Abstract
Directed cardiomyogenesis from human induced pluripotent stem cells (hiPSCs) has been greatly improved in the last decade but directed differentiation to pacemaking cardiomyocytes (CMs) remains incompletely understood. In this study, we demonstrated that inhibition of NODAL signaling by a specific NODAL inhibitor (SB431542) in the cardiac mesoderm differentiation stage downregulated PITX2c, a transcription factor that is known to inhibit the formation of the sinoatrial node in the left atrium during cardiac development. The resulting hiPSC-CMs were smaller in cell size, expressed higher pro-pacemaking transcription factors, TBX3 and TBX18, and exhibited pacemaking-like electrophysiological characteristics compared to control hiPSC-CMs differentiated from established Wnt-based protocol. The pacemaker-like subtype increased up to 2.4-fold in hiPSC-CMs differentiated with the addition of SB431542 relative to the control. Hence, Nodal inhibition in the cardiac mesoderm stage promoted pacemaker-like CM differentiation from hiPSCs. Improving the yield of human pacemaker-like CMs is a critical first step in the development of functional human cell-based biopacemakers.
Keywords: Cardiogenesis, Sinoatrial node, Pacemaker, Human induced pluripotent stem cells, Differentiation
1. Introduction
Cardiac rhythm is initiated and dictated by a group of pacemaking cardiomyocytes (CMs) forming the sinoatrial node (SAN). Since the 1960s, treatments of patients with bradycardia or SAN dysfunction often include implantation of an electronic pacemaker to restore normal heart rhythm (Cingolani et al., 2018). Although pacemakers have gone through iterations of improvements over the years, limitations associated with electronic devices still exist, including deficiency in sensitivity to autonomic regulation, interference from magnetic field, and limited battery life. A biopacemaker may be an alternative that can circumvent inadequacies intrinsic of electronics to improve quality of life for recipients.
The first step towards engineering biopacemakers is resolving the availability of a source of human pacemaking CMs. One potential de novo source is through differentiation from human pluripotent stem cells (hPSCs) such as human embryonic stem cells (hESCs) or human induced pluripotent stem cells (hiPSCs). Directed differentiation of CMs from hPSCs has greatly improved in recent years (Lian et al., 2012, 2013; Yang et al., 2008). Two well- established protocols first induce mesoderm formation, either through 1) Wingless/Integrase-1 (WNT) activation by a small molecule inhibiting glycogen synthase kinase (GSK)3 or 2) by activation of the bone morphogenetic protein (BMP) plus ACTIVIN A pathways in hPSCs, subsequently followed by inhibition of WNT pathways in cardiac mesodermal cells to induce CMs. Although these protocols produce CMs at high efficiency, the distribution of CM subtypes—pacemaking, atrial, and ventricular—is reminiscent of the population distribution in the heart, with most CMs exhibiting ventricular-like properties and only ~10% of which are the pacemaking-like subtype as assessed by action potential (AP) profiles (Yechikov et al., 2016). A few recent studies improved the specification of pacemaking CMs from hPSCs through inhibition of neuregulin or addition of retinoic acid in hPSC-derived cardiac mesodermal cells, however, the yield for this particular subtype remains low at no more than ~30% (Protze et al., 2017; Zhu et al., 2010). Therefore, the differentiation condition that promotes hiPSCs to pacemaking CMs remains incompletely understood.
Since CM differentiation from hPSCs generally recapitulates the cardiogenesis stages in a developing embryo, factors that affect the SAN development in embryogenesis may affect the differentiation of pacemaking CMs from hPSCs. Transcription factor pituitary homeobox (Pitx)2c has been shown to inhibit the development of the SAN during embryogenesis (Ammirabile et al., 2012). During cardiac development, Pitx2c is expressed in the left atrium but absent in the right atrium where the SAN is located. Interestingly, Pitx2c-knockout mice have been shown to develop a SAN in both the left and right atria, suggesting that Pitx2c may suppress the development of pacemaking CMs in vivo. Indeed, Pitx2c has been shown to downregulate the expression of transcription factors short-stature homeobox (Shox)2 by binding to the regulatory region of Shox2 (Wang et al., 2010). This has great ramification for the fate of pacemaking CMs, as Shox2 is known to be a promoter of the pacemaking phenotype by upregulating pro-pacemaking transcription factors, such as T-box (Tbx)3 and Islet (Isl)1, while inhibiting the phenotype of the working CMs (Clauss and Kaab, 2011; Espinoza-Lewis et al., 2009; Hu et al., 2018; Ye et al., 2015). Pitx2c expression has been shown to be regulated by Nodal, a cytokine of the transforming growth factor (Tgf)β superfamily (Clauss and Kaab, 2011). Therefore, in this study, we directly tested the effects of NODAL inhibition by small molecule SB431542 on the differentiation of pacemaking CMs from hiPSCs in three differentiation stages—mesoderm induction, cardiac mesoderm specification, and cardiac progenitor specification. Our data demonstrated that NODAL inhibition in the cardiac mesoderm stage downregulated expression of PITX2c in hiPSC-CMs and promoted pacemaking-like CMs.
2. Experimental procedures
2.1. hiPSC culture and cardiac differentiation
Feeder-free hiPSCs (iPS-DF19-9-7T and iPS-DF6-9-9T, WiCell) were cultured on hESC-qualified matrigel (Corning) with StemMACS iPSC-Brew XF media (Miltenyi Biotec). Conditions to differentiate hiPSCs into CMs in this work were tested against an established WNT-based cardiogenesis protocol (Lian et al., 2012, 2013). Briefly, confluent hiPSCs were differentiated in RPMI 1640 media supplemented with B-27 supplement minus insulin (ThermoFisher Scientific) from days 0–7. To promote cardiogenesis, RPMI media was supplemented with 6 μM GSK3 inhibitor CHIR99021 (Tocris), and 5 μM WNT inhibitor IWR1 (Tocris) from differentiation days 0–2 and 3–5, respectively. Small molecule Nodal inhibitor SB431542 (Tocris) was added either at days 0–2, 3–5 or 5–7 as specified in the results section.
2.2. RT-PCR for transcript expression assessment
Total RNA of CMs differentiated from iPS-DF6-9-9T and iPS-DF19-9-7T lines was extracted using TRIzol (ThermoFisher Scientific), chloroform (Sigma-Aldrich), and RNeasy Mini Kit (Qiagen). cDNA was synthesized using QuantiTect Reverse Transcription Kit (Qiagen) and quantified in triplicates for each sample by a ViiA 7 Real-Time PCR System (Applied Biosystems) using PowerUp SYBR Green Master Mix (ThermoFisher). GAPDH was used for normalization in analysis using the ΔΔCt method. Primer sequences are listed in Table S1.
2.3. Immunofluorescence staining for confocal microscopy
Paraformaldehyde-fixed hiPSC-derived CMs of day 14 post-differentiation were permeabilized with 0.05% Triton and blocked with 10% goat serum and 1% BSA for one hour. Cells were incubated overnight at 4 °C in primary antibodies—SHOX2 (Abcam, Cat# ab55740), ISL1 (Developmental Studies Hybridoma Bank, Cat# 39.4D5), TBX3 (ThermoFisher Scientific, Cat# 702055), TBX18 (Millipore-Sigma, Cat# SAB1412362), or TNT (ThermoFisher, Cat# PIMA512960). Then, cells were incubated with secondary antibodies for one hour at room temperature. Nuclei were counterstained with DAPI (ThermoFischer Scientific). Finally, cells were mounted in ProLong Gold Antifade Mountant (ThermoFisher Scientific) and allowed to cure overnight before imaging. Confocal fluorescent images were acquired using Zeiss LSM 700 laser scanning confocal microscope.
2.4. Flow cytometric analysis
To dissociate the hiPSC-CMs into single cells for flow cytometric analysis, hiPSC-CMs from day 14 post-differentiation were washed in phosphate-buffered saline (PBS) without Ca2+ and Mg2+, followed by a wash in Versene (ThermoFisher Scientific). Next, the hiPSC-CMs were incubated in 10X TrypLE (ThermoFisher Scientific) for 3 min at 37 °C in the incubator. Then, hiPSC-CMs were collected in a conical tube and further digested for an addition 7–10 min in a 37 °C water bath with a stir bar with constant stirring at 160 rpm. The cells were washed with PBS with ethylenediaminetetraacetic acid (EDTA) and 10% fetal bovine serum (FBS) two times and filtered with a 35-μm cell strainer before fixation with paraformaldehyde. Cells were permeabilized and stained as described for confocal microscopy above. Data was acquired using a BD Fortessa cytometer with FACSDiva acquisition software (Becton Dickinson) and analyzed using FlowJo (Becton Dickinson).
2.5. Electrophysiological assessment
Functional analyses were performed by two different experimenters who are blinded to the treatment groups using two assessment methods. Day-40 hiPSC-CMs were dissociated similarly as described for cells used for flow cytometry and plated on glass bottom dishes (MatTek Corporation) as single cells and allowed to recover for three days prior to assessment of APs on day 43–47 post-differentiation at 37 °C by either whole-cell patch-clamp using Axopatch 200A amplifier (Molecular Devices) at sampling frequency of 2 kHz or optically using voltage-sensitive fluorescent dye FluoVolt (ThermoFisher Scientific) at ~100 fps using Nipkow spinning disk microscope with a FITC filter set. For patch-clamp recordings, hiPSC-CMs were measured in Tyrode’s solution consisted of (mM): NaCl 138, KCl 4, MgCl2 1, CaCl2 2, NaH2PO4 0.33, glucose 10, and HEPES 10 at pH 7.4. The internal pipette solution back-filled with 200 μg/ml amphotericin consisted of (mM): K-glutamate 120, KCl 25, MgCl2 1, CaCl2 1, and HEPES 10 with pH 7.4. For optical recordings, hiPSC-CMs were incubated with FluoVolt for 20 min at 37 °C and then recorded in phenol-free RPMI 1640 media at 37 °C with a 488-nm excitation laser at ~0.5 mW (ThermoFisher Scientific). Both patch-clamp and optical recordings were acquired by experimenters from two different laboratories blinded to the differentiation conditions. Cluster analysis of AP parameters averaged from at least five APs recorded optically was performed to classify hiPSC-CMs into ventricular, atrial and pacemaking CM subtype as in our previous publications (Chang et al., 2019; Yechikov et al., 2016). The AP parameters were analyzed by cluster analysis using the Mclust package in R to determine the cutoff values for the decision tree to classify the CM subtypes. First, AP duration at 80% repolarization (APD80) values for all 941 CMs (from the two hiPSC lines and a total of four independently differentiated batches) were clustered to determine the cutoff value of 320 ms for the decision tree. Those that were < 320 ms were then clustered to determine the cutoff value for frequency (83 bpm). Those APs that were > 83 bpm for frequency of automaticity and < 320 ms for APD80 were classified as pacemaking CMs. The remaining cells were then pooled and clustered by a duration-slope-curvature (DSC) factor of the AP profile that is defined as the multiple of the APD50, the curvature between APD10 and APD30, and the slope between APD50 and APD80. Those APs with DSC < 1244 were classified as the atrial subtype and those that were ≥1244 were classified as the ventricular subtype.
2.6. Statistical analysis
qRT-PCR data with high expression differences were log transformed for better data representation. All data are reported as mean ± SEM. Statistical significance was determined using one-way ANOVA with Tukey post-hoc test for qPCR experiments with more than two test conditions and optically recorded AP recording comparison between CM subtypes. All other qPCR and flow cytometry experiments with only two test conditions were analyzed by t-test.
3. Results
3.1. Simultaneous inhibition of NODAL and WNT pathways in the cardiac mesoderm stage promotes cardiomyogenesis in hiPSCs
The cardiac mesoderm stage has been reported to be a key stage in determining the CM subtype (Karakikes et al., 2014; Protze et al., 2017; Weng et al., 2014). To determine the timing of cardiac mesoderm in hiPSC differentiation, we tracked the gene expression profile in differentiating hiPSCs induced by the IWR1 differentiation protocol (control protocol in Fig. 1A) from differentiation day 0 to 60 by assessing the transcript expression of pluripotency marker POU class 5 homeobox transcription factor (POU5F1) that encodes OCT4, mesodermal marker Brachyury T (T), cardiac mesodermal kinase insert domain receptor (KDR), first and second heart field progenitors NKX2-5 and ISL1, general CM marker troponin C (TNNC1), atrial CM myosin light chain (MYL)7 encoding myosin light chain (MLC)2a, and ventricular CM myosin light chain MYL2 encoding MLC2v (Fig. S1). This expression timeline follows the patterns reported in the literature (Burridge et al., 2014; Zhang et al., 2012). The cardiac mesoderm stage is defined as the period after the transient appearance of mesodermal marker T, at the start of the upregulation of cardiac mesodermal KDR and heart field progenitors NKX2-5 and ISL1, but prior to the expression of CM markers TNNC1. Based on our transcript data over time and reports in the literature, the cardiac mesoderm was determined to be day 3–5 post-differentiation, which corresponded to the time of IWR1 addition in the differentiation protocol.
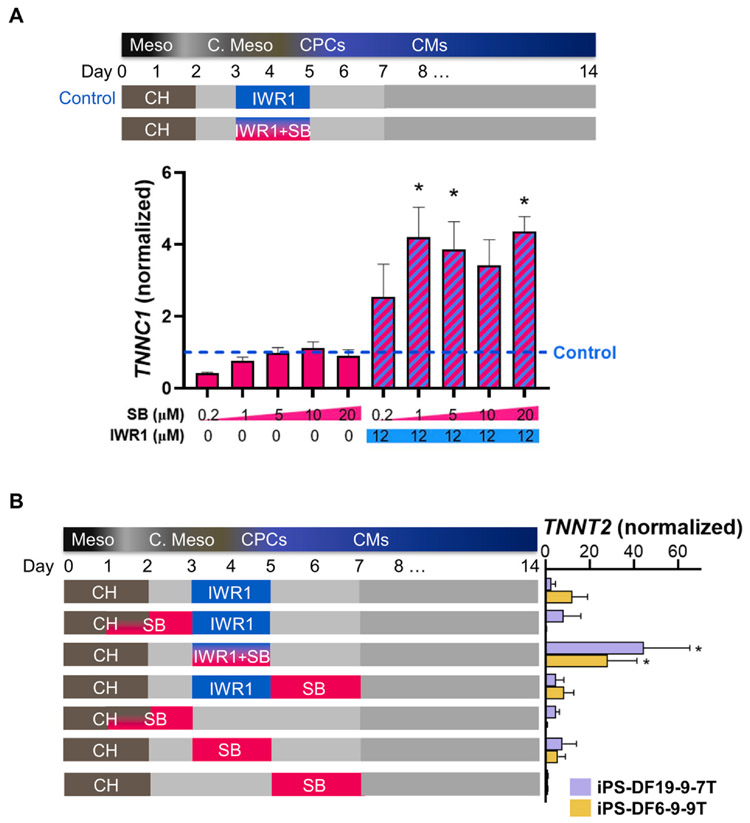
Fig. 1. Dose-dependence and temporal effects of Nodal inhibition on cardiomyocyte marker expression.
A) Troponin C (TNNC) expression of day 14 hiPSC-CMs differentiated with the addition of NODAL inhibitor SB431542 (SB) ranging from 0.2 to 20 μM with and without WNT inhibitor, IWR1, on differentiation day 3–5 normalized to the IWR1 only control (n = 3). B) Troponin T (TNNT2) transcript of day 20 hiPSC-CMs differentiated with the addition of NODAL inhibitor SB during mesodermal (Meso), cardiac mesodermal (C. Meso), and cardiac progenitor cell (CPCs) stages with and without IWR1. All experimental conditions were normalized to the control with no small molecule. Two hiPSC lines, iPS-DF19-9-7T and iPS-DF6-9-9T, were tested (n = 3). * denotes p < 0.05.
Once the cardiac mesoderm stage period was determined to be differentiation day 3–5, we first tested the concentration dependency of cardiomyogenesis in hiPSCs on a specific small molecule Nodal inhibitor, SB431542 (SB), in the cardiac mesoderm stage. A parallel culture was differentiated by an established cardiomyogenesis control protocol, which entailed the addition of 6 μM WNT activator or GSK3 inhibitor, CHIR99021, from differentiation days 0–2, followed by addition of 12 μM WNT inhibitor, IWR1, from differentiation day 3–5 (Fig. 1A, top). To inhibit the NODAL pathway, SB was added over a concentration range (1, 2, 5, 10, or 20 μM) either alone or in conjunction with IWR1 to differentiating hiPSC cultures from differentiation days 3–5. This differentiation stage was chosen for the concentration screen since WNT inhibition in the cardiac mesoderm stage has been shown to be critical for generating hiPSC-CMs. Cardiomyogenesis was assessed by the transcript level of cardiomyocyte marker troponin C (TNNC1) from differentiating hiPSCs of 14 days from the onset of differentiation (Fig. 1A, bottom). SB treatment alone at concentration of 5 μM or higher yielded comparable TNNC1 expression as the control differentiation protocol. Interestingly, IWR1 plus SB (IWR1+SB)-differentiated hiPSC-CM cultures expressed significantly higher TNNC1, which plateaued around 1 μM SB. Based on the concentration curve, 5 μM SB was selected as the concentration for optimal NODAL inhibition for subsequent experiments.
Each step of differentiation, from hiPSCs to mesodermal cells (differentiation day 0–2), to cardiac mesoderm (day 3–5), to cardiac progenitor cells (day 5–7), and finally to CMs (after day 7) can affect the resulting CM fate. To understand the impact of NODAL signaling during differentiation on the resulting CM yield, SB was tested in two hiPSC lines, iPS-DF19-9-7T and iPS-DF6-9-9T, at each stage of the differentiation timeline (Fig. 1B, left). No addition of inhibitors in the cardiac mesoderm stage for each respective hiPSC line served as the control condition. Cardiomyogenesis efficiency was assessed by the transcript level of cardiomyocyte marker troponin T (TNNT2) in day 14 hiPSC-CM cultures (Fig. 1B, right). Our results showed that addition of SB together with IWR1 to cardiac mesoderm from differentiation day 3–5 significantly increased TNNT2 expression over the standard IWR1 protocol in the same differentiation stage (iPS-DF19-9-7T: 44.4 ± 20.8 vs. 2.8 ± 1.8, n = 3; iPS-DF6-9-9T: 28.2 ± 13.1 vs. 12.3 ± 6.8, n = 3; p < 0.05), suggesting an additive effect of SB in improving cardiogenic efficiency when compared to the standard protocol without NODAL inhibitors. Interestingly, addition of SB to differentiating hiPSC cultures in any time period before or after this differentiation stage, with or without IWR1, consistently yielded lower TNNT2 expression levels for both hiPSC lines (Fig. 1B).
3.2. hiPSC-CMs differentiated from NODAL and WNT inhibited-cardiac mesoderm exhibit higher expression of pacemaking CM genes
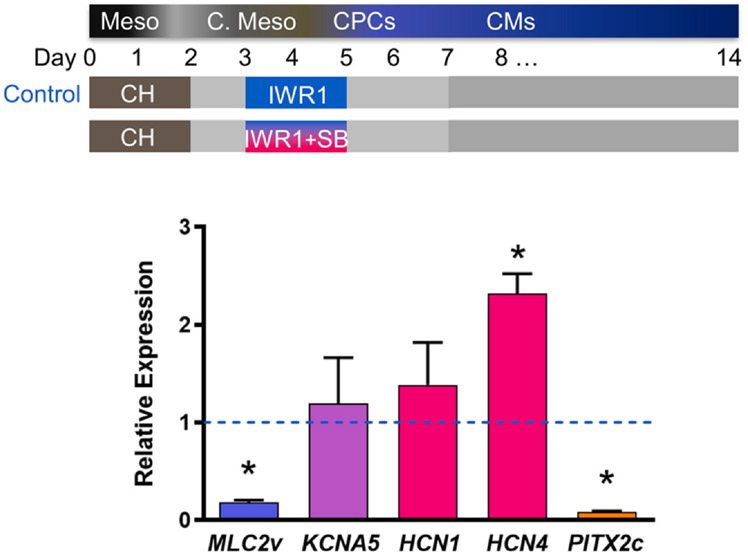
To determine whether WNT inactivation accompanied by NODAL inhibition in the cardiac mesoderm stage from IWR1+SB treatment may affect the differentiation of pacemaking CMs from hiPSCs, we examined gene expression using qRT-PCR analysis of CM subtype specific markers in day 20 unsorted hiPSC-CMs from IWR1+SB differentiation compared to the IWR1 standard treatment. Specifically, hiPSC-CMs were assessed for transcript expression of ventricular specific myosin light chain (MLC)2v, atrial specific voltage-gated K+ channel (KV)1.5, pacemaker CM-specific HCN1 and HCN4, and anti-sinoatrial node development transcription factor PITX2c (Fig. 2). hiPSC-CMs differentiated by IWR1 alone known to produce mostly ventricular CMs served as the control. MLC2v transcript expression in the IWR+SB-differentiated group was significantly lower by ~6-fold compared to IWR1-differentiated hiPSC-CMs, suggesting the resulting CM subtype may not be driven towards the ventricular subtype. Atrial voltage-gated KV1.5 channel (KCNA5) expression was upregulated in IWR1+SB-differentiated cultures compared to the control, but the increase was not significant. Both pacemaker channels HCN1 and HCN4 were also upregulated in the IWR1+SB-differentiated cultures compared to the control with significant upregulation of HCN4 expression by ~2-fold. Of note, PITX2c was significantly downregulated by ~12-fold in the IWR1+SB-differentiated cultures relative to the IWR1 control. The gene expression profile suggests the presence of more pacemaker-like CMs from the IWR1+SB differentiation protocol. Induction of more pacemaker-like CMs by the addition of IWR1+SB was consistent with the observed faster but less forceful contractions in the differentiation culture compared to the IWR1 control cells (Videos S1 and S2).
Fig. 2. Gene expression of hiPSC-CMs differentiated in the presence of Nodal inhibitor, SB431542.
Day-20 hiPSC-CMs post-differentiation that were differentiated in the presence of SB431542 during differentiation days 3–5 were assessed for ventricular marker (blue) myosin light chain (MLC)2v, atrial marker (purple) voltage-gated Kv1.5 channel (KCNA5), pacemaking CM markers (red) HCN1 and HCN4, and anti-sinoatrial node development pituitary homeobox (PITX)2c (orange). All expressions were normalized to control hiPSC-CMs differentiated without the addition of SB431542 (dotted line). Data are shown as mean ± SEM (n = 3). * indicates p < 0.05.
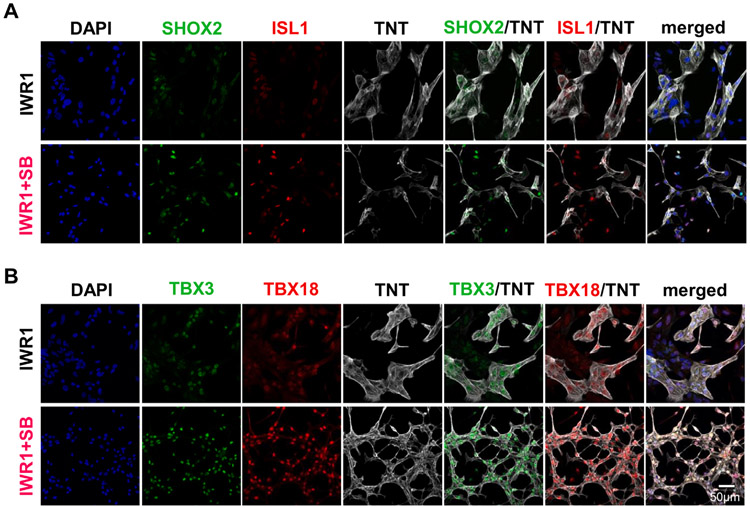
To assess the pro-pacemaking transcription factors in hiPSC-CMs, an earlier time point on day 14 post-differentiation was chosen because the pro-pacemaking transcription factors, such as TBX18, have been reported to downregulate over time and that high expression may only be needed during the development of the sinoatrial node (Wiese et al., 2009). In addition, tracking of the TBX3 expression from day 0 to 60 post-differentiation in IWR1-differentiated culture demonstrated that there was indeed a downregulation in its expression from day 10 to day 20 (Fig. S1), indicating that an earlier time point may maximize the differences in pro-pacemaking transcription factor expression in the hiPSC-CMs. To verify the pacemaker-specific protein expression in IWR1+SB-differentiated hiPSC-CMs, hiPSC-CMs on day 14 were replated and stained for pro-pacemaker transcription factors SHOX2 and ISL1 (Fig. 3A) or TBX3 and TBX18 (Fig. 3B) in conjunction with general cardiomyocyte marker troponin T (TNT) to enable assessment of transcription factor expression in TNT-positive CMs. ISL1 and SHOX2 staining exhibited slightly higher fluorescence intensity in IWR1+SB-than IWR1-differentiated hiPSC-CMs. Immunostainings of hiPSC-CMs for pro-pacemaking transcription factors TBX3 and TBX18 both presented much higher nuclear fluorescence in those CMs differentiated by IWR1+SB than the IWR1 control. Morphologically, IWR1+SB-differentiated hiPSC-CMs were smaller with spindle-like morphology compared to those from IWR1 differentiation. Both differentiated conditions resulted in hiPSC-CMs with myofilaments exhibiting sarcomeric pattern (Fig. S3).
Fig. 3. Morphology and protein expression of hiPSC-CMs differentiated by IWR1+SB.
A) SHOX2 and ISL1 staining in hiPSC-CMs of 14 days post-differentiation that were differentiated by the addition of IWR1+SB from day 3–5 compared to the IWR1 control. B) TBX3 and TBX18 staining in hiPSC-CMs from the IWR1+SB-differentiation relative to the IWR1 control. DAPI counterstain and TNT staining were included in all stained samples to allow identification of all cells and cardiomyocytes, respectively.
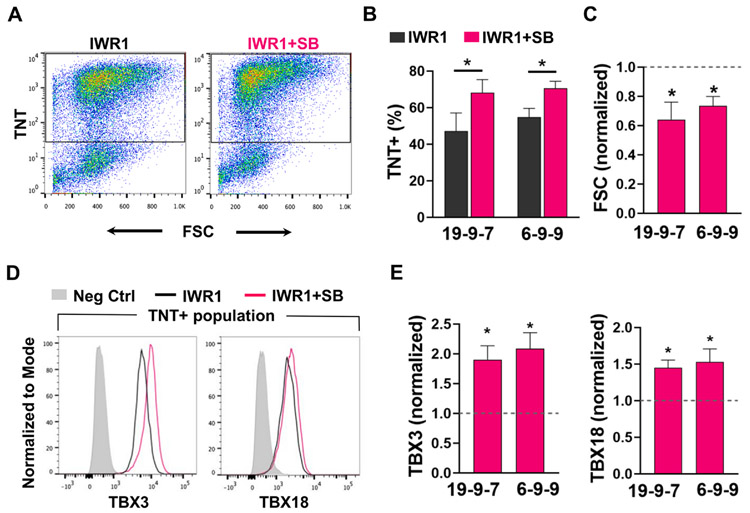
To further quantify the CM yield and the expression level of TBX3 and TBX18 per cell, hiPSC-CMs from day 14 post-differentiation were analyzed by flow cytometry. hiPSC-CMs were identified as TNT-positive cells (Fig. 4A). While there was a difference in cardiomyogenic efficiency between the hiPSC lines, the average CM yield for both hiPSC lines was statistically higher from IWR1+SB compared to IWR1 differentiation, at 68 ± 7% vs. 47 ± 10% of control for iPS-DF19-9-7T line and 71 ± 4% vs. 55 ± 5% of control for iPS-DF6-9–9 T line (Fig. 4B). The cell size of hiPSC-CMs, as indicated by forward scatter (FSC), was also smaller for IWR1+SB-differentiated hiPSC-CMs, with an average of ~ 64% and ~ 73% normalized FSC of IWR1-differentiated CMs for the iPS-DF19-9-7T and iPS-DF6-9-9T lines, respectively (Fig. 4C). The protein expression of TBX3 and TBX18 were both higher in IWR1+SB-differentiated CMs than the IWR1 control (Fig. 4D). For TBX3, the average median fluorescence change of independently differentiated batches was ~1.9 and ~2.1 times higher in hiPSC-CMs differentiated by IWR1+SB from iPS-DF19-9-7T (p = 0.03) and iPS-DF6-9-9T (p = 0.01) lines, respectively, compared to their respective IWR1 control (Fig. 4E). Similarly, the average median fluorescence for TBX18 was ~1.5 times higher for IWR1+SB-differentiated hiPSC-CMs than the IWR1 control for iPS-DF19-9-7T (p = 0.02) and iPS-DF6-9-9T (p = 0.03) lines (Fig. 4E). Expression of SHOX2 and ISL1 in hiPSC-CMs was also higher in the IWR1+SB-differentiated group than the IWR1 control as assessed by flow cytometric analysis (Fig. S2A-B), which is consistent with immunostaining of SHOX2 and ISL1 in hiPSC-CMs.
Fig. 4. CM yield and phenotype from the IWR1+SB differentiation.
A) Representative flow cytometry plots of troponin T (TNT) vs. forward scatter (FSC) for cells differentiated from the IWR1+SB vs. the IWR1 condition. B) Differentiation yield of TnT+ cells from the IWR1+SB and the control IWR1 differentiation of hiPSC lines iPS-DF19-9-7T (n = 9) and iPSF-DF6-9-9T (n = 17). C) FSC for hiPSC-CMs differentiated from the IWR1+SB condition normalized to the IWR1 control (dotted line) for two hiPSC lines. D) Representative histograms of TBX3 and TBX18 expression in the IWR1+SB- vs. IWR1-differentiated hiPSC-CMs (identified as TNT + cells). E) Bar graphs of TBX3 and TBX18 expression from CMs of independently differentiated batches by IWR1+SB relative to the IWR1 control (shown by dotted line) from two hiPSC lines (n = 4 for iPS-DF19-9-7T and n = 7 for iPS-DF6-9-9T). Data are shown as mean ± SEM. * denotes p < 0.05.
3.3. NODAL and WNT inhibition in the cardiac mesoderm stage generated hiPSC-CMs with pacemaking-like action potentials
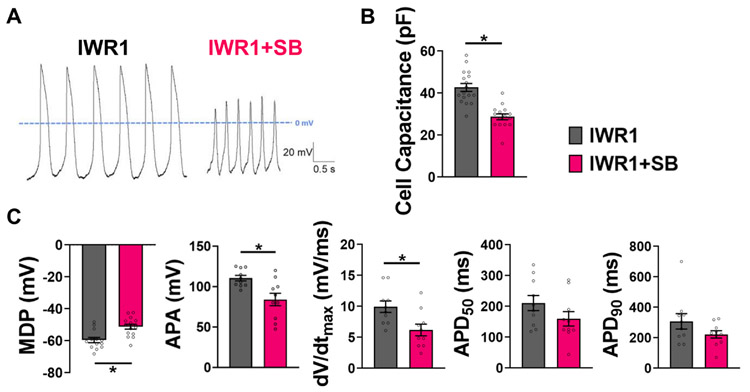
To assess functional differences, we had specifically chosen to assess the electrophysiology on more mature hiPSC-CMs after day 40 from onset of differentiation rather than day 14 because it is well known that all hiPSC-CMs start out with the ability to generate spontaneous APs, but this function is lost in the contractile CMs and only retained in the pacemaking CMs over time. Indeed, we have observed slowing of contraction frequency in the IWR1-differentiated culture by day 30 post-differentiation. Hence, assessing the automaticity in hiPSC-CMs after day 40 would minimize false-positive identification of pacemaking CMs. APs of day 43–47 hiPSC-CMs (re-plated as single cells for patch clamp on day 40) differentiated from IWR1+SB and control condition with IWR1 alone were recorded by perforated patch-clamp technique at 37 °C (Fig. 5A). Of note, the cell capacitance, a quantitative measure of cell size, was statistically smaller in IWR1+SB-differentiated hiPSC-CMs at 29 ± 1 pF compared to control CMs at 43 ± 2 pF (Fig. 5B). The AP recordings from the IWR1+SB-differentiated hiPSC-CMs showed characteristics that resembled more pacemaker-like CMs than the control (Fig. 5C). The maximum diastolic potential (MDP) on average was significantly more depolarized for CMs differentiated by IWR1+SB at −51 ± 2 mV compared to the control at −60 ± 2 mV. The AP amplitude (APA) was significantly smaller for IWR1+SB-differentiated CMs at 84 ± 8 mV compared to control CMs at 111 ± 3 mV. The maximum rate of rise (dV/dtmax) for the IWR1+SB group was 6 ± 1 mV/s and significantly slower than control hiPSC-CMs at 10 ± 1 mV/s. Following a similar pacemaker-like CM trend, the AP duration at 50% repolarization (APD50) was shorter, although not statistically significant, for the IWR1+SB-differentiated CMs at 160 ± 23 ms compared to the IWR1-differentiated control CMs at 211 ± 25 ms. The AP duration at 90% repolarization (APD90) was shorter for IWR1+SB-dfferentiated CMs at 308 ± 50 ms compared to control CMs at 222 ± 24 ms. APA, dV/dtmax, APD50, and APD90 were reported only for those CMs with stable APs.
Fig. 5. Patch-clamp recordings of action potentials from hiPSC-CMs differentiated by IWR1+SB.
A) Representative APs recorded using perforated patch-clamp in day-40 hiPSC-CMs post-differentiation for the IWR1+SB differentiation compared to the IWR1 control. B) Cell capacitance reflecting cell size of hiPSC-CMs differentiated from the IWR1+SB condition (n=17) compared to the IWR1 control (n=15). C) AP parameters—maximum diastolic potential (MDP; IWR1: n=13, IWR1+SB: n = 15), AP amplitude (APA), maximum rate of rise (dV/dtmax), AP duration at 50% repolarization (APD50), and APl duration at 90% repolarization (APD90)—quantified from patch-clamp recordings of hiPSC-CMs from the IWR1+SB-differentiated cultures relative to the control. Data are shown as mean ± SEM (n = 10, except for where indicated). * denotes p < 0.05.
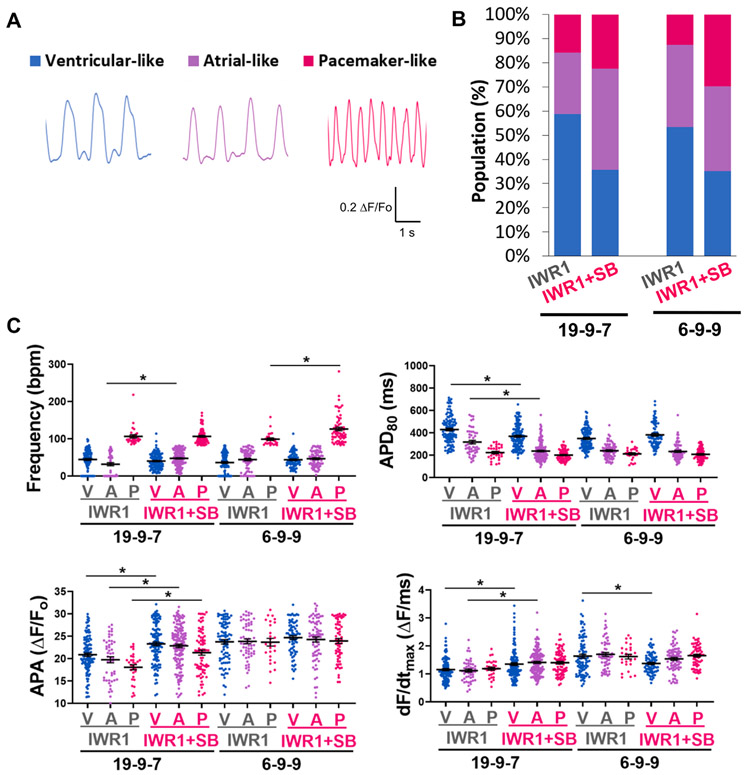
A non-biased approach of optical recordings was also performed to increase the AP assessment throughput and sample size. hiPSC-CMs of ~ 60 days post-differentiation generated from the IWR1+SB and IWR1 control differentiation protocols from two hiPSC lines (iPS-DF19-9-7T and iPS-DF6-9-9T) were assessed optically using voltage-sensitive dye, FluoVolt, to acquire AP profiles (Fig. 6A). In general, optically recorded AP profiles resembled those recorded by the whole-cell patch-clamp method. APs from optical recordings of hiPSC-CMs were quantified for AP parameters and classified into ventricular-, atrial-, and pacemaker-like CM subtype by cluster analysis of APD80, frequency of automaticity, and DSC factor DSC (Fig. 6B). The ventricular population was 59% and 53% for the two hiPSC lines differentiated by IWR1. The atrial population was 25% and 34% for the IWR1-differentiated CMs for the hiPSC lines. The lowest subtype yield from both hiPSC lines differentiated by IWR1 was the pacemaking population at 16% and 13%. Differentiation by IWR1+SB resulted in an increase in the pacemaking subtype by 1.4 and 2.4-fold compared to the IWR1 control in the iPS-DF19-9-7T and iPS-DF6-9-9T lines, respectively. The IWR1+SB-differentiated atrial fraction increased by 1.6-fold for the iPS-DF19-9-7 T line relative to the control, but the atrial yield was comparable for IWR1+SB and IWR1 differentiation in the iPS-DF6-9-9T line. The IWR1+SB-differentiated ventricular fraction decreased for both hiPSC lines by 0.6 to 0.7-fold.
Fig. 6. Electrophysiological phenotype of the IWR1+SB-differentiated hiPSC-CMs from optical recordings of action potentials.
A) Representative AP traces of ventricular-like, atrial-like, and pacemaker-like subtypes recorded optically using FluoVolt in day ~60 hiPSC-CMs. B) CM subtype distribution of IWR1+SB-differentiated cultures relative to the control for two hiPSC lines, iPS-DF19-9-7T (IWR1: n = 189, IWR1+SB: n = 338) and iPS-DF6-9-9T (IWR1: n = 206, IWR1+SB: n = 208), pooled from two independently differentiated batches for each line. C) Frequency of automaticity, AP duration at 80% repolarization (APD80), AP amplitude (APA), and maximum rate of rise (dF/dtmax) quantified from optically recorded APs of hiPSC-CMs differentiated by the addition of IWR1+SB compared to the IWR1 control from iPS-DF19-9-7T and iPS-DF6-9-9T lines. Data are shown as mean ± SEM. * denotes p < 0.05.
CM subtypes were compared for AP parameters between the IWR1+SB differentiation relative to the control (Fig. 6C). Frequency of automaticity was significantly faster for the pacemaking subtype from the IWR1+SB differentiation in the iPS-DF6-9–9 T line compared to the control, and the atrial subtype was slightly faster in the IWR1+SB differentiation for the iPS-DF19-9-7T line. APD80 was shortened in the atrial and ventricular subtypes of hiPSC-CMs differentiated by IWR1+SB compared to the control for the iPS-DF19-9-7T line. For iPS-DF19-9-7T line, the APA was increased in the IWR1+SB-differentiated hiPSC-CMs for all subtypes relative to the IWR1 counterpart. The maximum rate of rise (dF/dtmax) was also slightly faster for the IWR1+SB-differentiated atrial and ventricular subtypes than the IWR1 control for the iPS-DF19-9-7 T line. Therefore, although the distribution of the CM subtypes changed for the IWR1+SB-differentiated hiPSC-CMs relative to the control, there were only a few changes in the AP characteristics in each of the CM subtype between the differentiation conditions.
4. Discussion
An established monolayer differentiation protocol involving WNT activation followed by inactivation with small molecules CHIR99021 and IWR1, respectively, is a widely used protocol to differentiate hiPSC to CMs (Lian et al., 2012, 2013). This protocol has been shown to direct CMs toward the ventricular fate with minimal pacemaking fraction (Lian et al., 2012; Yechikov et al., 2016). Based on previous findings that downregulation of Pitx2c during cardiac development in embryogenesis enables the formation of the sinoatrial node in the left atrium (Ammirabile et al., 2012), we hypothesized that inhibition of PITX2c may promote the pacemaking CM fate. We took advantage of the fact that NODAL is a known upstream regulator of Pitx2c expression to test the effect of a NODAL inhibitor, SB431542, in a stage-specific manner on cardiomyogenesis. We demonstrated that the critical time to introduce SB431542 without adversely affecting cardiomyogenesis is in the cardiac mesoderm stage. This is in agreement with published reports that the addition of pro-ventricular CM factor, IWR1, in this particular differentiation stage results in higher ventricular CM yield (Karakikes et al., 2014; Weng et al., 2014). Indeed, the fate of the resulting CM subtype can be affected even prior to the cardiac progenitor stage.
4.1. Downregulation of PITX2c by the addition of SB during cardiac mesoderm stage
At optimized concentration and timing of SB introduction, the resulting CM phenotype from our modified IWR1+SB differentiation protocol was assessed. PITX2c was indeed significantly downregulated in the resulting CMs differentiated by the addition of IWR1+SB in the cardiac mesoderm stage compared to established IWR1 protocol. Pitx2c has been shown to downregulate pro-pacemaking transcription factor Shox2 (Ammirabile et al., 2012; Clauss and Kaab, 2011; Kirchhof et al., 2011; Wang et al., 2010). Shox2 promotes pacemaker-like CMs by upregulating other pro-pacemaking transcription factors, including Tbx3 and Tbx18 (Espinoza-Lewis et al., 2009; Hu et al., 2018; Ionta et al., 2015; Ye et al., 2015). Prior studies have shown Tbx3 to promote a pacemaking CM phenotype by inhibiting the atrial gene program, while Tbx18 is important for proper formation of the SAN head during cardiac development (Hoogaars et al., 2007, 2004; Wiese et al., 2009). We demonstrated in our study that the resulting IWR1+SB-differentiated hiPSC-CMs exhibited higher transcript and protein expression of TBX3, TBX18, SHOX2, and ISL1. Morphologically, IWR1+SB-differentiated CMs resembled those of pacemaker CMs from the sinoatrial node with spindle-like shape and smaller cell size, which was also confirmed by smaller cell capacitance measured by patch-clamp experiments and lower forward scatter assessed by flow cytometry. Most importantly, we demonstrated that the APs acquired by whole-cell patch-clamp recordings in these CMs functionally displayed electrophysiological characteristics that are more pacemaker-like than the control, with smaller APA, slower rate of rise, and shorter APD90.
4.2. Addition of IWR1+SB in the cardiac mesoderm stage induced an increase in the pacemaking fraction for both hiPSC lines
Previously, we had classified hiPSC-CMs into distinct ventricular-, atrial-, and pacemaker-like CM subtypes based on cluster analysis of optically acquired AP profiles (Chang et al., 2019; Yechikov et al., 2016). The optically recorded APs from IWR1 and IWR1+SB-differentiated hiPSC-CMs were similarly classified into CM subtypes. In this study, the IWR1-differentiated hiPSC-CMs on day ~60 yielded similar distribution of CM subtypes as our previous publication with majority of the CMs being the ventricular subtype and a small fraction of the pacemaking subtype at ~12% (Yechikov et al., 2016). Addition of IWR1+SB in the cardiac mesoderm stage indeed induced an increase in the pacemaking fraction for both hiPSC lines ranging from 1.4 to 2.4-fold. Hence, the faster spontaneous contractile frequency that we had observed in the IWR1+SB-differentiated monolayer cultures was likely due to an increase in the number of pacemaking-like CMs and not an overall increase in the frequency of automaticity in all CM subtypes. Although there were some differences in IWR1+SB induced differentiation between the hiPSC lines, the increase in expression of pro-pacemaking transcription factors TBX18 and TBX3 and an increase in the fraction of the pacemaking subtype as classified by AP profiles were consistently observed for both hiPSCs lines. Hence, an upregulation of pro-pacemaking TBX3 and TBX18 induced by the addition of IWR1+SB on differentiation days 3–5 is likely responsible for directing hiPSC-CMs towards a more pacemaking-like phenotype in hiPSC-CMs at day 60. It is important to note that the fraction of TBX3- and TBX18-positive cells does not directly correlate with the fraction of pacemaking CMs determined by the AP profile. The expression level is likely to be important for directing CM subtype, such that those CMs with a higher expression of TBX3 and TBX18 are likely to exhibit the pacemaking characteristics. This is support by the report that pro-pacemaking transcription factor can directly compete with pro-contractile transcription factor to determine the cell fate (Ye et al., 2015). A sufficiently high level of pro-pacemaking transcription factor will then be necessary to be competitive against transcription factors, such as NKX2.5, to drive the pacemaking fate.
From our classification of CM subtypes and qPCR data (downregulation of MLC2v expression), the increase in the pacemaking subtype was at the expense of a decrease in the ventricular fraction. The increase in the pacemaker-like fraction was not at the expense of a decrease in the atrial-like fraction as originally anticipated. Therefore, other factors besides relieving inhibition of the pacemaking gene program in hiPSC-derived atrial CMs by SB-induced PITX2C downregulation may be involved, by shifting the CM phenotype from ventricular-like to atrial-like and pacemaking-like subtypes. Although the presence of a low level of ACTIVIN (a ligand binding to the same receptors as NODAL) has been shown to drive hiPSCs towards pro-atrial mesodermal fate (Lee et al., 2017), the role of NODAL/ACTIVIN via SMAD signaling in directing CM subtype after cardiac mesoderm induction in hiPSCs remains to be elucidated.
The addition of SB to hPSC-derived cardiac progenitor cells has been reported to induce the downregulation of NK2 homeobox 5 (NKX2.5), a cardiac transcription factor that antagonizes SHOX2 (Birket et al., 2015; Ye et al., 2015). The observed pacemaking-like phenotype in our study may also be attributed to the SB-induced downregulation of NKX2.5, which promotes the contractile cardiomyocyte phenotype. Additionally, NKX2.5-knockout in zebrafish has been shown to shift the contractile cardiomyocytes from ventricular to the atrial fate (George et al., 2015). This may explain the observed SB-induced reduction in the ventricular hiPSC-derived cardiomyocytes without affecting the atrial fraction, such that there is a concurrent shift of the ventricular cardiomyocytes to the atrial fate and the atrial cardiomyocytes to the pacemaking subtype.
4.3. The roles of canonical and non-canonical WNT signaling during cardiac mesoderm stage
Of note, the WNT inhibitor added in conjunction with SB in this study was IWR1, which stabilizes Axin in the β-catenin destruction complex and consequently blocks only the canonical WNT signaling. In a recently reported differentiation protocol, WNT inhibitor IWP2 was added in the same differentiation stage (Protze et al., 2017). IWP2 acts by inhibiting the secretion of WNT via Porcupine (PORCN) inactivation and hence blocks both the canonical and non-canonical WNT signaling. We had specifically chosen IWR1 to block only the canonical WNT pathway to intentionally leave the non-canonical WNT pathways uninhibited as these pathways have been shown to be involved in cardiac development (Tian et al., 2010). Furthermore, recent publications reported activation of WNT in similar differentiation timing to promote pacemaker-like CMs in hPSCs. While one recent publication reported that canonical WNT3a activation promotes pacemaker-like CMs from hPSCs, another reported a typically non-canonical WNT5b activation that regulates the planar cell polarity pathway is responsible for inducing pacemaker-like CMs from hiPSCs (Liang et al., 2020; Ren et al., 2019). Based on the literature, the combination of NODAL inhibition by SB with canonical or non-canonical WNT activation in cardiac mesoderm on further improving the induction of pacemaker-like CMs from hiPSCs warrants additional studies.
4.4. Perspective and future studies
Although we have only tested the IWR1+SB differentiation protocol in hiPSCs, considering that most general cardiomyogenesis protocols and protocols that specifically drive the pacemaking, atrial, or ventricular CMs similarly work for both human embryonic stem cells (hESCs) and hiPSCs (Karakikes et al., 2014; Lee et al., 2017; Lian et al., 2012, 2013; Protze et al., 2017; Weng et al., 2014), we speculate that the IWR1+SB differentiation protocol should similarly improve the pacemaking yield in hESCs as well. Of note, the pacemaking CM yield was higher in the hiPSC line that had a higher CM yield. For the hESC lines that tend to have a higher CM yield than hiPSCs, the pacemaking CM yield in the CM fraction may also be higher. This notion is supported by Protze et al. that reported a higher pacemaking yield in hESCs that had a higher CM yield than the hiPSC line tested (Protze et al., 2017). Future studies are required to directly test the combination of WNT inhibition or activation in conjunction with SB differentiation in hESCs and additional lines of hiPSCs.
5. Conclusion
Collectively, our data suggest that the NODAL signaling pathway plays crucial roles in specifying pacemaker-like CMs from hiPSC-derived cardiac mesoderm. The directed differentiation of functional hiPSC-CMs with preserved pacemaking-like electrophysiology is a critical component and the first step towards the development of human cell-based biopacemakers.
Supplementary Material
Acknowledgments
Funding
This work is supported by California Institute of Regenerative Medicine (CIRM) Basic Biology Grant (DISC2-10120) to D.K.L. C.-W.C. is supported by a Postdoctoral Fellowship from National Institutes of Health T32 Training Program in Basic and Translational Science (NIH T32 HL086350). X.D.Z. is funded by NIH R56 HL138392. P.S. is supported by American Heart Association (AHA) Career Development Award (18CDA34110060), and Harold S. Geneen Charitable Trust Award. J.W.C. is supported by National Science Foundation (NSF) grants 1264776. NC is funded by NIH R01 HL085727, NIH R01 HL085844, NIH R01 HL137228, VA Merit Review Grant I01 BX000576 and I01 CX001490. The contents do not represent the views of the U.S. Department of Veterans Affairs or the United States Government. N.C. is the holder of the Roger Tatarian Endowed Professorship in Cardiovascular Medicine and a part-time staff physician at VA Northern California Health Care System, Mather, CA.
Footnotes
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Deborah Lieu is a financially compensated senior scientific consultant for Novoheart, Ltd.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2020.102043.
References
- Ammirabile G, Tessari A, Pignataro V, Szumska D, Sutera Sardo F, Benes J Jr., Balistreri M, Bhattacharya S, Sedmera D, Campione M, 2012. Pitx2 confers left morphological, molecular, and functional identity to the sinus venosus myocardium. Cardiovasc. Res 93, 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birket MJ, Ribeiro MC, Verkerk AO, Ward D, Leitoguinho AR, den Hartogh SC, Orlova VV, Devalla HD, Schwach V, Bellin M, Passier R, Mummery CL, 2015. Expansion and patterning of cardiovascular progenitors derived from human pluripotent stem cells. Nat. Biotechnol 33, 970–979. [DOI] [PubMed] [Google Scholar]
- Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, Plews JR, Abilez OJ, Cui B, Gold JD, Wu JC, 2014. Chemically defined generation of human cardiomyocytes. Nat. Methods 11, 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CW, Kao HKJ, Yechikov S, Lieu DK, Chan JW, 2019. An intrinsic, label-free signal for identifying stem cell-derived cardiomyocyte subtype. Stem Cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani E, Goldhaber JI, Marban E, 2018. Next-generation pacemakers: from small devices to biological pacemakers. Nat. Rev. Cardiol 15, 139–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauss S, Kaab S, 2011. Is Pitx2 growing up? Circ. Cardiovasc. Genet 4, 105–107. [DOI] [PubMed] [Google Scholar]
- Espinoza-Lewis RA, Yu L, He F, Liu H, Tang R, Shi J, Sun X, Martin JF, Wang D, Yang J, Chen Y, 2009. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev. Biol 327, 376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George V, Colombo S, Targoff KL, 2015. An early requirement for nkx2.5 ensures the first and second heart field ventricular identity and cardiac function into adulthood. Dev. Biol 400, 10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogaars WM, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LY, Bakker ML, Clout DE, Wakker V, Barnett P, Ravesloot JH, Moorman AF, Verheijck EE, Christoffels VM, 2007. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 21, 1098–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogaars WM, Tessari A, Moorman AF, de Boer PA, Hagoort J, Soufan AT, Campione M, Christoffels VM, 2004. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc. Res 62, 489–499. [DOI] [PubMed] [Google Scholar]
- Hu W, Xin Y, Zhao Y, Hu J, 2018. Shox2: the role in differentiation and development of cardiac conduction system. Tohoku J. Exp. Med 244, 177–186. [DOI] [PubMed] [Google Scholar]
- Ionta V, Liang W, Kim EH, Rafie R, Giacomello A, Marban E, Cho HC, 2015. SHOX2 overexpression favors differentiation of embryonic stem cells into cardiac pacemaker cells, improving biological pacing ability. Stem Cell Rep. 4, 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakikes I, Senyei GD, Hansen J, Kong CW, Azeloglu EU, Stillitano F, Lieu DK, Wang J, Ren L, Hulot JS, Iyengar R, Li RA, Hajjar RJ, 2014. Small molecule-mediated directed differentiation of human embryonic stem cells toward ventricular cardiomyocytes. Stem Cells Transl. Med 3, 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhof P, Kahr PC, Kaese S, Piccini I, Vokshi I, Scheld HH, Rotering H, Fortmueller L, Laakmann S, Verheule S, Schotten U, Fabritz L, Brown NA, 2011. PITX2c is expressed in the adult left atrium, and reducing Pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ. Cardiovasc. Genet 4, 123–133. [DOI] [PubMed] [Google Scholar]
- Lee JH, Protze SI, Laksman Z, Backx PH, Keller GM, 2017. Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell 21 (179–194), e4. [DOI] [PubMed] [Google Scholar]
- Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP, 2012. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. U.S.A 109, E1848–E1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP, 2013. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat. Protoc 8, 162–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W, Han P, Kim EH, Mak J, Zhang R, Torrente AG, Goldhaber JI, Marban E, Cho HC, 2020. Canonical Wnt signaling promotes pacemaker cell specification of cardiac mesodermal cells derived from mouse and human embryonic stem cells. Stem Cells 38, 352–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protze SI, Liu J, Nussinovitch U, Ohana L, Backx PH, Gepstein L, Keller GM, 2017. Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat. Biotechnol 35, 56–68. [DOI] [PubMed] [Google Scholar]
- Ren J, Han P, Ma X, Farah EN, Bloomekatz J, Zeng XI, Zhang R, Swim MM, Witty AD, Knight HG, Deshpande R, Xu W, Yelon D, Chen S, Chi NC, 2019. Canonical Wnt5b signaling directs outlying Nkx2.5+ mesoderm into pacemaker cardiomyocytes. Dev. Cell 50, 729–743 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Cohen ED, Morrisey EE, 2010. The importance of Wnt signaling in cardiovascular development. Pediatr. Cardiol 31, 342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, Martin JF, 2010. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc. Natl. Acad. Sci. U.S.A 107, 9753–9758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Z, Kong CW, Ren L, Karakikes I, Geng L, He J, Chow MZ, Mok CF, Keung W, Chow H, Leung AY, Hajjar RJ, Li RA, Chan CW, 2014. A simple, cost-effective but highly efficient system for deriving ventricular cardiomyocytes from human pluripotent stem cells. Stem Cells Dev. 23, 1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Grieskamp T, Airik R, Mommersteeg MT, Gardiwal A, de Gier-de VC, Schuster-Gossler K, Moorman AF, Kispert A, Christoffels VM, 2009. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ. Res 104, 388–397. [DOI] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, Keller GM, 2008. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature 453, 524–528. [DOI] [PubMed] [Google Scholar]
- Ye W, Wang J, Song Y, Yu D, Sun C, Liu C, Chen F, Zhang Y, Wang F, Harvey RP, Schrader L, Martin JF, Chen Y, 2015. A common Shox2-Nkx2-5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Development 142, 2521–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yechikov S, Copaciu R, Gluck JM, Deng W, Chiamvimonvat N, Chan JW, Lieu DK, 2016. Same-single-cell analysis of pacemaker-specific markers in human induced pluripotent stem cell-derived cardiomyocyte subtypes classified by electrophysiology. Stem Cells 34, 2670–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Klos M, Wilson GF, Herman AM, Lian X, Raval KK, Barron MR, Hou L, Soerens AG, Yu J, Palecek SP, Lyons GE, Thomson JA, Herron TJ, Jalife J, Kamp TJ, 2012. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ. Res 111, 1125–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu WZ, Xie Y, Moyes KW, Gold JD, Askari B, Laflamme MA, 2010. Neuregulin/ErbB signaling regulates cardiac subtype specification in differentiating human embryonic stem cells Circ. Res 107, 776–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.