the Dickeya dadantii hrpA promoter region in nptII‐gfp‐hrpA‐mCherry (Cui et al., 2018) was replaced with the promoter region of E. amylovora rprA. Constructs were cloned using the standard ligation‐dependent approach (Sambrook, 2001) or a ligation‐independent cloning approach (Li et al., 2011). The strains or plasmids used in this study are listed in Table 1. The oligonucleotide primers used are listed in Table 2. Constructs were transformed into E. coli Turbo cells using transformation and storage solution (TSS) (Chung et al., 1989) and/or into E. amylovora through electroporation.
TABLE 2.
Oligonucleotide primers used in this study
| Primer name | Sequence (5′‐3′) | Purpose |
|---|---|---|
| com_rprAF | TAGGAATTCGCAATAATCTGGCTTTACTGGA | Primers used for rprA complementation |
| com_rprAR | ATATCTAGATCGGTTCACCGATCGTCC | |
| oe_rprAF | GACGAATTCAGGATTTGAAATCTTCCCACTGA | Primer used for rprA overexpression |
| oe_rprAR | GACTCTAGACCGATCGTCCTTTTTTAAGGGC | |
| com_RcsBD_F | CCCGACTGGAAAGCGGGCAGTGCTAGCACAATTCACAAGGTTGG | Primer used for rcsB complementation |
| com_RcsBD_R | GTTGCGTCGCGGTGCATGGCTCCTAATGAACTGCCGCTACT | |
| backbone_pBBR1MCS5_F | CACTGCCCGCTTTCCAGTCGGG | |
| backbone_pBBR1MCS5_R | CCATGCACCGCGACGCAAC | |
| RcsB_D56E_F | CCAGGCATCGAAAGCTCGGTGACCAGCAC | Primers used for site‐directed mutagenesis of prcsB |
| RcsB_D56E_R | GTGCTGGTCACCGAGCTTTCGATGCCTGG | |
| RcsB_K154Q_F | GCGCAGAACTTCGCTCTCCTGCGGTGACAAACGCTTATC | |
| RcsB_K154Q_R | GATAAGCGTTTGTCACCGCAGGAGAGCGAAGTTCTGCGC | |
| hrpS_tsc_F | CGACCTGAATGGAAGCCGGCAGATTGTCTTTGCCGAGTACA | Primers used for transcriptional fusion constructions |
| hrpS_tsc_R | GAGCTCGGTACCCGGGGATCCTCAAAAAATTACCCCTGCCCTATC | |
| tsc_hrpL_F | CGACCTGAATGGAAGCCGGCTAAACGCGCATGCTGCGGAT | |
| tsc_hrpL_R | GAGCTCGGTACCCGGGGATCCTCGGCTTGCTCCGTTACTAAATCA | |
| tsc_dspE_F | CGACCTGAATGGAAGCCGGCCTGACTGTCAGACTGCGGAGTGG | |
| tsc_dspE_R | GAGCTCGGTACCCGGGGATCCTCGACCCGTTGCCCCCACCCTCT | |
| tsc_hrpA_F | CGACCTGAATGGAAGCCGGCCTGGTGAAGGCGCACCGGGAT | |
| tsc_hrpA_R | GAGCTCGGTACCCGGGGATCCTCATTAATCTCTCCAATTATTGAGGTTGTGTTCC | |
| tsc_lsc_F | CGACCTGAATGGAAGCCGGCAAGTGCACCTCCGCAAGGT | |
| tsc_lsc_R | GAGCTCGGTACCCGGGGATCCTC AAATATCCTCACAGGTTATTTCG | |
| backbone_pPROBE‐NT_F | GAGGATCCCCGGGTACCGAGCTC | |
| backbone_pPROBE‐NT_R | GCCGGCTTCCATTCAGGTCG | |
| tsc_amsG_F | CGACCTGAATGGAAGCCGGCCCTTAATGAGATGGTTGATAAATCCAT | |
| tsc_amsG_R | GAGCTCGGTACCCGGGGATCCTCAATTAGCTCTTAATTTTATCTCAGG | |
| tln_dspE_F | GAGATTGACATCCCTATCAGTGATAGAGATACTGAGCACAAAAATATCTAATGTTTACGGCAGAGG | Primers used for translational fusion constructs |
| tln_dspE_R | AGTTCTTCTCCTTTGCTCATGAATTCGCCAGAACCCTGCTGTAAGGCAACACC | |
| tln_hrpS129_F | GAGATTGACATCCCTATCAGTGATAGAGATACTGAGCACACAGCGTAAACTCAGAGTAAATA | |
| tln_hrpS227_F | GAGATTGACATCCCTATCAGTGATAGAGATACTGAGCACAATGTAGGGTAATCCCTACATTGC | |
| tln_hrpS_R | AGTTCTTCTCCTTTGCTCATGAATTCGCCAGAACCGATATCGATGGGTTGTTCTTCTGT | |
| backbone_pxg20_F | GAAGGTTCTGGCGAATTCATGAGCAAAGGAGAAGAACT | |
| backbone_pxg20_R | TGTGCTCAGTATCTCTATCACTGATAGGGATGTCAATCTC | |
| NptII‐gfp‐rprA‐mCherry_F | GTTGGATCCGCAATAATCTGGCTTTACTG | Primers used for generating pNptII‐gfp‐rprA‐mCherry |
| NptII‐gfp‐rprA‐mCherry_R | GGTGAGCTCGTAACCATAGTATGAAAAGGTG |
4.3. Bioinformatics
The secondary structure of RprA was predicted using the minimum free energy model of RNAfold (http://rna.tbi.univie.ac.at/cgi‐bin/RNAWebSuite/RNAfold.cgi). The genome‐wide targets of RprA were predicted using TargetRNA2 (http://cs.wellesley.edu/~btjaden/TargetRNA2/) with the default setting.
4.4. Quantification of amylovoran
The concentration of amylovoran was determined through a turbidity‐based assay using cetylpyrimidinium chloride (CPC) with modifications (Bellemann et al., 1994). Briefly, overnight E. amylovora cultures grown in LB medium were washed twice and resuspended in modified basal medium A (MBMA; Edmunds et al., 2013) supplemented with 1% galactose. Cultures were grown for 24 hr at 28 °C with shaking at 200 rpm. After centrifugation at 16,000 × g for 2 min, supernatant was harvested and mixed with 50 μl of 50 mg/ml CPC (Sigma‐Aldrich) per millilitre of culture supernatant. The mixtures were incubated at room temperature for 5 min and their turbidity at OD600 was measured using a spectrophotometer (Tecan) followed by normalization with the OD600 of the cultures.
4.5. Swimming motility assay
Swimming motility was examined following the method of Edmunds et al. (2013) with modifications. Briefly, 2 µl of overnight E. amylovora culture was stab‐inoculated into 0.3% agar LB plates and the inoculated plates were inoculated at 28 °C for 48 hr without any agitation. The radius of the motility area was determined for the subsequent statistical analysis.
4.6. Cellulose assay
Cellulose biosynthesis was assessed following a previously described method (Castiblanco & Sundin, 2018). In brief, 5 µl of E. amylovora overnight culture was spotted on NaCl‐free LB plates supplemented with Congo red (40 µg/ml). The inoculated plates were incubated for 48 hr at 28 °C without shaking. Red coloration of the E. amylovora colony is indicative of the production of cellulose that binds to Congo red.
4.7. Levansucrase activity
Levansucrase activity was quantified as described previously (Schachterle & Sundin, 2019). In brief, supernatants of E. amylovora overnight cultures were mixed with 0.5 × phosphate‐buffered saline (PBS) containing 2 M sucrose in a 1:1 ratio. The mixtures were incubated at 37 °C for 4 hr without shaking. The resultant turbidity from levan production, which is catalysed by the levansucrase enzyme, was measured at OD600 using a spectrophotometer (Tecan) followed by normalization with the OD600 values of the cultures.
4.8. Hypersensitive response assay
The HR assay followed the protocol of Zeng and Sundin (2014). In brief, overnight E. amylovora cultures were harvested, washed, and adjusted to the optical density OD600 = 0.05 in 0.5 × PBS. Around 100 μl cell suspension was infiltrated into N. benthamiana leaves of 10‐week‐old plants with a needleless syringe. The HR symptom was observed and image‐captured 16 hr postinfiltration.
4.9. Confocal microscopy
A FluoView 1000 (Olympus) laser scanning confocal microscope was used for examining E. amylovora cells expressing the nptII‐gfp‐rprA‐mCherry construct, which expressed gfp in a constitutive manner and mCherry under the control of the native promoter of rprA. To analyse rprA promoter activity in vitro, overnight cultures of E. amylovora Ea1189(nptII‐gfp‐rprA‐mCherry) were washed and resuspended in fresh LB medium with the OD600 adjusted to 1.0. To measure in vivo rprA promoter activity in pear ooze or pear flesh, E. amylovora Ea1189(nptII‐gfp‐rprA‐mCherry) cultures were inoculated on immature pears as described previously (Edmunds et al., 2013), except that the starting amount of inoculum was approximately 2 × 105 cfu to accelerate the infection and ooze emergence. Pear ooze was harvested using a sterile inoculation loop and was resuspended in 0.5 × PBS to OD600 = 1.0 immediately before imaging. Infected pear flesh tissue was dissected using a sterile razor blade immediately before imaging. The laser at 488 nm and the SDM560‐BA505‐525 emission filter were used for capturing the gfp fluorescence. The laser at 561 nm and the SDM640‐BA560‐620 emission filter were used for capturing the mCherry fluorescence. A sequential imaging recording method was used to avoid crosstalk between the fluorochromes.
4.10. Total RNA extraction and RT‐qPCR
Overnight E. amylovora cultures were washed and diluted to OD600 = 0.05 in fresh LB broth. The cultures were grown at 28 °C with shaking until the optical density reached OD600 = 0.5, corresponding to the exponential stage of bacterial growth. To examine the effect of environmental stressors on rprA expression, cultures of E. amylovora Ea1189 at OD600 = 0.5 were washed and resuspended in the same volume of the following media for 2 hr: LB medium, LB medium (pH 5.1), LB medium amended 0.4 M NaCl, and HrpMM (Huynh et al., 1989). To examine the effect of growth stage on rprA expression, late stationary phase cultures of E. amylovora were prepared by allowing cultures in the exponential phase to continue to grow for another 16 hr. To compare the in vivo and the in vitro expression of rprA and virulence factor genes in E. amylovora, E. amylovora Ea1189 was either inoculated into immature pears or grown overnight following the same procedures used for the confocal microscopic studies. To obtain enough E. amylovora cells from pear ooze and also to control the possible variations of E. amylovora gene expression due to the physiological differences of the pears inoculated, ooze from six inoculated pears was pooled together as one biological replicate and three biological replicates were investigated in this study, “Ooze 1”, “Ooze 2”, and “Ooze 3.” Crude total RNA was extracted following a previously reported method (Rivas et al., 2001). Crude total RNA was purified using the RNA Clean & Concentrator‐25 kit (Zymo Research) following the manufacturer’s instructions. Contaminating genomic DNA was eliminated using the TURBO DNA‐free kit (Thermo Fisher Scientific) according to the manufacturer's instructions. First‐strand cDNA was synthesized with the High Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific) according to the manufacturer's instructions. Expression levels of RprA were quantified routinely using a StepOne Plus Real‐Time PCR system (Applied Biosystems) (Zeng et al., 2013). The reference gene recA was used as an endogenous control (Zeng et al., 2013). Relative expression values were calculated through the 2−ΔΔCt method.
4.11. Biofilm assay
Overnight cultures of E. amylovora Ea1189(pHM‐tac) and Ea1189(pOE‐rprA) were washed and resuspended in 0.5 × LB medium with the OD600 adjusted to 1.0. To enhance attachment of E. amylovora cells, wells of 96‐well round‐bottom microplates were etched with 200 µl acetone for 20 s to increase the roughness of the surface (Davies & Marques, 2009). After complete drying of the plates, resuspended E. amylovora cultures in 200 µl per well were inoculated and incubated at room temperature for 48 hr with light horizontal shaking. After depletion of planktonic cultures, biofilm cells were stained by adding 250 µl of 10% crystal violet solution for 1 hr. Stained plates were washed twice by water and dried. A total of 300 μl of destaining solution (40% methanol and 10% acetic acid) was added into each well incubated for 1 hr at room temperature with light shaking. The A594 values of the suspensions were measured using a Tecan spectrophotometer.
4.12. Biofilm dispersal assay
Two millilitres of washed overnight cultures at OD600 of 1 in 0.5 × LB medium were inoculated into 12‐well plates without adding any IPTG. One polystyrene bead (7 mm) was added to each inoculated well. The inoculated plates were incubated at room temperature for 48 hr with light shaking to allow even formation of biofilms on the beads. Each bead covered with biofilm cells was washed six times by 10 ml of 0.5 × PBS to remove any planktonic cells on the surface of the bead. Washed beads were transferred to wells in 2 ml of fresh 0.5 × LB medium and 1 mM IPTG and incubated at room temperature without any shaking. Dispersed cells were quantified by withdrawing 10 µl of the culture suspension at 1‐hr intervals, and the cfus were determined by dilution plating. To count the starting number of biofilm cells on the beads, beads with biofilm cells attached were added into 1.7‐ml Eppendorf centrifuge tubes containing 1 ml of 0.5 × PBS followed by sonication for 5 min to release the attached cells. The cfus of the suspension were counted through dilution plating.
4.13. Statistical analyses
Results represent the means of at least three replications, and error bars represent the standard deviations. Statistical analyses of Student's t test or Tukey's HSD test were performed using JMP Pro 14 statistical software.
AUTHOR CONTRIBUTIONS
J.P., J.K.S., and G.W.S. designed the experiments. J.P. and J.K.S. performed the experiments. J.P. and G.W.S. wrote the manuscript. All authors contributed to the revisions.
Supporting information
FIGURE S1 The Rcs binding site in the promoter of rprA. The rprA gene sequence is in bold font. The −10 and −35 promoter regions and the “Rcs box” region are underlined
TABLE S1 Genome‐wide prediction of the targets of RprA using TargetRNA2
ACKNOWLEDGEMENTS
This project was supported by funds from the Agriculture and Food Research Initiative Competitive Grants Program grant no. 2015‐67013‐23068 from the USDA National Institute of Food and Agriculture and by Michigan State University AgBioResearch. We declare no conflict of interest.
Peng J, Schachterle JK, Sundin GW. Orchestration of virulence factor expression and modulation of biofilm dispersal in Erwinia amylovora through activation of the Hfq‐dependent small RNA RprA. Mol Plant Pathol. 2021;22:255–270. 10.1111/mpp.13024
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Ancona, V. , Chatnaparat, T. & Zhao, Y. (2015) Conserved aspartate and lysine residues of RcsB are required for amylovoran biosynthesis, virulence, and DNA binding in Erwinia amylovora . Molecular Genetics and Genomics, 290, 1265–1276. [DOI] [PubMed] [Google Scholar]
- Andreassen, P.R. , Pettersen, J.S. , Szczerba, M. , Valentin‐Hansen, P. , Møller‐Jensen, J. & Jørgensen, M.G. (2018) sRNA‐dependent control of curli biosynthesis in Escherichia coli: McaS directs endonucleolytic cleavage of csgD mRNA. Nucleic Acids Research, 46, 6746–6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayot, R.G. & Ries, S.M. (1986) Role of motility in apple blossom infection by Erwinia amylovora and studies of fire blight control with attractant and repellent compounds. Phytopathology, 76, 441–445. [Google Scholar]
- Bellemann, P. , Bereswill, S. , Berger, S. & Geider, K. (1994) Visualization of capsule formation by Erwinia amylovora and assays to determine amylovoran synthesis. International Journal of Biological Macromolecules, 16, 290–296. [DOI] [PubMed] [Google Scholar]
- Bellemann, P. & Geider, K. (1992) Localization of transposon insertions in pathogenicity mutants of Erwinia amylovora and their biochemical characterization. Microbiology, 138, 931–940. [DOI] [PubMed] [Google Scholar]
- Bogdanove, A.J. , Bauer, D.W. & Beer, S.V. (1998) Erwinia amylovora secretes DspE, a pathogenicity factor and functional AvrE homolog, through the Hrp (type III secretion) pathway. Journal of Bacteriology, 180, 2244–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs, J. , Bruchmüller, I. , Erbar, C. & Geider, K. (1998) Colonization of host plants by the fire blight pathogen Erwinia amylovora marked with genes for bioluminescence and fluorescence. Phytopathology, 88, 416–421. [DOI] [PubMed] [Google Scholar]
- Brennan, R.G. & Link, T.M. (2007) Hfq structure, function and ligand binding. Current Opinion in Microbiology, 10, 125–133. [DOI] [PubMed] [Google Scholar]
- Bugert, P. & Geider, K. (1995) Molecular analysis of the ams operon required for exopolysaccharide synthesis of Erwinia amylovora . Molecular Microbiology, 15, 917–933. [DOI] [PubMed] [Google Scholar]
- Castiblanco, L.F. & Sundin, G.W. (2016) New insights on molecular regulation of biofilm formation in plant‐associated bacteria. Journal of Integrative Plant Biology, 58, 362–372. [DOI] [PubMed] [Google Scholar]
- Castiblanco, L.F. & Sundin, G.W. (2018) Cellulose production, activated by cyclic di‐GMP through BcsA and BcsZ, is a virulence factor and an essential determinant of the three‐dimensional architectures of biofilms formed by Erwinia amylovora Ea1189. Molecular Plant Pathology, 19, 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, C.T. , Niemela, S.L. & Miller, R.H. (1989) One‐step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proceedings of the National Academy of Sciences of the United States of America, 86, 2172–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, Z. , Yuan, X. , Yang, C.H. , Huntley, R.B. , Sun, W. , Wang, J. et al. (2018) Development of a method to monitor gene expression in single bacterial cells during the interaction with plants and use to study the expression of the type III secretion system in single cells of Dickeya dadantii in potato. Frontiers in Microbiology, 9, 1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, D.G. & Marques, C.N. (2009) A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. Journal of Bacteriology, 191, 1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds, A.C. , Castiblanco, L.F. , Sundin, G.W. & Waters, C.M. (2013) Cyclic Di‐GMP modulates the disease progression of Erwinia amylovora . Journal of Bacteriology, 195, 2155–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier, G. & Geider, K. (1993) Characterization and influence on virulence of the levansucrase gene from the fireblight pathogen Erwinia amylovora . Physiological and Molecular Plant Pathology, 42, 387–404. [Google Scholar]
- Goodman, R.N. , Huang, J.S. & Huang, P.‐Y. (1974) Host‐specific phytotoxic polysaccharide from apple tissue infected by Erwinia amylovora . Science, 183, 1081–1082. [DOI] [PubMed] [Google Scholar]
- Gross, M. , Geier, G. , Rudolph, K. & Geider, K. (1992) Levan and levansucrase synthesized by the fireblight pathogen Erwinia amylovora . Physiological and Molecular Plant Pathology, 40, 371–381. [Google Scholar]
- Hu, L.I. , Chi, B.K. , Kuhn, M.L. , Filippova, E.V. , Walker‐Peddakotla, A.J. , Bäsell, K. et al. (2013) Acetylation of the response regulator RcsB controls transcription from a small RNA promoter. Journal of Bacteriology, 195, 4174–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y.H. , Ferrières, L. & Clarke, D.J. (2006) The role of the Rcs phosphorelay in Enterobacteriaceae . Research in Microbiology, 157, 206–212. [DOI] [PubMed] [Google Scholar]
- Huynh, T.V. , Dahlbeck, D. & Staskawicz, B.J. (1989) Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science, 245, 1374–1377. [DOI] [PubMed] [Google Scholar]
- Jackson, D.W. , Suzuki, K. , Oakford, L. , Simecka, J.W. , Hart, M.E. & Romeo, T. (2002) Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli . Journal of Bacteriology, 184, 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kery, M.B. , Feldman, M. , Livny, J. & Tjaden, B. (2014) TargetRNA2: identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Research, 42, W124–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharadi, R.R. & Sundin, G.W. (2020) Dissecting the process of xylem colonization through biofilm formation in Erwinia amylovora . Journal of Plant Pathology. 10.1007/s42161-020-00635-x [DOI] [Google Scholar]
- Kim, J. , Mannaa, M. , Kim, N. , Lee, C. , Kim, J. , Park, J. et al. (2018) The roles of two hfq genes in the virulence and stress resistance of Burkholderia glumae . The Plant Pathology Journal, 34, 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.F. , Wei, Z.M. & Beer, S.V. (1997) The hrpA and hrpC operons of Erwinia amylovora encode components of a type III pathway that secretes harpin. Journal of Bacteriology, 179, 1690–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koczan, J.M. , Lenneman, B.R. , McGrath, M.J. & Sundin, G.W. (2011) Cell surface attachment structures contribute to biofilm formation and xylem colonization by Erwinia amylovora . Applied and Environmental Microbiology, 77, 7031–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koczan, J.M. , McGrath, M.J. , Zhao, Y. & Sundin, G.W. (2009) Contribution of Erwinia amylovora exopolysaccharides amylovoran and levan to biofilm formation: implications in pathogenicity. Phytopathology, 99, 1237–1244. [DOI] [PubMed] [Google Scholar]
- Kovach, M.E. , Elzer, P.H. , Hill, D.S. , Robertson, G.T. , Farris, M.A. , Roop, R.M. 2nd et al. (1995) Four new derivatives of the broad‐host‐range cloning vector pBBR1MCS, carrying different antibiotic‐resistance cassettes. Gene, 166, 175–176. [DOI] [PubMed] [Google Scholar]
- Lai, J.‐L. , Tang, D.‐J. , Liang, Y.‐W. , Zhang, R. , Chen, Q. , Qin, Z.‐P. et al. (2018) The RNA chaperone Hfq is important for the virulence, motility and stress tolerance in the phytopathogen Xanthomonas campestris . Environmental Microbiology Reports, 10, 542–554. [DOI] [PubMed] [Google Scholar]
- Lee, J.H. , Sundin, G.W. & Zhao, Y. (2016) Identification of the HrpS binding site in the hrpL promoter and effect of the RpoN binding site of HrpS on the regulation of the type III secretion system in Erwinia amylovora . Molecular Plant Pathology, 17, 691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.H. & Zhao, Y. (2018) Integration of multiple stimuli‐sensing systems to regulate HrpS and type III secretion system in Erwinia amylovora . Molecular Genetics and Genomics, 293, 187–196. [DOI] [PubMed] [Google Scholar]
- Li, C. , Wen, A. , Shen, B. , Lu, J. , Huang, Y. & Chang, Y. (2011) FastCloning: a highly simplified, purification‐free, sequence‐ and ligation‐independent PCR cloning method. BMC Biotechnology, 11, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhugiri, R. , Basineni, S.R. & Klug, G. (2010) Turn‐over of the small non‐coding RNA RprA in E. coli is influenced by osmolarity. Molecular Genetics and Genomics, 284, 307–318. [DOI] [PubMed] [Google Scholar]
- Majdalani, N. , Chen, S. , Murrow, J. , St John, K. & Gottesman, S. (2001) Regulation of RpoS by a novel small RNA: the characterization of RprA. Molecular Microbiology, 39, 1382–1394. [DOI] [PubMed] [Google Scholar]
- Majdalani, N. , Heck, M. , Stout, V. & Gottesman, S. (2005) Role of RcsF in signaling to the Rcs phosphorelay pathway in Escherichia coli . Journal of Bacteriology, 187, 6770–6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani, N. , Hernandez, D. & Gottesman, S. (2002) Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Molecular Microbiology, 46, 813–826. [DOI] [PubMed] [Google Scholar]
- Malnoy, M. , Martens, S. , Norelli, J.L. , Barny, M.A. , Sundin, G.W. , Smits, T.H. et al. (2012) Fire blight: applied genomic insights of the pathogen and host. Annual Review of Phytopathology, 50, 475–494. [DOI] [PubMed] [Google Scholar]
- McCullen, C.A. , Benhammou, J.N. , Majdalani, N. & Gottesman, S. (2010) Mechanism of positive regulation by DsrA and RprA small noncoding RNAs: pairing increases translation and protects mRNA from degradation. Journal of Bacteriology, 192, 5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, R.R. , Toth, I.K. , Cock, P.J. , Pritchard, L. , Hedley, P.E. , Morris, J.A. et al. (2012) Genetic characterization of the HrpL regulon of the fire blight pathogen Erwinia amylovora reveals novel virulence factors. Molecular Plant Pathology, 13, 160–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika, F. , Busse, S. , Possling, A. , Berkholz, J. , Tschowri, N. , Sommerfeldt, N. et al. (2012) Targeting of csgD by the small regulatory RNA RprA links stationary phase, biofilm formation and cell envelope stress in Escherichia coli . Molecular Microbiology, 84, 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, W.G. , Leveau, J.H. & Lindow, S.E. (2000) Improved gfp and inaZ broad‐host‐range promoter‐probe vectors. Molecular Plant‐Microbe Interactions, 13, 1243–1250. [DOI] [PubMed] [Google Scholar]
- Mina, I.R. , Jana, N.P. , Criollo, J.E. & Castiollo, J.A. (2019) The critical role of biofilms in bacterial vascular plant pathogenesis. Plant Pathology, 68, 1439–1447. [Google Scholar]
- Nimtz, M. , Mort, A. , Domke, T. , Wray, V. , Zhang, Y. , Qiu, F. et al. (1996) Structure of amylovoran, the capsular exopolysaccharide from the fire blight pathogen Erwinia amylovora . Carbohydrate Research, 287, 59–76. [DOI] [PubMed] [Google Scholar]
- Norelli, J.L. , Jones, A.L. & Aldwinckle, H.S. (2003) Fire blight management in the twenty‐first century: using new technologies that enhance host resistance in apple. Plant Disease, 87, 756–765. [DOI] [PubMed] [Google Scholar]
- Oh, C.S. , Kim, J.F. & Beer, S.V. (2005) The Hrp pathogenicity island of Erwinia amylovora and identification of three novel genes required for systemic infection. Molecular Plant Pathology, 6, 125–138. [DOI] [PubMed] [Google Scholar]
- Park, H. , Bak, G. , Kim, S.C. & Lee, Y. (2013) Exploring sRNA‐mediated gene silencing mechanisms using artificial small RNAs derived from a natural RNA scaffold in Escherichia coli . Nucleic Acids Research, 41, 3787–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purevdorj‐Gage, B. , Costerton, W.J. & Stoodley, P. (2005) Phenotypic differentiation and seeding dispersal in non‐mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology, 151, 1569–1576. [DOI] [PubMed] [Google Scholar]
- Rivas, R. , Vizcaíno, N. , Buey, R.M. , Mateos, P.F. , Martínez‐Molina, E. & Velázquez, E. (2001) An effective, rapid and simple method for total RNA extraction from bacteria and yeast. Journal of Microbiological Methods, 47, 59–63. [DOI] [PubMed] [Google Scholar]
- Rumbaugh, K.P. & Sauer, K. (2020) Biofilm dispersion. Nature Reviews Microbiology, 18, 571–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. (2001) Molecular cloning: a laboratory manual. 3rd edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sauer, K. , Camper, A.K. , Ehrlich, G.D. , Costerton, J.W. & Davies, D.G. (2002) Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. Journal of Bacteriology, 184, 1140–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachterle, J.K. & Sundin, G.W. (2019) The leucine‐responsive regulatory protein Lrp participates in virulence regulation downstream of small RNA ArcZ in Erwinia amylovora . mBio, 10, e00757‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachterle, J.S. , Zeng, Q. & Sundin, G.W. (2019) Three Hfq‐dependent small RNAs regulate flagellar motility in the fire blight pathogen Erwinia amylovora . Molecular Microbiology, 111, 1476–1492. [DOI] [PubMed] [Google Scholar]
- Sjulin, T.M. & Beer, S.V. (1978) Mechanism of wilt induction by amylovorin in cotoneaster shoots and Its relation to wilting of shoots infected by Erwinia amylovora . Phytopathology, 68, 89–94. [Google Scholar]
- Slack, S.M. , Zeng, Q. , Outwater, C.A. & Sundin, G.W. (2017) Microbiological examination of Erwinia amylovora exopolysaccharide ooze. Phytopathology, 107, 403–411. [DOI] [PubMed] [Google Scholar]
- Srikumar, S. , Kröger, C. , Hébrard, M. , Colgan, A. , Owen, S.V. , Sivasankaran, S.K. et al. (2015) RNA‐seq brings new insights to the intra‐macrophage transcriptome of Salmonella Typhimurium . PLoS Pathogens, 11, e1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny, M. , Beloin, C. & Ghigo, J.M. (2018) Increased osmolarity in biofilm triggers RcsB‐dependent lipid A palmitoylation in Escherichia coli . mBio, 9, e01415‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, S.V. (2000) Epidemiology of fire blight In: Vanneste J. (Ed.) Fire blight: the disease and its causative agent, Erwinia amylovora. New York: CABI Publishing, pp. 9–36. [Google Scholar]
- Triplett, L.R. , Melotto, M. & Sundin, G.W. (2009) Functional analysis of the N terminus of the Erwinia amylovora secreted effector DspA/E reveals features required for secretion, translocation, and binding to the chaperone DspB/F. Molecular Plant‐Microbe Interactions, 22, 1282–1292. [DOI] [PubMed] [Google Scholar]
- Urban, J.H. & Vogel, J. (2007) Translational control and target recognition by Escherichia coli small RNAs in vivo . Nucleic Acids Research, 35, 1018–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste, J.L. (1995) Erwinia amylovora In: Singh U.S., Singh R.P., & Kohmoto K. (Eds.) Pathogenesis and host specificity in plant diseases: histopathological, biochemical, genetic and molecular bases, 21–46. Oxford and London: Pergammon Press. [Google Scholar]
- Vogel, J. & Luisi, B.F. (2011) Hfq and its constellation of RNA. Nature Reviews Microbiology, 9, 578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Pu, T. , Lou, W. , Wang, Y. , Gao, Z. , Hu, B. et al. (2018) Hfq, a RNA chaperone, contributes to virulence by regulating plant cell wall–degrading enzyme production, type VI secretion system expression, bacterial competition, and suppressing host defense response in Pectobacterium carotovorum . Molecular Plant‐Microbe Interactions, 31, 1166–1178. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Korban, S.S. & Zhao, Y. (2009) The Rcs phosphorelay system is essential for pathogenicity in Erwinia amylovora . Molecular Plant Pathology, 10, 277–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Korban, S.S. , Pusey, P.L. & Zhao, Y. (2012a) AmyR is a novel negative regulator of amylovoran production in Erwinia amylovora . PLoS One, 7, e45038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Qi, M. , Calla, B. , Korban, S.S. , Clough, S.J. , Cock, P.J.A. et al. (2012b) Genome‐wide identification of genes regulated by the Rcs phosphorelay system in Erwinia amylovora . Molecular Plant‐Microbe Interactions, 25, 6–17. [DOI] [PubMed] [Google Scholar]
- Wei, Z.M. & Beer, S.V. (1995) hrpL activates Erwinia amylovora hrp gene transcription and is a member of the ECF subfamily of sigma factors. Journal of Bacteriology, 177, 6201–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Z. , Kim, J.F. & Beer, S.V. (2000) Regulation of hrp genes and type III protein secretion in Erwinia amylovora by HrpX/HrpY, a novel two‐component system, and HrpS. Molecular Plant‐Microbe Interactions, 13, 1251–1262. [DOI] [PubMed] [Google Scholar]
- Wilms, I. , Möller, P. , Stock, A.‐M. , Gurski, R. , Lai, E.‐M. & Narberhaus, F. (2012) Hfq influences multiple transport systems and virulence in the plant pathogen Agrobacterium tumefaciens . Journal of Bacteriology, 194, 5209–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, M. , Singh, J. , Khan, A. , Sundin, G.W. & Zhao, Y.F. (2020) Complete genome sequence of the fire blight pathogen Erwinia amylovora strain Ea1189. Molecular Plant‐Microbe Interactions, 33, 1277–1279. 10.1094/MPMI-06-20-0158-A. [DOI] [PubMed] [Google Scholar]
- Yuan, X. , Zeng, Q. , Khokhani, D. , Tian, F. , Severin, G.B. , Waters, C.M. et al. (2019) A feed‐forward signaling circuit controls bacterial virulence through linking cyclic di‐GMP and two mechanistically distinct sRNAs, ArcZ and RsmB. Environmental Microbiology, 21, 2755–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Q. , McNally, R.R. & Sundin, G.W. (2013) Global small RNA chaperone Hfq and regulatory small RNAs are important virulence regulators in Erwinia amylovora . Journal of Bacteriology, 195, 1706–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Q. & Sundin, G.W. (2014) Genome‐wide identification of Hfq‐regulated small RNAs in the fire blight pathogen Erwinia amylovora discovered small RNAs with virulence regulatory function. BMC Genomics, 15, 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Sundin, G.W. & Wang, D. (2009a) Construction and analysis of pathogenicity island deletion mutants of Erwinia amylovora . Canadian Journal of Microbiology, 55, 457–464. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Wang, D. , Nakka, S. , Sundin, G.W. & Korban, S.S. (2009b) Systems level analysis of two‐component signal transduction systems in Erwinia amylovora: Role in virulence, regulation of amylovoran biosynthesis and swarming motility. BMC Genomics, 10, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 The Rcs binding site in the promoter of rprA. The rprA gene sequence is in bold font. The −10 and −35 promoter regions and the “Rcs box” region are underlined
TABLE S1 Genome‐wide prediction of the targets of RprA using TargetRNA2
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


