Abstract
Remarkable progress has been made in elucidating important roles of plant non-coding RNAs. Among these RNAs, long noncoding RNAs (lncRNAs) have gained widespread attention, especially their role in plant environmental stress responses. LncRNAs act at different levels of gene expression regulation, and one of these mechanisms is by recruitment of DNA methyltransferases or demethylases to regulate the target gene transcription. In this mini-review, we highlight the function of lncRNAs, including their potential role in RNA-directed DNA Methylation (RdDM) silencing pathway and their potential function under abiotic stresses conditions. Moreover, we also present and discuss studies of lncRNAs in crops. Finally, we propose a path outlook for future research that may be important for plant breeding.
Keywords: epigenetic, non-coding RNAs, gene regulation, environmental stresses, plant breeding
Introduction
In 1970, the central dogma of molecular biology was proposed, suggesting that the flow of information would follow the DNA to RNA to Protein (Crick, 1970). With the sequencing of the human genome, it was found that only about 3% of the genomic DNA encoded proteins and the rest was composed of the so-called “junk” DNA, including transposable elements (TEs) and highly repetitive DNA (Nowak, 1994). They also show that despite of not encoding proteins, the vast majority of human genome is transcribed into RNA. This also occurs in plant genomes. For instance, in Arabidopsis thaliana, the minority of its genome has the capacity of encoding proteins (Yamada et al., 2003). Nowadays, what initially was considered trash DNA became the luxury, as researchers are unraveling important roles out of the genomic non-coding sequences.
Non-coding RNAs (ncRNAs) include a huge variety of RNAs. The regulatory ncRNAs contain small RNAs (sRNAs) and long non-coding RNAs (lncRNAs) that do not encode proteins, but can generate small peptides (BenAmor et al., 2009). The best characterized are the sRNAs: microRNAs (miRNAs) and small interference RNAs (siRNAs). Several studies have highlighted the important role of sRNAs in transcriptional and post-transcriptional regulation of gene expression in plants. Although lncRNAs were previously considered to be “transcriptional noise,” emerging plant studies have Also, revealed the crucial involvement of lncRNAs in various biological processes including flowering (Fan et al., 2016), development (Zhang and Chen, 2013) and stresses responses (Sun et al., 2020).
LncRNAs are classified as ncRNAs longer than 200 nt (Ma et al., 2013). The first lncRNAs with regulatory function identified in plants was the enod40 (early nodulin 40) in Medicago, a “riboregulator” involved in plant growth (Crespi et al., 1994). With the advance of computational methods, 503 mRNA (messenger RNA)-like transcripts that appear to not encode proteins were identified in Medicago (Wen et al., 2007). Then, an increasing number of lncRNAs have been found by computational approach in different plant species (Vieira et al., 2017; Danilevicz et al., 2018). CANTATAdb1 is one database created to deposit these sequences, actually it collects 239,631 lncRNAs predicted in 39 species (Szcześniak et al., 2015). Although, the sequences of most lncRNAs are much less conserved than those of mRNAs, analysis of primary sequence conservation using 10 plant species revealed that the majority of lncRNAs had high sequence conservation at the intra-species and sub-species levels, in contrast to the highly diverged inter-species level (Deng et al., 2018). Moreover, lncRNAs are less expressed than mRNAs, which requires high sensitivity techniques such as RNA fluorescence in situ hybridization (RNA FISH), and real-time quantitative polymerase chain reaction (qRT-PCR) for the analysis of expression (Wu et al., 2020). Another feature of lncRNAs is their genomic localization, that can be located in intergenic, intronic, or coding regions, both at the sense and antisense directions (Wu et al., 2020). Interestingly, lncRNAs are regulated in response to various stimulus. Analysis of 76 lncRNAs in Arabidopsis revealed that 22 lncRNAs showed altered expression under abiotic stress (BenAmor et al., 2009). For instance, npc60 showed to be 100 times more expressed under salt stress. In cotton, lncRNA973 was increased by salt treatments and analysis by in situ hybridization showed that it was localized mainly to the nucleus (Zhang et al., 2019). Some studies use the subcellular localization of lncRNAs to infer their functions, since it can act both in the nucleus and cytoplasm (Karlik et al., 2019).
Although plant lncRNAs have a potential role in regulating plant responses to environmental conditions, their mechanism of function in gene regulation is poorly understood. Here, we highlight some studies that have been analyzing the importance of lncRNAs in plants. First, we included the potential roles of lncRNAs on RNA-directed DNA methylation (RdDM) silencing pathway, since many genes are methylated in response to abiotic stress. Despite showing studies on model plants, we also discuss studies of lncRNAs carried on crops, with the potential used as tools for biotechnological improvement of plants.
LncRNAs as Precursors in RdDM Silencing Pathway
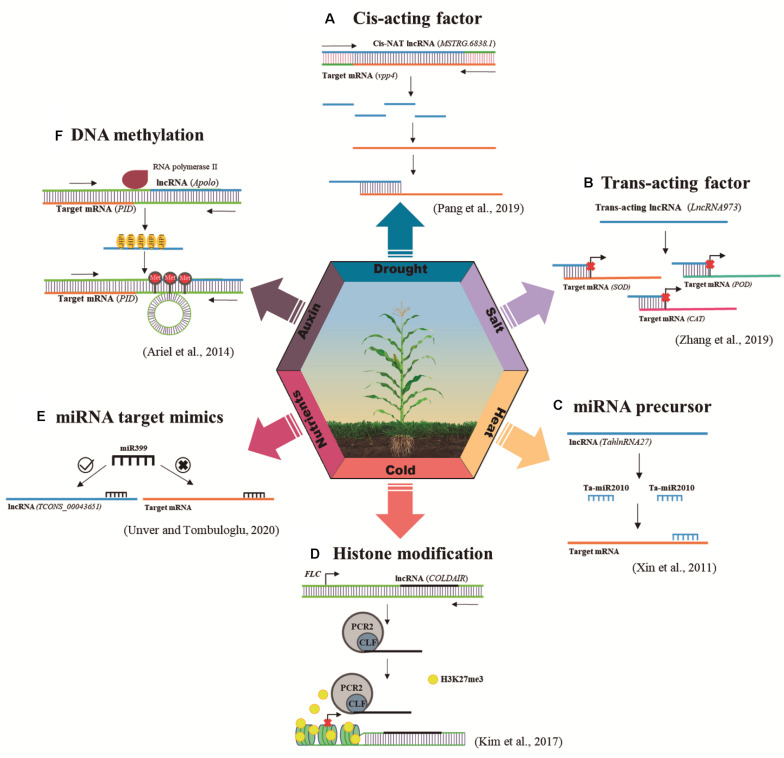
LncRNAs can act as key genetic and epigenetic regulators of gene expression (Karlik et al., 2019). They may function as cis-acting elements by working near the site of RNA synthesis, acting directly on consecutive genes on the same strand (Zhao et al., 2020; Figure 1A); or as trans-acting factors by operating far from the site of synthesis (Suksamran et al., 2020; Figure 1B). LncRNAs may interfere with the binding of transcription factors to promoter regions (Csorba et al., 2014). Moreover, they can also function as miRNAs and trans-acting small interfering RNA (tasiRNA) precursors (Zhang et al., 2014; Fukuda et al., 2019; Figure 1C), miRNA target mimics (Shuai et al., 2014; Figure 1E) and can be processed in siRNA (Wunderlich et al., 2014). Curiously, similar to what occurs in mRNA biogenesis, the RNA polymerase II (Pol II) transcribes the majority of lncRNAs. Other RNA polymerases, such as Pol IV and Pol V that are exclusive to plants, can also act in the lncRNA generation, participating mainly in the epigenetic regulation mediated by RdDM (Wierzbick et al., 2008; Li et al., 2014). Furthermore, epigenetic mechanisms including DNA methylation (Ariel et al., 2014; Figure 1F) and histone modification (Heo and Sung, 2011; Figure 1D) are usually reported to be regulated by lncRNAs.
FIGURE 1.
Regulatory mechanisms of plant lncRNAs in response to abiotic stresses. The main mechanisms of action triggered by lncRNAs responsive to abiotic stresses are miRNA precursor, histone modification, target mimicry, RdDM, cis-acting factor and trans-acting factor. This figure illustrates one example of each of these mechanisms. (A) Cis-acting factor: vpp4 encoding a vacuolar (H+)-pumping ATPase subunit was identified as a putative target of an adjacent lncRNA MSTRG.6838.1. The expressions of vpp4 and MSTRG.6838.1 were significantly correlated in many tissues and development stages, being both repressed under drought stress, which indicates that MSTRG.6838.1 and vpp4 could be a promising cis-acting pair (Pang et al., 2019). (B) Trans-acting factor: LncRNA973 corresponds to a trans-acting lncRNA responsive to salt stress, which regulates plant stress responses by modulating the expression of a series of key salt-related genes, as superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) (Zhang et al., 2019). (C) miRNA precursor: TahlnRNA27, a heat-induced lncRNA, can act as a miRNA precursor since it presents Ta-miR2010 family sequences. After 1 h of heat-treatment, TahlnRNA27 expression was induced as well as Ta-miR2010 expression. The secondary structure and the corresponding expression pattern indicate that TahlnRNA27 might be the precursor of Ta-miR2010 (Xin et al., 2011). (D) Histone modification: The repression of FLC by vernalization is accompanied by a series of changes in histone modifications at FLC chromatin, including the deposition of repressive histone markers, such as Histone H3 Lys 27 (H3K27me3). COLDAIR is up-regulated in response to cold, physically interacting with a component of PRC2, CURLY LEAF (CLF), for the increased enrichment of PRC2 at FLC chromatin to promote H3K27me3 accumulation at FLC (Kim et al., 2017). (E) miRNA target mimics: TCONS_00043651 function as a potential natural miRNA sponge of miR399 sequence in response to boron-stress. Results obtained from barley roots analysis showed that TCONS_00043651 was up-regulated (three times than that of control) upon boron-exposure, meanwhile miR399 expression was repressed (three times down-regulated) in the same stress conditions (Unver and Tombuloglu, 2020). (F) DNA methylation: APOLO can trigger RdDM in response to an auxin stimulus. In response to auxin, Pol II APOLO transcripts gradually recruit LHP1 to mediate loop formation, whereas Pol IV/V transcription triggers DNA methylation. Then, Pol II APOLO-LHP1 mediated loop is conformed and maintained by Pol IV/V-dependent DNA methylation to repress PID expression (Ariel et al., 2014).
Plant lncRNAs play a key role in the RdDM silencing pathway. This regulatory route is based on the performance of Pol IV-dependent RNAs (P4RNAs) transcribed by Pol IV (Blevins et al., 2015; Zhai et al., 2015; Yang et al., 2016). These precursor RNAs are processed by RNA-dependent RNA Polymerase 2 (RDR2) to form double-stranded RNAs (dsRNAs), which are primarily cleaved by Dicer-like 3 (DCL3) to produce 24-nt siRNAs (Xie et al., 2004). These siRNAs are associated with Argonaute 4 (AGO4), forming AGO-siRNA complex (Holoch and Moazed, 2015). Simultaneously, lncRNAs transcribed by Pol V work as scaffold RNAs being recognized by the siRNA-AGO complex through sequence complementarity (Böhmdorfer et al., 2016). Once AGO4–siRNA–lncRNA complex is formed, it is driven to the chromatin target site together with a DNA methylation enzyme, the DNA methyltransferase domains rearranged methyltransferase 2 (DRM2) (Gao et al., 2010). This methyltransferase mediates de novo methylation of cytosines in all classes of sequence contexts at the target region to initiate gene silencing (Wierzbick et al., 2008). Therefore, RdDM correspond to a plant-specific de novo DNA methylation mechanism that requires lncRNAs as scaffold to define target genomic loci (Wierzbicki et al., 2009).
The understanding of lncRNAs role as precursors in epigenetic silencing via RdDM have received remarkable contributions (Chen et al., 2018, 2019). Several reports have suggested that plant lncRNAs are involved with DNA methylation performing different developmental functions such as in the regulation of embryogenesis (Chen et al., 2018), root organogenesis (Chen et al., 2019), reproduction (Ding et al., 2012), and gene silencing (Yan et al., 2018). Besides that, researchers have explored the potential of stress-regulated lncRNAs to trigger DNA methylation in response to environmental conditions. The well-characterized AUXIN REGULATED PROMOTER LOOP (APOLO) was identified as an auxin-induced lncRNA in Arabidopsis (Ariel et al., 2014). The double transcription of APOLO by Pol II and V was reported as responsible for originating a chromatin loop, which encompasses the promoter of its neighboring gene PINOID (PID), a key regulator of polar auxin transport, leading to downregulation of its transcripts. Alternatively, it was proposed that APOLO also recognizes distant non-associated loci by R-loop formation. APOLO-mediated LIKE HETEROCHROMATIC PROTEIN 1 (LHP1) decoy may trigger the transcription of the target loci modulating local chromatin conformation, co-regulating auxin-responsive genes (Ariel et al., 2020). A systematic methylome study (Song et al., 2016) evaluated DNA methylation changes in Populus simonii submitted to salinity, osmotic and temperature stress, suggesting that, in association with miRNAs and lncRNAs, this regulatory mechanism can act in response to abiotic stresses in poplar. Ultimately, analysis in soybean roots continuously treated with high salinity solutions revealed that more than 75% of the lncRNAs identified were activated or induced in transcriptome sequencing (Chen et al., 2019).
The RdDM pathway constitutes an impressive extension of the transcriptional capacity of eukaryotic organisms, being considered the main epigenetic pathway mediated by siRNA in plants (Matzke and Mosher, 2014). The canonical RdDM pathway involves the recruitment of Pol IV to transcribe single-stranded RNAs (ssRNAs) at its target loci. The RDR2 copies the ssRNAs to produce dsRNAs. DCL3 processes dsRNAs to 24-nt siRNAs. Finally, de novo methylation occurs, which requires Pol V–dependent scaffold RNAs, AGO4-bound 24-nt siRNAs, and DRM2 (Mosher et al., 2008). Meanwhile, non-canonical RdDM pathways provides a link between PTGS of transposon transcripts and de novo methylation of transposon DNA, since it was reported that tasiRNAs and transposons are initially transcribed by Pol II, copied by RDR6 and processed by DCL2 and DCL4 into 21–22-nt siRNAs (Matzke et al., 2015). Additionally, experiments conducted in Arabidopsis dcl1/2/3/4 mutants by Yang et al. (2016) demonstrated that DNA methylation at many of the RdDM target loci did not correlate with 24-nt siRNAs and it was completely independent of DCLs. Instead, it was observed that 25–50 nt RNAs were the main class of sRNAs generated from most RdDM loci in dcl plants. Interesting studies have contributed to broaden our understanding about RdDM biological functions of RdDM, reporting its involvement in regulating transposon silencing (La et al., 2011), gene expression (Lang et al., 2017), plant development (Kawakatsu et al., 2017), and biotic interactions (Satgé et al., 2016). Special attention has been given to the potential roles of DNA methylation in plant responses to a wide range of abiotic stresses, such as nutritional deficit (Secco et al., 2015), temperature (Liu et al., 2018), high salinity (Lira-Medeiros et al., 2010), and drought (Wang et al., 2016). Despite great efforts, issues such as the mechanism, biological roles and evolutionary importance of RdDM still remains to be fully elucidated, as well as the fundamental role that lncRNAs may be playing in regulating this silencing mechanism.
Functional investigations suggested the contributions of lncRNAs as essential modulators in plant responses to stresses (Figure 1). A growing body of evidence points to the great potential role for plant lncRNAs in responses to abiotic stresses via RdDM (Ariel et al., 2014; Yong-Villalobos et al., 2015). Given the limited number of studies, it is assumed that there is a great potential for RdDM-associated lncRNAs to be studied.
LncRNAs as Precursors to Abiotic Stress Responses
Here, we briefly summarize recent examples of lncRNAs responsive to abiotic stresses in different plant species, with an emphasis on crop species, providing details of other mechanisms of action, in addition to the aforementioned epigenetic silencing via RdDM (Table 1).
TABLE 1.
Summary of studies with abiotic stress-responsive lncRNAs in plants.
| LncRNA | Stress | Plant species | Regulation mechanism | Expression | References |
| IPS1 | Phosphate deficiency | A. thaliana | Target mimicry | Induced | Franco-Zorrilla et al., 2007 |
| npc536 | Salt stress | A. thaliana | Nat. antisense siRNAs | Induced | BenAmor et al., 2009 |
| npc60 | Salt stress | A. thaliana | Nat. antisense siRNAs | Induced | BenAmor et al., 2009 |
| COLDAIR | Cold stress | A. thaliana | Histone modification | Induced | Heo and Sung, 2011 |
| TahlnRNA27 | Heat stress | T. aestivum | miRNA precursor | Induced | Xin et al., 2011 |
| TalnRNA5 | Heat stress | T. aestivum | miRNA precursor | Induced | Xin et al., 2011 |
| AtR8 | Hypoxic stress | A. thaliana | Trans-acting factor | Repressed | Wu et al., 2012 |
| Cis-NAT PHO1;2 | Phosphate deficiency | O. sativa | Translation enhancer | Induced | Jabnoune et al., 2013 |
| Si NAT 80 | Drought stress | S. italica | Cis-acting factor | Induced | Qi et al., 2013 |
| APOLO | Auxin | A. thaliana | DNA demethylation | Induced | Ariel et al., 2014 |
| asHSFB2a | Heat stress | A. thaliana | Nat. antisense siRNAs | Induced | Wunderlich et al., 2014 |
| COOLAIR | Cold stress | A. thaliana | Histone modification | Induced | Csorba et al., 2014; Marquardt et al., 2014 |
| Lnc-173 | High-light stress | A. thaliana | Cis-acting factor | Induced | Di et al., 2014 |
| Lnc-225 | High-light stress | A. thaliana | Cis-acting factor | Induced | Di et al., 2014 |
| LincRNA1128 | Drought stress | P. trichocarpa | Target mimicry | Repressed | Shuai et al., 2014 |
| LincRNA2962 | Drought stress | P. trichocarpa | Target mimicry | Induced | Shuai et al., 2014 |
| LincRNA1039 | Drought stress | P. trichocarpa | Target mimicry | Induced | Shuai et al., 2014 |
| LincRNA20 | Drought stress | P. trichocarpa | Target mimicry | Induced | Shuai et al., 2014 |
| LincRNA2752 | Drought stress | P. trichocarpa | Target mimicry | Induced | Shuai et al., 2014 |
| LincRNA2623 | Drought stress | P. trichocarpa | Target mimicry | Repressed | Shuai et al., 2014 |
| TCONS_00056395 | Drought stress | Z. mays | miRNA precursor | Induced | Zhang et al., 2014 |
| TCONS_00082174 | Drought stress | Z. mays | miRNA precursor | Induced | Zhang et al., 2014 |
| GRMZM2G088590_T04 | Drought stress | Z. mays | miRNA precursor | Induced | Zhang et al., 2014 |
| TCONS_00037470 | Drought stress | Z. mays | miRNA precursor | Induced | Zhang et al., 2014 |
| TCONS_00012768 | Drought stress | Z. mays | miRNA precursor | Induced | Zhang et al., 2014 |
| XLOC_011965 | Cadmium stress | O. sativa | Unknown | Induced | He et al., 2015 |
| XLOC_054416 | Cadmium stress | O. sativa | Unknown | Induced | He et al., 2015 |
| XLOC_001126 | Cadmium stress | O. sativa | Unknown | Repressed | He et al., 2015 |
| XLOC_048220 | Cadmium stress | O. sativa | Unknown | Repressed | He et al., 2015 |
| TCONS_00046739 | Salt stress | M. truncatula | Unknown | Induced | Wang et al., 2015 |
| TCONS_00100258 | Salt stress | M. truncatula | Unknown | Induced | Wang et al., 2015 |
| TCONS_00118328 | Salt stress | M. truncatula | Unknown | Induced | Wang et al., 2015 |
| Os02g0250700-01 | Drought stress | O. sativa | Nat. antisense transcript | Repressed | Chung et al., 2016 |
| Os02g0180800-01 | Drought stress | O. sativa | Nat. antisense transcript | Repressed | Chung et al., 2016 |
| TCONS_00052316 | Low-nitrogen stress | P. tomentosa | Target mimicry | Repressed | Chen et al., 2016 |
| TCONS_00069233 | Low-nitrogen stress | P. tomentosa | Target mimicry | Repressed | Chen et al., 2016 |
| TCONS_00052315 | Low-nitrogen stress | P. tomentosa | Target mimicry | Repressed | Chen et al., 2016 |
| TCONS_00064021 | Low-nitrogen stress | P. tomentosa | Cis-acting factor | Repressed | Chen et al., 2016 |
| TCONS_00049805 | Low-nitrogen stress | P. tomentosa | Cis-acting factor | Repressed | Chen et al., 2016 |
| TCONS_00017288 | Low-nitrogen stress | P. tomentosa | Unknown | Induced | Chen et al., 2016 |
| TCONS_0002186 | Low-nitrogen stress | P. tomentosa | Cis-acting factor | Induced | Chen et al., 2016 |
| TCONS_00021860 | Low-nitrogen stress | P. tomentosa | Unknown | Induced | Chen et al., 2016 |
| c70772_g2_i1 | Drought stress | T. turgidum | Target mimicry | Induced | Cagirici et al., 2017 |
| c90557_g1_i1 | Drought stress | T. turgidum | Target mimicry | Induced | Cagirici et al., 2017 |
| TCONS_00043651 | Boron stress | H. vulgare | Target mimicry | Induced | Karakulah and Unver, 2017 |
| DRIR | Drought and salt stress | A. thaliana | Unknown | Induced | Qin et al., 2017 |
| AK370814 | Salt stress | H. vulgare | Cis-acting factor | Induced | Karlik and Gozukirmizi, 2018 |
| LncRNA_082364 | Ca2+ -channel blocking | T. aestivum | Trans-acting factor | Induced | Ma et al., 2018 |
| LncRNA_047461 | Ca2+ -channel blocking | T. aestivum | Trans-acting factor | Induced | Ma et al., 2018 |
| LncRNA_074658 | Ca2+ -channel blocking | T. aestivum | Trans-acting factor | Repressed | Ma et al., 2018 |
| LncRNA_000823 | Ca2+ -channel blocking | T. aestivum | Trans-acting factor | Repressed | Ma et al., 2018 |
| LncRNA_058136 | Ca2+ -channel blocking | T. aestivum | Trans-acting factor | Repressed | Ma et al., 2018 |
| LncRNA_008977 | Ca2+ -channel blocking | T. aestivum | Trans-acting factor | Induced | Ma et al., 2018 |
| LncRNA_061738 | Ca2+ -channel blocking | T. aestivum | Trans-acting factor | Induced | Ma et al., 2018 |
| LncRNA_018111 | Ca2+ -channel blocking | T. aestivum | Trans-acting factor | Induced | Ma et al., 2018 |
| MSTRG.4636 | Heat stress | Z. mays | Unknown | Repressed | Lv et al., 2019 |
| MSTRG.38321 | Heat stress | Z. mays | Unknown | Repressed | Lv et al., 2019 |
| MSTRG.11125 | Heat stress | Z. mays | Unknown | Induced | Lv et al., 2019 |
| MSTRG.15555 | Heat stress | Z. mays | Unknown | Induced | Lv et al., 2019 |
| MSTRG.31362 | Heat stress | Z. mays | Unknown | Induced | Lv et al., 2019 |
| MSTRG.63799 | Heat stress | Z. mays | Unknown | Repressed | Lv et al., 2019 |
| AT1G34844 | Cold stress | A. thaliana | Nat. antisense transcript | Induced | Calixto et al., 2019 |
| AT3G26612 l | Cold stress | A. thaliana | Nat. antisense transcript | Induced | Calixto et al., 2019 |
| TAS3 | Low-nitrogen stress | A. thaliana | Trans-acting factor | Repressed | Fukuda et al., 2019 |
| LncRNA-tomato_535 | Drought stress | S. lycopersicum | Target mimicry | Induced | Eom et al., 2019 |
| LncRNA-tomato_146 | Drought stress | S. lycopersicum | Target mimicry | Induced | Eom et al., 2019 |
| LncRNA-tomato_178 | Drought stress | S. lycopersicum | Target mimicry | Induced | Eom et al., 2019 |
| LncRNA_tomato_467 | Drought stress | S. lycopersicum | Unknown | Induced | Eom et al., 2019 |
| MSTRG.6838.1 | Drought stress | Z. mays | Cis-acting factor | Repressed | Pang et al., 2019 |
| VIT_216s0100n00030 | Cold stress | V. vinifera | Cis-acting factor | Induced | Wang et al., 2019 |
| LXLOC_027751 | Cold stress | V. vinifera | Cis-acting factor | Induced | Wang et al., 2019 |
| LXLOC_010422 | Cold stress | V. vinifera | Cis-acting factor | Induced | Wang et al., 2019 |
| VIT_202s0025n00100 | Cold stress | V. vinifera | Cis-acting factor | Induced | Wang et al., 2019 |
| VIT_200s0225n00020 | Cold stress | V. vinifera | Trans-acting factor | Repressed | Wang et al., 2019 |
| MSTRG.43964.1 | Drought stress | C. songorica | Target mimicry | Induced | Yan et al., 2019 |
| MSTRG.4400.2 | Drought stress | C. songorica | Target mimicry | Induced | Yan et al., 2019 |
| LncRNA973 | Salt stress | G. hirsutum | Trans-acting factor | Induced | Zhang et al., 2019 |
| TCONS_00024229 | Salt stress | S. polyrhiza | Cis-acting factor | Induced | Fu et al., 2020 |
| TCONS_00057092 | Salt stress | S. polyrhiza | Cis-acting factor | Induced | Fu et al., 2020 |
| TCONS_00018576 | Salt stress | S. polyrhiza | Cis-acting factor | Induced | Fu et al., 2020 |
| TCONS_00023928 | Salt stress | S. polyrhiza | Cis-acting factor | Induced | Fu et al., 2020 |
| TCONS_00045028 | Salt stress | S. polyrhiza | Cis-acting factor | Induced | Fu et al., 2020 |
| TCONS_00033722 | Salt stress | S. polyrhiza | Target mimicry | Induced | Fu et al., 2020 |
| TCONS_00018793 | Salt stress | S. polyrhiza | Target mimicry | Induced | Fu et al., 2020 |
| TCONS_ 00045706 | Salt stress | S. polyrhiza | Target mimicry | Induced | Fu et al., 2020 |
| TCONS_00057092 | Salt stress | S. polyrhiza | Target mimicry | Induced | Fu et al., 2020 |
| TCONS_ 00045512 | Salt stress | S. polyrhiza | Target mimicry | Induced | Fu et al., 2020 |
| TCONS_00051908 | Heat stress | B. juncea | Unknown | Induced | Bhatia et al., 2020 |
| TCONS_00088973 | Drought stress | B. juncea | Unknown | Induced | Bhatia et al., 2020 |
| NcM9574 | Cold stress | M. esculenta | Cis-acting factor | Induced | Suksamran et al., 2020 |
| NcP12248 | Cold stress | M. esculenta | Cis-acting factor | Repressed | Suksamran et al., 2020 |
| NcM17949 | Drought stress | M. esculenta | Cis-acting factor | Induced | Suksamran et al., 2020 |
| NcP456 | Cold stress | M. esculenta | Trans-acting factor | Repressed | Suksamran et al., 2020 |
| NcP12197 | Drought stress | M. esculenta | Trans-acting factor | Induced | Suksamran et al., 2020 |
| NcM15664 | Drought stress | M. esculenta | Trans-acting factor | Repressed | Suksamran et al., 2020 |
| LncRNA13472 | Salt stress | S. bicolor | Target mimicry | Induced | Sun et al., 2020 |
| LncRNA14798 | Salt stress | S. bicolor | Target mimicry | Repressed | Sun et al., 2020 |
| LncRNA11310 | Salt stress | S. bicolor | Target mimicry | Repressed | Sun et al., 2020 |
| LncRNA2846 | Salt stress | S. bicolor | Target mimicry | Repressed | Sun et al., 2020 |
| LncRNA26929 | Salt stress | S. bicolor | Target mimicry | Repressed | Sun et al., 2020 |
| XLOC_012868 | Drought stress | B. napus | Unknown | Repressed | Tan et al., 2020 |
| XLOC_052298 | Drought stress | B. napus | Unknown | Induced | Tan et al., 2020 |
| XLOC_094954 | Drought stress | B. napus | Unknown | Induced | Tan et al., 2020 |
| TCONS_00043651 | Boron stress | H. vulgare | Target mimicry | Induced | Unver and Tombuloglu, 2020 |
| TCONS_00061958 | Boron stress | H. vulgare | Cis-acting factor | Induced | Unver and Tombuloglu, 2020 |
| MtCIR1 | Cold stress | M. truncatula | Cis-acting factor | Induced | Zhao et al., 2020 |
A genome-wide study by Fukuda et al. (2019) reported lncRNAs that are involved in the response to low availability of nutrients in Arabidopsis, allowing the identification of 60 differentially expressed lncRNAs. Among them, TAS3 was revealed as repressed under low-nitrogen conditions with high affinity to target nitrate transporter 2.4 (NRT2.4). Similarly, a genome-wide strategy was used to identify lncRNAs differentially expressed in response to nutritional stress in poplar (Chen et al., 2016) and Arabidopsis (Franco-Zorrilla et al., 2007).
Extreme temperatures can also alter plants lncRNAs expression. In Arabidopsis, HSFB2a is a heat shock gene required for the gametophytic development, controlled by an antisense heat-inducible lncRNA, asHSFB2a (Wunderlich et al., 2014). Intriguingly, the overexpression of asHSFB2a represses HSFB2a RNA accumulation and overexpression of HSFB2a has a similar negative effect on asHSFB2a expression. Despite the lack of knowledge of the molecular mechanisms involved in this “Yin–Yang” control of sense and antisense RNA expression, the study by Wunderlich et al. (2014) showed that the vegetative and gametophytic development are impacted by this regulation of gene expression at the HSFB2a locus. Meanwhile, 1,614 lncRNAs were found to be differentially expressed in Brassica juncea under heat and drought stress conditions (Bhatia et al., 2020). Cold-responsive lncRNAs have been identified in plants such as grape (Wang et al., 2019) and Arabidopsis (Calixto et al., 2019). Both COLDAIR and COOLAIR are well-characterized examples of cold-induced lncRNAs that have been detected as regulating the vernalization process through silencing of FLOWERING LOCUS C (FLC). FLC encodes a MADS box transcription regulator of flowering time, repressing the induction of flowering (Heo and Sung, 2011; Marquardt et al., 2014). COLDAIR is transcribed from the first intron of FLC and physically interacts with a component of Polycomb Repressive Complex 2 (PRC2) to promote H3K27me3 accumulation at the FLC locus (Kim et al., 2017). COOLAIR is an FLC antisense transcript, involved in FLC repression by both autonomous (Tian et al., 2019) and vernalization pathways (Csorba et al., 2014), inducing H3K27me3 by recruiting plant homeo-domain (PHD)-PRC2 (Swiezewski et al., 2009).
Drought and high salinity are the main environmental conditions that adversely affect plant productivity and both can perform the same effects by overlapping genetic regulatory mechanisms. For instance, Drought Induced lncRNA (DRIR) was reported in Arabidopsis as a positive regulator of plant responses to drought and salt stress (Qin et al., 2017). Previous work identified 3 up-regulated lncRNAs under NaCl treatment (BenAmor et al., 2009) and 2,815 novel salt-responsive lncRNAs were reported in Spirodela polyrhiza (Fu et al., 2020). Drought-responsive lncRNAs were investigated in poplars submitted to a water deficit (Shuai et al., 2014). For example, drought induced lincRNA2752 is a target mimic of ptc-miR169, a NF-YA transcription factor regulator. Similar results were found in drought-responsive lncRNAs identified in Cleistogenes songorica (Yan et al., 2019) and B. napus (Tan et al., 2020).
LncRNA in Crop Plants
All findings reporting lncRNAs involvement in response to environmental stresses are particularly important in the context of crop species, since abiotic stresses are a major constraint to improve agriculture yields (Halford et al., 2015). Identification of lncRNAs during crop stress responses remains largely premature, presenting few examples (Karakulah and Unver, 2017; Pang et al., 2019).
LncRNAs have been identified as involved in nutritional homeostasis in crops such as rice (Jabnoune et al., 2013; He et al., 2015) and wheat (Ma et al., 2018). Recent reports demonstrated roles of barley lncRNAs upon excessive boron-treatment (Karakulah and Unver, 2017; Unver and Tombuloglu, 2020). Both studies suggest that boron-regulation can be cooperatively controlled by the interaction of miRNA-lncRNA-coding target transcript modules. For instance, TCONS_00043651, a potential miRNA sponge of miR399, was positively regulated under boron-exposure (Unver and Tombuloglu, 2020). Oppositely, miR399 expression was repressed under this stress condition.
Whereas changes in temperature often causes yield loss, heat-responsive lncRNAs were identified in wheat (Xin et al., 2011) and maize (Lv et al., 2019). The lncRNA TahlnRNA27 was induced under heat treatment and characterized as putative miRNA precursor by presenting Ta-miR2010 family sequences (Xin et al., 2011). Similarly, 182 novel cold-responsive lncRNAs are known to be differentially expressed in cassava (Suksamran et al., 2020); whereas 2,271 lncRNAs were cold-responsive in alfalfa (Zhao et al., 2020).
Salinity stress is currently an environmental factor that most constraints agricultural productivity (Song and Wang, 2015). Studies have attempted to expand knowledge about functional mechanisms of lncRNAs in response to salt stress as well as in alfalfa (Wang et al., 2015); barley (Karlik and Gozukirmizi, 2018); cotton (Zhang et al., 2019); and sorghum (Sun et al., 2020). In particular, the lncRNA973 overexpression had increased salt tolerance, modulating the expression of cotton salt stress-related genes (Zhang et al., 2019).
To improve crop performance in regions limited by water deficit, studies have been conducted to investigate the drought-responsive lncRNAs in crop species including foxtail millet (Qi et al., 2013); maize (Zhang et al., 2014); rice (Chung et al., 2016); wheat (Cagirici et al., 2017); tomato (Eom et al., 2019); and cassava (Suksamran et al., 2020). A recent work carried out with maize identified 124 drought-responsive lncRNAs characterized as cis-acting factors (Pang et al., 2019). The repressed expression correlation between vpp4, encoding a vacuolar (H+)-pumping ATPase subunit, and its adjacent lncRNA MSTRG.6838.1 provides the idea that both could be a promising cis-acting pair.
Conclusion and Perspective
Due to the rapid progress in high-throughput sequencing, several findings have significantly expanded our knowledge of lncRNA biology. However, despite the relevant results reported recently, the biological role and mechanisms of action of plant lncRNAs remain poorly understood. Further studies on lncRNAs responsive to abiotic stresses in crop species will open paths for a better understanding of their function in various processes of plant development and management of stress. It is notable in Table 1 that several lncRNAs regulated in response to abiotic stress have unknown regulation mechanisms. Remarkable progress has been made in elucidating the roles of plant lncRNAs in RdDM silencing pathway. The complexity of RdDM and its involvement in the activation of stress-responsive genes are undeniable, although more efforts are needed to understand RNA-induced DNA methylation and its function in plants, especially during abiotic stresses.
MiRNAs and lncRNAs are regulatory genes that can be targets for improving crop tolerance to abiotic stresses by using the currently advanced genome editing tools, as clustered regularly interspaced short palindromic repeats associated nucleases (CRISPR/Cas) (Zhang et al., 2020). A few successful reports on CRISPR/Cas9-based gene editing for miRNAs were published recently (Li et al., 2016; Zhou et al., 2017). The short sequences of miRNAs make it difficult to find a PAM sequence that is required for CRISPR/Cas genome editing. As more diversity of Cas proteins are identified and current Cas proteins are being continuously modified, the PAM requirement will be relaxed, and more genetic loci will become accessible by CRISPR/Cas system (Zhang and Zhang, 2020), including lncRNAs once they are already longer than miRNAs.
As the regulation for the use of genetically modified organisms (GMOs) and CRISPR-gene editing is still very tight in several countries, alternative approaches for crop breeding should be considered, such as the exogenous application of RNA molecules (Dalakouras et al., 2020). Based on successful examples of delivery of RNAs with the potential to trigger RNAi in plants (Cagliari et al., 2019; Werner et al., 2020), possible shortcomings of these methods might include optimization in application of several other types of RNA molecules, including lncRNAs, as well as grouped components of CRISPR/Cas to promote GMO independent editing events in lncRNA sequences.
Author Contributions
MU and FT wrote the manuscript. AH reviewed the manuscript. PF contributed to design of this mini-review. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to researchers whose work has not been included in this manuscript owing to a limit in space.
Funding. The FAPERJ (Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro), the CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), and the CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) supported this work. The CAPES and the CNPq support postgraduate fellowships of FT and Ph.D. fellowship of MU, respectively. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Ariel F., Jegu T., Latrasse D., Romero-Barrios N., Christ A., Benhamed M., et al. (2014). Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell 55 383–396. 10.1016/j.molcel.2014.06.011 [DOI] [PubMed] [Google Scholar]
- Ariel F., Lucero L., Christ A., Mammarella M. F., Jegu T., Veluchamy A., et al. (2020). R-Loop mediated trans action of the APOLO long noncoding RNA. Mol. Cell 77 1055–1065. 10.1016/j.molcel.2019.12.015 [DOI] [PubMed] [Google Scholar]
- BenAmor B., Wirth S., Merchan F., Laporte P., D’Aubenton-Carafa Y., Hirsch J., et al. (2009). Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 19 57–69. 10.1101/gr.080275.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia G., Singh A., Verma D., Sharma S., Singh K. (2020). Genome-wide investigation of regulatory roles of lncRNAs in response to heat and drought stress in Brassica juncea (Indian mustard). Environ. Exp. Bot. 171:103922 10.1016/j.envexpbot.2019.103922 [DOI] [Google Scholar]
- Blevins T., Podicheti R., Mishra V., Marasco M., Wang J., Rusch D., et al. (2015). Identification of pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 4 1–22. 10.7554/eLife.09591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhmdorfer G., Sethuraman S., Jordan Rowley M., Krzyszton M., Hafiz Rothi M., Bouzit L., et al. (2016). Long non-coding RNA produced by RNA polymerase V determines boundaries of heterochromatin. eLife 5 1–24. 10.7554/eLife.19092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagirici H. B., Alptekin B., Budak H. (2017). RNA sequencing and co-expressed long non-coding RNA in modern and wild Wheats. Sci. Rep. 7 1–16. 10.1038/s41598-017-11170-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliari D., Dias N. P., Galdeano D. M., dos Santos E. Á, Smagghe G., Zotti M. J. (2019). Management of pest insects and plant diseases by non-transformative RNAi. Front. Plant Sci. 10:1319. 10.3389/fpls.2019.01319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calixto C. P. G., Tzioutziou N. A., James A. B., Hornyik C., Guo W., Zhang R., et al. (2019). Cold-dependent expression and alternative splicing of Arabidopsis long non-coding RNAs. Front. Plant Sci. 10:235. 10.3389/fpls.2019.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Wang C., Bao H., Chen H., Wang Y. (2016). Genome-wide identification and characterization of novel lncRNAs in Populus under nitrogen deficiency. Mol. Genet. Genomics 291 1663–1680. 10.1007/s00438-016-1210-3 [DOI] [PubMed] [Google Scholar]
- Chen R., Li M., Zhang H., Duan L., Sun X., Jiang Q., et al. (2019). Continuous salt stress-induced long non-coding RNAs and DNA methylation patterns in soybean roots. BMC Genomics 20:730. 10.1186/s12864-019-6101-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Li X., Su L., Chen X., Zhang S., Xu X., et al. (2018). Genome-wide identification and characterization of long non-coding RNAs involved in the early somatic embryogenesis in Dimocarpus longan Lour. BMC Genomics 19:805. 10.1186/s12864-018-5158-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung P. J., Jung H., Jeong D. H., Ha S. H., Choi Y. D., Kim J. K. (2016). Transcriptome profiling of drought responsive noncoding RNAs and their target genes in rice. BMC Genomics 17:563. 10.1186/s12864-016-2997-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi M. D., Jurkevitch E., Poiret M., D’Aubenton-Carafa Y., Petrovics G., Kondorosi E., et al. (1994). enod4O,a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 13 5099–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. (1970). Central dogma of molecular biology. Nature 227 561–563. 10.1038/227561a0 [DOI] [PubMed] [Google Scholar]
- Csorba T., Questa J. I., Sun Q., Dean C. (2014). Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. U.S.A. 111 16160–16165. 10.1073/pnas.1419030111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalakouras A., Wassenegger M., Dadami E., Ganopoulos I., Pappas M. L., Papadopoulou K. (2020). Genetically modified organism-free RNA interference: exogenous application of RNA molecules in plants. Plant Physiol. 182 38–50. 10.1104/pp.19.00570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevicz M. F., Moharana K. C., Venancio T. M., Franco L. O., Cardoso S. R. S., Cardoso M., et al. (2018). Copaifera langsdorffii novel putative long non-coding RNAs: interspecies conservation analysis in adaptive response to different biomes. Non Coding RNA 4 1–21. 10.3390/ncrna4040027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng P., Liu S., Nie X., Weining S., Wu L. (2018). Conservation analysis of long non-coding RNAs in plants. Sci. China Life Sci. 61 190–198. 10.1007/s11427-017-9174-9 [DOI] [PubMed] [Google Scholar]
- Di C., Yuan J., Wu Y., Li J., Lin H., Hu L., et al. (2014). Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 80, 848–861. 10.1111/tpj.12679 [DOI] [PubMed] [Google Scholar]
- Ding J., Shen J., Mao H., Xie W., Li X., Zhang Q. (2012). RNA-directed DNA methylation is involved in regulating photoperiod- sensitive male sterility in rice. Mol. Plant 5 1210–1216. 10.1093/mp/sss095 [DOI] [PubMed] [Google Scholar]
- Eom S. H., Lee H. J., Lee J. H., Wi S. H., Kim S. K., Hyun T. K. (2019). Identification and functional prediction of drought-responsive long non-coding RNA in tomato. Agronomy 9 629 10.3390/agronomy9100629 [DOI] [Google Scholar]
- Fan Y., Yang J., Mathioni S. M., Yu J., Shen J., Yang X., et al. (2016). PMS1T, producing Phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice. Proc. Natl. Acad. Sci. U.S.A. 113 15144–15149. 10.1073/pnas.1619159114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J. M., Valli A., Todesco M., Mateos I., Puga M. I., Rubio-Somoza I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39 1033–1037. 10.1038/ng2079 [DOI] [PubMed] [Google Scholar]
- Fu L., Ding Z., Tan D., Han B., Sun X., Zhang J. (2020). Genome-wide discovery and functional prediction of salt-responsive lncRNAs in duckweed. BMC Genomics 21:212. 10.1186/s12864-020-6633-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., Nishida S., Kakei Y., Shimada Y., Fujiwara T. (2019). Genome-wide analysis of long intergenic noncoding RNAs responding to low-nutrient conditions in Arabidopsis thaliana: Possible involvement of trans-acting siRNA3 in response to low nitrogen. Plant Cell Physiol. 60 1961–1973. [DOI] [PubMed] [Google Scholar]
- Gao Z., Liu H. L., Daxinger L., Pontes O., He X., Qian W., et al. (2010). An RNA polymerase II-and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 465 106–109. 10.1038/nature09025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford N. G., Curtis T. Y., Chen Z., Huang J. (2015). Effects of abiotic stress and crop management on cereal grain composition: implications for food quality and safety. J. Exp. Bot. 66 1145–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Liu Q., Zheng L., Cui Y., Shen Z., Zheng L. (2015). Rna-seq analysis of rice roots reveals the involvement of post-transcriptional regulation in response to cadmium stress. Front. Plant Sci. 6:1136. 10.3389/fpls.2015.01136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo J. B., Sung S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331 76–79. 10.1126/science.1197349 [DOI] [PubMed] [Google Scholar]
- Holoch D., Moazed D. (2015). RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16 71–84. 10.1038/nrg3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabnoune M., Secco D., Lecampion C., Robaglia C., Shu Q. Y., Poirier Y. (2013). A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 25 4166–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakulah G., Unver T. (2017). Barley long non-coding RNAs and their tissue-specific co-expression pattern with coding-transcripts. bioRxiv[Preprint] 10.1101/229559 [DOI] [Google Scholar]
- Karlik E., Ari S., Gozukirmizi N. (2019). LncRNAs: genetic and epigenetic effects in plants. Biotechnol. Biotechnol. Equip. 33 429–439. 10.1080/13102818.2019.1581085 [DOI] [Google Scholar]
- Karlik E., Gozukirmizi N. (2018). Expression analysis of lncRNA AK370814 involved in the barley vitamin B6 salvage pathway under salinity. Mol. Biol. Rep. 45 1597–1609. 10.1007/s11033-018-4289-2 [DOI] [PubMed] [Google Scholar]
- Kawakatsu T., Nery J. R., Castanon R., Ecker J. R. (2017). Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol. 18 1–12. 10.1186/s13059-017-1251-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H., Xi Y., Sung S. (2017). Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet. 13:e1006939. 10.1371/journal.pgen.1006939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La H., Ding B., Mishra G. P., Zhou B., Yang H., Bellizzi M. D. R., et al. (2011). A 5-methylcytosine DNA glycosylase/lyase demethylates the retrotransposon Tos17 and promotes its transposition in rice. Proc. Natl. Acad. Sci. U.S.A. 108 15498–15503. 10.1073/pnas.1112704108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang Z., Wang Y., Tang K., Tang D., Datsenka T., Cheng J., et al. (2017). Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. U.S.A. 114 E4511–E4519. 10.1073/pnas.1705233114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Eichten S. R., Shimizu R., Petsch K., Yeh C. T., Wu W., et al. (2014). Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol. 15 1–15. 10.1186/gb-2014-15-2-r40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Li X., Zhou Z., Wu P., Fang M., Pan X., et al. (2016). Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 7:377. 10.3389/fpls.2016.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira-Medeiros C. F., Parisod C., Fernandes R. A., Mata C. S., Cardoso M. A., Ferreira P. C. G. (2010). Epigenetic variation in mangrove plants occurring in contrasting natural environment. PLoS One 5:e10326. 10.1371/journal.pone.0010326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Xia Y., Liu T., Dai S., Hou X. (2018). The DNA methylome and association of differentially methylated regions with differential gene expression during heat stress in brassica rapa. Int. J. Mol. Sci. 19 1414. 10.3390/ijms19051414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y., Hu F., Zhou Y., Wu F., Gaut B. S. (2019). Maize transposable elements contribute to long non-coding RNAs that are regulatory hubs for abiotic stress response. BMC Genomics 20:864. 10.1186/s12864-019-6245-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K., Shi W., Xu M., Liu J., Zhang F. (2018). Genome-wide identification and characterization of long non-coding RNA in wheat roots in response to Ca2+ channel blocker. Front. Plant Sci. 9:244. 10.3389/fpls.2018.00244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Bajic V. B., Zhang Z. (2013). On the classification of long non-coding RNAs. RNA Biol. 10 924–933. 10.4161/rna.24604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S., Raitskin O., Wu Z., Liu F., Sun Q., Dean C. (2014). Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol. Cell. 54 156–165. 10.1016/j.molcel.2014.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M. A., Kanno T., Matzke A. J. M. (2015). RNA-directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu. Rev. Plant Biol. 266 243–267. 10.1146/annurev-arplant-043014-114633 [DOI] [PubMed] [Google Scholar]
- Matzke M. A., Mosher R. A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15 394–408. 10.1038/nrg3683 [DOI] [PubMed] [Google Scholar]
- Mosher R. A., Schwach F., Studholme D., Baulcombe D. C. (2008). PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 105 3145–3150. 10.1073/pnas.0709632105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak R. (1994). Mining treasures from “junk DNA.”. Science 263 608–610. 10.1126/science.7508142 [DOI] [PubMed] [Google Scholar]
- Pang J., Zhang X., Ma X., Zhao J. (2019). Spatio-temporal transcriptional dynamics of maize long non-coding RNAs responsive to drought stress. Genes (Basel) 10 138. 10.3390/genes10020138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Xie S., Liu Y., Yi F., Yu J. (2013). Genome-wide annotation of genes and noncoding RNAs of foxtail millet in response to simulated drought stress by deep sequencing. Plant Mol. Biol. 83 459–473. 10.1007/s11103-013-0104-6 [DOI] [PubMed] [Google Scholar]
- Qin T., Zhao H., Cui P., Albesher N., Xionga L. (2017). A nucleus-localized long non-coding rna enhances drought and salt stress tolerance. Plant Physiol. 175 1321–1336. 10.1104/pp.17.00574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satgé C., Moreau S., Sallet E., Lefort G., Auriac M.-C., Remblière C., et al. (2016). Reprogramming of DNA methylation is critical for nodule development in Medicago truncatula. Nat. Plants 2 16166. [DOI] [PubMed] [Google Scholar]
- Secco D., Wang C., Shou H., Schultz M. D., Chiarenza S., Nussaume L., et al. (2015). Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife 4 1–26. 10.7554/elife.09343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai P., Liang D., Tang S., Zhang Z., Ye C. Y., Su Y., et al. (2014). Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J. Exp. Bot. 65 4975–4983. 10.1093/jxb/eru256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Wang B. (2015). Using euhalophytes to understand salt tolerance and to develop saline agriculture: Suaeda salsa as a promising model. Ann. Bot. 115 541–553. 10.1093/aob/mcu194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Ci D., Tian M., Zhang D. (2016). Stable methylation of a non-coding RNA gene regulates gene expression in response to abiotic stress in Populus simonii. J. Exp. Bot. 67 1477–1492. 10.1093/jxb/erv543 [DOI] [PubMed] [Google Scholar]
- Suksamran R., Saithong T., Thammarongtham C., Kalapanulak S. (2020). Genomic and transcriptomic analysis identified novel putative cassava lncRNAs involved in cold and drought stress. Genes (Basel) 11 366. 10.3390/genes11040366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Zheng H., Li J., Liu L., Zhang X., Sui N. (2020). Comparative transcriptome analysis reveals new lncRNAs responding to salt stress in sweet sorghum. Front. Bioeng. Biotechnol. 8:331. 10.3389/fbioe.2020.00331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S., Liu F., Magusin A., Dean C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis polycomb target. Nature 462 799–802. 10.1038/nature08618 [DOI] [PubMed] [Google Scholar]
- Szcześniak M. W., Rosikiewicz W., Makalowska I. (2015). CANTATAdb: A collection of plant long non-coding RNAs. Plant Cell Physiol. 57 e8. 10.1093/pcp/pcv201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Li S., Hu L., Zhang C. (2020). Genome-wide analysis of long non-coding RNAs (lncRNAs) in two contrasting rapeseed (Brassica napus L.) genotypes subjected to drought stress and re-watering. BMC Plant Biol. 20:81 10.1186/s12870-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Zheng H., Zhang F., Wang S., Ji X., Xu C., et al. (2019). PRC2 recruitment and H3K27me3 deposition at FLC require FCA binding of COOLAIR. Sci. Adv. 5 1–13. 10.1126/sciadv.aau7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unver T., Tombuloglu H. (2020). Barley long non-coding RNAs (lncRNA) responsive to excess boron. Genomics 112 1947–1955. 10.1016/j.ygeno.2019.11.007 [DOI] [PubMed] [Google Scholar]
- Vieira L. L. M., Grativol C., Thiebaut F., Carvalho T. G. T., Hardoim P. P. R., Hemerly A., et al. (2017). PlantRNA_Sniffer: a SVM-based workflow to predict long intergenic non-coding RNAs in plants. Non Coding RNA 3 11. 10.3390/ncrna3010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Dai L., Ai J., Wang Y., Ren F. (2019). Identification and functional prediction of cold-related long non-coding RNA (lncRNA) in grapevine. Sci. Rep. 9 1–15. 10.1038/s41598-019-43269-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. Z., Liu M., Zhao M. G., Chen R., Zhang W. H. (2015). Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing. BMC Plant Biol. 15:131. 10.1186/s12870-015-0530-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Qin Q., Sun F., Wang Y., Xu D., Li Z., et al. (2016). Genome-wide differences in DNA methylation changes in two contrasting rice genotypes in response to drought conditions. Front. Plant Sci. 7:1675. 10.3389/fpls.2016.01675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Parker B. J., Weiller G. F. (2007). In Silico identification and characterization of mRNA-like noncoding transcripts in Medicago truncatula. In Silico Biol. 4 485–505. [PubMed] [Google Scholar]
- Werner B. T., Gaffar F. Y., Schuemann J., Biedenkopf D., Koch A. M. (2020). RNA-spray-mediated silencing of Fusarium graminearum AGO and DCL genes improve barley disease resistance. Front. Plant Sci. 11:476. 10.3389/fpls.2020.00476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbick A. T., Haag J. R., Pikaard C. S. (2008). Noncoding transcription by RNA Polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135 635–648. 10.1016/j.cell.2008.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A. T., Ream T. S., Haag J. R., Pikaard C. S. (2009). RNA polymerase v transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 41 630–634. 10.1038/ng.365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Okada T., Fukushima T., Tsudzuki T., Sugiura M., Yukawa Y. (2012). A novel hypoxic stress-responsive long non-coding RNA transcribed by RNA polymerase III in Arabidopsis. RNA Biol. 9, 302–313. 10.4161/rna.19101 [DOI] [PubMed] [Google Scholar]
- Wu L., Liu S., Qi H., Cai H., Xu M. (2020). Research progress on plant long non-coding RNA. Plants 9 1–10. 10.3390/plants9040408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich M., Groß-Hardt R., Schöffl F. (2014). Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol. Biol. 85 541–550. 10.1007/s11103-014-0202-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Johansen L. K., Gustafson A. M., Kasschau K. D., Lellis A. D., Zilberman D., et al. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2:642. 10.1371/journal.pbio.0020104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M., Wang Y., Yao Y., Song N., Hu Z., Qin D., et al. (2011). Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 11:61. 10.1186/1471-2229-11-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Lim J., Dale J. M., Chen H., Shinn P., Palm C. J., et al. (2003). Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302 842–846. 10.1126/science.1088305 [DOI] [PubMed] [Google Scholar]
- Yan H., Bombarely A., Xu B., Frazier T. P., Wang C., Chen P., et al. (2018). SiRNAs regulate DNA methylation and interfere with gene and lncRNA expression in the heterozygous polyploid switchgrass. Biotechnol. Biofuels 11 1–19. 10.1186/s13068-018-1202-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q., Wu F., Yan Z., Li J., Ma T., Zhang Y., et al. (2019). Differential co-expression networks of long non-coding RNAs and mRNAs in Cleistogenes songorica under water stress and during recovery. BMC Plant Biol. 19:23. 10.1186/s12870-018-1626-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D. L., Zhang G., Tang K., Li J., Yang L., Huang H., et al. (2016). Dicer-independent RNA-directed DNA methylation in Arabidopsis. Cell Res. 26 66–82. 10.1038/cr.2015.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong-Villalobos L., González-Morales S. I., Wrobel K., Gutiérrez-Alanis D., Cervantes-Peréz S. A., Hayano-Kanashiro C., et al. (2015). Methylome analysis reveals an important role for epigenetic changes in the regulation of the Arabidopsis response to phosphate starvation. Proc. Natl. Acad. Sci. U.S.A. 112 E7293–E7302. 10.1073/pnas.1522301112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., Bischof S., Wang H., Feng S., Lee T., Chen X., et al. (2015). One precursor One siRNA model for Pol IV- dependent siRNAs Biogenesis. Cell 163 1–25. 10.1016/j.cell.2015.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Zhang B. (2020). SpRY: engineered CRISPR/Cas9 harnesses new genome-editing power. Trends Genet. 36 546–548. 10.1016/j.tig.2020.05.001 [DOI] [PubMed] [Google Scholar]
- Zhang D., Zhang Z., Unver T., Zhang B. (2020). CRISPR/Cas: a powerful tool for gene function study and crop improvement. J. Adv. Res. 10.1016/j.jare.2020.10.003 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Han Z., Guo Q., Liu Y., Zheng Y., Wu F., et al. (2014). Identification of maize long non-coding RNAs responsive to drought stress. PLoS One 9:e98958. 10.1371/journal.pone.0098958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Dong J., Deng F., Wang W., Cheng Y., Song L., et al. (2019). The long non-coding RNA lncRNA973 is involved in cotton response to salt stress. BMC Plant Biol. 19:459. 10.1186/s12870-019-2088-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen Y. (2013). Biochemical and biophysical research communications long noncoding RNAs: new regulators in plant development. Biochem. Biophys. Res. Commun. 436 111–114. 10.1016/j.bbrc.2013.05.086 [DOI] [PubMed] [Google Scholar]
- Zhao M., Wang T., Sun T., Yu X., Tian R., Zhang W. H. (2020). Identification of tissue-specific and cold-responsive lncRNAs in Medicago truncatula by high-throughput RNA sequencing. BMC Plant Biol. 20:99. 10.1186/s12870-020-2301-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Deng K., Cheng Y., Zhong Z., Tian L., Tang X., et al. (2017). CRISPR-cas9 based genome editing reveals new insights into microRNA function and regulation in rice. Front. Plant Sci. 8:1598. 10.3389/fpls.2017.01598 [DOI] [PMC free article] [PubMed] [Google Scholar]