Abstract
Most mammalian cells possess a single, non-motile primary cilium that plays an important role in mediating cellular signaling pathways, such as Hedgehog (Hh) signaling. Primary cilia are present on testicular somatic cells and demonstrate a temporal expression during development; however, their role in testicular morphogenesis is not well characterized. To investigate the role of primary cilia and Hh signaling in Sertoli cells on morphogenesis, we inhibited assembly of primary cilia through CRISPR Cas9–mediated gene editing of ODF2, a structural component of primary cilia and siRNA-mediated gene silencing of IFT88, a functional component of the intraflagellar transport system. Knockdown of ODF2 and IFT88 resulted in a 50% reduction in the number of cells with primary cilia and significant shortening of the remaining cilia. The expression of GLI1, a downstream target of Hh signaling, was significantly reduced when IFT88 but not ODF2, was downregulated. When morphogenesis was examined using tubule formation in vitro and a novel testicular organoid system, loss of cilia after knockdown of both targets affected cellular assembly and organization. While the Hh pathway was found to be active during morphogenesis in vitro, addition of the Hh antagonist cyclopamine did not affect morphogenesis in either in vitro system. These results indicate that primary cilia are important for morphogenesis in vitro but Hh signaling is not the cilia-mediated pathway responsible for orchestrating morphogenic organization.
Keywords: Primary cilia, Morphogenesis, Hedgehog signaling, Sertoli cells, Organoids
Introduction
The primary cilium was once considered a vestigial structure with little to no important function within the body; however, it has been proven to be a critical organelle with a wide variety of cellular functions. Almost all cell types possess a primary cilium, including terminally differentiated cells, differentiating cells, embryonic stem cells and some adult stem cells (Kiprilov et al. 2008; He et al. 2014). Primary cilia have the ability to respond to a wide variety of stimuli including changes in light, temperature, and signaling molecules (Christensen et al. 2005; Insinna and Besharse 2008; Kihara et al. 2008; Moorman and Shorr 2008). These organelles are important in the determination of cell fate and directed differentiation and play a critical role in embryogenesis, where they can be found on epiblast cells as early as embryonic day 6.0 to assist with patterning of the mouse embryo (Bangs et al. 2015). The importance of the primary cilium is further emphasized by its connection to a wide variety of diseases, collectively termed ciliopathies, which occur when one or more genes implicated in ciliary structure or function are disrupted.
The structure of the primary cilium is granted by the central axoneme, composed of microtubule doublets in a 9+0 pattern (Satir and Christensen 2007). The axoneme emanates from the mother centriole (now termed basal body) that is anchored to the cellular membrane by distal and subdistal appendages (Szymaska and Johnson 2012; Tanos et al. 2013). While better known for its role in the sperm tail, Outer Dense Fibre Protein 2 (ODF2) is localized at the distal appendages of the basal body at the base of the cilium (Nakagawa et al. 2001; Donkor et al. 2004). When both alleles of ODF2 were deleted in F9 teratocarcinoma cells, primary cilia failed to form, implicating ODF2 as an important contributor to ciliary stability (Ishikawa et al. 2005).
The primary cilium lacks the cellular machinery for protein synthesis and therefore relies on the intraflagellar transport (IFT) system to import and export proteins important for assembly and signaling. Cargo is moved by two cellular motors, kinesin-2 and cytoplasmic dynein 2, in the antero- and retrograde directions, respectively (Ishikawa and Marshall 2017). Both motors are essential for ciliary assembly and cilia fail to form if some motor subunits are absent (Ishikawa and Marshall 2017). Two IFT complexes, termed A and B, link the cargo being transported to the cellular motors. Both complexes are composed of a variety of IFT proteins whose individual loss can negatively affect, or even completely disrupt, the entire transport system (Cole et al. 1998; Pazour et al. 2000). IFT complex B acts in conjunction with kinesin-2 in the anterograde direction and IFT complex A works with dynein in the retrograde direction (Pazour et al. 2000; Iomini et al. 2009).
Primary cilia function as a cellular antenna and therefore possess the machinery necessary for a variety of different signaling pathways, including Hedgehog (Hh), PDGFRα, and Wnt (Satir, Pedersen and Christensen 2010; Wheway, Nazlamova and Hancock 2018). Currently, the best described signaling pathway mediated by the primary cilium is the Hh pathway that is entirely reliant upon primary cilia to facilitate signaling (Huangfu et al. 2003; Bangs and Anderson 2017). Hh signaling is critical in processes such as embryogenesis, body axis patterning, homeostasis, cellular migration, and stem cell maintenance (Bangs and Anderson 2017). Briefly, in the presence of Hh ligand, the transmembrane protein Patched1 no longer represses the movement of another transmembrane protein Smo, allowing it to enter the ciliary compartment and inhibit proteins that sequester GLI transcription factors that activate genes downstream of this pathway (Rohatgi, Milenkovic and Scott 2007). There are a large variety of genes downstream of the Hh pathway whose expression can be variable depending on the developmental stage of the cell, cell type and cell cycle.
Primary cilia are present on many of the cells in the male reproductive tract, including those within the testis (reviewed by Girardet et al. 2019). We previously demonstrated that loss of primary cilia in testicular somatic cells via the cytoplasmic dynein antagonist Ciliobrevin D resulted in disruption of tubular morphogenesis in vitro (Firestone et al. 2012; Dores and Dobrinski 2014). As Ciliobrevin D does not permanently inhibit cilia formation, here we employed two different genetic disruption techniques, CRISPR-Cas9 and siRNA, of two different ciliary targets, ODF2 and IFT88, to study the effect of their loss on primary cilia formation and morphogenesis. We investigated morphogenesis in testicular tubules formed in vitro and in testicular organoids, as organoids provide a highly reproducible and quantifiable culture system (Sakib et al., 2019). We also investigated Hh signaling upon loss of primary cilia and during in vitro morphogenesis in both systems.
Materials and methods
Enzymatic digestion
Porcine testes were donated from a commercial Alberta farm following castration of 1-week old pigs. Sourcing of testes was approved by the Animal Care Committee at the University of Calgary. The epididymis and tunica vaginalis were removed, the testes decapsulated and the mediastinum discarded. Tissue was minced and then subjected to a two-step enzymatic digestion (Sakib et al. 2019). Briefly, collagenase IV (2 mg/mL; Sigma and Worthington) was used to isolate tubules and trypsin 0.25% EDTA (Gibco) was then added to separate the cells within the tubules (Sertoli and germ cells).
Differential plating
Sertoli cells were separated from germ cells through their differential adhesion properties (Luo et al. 2006). Testicular cells were plated at a concentration of 25 x 106 cells per 100-mm tissue culture dishes (Corning) in 8 mL DMEM F12 (Sigma) with 10% fetal bovine serum (FBS) (Gibco), hereby referred to as Sertoli cell medium and incubated at 37 °C and 5% CO2 for 90 min. During this time Sertoli cells adhere, while germ cells remain in suspension. Then media containing the non-adherent germ cells and cellular debris was removed and replaced with new Sertoli cell medium and cells were placed at 37 °C and 5% CO2 overnight for collection the following morning.
Cell characterization
The collected cells were characterized for Sertoli cell number. From each cell preparation, 5 × 106 cells were permeabilized in 0.1% Triton-X (EMD) in DPBS for 10 min, washed in 2 mL of 1% BSA in DPBS and centrifuged at 500×g for 5 min. Samples were blocked for 15 min with CAS Block (Thermo Fisher Scientific 008120) and then incubated with a primary antibody to GATA 4 to identify Sertoli cells (Tevosian et al. 2002) (1:200, Santacruz Biotechnology) or an isotype control overnight at 4 °C. Cells were washed again with 2 mL of 1% BSA in DPBS and secondary antibodies were added and incubated for 1 h at room temperature in the dark. Following another wash and centrifugation, the supernatant was removed and 200 μL of 1% BSA in DPBS was added and cells were transferred to 5 mL round-bottom FACS tubes for analysis on a BD FACSAria flow cytometer.
Gene targeting constructs
Three different CRISPR-Cas9 guide RNA sequences were designed based on the predicted porcine ODF2 sequence (Table S1). The gRNA locations were chosen based on their proximity to the first functional (coiled coil) domain of ODF2 and their lack of off target effects. The gRNAs, Cas9 protein and Cy3-labelled tracr-RNA were obtained from Integrated DNA Technologies (IDT) as part of the IDT Alt-R system and a non-targeting gRNA was used as a control. Custom siRNA (Dharmacon) was designed based on a published siRNA sequence for porcine IFT88 that was successfully used to knockdown IFT88 in a porcine kidney cell line (Table S1) (Delaval et al. 2011). The siRNA construct was linked to DY647 to allow for FACS sorting of transfected cells. A scrambled siRNA was used as a control (Invitrogen).
Transfection
gRNAs and Cas9 were delivered using lipofection as per IDT delivery protocol. Cells and complexes were plated on 6-well plates (Corning 353846) and left for 48 h at 37 °C. IFT88 siRNA was delivered using Lipofectamine 3000 (Thermo Fisher), following the manufacturers protocol to 106 adherent cells in OptiMEM on 6-well plates. Cells were incubated overnight and collected for FACS the following morning.
FACS
After 48 h of culture in Sertoli cell medium, CRISPR-transfected cells were collected. After overnight culture in OptiMEM, siRNA transfected cells were collected. All cells were subjected to fluorescence-activated cell sorting (FACS), on a BD FACSAria and were analyzed and gated using FACSDiva software. Only Cy3- or DY647-positive cells were collected. Cells were sorted directly into Sertoli cell media for further culture or Trizol (Ambion) for immediate RNA isolation.
RNA isolation and cDNA synthesis
RNA was isolated using the Qiagen RNeasy microkit protocol. Following elution, RNA concentration was assessed on a nanophotometer and RNA was stored at − 80 °C until it was converted to cDNA using the Applied Biosystems high capacity cDNA reverse transcription kit with RNase inhibitor (LS4374966) following the manufacturers protocol. cDNA was diluted to a final concentration of 100 pg/μL.
RT-qPCR
qPCR was performed using the Applied Biosystems 7500 Fast Real-Time PCR system. cDNA was used at a concentration of 0.01 ng/μL. A list of genes and primers investigated is included in Table S2. Primers were designed to span exon-exon junctions to avoid amplification of genomic DNA. Relative expression is calculated using 2−ΔΔCT.
Hedgehog agonist treatment
Sorted cells were seeded onto 6-well plates and allowed 24 h recovery in Sertoli cell media. The following day, Sertoli cell media was removed and replaced with serum-free media (DMEM F12, 1× P/S) supplemented with 10 μM SAG (EMD Millipore) or DMSO (Sigma) as a vehicle control. Cells were treated for 24 h, the media removed and cells were lysed directly on the plate with Buffer RLT from a Qiagen microkit. Lysate was moved to a 15-mL conical tube and the manufacturers RNA extraction protocol was followed.
Assessment of editing
Sorted cells were collected after FACS, washed in PBS, pelleted at 500×g and stored at − 80 °C until analysis of cleavage by CelI assay. PCR flanking the targeted sites (primer sequences included Table S2) was conducted using AccuStart™ Taq DNA Polymerase HiFi (Quanta Biosciences) with 100 ng of template DNA according to the manufacturer’s recommendations. The frequency of mutation in a population was analyzed with the Surveyor mutation detection kit (Transgenomic) according to the manufacturer’s recommendations, using 10 μL of the PCR product as described above. Surveyor reactions were resolved on a 10% TBE polyacrylamide gel and visualized by ethidium bromide staining. Densitometry measurements of the bands were performed using ImageJ and the mutation rate of Surveyor reactions was calculated as described previously (Guschi et al. 2010). Extracted DNA from edited cells was analyzed by Sanger sequencing followed by ICE analysis (Synthego) to generate ICE and KO scores to assess editing and knockout efficiencies.
Immunocytochemistry for primary cilia
Cells were grown on glass cover slips (Fisher) in 24-well plates for immunocytochemistry. Cells were serum starved for 48 h before fixation in 4% PFA for 15 min at room temperature. Cells were permeabilized in 0.2% Triton-X100 in PBS, washed in PBS three times for 5 min, and blocked with CasBlock (Life Technologies) for 15 min at room temperature. An ARL13B antibody (1:500, Proteintech) was diluted in 10% CasBlock in PBS, added to the cells and left at 4 °C overnight. Cells were then washed with PBS two times for 5 min and incubated with donkey anti-rabbit Alexafluor 555 (1:500, Invitrogen) for 1–2 h at room temperature. Following 3 washes in PBS and one wash in Milli-Q H2O, Vectashield with DAPI (Vector) was added and glass coverslips were placed on slides for imaging using a Zeiss ImagerM.2 microscope with Zeiss AxioVision Rel.4.8 software. Quantification of cells in immunofluorescence images was performed with ImageJ software (Version 2.0.0). Ciliary length was quantified using ImageJ software after immunocytochemistry. Slides were imaged in one plane and all cilia were measured from point of origin to tip as visualized by ARL13B.
In vitro tubule formation assay
Glass cover slips were added to the bottom of a 24-well plate, and 300 μL of 1:1 DMEM:Growth factor reduced Matrigel (Corning) was added and allowed to solidify for 30 min at 37 °C. A total of 106 cells were then seeded onto the solid Matrigel in serum-free media (DMEM F12, 1× P/S, 1× ITS) and incubated at 37 °C for 1 to 5 days to allow for tubules to form (Dores et al. 2017; adapted from Gassei et al. 2006). Tubules were imaged at various times during culture.
Cell proliferation assay
Cells were pulsed with EdU (5-ethynyl-2′-deoxyuridine), a nucleoside analog incorporated into DNA during replication, for 3 h at 37 °C. EdU uptake was visualized using Click-iT Edu Alexa Fluor 488 imaging kit as per manufacturers protocol (Invitrogen). Cells were imaged on a fluorescence microscope for EdU incorporation.
Organoid formation assay
Organoids were formed in microwells as described (Sakib et al. 2019). Briefly, each well was seeded with 1.2 × 106 Sertoli cells and centrifuged at 500×g for 5 min. After 48 h in culture, organoids were fixed in 4% paraformaldehyde (PFA) for 15 min at room temperature and expression of Sertoli cell and basement membrane markers were examined using GATA4 (1:200, Santa Cruz Biotechnology) and collagen IV (1:500, Abcam) antibodies as previously described (Sakib et al. 2019). Organoids were imaged using confocal microscopy and scored using a binary system for presence or absence of a defined basement membrane.
Hedgehog antagonist treatment
Cells in both in vitro systems were treated with 10 μM of cyclopamine (Sigma-Aldrich) added directly to media upon cell seeding. Cultures underwent half media changes every 48 h. Tubules remained in culture for 6 days and organoids remained in culture for 4 days. Structures were collected for RNA extraction using a Qiagen RNeasy microkit.
Statistical analysis
qPCR data were normalized to housekeeping genes and are displayed as average RQ ± average minimum and maximum RQ relative to controls. T-tests were performed when there were two data sets and two-way ANOVAs with Tukey’s multiple comparison test were used when there were three or more data sets. Organoid scoring values were analyzed by chisquared test. Significance was set at p < 0.05.
Results
The cells used for all experiments contained 84.5 ± 2.3% Sertoli cells (n = 9 cell preparations, 5 × 106 cells/sample). The remaining cells were peritubular myoid cells 15.5 ± 2.5%; n = 5 cell preparations, 1 × 105 cells/sample.
Loss of ODF2 or IFT88 results in reduction in number and length of primary cilia
To examine the role of ODF2 in cilia development in Sertoli cells, we disrupted gene expression by transient transfection of CRISPR/Cas9 targeted to ODF2. The efficiency of gene editing was calculated to be 61% with a KO score of 52.3% (SFig. 1) leading to a 95% decrease (p < 0.001) in the relative expression of ODF2 mRNA in edited cells (ODF2e) (n = 3) (Fig. 1a). ODF2e cells showed a significant reduction in the number of cells that possessed primary cilia (38.3 ± 2.9% of ODF2e cells vs. 67.8 ± 2.3% of control cells had primary cilia; n = 6 for control, n = 4 for ODF2e cells; p < 0.0001; Fig. 1b). The remaining cilia on these cells were shorter (1.6 ± 0.04 μm in ODF2e cells vs. 2.5 ± 0.05 μm in controls; n = 493 cells for controls, n = 483 cells for ODF2ecells; p < 0.0001; Fig. 1c).
Fig. 1.
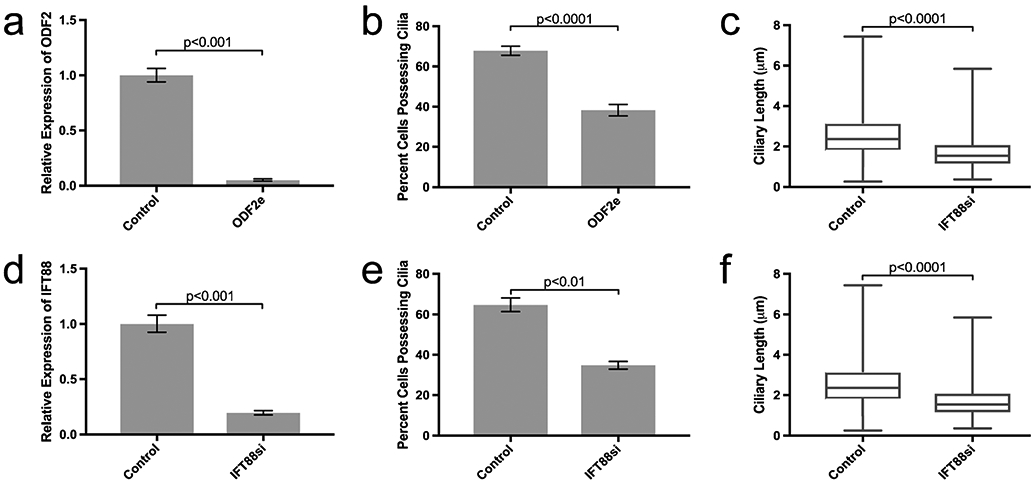
Editing of ODF2 and silencing of IFT88 reduces the number and length of primary cilia. a, d Relative expression of target mRNA. In a, n = 3. In d, n = 5. Bars indicate minimum and maximum RQ. b, e Percent of cells possessing primary cilia. In b, control n = 6, ODF2e n = 4. In e, control n = 7, IFT88si n = 6. Bars indicate ± SEM. c, f Length of primary cilia. In c, control n = 493, ODF2e n = 483. In f, control n = 371, IFT88si n = 400 cells counted. Bars indicate minimum and maximum ciliary lengths measured; center line is median length
Relative expression of IFT88 mRNA decreased by 81% upon silencing with siRNA (IFT88si) (n = 5; p < 0.001; Fig. 1d)The number of cells with primary cilia was also reduced (34.8 ± 1.9% of IFT88si cells vs. 64.8 ± 3.4% of control cells possessing primary cilia; n = 7 for control, n = 6 for IFT88si cells; p < 0.01; Fig. 1e). The remaining IFT88si cells that retained their cilia had shorter cilia (1.7 ± 0.04 μm in IFT88si cells vs. 2.6 ± 0.06 μm in controls; n = 371 for controls, n = 400 for IFT88si cells; p < 0.0001; Fig. 1f).
Loss of IFT88 but not ODF2 affects cellular proliferation
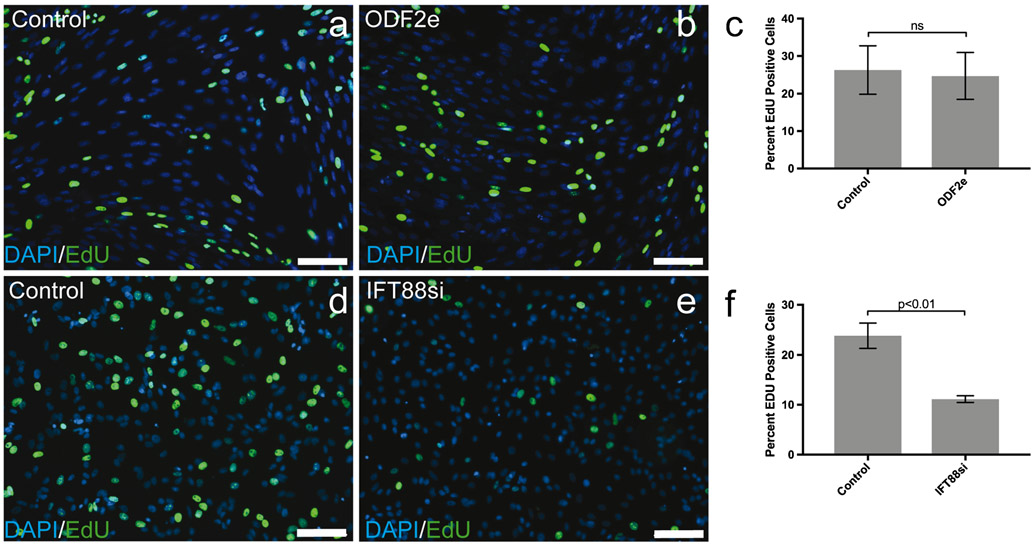
There was no difference in cellular proliferation between ODF2e cells and control as determined by EdU uptake (n = 3) (Fig. 2a-c). By contrast, IFT88si showed less proliferation (p < 0.01) than control cells (n = 3) (Fig. 2d-f).
Fig. 2.
Knockdown of IFT88 but not ODF2 affects cellular proliferation. a, b, d, e EdU-positive cells in control and treated populations. Scale bars 100 μm. c, f Quantification of EdU-positive cells in control and treated populations (n = 3). Bars indicate ± SEM
Loss of IFT88 but not ODF2 affects hedgehog signaling
Relative expression of GLI1 increased in ODF2e cells from 0.98 to 3.91 vs. 1 to 3.6 in controls (n = 3) upon treatment with the Hh agonist SAG (Fig. 3a). In contrast, IFT88si cells did not upregulate GLI1 expression upon treatment with SAG. Relative expression of GLI1 increased in control cells from 1 to 3.03, while relative expression in IFT88si cells was 0.58 and 0.87 for untreated and treated cells, respectively (n = 4) (Fig. 3b).
Fig. 3.
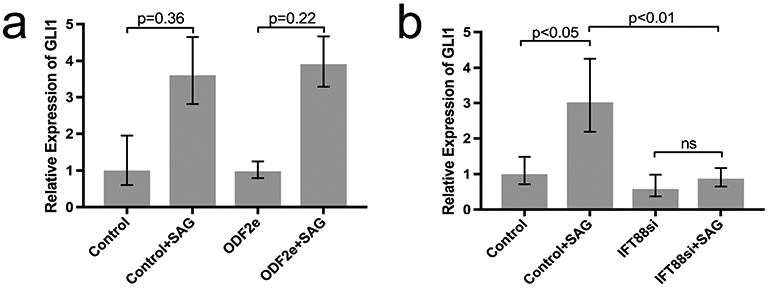
Effect of targeting ODF2 and IFT88 on GLI1 expression upon treatment with SAG. a Control and ODF2e cells, n = 3. b Control and IFT88si cells, n = 4. Bars indicate minimum and maximum RQ
Reduction in primary cilia affects in vitro morphogenesis
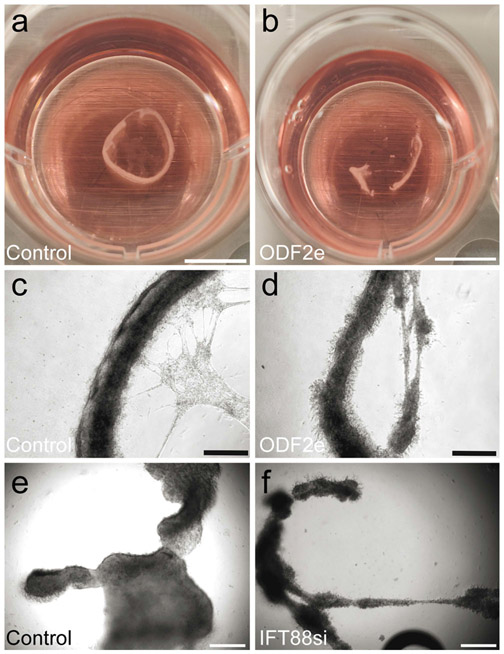
Cells were investigated using in vitro testicular tubule culture as described (Dores et al. 2017). Both ODF2e and IFT88si cells formed tubules in vitro that were generally more disorganized when compared with respective controls (Fig. 4). However, results were variable in size and shape of tubules between replicates.
Fig. 4.
Reduction of primary cilia impairs formation of tubules in vitro. a, b Tubules made from control and ODF2e cells. Scale bars 5 mm. c, d Higher magnification of tubules in a and b. Scale bars 500 μm. e, f Tubules formed from control and IFT88si cells. Scale bars 500 μm
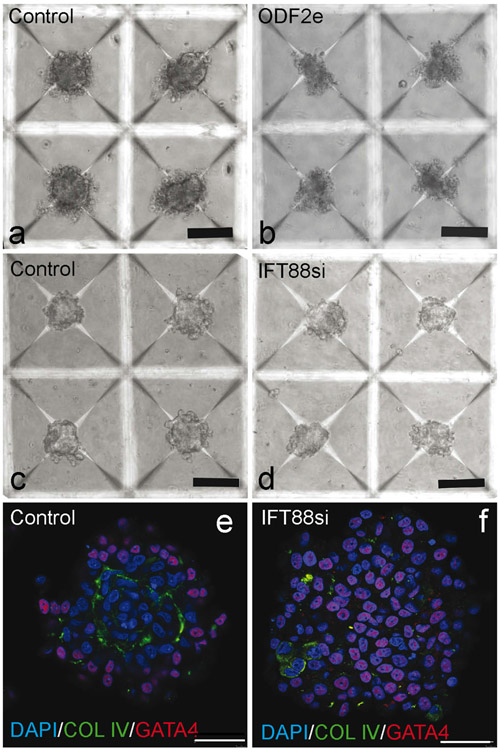
To decrease replicate variability, the testicular organoid system was used to investigate morphogenesis (Sakib et al. 2019). ODF2e and IFT88si cells failed to create a defined basement membrane structure, distinctive of a formed organoid. Organoids created from ODF2e cells dissociated upon removal from microwells and could not be analyzed further (Fig. 5a, b). When examined with brightfield microscopy, organoids created from IFT88sicells did not appear to be strikingly different from controls (Fig. 5c, d). However, when examined using immunofluorescence, these organoids lacked the distinct basement membrane that was present in controls (Fig. 5e, f). In controls, 128 of 150 organoids had a distinct basement membrane, while only 5 of 150 organoids formed from IFT88si cells formed a basement membrane.
Fig. 5.
Reduction of primary cilia impairs formation of organoids in vitro. a, b Organoids formed from control and ODF2e cells. Scale bars 50 μm. c, d Organoids formed from control and IFT88si cells. Scale bars 50 μm. e, f Basement membrane deposition and cellular organization of organoids from control and IFT88si cells. Scale bars 40 μm
Cyclopamine does not affect in vitro morphogenesis
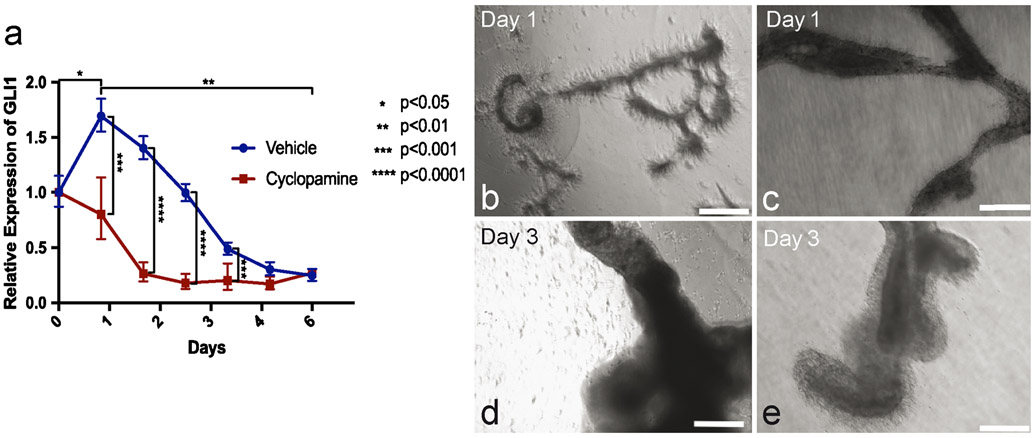
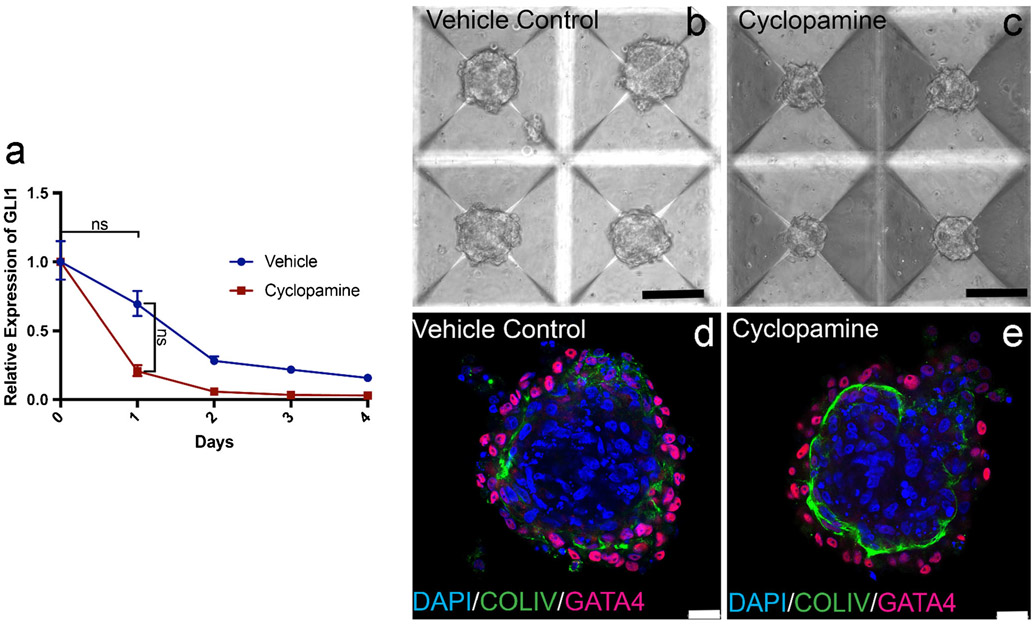
Hh signaling was active during tubule formation in vitro, with GLI1 mRNA expression significantly increasing after 1 day in culture. Over the next 5 days, GLI1 expression progressively decreased with a significant difference in expression between 1 and 7 days in culture (Fig. 6a). Tubules at 1 day in culture were beginning assembly, while at 7 days, they had fully assembled into a tubular structure (Fig. 6b). Tubules that were treated with cyclopamine showed attenuation of GLI1 expression over 7 days but were still able to form, with a similar formation pattern at 1 and 7 days as controls (Fig. 6c).
Fig. 6.
Hedgehog signaling is active but not essential for tubule morphogenesis in vitro. a GLI1 expression (blue line) over the course of 6 days in in vitro tubules. Day 0, n = 7; day 1, n = 5; days 2–6, n = 4. GLI1 expression upon treatment with cyclopamine (red line). n = 3. Bars indicate minimum and maximum RQ. b Tubule formation at 1 and 3 days. Scale bars 500 μm. c Tubule formation at 1 and 3 days upon treatment with cyclopamine. Scale bars 500 μm
GLI1 expression also decreased in organoids over time but there was no significant difference between time points and no distinctive peak at 1 day in culture (Fig. 7a). There was no significant difference in GLI1 expression between cyclopamine treated and untreated organoids and while cyclopamine treated organoids appeared smaller with brightfield microscopy, they were still able to form a distinct basement membrane similar to controls (Fig. 7b-e).
Fig. 7.
Hedgehog signaling in organoids was not essential for morphogenesis. a GLI1 expression over 4 days (blue line) in organoids. n = 3. GLI1 expression upon treatment with cyclopamine (red line). n = 3. Bars indicate minimum and maximum RQ. b, c Appearance of organoids treated with DMSO vehicle control or cyclopamine. Scale bars 50 μm. d, e Basement membrane deposition and cellular organization of control organoid and organoid formed from cells treated with cyclopamine. Scale bars 25 μm
Discussion
Targeting ODF2, a structural component of the primary cilia, or IFT88, a functional component of the IFT system, in Sertoli cells resulted in a reduction in the number of cells with primary cilia. Both the number of cells with cilia and the length of the remaining primary cilia was significantly reduced upon targeting, corroborating other studies that underscore their importance to the primary cilium (Pazour et al. 2000; Ishikawa et al. 2005). The distal appendages serve as a recruitment point for proteins to be transported up the axoneme during ciliary assembly, so the shortened cilia that resulted upon editing of ODF2 could be due to improper protein localization of cargo at this location (Yang et al. 2018). In the case of IFT88 silencing, the intraflagellar transport system is paramount in transporting tubulin monomers to the growing ciliary tip and loss of a key component of complex B could severely disrupt the ability of these building blocks to be transported (Cole et al. 1998). It is important to note that despite losing nearly all expression of mRNA for both targets, some cilia, while shortened, were still present. The percentage of cells that lost primary cilia (56.5%) is comparable to the KO score (52.3%), indicating that not all indels created at this locus were sufficient to disrupt protein function. It is also possible that other transcript variants of ODF2, collectively termed Cenexins, were still present upon editing of exon 2 and that they contribute to the structure of the cilium via distal appendages as well (Chen and Megraw 2013). ODF2 could also be a very stable protein and despite cell proliferation, unedited ODF2 remained active in some cells. As IFT88 was targeted using siRNA, the transient nature of this system could have allowed some cilia to assemble. Loss of IFT88 in Sertoli cells affected cellular proliferation, as previously reported in other cell types (Delaval et al. 2011).
Interestingly, only the loss of IFT88 resulted in a change in Hh signaling, as measured by GLI1 upregulation upon treatment with SAG. As the Hh signaling pathway relies on the intraflagellar transport system to transport GLI proteins down the axoneme to become transcriptional activators, perturbation in this cellular transport system would affect the signaling process (May et al. 2005). Since IFT88 is a component of complex B, its loss would negatively affect the migration of components such as SMO to the tip of the cilium and prevent the release of GLI proteins from the ciliary tip. On the other hand, ODF2 acts as a structural protein and is not involved in transport and its loss is not expected to affect the movement of Hh components up and down the axoneme.
Loss of either ODF2 or IFT88 and the reduction in primary cilia, affected morphogenesis in vitro. In the tubule system, reduction of cilia created more disorganized tubules than controls. This disrupted morphogenesis is unlikely to be due to impaired proliferation because targeting of both ODF2 and IFT88 show the same outcome despite the two targeted cell populations having different proliferative ability. Tubule width and length parameters could not be reliably quantified as replicates were variable in size and shape even in controls. The novel testicular organoid system provides a more quantifiable and reproducible in vitro model of morphogenesis and it was primarily used to examine the effect of ciliary loss. Reduction of primary cilia impaired organoid formation and the deposition of collagen IV in the basement membrane, possibly due to aberrant cell-cell interaction. Hence, primary cilia are essential for morphogenesis of testicular tubules in vitro.
During in vitro morphogenesis, the Hh pathway was active in in vitro forming tubules and to a lesser degree in organoids. In tubules, GLI1 expression peaked coincident with cell migration to form initial tubule structures, indicating that Hh signaling may be involved in this initial migration step of morphogenesis. This result, alongside the morphogenesis results of cells with and without impaired proliferation, points towards the initial morphogenesis being less dependent on proliferation than cellular migration. However, when Hh signaling was disrupted by addition of cyclopamine and GLI1 expression was attenuated, tubules were still able to form comparably to controls. Therefore, while Hh signaling is active during in vitro morphogenesis, it is not essential. There was no distinct peak of GLI1 expression during organoid formation but GLI1 expression decreased over time. Absence of a distinct Hh signaling peak can perhaps be attributed to a lack of cellular migration during organoid formation, as cells are forced into contact with each other immediately upon introduction to the microwell system. GLI1 expression was attenuated when cells in organoids were treated with cyclopamine but morphogenesis was not affected. Therefore, cilia-mediated signaling pathways other than Hh signaling appear to be involved in morphogenesis in vitro. One candidate pathway would be PDGFα signaling, which is also regulated by the primary cilium and is important in directing cellular migration events such as those found in wound healing (Schneider et al., 2010).
In conclusion, targeting of either the structural cilia target ODF2 or the functional transport target IFT88 led to a reduction in number and length of primary cilia on porcine Sertoli cells. Reduction of primary cilia resulted in the loss of organized assembly of cells during in vitro tubular morphogenesis. The Hh pathway is dependent on primary cilia and was active during structural assembly of testicular cells in vitro; however, it was dispensable for morphogenesis. Other cilia-mediated pathways must be active during morphogenesis and their perturbation contributes to disruption of morphogenesis. The approaches presented here will allow identification of other primary cilia-mediated signaling pathways in Sertoli cells and their role in testicular morphogenesis.
Supplementary Material
Acknowledgments
Funding information This study was funded by NIH/ORIP OD016575 to ID.
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
Ethical approval All applicable international, national and/or institutional guidelines for the care and use of animals were followed.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00441-019-03121-8) contains supplementary material, which is available to authorized users.
References
- Bangs FK, Anderson KV (2017) Primary cilia and mammalian hedgehog signaling. Cold Spring Harb Perspect Biol 9:48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs FK, Shrode N, Hadjantonakis AK, Anderson KV (2015) Lineage specificity of primary cilia in the mouse embryo. Nat Cell Biol 17: 112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JV, Megraw TL (2013) Cenexin1 and Odf2: splice variants with diverged cilium functions. Cell Cycle 12:869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen ST, Voss JW, Teilmann S, Lambert IH (2005) High expression of the taurine transporter taut in primary cilia of NIH3T3 fibroblasts. Cell Biol Int 29:347–351 [DOI] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL (1998) Chlamydomonas kinesin-II dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol 141:993–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaval B, Bright A, Lawson ND, Doxsey S (2011) The cilia protein IFT88 is required for spindle orientation in mitosis. Nat Cell Biol 13:461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkor FF, Monnich M, Czirr E, Hollemann T, Hoyer-Fender S (2004) Outer dense fibre protein 2 (ODF2) is a self-interacting centrosomal protein with affinity for microtubules. J Cell Sci 117:4643–4651 [DOI] [PubMed] [Google Scholar]
- Dores C, Dobrinski I (2014) De novo morphogenesis of testis tissue: an improved bioassay to investigate the role of VEGF-165 during testis formation. Reproduction 148:109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dores C, Alpaugh W, Su L, Biernaskie J, Dobrinski I (2017) Primary cilia on porcine testicular somatic cells and their role in hedgehog signaling and tubular morphogenesis in vitro. Cell Tissue Res 368:215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestone AJ, Weigner JS, Maldonado M, Barlan K, Langston LD, O’Donnell M, Gelfand VI, Kapoor TM, Chen JK (2012) Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature 484:15–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassei K, Schlatt S, Ehmcke J (2006) De novo morphogenesis of seminiferous tubules from dissociated immature rat testicular cells in xenografts. J Androl 27:611–618 [DOI] [PubMed] [Google Scholar]
- Girardet L, Augiére C, Asselin M, Belleannée C (2019) Primary cilia: biosensors of the male reproductive tract. Andrology 7:588–602 [DOI] [PubMed] [Google Scholar]
- Guschi GY, Waite AJ, Katibah GE, Miller JC, Holmes MS, Rebar EJ (2010) A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol 649:247–256 [DOI] [PubMed] [Google Scholar]
- He Q, Wang G, Wakade S, Dasgupta S, Dinkins M, Kong JN, Spassieva SD, Bieberich E (2014) Primary cilia in stem cells and neural progenitors are regulated by neutral sphingomyelinase 2 and ceramide. Mol Biol Cell 25:1714–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV (2003) Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426:83–87 [DOI] [PubMed] [Google Scholar]
- Insinna C, Besharse JC (2008) Intraflagellar transport and the sensory outer segment of vertebrate photoreceptors. Dev Dyn 237:1982–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C, Li L, Esparza JM, Dutcher SK (2009) Retrograde intraflagellar transport mutants identify complex A proteins with multiple genetics interactions in Chlamydomonas reinhardtii. Genetics 183:885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF (2017) Intraflagellar transport and ciliary dynamics. Cold Spring Harb Perspect Biol 9:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Kubo A, Tsukita S, Tsukita S (2005) Odf2 deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol 7:517–524 [DOI] [PubMed] [Google Scholar]
- Kihara A, Okmura M, Kimata T, Tanizawa Y, Takano R, Kimura KD, Inada H, Matsumoto K, Mori I (2008) Temperature sensing by an olfactory neuron in a circuit controlling behavior of C. elegans. Science 320:803–808 [DOI] [PubMed] [Google Scholar]
- Kiprilov EN, Awan A, Desprat R, Velho M, Clement CA, Byskov AG, Cy A, Satir P, Bouhassira EE, Chistensen ST, Hirsh RE (2008) Human embryonic stem cells in culture possess primary cilia with hedgehog signaling machinery. J Cell Biol 180:897–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Megee S, Rathi R, Dobrinski I (2006) Protein gene product 9.5 is a spermatogonia-specific marker in pig testis: application to enrichment and culture of porcine spermatogonia. Mol Reprod Dev 73: 1531–1540 [DOI] [PubMed] [Google Scholar]
- Moorman SJ, Shorr AZ (2008) The primary cilium as a gravitational force transducer and a regulator of transcriptional noise. Dev Dyn 237:1955–1959 [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Yamane Y, Okanoue T, Tsukita S, Tsukita S (2001) Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol Biol Cell 12:1687–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Vucica Y, Es S, Rosenbaum JL, Witman GB, Cole DG (2000) Chlamydomonas IFT88 and its mouse homologue polycystic kidney disease gene Tg737 are required for assembly of cilia and flagella. J Cell Biol 151:709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP (2007) Patched1 regulates hedgehog signaling at the primary cilium. Science 317:372–376 [DOI] [PubMed] [Google Scholar]
- Satir P, Christensen S (2007) Overview of structure and function of mammalian cilia. Annu Rev Physiol 69:377–400 [DOI] [PubMed] [Google Scholar]
- Satir P, Pedersen LB, Christensen ST (2010) The primary cilium at a glance. J Cell Sci 123:499–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider L, Cammer M, Lehman J, Nielsen SK, Guerra CF, Veland IR, Stock C, Hoffman EK, Yoder BK, Schwab A, Satir P, Christensen ST (2010) Direction cell migration and chemotaxis in wound healing response to PDGF-AA are coordinated by the primary cilium in fibroblasts. Cell Physiol Biochem 25:279–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakib S, Uchida A, Valenzuela-Leon P, Yu Y, Valli-Pulaski H, Orwig K, Ungrin M, Dobrinski I (2019) Formation of organotypic testicular organoids in microwell culture. Biol Reprod. 10.1093/biolre/ioz053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May S, Ashique AM, Karlen M, Wang B, Shen Y, Zarbalis K, Reiter J, Ericson J, Peterson AS (2005) Loss of retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev Biol 287:378–389 [DOI] [PubMed] [Google Scholar]
- Szymaska K, Johnson CA (2012) The transition zone: an essential functional compartment of cilia. Cilia 1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos BE, Yang HJ, Soni R, Wang WJ, Macaluso FP, Asara JM, Tsou MF (2013) Centriole distal appendages promote membrane docking leading to cilia initiation. Genes Dev 27:163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevosian SG, Albrecht KH, Crispino JD, Fujiwara Y, Eicher EM, Orkin SH (2002) Gonadal differentiation, sex determination and normal Sry expression in mice require direct interaction between transcription partners GATA4 and FOG2. Development 129:4627–4634 [DOI] [PubMed] [Google Scholar]
- Wheway G, Nazlamova L, Hancock JT (2018) Signaling through the primary cilium. Front Cell Dev Biol. 10.3389/fcell.2018.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TT, Chong WM, Wang WJ, Mazo G, Tanos B, Chen Z, Tran TMN, Chen YD, Weng RR, Huang CE, Jane WN, Tsou MB, Liao JC (2018) Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat Commun 9:2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.