Abstract
Maternal infection (i.e., ascending infection) and the resulting host inflammatory response are risk factors associated with spontaneous preterm birth (PTB), a major pregnancy complication. However, the path of infection and its propagation from the maternal side to the fetal side have been difficult to study due to lack of appropriate in vitro models and limitations of animal models. A better understanding of the propagation kinetics of infectious agents and development of the host inflammatory response at the feto-maternal (amniochorion-decidua, respectively) interface (FMi) is critical in curtailing host inflammatory responses that can lead to PTB. To model ascending infection and determine inflammatory responses at the FMi, we developed a microfluidic organ-on-chip (OOC) device containing primary cells from the FMi (decidua, chorion, and amnion [mesenchyme and epithelium]) and collagen matrix harvested from primary tissue. The FMi-OOC is composed of four concentric circular cell/collagen chambers designed to mimic the thickness and cell density of the FMi in vivo. Each layer is connected by arrays of microchannels filled with type IV collagen to recreate the basement membrane of the amniochorion. Cellular characteristics (viability, morphology, production of nascent collagen, cellular transitions, and migration) in the OOC were similar to those seen in utero, validating the physiological relevance and utility of the developed FMi-OOC. The ascending infection model of the FMi-OOC, triggered by exposing the maternal (decidua) side of the OOC to lipopolysaccharide (LPS, 100 ng/mL), shows that LPS propagated through the chorion, amnion mesenchyme, and reached the fetal amnion within 72 h. LPS induced time-dependent and cell-type-specific pro-inflammatory cytokine production (24 h decidua: IL-6, 48 h chorion: GM-CSF and IL-6, and 72 h amnion mesenchyme and epithelium: GM-CSF and IL-6). Collectively, this OOC model and study successfully modeled ascending infection, its propagation, and distinct inflammatory response at the FMi indicative of pathologic pathways of PTB. This OOC model provides a novel platform to study physiological and pathological cell status at the FMi, and is expected to have broad utility in the field of obstetrics.
Keywords: microfluidics, organ-on-a-chip, fetal membrane, feto-maternal interface, inflammation, decidua, preterm birth, infection
Introduction
Spontaneous preterm birth (PTB), or birth before the 37th week of gestation, is a major pregnancy pathology contributing to high neonatal mortality and morbidity 1, 2. Although the etiology of PTB is not clearly understood, infection and infection-associated host inflammatory response are significant risk factors associated with PTB 1, 3. Approximately 50% of all cases of PTB have an infectious etiology 2. Although the development of maternal infection is a considerable risk factor, inflammation induced within the feto-maternal tissues is a major effector of preterm labor pathways 4-6. It is currently hypothesized that host inflammatory responses are amplified at the feto-maternal interface (FMi), and this amplification compromises the immune homeostasis of pregnancy or promotes premature parturition-related changes 7-9.
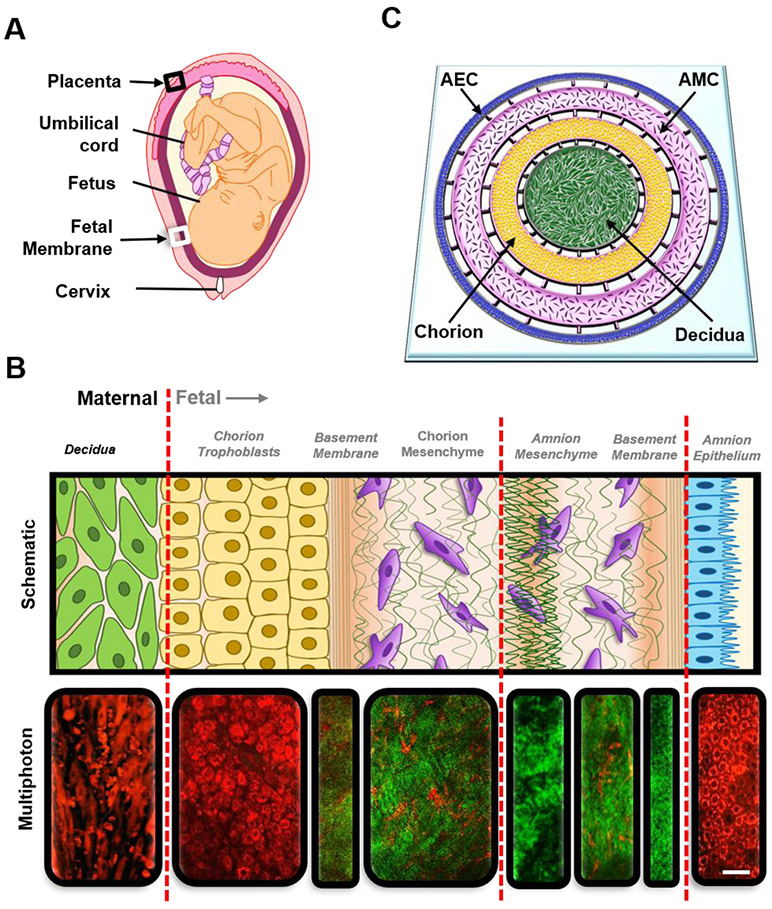
There are two FMis in pregnancy (Figure 1A): 1) between the placenta and decidua basalis (black box), and 2) between the fetal membrane (amniochorion) and decidua parietalis (white box). Structurally and functionally, these two interfaces are very different; however, the selective passage of various materials, including infectious agents or inflammatory cytokines, are likely through both interfaces 10, 11. The placenta interface is the better studied interface 10, 12, 13, whereas the fetal membrane-decidua interface is not well studied 14, 15. In this report, we focus on the less studied fetal membrane-decidual interface (referred to as the FMi in the rest of the manuscript) to study ascending infection through the development of an FMi organ-on-chip (OOC) modeling this interface. The FMi is a multi-cellular multi-layer structure, composed of decidualized maternal endometrial stromal cells (Figure 1B black text) connected to the fetal-derived amniochorionic membrane (Figure 1B gray text) 16. The amniochorion protects the fetus throughout gestation by acting as a mechanical and immune barrier 17. The human amniochorionic membrane has two components: the amnion layer that lines the intra-amniotic cavity and the chorion that forms the interface with the decidua. The amnion layer is composed of two different cell types: the amnion mesenchymal cells (AMCs) and the amnion epithelial cells (AECs). The chorion is also composed of two different cell types: the chorion trophoblasts (CTs) and the chorion mesenchymal cells (CMCs). Altogether, this interface is composed of five distinct cellular layers, each having unique phenotypes and characteristics. This complex multi-cellular multi-layer structure is what makes it challenging to develop an in vitro model system 16, 17.
Figure 1: Recreating the FMi with organ-on-chip technology.
A) An illustration of the anatomy of the intrauterine tissue showing maternal and fetal components and both FMis (black arrows [black box: placenta-decidua basalis; white box: fetal membrane – decidua parientalis]). Maternal tissues are comprised of the uterus (myometrium) and cervix, while the fetal tissues include the placenta, umbilical cord, fetus, and fetal membrane (amniochorionic membrane).
B) Region of the decidua and amniochorionic membrane indicated as a white box in A, showing the FMi of interest. The FMi is divided by maternal decidual cells (black text) and fetal-derived amniochorionic cells (gray text). The top row shows a schematic of the amniochorionic membrane and maternal decidua. From left to right, the description starts from the outermost layer (the maternal decidua) and ends at the fetal amnion. The chorion interfaces with the maternal decidua (green), thus connecting the fetal layers to the maternal compartment of the uterus. The chorion (yellow) is connected to the ECM through a type IV collagen basement membrane (green stripes) and is made up of chorion trophoblasts (CTs). The fibroblast, spongy, and reticular layers of the ECM harbor amnion mesenchymal cells (AMCs) and chorion mesenchymal cells (CMCs) (purple). The AECs (blue) are connected to the ECM through the basement membrane/compact layer (green stripes). The bottom row shows corresponding multiphoton microscopy images of each section, where cells are stained in red color and the collagen layers are stained in green color. The red lines divide the four different compartments of the FMi that are represented in the FMi-OOC. Scale bar = 30 μm.
C) Schematic of the FMi-OOC device designed to mimic the FMi. The FMi-OOC is composed of four concentric circular cell culture chambers separated by arrays of microchannels. The cells are seeded as follows, from the center to the outside: decidua cells (green), CMCs/CTs (yellow), AMCs – amnion mesenchyme (purple), and AECs – amnion epithelium (blue), respectively. Primary fetal membrane collagen and Matrigel (pink) can enable culturing AMCs and CMC/CTs in a 3D format. To recreate cell-collagen interfaces, the microchannels are filled with type IV collagen (pink) to mimic the basement membrane of the amnion and chorion layers. In contrast, the choriodecidua interface was not filled with collagen (gray).
It is hypothesized that vaginal dysbiosis, due to an imbalance in the vaginal microbiome representing a pathogenic state or the introduction of an infectious microbe, induces localized inflammation that disrupts tissue homeostasis within the vagina and cervix 18-20. Tissue damage allows microbes to ascend through the vagina (vaginitis), transverse the cervix (cervicitis), infect the decidua (deciduaitis), and cross the amniochorionic membrane (chorioamnionitis), leading to microbial invasion of the intra-amniotic cavity (MIAC) 21 and causing intra-amniotic inflammation (IAI) 22. Collectively, this is referred to as intra-amniotic invasion, infection, and inflammation. Inflammation at different tissue levels is not a prerequisite for MIAC or IAI, as localized inflammation could be dependent on the type of pathogen. Inflammation in these tissues can also occur in the absence of infection due to various other pregnancy risk factors (sterile inflammation) 1, 5, 23.
Ascending infections are hypothesized to be common, but have not been adequately modeled either through in vitro or animal models. Large (non-human primate [NHP]) and small (mice or rabbit) animal models have been used routinely to test ascending infection-induced PTB 24-26. Many of these models inject a non-physiological load of either live or attenuated bacteria (e.g., group B streptococcus [GBS], Escherichia coli [E. coli], Ureaplasma parvum, and Gardnerella vaginalis), viral factors (e.g., DNA isolates of adenovirus, cytomegalovirus, and enterovirus), proxies for infection (e.g., lipopolysaccharide [LPS] and polyinosinic-polycytidylic acid [poly I:C]), or infectious inflammatory chemicals (e.g., tumor necrosis factor [TNF]-α) into the cervix of animals or with intrauterine injections 22, 24, 26. However, the anatomy and functionality of intrauterine organs of small animal models 26, 27, especially the FMi, neither mimic human pregnancy nor do these models naturally undergo PTB 27, limiting the degree of information that can be gained from small animal studies. NHPs are anatomically and functionally the most similar to humans, thus they serve as the best model; however, they are incredibly costly, difficult to handle, and require a dedicated facility to conduct experiments. Therefore, to move this field forward, the development of advanced cell-based in vitro models that can mimic the physiology of human pregnancy and can be used to study ascending infection-induced preterm labor is critical.
Understanding the role of the FMi during pregnancy and parturition is vital to advancing basic and clinical research in the field of obstetrics. The emergence of in vivo-like in vitro models, such as organ-on-chip (OOC) that can better recapitulate in vivo functions and responses, has the potential to move the field of obstetrics forward significantly. Many OOCs mimicking diverse ranges of organ systems have been developed in the past decade 28-32. These multi-cellular multi-layer devices often have distinct cell culture compartments that are interconnected to allow communication via biomolecular signals and cell-to-cell contact, while allowing control over their microenvironments. In the field of obstetrics, several basic OOC models of the placenta have been developed 33-37. Recently, OOC models of the fetal membrane (2-chamber co-culture model) 38-40 and the amnion membrane (2-chamber co-culture model) have been also developed. However, none of these current OOC models fully recreate the cellular and collagen components of the decidua-amniochorionic membrane interface as shown in Figure 1A-B. To fill this major gap, here we present the first FMi-OOC model that recreates both the fetal and maternal sides of the human FMi (amniochorion-decidua) in an OOC platform (Figure 1C).
This FMi-OOC was designed to contain four primary human cell types and collagen to represent both the maternal side (decidua, Figure 1C green) and fetal side (chorion, [Figure 1C yellow], amnion cells [AMC purple; AEC blue]) of the FMi (decidua [green], chorion [yellow], and amnion cells [AMCs-purple; AECs-blue]) (Figure 1C). This device was fabricated, validated, and utilized to create a model of ascending infection across the FMi.
Methods
Institutional review board approval:
Placentas for this study were collected from John Sealy Hospital, University of Texas Medical Branch (UTMB) at Galveston, Texas, according to the inclusion and exclusion criteria described below. As discarded placentas after delivery were used for the study, subject recruitment or consenting was not required. The Institutional Review Board (IRB) at UTMB approved the study protocol (UTMB 11-251), and placentas were collected according to the regulations of the IRB as an exempt protocol that allowed the use of discarded placentas for fetal membrane and placental research.
Inclusion criteria for term categories:
Normal term births were women with term, not-in-labor, delivery (> 390/7 weeks), and without any pregnancy-related complications.
Exclusion criteria for term categories:
Subjects with multiple gestations, placenta previa, fetal anomalies, and/or medical or surgical intervention (for clinical conditions that are not linked to pregnancy) during pregnancy were excluded. Severe cases of preeclampsia or persistent symptoms (e.g., headache, vision changes, or right upper quadrant pain), abnormal laboratory findings (thrombocytopenia, repeated abnormal liver function tests, creatinine doubling or > 1.2, or hemolysis, elevated liver enzymes, low platelet count [HELLP] syndrome), and abnormal clinical findings (pulmonary edema or eclampsia) were all excluded. Subjects who had any surgical procedures during pregnancy or who were treated for hypertension, preterm labor, or have suspected clinical chorioamnionitis (reports of foul-smelling vaginal discharge or fetal tachycardia), positive GBS screening or diagnosis of bacterial vaginosis, and behavioral issues (cigarette smoking, drug or alcohol abuse) were also excluded.
Cell preparation and culture:
Maternal and fetal cells were collected from the same fetal membrane from term, not-in-labor, and Cesarean deliveries for each experiment to reduce patient-to-patient variability when studying the responses of each cell type within the FMi-OOC (n = 6).
Decidua cell preparation:
Decidua was separated from the chorion by blunt dissection with forceps and a scalpel and minced by cross-cutting with scalpel blades 41. The tissues were processed in a digestion buffer containing 0.125% trypsin (Cat# 85450c, Sigma), 0.2% collagenase (Cat# C0130, Sigma), 0.02% DNase I (Cat# DN25, Sigma), and incubated at 37°C for 60–90 min. Samples were subsequently neutralized with complete media (1:1 mixture of Ham’s F12/DMEM, supplemented with 5% heat-inactivated FBS, 10 ng/mL epidermal growth factor, 100 U/mL penicillin G, and 100 mg/mL streptomycin) (Cat# 30-001-CI, Corning). After filtration, the cell solution was centrifuged at 3,000 rpm for 10 min. A cell-separation gradient was prepared using an Optiprep column (Axis-Shield), with steps ranging from 4% to 40% of 4 mL each (4%, 6%, 8%, 10%, 20%, 30%, and 40%). Processed decidual cells were added to the top of the gradient and centrifuged (3,000 rpm) at room temperature for 35 min. Cell densities of 1.027-1.038 g/mL could be obtained through this procedure from the decidua layer. Cell viability was tested using the trypan blue exclusion method and microscopy.
Chorion cell preparation:
Separation of the chorion and decidual layers involved blunt and gentle scraping with a scalpel 41. Tissues were first processed in a digestion buffer containing 2.4 U/mL dispase (Cat# D4693, Sigma) and incubated at 37°C for 8 min. Tissues were then allowed to rest for 5-10 min at room temperature in complete media (1:1 mixture of Ham's F12/DMEM, supplemented with 10% FBS, 100 U/mL penicillin G, and 100 mg/mL streptomycin). The dispase incubation and rest cycle were repeated once more. Subsequently, the tissue was incubated in a digestion buffer containing 0.75 mg/mL collagenase (Cat# C0130, Sigma) and 0.02% DNase I (Cat# DN25, Sigma) with rotation at 37°C for 3 h. The remaining tissue was incubated in a solution containing 0.25% trypsin (Cat# 85450c, Sigma) and 0.02% DNase I (Cat# DN25, Sigma) at 37°C for 5 min. The digested solution was then filtered through a 70 μm cell strainer for cell collection. After filtration, the cell solution was centrifuged at 3,000 rpm for 10 min. Pelleted cells were resuspended in complete media and plated. The resulting culture contained a mixture of chorion mesenchymal and trophoblast cells. Cell viability was tested using the trypan blue exclusion method and microscopy.
AMC preparation:
AMCs were isolated from amnion membranes, as previously described by Kendal-Wright et al. 42 (but with slight modifications). The reflected amnion (~10 g) was peeled from the chorion layer and rinsed 3-4 times in sterile HBSS (21-021-CV; Corning, Corning, NY, USA) to remove blood debris. The sample was then incubated with 0.05% trypsin-EDTA (25-053-CI; Corning) at 37°C for 1 h to disperse the cells and remove the epithelial cell layer. The membrane pieces were then washed 3-4 times using cold HBSS to inactivate the enzyme. The washed membrane was transferred into a second digestion buffer containing Eagle's minimum essential medium (10-010-CV; Corning), 1 mg/mL collagenase type IV, and 25 μg/mL DNase I and incubated in a rotator at 37°C for 1 h. The digested membrane solution was neutralized using complete DMEM-F12 medium (10-092-CV; Corning) that was filtered using a 70 μm cell strainer and centrifuged at 3,000 rpm for 10 min. The cell pellet was resuspended in complete DMEM-F12 medium supplemented with 5% heat-inactivated fetal bovine serum (35-010-CV; Corning), 100 U/mL penicillin G, and 100 mg/mL streptomycin (30-001-CI; Corning). The resuspended cells were subsequently seeded at a density of 3–5 million cells per T75 flask and incubated at 37°C with 5% CO2 until 80–90% confluency was achieved.
AEC preparation:
Approximately 10 g of the amniotic membrane, peeled from the chorion layer, was dispersed by successive treatments with 0.125% collagenase and 1.2% trypsin. All cell culture reagents were purchased from Millipore Sigma (Burlington, MA, USA). Details of AEC isolation protocols can be found in our previous report 43. Briefly, the dispersed cells were plated in a 1:1 mixture of Ham's F12-DMEM, supplemented with 10% heat-inactivated fetal bovine serum, 10 ng/mL epidermal growth factor, 2 mM L-glutamine, 100 U/mL penicillin G, and 100 mg/mL streptomycin at a density of 3–5 million cells per T75 flask, and then incubated at 37°C with 5% CO2 until 80–90% confluency was achieved.
Microfluidic FMi-OOC design:
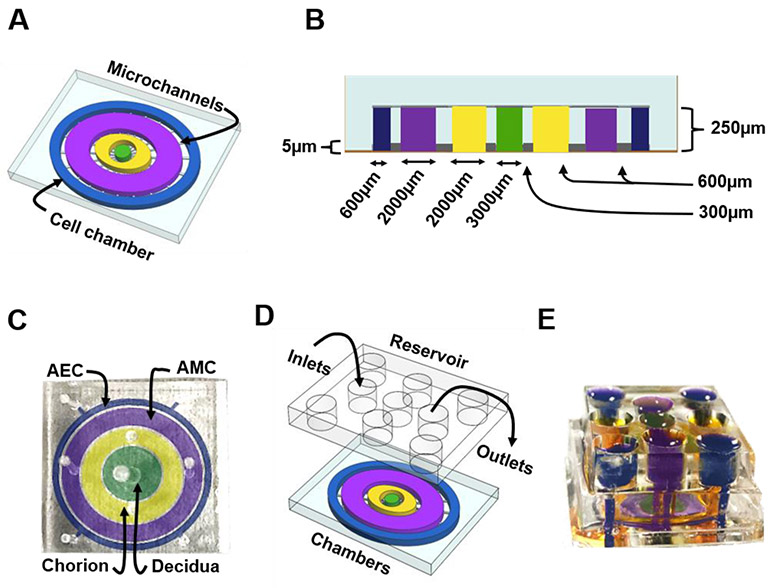
The microfluidic FMi chip is composed of four poly(dimethylsiloxane) (PDMS) concentric circles that each form a cell culture chamber (Figure 2A). Chamber 1, the center chamber, contains maternal decidual cells (green), chamber 2 contains fetal CTs and CMCs (yellow), chamber 3 contains AMCs (purple), and chamber 4 contains the outermost layer containing AECs (blue) (Figure 2A). Each cell chamber was 250 μm in height, and the width of each chamber was designed to mimic the thickness of each maternal and fetal layer as seen in utero (maternal: decidua – 3,000 μm [green]; fetal: chorion – 2,000 μm [yellow], AMC – 2,000 μm [purple], and AEC – 600 μm [blue]) (Figure 2B-C) 44, 45. Thus, this design allowed the four different cell types to be cultured in four separate microenvironments (e.g., different culture medium). The chambers were interconnected through an array of 24 microchannels (5 μm in height, 30 μm in width, and 300-600 μm in length) (Figure 2A-B). The microchannel arrays perform multiple independent functions, including: 1) preventing the flow of cells between compartments during the initial cell loading process, 2) allowing localized treatment of each cell layer with infectious or other stimulants while limiting their diffusion to the neighboring chambers, 3) enabling independent elution of supernatant from each cell chamber, and 4) allowing biochemicals to diffuse between chambers in a time-dependent way and also permit active cell migration that may involve cellular transitions 39, 40, 46. The device also contains an on-chip reservoir block, having multiple 4mm diameter and 2mm deep reservoirs where each reservoir is aligned in such a way that they are placed on top of the inlets and outlets of each chamber in the main cell culture layer (Figure 2D-E). This on-chip reservoir block provides sufficient cell culture media volume for culture, allows hydrostatic pressure difference-based diffusion control between the cell culture compartments by adjusting the culture volume fill height, as well as allows easy cell loading, treatment, and effluent access throughout the culture period using a simple pipette operation.
Figure 2: Design and images of the FMi-OOC model.
A) Schematic illustration of the FMi-OOC highlighting the four cell culture chambers connected by arrays of 24 microchannels.
B) Cross-sectional view of the cell culture chambers showing the chamber height (250 μm), individual chamber diameters (mimicking in utero thickness), and the interconnecting microchannel lengths.
C) Image of the microfabricated FMi-OOC where each cell culture chamber were filled with different color dye for easy visualization, namely the choriodecidua interface (decidua – green; chorion – yellow) and the amniochorionic interface (chorion – yellow; amnion – AMC [purple] and AEC [blue]).
D) Design of an on-chip media reservoir layer that was aligned and placed on top of the cell loading inlets and outlets (seen in image C) of the main cell culture layer, both to provide sufficient media for culture and also to control the diffusion between the cell culture layer through controlling the hydrostatic pressure between the chambers by filling the reservoirs to different height. I addition, localized chemical treatments can be performed as well as effluents collected from each culture chamber through these reservoirs. The center decidua layer has one reservoir, the chorion and AMC have two each, while the AEC chamber has four.
E) The FMi-OOC device with the integrated media reservoir filled with color dye in each of the corresponding cell culture layers.
The designed platform was fabricated in PDMS using a two-step photolithography master mold fabrication process, followed by a soft lithography process of replica molding the final PDMS device from the master mold. First, to create the master mold, two layers of photosensitive epoxy (SU-8; MicroChem, Westborough, MA, USA) with different thicknesses were sequentially patterned on a 3 inch diameter silicon substrate. The first layer forming the 5 μm deep microchannels (Figure 2A-B) was obtained by spin coating SU-8™ 3005 at 4,000 rpm and soft baking at 95°C for 5 min. It was then exposed to UV light through a photomask, followed by a postexposure bake at 95°C for another 5 min. The second layer forming the cell culture chambers was 250 μm thick (Figure 2A-B) and patterned by spin coating SU-8™ 3050 at 1,000 rpm, soft-baked first at 65°C for 24 h and then at 95°C for 40 min, exposed to UV through a second photomask, and then post-exposure baked at 65°C for 4 h.
The master mold was then coated with (tridecafluoro-1,1,2,2-tetrahydro octyl) trichlorosilane (United Chemical Technologies, Bristol, PA, USA) to facilitate PDMS release from the master mold after replication. The PDMS devices were replicated from the master mold by pouring PDMS pre-polymer (1:5 mixture, Sylgard 184; DowDuPont, Midland, MI, USA) on the mold, followed by curing at 85°C for 45–60 min. The reservoirs to hold the culture medium were punched out of a 4.5–5 mm thick PDMS block using a 4 mm diameter drill punch (Figure 2D-E). To improve the bonding of the PDMS layer onto the glass substrate and to make the device hydrophilic for easy cell and culture medium loading, the PDMS layers were treated with oxygen plasma (Harrick Plasma, Ithaca, NY, USA) for 90 sec, followed by bonding onto a glass substrate. This process was repeated to bond the PDMS reservoir layer on top of the device. The assembled device was then stored dry (for up to 1 month), then sterilization with 70% ethanol for 15 min before use.
Microfluidic FMi-OOC device preparation for type IV collagen Matrigel filling of microchannels:
Before using the FMi-OOC, the devices were washed three times with PBS, filled with type IV basement membrane collagen Matrigel (Corning Matrigel Basement Membrane Matrix, DEV-free; 1:25 in media), and incubated at 37°C with 5% CO2 overnight. Diluted type IV basement membrane Matrigel was used to fill the microchannels connecting the AECs to AMCs and AMCs to chorion, which mimics the amnion and chorion basement membranes in utero. After this process, cell chambers were rinsed with PBS to remove extra Matrigel, and the devices were then ready for cell loading.
Cell seeding and culture in the FMi-OOC:
One day after the Matrigel coating, the devices were washed two times with complete DMEM-F12 medium before cell seeding. Primary cells were then trypsinized and loaded starting from the inside chamber to the outside (65,000 decidual cells for chamber 1, 250,000 chorion cells + 5% primary collagen + 25% Matrigel for chamber 2, 62,500 AMCs + 20% primary collagen + 25% Matrigel for chamber 3, and 120,000 AECs for chamber 4), similar to the concentration ratios seen in utero (AEC: 10 cells, AMC: 1 cell, CMC/CT: 20 cells, and decidua: 1.7 cells) 44. Following cell seeding, the FMi-OOCs were incubated at 37°C with 5% CO2 for 24 h before LPS treatment in specific cell chambers.
Collection of primary amnion collagen:
Methods for cell-free collagen preparation from amniotic membrane ECM following a protocol from Predictive Biotech, Inc. (Salt Lake City, UT) with some modifications. Primary collagen samples were isolated from amnion membranes obtained from fetal membranes from term, not-in-labor, and Cesarean deliveries. Approximately 10 g of amniotic membrane, peeled from the chorion layer, was washed with PBS three times thoroughly to remove any blood clots, then rinsed three more times in HBSS media. The amniotic membranes (1 g each) were then placed in a Dounce homogenizer and processed until the tissue was completely homogenized. The sample was then tested for cellular components by Masson trichome stain to confirm that collagen preparation was decellularized. Furthermore, we tested for type I collagen concentrations using an Enzyme-Linked Immunoassay (ELISA) (data not shown). Primary collagen for all experiments were prepared from a single placental sample to minimize patient-to-patient variability.
Creating an ascending infection model using the FMi-OOC:
To create an ascending infection model (i.e., maternal infection), as expected in subjects with preterm labor, cells in each FMi-OOC were treated with one of the following conditions for up to 72 h: 1) normal cell culture conditions with media specific for each type of cell (referred to in the rest of the manuscript as “control”) and 2) LPS treatment (100 ng/mL) of decidual cells in the FMi-OOC. To verify the propagation of LPS through the microchannels interconnecting each circular cell culture layer, a batch of FMi-OOCs were fabricated without microchannels and treated with LPS only on the AEC chamber. LPS from E. coli 055:B5 (Sigma-Aldrich #L2880, St. Louis, MO) was reconstituted at a concentration of 1 mg/mL. Concentrated LPS was then diluted to 100 ng/mL in culture media prior to use. This dose of LPS is within the range reported in the amniotic fluid of women with intra-amniotic infection and inflammation and as reported in our prior publications 47, 48. Once cells reached 70–80% confluency in the FMi-OOCs, each device was rinsed with sterile 1× PBS, serum-starved for 1 h, treated with the respective conditions, and then incubated at 37°C, 5% CO2, and 95% humidity for up to 72 h. To maintain low levels of perfusion between the cell culture chambers, the reservoirs on top of the device were filled with the following volumes of culture media every 24 h: decidua – 90 μl, chorion – 60 μl, AMC – 60 μl, and AEC – 50 μl.
Analysis of molecular diffusion in the FMi-OOC:
To determine the extent and time of LPS loaded into the center decidua chamber diffusing to the outer cell culture chambers through the interconnected microfluidic channel array, we conducted a set of perfusion studies. For this, fluorescein isothiocyanate (FITC) labeled LPS (1 mg/mL) was loaded into the center decidua chamber, and microscopy images of the device were taken over a 72 h period. Fluorescence intensity was used to measure the degree of diffusion from chamber to chamber. ImageJ software (National Institutes of Health, Bethesda, MD, USA) was used to analyze the images.
Cell staining
Masson trichrome staining for matrix collagen and cell imaging:
Before and after the FMi-OOC experiments, devices were stained with Masson trichrome stain to image type IV collagen inside the microchannels. To show that our matrix loading into the microchannels was evenly distributed, devices were rinsed with PBS and fixed at room temperature with 4% paraformaldehyde for 20 min. The devices were then stained with Biebrich scarlet-acid fuchsin for 10 min and then rinsed with water three times. This process stained all the cells and collagen a red color. Next, phosphomolybdic-phosphotungstic acid was applied for 15 min, which removed the red stain from the collagen. Aniline blue solution was then added for 10 min to stain the collagen a blue color. Once the device was stained, it was rinsed three times with water and imaged. This procedure was also carried out on some devices after 72 h of cell culture to monitor the collagen degradation caused by cell migration.
Staining for live and dead cells:
Before cell fixation, media were removed from all cell culture chambers, and each chamber was washed with 1x PBS. A combination of conjugated live and dead cell staining antibodies, calcein AM (for live cells; green; 4 mM) and ethidium homodimer-1 (for dead cells; red; 2 mM) (Thermo Fisher Scientific; L3224), were added into the cell culture chambers and incubated for 30 min in the dark with gentle rocking. The chambers were then washed with PBS, treated with NucBlue Live ReadyProbes Reagent (Thermo Fisher Scientific), and imaged as described below.
Crystal violet staining for cell viability:
After 72 h under control and LPS treatment conditions, all cell culture chambers were washed with 1x PBS, fixed with 4% paraformaldehyde for 15 min, washed with 1x water, and stained with 0.1% crystal violet dye for 20 min. After 20 min, cells were washed, allowed to dry, and imaged with bright field microscopy. Viable cells retained a uniform amount of violet dye.
Immunocytochemical staining of intermediate filaments cytokeratin-18 and vimentin:
Immunocytochemical staining for vimentin (3.7 μl/mL; ab92547; Abcam, Cambridge, MA, USA) and cytokeratin-18 (CK-18; 1 μl/mL; ab668; Abcam) were performed after 72 h, as previously reported 49-51. Manufacturers' instructions were followed to determine optimum dilution to ensure uniform staining. After 72 h, cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X, and blocked with 3% bovine serum albumin in PBS prior to incubation with primary antibodies overnight at 4°C. After washing with PBS, the FMi-OOCs were incubated with Alexa Fluor 488- and 594-conjugated secondary antibodies (Thermo Fisher Scientific) and diluted 1:1000 in 3% bovine serum albumin for 1 h in the dark. The devices were washed with PBS, treated with NucBlue Live ReadyProbes Reagent (Thermo Fisher Scientific), and imaged as described below.
Immunocytochemical staining for NF-kB:
Immunocytochemical staining for NF-kB (p65;pS529) (1:100; BD Biosciences - BDB558422) was performed after 15 min of LPS treatment. Decidua and AECs inside the device were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X, and blocked with 3% bovine serum albumin in PBS prior to incubation with 594-conjugated NF-kB antibody for 1 h at room temperature in the dark. The devices were washed with PBS, treated with NucBlue Live ReadyProbes Reagent (Thermo Fisher Scientific), and imaged as described below.
Immunocytochemical staining for LPS:
Immunocytochemical staining for biotin-linked LPS (30 μg/mL; My Bioscource MBS2012722) was performed after 24 h, 48 h, and 72 h. After each time point, cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X, and blocked with 3% bovine serum albumin in PBS prior to incubation with primary antibodies overnight at 4°C. After washing with PBS, the devices were incubated with streptavidin 594-conjugated secondary antibodies (BioLegend 405204) at a concentration of 0.0012 μg/uL in 3% bovine serum albumin for 1 h in the dark. The devices were washed with PBS, treated with NucBlue Live ReadyProbes Reagent (Thermo Fisher Scientific), and imaged as described below.
Microscopy:
After 24 h, 48 h, or 72 h of culture/treatment, bright field microscopy was performed (Nikon Eclipse TS100 microscope; ×4 and ×10 magnification, or Keyence All-in-one Fluorescence BZ-X810 microscope, ×4, ×10, and ×40 magnification) to determine cell morphology, nascent collagen production by cells, intermediate filament expression, cell migration, and LPS propagation.
Image analysis:
Two random regions of interest per device were used to determine overall CK-18, vimentin, and fluorescent LPS expression. Laser settings, brightness, contrast, and collection settings were all uniform for all images collected. Images were not modified (brightness, contrast, and smoothing) for intensity analysis. ImageJ software was used to measure LPS staining intensity from two different regions per treatment condition, per cell type, and at each time point.
Multiplex assays for pro- and anti-inflammatory cytokines:
Supernatants were collected from the reservoirs of all cell culture chambers after 24 h, 48 h, and 72 h of treatment. Multiplex assays were performed with cytokine granulocyte-macrophage colony-stimulating factor (GM-CSF), tumor necrosis factor (TNF)-α, interleukin (IL)-6 and IL-10 antibody-coated beads (Merck, Darmstadt, Germany), as indicators of inflammation in the cell culture supernatant. Standard curves were developed with duplicate samples of known quantities of recombinant proteins that were provided by the manufacturer. Sample concentrations were determined by relating the absorbance values that were obtained to the standard curve by linear regression analysis.
Statistical analyses:
All experiments were conducted in triplicate and graphs plotted using Prism 8 software (GraphPad Software, La Jolla, CA, USA). One-way ANOVA and independent samples Student's t-test were used, and p < 0.05 was considered significant. Bar graph values are expressed as mean intensities/concentrations ± standard error of the mean (SEM).
Results
Development of the FMi-OOC
Figure 2C-E shows images of the developed FMi-OOC device, where four different color dyes were loaded into each cell culture chamber for easy visualization. From the center to outside, the center chamber contains maternal decidual cells (green), the next chamber contains fetal CTs and CMCs (yellow), the third chamber contains AMCs (purple), and the outermost chamber containing AECs (blue). The circular chambers are connected through arrays of 24 microchannels (5 μm in height, 30 μm in width, and 300-600 μm in length). The cell media reservoir layer is shown placed on top of the main device layer after alignment (Figure 2D showing the schematic). Figure 2E shows the media reservoirs filled with color dye for visualization. Taken together, the internal FMi-OOC structure is similar to having distinct cellular layers, as seen in the human decidua-amniochorionic membrane (i.e., the FMi).
Characteristics of co-cultured amniochorionic and decidua cells in the FMi-OOC
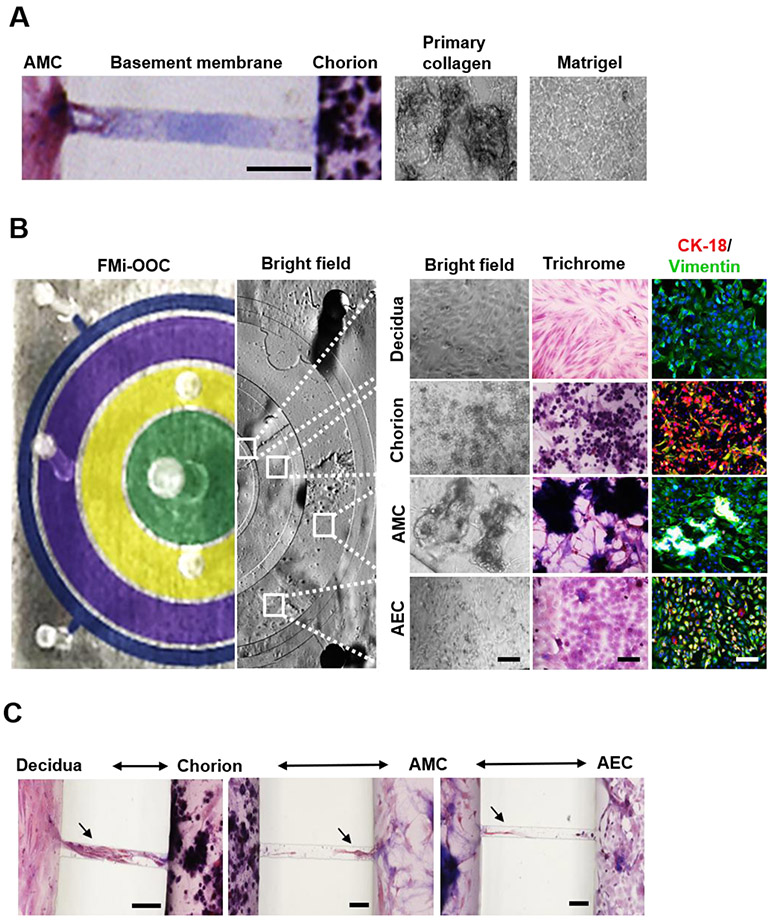
After sterilization, each device was placed inside a well of a conventional 6-well plate, and the microchannels were filled with type IV collagen (stained blue using Masson trichome stain), mimicking the basement membrane seen in utero between the amnion (AEC-AMC) and chorion layers (AMC-CT/CMC) (Figure 3A) 45. Microchannels are permeable to biochemicals but do not allow free movement of cells, except for supporting active cell migration, as we have previously described 50.
Figure 3: Characterization of decidua and amniochorionic derived cells cultured within the FMi-OOC.
A) Microchannels between the AEC and AMC, as well as the AMC and CTs/CMCs, chambers are filled with type IV collagen to recreate the two basement membranes. Collagen was stained with Masson trichome for visualization (blue color, left image). Scale bar = 100 μm. Additionally, AMCs and CTs/CMCs were cultured with decellularized primary collagen (middle image) harvested from the membrane of the amnion to provide biochemical factors necessary to recreate stromal layers, as well as function as and matrix to provide a scaffold for the cells to grow in 3D (right image).
B) On the left is a cross-sectional view of the four-chamber FMi-OOC device, highlighting the decidua chamber as green, the chorion chamber as yellow, the AMC chamber as purple, and the AEC chamber as blue. The right section of this image shows bright field microscopy images of cells growing in each cell culture chamber. A variety of in utero characteristics were measured to determine if cells grown within the FMi-OOC retained their in vivo characteristics (n = 3). These measurements included cell morphology, collagen production (Masson trichome staining [non-collagen producing cells show up as red, collagen-producing cells show up as purple, and collagen components show up as dark purple/blue]), cellular transition status (epithelial [CK-18; red] and mesenchymal [vimentin; green]) intermediate filament expression, and migratory potential. Scale bar = 50 μm.
C) Visualization of cell migration between compartments through the use of Masson trichome stain as a counter stain. Cellular migration between the chorion and amnion chambers were observed (see black arrowhead), as well as degradation of type IV collagen in the microchannel within 72 h. Scale bar = 100 μm.
The FMi-OOC was validated first by characterizing whether in utero cellular and matrix characteristics are maintained during culture. For this, we determined cell morphology, nascent collagen production, cellular transitions, collagen degradation, cell migration, and viability (Figure 3A-B; Supplemental Figure 1). AECs were cultured in 2D monolayers, whereas the AMCs and CMCs were grown in 3D using primary fetal membrane collagen as a scaffold to recreate the 3D microenvironment seen in utero (Figure 3A). Starting with the maternal side (innermost circular chamber), decidual cells cultured in 2D within the FMi-OOC retained their elongated morphology, did not produce nascent collagen (pink cells when stained with Masson trichome stain), and expressed the mesenchymal marker vimentin (green) (Figure 3B top row). Adjacent to the decidual chamber, chorion cells grown in primary collagen matrix formed 3D growths of cuboidal CK-18 (red) positive CTs and fibroblastoid vimentin (green) positive CMCs. Both cell types showed up as purple with Masson trichome stain, suggesting nascent collagen production inside the 3D collagen matrix (Figure 3B second row). In the next chamber, AMCs cultured with primary collagen showed 3D growth characteristics. AMCs retained their fibroblastoid morphology, produced nascent collagen (purple cells with Masson trichome stain), and expressed the mesenchymal marker vimentin (Figure 3B third row). In the outermost chamber, AECs grown in a 2D monolayer retained their cuboidal epithelial shape, while nascent collagen production was random with a mixture of purple and pink cells stained with trichome. Overall, the AEC population remained in a 'metastate' (as we have previously reported) 49, 50, co-expressing both epithelial (CK-18) and mesenchymal (vimentin) markers (Figure 3B bottom row). Further characterization of the FMi-OOC device showed that the addition of an on-chip reservoir layer provided consistent media availability to maintain cell viability (calcein AM+ [green] and ethidium homodimer-1− cells [red]) in each cell chamber of the FMi-OOC for 72 h (Supplemental Figure 1A-B).
Next, cell migration between the culture chambers was documented using trichome stain. Cell migration is an essential characteristic of amniochorion membrane remodeling commonly seen in utero 50-52, especially between AECs and AMCs. Here, the choriodecidua chambers formed a tight interface (cell-cell contact) within the microchannels, as seen in vivo (Figure 3C). After 72 h, cell migration between the chorion and amnion chambers were observed (Figure 3C arrowhead as examples), where this migration was associated with degradation of type IV collagen, as visualized by the absence of collagen (Figure 1C no purple color when stained with trichrome stain). Taken together, these results validate that the FMi-OOC model retains key cellular characteristics seen in the in vivo FMi. Table 1 summarizes key cellular characteristics of the FMi-OOC, comparing them to 2D cell culture, 3D cell culture, and those seen in humans.
Table 1:
Characteristics of cell cultured in the developed FMi-OOC compared to those seen in conventionally used in vitro models, as well as to the human in vivo conditions (* Red font highlights key differences that do not recapitulate those of human in vivo environment).
| Characteristics | Cell Type | 2D Cell Culture | 3D Cell Culture |
FMi-OOC | Human |
|---|---|---|---|---|---|
| Morphology | AEC | Cuboidal/Fibroblastoid | Fibroblastoid | Cuboidal/Fibroblastoid | Cuboidal/Fibroblastoid |
| AMC | Fibroblastoid | Cuboidal/Fibroblastoid | Fibroblastoid | Fibroblastoid | |
| CMC/CT | Fibroblastoid/Cuboidal | Fibroblastoid/Cuboidal | Fibroblastoid/Cuboidal | Fibroblastoid/Cuboidal | |
| Decidua | Fibroblastoid | Fibroblastoid | Fibroblastoid | Fibroblastoid | |
| Collagen production | AEC | low | high | high | high |
| AMC | high | high | high | high | |
| CMC/CT | low | high | high | high | |
| Decidua | low | low | low | low | |
| Intermediate filament expression | AEC | Metastate | Mesenchymal | Metastate | Metastate |
| AMC | Mesenchymal | Metastate | Mesenchymal | Mesenchymal | |
| CMC/CT | Mesenchymal/Epithelial | Mesenchymal/Epithelial | Mesenchymal/Epithelial | Mesenchymal/Epithelial | |
| Decidua | Mesenchymal | Mesenchymal | Mesenchymal | Mesenchymal |
Fluid diffusion over time between the culture chambers in the FMi-OOC
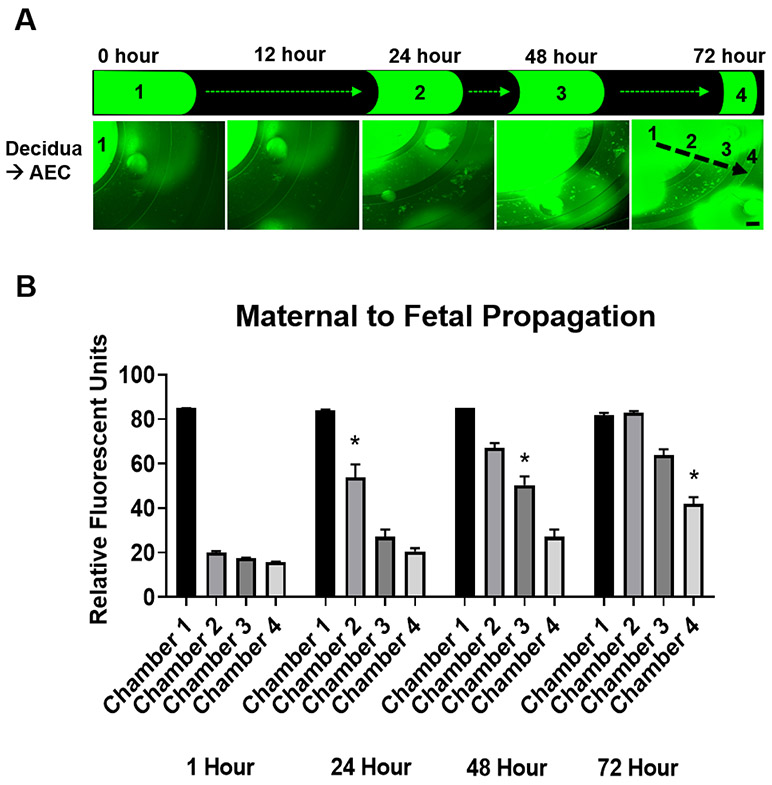
In the multi-cellular multi-compartment FMi-OOC system, fluid diffusion characteristics between each of the four chambers are important to understand how stimulants and biochemicals produced by cells diffuse from one chamber to the other chamber. This allows the assessment of feto-maternal cells in their respective compartments while maintaining intracellular and intercellular interactions. The efficiency to maintain fluidic separation between the four chambers was tested using a FITC labeled LPS (1 mg/mL) perfusion assay by creating a minute fluidic level difference between the decidua (chamber 1 into which FITC-LPS was loaded) and AEC (chamber 4) chamber, which results in hydrostatic pressure differences. The effects of such a fluid level difference on diffusion and fluidic isolation is well studied 53, 54. Figure 4A shows that the FITC intensity in chambers 2-4 increased over the 72 h period as FITC-LPS diffused between chambers, reaching chorion in 24 h, AMCs in 48 h, and AECs in 72 h (Figure 4B). These data suggest that FITC-LPS can gradually diffuse from the decidua to the AEC chamber within 72 h, taking approximately 24 h for sufficient diffusion to occur from one chamber to the next neighboring chamber. This characterization allowed identifying the ideal time points for studying the propagation of infection or inflammation within the FMi-OOC device, as shown in the subsequent section.
Figure 4: Fluidic isolation and diffusion of florescent LPS over time between the FMi-OOC chambers.
A) FITC-LPS was tracked every 12 h for 72 h to monitor its diffusion from the maternal decidua side (chamber 1) to the fetal AEC side (chamber 4). Scale bar = 600 μm.
B) FITC-LPS significantly propagated between each chamber in the 24 h time increment: chamber 1 (1 h [85±0 RFU] → 24 h [83±0.5 RFU]), chamber 2 (1 h [20±0.6 RFU] → 24 h [53±6 RFU] [P<0.0001]), chamber 3 (24 h [27±3 RFU] → 48 h [50±4 RFU] [P<0.0001]), and chamber 4 (48 h [27±3 RFU] → 72 h [42±3 RFU] [P=0.0074]) (n = 3). Values are expressed as mean intensities ± SEM.
Kinetics of the ascending infection signals in the FMi-OOC
PDMS has been documented to absorb various cell-secreted factors and other biochemicals 55. To first characterize how much cytokines may be adsorbed to the PDMS surface, which can prevent accurate measurement of cell-secreted inflammatory cytokines, we tested the degree of IL-6 adsorption in the FMi-OOC device. Supplemental Figure 1C shows that when the device was loaded with 10 ng/mL of IL-6 and sampled after 24 h and 72 h, no noticeable difference in IL-6 concentration was observed. This result shows that our cytokine adsorption to the PDMS device is not a problem, and that the PDMS-based FMi-OOC can be utilized to study inflammatory mediators of the FMi.
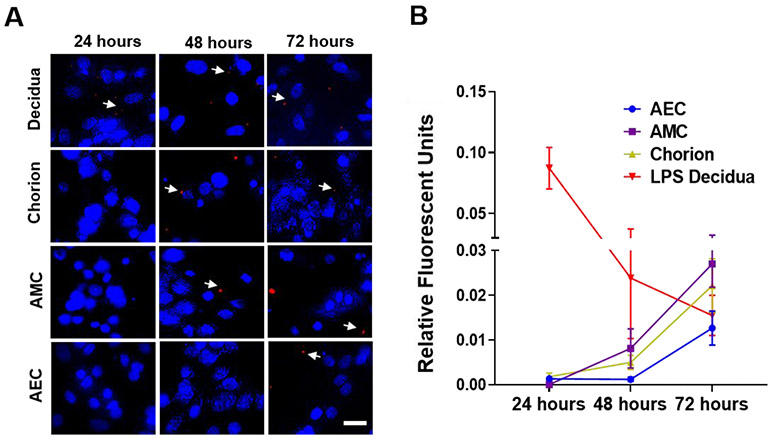
For the LPS stimulation experiments using the FMi-OOC, we first confirmed that 100 ng/mL of LPS did not impact the viability of cells in any of the chambers (Supplemental Figure 2A). Fluorescent microscopy showed that LPS positive decidual cells (red spots; white arrows) were seen within 24 h and remained positive throughout the 72 h testing period (Figure 5A first row). This LPS propagated from the decidua to the fetal AMC chamber within 48 h, and then to the AEC chamber by 72 h. Quantitation of LPS florescent intensity further confirmed the propagation of this infectious stimulant from the maternal compartment to the fetal compartment of the FMi-OOC (Figure 5B). Additionally, LPS treatment induced canonical NF-kB activation and decidual/AEC cell nuclear translocations (Supplemental Figure 2B). This suggests that the propagation of an infectious stimulant resulted in pro-inflammatory transcription factor activation across cells within the FMi-OOC.
Figure 5: Propagation of LPS in the FMi-OOC.
Immunocytochemistry followed by fluorescent microscopy shows LPS (red dots; white arrows) propagation from the decidua (24 h – 0.09±0.02 RFI), to chorion (24 h – 0.005±0.001 RFI), and then to AMCs (48 h – 0.009±0.006 RFI) within 48 h, and finally to AECs within 72 h (72 h – 0.01±0.003 RFI) (n = 3). Scale bar = 10 μm. Blue – DAPI, Red – LPS. Values are expressed as mean intensities ± SEM.
Propagation of inflammatory mediators in the FMi-OOC
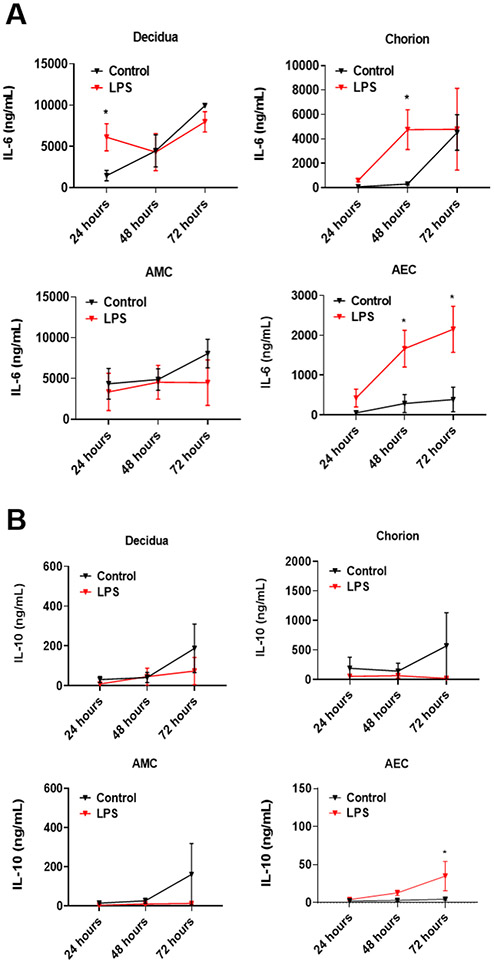
Concentrations of pro- and anti-inflammatory cytokines, IL-6 and IL-10, respectively, were measured after 24 h, 48 h, and 72 h to determine whether LPS-induced inflammation remained localized to the maternal chamber or whether propagation of LPS and NF-kB activation resulted in an immune response in the fetal chambers (Figure 6). LPS-treated decidual cells significantly increased IL-6 production within 24 h (6115±1641 ng/mL) inside the FMi-OOC compared to control (1469±629 ng/mL; P=0.028). IL-6 was also significantly increased in the chorion (LPS - 4765±1624 ng/mL; controls - 313±94 ng/mL; P=0.026) and AEC chambers by 48 h (LPS - 1665±459 ng/mL; controls - 285±226 ng/mL; P=0.016). However, by 72 h, IL-6 levels increased in untreated control decidua, chorion, and AMC chambers, suggesting a culture condition-induced effect. In the AEC chamber, interestingly, IL-6 continued to increase by 72 h, while almost no increase was observed for the case of controls (LPS - 2151±576 ng/mL; controls - 387±309 ng/mL; P=0.027). This data shows the ability of LPS to propagate from the maternal side to the fetal side and subsequently cause inflammation in the fetal cell chambers. While we cannot rule out the confounding effect of inflammatory mediators diffusing through the microchannels between the compartments, such paracrine interactions are also expected in utero. Therefore, we presume that such an effect is successfully recreated in our FMi-OOC model.
Figure 6: Production and propagation of pro- and anti-inflammatory cytokines in the FMi-OOC device.
A) Increased IL-6 level in decidual cells after 24-h LPS treatment can be observed compared to controls (P=0.0287). The chorion and AECs also secreted significant levels of IL-6 after 48 h of LPS treatment (P=0.0261 and P=0.0161, respectively). This trend continued to increase in the AEC compartment after 72 h (P=0.0376), showing a progressive induction of inflammatory mediators from the decidua to AEC (n = 4).
B) LPS decreased the production of the anti-inflammatory IL-10 in the decidua, chorion, and AMCs at all time points compared to controls. However, LPS significantly increased IL-10 levels in AECs after 72 h (P=0.0376) (n = 4). For all graphs, values are expressed as mean values ± SEM.
Although not significant, LPS treatment resulted in lower levels of anti-inflammatory IL-10 levels compared to controls in the decidual and the two fetal cell chambers (chorion and AMCs), regardless of the time points. After 72 h, the IL-10 concentration was 2-fold lower for each of these cell types after LPS treatment compared to controls (Figure 6B). However, in the case of AECs, a significant increase in IL-10 was determined after 72 h of LPS treatment (LPS - 35±19 ng/mL, controls - 4±1 ng/mL; P=0.037).
The ratio between IL-10 and IL-6 concentrations was determined to test whether the balance between pro- and anti-inflammatory mediators shifted in our FMi-OOC model system. However, even with an increase in the IL-10 level, we did not see a balanced immune response in the AEC chamber (72 h IL-10:IL-6 ratio; P=0.226) (Supplemental Figure 3A). This suggests that AECs innately respond by producing IL-10; however, this was insufficient to overcome the pro-inflammatory impact of LPS. Besides, LPS induced immune imbalance in all maternal and fetal cell chambers within the FMi-OOC (Supplemental Figure 3A).
Discussion
Structural, biological, and physiological differences in pregnancies between human and animal models, along with the limitations of existing in vitro cell culture models, have led to the development of advanced human physiology-mimicking in vitro models, such as OOCs, to be developed and utilized in the field of obstetrics 35, 46, 56, 57. Herein, we describe a FMi-OOC device that contains cells from both fetal and maternal primary human tissue. The FMi-OOC developed and validated in this study revealed that decidua and amniochorionic membrane cells cultured in the OOC model could preserve key characteristics observed during pregnancy in utero. These include the ability to: 1) proliferate and maintain epithelial or mesenchymal cellular transition status as well as migration capabilities; 2) produce nascent collagen; 3) maintain viability; and 4) produce localized inflammation to degrade type IV collagen and to facilitate cellular migration between the culture chambers. These findings mimic the in utero FMi, showing classical signs of tissue remodeling and/or repair 49, 58; thus, validating the physiological relevance of the developed FMi-OOC model. Utilizing the developed FMi-OOC, we created an ascending infection model using LPS as a stimulant that propagated from the decidua to the amnion within 72 h. Data from this model demonstrated: 1) propagation of the infectious stimulus from the maternal chamber to the fetal chambers, 2) maternal inflammatory activation (cytokine release) that was subsequently observed in fetal cells, and 3) inflammatory response in both the maternal and fetal sides upon infectious stimulation of the maternal side, a phenomenon that is similar to the immune imbalance observed at the FMi in utero. In summary, the developed FMi-OOC presented here provides a model that can be used to study ascending infection and associated feto-maternal inflammatory responses.
Typically, to answer physiological or pathological questions related to the FMi in vitro, a standard 2D cell culture model 59, transwell co-culture model 60, or 3D cell spheroid or cell sheet models 61 are normally used. Each of these methods have advantages and limitations 46. However, the loss of intercellular interactions and cell-collagen interactions between the decidua-amniochorionic membrane, as well as most co-culture models allowing only two different cell types to be tested, are some of their limitations. Studying individual cellular components does not address tissue-level complexity that can provide a comprehensive and physiologically relevant answer when studying the multi-cellular, multi-organ FMi. To some extent, cellular interaction studies can be conducted using in vitro explant culture methods 62. However, in this model, intercellular interactions and contributions by individual cellular layers are difficult to assess, since controlling and accessing each cellular layer is often not possible. Tissue explants also lack cell division, have difficult to determine the proliferative properties of cells (as well as their transition and migration), and are limited in their in vitro lifespan, as their responsiveness is not reliable beyond 48 h 62. The FMi-OOC overcomes many of the above listed limitations, highlighting its usefulness as an alternative model for studying ascending infection.
Specifically, the four-chamber FMi-OOC allowed us to make several key observations, validating the model and providing insights into the physiological state of the FMi in utero. For example, AMCs are sparse during normal gestation in the membrane ECM, and the ratio between AECs and AMCs is approximately 10:1. However, at term, AECs undergo a terminal state of epithelial-to-mesenchymal transition (EMT), leading to the accumulation of AMCs. Massive inflammation derived from AMCs can cause ECM damage and membrane weakening. Under normal culture conditions in the FMi-OOC, we were able to maintain this ratio, suggesting a normal physiological state (Figure 3). Moreover, an increase in inflammatory cytokines was also not seen after LPS treatment in the AMC chamber, which is likely due to the lack of any changes in the AMC numbers (Figure 6). To note, the number of AMCs were also not increased, since LPS is a poor inducer of EMT (AECs becoming AMCs in response to a stimulant). It is also indicative of an innate resistance within the membranes to support FMi integrity in the wake of an infection. Interestingly, inflammation was noted in the AEC chambers (within 24 h) prior to LPS reaching this chamber (Figure 6), supporting the gradual propagation of stimulants.
Additionally, the role of the chorion in physiological and pathological pregnancies is relatively unknown due to the difficulty in culturing primary fetal membrane-derived chorion trophoblast cells. Furthermore, the interaction between the decidua and chorion (maternal-fetal barrier) is even more elusive. Besides monitoring IL-6 and IL-10 levels, we also monitored cytokines, such as GM-CSF and TNF-α, that are more cell-type-specific responders. Although not significant, the chorion showed an increased level of TNF-α secretion in this model, as seen in previous studies 63, 64 (Supplemental Figure 3). Chorion is susceptible to TNF-α-induced apoptosis 65-67. An increase in this cytotoxic cytokine is indicative of cellular- and immune-level compromise that may facilitate ascending infection. As noted in Supplemental Figure 3, the cytokine response in each cell layer is different, which is indicative of distinct immune responses (Table 2). This data supports the heterogeneity in inflammatory biomarkers reported during MIAC and IAI 64, 68.
Table 2:
Propagation of LPS-induced cytokine production (*Green color indicates increasing levels of pro-inflammatory cytokines, while red indicates decreasing levels of anti-inflammatory cytokines. This was determined by a greater than 2-fold difference).
| Cytokine | Tissue compartment |
|||
|---|---|---|---|---|
| 24 hours | 48 hours | 72 hours | ||
| IL-10 | Decidua | ↓ | ←→ | ↓ |
| Chorion | ↓ | ↓ | ↓ | |
| AMC | ↓ | ↓ | ↓ | |
| AEC | ←→ | ↑ | ↑ | |
| IL-6 | Decidua | ↑ | ←→ | ←→ |
| Chorion | ↑ | ↑ | ←→ | |
| AMC | ←→ | ←→ | ←→ | |
| AEC | ↑ | ↑ | ↑ | |
| IL-8 | Decidua | ←→ | ↓ | ←→ |
| Chorion | ←→ | ←→ | ←→ | |
| AMC | ←→ | ←→ | ←→ | |
| AEC | ↑ | ↑ | ←→ | |
| TNF-α | Decidua | ←→ | ←→ | ←→ |
| Chorion | ↑ | ↑ | ←→ | |
| AMC | ←→ | ←→ | ←→ | |
| AEC | ←→ | ↑ | ←→ | |
| GM-CSF | Decidua | ↑ | ←→ | ←→ |
| Chorion | ↓ | ↑ | ←→ | |
| AMC | ←→ | ←→ | ↑ | |
| AEC | ←→ | ↑ | ←→ | |
Conclusion
The FMi-OOC model developed here helped to visualize cellular and collagen components throughout the culture, as well as tracking the propagation of infection and inflammation across the FMi. However, the current OOC has a few biological, device-related, and experimental limitations that we plan to overcome in the future. From a biology side, although we included decidua, chorion, and amnion cells in our model and established a baseline characteristic for studying ascending infection, this model lacks immune cells (e.g., natural killer cells, macrophages, and T-cells) that are also critical components of the feto-maternal immune system 11. Future studies will test their contributions during ascending infection. Additionally, the effect of infection and inflammation on neither the cervix nor placenta could be studied in this model, even though they are vital components of the intrauterine cavity, which we plan to include as additional cell culture compartments in future studies. On the microfabrication side, the FMi-OOC was not designed to induce biomechanical stressors to the cellular layers, such as physical stretch or dynamic shear stress that cells in utero may experience. Fetal membrane cells throughout gestation must adapt and remodel the membrane to accommodate these biomechanical stressors. Recreating these components could add to the physiological relevance of the FMi-OOC device in the future. From the experimental side, there are also a few limitations that we plan to address in our future studies. The current study examined maternal to fetal transmission of infectious agents (simulated by LPS treatment) and the development of inflammation. Although systemically introduced amniotic infection is rare, it can still occur; However, we did not examine the fetal to maternal propagation of infectious stimuli and production of inflammation. Besides, mechanistic mediators and signalers (e.g., toll-like receptor-4/ myeloid differentiation primary response-88) of LPS action were also not examined. Cytokines analyzed in this work showed an increased level (over 2-fold), but many did not reach to a significant level. This could be due to multiple reasons, including statistical power was likely insufficient to detect significance and the duration of culture (72 h) might be insufficient to produce significant changes. However, in this work our primary goal was not to document significant changes in any specific biochemical marker, but rather to determine the overall shift in a specific cell's inflammatory status in response to the propagating of infection. Therefore, a power analysis was not conducted prior to the initiation of this study. Despite this limitation, we were still able to report time- and cell type-dependent changes in the cytokine profiles and shifts indicative of immune imbalance with respect to the two cytokines tested (IL-6 and IL-10). In summary, future designs of the FMi-OOC will include other critical cellular components (e.g., fetal and maternal immune cells) and biomechanical stressors (e.g., physical stretch and shear stress) that will allow researchers to address new questions regarding the function of the FMi during normal and pathological pregnancies.
The FMi-OOC system developed and presented here allowed us to overcome several limitations of traditional in vitro models (2D and 3D culture systems) and the simplistic two-chamber OOC design while investigating the propagation of infection and inflammation across the FMi. Maternal infection was able to propagate through the fetal chambers and induce cell-specific inflammatory cytokine profiles, suggesting that each cellular layer responds differentially and induces infection-related cell-cell communication. In the future, elucidation of these cellular responses could identify novel points of treatment for limiting maternal infection-induced fetal inflammation. Taken together, the FMi-OOC provides a robust model to study normal pregnancy as well as the mechanism and potential treatment of maternal infection-induced preterm birth.
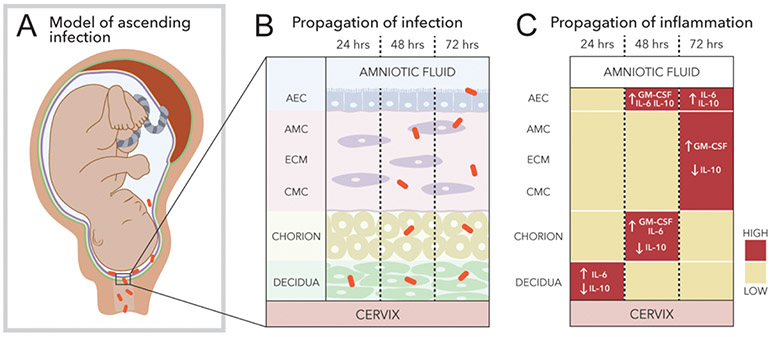
In conclusion, this study showed the disruption of the FMi integrity during ascending infection due to immune marker-associated dysregulation at both the maternal and fetal sides. As summarized in Figure 7, a functional ascending infection model and host inflammation characteristics that could contribute to PTB are presented. During ascending infection, infectious microbes (illustrated as rod-shaped and orange colored) transverse the cervix (Figure 7A) and propagate across decidua to reach the fetal amnion epithelium in a stepwise fashion, crossing both cellular and collagen layers (Figure 7B). Propagation of infection coincides with an increased pro-inflammatory profile across the FMi (red boxes; Figure 7C) (infectious inflammation). Shifts in the inflammatory profile at this critical interface can induce immune imbalance (red boxes; Figure 7C), contributing to membrane weakening and leading to PTB.
Figure 7: Schematic showing the propagation of infection and inflammation across the FMi observed through the FMi-OOC model.
Using the developed FMi-OOC model, an ascending infection model (decidua → amnion) of microbial invasion of the amniotic cavity was recreated. Ascending infection was shown associated with a pro-inflammatory shift, compromising the FMi immune homeostasis that can eventually result in intra-amniotic inflammation predisposing subjects to preterm labor-associated feto-maternal inflammatory responses.
A) Illustration of ascending infection during pregnancy that the developed model recapitulates.
B) The FMi (decidua-fetal amniochorionic interface) interconnects the cervix and intra-amniotic cavity. During ascending infection, infectious microbes transverse through the cervix, propagate from the decidua to reach AECs, and initiate inflammation.
C) Propagation of infection coincides with an increased pro-inflammatory environment (high IL-6 and GM-CSF levels; low IL-10 level), causing an immune imbalance.
Supplementary Material
Acknowledgments
Funding: This study was supported by funding from the NIH/NICHD grant R01 HD100729 and NCATS grant UG3TR003283 to R. Menon and A. Han. L. Richardson was supported by the Jeane B. Kempner Fellowship of the UTMB (2019–2020) and by a postdoctoral fellowship through the Regulatory Science in Environmental Health and Toxicology Training Grant (T32 ES026568) from the National Institute of Environmental Health Sciences (NIEHS) of the National Institutes of Health (NIH).
Footnotes
Conflict of interest statement: A patent application describing the feto-maternal interface organ-on-a-chip system has been submitted.
Data availability:
Data will be made available upon request.
References
- 1.Keelan JA, J Reprod Immunol, 2018, 125, 89–99. [DOI] [PubMed] [Google Scholar]
- 2.Romero R, Dey SK and Fisher SJ, Science, 2014, 345, 760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Menon R and Fortunato SJ, Best Pract Res Clin Obstet Gynaecol, 2007, 21, 467–478. [DOI] [PubMed] [Google Scholar]
- 4.Bastek JA, Gomez LM and Elovitz MA, Clin. Perinatol, 2011, 38, 385–406. [DOI] [PubMed] [Google Scholar]
- 5.Behnia F, Sheller S and Menon R, Am. J Reprod Immunol, 2016, DOI: 10.1111/aji.12496 [doi]. [DOI] [PubMed] [Google Scholar]
- 6.Bonney EA and Johnson MR, Placenta, 2019, 79, 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abrahams VM, Potter JA, Bhat G, Peltier MR, Saade G and Menon R, Am J Reprod Immunol, 2013, 69, 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menon R and Taylor BD, Obstet Gynecol, 2019, 134, 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menon R, Dunlop AL, Kramer MR, Fortunato SJ and Hogue CJ, Acta Obstet. Gynecol. Scand, 2011, 90, 1325–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ander SE, Diamond MS and Coyne CB, Sci Immunol, 2019, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park JE, Stephenson E, Polanski K, Goncalves A, Gardner L, Holmqvist S, Henriksson J, Zou A, Sharkey AM, Millar B, Innes B, Wood L, Wilbrey-Clark A, Payne RP, Ivarsson MA, Lisgo S, Filby A, Rowitch DH, Bulmer JN, Wright GJ, Stubbington MJT, Haniffa M, Moffett A and Teichmann SA, Nature, 2018, 563, 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erlebacher A, Annu Rev Immunol, 2013, 31, 387–411. [DOI] [PubMed] [Google Scholar]
- 13.Rinaldi SF, Makieva S, Saunders PT, Rossi AG and Norman JE, Mol Hum Reprod, 2017, 23, 708–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez-Lopez N, Vadillo-Perez L, Hernandez-Carbajal A, Godines-Enriquez M, Olson DM and Vadillo-Ortega F, Am J Obstet Gynecol, 2011, 205, 235 e215–224. [DOI] [PubMed] [Google Scholar]
- 15.Norwitz ER, Bonney EA, Snegovskikh VV, Williams MA, Phillippe M, Park JS and Abrahams VM, Cold Spring Harb Perspect Med, 2015, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menon R, Richardson LS and Lappas M, Placenta, 2018, DOI: 10.1016/j.placenta.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menon R and Moore JJ, Obstet Gynecol Clin North Am, 2020, 47, 147–162. [DOI] [PubMed] [Google Scholar]
- 18.Gerson KD, McCarthy C, Elovitz MA, Ravel J, Sammel MD and Burris HH, Am J Obstet Gynecol, 2020, 222, 491 e491–491 e498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donders GG, Van CK, Bellen G, Reybrouck R, Van den Bosch T, Riphagen I and Van LS, BJOG, 2009, 116, 1315–1324. [DOI] [PubMed] [Google Scholar]
- 20.Donders GGG, Bellen G, Grinceviciene S, Ruban K and Vieira-Baptista P, Res Microbiol, 2017, 168, 845–858. [DOI] [PubMed] [Google Scholar]
- 21.Romero R, Gomez-Lopez N, Winters AD, Jung E, Shaman M, Bieda J, Panaitescu B, Pacora P, Erez O, Greenberg JM, Ahmad MM, Hsu CD and Theis KR, Journal of perinatal medicine, 2019, 47, 915–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Racicot K, Cardenas I, Wunsche V, Aldo P, Guller S, Means RE, Romero R and Mor G, J Immunol, 2013, 191, 934–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomez-Lopez N, Romero R, Plazyo O, Schwenkel G, Garcia-Flores V, Unkel R, Xu Y, Leng Y, Hassan SS, Panaitescu B, Cha J and Dey SK, Am J Obstet Gynecol, 2017, DOI: 10.1016/j.ajog.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armistead B, Oler E, Adams Waldorf K and Rajagopal L, J Mol Biol, 2019, 431, 2914–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gravett MG, Haluska GJ, Cook MJ and Novy MJ, Am J Obstet Gynecol, 1996, 174, 1725–1731; discussion 1731-1723. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen BW, Bonney EA, Pearce BD, Donahue LR, Sarkar IN and Preterm Birth International C, Reprod Sci, 2016, 23, 482–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarthy R, Martin-Fairey C, Sojka DK, Herzog ED, Jungheim ES, Stout MJ, Fay JC, Mahendroo M, Reese J, Herington JL, Plosa EJ, Shelton EL and England SK, Biol Reprod, 2018, 99, 922–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haderspeck JC, Chuchuy J, Kustermann S, Liebau S and Loskill P, Expert Opin Drug Discov, 2019, 14, 47–57. [DOI] [PubMed] [Google Scholar]
- 29.Jodat YA, Kang MG, Kiaee K, Kim GJ, Martinez AFH, Rosenkranz A, Bae H and Shin SR, Curr Pharm Des, 2018, 24, 5471–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.An F, Qu Y, Liu X, Zhong R and Luo Y, Anal Chem Insights, 2015, 10, 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balijepalli A and Sivaramakrishan V, Drug Discov Today, 2017, 22, 397–403. [DOI] [PubMed] [Google Scholar]
- 32.Bhatia SN and Ingber DE, Nat Biotechnol, 2014, 32, 760–772. [DOI] [PubMed] [Google Scholar]
- 33.Blundell C, Yi YS, Ma L, Tess ER, Farrell MJ, Georgescu A, Aleksunes LM and Huh D, Adv Healthc Mater, 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JS, Romero R, Han YM, Kim HC, Kim CJ, Hong JS and Huh D, J Matern Fetal Neonatal Med, 2016, 29, 1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pemathilaka RL, Reynolds DE and Hashemi NN, Interface Focus, 2019, 9, 20190031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pemathilaka RL, Caplin JD, Aykar SS, Montazami R and Hashemi NN, Glob Chall, 2019, 3, 1800112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin F, Zhu Y, Zhang M, Yu H, Chen W and Qin J, Toxicol In Vitro, 2019, 54, 105–113. [DOI] [PubMed] [Google Scholar]
- 38.Gnecco JS, Anders AP, Cliffel D, Pensabene V, Rogers LM, Osteen K and Aronoff DM, Curr Pharm Des, 2017, DOI: 10.2174/1381612823666170825142649. [DOI] [PubMed] [Google Scholar]
- 39.Richardson L, Gnecco J, Ding T, Osteen K, Rogers LM, Aronoff DM and Menon R, Reprod Sci, 2019, DOI: 10.1177/1933719119828084, 1933719119828084. [DOI] [PubMed] [Google Scholar]
- 40.Richardson L, Jeong S, Kim S, Han A and Menon R, FASEB J, 2019, 33, 8945–8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin J, Richardson L, Sheller-Miller S, Zhong N and Menon R, Placenta, 2018, 67, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato BL, Collier ES, Vermudez SA, Junker AD and Kendal-Wright CE, Placenta, 2016, 44, 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menon R, Boldogh I, Urrabaz-Garza R, Polettini J, Syed TA, Saade GR, Papaconstantinou J and Taylor RN, PLoS ONE [Electronic Resource], 2013, 8, e83416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gnecco JS, Anders AP, Cliffel D, Pensabene V, Rogers LM, Osteen K and Aronoff DM, Curr Pharm Des, 2017, 23, 6115–6124. [DOI] [PubMed] [Google Scholar]
- 45.Richardson Lauren, Vargas Gracie, Brown Tyra, Ochoa Lorenzo, Trivedi J, Kacerovský Marian, Lappas Martha and Menon R, 2017, 53. [DOI] [PubMed] [Google Scholar]
- 46.Richardson L, Kim S, Menon R and Han A, Front Physiol, 2020, 11, 715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Romero R, Avila C, Santhanam U and Sehgal PB, J Clin Invest, 1990, 85, 1392–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dixon CL, Richardson L, Sheller-Miller S, Saade G and Menon R, Am J Reprod Immunol, 2017, DOI: 10.1111/aji.12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richardson L and Menon R, Am J Pathol, 2018, DOI: 10.1016/j.ajpath.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richardson L, Jeong S, Kim S, Han A and Menon R, Faseb j, 2019, DOI: 10.1096/fj.201900020RR, fj201900020RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richardson LS, Taylor RN and Menon R, Sci Signal, 2020, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richardson LS, Vargas G, Brown T, Ochoa L, Sheller-Miller S, Saade GR, Taylor RN and Menon R, The American Journal of Pathology, DOI: 10.1016/j.ajpath.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park J, Koito H, Li J and Han A, Biomed Microdevices, 2009, 11, 1145–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park J, Koito H, Li J and Han A, Lab Chip, 2012, 12, 3296–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang JD, Douville NJ, Takayama S and ElSayed M, Ann Biomed Eng, 2012, 40, 1862–1873. [DOI] [PubMed] [Google Scholar]
- 56.Aronoff DM, Trans Am Clin Climatol Assoc, 2020, 131, 72–79. [PMC free article] [PubMed] [Google Scholar]
- 57.Horii M, Touma O, Bui T and Parast MM, Reproduction, 2020, 160, R1–r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Richardson LS, Radnaa E, Urrabaz-Garza R, Lavu N and Menon R, Placenta, 2020, 99, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feng L, Allen TK, Marinello WP and Murtha AP, Reprod Sci, 2018, DOI: 10.1177/1933719118776790, 1933719118776790. [DOI] [Google Scholar]
- 60.Zaga-Clavellina V, López GG, Estrada-Gutierrez G, Martinez-Flores A, Maida-Claros R, Beltran-Montoya J and Vadillo-Ortega F, Mycoses, 2006, 49, 6–13. [DOI] [PubMed] [Google Scholar]
- 61.Richardson LS, Menon PR and Menon R, Placenta, 2020, 90, 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fortunato SJ, Menon R, Swan KF and Lyden TW, Am J Reprod Immunol, 1994, 32, 184–187. [DOI] [PubMed] [Google Scholar]
- 63.Arechavaleta-Velasco F, Ogando D, Parry S and Vadillo-Ortega F, Biol Reprod, 2002, 67, 1952–1958. [DOI] [PubMed] [Google Scholar]
- 64.Hadley EE, Richardson LS, Torloni MR and Menon R, Am J Reprod Immunol, 2017, DOI: 10.1111/aji.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Menon R, Fortunato SJ, Yu J, Milne GL, Sanchez S, Drobek CO, Lappas M and Taylor RN, Placenta, 2011, 32, 317–322. [DOI] [PubMed] [Google Scholar]
- 66.Fortunato SJ, Menon R and Lombardi SJ, Am J Obstet Gynecol, 2001, 184, 1392–1397; discussion 1397-1398. [DOI] [PubMed] [Google Scholar]
- 67.Fortunato SJ, Menon R, Bryant C and Lombardi SJ, Am J Obstet Gynecol, 2000, 182, 1468–1476. [DOI] [PubMed] [Google Scholar]
- 68.Menon R, Torloni MR, Voltolini C, Torricelli M, Merialdi M, Betran AP, Widmer M, Allen T, Davydova I, Khodjaeva Z, Thorsen P, Kacerovsky M, Tambor V, Massinen T, Nace J and Arora C, Reprod Sci, 2011, 18, 1046–1070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available upon request.