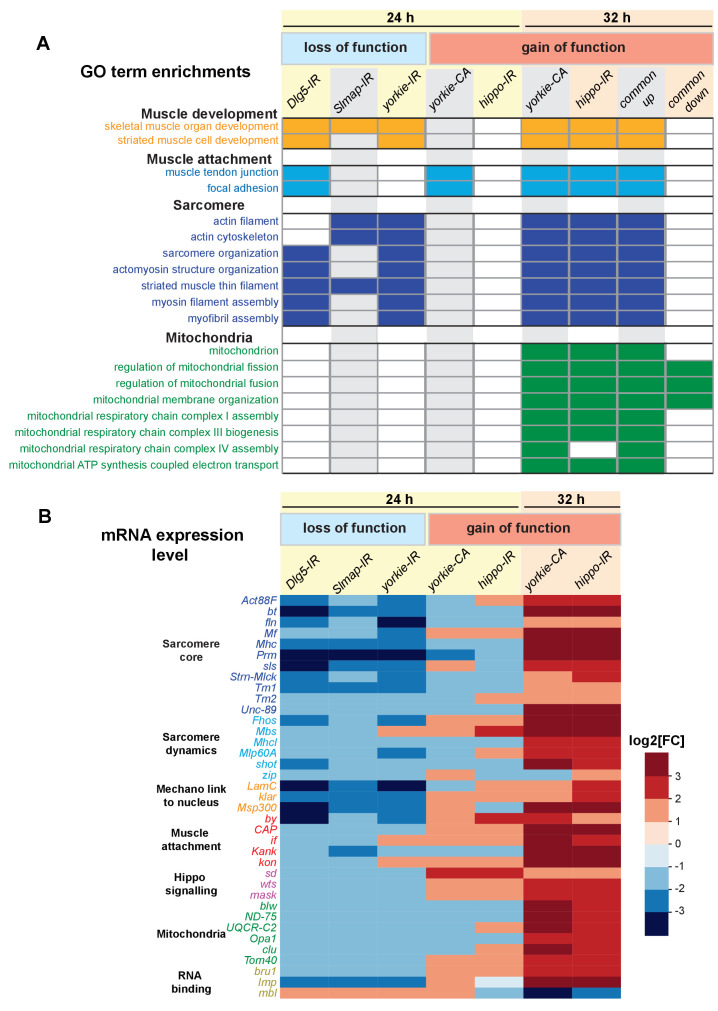
Figure 8. Yorkie transcriptionally controls sarcomeric and mitochondrial genes expression.
(A) Gene ontology (GO)-term enrichments in genes lists that are significantly changed in the various loss (Dlg5-IR, Slmap-IR, yorkie-IR) and gain-of-function (yorkie-CA, hippo-IR) yorkie conditions at 24 hr and 32 hr after puparium formation (APF) compared to wild-type controls (see Supplementary file 3). Note the strong enrichment of sarcomere related GO-terms in the 24 hr APF loss-of-function and the 32 hr APF gain-of-function condition. Thirty-two hr APF gain of function is also strongly enriched for mitochondrial GO-terms. (B) Plot displaying the log2-fold change of transcript levels for individual genes of the above genotypes compared to control (up in red, down in blue). Note the strong downregulation of the sarcomeric genes in the 24 hr APF loss-of-function conditions, in particular the titin homologs bt and sls, as well as Mhc, Prm and Act88F.