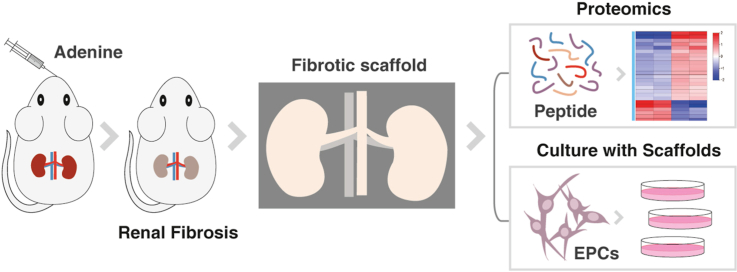
Abstract
Chronic kidney disease has been recognized as a major public health problem worldwide and renal fibrosis is a common pathological process occurring in chronic renal failure. It is very promising to find the strategies to slow or even prevent the progression of fibrosis. This study focused on whether renal fibrosis decellularized scaffolds has the potential to be a model of cellular mechanisms of tissue fibrosis or donors for tissue engineering. In order to evaluate the feasibility of decellularized scaffolds derived from pathological kidneys, histology, proteomics and ELISA will be used to analysis the changes in the structure and main components of fibrotic tissue. The fibrosis model in this paper was induced by adenine-fed and the results showed that the structure of fibrotic scaffold was changed and some protein were up-regulated or down-regulated, but the cytokines associated with renal regeneration after injury were remained. In cell experiments, endothelial progenitor cells proliferated well, which proved that the fibrotic scaffolds have non-cytotoxic. All these conclusions indicate that the renal fibrosis decellularized scaffolds model has the ability to study fibrosis mechanism and the potential to be engineering donors as well as normal scaffolds.
Keywords: Extracellular matrix, Proteomics, Growth factor, Tissue engineering, Chronic renal diseases
Graphical abstract
Highlights
-
•
A study model of the mechanism of renal fibrosis from natural sources.
-
•
Adenine-induced renal fibrosis with a cytopathic scaffold preserves important cytokines.
-
•
There was no significant difference in cytokine content with natural scaffolds.
-
•
Heat maps of differentially expressed proteins from subcellular localization.
1. Introduction
Chronic kidney disease (CKD) has been recognized as a major public health problem worldwide, the global prevalence of which had reached 9.1% [1]. Developing countries such as China had a higher incidence of 10.8% [2]. CKD will eventually progress to end-stage renal disease (ESRD) which is a serious disease threatens human life, and the best treatment for most patients with ESRD to improve survival rate and the quality of life is kidney transplantation. However, the huge donor gap and the high cost make it impossible for many patients to receive treatment, making it crucial to find more strategies to slow or even prevent the progression of CKD to ESRD.
The progression of CKD is the progressive accumulation of renal fibrosis, characterized by excessive extracellular matrix (ECM) deposition and myofibroblast accumulation [3]. At the same time, glomerular sclerosis, tubule atrophy and irreversible damage of parenchymal cells lead to progressive deterioration of renal function, and eventually to ESRD. In general, renal fibrosis is thought to be a failed wound healing process [4]. They usually begin at a specific focal point, where interstitial fibroblasts are activated, proliferate, and produce a large amount of ECM components [5,6]. Based on these events, the pathogenesis of renal fibrosis can be artificially divided into four overlapping phases: priming, activation, execution and progression [7,8]. In the dynamic process of renal fibrosis, many of these events happened at the same time, but in the end, excessive accumulation of connective tissue, including interstitial collagen (type Ⅰ, Ⅲ and Ⅳ) fibronectin and some glycoprotein, leading to changes in normal renal structure. Due to the immense complexity of renal fibrosis and the low repeatability of in vivo models, there is a need to develop an in vitro biological platform to provide a reliable test means for clinical transformation of drugs, biomaterials and tissue engineering products [[9], [10], [11]].
Recently, decellularized scaffolds derived from animal tissues have been provide a new platform for detecting cell function and differentiation [[12], [13], [14], [15]], and fabricating tissue engineered organs [16,17]. Traditional cell culture did not mimic the way cells grow in vivo, while decellularized scaffolds provide a contact surface and a room for cell growth and migration as a three-dimension culture system. In previous studies, researchers reported the application of human adipose tissue-derived extracellular matrix scaffolds or decellularized lung scaffolds for the investigation of the growth, migration/invasion, morphology, and drug response of breast cancer cells [18,19]. Even more remarkable is that tissue scaffolds with intact blood vessels and ECM structure have a great therapeutic prospect in the field of organ reconstruction. Previous study had reported that they successfully cultured rat donor kidneys induced by acellular scaffolds and successfully transplanted them into rats with certain physiological functions [20]. However, the current research is mostly limited to healthy kidneys, and there is not much research on kidneys under pathological conditions [9]. In fact, most of the donors obtained from the clinic were older or already have kidney disease. Thus, it is of great concern that scaffolds in pathological conditions can be used not only as models for disease research but also to study whether diseased organs have the potential to be tissue engineered kidney donors. Studies have showed that matrix scaffold prepared from diseased kidney retained diseased ECM characteristics and promoted greater ex vivo fibroblast proliferation than did scaffolds prepared from healthy kidney [21].
Currently, there were many methods for the preparation of kidney failure models [22]. The main methods to induce renal fibrosis are drugs or poisons (mercuric chloride, cyclosporine A, nephrotoxic drugs, aminoglycoside drugs, etc.), surgery (unilateral ureteral ligation, 5-stroke 6 nephrectomy model, renal ischemia-reperfusion, etc.), unilateral nephrectomy combined with AngII and transgenic model, etc. An excessive adenine feeding to rats (Yokozawa Method) was an excellent method to make the model of renal fibrosis because of the similar metabolic abnormalities in human chronic renal failure compared with the rat's [23]. However, there have been no reports of scaffolds using this fibrous kidney model.
The goal of the present study was to engineer decellularized fibrotic kidney scaffolds and analyzed the changes in structure and major protein component of fibrotic scaffold. We hypothesized that the fibrotic scaffolds was a good model to study mechanism of renal fibrosis and a potential source of tissue-engineered donors.
2. Materials and methods
2.1. Preparation of renal fibrosis rat model
All procedures involving animal use, housing, and surgery were approved by the administration of Wenzhou Medical University. Forty male Sprague Dawley (SD) rats initially weighing 200 g were used, with a certification number of SYXK (Zhejiang) 2019-0009. The animals were divided into two groups randomly (n = 20). The first group was prepared for renal fibrosis model as the experimental group, whereas the second one was the normal group. According to the Yokozawa method [23], rats were given 200 mg/(kg · d) adenine (Shanghai Bojing Chemical Co., Ltd., China) suspension (freshly dissolved in distilled 0.9% saline) by gavage continuously for 4 weeks to establish models of renal injury, while the normal group were treated with an equal volume of 0.9% saline at the same time.
2.2. Preparation of decellularized kidney scaffolds
The method of decellularizing the kidneys comes from previous research [24]. Animals were anesthetized with an intraperitoneal injection of 10% chloral hydrate (3.5 ml/kg body weight) (Beijing Solarbio Science & Technology Co., Ltd., China). Venous blood collected with anticoagulation tube from lower cavity veins and the serum obtained by centrifugation (3000 g, 10 min) will then be measured blood urea nitrogen (BUN) and creatinine (Cr).
Decellularization program were as follows: inserting 24 G cannula into the abdominal aorta near the lower pole of kidneys; injecting heparinized in saline to wash the residual blood; finally perfusing heparinized phosphate buffer saline (PBS), 1% TritonX-100 (Sigma, USA), PBS 500 ml each, 0.8% SDS (Sigma, USA) 1000 ml, PBS +1% penicillin 2000 ml in sequence at 37 °C (Deionized water was autoclaved by high-pressure steam). The content of deoxyribonucleic acid (DNA) was detected by spectrophotometry (Nano Drop One).
2.3. Histological examination
Tissue samples were excised from native and fibrotic kidneys and their decellularized scaffolds for morphological analysis. Tissues of a proper size were fixed in 4% paraformaldehyde for 24–48 h and partly dehydrated in 50% ethanol prior to dehydration in a graded series of alcohol concentrations. Samples were then immersed in xylene and embedded in paraffin. Paraffin blocks were cut into 4-μm. Slides were cleared in xylene and rehydrated before staining with either haematoxylin (HE) and eosin, Alcian blue and periodic acid-Schiff (AB-PAS) or with Masson's Trichrome according to the manufacturer's recommended procedure. All images were captured on a Leica upright microscope. Collagen fibers were stained blue with a cord-like structure in Masson trichrome staining and collagen volume fraction (CVF) was calculated using the Image J software (National Institute of Health, NIH). For AB-PAS staining, acidic mucin and proteoglycan were stained blue, the glycogen was stained purple, and the areal density in blue and purple were calculated by Image J software.
2.4. Immunofluorescence analysis
For immunofluorescence, tissue sections were incubated with the following primary antibodies: mouse anti-α-smooth muscle actin (α-SMA; 1:5000; Abcam, UK), rabbit anti-collagen Ⅳ (Col Ⅳ; 1:200; Abcam, UK), mouse anti-laminin (LN; 1:200; Abcam, UK). After being washed in PBS, sections were then incubated in 488- or 594-conjugated species-specific secondary antibody (1:1000; Chemicon, USA) for 2 h at room temperature. Slides were observed with an Olympus fluorescent microscope and images were captured using Olympus image viewer.
2.5. Immunohistochemical analysis
For immunohistochemical staining, tissue sections were incubated with the following antibodies: mouse anti-α-smooth muscle actin (α-SMA; 0.034 μg/ml; Abcam, UK), rabbit anti-collagen Ⅳ (Col Ⅳ; 1:2000; Abcam, UK), mouse anti-laminin (LN; 1:2000; Abcam, UK).The stained sections were scanned on a microscope (Leica Microsystems, Wetzlar, Germany).
Areal density was used for analysis of α-SMA, Col IV and LN. Five images were randomly taken for analysis in one section of each animal. The brown yellow was used as the standard to define the positive staining via the Image J software. The pixel area were measured in each photo.
2.6. Scanning electron microscopy (SEM)
To observe the structure of scaffolds, we firstly put the specimens in 2.5% glutaraldehyde fixation fixed in 4 °C for 24 h. After dehydration through an ethanol gradient (30, 50, 70, 90, and 100% ethanol for 15 min each), the samples were freeze-dried in a vacuum dryer. Subsequently, dry samples were coated under vacuum with platinum alloy at a thickness of 25 nm. Gently with forceps clip sample profile, the samples were then observed under a scanning electron microscope.
2.7. Enzyme-linked immunosorbent assay (ELISA)
Samples were collected for tissue homogenization, and the supernatant was obtained by centrifugation (5000 rpm/min, 10 min) to detect cytokine levels by ELISA. The content of transforming growth factor-β (TGF-β), platelet derived growth factor (PDGF), hepatocyte growth factor (HGF), Vascular endothelial growth factor (VEGF) and fibroblast growth factor (FGF) (n = 5) were measured according to the manufacturer's instructions of commercial ELISA kits (R&D Systems, Minnesota, USA).
2.8. Proteomics analysis
2.8.1. Sample preparation and trypsin digestion
The samples were grinded separately in liquid nitrogen and lysed in 8 M urea, 1% Triton X-100, 65 mM dithiothreitol (DTT), 1% protease inhibitor, 3 μM trichostatin A (TSA), 50 mM Nicotinamide (NAM) and 2 mM ethylenediaminetetraacetic acid (EDTA). The remaining debris was removed by centrifugation at 20,000 g at 4 °C for 10 min. Finally, the protein was precipitated with cold 15% TCA for 2 h at −20 °C. After centrifugation at 4 °C for 10 min, the supernatant was discarded. The remaining precipitate was washed with cold acetone for three times. The protein was redissolved in buffer (8 M urea, 100 mM TEAB, pH 8.0) and the protein concentration was determined with 2-D Quant kit (GE Healthcare, Fairfield, Connecticut, USA) according to the manufacturer's instructions. For digestion, the protein solution was reduced with 10 mM DTT for 1 h at 37 °C and alkylated with 20 mM IAA for 45 min at room temperature in darkness. For trypsin digestion, the protein sample was diluted by adding 100 mM the triethylammonium bicarbonate (TEAB) to urea concentration less than 2 M. Finally, trypsin was added at 1:50 trypsin-to-protein mass ratio for the first digestion overnight and 1:100 trypsin-to-protein mass ratio for a second 4 h-digestion. Approximately 100 μg protein for each sample was digested with trypsin for the following experiments.
2.8.2. iTRAQ/TMT labeling
After trypsin digestion, peptide was desalted by Strata X C18 SPE column (Phenomenex) and vacuum-dried. The peptide was reconstituted in 0.5 M TEAB and processed according to the manufacturer's protocol for the iTRAQ/TMT kit (AB Sciex, USA). Briefly, iTRAQ/TMT reagent (defined as the amount of reagent required to label 100 μg protein) was thawed and reconstituted in 24 μl acetonitrile. Peptides were labeled with iTRAQ/TMT reagent by incubation at room temperature for 2 h and then freeze-dried under vacuum centrifugation.
2.8.3. Fractionation based on liquid chromatography-mass spectrometry
The labeled peptide was then fractionated by high pH reverse-phase high performance liquid chromatography with Agilent 300 Extend C18 column (5 μm particles, 4.6 mm ID, and 250 mm length). Briefly, the peptide gradient was from 2% to 60% acetonitrile in 10 mM ammonium bicarbonate pH 10 over 80 min into 80 fractions. Then, the peptides were combined into 18 fractions and dried by vacuum centrifuging. Finally, Thermo Scientific™ Q Exactive™ Plus mass spectrometer was used for proteomics analysis.
2.9. Cell culture and proliferation assay
Endothelial progenitor cells (EPCs) isolated from rats’ bone marrow were cultured in endothelial cell growth medium-2, and then labeled with CD133 and kinase insert domain receptor (KDR). For better culture together with cells, the samples were cut into 100 μm thick and following attached to 12 mm cell climbing (Hong-da Medical Equipment Co., China). In addition, the control group with only an equal number of cells and no scaffold was set. The scaffolds in experiment group were sterilized before seeding the cells, using the previous protocol [25]. EPCs were seeded into a 24-well plate at a density of 2.0 × 104 cells/cm2.
Cell proliferation was measured by Cell Counting kit-8 (CCK-8) (Beyotime, Shanghai, China) according to the manufacturer's instructions. Cells were incubated in 10% CCK-8 that was diluted in normal culture medium at 37 °C until the visual color conversion occurred. Proliferation rates were determined at 1, 3, 5, 7 days after cells were cultured with DC kidney scaffolds.
2.10. Statistical analysis
Quantitative results were reported as mean ± standard deviation. Independent samples t-test was used to compare the levels of two experimental groups, and One-Way ANOVA was used to show a comparison over two groups. The significance level was set at 0.05 and SPSS 18.0 (SPSS Inc., Chicago, USA) was used for statistical analysis.
3. Results
3.1. Preparation of renal fibrosis model
Rats were administered with adenine according to the Yokozawa method [23]. In fact, when 20 rats were used to prepare renal fibrosis models, eight of them sacrificed, and the 12 alive rats were randomly divided into two groups: the fibrotic kidney group (FK) and the fibrotic scaffold group (FS). The same number of rats were then taken from the normal control group as the normal kidney group (NK) and the normal scaffold group (NS). The rest of rats will be used in other experiments.
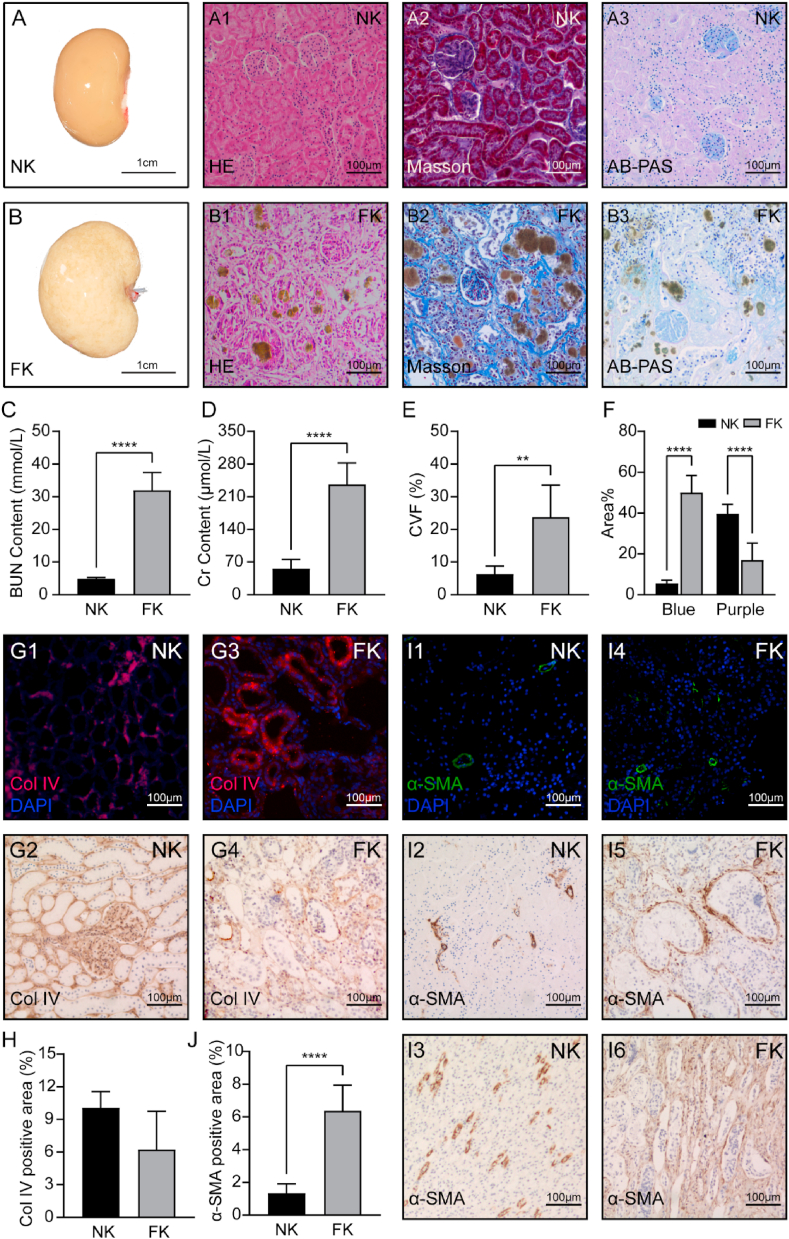
The degree of renal function was determined by the evaluation of blood urea nitrogen (BUN) and creatinine (Cr). BUN levels in FK (31.93 ± 5.46 mmol/L) markedly higher than NK (4.83 ± 0.46 mmol/L) (Fig. 1C), and the Cr contents in FK (236.34 ± 45.9 μmol/L) were also significantly increased (NK, 55.77 ± 19.76 μmol/L) (Fig. 1D). From the general, the fibrotic kidney showed marked hypertrophy and visible spots on the surface (Fig. 1B). HE staining, Masson Trichrome staining and AB-PAS staining showed that the detailed histological alterations were observed in FK (Fig. 1B1-B3). Although the glomeruli were structurally normal, many brown black crystals (2, 8-dihydroxyadenine) of various sizes were deposited in renal tubules which were observed in renal cortex area and the tubular dilation was a result of this. Masson trichrome staining showed that the extracellular matrix increased considerably and the collagen fibers deposited in the renal interstitium (Fig. 1B2). CVF calculated in FK were significantly higher than that in NK (Fig. 1E). AB-PAS staining displayed the approximate content of mucin, proteoglycan and glycogen (Fig. 1B2). The purple area decreased while the blue area increased. This could be a massive loss of glycogen.
Fig. 1.
Preparation of renal fibrosis model, detection of renal function and characteristics. (A) (B) Macroscopic appearance of normal and fibrotic kidneys after blood removal. Representative photographs of haematoxylin and eosin staining (HE), Masson, AB-PAS staining results in NK and FK group. The brownish-yellow granules were crystallized adenine and deposited in renal tubules. (C) (D) BUN and Cr test results. The levels of serum BUN and Cr of the fibrotic group were much higher than the normal group. **, P<0.01; ****, P<0.0001. (E) The content of collagen volume fraction (CVF) in NK and FK. (F) the areal density of AB-PAS staining. (G) (I) the immunofluorescence and immunohistochemical micrograph of collagen Ⅳ and α-SMA. G1-G2: collagen Ⅳ, NK; G3-G4: collagen Ⅳ, FK. I1-I3: α-SMA, NK; I4-I6: α-SMA, FK. Especially, I2 and I5 were in cortex and I3, I6 was in the medulla. (H) (J) the positive area contents of collagen Ⅳ and α-SMA. NK: Normal kidneys; FK: Fibrotic kidneys.
The immunofluorescence and immunohistochemical staining results showed that the expression of collagen Ⅳ, which were the important component of extracellular matrix, markedly no difference between NK and FK (Fig. 1G and H). However, the proliferation and differentiation of fibroblasts into myofibroblasts is the central cellular event in renal fibrosis and α-SMA is a marker protein for myofibroblasts [26]. Generally, the expression of α-SMA was mainly located in the small arteries (Fig. 1, Fig. 2, Fig. 3). Nevertheless, there were significantly positive in renal interstitial in FK, especially in the medulla area (Fig. 1I5, I6). These results demonstrate the successfully establishment of renal fibrosis model by using adenine-fed methods.
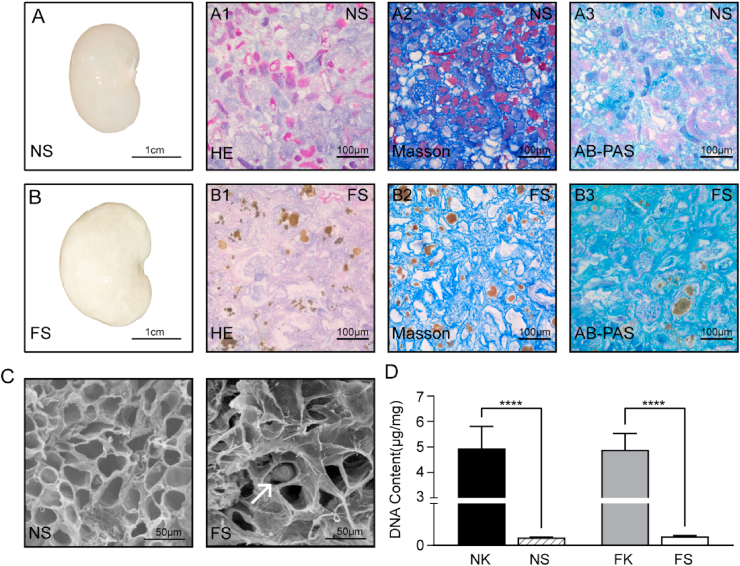
Fig. 2.
Characteristic of normal and fibrotic scaffolds. (A) (B) Macroscopic appearance of normal and fibrotic scaffolds after decellularization. Representative photographs of haematoxylin and eosin staining (HE), Masson, AB-PAS staining results in NS and FS group. The brownish-yellow granules were crystallized adenine and deposited in renal tubules. (C) SEM images showing the three-dimensional microarchitectures of the NS and FS. The white arrow points to crystals. (D) Comparison of DNA content in normal and fibrotic renal tissue before and after decellularization. ****, P<0.0001. NK: Normal kidneys; FK: Fibrotic kidneys; NS: Normal Scaffolds; FS: Fibrotic Scaffolds.
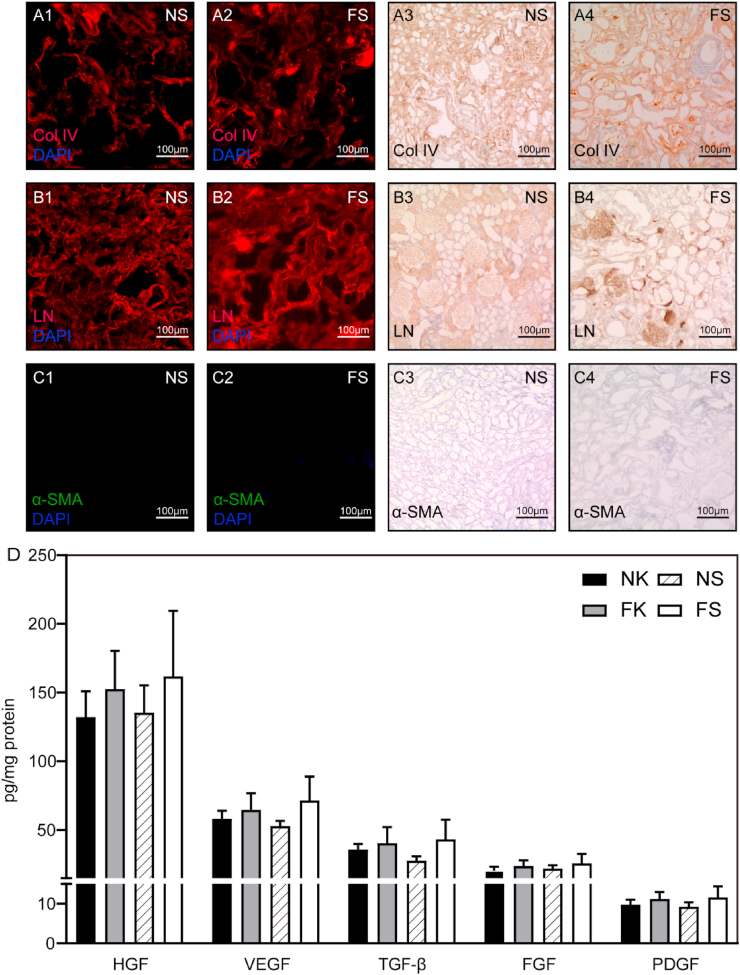
Fig. 3.
(A–C) the immunofluorescence and immunohistochemical micrograph of collagen Ⅳ, LN and α-SMA. (D) Quantitative assay of cytokines in NK, FK, NS and FS. The level of HGF, VEGF, TGF-β, FGF and PDGF in four groups in the legend were determined. NK: Normal kidneys; FK: Fibrotic kidneys; NS: Normal Scaffolds; FS: Fibrotic Scaffolds.
3.2. Characterization of fibrotic scaffolds
Normal and fibrotic kidneys were decellularized to remove cellular components. Post decellularization, normal scaffolds (NS) had a transparent appearance (Fig. 2A), while fibrotic scaffolds (FS) were milk-white (Fig. 2B). HE staining showed that no blue-stained nuclei was observed in either NS or FS (Fig. 2A1, B1). DNA content determination results showed that the DNA removal rate of both FS and NS reached more than 97.72% (Fig. 2D).
The morphology of the glomeruli in fibrotic scaffolds kept well, the dilated renal tubules were noticeable, and the 2, 8-dihydroxyadenine crystals deposited in the renal tubules were not removed post decellularization. Masson trichrome staining and AB-PAS staining of paraffin-embedded tissues showed collagen, mucin, proteoglycan and the glycogen observed was kept but disordered (Fig. 2A and B). By scanning electron microscopy, the extracellular matrix structure of NS was smooth, round and uniform, while the FS was rough and angular with disordered three-dimensional structure (Fig. 2C). All of these can be indicated that our decellularization protocol maintained the structural excellular matrix components intact.
To clarify the several main ECM structural proteins in both NS and FS, immunofluorescence staining and immunohistochemical staining was performed. The collagen Ⅳ (Col Ⅳ) and laminin both locating in the glomerular basement membrane was remained both in NS and FS (Fig. 3A and B). As for α-SMA, there was no positive signal on either scaffold because only the well-structured extracellular matrix was present (Fig. 3C). To assess the difference of biological activity between NS and FS, we performed an ELISA analysis to quantify various growth factors. As shown in Fig. 3D, the cell factors including HGF, TGF-β, FGF, VEGF and PDGF were found to be retained in the decellularized scaffolds. Thus, although the rat kidneys had been completely decellularized, some cell factors were remained within the scaffold which might be sufficient to contribute to renal regeneration after injury. Based on the above results, this decellularization method did not negatively affect the amount of retained protein.
3.3. Differential proteomics of normal scaffolds and fibrotic scaffolds
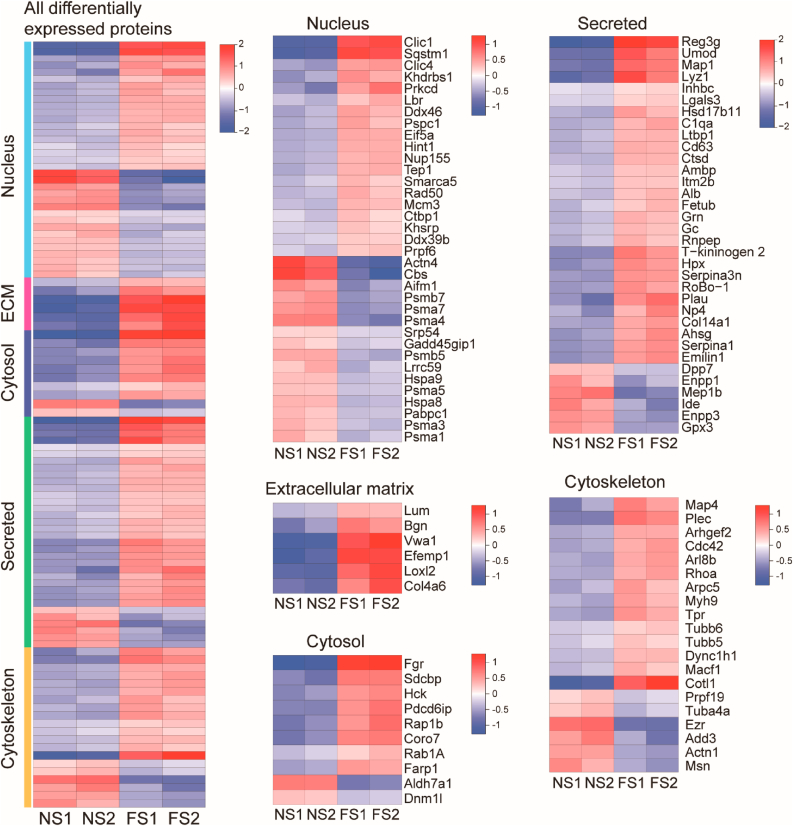
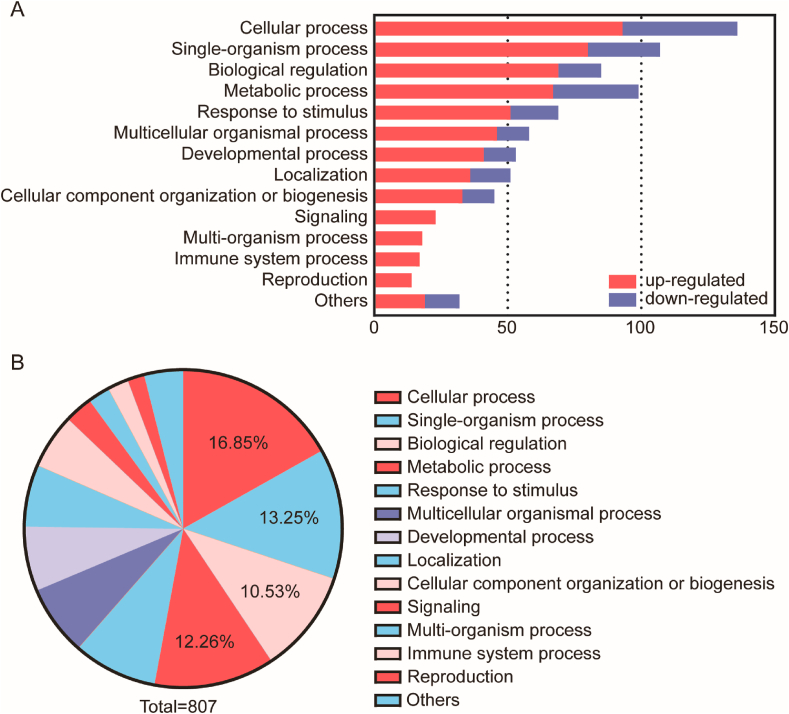
We hypothesized residual protein content as determined by mass spectrometry would differ between normal and fibrotic scaffolds. In iTRAQ/TMT high-throughput proteomics studies, we analyzed extracellular matrix-related proteins, secreted protein-related proteins, cytoskeleton-related proteins, nuclear-related proteins, cytoplasm-related proteins, and cell membrane-related proteins in normal and fibrotic scaffolds. The majority of statistically significant differences appeared between groups in secreted protein-related proteins and cell membrane-related proteins (Table 1). The heat map results of specific protein up or down-regulation were shown in Fig. 4. Compared with normal groups, 81 proteins were up regulated and 31 proteins were down regulated in FS (Table S1). Gene Ontology (GO) distribution was used to analyze the differentially express protein. Half of all differential proteins were related to cellular process, single organism processes, biological regulation and metabolic process (Fig. 5). Most of the down-regulated proteins were associated with the body's energy metabolism among the rest. For example, enzymes related to glycolytic amino acid metabolism were obviously reduced, which also explained the slower weight gain in fibrotic groups compared to normal ones. Differences in these differential proteins may account for the structural and functional differences between normal and fibrotic scaffolds. These up-regulated or down-regulated proteins can be studied to explore the signaling pathways that cause renal fibrosis. Up-regulated protein (integrin) were mainly related to cell adhesion and migration. The results showed that the Cateninβ-1, an important non-biodegradable protein associated with fibrosis process, was significantly rise. Studies have shown that the expression of Cateninβ-1 can lead to spontaneous skin fibrosis in mice, accompanied by fibroblast proliferation [27,28]. In addition, inhibition of the Wnt/catenin pathway could prevents renal and dermal fibrosis [29,30].
Table 1.
Subcellular localization categories statistics of identified proteins and differentially expressed proteins. All proteins identified by mass spectrometer were classified six categories according to UniprotKB Subcellular localization database. If a protein was matched to more than one category, its predominant subcellular location was used for functional grouping.
| Subcellular localization | Number of proteins |
Percent |
Fisher's exact test P value | ||
|---|---|---|---|---|---|
| Identified | Different | Identified | Different | ||
| ECM | 52 | 6 | 1.59% | 1.34% | 7.37E-01 |
| Secreted | 146 | 34 | 4.47% | 7.59% | 9.26E-04 |
| Cytoskeleton | 157 | 20 | 4.80% | 4.46% | 6.77E-01 |
| Nucleus | 350 | 36 | 10.71% | 8.04% | 9.83E-01 |
| Cytosol | 51 | 10 | 1.56% | 2.23% | 1.51E-01 |
| Membrane | 835 | 150 | 25.54% | 33.48% | 3.20E-05 |
Fig. 4.
Heat maps of differentially expressed proteins from six subcellular localization categories (ECM, Secreted, Cytoskeleton, Nucleus, and Cytosol). NS1, NS2 was the normal scaffold and FS1, FS2 was the fibrotic scaffold. The combined heat map and specific heat maps for each of the designated protein categories were shown in the figures and the color scheme represents the log2 order transformation value, which depicts the relative abundance of each protein compared to the average strength.
Fig. 5.
(A) Distribution of down-regulated and up-regulated proteins in GO secondary annotation in the comparison of NS vs. FS. The horizontal axis represented the number of enriched proteins, and the left y-axis represented different biological process. The red bars were up-regulated proteins and the blue bars were down-regulated proteins. (B) The relative abundance of matrix proteins was shown in a pie chart after the proteins were classified. GO: Gene ontology.
3.4. Cytocompatibility of fibrotic scaffolds
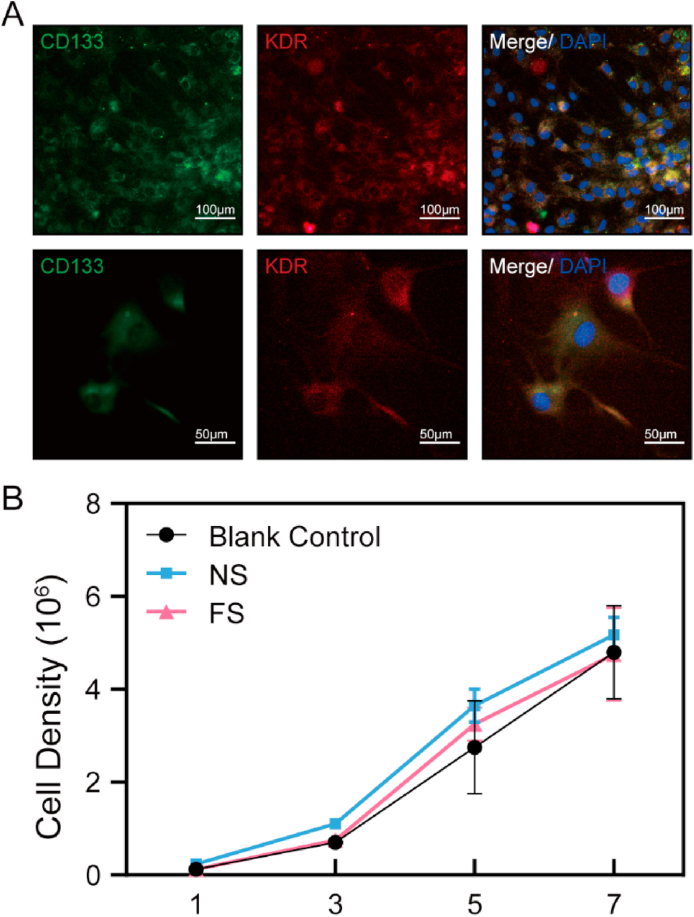
Scaffolds were cut into 100 μm-thick sections, and EPCs were seeded onto the scaffolds-coated cover slips and cultured to assess the cytocompatibility of FS. Double labeling of CD133 and kinase insert domain receptor (KDR) identified the cells after extraction, and the double positives indicated that the cells were indeed EPCs (Fig. 6A). CCK-8 was used to detect the proliferation and apoptosis of EPCs. The results showed that the cells in blank control group, NS and FS have no cytotoxicity to affect cell proliferation (Fig. 6B).
Fig. 6.
Identification and proliferation of EPCs. (A) EPCs labeled by CD133 and KDR. The cells like triangles were EPCs and almost all cells in the field of view showed double positive, which were CD133 (green) and KDR (red). Scale bar = 100 μm (top), 50 μm (bottom) (B) The result of CCK-8 cell proliferation. The cells in the normal group, the fibrotic group and the control group had proliferated. NS: Normal Scaffolds; FS: Fibrotic Scaffolds.
4. Discussion
Our previous studies had reported decellularized renal scaffolds could mediate renal regeneration for repairing of partially resected kidneys [31,32], since ECM is not only a good biological scaffold, but also an important signaling molecule that regulates cell growth, metabolism and differentiation. At present, the preparation of renal scaffolds under pathological conditions was deficient. During the development process of renal fibrosis, the typical features of renal fibrosis is the excessive deposition of ECM proteins, and the rate of ECM production exceeds its degradation rate [33]. Both of the preparation of two groups used the same protocol [24], which removed renal cells, retained the normal vascular tree and continuous extracellular matrix and preserved the three dimensional structure of the cells.
Due to the complexity of renal fibrosis, the renal fibrosis model was prepared by unilateral ischemia-reperfusion injury [34] or ureteral obstruction [21]. Adenine induced renal failure can be progressively aggravated with the prolongation of feeding time. The fibrotic kidney induced by an adenine rich diet was observed the destruction and obstruction of renal tubules, and eventually the loss of a large number of units. Four weeks of modeling is a suitable time to achieve a certain degree of fibrosis and the livability of animals was guaranteed. Besides, although some cytokines may be lost during fluid perfusion, the cytokines affecting cell proliferation and differentiation were reserved in our scaffold. For instance, the substantial retention of TGF-β in fibrotic scaffolds can not only increase cell adhesion to ECM but also regulate a variety of cytokines, which is an important regulator of renal fibrosis. Previous study indicated that VEGF receptor exists in renal tubular epithelial cells, and proved that VEGF can promote the proliferation of renal tubular epithelial cells and improve their anti-apoptosis ability, which is an important cytokine for the survival of renal tubular epithelial cells [35]. Moreover, both VEGF and FGF can promote the formation of new blood vessels. HGF has a wide range of biological functions including kidney development, acute injury and regeneration. The retention of these important cytokines provided a more realistic microenvironment for cell growth, whereas the artificial model in vitro did not. The cell experiments of EPCs also confirmed this. The reason why we used EPCs to judge the biocompatibility of fibrotic scaffolds because EPCs can form new blood vessels in the ischemic injury area of body under certain conditions, promote blood flow recovery and repair wound. In this study, we used the natural spatial structure of extracellular matrix for cell growth and assessed the cytotoxicity of residual crystals in scaffolds.
Proteomics was used to better understand the changes in extracellular matrix protein caused by renal fibrosis and screen out proteins with different expression levels, for laying a foundation for further study on the pathogenesis of renal fibrosis. After KEGG pathway enrichment of the up or down-regulated proteins, the glycolysis/gluconeogenesis, glycine, serine and threonine metabolism, sulfur metabolism and biosynthesis of amino acids pathways were mainly affected. For example, the protein domain enrichment analysis of up-regulated proteins (FS/NS) showed that the catalytic domain of tyrosine-protein kinase was the most obvious differences. In addition, the expression of Catenin-β1 and integrin associated with Wnt/Catenin and TGF-β1 pathways were up-regulated. Several studies have explored the key role of the Wnt/catenin signaling pathway in the fibrosis process [36] and TGF-β1 could regulates the expression of integrin by increasing the adhesion of cells to the matrix. Some scholars have showed that the increased concentration of TGF-β1 in hepatic fibrosis patients was correlated with the severity of liver fibrosis [37]. The further analysis of signal pathways and the verification of landmark proteins need a lot of work in future research.
The high proliferation potential of EPC cells is a promising source of seed cells for tissue engineering blood vessels. An important indicator of tissue regeneration is the regeneration of blood vessels. If EPC cells can adhere and proliferate well on scaffolds, it will provide help for future research on tissue engineering kidneys, and may expand the application scope of fibrotic renal scaffolds. Moreover, there were no significant changes in cell proliferation. we hypothesized that the factors affecting cell proliferation were not only cytokines but also spatial structure. The spatial structure of the fibrotic kidney has changed significantly compared with that of the normal kidney. In addition, the hardness of the tissue has an effect on the growth of cells. The factors affecting cell growth are very complex, so it is necessary to use the biological origin of the original spatial structure and the main cytokine model, to study the process of the occurrence and development of fibrosis. However, our research was still in its infancy, whether more stable, comprehensive and positive results can be obtained remains to be further studied.
5. Conclusion
In summary, we prepared and detected the decellularized adenine-fed renal fibrosis model, and analyzed the difference between normal and fibrotic scaffolds through proteomics and cultivation with cells. The fibrotic scaffolds was a good model to study mechanism of renal fibrosis and a potential source of tissue-engineered donors.
Funding sources
National Natural Science Foundation of China (grant No. 81970653); Zhejiang Medical and Health Technology Project (grant No. 2018248656); Zhejiang pharmacy Top Key Project (grant No. YKFJ2-008).
CRediT authorship contribution statement
Rui Zhang: Writing - original draft, Investigation, Methodology, Formal analysis, Visualization. Junqun Jiang: Writing - original draft, Methodology, Investigation, Conceptualization. Yaling Yu: Writing - original draft, Investigation, Validation. Fangfang Wang: Investigation, Formal analysis. Niuniu Gao: Date curation. Yingjie Zhou: Date curation, Formal analysis. Xinlong Wan: Date curation. Zhibin Wang: Resources. Peng Wei: Supervision, Writing - review & editing, Project administration. Jin Mei: Conceptualization, Writing - review & editing, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare no competing financial interest.
Acknowledgment
The authors would like to acknowledge the Institute of Bioscaffold Transplantation and Immunology whose research was funded by the National Natural Science Foundation of China (grant NO. 81970653). And the Medical Research Center of Ningbo City First Hospital whose research was funded by Zhejiang Medical and Health Technology Project (grant No. 2018248656) and Zhejiang pharmacy Top Key Project (grant No. YKFJ2-008).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2020.12.028.
Contributor Information
Peng Wei, Email: weipeng@nbu.edu.cn.
Jin Mei, Email: meijin@wmu.edu.cn.
Abbreviations
- iTRAQ
isobaric tag for relative and absolute quantitation
- TMT
tandem mass tag
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bikbov B., Purcell C.A., Levey A.S., Smith M., Abdoli A., Abebe M., Adebayo O.M., Afarideh M., Agarwal S.K., Agudelo-Botero M., Ahmadian E., Al-Aly Z., Alipour V., Almasi-Hashiani A., Al-Raddadi R.M., Alvis-Guzman N., Amini S., Andrei T., Andrei C.L., Andualem Z., Anjomshoa M., Arabloo J., Ashagre A.F., Asmelash D., Ataro Z., Atout M.M.'d.W., Ayanore M.A., Badawi A., Bakhtiari A., Ballew S.H., Balouchi A., Banach M., Barquera S., Basu S., Bayih M.T., Bedi N., Bello A.K., Bensenor I.M., Bijani A., Boloor A., Borzì A.M., Cámera L.A., Carrero J.J., Carvalho F., Castro F., Catalá-López F., Chang A.R., Chin K.L., Chung S.-C., Cirillo M., Cousin E., Dandona L., Dandona R., Daryani A., Das Gupta R., Demeke F.M., Demoz G.T., Desta D.M., Do H.P., Duncan B.B., Eftekhari A., Esteghamati A., Fatima S.S., Fernandes J.C., Fernandes E., Fischer F., Freitas M., Gad M.M., Gebremeskel G.G., Gebresillassie B.M., Geta B., Ghafourifard M., Ghajar A., Ghith N., Gill P.S., Ginawi I.A., Gupta R., Hafezi-Nejad N., Haj-Mirzaian A., Haj-Mirzaian A., Hariyani N., Hasan M., Hasankhani M., Hasanzadeh A., Hassen H.Y., Hay S.I., Heidari B., Herteliu C., Hoang C.L., Hosseini M., Hostiuc M., Irvani S.S.N., Islam S.M.S., Jafari Balalami N., James S.L., Jassal S.K., Jha V., Jonas J.B., Joukar F., Jozwiak J.J., Kabir A., Kahsay A., Kasaeian A., Kassa T.D., Kassaye H.G., Khader Y.S., Khalilov R., Khan E.A., Khan M.S., Khang Y.-H., Kisa A., Kovesdy C.P., Kuate Defo B., Kumar G.A., Larsson A.O., Lim L.-L., Lopez A.D., Lotufo P.A., Majeed A., Malekzadeh R., März W., Masaka A., Meheretu H.A.A., Miazgowski T., Mirica A., Mirrakhimov E.M., Mithra P., Moazen B., Mohammad D.K., Mohammadpourhodki R., Mohammed S., Mokdad A.H., Morales L., Moreno Velasquez I., Mousavi S.M., Mukhopadhyay S., Nachega J.B., Nadkarni G.N., Nansseu J.R., Natarajan G., Nazari J., Neal B., Negoi R.I., Nguyen C.T., Nikbakhsh R., Noubiap J.J., Nowak C., Olagunju A.T., Ortiz A., Owolabi M.O., Palladino R., Pathak M., Poustchi H., Prakash S., Prasad N., Rafiei A., Raju S.B., Ramezanzadeh K., Rawaf S., Rawaf D.L., Rawal L., Reiner R.C., Rezapour A., Ribeiro D.C., Roever L., Rothenbacher D., Rwegerera G.M., Saadatagah S., Safari S., Sahle B.W., Salem H., Sanabria J., Santos I.S., Sarveazad A., Sawhney M., Schaeffner E., Schmidt M.I., Schutte A.E., Sepanlou S.G., Shaikh M.A., Sharafi Z., Sharif M., Sharifi A., Silva D.A.S., Singh J.A., Singh N.P., Sisay M.M.M., Soheili A., Sutradhar I., Teklehaimanot B.F., Tesfay B.e., Teshome G.F., Thakur J.S., Tonelli M., Tran K.B., Tran B.X., Tran Ngoc C., Ullah I., Valdez P.R., Varughese S., Vos T., Vu L.G., Waheed Y., Werdecker A., Wolde H.F., Wondmieneh A.B., Wulf Hanson S., Yamada T., Yeshaw Y., Yonemoto N., Yusefzadeh H., Zaidi Z., Zaki L., Zaman S.B., Zamora N., Zarghi A., Zewdie K.A., Ärnlöv J., Coresh J., Perico N., Remuzzi G., Murray C.J.L. Global, regional, and national burden of chronic kidney disease, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2020;395:709–733. doi: 10.1016/S0140-6736(20)30045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L., Wang F., Wang L., Wang W., Liu B., Liu J., Chen M., He Q., Liao Y., Yu X., Chen N., Zhang J.-e., Hu Z., Liu F., Hong D., Ma L., Liu H., Zhou X., Chen J., Pan L., Chen W., Wang W., Li X., Wang H. Prevalence of chronic kidney disease in China: a cross-sectional survey. Lancet. 2012;379:815–822. doi: 10.1016/S0140-6736(12)60033-6. [DOI] [PubMed] [Google Scholar]
- 3.Meng X.-M., Nikolic-Paterson D.J., Lan H.Y. TGF-β: the master regulator of fibrosis. Nat. Rev. Nephrol. 2016;12:325–338. doi: 10.1038/nrneph.2016.48. [DOI] [PubMed] [Google Scholar]
- 4.Wynn T.A. Cellular and molecular mechanisms of fibrosis. J. Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farris A.B., Alpers C.E. What is the best way to measure renal fibrosis?: a pathologist's perspective. Kidney Int. Suppl. 2011;4:9–15. doi: 10.1038/kisup.2014.3. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farris A.B., Adams C.D., Brousaides N., Della Pelle P.A., Collins A.B., Moradi E., Smith R.N., Grimm P.C., Colvin R.B. Morphometric and visual evaluation of fibrosis in renal biopsies. J. Am. Soc. Nephrol. 2011;22:176–186. doi: 10.1681/ASN.2009091005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y. Cellular and molecular mechanisms of renal fibrosis. Nat. Rev. Nephrol. 2011;7:684–696. doi: 10.1038/nrneph.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y. Renal fibrosis: new insights into the pathogenesis and therapeutics. Kidney Int. 2006;69:213–217. doi: 10.1038/sj.ki.5000054. [DOI] [PubMed] [Google Scholar]
- 9.McCrary M.W., Bousalis D., Mobini S., Song Y.H., Schmidt C.E. Decellularized tissues as platforms for in vitro modeling of healthy and diseased tissues. Acta Biomater. 2020;111:1–19. doi: 10.1016/j.actbio.2020.05.031. [DOI] [PubMed] [Google Scholar]
- 10.Boor P., Ostendorf T., Floege J. Renal fibrosis: novel insights into mechanisms and therapeutic targets. Nat. Rev. Nephrol. 2010;6:643–656. doi: 10.1038/nrneph.2010.120. [DOI] [PubMed] [Google Scholar]
- 11.Zeisberg M., Neilson E.G. Mechanisms of tubulointerstitial fibrosis. J. Am. Soc. Nephrol. 2010;21:1819–1834. doi: 10.1681/ASN.2010080793. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y., He Y., Bharadwaj S., Hammam N., Carnagey K., Myers R., Atala A., van Dyke M. Tissue-specific extracellular matrix coatings for the promotion of cell proliferation and maintenance of cell phenotype. Biomaterials. 2009;30:4021–4028. doi: 10.1016/j.biomaterials.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hussein K.H., Park K.M., Ghim J.H., Yang S.R., Woo H.M. Three dimensional culture of HepG2 liver cells on a rat decellularized liver matrix for pharmacological studies. J. Biomed. Mater. Res. B Appl. Biomater. 2016;104:263–273. doi: 10.1002/jbm.b.33384. [DOI] [PubMed] [Google Scholar]
- 14.Park K.M., Hussein K.H., Hong S.H., Ahn C., Yang S.R., Park S.M., Kweon O.K., Kim B.M., Woo H.M. Decellularized liver extracellular matrix as promising tools for transplantable bioengineered liver promotes hepatic lineage commitments of induced pluripotent stem cells. Tissue Eng. 2016;22:449–460. doi: 10.1089/ten.TEA.2015.0313. [DOI] [PubMed] [Google Scholar]
- 15.Hoshiba T., Chen G., Endo C., Maruyama H., Wakui M., Nemoto E., Kawazoe N., Tanaka M. Decellularized extracellular matrix as an in vitro model to study the comprehensive roles of the ECM in stem cell differentiation. Stem Cell. Int. 2016;2016 doi: 10.1155/2016/6397820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert T.W., Sellaro T.L., Badylak S.F. Decellularization of tissues and organs. Biomaterials. 2006;27:3675–3683. doi: 10.1016/j.biomaterials.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 17.Arenas-Herrera J.E., Ko I.K., Atala A., Yoo J.J. Decellularization for whole organ bioengineering. Biomed. Mater. 2013;8 doi: 10.1088/1748-6041/8/1/014106. [DOI] [PubMed] [Google Scholar]
- 18.Xiong G., Flynn T.J., Chen J., Trinkle C., Xu R. Development of an ex vivo breast cancer lung colonization model utilizing a decellularized lung matrix. Integr. Biol. (Camb) 2015;7:1518–1525. doi: 10.1039/c5ib00157a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunne L.W., Huang Z., Meng W., Fan X., Zhang N., Zhang Q., An Z. Human decellularized adipose tissue scaffold as a model for breast cancer cell growth and drug treatments. Biomaterials. 2014;35:4940–4949. doi: 10.1016/j.biomaterials.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Song J.J., Guyette J.P., Gilpin S.E., Gonzalez G., Vacanti J.P., Ott H.C. Regeneration and experimental orthotopic transplantation of a bioengineered kidney. Nat. Med. 2013;19:646–651. doi: 10.1038/nm.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu H., Tian Y., Zhou L., Zhou D., Tan R.J., Stolz D.B., Liu Y. Tenascin-C is a major component of the fibrogenic niche in kidney fibrosis. J. Am. Soc. Nephrol. 2017;28:785–801. doi: 10.1681/ASN.2016020165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nogueira A., Pires M.J., Oliveira P.A. Pathophysiological mechanisms of renal fibrosis: a review of animal models and therapeutic strategies. In Vivo. 2017;31:1–22. doi: 10.21873/invivo.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yokozawa T., Zheng P.D., Oura H., Koizumi F. Animal model of adenine-induced chronic renal failure in rats. Nephron. 1986;44:230–234. doi: 10.1159/000183992. [DOI] [PubMed] [Google Scholar]
- 24.Sullivan D.C., Mirmalek-Sani S.-H., Deegan D.B., Baptista P.M., Aboushwareb T., Atala A., Yoo J.J. Decellularization methods of porcine kidneys for whole organ engineering using a high-throughput system. Biomaterials. 2012;33:7756–7764. doi: 10.1016/j.biomaterials.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 25.Wang X., Yu Y., Li M., Alkhawaji A., Chen C., Liu X., Jiang J., Zhang J., Wang Z., Li T., Zhang W., Mei J. EPCs enhance angiogenesis in renal regeneration. Oncotarget. 2016;7:44941–44949. doi: 10.18632/oncotarget.10377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LeBleu V.S., Taduri G., O'Connell J., Teng Y., Cooke V.G., Woda C., Sugimoto H., Kalluri R. Origin and function of myofibroblasts in kidney fibrosis. Nat. Med. 2013;19:1047–1053. doi: 10.1038/nm.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akhmetshina A., Palumbo K., Dees C., Bergmann C., Venalis P., Zerr P., Horn A., Kireva T., Beyer C., Zwerina J., Schneider H., Sadowski A., Riener M.-O., MacDougald O.A., Distler O., Schett G., Distler J.H.W. Activation of canonical Wnt signalling is required for TGF-β-mediated fibrosis. Nat. Commun. 2012;3:735. doi: 10.1038/ncomms1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamburg E.J., Atit R.P. Sustained β-catenin activity in dermal fibroblasts is sufficient for skin fibrosis. J. Invest. Dermatol. 2012;132:2469–2472. doi: 10.1038/jid.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beyer C., Reichert H., Akan H., Mallano T., Schramm A., Dees C., Palumbo-Zerr K., Lin N.Y., Distler A., Gelse K., Varga J., Distler O., Schett G., Distler J.H.W. Blockade of canonical Wnt signalling ameliorates experimental dermal fibrosis. Ann. Rheum. Dis. 2013;72:1255–1258. doi: 10.1136/annrheumdis-2012-202544. [DOI] [PubMed] [Google Scholar]
- 30.Surendran K., Schiavi S., Hruska K.A. Wnt-dependent beta-catenin signaling is activated after unilateral ureteral obstruction, and recombinant secreted frizzled-related protein 4 alters the progression of renal fibrosis. J. Am. Soc. Nephrol. 2005;16:2373–2384. doi: 10.1681/ASN.2004110949. [DOI] [PubMed] [Google Scholar]
- 31.Yu Y., Cui H., Chen C., Wen G., Xu J., Zheng B., Zhang J., Wang C., Chai Y., Mei J. Hypoxia-inducible Factor-1α directs renal regeneration induced by decellularized scaffolds. Biomaterials. 2018;165:48–55. doi: 10.1016/j.biomaterials.2018.02.045. [DOI] [PubMed] [Google Scholar]
- 32.Yu Y.L., Shao Y.K., Ding Y.Q., Lin K.Z., Chen B., Zhang H.Z., Zhao L.N., Wang Z.B., Zhang J.S., Tang M.L., Mei J. Decellularized kidney scaffold-mediated renal regeneration. Biomaterials. 2014;35:6822–6828. doi: 10.1016/j.biomaterials.2014.04.074. [DOI] [PubMed] [Google Scholar]
- 33.Eddy A.A. Molecular basis of renal fibrosis. Pediatr. Nephrol. 2000;15:290–301. doi: 10.1007/s004670000461. [DOI] [PubMed] [Google Scholar]
- 34.Zhu H., Liao J., Zhou X., Hong X., Song D., Hou F.F., Liu Y., Fu H. Tenascin-C promotes acute kidney injury to chronic kidney disease progression by impairing tubular integrity via αvβ6 integrin signaling. Kidney Int. 2020;97:1017–1031. doi: 10.1016/j.kint.2020.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.John Kanellis, Scott Fraser, Marina Katerelos, and David A. Power, Vascular Endothelial Growth Factor Is a Survival Factor for Renal Tubular Epithelial Cells.. [DOI] [PubMed]
- 36.Wei J., Fang F., Lam A.P., Sargent J.L., Hamburg E., Hinchcliff M.E., Gottardi C.J., Atit R., Whitfield M.L., Varga J. Wnt/β-catenin signaling is hyperactivated in systemic sclerosis and induces Smad-dependent fibrotic responses in mesenchymal cells. Arthritis Rheum. 2012;64:2734–2745. doi: 10.1002/art.34424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee U.E., Friedman S.L. Mechanisms of hepatic fibrogenesis. Best Pract. Res. Clin. Gastroenterol. 2011;25:195–206. doi: 10.1016/j.bpg.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.