Abstract
Introduction
Multiple myeloma (MM) remains an incurable disease, and patient survival requires a better understanding of this malignancy’s molecular aspects. Heparanase (HPSE) is highly expressed in aggressive MM cells and related to tumor growth, metastasis, and bortezomib (BTZ) resistance. Thus, targeting HPSE seems to be a promising approach for MM treatment, and because microRNAs (miRNAs) have emerged as potential regulators of HPSE expression, the use of extracellular vesicles (EVs) can allow the efficient delivery of therapeutic miRNAs.
Methods
We used prediction algorithms to identify potential miRNAs that regulate negatively HPSE expression. RT-qPCR was performed to assess miRNAs and HPSE expression in MM lines (U266 and RPMI-8226). Synthetic miRNA mimics were electroporated in MM cells to understand the miRNA contribution in HPSE expression, glycosaminoglycans (GAGs) profile, cell proliferation, and cell death induced by BTZ. EVs derived from HEK293T cells were engineered with miRNAs to evaluate their therapeutic potential combined with BTZ.
Results
It revealed a direct association between BTZ sensitivity, HPSE, and miR-1252-5p expressions. Moreover, overexpression of miR-1252-5p significantly reduced HPSE expression and HPSE enzymatic activity in MM cells. The higher level of miR-1252-5p was correlated with a reduction of cell viability and higher sensitivity to BTZ. Further, EVs carrying miR-1252-5p increased MM cells' sensitivity to BTZ treatment.
Conclusion
These results showed that miR-1252-5p could negatively regulate HPSE in MM, indicating the use of EVs carrying miR-1252-5p as a potential novel BTZ sensitization approach in MM cells.
Keywords: bortezomib, cancer, extracellular vesicles, exosomes, heparanase, multiple myeloma, microRNA
Introduction
Multiple myeloma (MM) is a B cell malignancy, representing the second most common hematological neoplasia (after non‑Hodgkin’s lymphoma), characterized by destructive lesions in the bone marrow (BM), chemoresistance, and disease relapses with a fatal outcome.1,2 Although MM is incurable, there was significant progress in managing the disease, mainly because of new therapeutic agents, such as proteasome inhibitors (PIs).3–5 Nonetheless, responses to therapy are transient, and patients treated with PIs frequently develop resistance and become refractory to their treatment regimens, contributing to a poor patient outcome.6,7 Hence, different therapeutic strategies are being explored to address the increment of treatment efficacy, reducing target relapse and improving MM patient outcome.8
Among the pathways involved in MM carcinogenesis, heparanase (HPSE) signaling promotes MM progression through tumor microenvironment regulation, driving angiogenesis, tumor growth, and metastasis.9–11 HPSE was previously reported highly expressed in cell fractions of the bone marrow microenvironment, especially monocytes and osteoclasts, and correlated with shorter event-free survival (EFS), and overall survival (OS) of MM patients.12 HPSE is an endo-β-D-glucuronidase that cleaves the heparan sulfate (HS) chains of proteoglycans in the tumor microenvironment, and cells with higher expression of this protein are more resistant to PIs treatment, including bortezomib (BTZ).13–15 In accordance with this, it was revealed that therapies targeting HPSE could sensitize MM cells to PIs cytotoxic effects.15
Although few studies had observed the role of microRNAs (miRNAs) regulating HPSE expression in cancers such as breast, melanoma, and hepatocellular carcinoma, it still has not been reported miRNAs regulating HPSE expression in MM cells.16–18 MicroRNAs are non-coding RNAs with 18–25 nucleotides that interact mainly with the 3′-untranslated regions (UTR) of specific messenger RNAs (mRNA), which induce their degradation or inhibit their translation, and are frequently dysregulated in cancer.19,20 The amount of data regarding miRNAs role in MM pathogenesis has increased, including the use of miRNA mimics or antimiRs to modulate the expression of specific miRNAs, providing potential new tools in clinical interventions for this dyscrasia.21,22 However, most of the methods applied for RNAs drug delivery (eg, viruses, lipid transfection, and lipid nanoparticles) are usually immunogenic and/or cytotoxic.23,24 Hence, the use of extracellular vesicles (EVs), which are natural lipid bilayer membrane nanovesicles (eg, exosomes, microvesicles, and apoptotic bodies) carrying proteins, lipids, nucleic acids, and sugars, could circumvent the delivery dilemma becoming an efficient vehicle for therapeutic miRNAs.24–26 Based on computational target prediction analysis to identify putative miRNAs that could negatively regulate HPSE expression, we identified interesting candidates to target HPSE transcripts in MM. Therefore, this study was performed to gain an understanding of miRNAs biological function in MM tumorigenesis, as well as to evaluate its contribution to HPSE regulation and BTZ treatment response.
Materials and Methods
Cell Line Culture
The MM cell lines RPMI-8226 (ATCC® CCL-155TM) and U266 (ATCC® TIB-196) were maintained in suspension with RPMI-1640 medium, while the kidney cell line HEK293T (ATCC® CRL-11268TM) was maintained with DMEM medium, and all cells were supplemented with 10% fetal bovine serum (FBS), 1% non‑essential amino acids and 0.01 μg/mL of penicillin‑streptomycin (cell culture reagents were purchased from ThermoFisher Scientific, USA). These cells were grown in an incubator at 37°C with 5% CO2.
Computational Target Prediction of miRNAs Targeting HPSE and miRNA Pathway Analysis
To predict potential miRNAs that target HPSE 3ʹUTR fraction by the presence of conserved 8mer, 7mer, and 6mer sites that match the seed region of each miRNA, we used TargetScan v.7.0 (http://www.targetscan.org/).27 Further, to understand the biological role of these miRNAs, Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathways enrichment analyses were performed for the selected miRNAs using DIANA miRPath v.3.0 software (http://snf‐515788.vm.okeanos.grnet.gr/) with microT-CDS.28 The p-values were calculated by DIANA software.
RNA Extraction, cDNA Synthesis, and RT-qPCR
RNA extraction and RT-qPCR were performed as previously described.8 Relative transcript levels of HPSE were determined by RT-qPCR using glyceraldehyde 3-phosphate dehydrogenase (GAPDH, F: 5ʹTTGAGGACCTCTGTGTATTTGTCAA 3ʹ and R: 5ʹ CCTGGAGGAGAAGAGGAAAGAGA 3ʹ and ribosomal protein L13a (RPL13a, F: 5ʹ TCGACAGTCAGCCGCATCTTCTTT 3ʹ and R: 5ʹ GCCCAATACGACCAAATCCGTTGA 3ʹ) to normalize the expression. The primer sequences for HPSE were F: 5ʹ TGGCAAGAAGGTCTGGTTAGGAGA 3ʹ and R: 5ʹ GCAAAGGTGTCGGATAGCAAGGG 3ʹ. The microRNAs expression assays were conducted using the TaqMan MicroRNA Reverse Transcription kit (ThermoFisher Scientific, USA) specific for each selected microRNAs, according to the manufacture’s protocols. Briefly, 10ng of total RNA was reverse-transcribed to cDNA and quantified on a 7500 Fast Real-Time PCR System (Applied Biosystems, USA). U6 small nuclear RNA (U6) and U47 small nuclear RNA (U47) were used as internal controls. All reactions were performed in triplicates and the 2−ΔΔCt method was employed to evaluate the transcript expressions.
MM Cells Transfection
The U266 and RPMI-8226 wild type (WT) cells were transfected with 30 pmol of synthetic hsa-miR-1252-5p mimic (miR-1252-5p mimic; #4464066, mirVanaTM miRNA mimic, ThermoFisher Scientific, USA) or with 30 pmol of miRNA mimic negative control (miRctrl; #4464058, ThermoFisher Scientific, USA). Electroporation was performed using Nucleofector 2b, following the manufacturer’s recommendations (Cell Line Nucleofector kit C, program X‑005; Lonza, Switzerland), as previously described.8 Upon 48 h after transfection, cells were collected and subjected to downstream analysis.
HPSE Expression by Flow Cytometry
HPSE expression was measured by flow cytometry. Transfected MM cells were centrifuged, washed, and incubated with anti-HPSE (HPA1 C-20: Sc26137, Santa Cruz Biotechnology, USA) and secondary antibody IgG labeled with AlexaFluor 488 (ThermoFisher Scientific, USA) in PBS for 60 min. Flow cytometry evaluation was performed in a FACS Aria III (BD Biosciences, USA). Data were analyzed using the FlowJo software.
Degradation of Biotinylated Heparan Sulfate by HPSE
HPSE action was measured by an ELISA-like method using HS biotinylated, as previously described.29 Transfected MM cells (1×105 per well) were cultured for 3 days in 60-mm plates. The cells were collected by centrifugation and resuspended in 500 μL of sodium acetate 25 mM, pH 5.0, containing protease inhibitors (ThermoFisher Scientific, USA), and 50 μL of cell extract was incubated with the pre-coated plate, revealed with europium-labeled streptavidin, washed and submitted to 200 μL of enhancement solution (PerkinElmer Life Sciences-Wallac Oy, Finland). Free europium was measured and the data analyzed in the MultiCalc software (PerkinElmer Life Sciences-Wallac Oy, Finland). The product obtained by HPSE incubation was expressed by the ratio of degraded HS and total protein from the cellular fraction (μg of HS/μg of total protein).
Cell Viability and Death Assays
Cell viability was quantified using PrestoBlue Cell Viability Reagent (ThermoFisher Scientific, USA), according to the manufacturer’s protocol, as previously described.8 Briefly, the IC50 values for BTZ were calculated from WT cell viability plots. Hence, 48 hours after transfections, RPMI-8226 and U266 cells were treated for 48 h with 5.0 nM and 20 nM of BTZ, respectively. The relative luminescence units from treated cells were normalized against DMSO control cells and expressed as a percentage of viable cells. Cell death assays were performed as previously described by quantifying Annexin V (AV)/7-AAD (7-aminoactinomycin D) levels, according to the manufacturer’s protocol (#1006; Biovision, USA).30 All treatments were performed in triplicate, with three separate biological replicates for each assay.
EVs Isolation and Characterization
The EVs isolation from HEK293T cells was performed as previously described.30 Briefly, the cells were incubated for 72 h in phenol-red free DMEM (#31053, Gibco, USA), supplemented with 5% Insulin-Transferrin-Selenium-Ethanolamine (#51500-056, ThermoFisher, USA), 10mM of Non Essential Amino Acids (#11140-050, ThermoFisher, USA), 500μg of fibroblast growth factor-basic (#13256-029, ThermoFisher, USA), 100mM of Sodium Pyruvate and 55mM of β-Mercaptoethanol (#21985-023, ThermoFisher, USA). The FBS-free medium was collected and centrifuged at 250 x g for 5 min to remove the cell debris. The supernatant was filtered on a 0.22-μm filter and further concentrated 20X by tangential flow filtration on a 50 kDa Ultra-15 Centrifugal Filter (Amicon; Merck, USA) by centrifugation at 1200 x g for 20 min. This concentrated fraction media enriched on EVs is the vesicular secretome fraction (VSF). Total protein contents of VSF was measured by Qubit™ Protein Assay Kit (#Q33212, ThermoFisher, USA). Further, to visualize EVs through transmission electron microscopy (TEM), the VSF preparation was incubated with the uranyl-oxalate solution for 5 min, as previously described, and the images were acquired on a JEM 1200 EX II (JEOL, Japan) at 80 kV at UNIFESP – Electron Microscopy Center (CEME).31 Western blots were performed as previously described and the antibodies used were purchased from Santa Cruz Biotechnology (USA): anti-ALIX (#SC-53540) and anti-CD81 (#SC-7637).8 The size distribution of EVs present in the VSF was measured using the NanoSight system (Malvern Panalytical, UK) for nanoparticle tracking analysis (NTA). To identify EVs’ size, the NanoSight system was equipped with a 405 nm laser and NTA 3.1 analytic software. The average diameter of the EVs was used in the statistical analyses.
EVs Loading Protocol and Effects on MM Cells
Subsequently to VSF characterization, 100 µL of VSF (containing around 5×109 nanoparticles) and 30 pmol of miR-1252-5p mimic or miRctrl were mixed to the final volume of 110 µL. Then, the EVs were engineered using electroporation on a Nucleofector 2b, program Q‑001 (Lonza, Switzerland), and incubated for 30 min at 37°C. To access the efficiency of miRNAs loading into the EVs, the VSF-miRNA mixtures were treated with RNase, as previously described.26 Briefly, the VSF-miRNA electroporated or unelectroporated mixtures were incubated with 0.2 mg/mL of RNase A (ThermoFisher Scientific, USA) for 30 min at 37°C. To stop the reaction, the RNase inhibitor (ThermoFisher Scientific, USA) was applied for 30 min at 37°C, as described by the manufacturer’s protocol. Upon RNase treatment, Trizol-LS (ThermoFisher Scientific, USA) was added into each sample for total RNA extraction.26 Further, the VSF-miRNA mixtures were subjected to miRNA expression analyses by RT-qPCR, as described above. Finally, to perform the VSF co-treatment with BTZ [IC50], MM cells were seeded in a 96 well plate at a density of 103 cells in an EV-free medium.30 Then, 10 µL of 1) pure VSF, 2) VSF electroporated with miR-1252-5p mimic (eVSF), or 3) VSF electroporated with miRctrl (eVSFctrl) were added. Simultaneously to the addition of the VSFs, BTZ [IC50] was also supplied. Succeeding 48 h of co-treatment, cytotoxic assays were performed as described above.
Statistical Analyses
Comparisons of the values obtained in this study were performed using Mann–Whitney or one-way ANOVA when appropriate. Multiple paired comparisons were conducted employing Bonferroni’s post-test method to maintain the 5% significance level.
Results
HPSE Transcripts Have miR-20b-5p and miR-1252-5p as Potential Target Regulators
To identify putative miRNAs regulating HPSE expression, TargetScan prediction algorithm27 was used, and a panel of miRNAs that could regulate HPSE negatively was revealed (data not shown). From this screen, hsa-miR-20b-5p and hsa-miR-1252-5p appear with a high probability of directly regulate HPSE transcripts negatively through binding the 3ʹ-UTR (TargetScan: Supplementary Figure 1). To assess the biological significance of miR-20b-5p and miR-1252-5p, we conducted an in silico analysis to predict potential pathways regulated by these miRNAs (Table 1). Predicted genes were functionally clustered into key pathways using the DIANA miRPath software v.3.0 with microT-CDS.28 Thus, 14 pathways were significantly associated with miR-20b-5p in the Kyoto Encyclopedia of Genes and Genomes (KEGG). The most prominent cancer networks targeted by this microRNA included transcriptional misregulation, chronic myeloid leukemia, proteoglycans, MAPK signaling, renal cell carcinoma, acute myeloid leukemia, choline metabolism, and ubiquitin-mediated proteolysis (Table 1). In addition, of the seven KEGG pathways identified as putative targets for miR-1252-5p, the pathways related to tumorigenesis signaling were linked to lysine degradation, transcriptional misregulation, glycosphingolipid biosynthesis, and also proteoglycans (Table 1). Therefore, miR-20b-5p and miR-1252-5p were chosen due to the relevance they might play by regulating tumorigenesis pathways and because none of these miRNAs had been evaluated in MM.
Table 1.
List of Significant Predicted Pathways Identified Through DIANA miRPath V.3 Software Database with microT-CDS.
| KEGG Pathway | p-Value | No. of Related Genes |
|---|---|---|
| hsa-miR-20b-5p | ||
| Transcriptional misregulation in cancer | 0.00251 | 5 |
| Pathways in cancer | 0.00347 | 11 |
| Neurotrophin signaling pathway | 0.00534 | 7 |
| Hepatitis B | 0.00786 | 4 |
| Sphingolipid signaling pathway | 0.01025 | 6 |
| Axon guidance | 0.01727 | 6 |
| Chronic myeloid leukemia | 0.01831 | 4 |
| Proteoglycans in cancer | 0.01846 | 8 |
| MAPK signaling pathway | 0.02168 | 9 |
| Renal cell carcinoma | 0.02314 | 4 |
| Hepatitis C | 0.02906 | 6 |
| Ubiquitin mediated proteolysis | 0.03487 | 6 |
| Acute myeloid leukemia | 0.04346 | 3 |
| Choline metabolism in cancer | 0.04696 | 5 |
| hsa-miR-1252-5p | ||
| Metabolism of xenobiotics by cytochrome P450 | 1.38E+03 | 4 |
| Lysine degradation | 1.91E+06 | 6 |
| Transcriptional misregulation in cancer | 0.00010 | 13 |
| Mucin type O-glycan biosynthesis | 0.01004 | 3 |
| Glycosphingolipid biosynthesis - globo series | 0.01154 | 3 |
| Glycosphingolipid biosynthesis - ganglio series | 0.01174 | 2 |
| Proteoglycans in cancer | 0.02550 | 13 |
Note: Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways Related to the Putative Genes That miR-20b-5p or miR-1252-5p May Be Involved.
HPSE and miRNA Levels in MM Cells
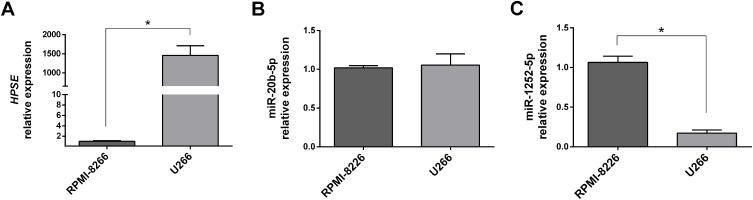
Our previous data showed that U266 cells were more resistant to BTZ treatment than RPMI-8226.8 To evaluate the correlation between HPSE levels and BTZ resistance, and trying to connect the HPSE levels with miR-20b-5p and miR-1252-5p expressions, we performed RT-qPCR analysis in these two MM cell lines. This analysis showed that HPSE expression level in U266 was significantly higher than in RPMI-8226 (almost 1500×; Figure 1A). Besides this, no significant difference in the expression levels of miR-20b-5p was observed between both MM cell lines tested (Figure 1B). On the other hand, it was observed an 82.6%-reduction in the miR-1252-5p expression in U266 cells compared to RPMI-8226 cells, suggesting that this miRNA might contribute to the regulation of HPSE expression in MM cells (Figure 1C).
Figure 1.
HPSE, miR-20b-5p and miR-1252-5p expression in MM cells. The RT-qPCR analysis was used to evaluate the relative expression level of HPSE (A), miR-20b-5p (B) and miR-1252-5p (C) in RPMI-8226 and U266 MM cell lines. All the relative expression levels were calculated using the 2−ΔΔCt method, based on RPMI-8226 levels. The standard deviation observed in three independent experiments was indicated. For all bar graphs, p-values denoted as *p < 0.05 by Mann–Whitney test.
miR-1252-5p Overexpression Regulates Negatively HPSE Expression and Its Enzymatic Activity in MM Cells
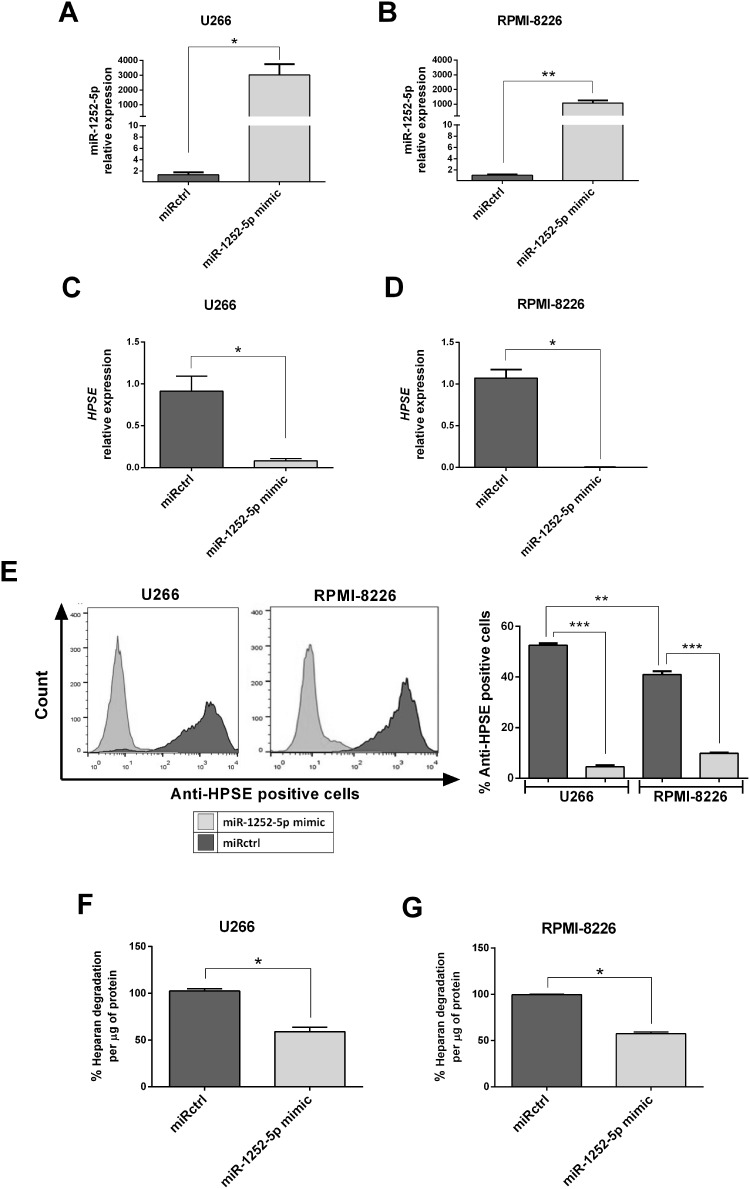
The hsa-miR-1252-5p mimic (miR-1252-5p mimic) was overexpressed in the two MM cell lines to better understand this miRNA functional role in MM tumorigenesis and to assess its impact in the regulation of HPSE expression. Thus, the presence of miR-1252-5p mimic promoted a high expression of this microRNA in U266 (Figure 2A) and RPMI-8226 cells (Figure 2B). The negative regulation of HPSE expression induced by the miR-1252-5p mimic was confirmed by a significant reduction in HPSE transcript levels in U266 (91.96%; Figure 2C) and RPMI-8226 (99.8%; Figure 2D). As expected, according to the HPSE expression data, U266 significantly expressed higher levels of HPSE in comparison to RPMI-8226. The miR-1252-5p induced a drastic reduction of this protein in both cell lines (91.37% for U266 and 75.88% for RPMI-8226) in contrast to the cells transfected with miRNA mimic negative control (miRctrl) (Figure 2E). In addition, U266 (Figure 2F) and RPMI-8226 (Figure 2G) cells overexpressing miR-1252-5p mimic presented a significant reduction in the HPSE activity (41.01% for U266 and 42.55% for RPMI-8226), as attested by evaluation of the level of heparan sulfate degraded by those cells, which corroborates with the low mRNA and protein expression level of such enzyme in transfected MM cells.
Figure 2.
Overexpression of miR-1252-5p regulates negatively HPSE expression and activity in MM cells. The MM cells (A; U266 and B; RPMI-8226) were transfected with synthetic miR-1252-5p mimic and with a miRNA mimic negative control (miRctrl). The levels of HPSE mRNA (C; U266 and D; RPMI-8226) and protein (E) were checked by RT-qPCR and flow cytometry, respectively. HPSE enzymatic activity was measured by an ELISA-like method using HS biotinylated (F; U266 and G; RPMI-8226). The standard deviation observed in three independent experiments was indicated. Asterisks denote significance as determined by Mann–Whitney test; *p < 0.05, **p < 0.01, ***p < 0.0001.
EVs-miR-1252-5p Decreases Cell Viability and Enhances BTZ Sensitivity in MM Cells
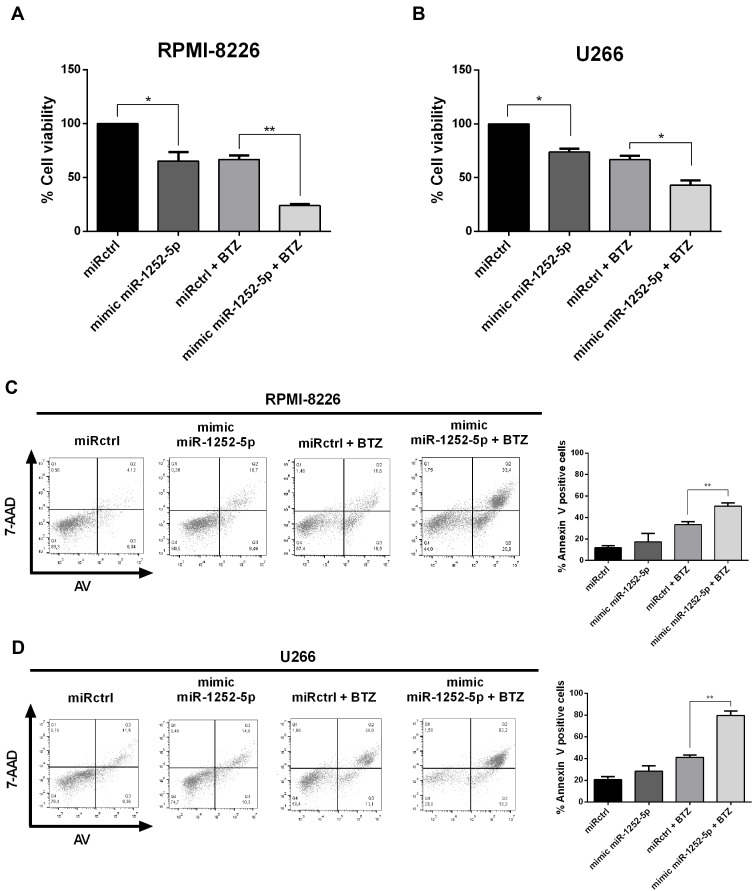
Firstly, the IC50 value for BTZ in both cell lines was established by 48-h treatment with increasing concentrations of this drug resulting in dose-dependent reduction of cell viability in both cell lines (Supplementary Figure 2). According to PrestoBlue assays, the BTZ half-maximal inhibition value for U266 WT was 21.43 ± 2.36 nM, while the IC50 for RPMI-8226 WT was 4.45 ± 1.24 nM. Further, we assessed the effect of miR-1252-5p in the viability of MM cells. The overexpression of miR-1252-5p reduced the MM cell viability (34.81% in RPMI-8226 and 26.22% in U266), compared to the respective miRctrl transfected cells (Figure 3A; RPMI-8226 and Figure 3B; U266). In turn, upon 48 h of treatment with BTZ (5.0 nM for RPMI-8226 and 20.0 nM for U266), the HPSE blockage by miR-1252-5p mimic promoted a significant increment of drug sensitivity in RPMI-8226 (64.08%; Figure 3A) and in U266 cells (35.45%; Figure 3B), when compared to BTZ response in the respective MM cells transfected with miRctrl. To confirm the cell death induced by BTZ, after a 48h-BTZ treatment, an Annexin V-7AAD dual staining assay was conducted. It is essential to mention that, although there was no significant difference, we observed an increment tendency in the percentage of cell death in MM cells overexpressing miR-1252-5p (5.6% in RPMI-8226 and 7.7% in U266) in comparison to the respective cells transfected with miRctrl (Figure 3C; RPMI-8226 and Figure 3D; U266). Besides, as shown for both MM cell lines, the overexpression of miR-1252-5p induced a significant increase in the number of dead cells subsequently to BTZ treatment in comparison to cells transfected with miRctrl (33.4% vs 50.7% for RPMI-8226 and 41.2% vs 79.6% for U266) (Figure 3C; RPMI-8226 and Figure 3D; U266). Taken collectively, these data allow us to hypothesize that miR-1252-5p could be able to modulate the MM response to BTZ by targeting HPSE transcripts and, consequently, reducing the HPSE activity.
Figure 3.
Overexpression of miR-1252-5p enhanced the BTZ effect on MM cells. MM cell viability was quantified using PrestoBlue assay (A; RPMI-8226 and B; U266), while cell death was assayed by Annexin V – 7-AAD staining after BTZ treatment (C; RPMI-8226, 5.0 nM and D; U266, 20 nM). The standard deviation observed in three independent experiments was indicated. Asterisks denote significance as determined by one-way ANOVA, multiple comparisons followed by Bonferroni test (post-test) (*p < 0.001 and **p < 0.0001).
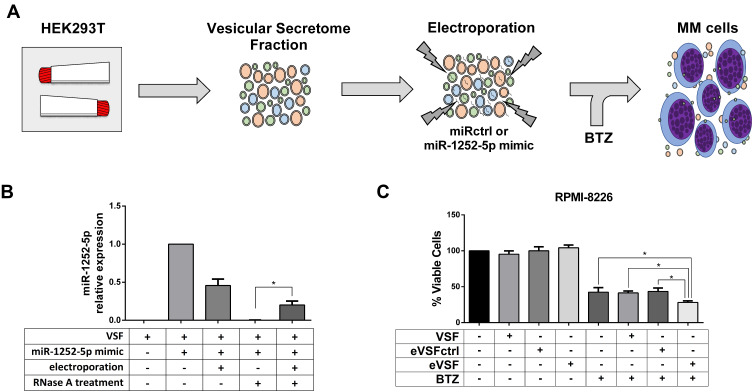
Afterwards, we collected by filtration the EVs released by HEK293T, and electroporated the vesicular secretome fraction (VSF) with miRctrl or synthetic miR-1252-5p to assess the co-treatment effect of these modified EVs in BTZ response of MM cells (Figure 4A). To this end, the EVs presented in the VSF were characterized by transmission electron microscopy (TEM). As expected, vesicles-like nanoparticles of 100–200 nm diameter-size were observed (Supplementary Figure 3A). We also certified the presence of EVs in the HEK293T VSF by detecting the expression of EVs markers such as CD81 and ALIX (Supplementary Figure 3B). Before and after electroporation, the NTA analysis of the VSF showed EVs with similar size to that observed by TEM. Both VSF and eVSF (electroporated VSF with synthetic miR-1252-5p) were rich in nanoparticles with a mean size of 119.63 ± 13.01 nm and 137.4 ± 9.24 nm, respectively. Therefore, we reasoned that the electroporation protocol applied did not induce any significant change in EVs size distribution (Supplementary Figure 3C).
Figure 4.
Engineered EVs carrying miR-1252-5p sensitizes MM cells to BTZ. (A) Experimental strategy of miR-1252-5p delivery by HEK293T EVs. (B) Relative levels of miR-1252-5p in untreated HEK293T VSF, incubated or electroporated with 30 pmol of synthetic miR-1252-5p, and treated with RNase was measured by RT-qPCR. (C) RPMI-8226 WT cells were treated with HEK293T VSF, eVSFctrl, or eVSF, on the presence of 5.0 nM BTZ and cell viability was quantified using PrestoBlue assay. Asterisks denote significance as determined by one-way ANOVA, multiple comparisons followed by Bonferroni test (post-test) (*p < 0.0001).
Moreover, since the analysis of miR-1252-5p expression in the total RNA of HEK293T cells did not detect the presence of this miRNA (Supplementary Figure 4), it was once expected no levels of miR-1252-5p in the fresh HEK293T-VSF. Hence, to quantify the loading of miR-1252-5p into EVs, the VSF was mixed with 30 pmol of synthetic miR-1252-5p and, as was foreseeable, we were able to detect the presence of this miRNA in the mixture before electroporation (this amount detected was considered as 100% for the further RT-qPCR relative expression analysis). Subsequently to electroporation, we observed a reduction of 54.4% in the level of the miR-1252-5p in the VSF. RNase treatment was able to eliminate all miR-1252-5p in the VSF (outside the EVs). Unsurprisingly, it was not possible to detect miRNA in the VSF not submitted to electroporation. However, 20.2% of miR-1252-5p could be distinguished in the electroporated VSF even after the RNase treatment. In light of these facts, it can be assumed that about a fifth of the miR-1252-5p mixed with the VSF was protected to RNase degradation because it was loaded in the EVs by the electroporation process (Figure 4B).
Lastly, we tested the effect of EVs carrying miR-1252-5p in the response of RPMI-8226 cells to BTZ treatment. Accordingly, VSF, eVSF, or eVSFctrl were added to RPMI-8226 in the presence of 5.0 nM of BTZ. Consolidating our study, there was a significant decrease in the percentage of viable RPMI-8226 cells treated for 48 h with eVSF and BTZ (28.2%) in comparison to the BTZ treatment alone (42.4%) or VSF alone (without miR-1252-5p, 41.7%), or with eVSF+miRctrl (43.5%) (Figure 4C). These findings indicated that EVs containing miR-1252-5p were able to sensitize MM cells to BTZ treatment.
Discussion
MM is an incurable plasma dyscrasia and one of the deadliest malignancies worldwide.2 Meanwhile, nowadays, the main goal of MM therapy is to prolong as much as possible the overall survival of the patients.32 Although the insertion of BTZ in all treatment phases significantly improved MM patients’ prognosis, including benefits to the progression-free survival and all response rates, most patients develop resistance and become refractory to their treatment regimens.4 Therefore, there is a critical unmet need for the discovery of new approaches to increase BTZ sensitivity that could efficiently destroy MM cells.
Several groups have made attempts to identify factors that predict BTZ response based on biological hypotheses. HPSE is an enzyme highly expressed in the tumor microenvironment and associated with poor survival and treatment resistance in the MM context.11,12,33 Of note, Ramani et al15 demonstrated that HPSE could promote BTZ resistance and relapse in MM cells via extracellular-signal-regulated kinase (ERK) signaling, and the HPSE inhibitor (Roneparstat) was able to enhance chemotherapy effect on MM cells. Moreover, it was observed that HPSE expression was induced by anti-MM drugs in cells expressing little or no HPSE, and extracellular vesicles shed by treated MM cells could transfer HPSE, mediating tumor chemoresistance and eventual post-therapy relapse to the recipient cells through the activation of cell signaling, cell migration, and cytokine expression.34,35 Therefore, anti-HPSE therapies could be a novel sensitization approach in MM, intending to enhance BTZ response and to prevent tumor relapse.
The use of miRNA as potential therapy has provided new applicable tools to increase BTZ-induced cytotoxicity on MM cells.21 MiR-145-3p was recently related to lead to autophagic flux through the regulation of histone deacetylase 4, enhancing BTZ activity on both in vitro and in vivo MM models.36 Moreover, miR-520g and miR-520h triggered the inhibition of DNA repair system through downregulation of human apurinic/apyrimidinic endonuclease 1 (APE1), while miR‑497 targeted B‑cell lymphoma 2 (BCL2) expression, contributing to MM cell apoptosis through cell cycle arrest, and these miRNAs overcame BTZ resistance in MM cell lines.37,38 Nonetheless, few data providing evidence of miRNAs regulating HPSE activity negatively are available. The miR-1258 was described as modulating HPSE expression in gastric, breast, and brain tumors.16–18 Likewise, it was demonstrated that miR-30 in melanoma metastasis as well as miR-429 and miR-299-3p in gastric cancer could also target HPSE, promoting its downregulation.39–41 Conversely, the role of miRNA regulating HPSE expression in MM and, consequently, its impact on sensitivity to BTZ has not yet been reported.
Analysis using in silico prediction tools could indicate several microRNAs that can regulate HPSE expression, including miR-20b-5p and miR-1252-5p, and these miRNAs were also associated with tumorigenesis signaling pathways. However, it is not fully understood the role of miR-1252-5p. This miRNA was reported as an oncomiR in lung cancer due to the negative regulation of Itchy E3 ubiquitin-protein ligase,42 which inhibits cell proliferation, and as an anti-oncomiR by targeting FOXR2 and suppressing tumor growth.43 On the other hand, miR-20b seems to play a role as a tumor suppressor inhibiting cell proliferation, migration, and invasion of bladder cancer cells by targeting matrix metalloproteinase-2 expression,44 and suppressing the levels of a desintegrin and metalloprotease 9 and consequently decreasing the 5-FU resistance in colon cancer cells.45 Furthermore, there is no evidence of a biological function of these microRNAs on MM or targeting HPSE.
Consistently with our previous data reporting, U266 cells are less susceptible to BTZ therapy than RPMI-8226.8 In light of this fact, we sought to evaluate if there was any correlation among the expression levels of miRNAs (miR-20b-5p and miR-1252-5p) and the quantity of HPSE transcripts. Our data indicated that U266 had higher levels of HPSE transcripts than RPMI-8226. Because of the lack of significant correlation between the expression levels of miR-20b-5p and HPSE expression in both MM lines, we suspected that this miRNA should not play a meaningful role regulating HPSE expression. Interestingly, we detected higher levels of miR-1252-5p in RPMI-8226 cells than in U266 cells, which present elevated levels of HPSE transcript and are more resistant to BTZ treatment, as reported previously.8 Together, these findings suggest an association within BTZ sensitivity, miR-1252-5p expression level, and HPSE expression.
To better understand the role of miR-1252-5p in MM tumorigenesis, we overexpressed a synthetic mimic of miR-1252-5p in the MM cells. Our data revealed a dramatic reduction in the levels of HPSE transcripts, protein, and enzymatic activity in MM cells overexpressing miR-1252-5p, suggesting that this microRNA could promote the downregulation of this gene. Hereinafter, our results propose that synthetic miR-1252-5p could decrease the viability of MM cells and potentiate BTZ cytotoxic effect by targeting HPSE transcripts, enhancing the cell death rate in MM cells. These findings follow Ramani et al15 who proposed that HPSE absence was associated with better response to BTZ in MM cells. These results point out a biological rationale using synthetic miR-1252-5p mimic, in combination with BTZ, as a novel therapeutic strategy for the treatment of multiple myeloma. However, although it was not evaluated in this study, miR-1252-5p might also regulate other phenotypes, such as the cell cycle control or induce changes in the cytoskeleton and oxidative stress that end up altering cell viability, but not cell death.
Even though some of the synthetic RNAs reached clinical trials, the use of miRNA as theranostic is limited because most of the miRNA delivery methods are known to induce immunogenic or cytotoxic responses.23 Extracellular vesicles have been implicated in physiological intercellular communication mediated by its cargo and are considered natural carriers of coding and noncoding RNAs, overcoming most cellular barriers, such as RNase susceptibility and cytotoxicity.25,46 In this scenario, EVs represent an emerging vehicle for therapeutic miRNA delivery, with great potential clinical relevance for blood tumors, including MM.47–49 Currently, electroporation has been established as a standard method for loading RNAs into EVs.24–26 Although it was described that electric fields can induce EVs aggregation, disturbing its morphology and function,50 our data showed no relevant difference in the size distribution between control and electroporated EVs. Hence, we sought to use these modified HEK293T-derived EVs, which are considered safe EVs from immortalized cells that do not induce toxicity or immune response in animal models,51 carrying miR-1252-5p to increase the sensitivity of MM cells to BTZ treatment. Our results indicate that EVs positively carrying miR-1252-5p are sufficient to sensitize MM cells to BTZ treatment.
Notwithstanding, one of the major technical issues in EVs field is the reliability and reproducibility of techniques to isolate biologically relevant EVs. Therefore, further efforts are still required to optimize a clinical protocol, from the EVs isolation and electroporation to provide an efficient transfer of therapeutic miRNA to the target cell. Moreover, there are some pitfalls in this study, including the lack of in vivo models for MM treated with BTZ under the conditions presented in this study, as well as the use of an anti-miR-1252-5p. Besides, it does not go unnoticed that mutation assays or pull-down assays should confirm that miR-1252-5p directly binds to HPSE 3ʹUTR. Indeed, a better molecular characterization of miR-1252-5p mechanism of action is relevant, especially by identifying other pathways regulated by this microRNA on MM cells, that might increase BTZ sensitivity in MM.
In summary, our results indicate for the first time that miR-1252-5p downregulated HPSE expression in MM that possibly affects the degradation of heparan sulfate chains and, consequently, proteoglycans signaling in carcinogenesis, contributing to the reduction of cell viability, and improving cell death rates prompted by BTZ. Based on these results, we suggest that synthetic miR-1252-5p mimics loaded into EVs could be a novel sensitization approach for MM cells regardless of the HPSE levels. We also speculate that patients expressing miR-1252-5p may have better response and lower relapse rates for BTZ-based therapy approaches, although further studies are required to confirm this hypothesis.
Acknowledgments
DMR-Jr acknowledges the support of São Paulo Research Foundation (FAPESP #2015/21420-3).
Funding Statement
This study was supported by grants from São Paulo Research Foundation (FAPESP #2015/09182-0; #2015/03964-6; and #2016/01357-8).
Abbreviations
7-AAD, 7-aminoactinomycin D; AV, annexin V; Bcl-2, B‑cell lymphoma 2; BM, bone marrow; BTZ, bortezomib; EFS, event-free survival; ERK, extracellular-signal-regulated kinase; EV, extracellular vesicle; eVSF, electroporated VSF with miR-1252-5p; FBS, fetal bovine serum; GAGs, glycosaminoglycans; HPSE, heparanase; HS, heparan sulfate; KEGG, Kyoto Encyclopedia of Genes and Genomes; miRNA, microRNA; MM, multiple myeloma; mRNA, messenger RNAs; NTA, nanoparticle tracking analysis; OS, overall survival; PDA, propylene–diamine–acetate; PIs, proteasome inhibitors; RT-qPCR, quantitative reverse transcription–polymerase chain reaction; TEM, transmission electron microscopy; UTR, untranslated regions; VSF, vesicular secretome fraction.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author (DMR-Jr) on reasonable request.
Ethics Approval and Consent to Participate
This study was approved by the institutional ethics committees (CEP-UNIFESP: 0045/2015).
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
Maria Aparecida Silva Pinhal and André Luiz Vettore report grants from FAPESP - São Paulo Research Foundation (Brazil), during the conduct of the study. The authors declare that they have no other potential conflicts of interest for this work.
References
- 1.Kyle RA, Rajkumar S. Criteria for diagnosis, staging, risk stratification and response assessment of multiple myeloma. Leukemia. 2009;23(1):3–9. doi: 10.1038/leu.2008.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364(11):1046–1060. doi: 10.1056/NEJMra1011442 [DOI] [PubMed] [Google Scholar]
- 3.Mateos MV, Ocio EM, San Miguel JF. Novel generation of agents with proven clinical activity in multiple myeloma. Semin Oncol. 2013;40(5):618–633. doi: 10.1053/j.seminoncol.2013.07.005 [DOI] [PubMed] [Google Scholar]
- 4.Moreau P, Richardson PG, Cavo M, et al. Proteasome inhibitors in myeloma: 10 years later. Blood. 2012;120(5):947–959. doi: 10.1182/blood-2012-04-403733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grosicki S, Barchnicka A, Jurczyszyn A, Grosicka A. Bortezomib for the treatment of multiple myeloma. Expert Rev Hematol. 2014;7(2):173–185. doi: 10.1586/17474086.2014.899144 [DOI] [PubMed] [Google Scholar]
- 6.Hostenkamp G, Lichtenberg FR. The impact of recent chemotherapy innovation on the longevity of myeloma patients: US and international evidence. Soc Sci Med. 2015;130:162–171. doi: 10.1016/j.socscimed.2015.02.003 [DOI] [PubMed] [Google Scholar]
- 7.Pulte D, Redaniel MT, Brenner H, Jansen L, Jeffreys M. Recent improvement in survival of patients with multiple myeloma: variation by ethnicity. Leuk Lymphoma. 2014;55(5):1083–1089. doi: 10.3109/10428194.2013.827188 [DOI] [PubMed] [Google Scholar]
- 8.Rodrigues-Junior DM, Biassi TP, de Albuquerque GE, et al. Downregulation of DCC sensitizes multiple myeloma cells to bortezomib treatment. Mol Med Rep. 2019;19(6):5023–5029. doi: 10.3892/mmr.2019.10142 [DOI] [PubMed] [Google Scholar]
- 9.Kelly T, Miao HQ, Yang Y, et al. High heparanase activity in multiple myeloma is associated with elevated microvessel density. Cancer Res. 2003;63(24):8749–8756. [PubMed] [Google Scholar]
- 10.Sanderson RD, Yang Y, Suva LJ, Kelly T. Heparan sulfate proteoglycans and heparanase—partners in osteolytic tumor growth and metastasis. Matrix Biol. 2004;23(6):341–352. doi: 10.1016/j.matbio.2004.08.004 [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Macleod V, Bendre M, et al. Heparanase promotes the spontaneous metastasis of myeloma cells to bone. Blood. 2005;105(3):1303–1309. doi: 10.1182/blood-2004-06-2141 [DOI] [PubMed] [Google Scholar]
- 12.Mahtouk K, Hose D, Raynaud P, et al. Heparanase influences expression and shedding of syndecan-1, and its expression by the bone marrow environment is a bad prognostic factor in multiple myeloma. Blood. 2007;109(11):4914–4923. doi: 10.1182/blood-2006-08-043232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hulett MD, Freeman C, Hamdorf BJ, Baker RT, Harris MJ, Parish CR. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis. Nat Med. 1999;5(7):803–809. doi: 10.1038/10525 [DOI] [PubMed] [Google Scholar]
- 14.Toyoshima M, Nakajima M. Human heparanase: purification, characterization, cloning, and expression. J Biol Chem. 1999;274(34):24153–24160. doi: 10.1074/jbc.274.34.24153 [DOI] [PubMed] [Google Scholar]
- 15.Ramani VC, Zhan F, He J, et al. Targeting heparanase overcomes chemoresistance and diminishes relapse in myeloma. Oncotarget. 2016;7(2):1598–1607. doi: 10.18632/oncotarget.6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Sullivan PS, Goodman JC, Gunaratne PH, Marchetti D. MicroRNA-1258 suppresses breast cancer brain metastasis by targeting heparanase. Cancer Res. 2011;71(3):645–654. doi: 10.1158/0008-5472.CAN-10-1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang D, Zhang Q, Zhao S, et al. The expression and clinical significance of microRNA-1258 and heparanase in human breast cancer. Clin Biochem. 2013;46(10–11):926–932. doi: 10.1016/j.clinbiochem.2013.01.027 [DOI] [PubMed] [Google Scholar]
- 18.Shi J, Chen P, Sun J, et al. MicroRNA-1258: an invasion and metastasis regulator that targets heparanase in gastric cancer. Oncol Lett. 2017;13(5):3739–3745. doi: 10.3892/ol.2017.5886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- 20.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- 21.Ahmad N, Haider S, Jagannathan S, Anaissie E, Driscoll JJ. MicroRNA theragnostics for the clinical management of multiple myeloma. Leukemia. 2014;28(4):732–738. doi: 10.1038/leu.2013.262 [DOI] [PubMed] [Google Scholar]
- 22.Abdi J, Rastgoo N, Li L, Chen W, Chang H. Role of tumor suppressor p53 and micro-RNA interplay in multiple myeloma pathogenesis. J Hematol Oncol. 2017;10(1):169–180. doi: 10.1186/s13045-017-0538-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juliano RL. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44(14):6518–6548. doi: 10.1093/nar/gkw236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Usman WM, Pham TC, Kwok YY, et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat Commun. 2018;9(1):2359. doi: 10.1038/s41467-018-04791-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Andaloussi S, Lee Y, Lakhal-Littleton S, et al. Exosome-mediated delivery of siRNA in vitro and in vivo. Nat Protoc. 2012;7(12):2112–2126. doi: 10.1038/nprot.2012.131 [DOI] [PubMed] [Google Scholar]
- 26.Pomatto MAC, Bussolati B, D’Antico S, et al. Improved loading of plasma-derived extracellular vesicles to encapsulate antitumor miRNAs. Mol Ther Methods Clin Dev. 2019;13:133–144. doi: 10.1016/j.omtm.2019.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are MicroRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 28.Vlachos IS, Zagganas K, Paraskevopoulou MD, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43(W1):W460–6. doi: 10.1093/nar/gkv403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouças RI, Trindade ES, Tersariol IL, Dietrich CP, Nader HB. Development of an enzyme-linked immunosorbent assay (ELISA)-like fluorescence assay to investigate the interactions of glycosaminoglycans to cells. Anal Chim Acta. 2008;618(2):218–226. doi: 10.1016/j.aca.2008.04.059 [DOI] [PubMed] [Google Scholar]
- 30.Rodrigues-Junior DM, Tan SS, Lim SK, et al. Circulating extracellular vesicle-associated TGFβ3 modulates response to cytotoxic therapy in head and neck squamous cell carcinoma. Carcinogenesis. 2019;40(12):1452–1461. doi: 10.1093/carcin/bgz148 [DOI] [PubMed] [Google Scholar]
- 31.Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;30(1). doi: 10.1002/0471143030.cb0322s30 [DOI] [PubMed] [Google Scholar]
- 32.Röllig C, Knop S, Bornhäuser M. Multiple myeloma. Lancet. 2015;385(9983):2197–2208. doi: 10.1016/S0140-6736(14)60493-1 [DOI] [PubMed] [Google Scholar]
- 33.Ritchie JP, Ramani VC, Ren Y, et al. SST0001, a chemically modified heparin, inhibits myeloma growth and angiogenesis via disruption of the heparanase/syndecan-1 axis. Clin Cancer Res. 2011;17(6):1382–1393. doi: 10.1158/1078-0432.CCR-10-2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramani VC, Vlodavsky I, Ng M, et al. Chemotherapy induces expression and release of heparanase leading to changes associated with an aggressive tumor phenotype. Matrix Biol. 2016;55:22–34. doi: 10.1016/j.matbio.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bandari SK, Purushothaman A, Ramani VC, et al. Chemotherapy induces secretion of exosomes loaded with heparanase that degrades extracellular matrix and impacts tumor and host cell behavior. Matrix Biol. 2018;65:104–118. doi: 10.1016/j.matbio.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu H, Liu C, Yang Q, et al. MIR145-3p promotes autophagy and enhances bortezomib sensitivity in multiple myeloma by targeting HDAC4. Autophagy. 2019;4:1–15. doi: 10.1080/15548627.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian F, Zhan Y, Zhu W, et al. MicroRNA-497 inhibits multiple myeloma growth and increases susceptibility to bortezomib by targeting Bcl-2. Int J Mol Med. 2019;43(2):1058–1066. doi: 10.3892/ijmm.2018.4019. [DOI] [PubMed] [Google Scholar]
- 38.Yuan X, Ma R, Yang S, et al. miR-520g and miR-520h overcome bortezomib resistance in multiple myeloma via suppressing APE1. Cell Cycle. 2019;18(14):1660–1669. doi: 10.1080/15384101.2019.1632138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu XY, Tang QS, Chen HC, Jiang XL, Fang H. Lentiviral miR30-based RNA interference against heparanase suppresses melanoma metastasis with lower liver and lung toxicity. Int J Biol Sci. 2013;9(6):564–577. doi: 10.7150/ijbs.5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheng N, Zhang L, Yang S. MicroRNA-429 decreases the invasion ability of gastric cancer cell line BGC-823 by downregulating the expression of heparanase. Exp Ther Med. 2018;15(2):1927–1933. doi: 10.3892/etm.2017.5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou X, Hu M, Ge Z. Tumor suppressive miR 299 3p inhibits gastric cancer cell invasion by targeting heparanase. Mol Med Rep. 2019;20(3):2151–2158. doi: 10.3892/mmr.2019.10436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian F, Yu CT, Ye WD, Wang Q. Cinnamaldehyde induces cell apoptosis mediated by a novel circular RNA hsa_circ_0043256 in non-small cell lung cancer. Biochem Biophys Res Commun. 2017;493(3):1260–1266. doi: 10.1016/j.bbrc.2017.09.136. [DOI] [PubMed] [Google Scholar]
- 43.Tian X, Zhang L, Jiao Y, Chen J, Shan Y, Yang W. CircABCB10 promotes nonsmall cell lung cancer cell proliferation and migration by regulating the miR-1252/FOXR2 axis. J Cell Biochem. 2018;120(3):3765–3772. doi: 10.1002/jcb.27657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park SL, Cho TM, Won SY, et al. MicroRNA-20b inhibits the proliferation, migration and invasion of bladder cancer EJ cells via the targeting of cell cycle regulation and Sp-1-mediated MMP-2 expression. Oncol Rep. 2015;34(3):1605–1612. doi: 10.3892/or.2015.4119. [DOI] [PubMed] [Google Scholar]
- 45.Fu Q, Cheng J, Zhang J, et al. miR-20b reduces 5-FU resistance by suppressing the ADAM9/EGFR signaling pathway in colon cancer. Oncol Rep. 2017;37(1):123–130. doi: 10.3892/or.2016.5259. [DOI] [PubMed] [Google Scholar]
- 46.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is anovel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 47.Moloudizargari M, Abdollahi M, Asghari MH, et al. The emerging role of exosomes in multiple myeloma. Blood Rev. 2019;38:100595. doi: 10.1016/j.blre.2019.100595 [DOI] [PubMed] [Google Scholar]
- 48.Gargiulo E, Morande PE, Largeot A, et al. Diagnostic and therapeutic potential of extracellular vesicles in b-cell malignancies. Front Oncol. 2020;10:580874. doi: 10.3389/fonc.2020.580874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desantis V, Saltarella I, Lamanuzzi A, et al. MicroRNAs-based nano-strategies as new therapeutic approach in multiple myeloma to overcome disease progression and drug resistance. Int J Mol Sci. 2020;21(9):3084. doi: 10.3390/ijms21093084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnsen KB, Gudbergsson JM, Skov MN, et al. Evaluation of electroporation-induced adverse effects on adipose-derived stem cell exosomes. Cytotechnology. 2016;68(5):2125–2138. doi: 10.1007/s10616-016-9952-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu X, Badawi M, Pomeroy S, et al. Comprehensive toxicity and immunogenicity studies reveal minimal effects in mice following sustained dosing of extracellular vesicles derived from HEK293T cells. J Extracell Vesicles. 2017;6(1):1324730. doi: 10.1080/20013078.2017.1324730. [DOI] [PMC free article] [PubMed] [Google Scholar]