Abstract
Background
Spike protein is the surface glycoprotein of the severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) necessary for the entry of the virus via the transmembrane receptors of the human respiratory cells causing COVID-19 disease.
Aim
Here, we aimed to predict the three-dimensional monomer structure of spike protein of SARS-CoV-2 from 20 Jordanian nasopharyngeal samples and to determine the percentage of single amino acid variants (SAV) in the spike protein of SARS-CoV-2.
Methods
The output of the Protein Homology/analogY Recognition Engine V 2.0 (Phyre2) found four single amino acid variants in the spike gene.
Results
The first variant represented by 5% of samples that showed tyrosine deletion at Y144 located in the N terminal domain. The second and the dominant variant, represented by 62%, showed aspartate a coil amino acid substitution to glycine an extracellular amino acid at D614G located in the spike recognition binding site. The third variant, represented by 5%, showed aspartate substitution to tyrosine at D1139Y, and the fourth variant, represented by 5% glycine substitution to serine at G1167S.
Conclusion
Our results have shown low mutational sensitivity in all variants except to D614G the one with the most likely neutral mutational sensitivity that all variants might not explicitly affect the function of spike glycoprotein. However, D614G might change the viral conformational plasticity and hence a potential viral fitness gain but one must be cautious about drawing any concrete conclusions about the severity of symptoms and viral transmission from genomic data only.
General significance
Studying mutations such as D614G in deep is essential to control the pandemic in terms of immune systems, antibodies, or even vaccines.
Keywords: COVID-19, SARS-CoV-2, Spike, D614G & Mutation
Highlights
-
•
Protein Homology/AnalogY Recognition Engine V 2.0 (Phyre2) found four single amino acid variants in the spike gene.
-
•
The dominant variant (62%) showed aspartate substitution to glycine at D614G in the spike recognition-binding site.
-
•
D (aspartate) to G (glycine) has changed from a coil amino acid to anextracellular amino acid.
-
•
D614G might change the viral conformational plasticity and hence a potential viral fitness gain.
-
•
Most likely neutral mutational sensitivity has been shown in the variant D614G in the spike glycoprotein.
-
•
Studying mutations such as D614G in deep is essential to control the pandemic in terms of immune systems, or vaccines.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused an outbreak in Wuhan city, China, at the beginning of December 2019 that rapidly spread across the country and to other nations around the world and characterized as a pandemic by the World Health Organization WHO [1,2]. The first case of SARS-CoV-2 in Jordan was reported to the Ministry of health on March 2, 2020 for a citizen who returned from Italy. To the date, there are 2945 confirmed cases, 2084 recovered, and 21 deaths of COVID-19 in Jordan, according to the official web site launched by the Jordanian Ministry of health as a unified source of information about coronavirus (https://corona.moh.gov.jo/en).
SARS-CoV-2 has a positive, single-strand RNA genome that is over 29 kilobases in length, which belongs to one of the four genera of Orthocoronaviridae, the beta-coronavirus [3]. Moreover, SARS-CoV-2 encodes four major structural proteins, the envelope (E), membrane (M), nucleocapsid (N), and spike (S) proteins. Spike protein (approx. 180 kDa) is the surface glycoprotein of the severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) [4]. Spike glycoprotein is necessary for the interaction of the virus with human cell receptors for a sequential combination of the viral encompass with the cell membrane to be engulfed and permit COVID-19 disease by binding with the angiotensin-converting enzyme 2 (ACE2) [5,6] after an evident activation by type II transmembrane protease TMPRSS2 [7]. Spike protein has two domains, the S1 that has the receptor-binding domain (RBD) to bind with the trans-membrane ACE2 receptor at the cell host to facilitate the virus fusion and the fusion facilitator S2 domain [8]. The alteration within spike protein is crucial, as it may suggest the production of novel SARS-CoV-2 strains that changes its transmissibility or pathogenicity. The S protein is a highly glycosylated trimeric protein that fuses with ACE2 to mediate entrance into host cells, which explains why S protein is a prime target for to be designed therapies against COVID-19 [9].
Mutations and amino acid variants analysis are essential in understanding the mechanism of binding of spike protein with its receptor to have insights on possibilities to design a peptide or nucleotide-based vaccine for COVID-19. To understand the early steps of COVID-19 infection, in this study, we determined the single amino acid variant (SAV) percentage of the spike protein of SARS-CoV-2 in Jordan. One of the most interesting amino acid variants in the spike protein of SARS-Cov-2 is the D614G due to a missense mutation. The D614G mutation became an alarm due to it has been increasingly prevalent all over the world, which might be the new transmissible form of SARS-Cov-2, which has dismayed many scientists according to previous reports [10,11]. Studying such mutations in deep is essential to control the pandemic in terms of immune systems, antibodies, or even vaccines.
Besides, we predicted a three-dimensional structure of the spike glycoprotein of SARS-CoV-2 from positive nasopharyngeal specimens collected in Jordan. The samples were sequenced by Biolab Diagnostic Laboratories (Jordan) & Andersen lab at Scripps Research (USA), who published sequences were retrieved from GISAID, a maintained global database based in Germany. The insight in this work is helpful for scientists to understand different molecular and cytological approaches involved in vaccine development for COVID-19. To the best of our knowledge, what is unique about this study is that it is the first of its kind in Jordan or even the Middle East to investigate the exact percentages of newly missense mutations and its predicted mutational sensitivity in the spike protein.
2. Materials and methods
2.1. Genomic sequence retrieval
Twenty whole-genome sequences of SARS-CoV-2 collected in Jordan were retrieved from the GISAID database. From the above-mentioned genome sequences, the amino sequence of the spike glycoprotein were generated and analyzed. The database showed that the twenty nasopharyngeal specimens were collected over March 2020 and submitted GISAID sequential accession number from EPI_ISL_429992 to EPI_ISL_4300015.
2.2. Submitting sequence in FASTA format and Multiple Alignment using Fast Fourier Transform
The FASTA formats of the spike gene were aligned (Appendix A), isolated, and translated into 1273 amino acids from the whole genome 20 sequences versus 1 reference sequence (accession number YP_009724390.1) of the SARS-CoV-2 by using open-source functions developed by The University of Alcalá, Madrid, Spain at (http://biomodel.uah.es/en/lab/cybertory/analysis/trans.htm). The BLAST function at the NCBI, a web-based service, in addition to Multiple Alignment using Fast Fourier Transform (MAFFT) [12] and viewed by Jalview [13] of Dundee University Scotland. Then the FASTA format of an amino acid sequence of spike protein was submitted to I-TASSER (Iterative Threading ASSEmbly Refinement), which is a hierarchical approach to predict protein structure and function.
2.3. Spike protein secondary structure by position-specific scoring matrices (PSIPRED)
The secondary structure of the wild type and the D614G spike protein was predicted by utilizing the PSIPRED. PSIPRED is a protein structure analysis workbench based on two-stage neural network to predicts the protein secondary structure by generating position-specific scoring matrices. PSIPRED was developed by the bioinformatics aggregate of groups headed by Professor David Jones at the University College London [14]. The PSIPRED was adopted in this work due to its superior benchmarking results of protein secondary structure prediction when compared with other methods according to the globally well-known critical assessment for protein structure (CAS3).
2.4. Iterative Threading ASSEmbly refinement (I-TASSER)
In recent community-wide studies [15] of critical assessment of protein structure prediction or (CASP), particularly in CASP7, CASP8, CASP9, CASP10, CaSP11, CASP12 and CASP13, I-TASSER, as ‘Zhang-Server’, were listed as No. 1 protein structure prediction node which has motivated us to adopt it in this study. It was also ranked highest in CASP9 for the estimation of functions. The system is in active development to utilize state-of-the-art algorithms offering the most reliable protein structure and feature predictions (See appendix for the submitted Sequence in FASTA format). Initially, the I-TASSER was utilized to recognize the basic templates from the PDB by multiple threading approach LOMETS, with full-length atomic models produced by iterative fragment assembly simulations based on templates. Function insights of the targeted molecule are then obtained by rethreading the three-dimensional models via the BioLiP database of protein functions. To produce a predicted three-dimensional structure/model for the S-protein of SARS-CoV-2 collected in Jordan as a PDB file, a hierarchical approach to protein structure and function prediction known as I-TASSER server was used. The I-TASSER pipeline consists of three steps: 1) identification of models, 2) assembly of full-length structures, and 3) annotation of structure-based functions. The reliability of each model is evaluated quantitatively by a C-score based on the value of threaded prototype alignments and the parameters of convergence of structural mounting simulations. C-score is usually [-5, 2], where a higher-value C-score means a more positive and vice versa scale. Following the association observed between these attributes, the TM-score and RMSD are calculated using the C and the protein frequency. Since the group size classes the top 5 models, in some situations, a higher C-score is possible for the lower-ranking models. While the first model is better in most cases, as seen in our research (Fig. 4), lower-level models can also be better than higher-level models. If the I-TASSER simulations converge, less than 5 clusters can have been generated; it usually shows that because of the combined simulations, the models have good quality.
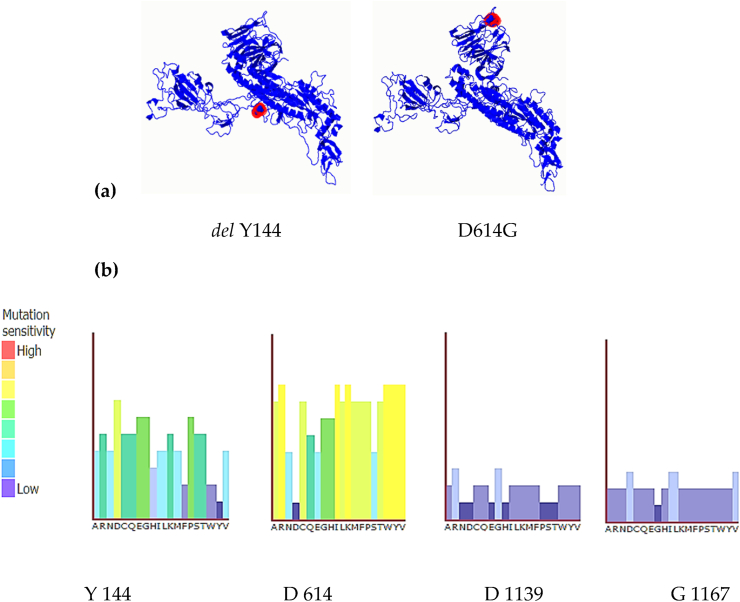
Fig. 4.
(Phyre2) Mutational sensitivity (a) the locations of single amino acid variants del Y144 (left side) & D614G (right side) on spike protein as they have the highest mutation sensitivity scores. (b) The mutation sensitivity histograms for all four single amino acid variants on spike protein.
2.5. Single amino acid variant (SAV), phenotype and mutation analysis
Surface glycoprotein [Severe acute respiratory syndrome coronavirus-2] with accession number YP_009724390.1 was used as a reference sequence to compare with, and it was downloaded from the link: https://www.ncbi.nlm.nih.gov/protein/YP_009724390.1?report=fasta.
Four Amino Acid Variants (SAV) were found from 20 spike glycoprotein sequences submitted and retrieved by a server available on the web called Phyre2. The Phyre2 is a user–friendly suite of bioinformatics tools that are available online to predict and analyze the structure, function, and mutations of proteins [[16], [17], [18]]. At a particular position, color-coded bars of residue preference out of the 20 amino acids represent the Phyre2 mutational sensitivity results for any protein sequence, were tall red bars represent the highest favorable residues to have a phenotypic effect than shorter blue bars. The bar tallness is the values of the position-specific scoring matrix (PSSM) calculated by Position-Specific Iterated Basic Local Alignment (PSI-BLAST) [18].
3. Results
3.1. Spike protein secondary structure by position-specific scoring matrices (PSIPRED)
The prediction output of the secondary structure of the wild type spike glycoprotein versus the D614G variant showed that the substitution from D (aspartate) to G (glycine). D (aspartate) to G (glycine) has changed from a coil amino acid in the light green box to an extracellular amino acid in the orange box according to the color-coding of the PSIPRED, as shown in Fig. 1.
Fig. 1.
Spike protein secondary structure by position-specific scoring matrices (PSIPRED). The upper box shows the wild type D614 as a coil amino acid in light green color, and the lower box shows the G614 as an extracellular amino acid in orange color. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. The final predicted model of the monomer of spike protein by I-TASSER
At first, it was not feasible to get the multimer (trimer) form of the spike protein, as the intricate building is one of the tool limitations of such kind of a computational model prediction. For each target, I-TASSER simulations called decoys generate an extensive collection of structural conformations. I-TASSER uses the SPICKER to cluster all architectural structures based on the pair-sided similarity and records up to 5 models corresponding to the five largest structural clusters.
Recently, the experimental cryo-EM structures of the glycoprotein of SARS-CoV-2 have been reported by two groups; Wrapp et al. and Walls et al., and the structures are available in the Protein Data Bank, e.g., 6vsb, 6vxx [4,19]. While the deposited structures are not full-length and not solved by crystallography, but cryo-EM, the resolution of those structures is close to the crystal structures. Zhang's group (zhanglab.ccmb.med.umich.edu/COVID-19/) has modeled other structural models of SARS-CoV-2, including the surface glycoprotein, with I-TASSAR. We found no difference between the recently reported experimental structures and the in silico models we generated, even with the new single amino variants in terms of the general 3D-structures (Fig. 2).
Fig. 2.
(a) The final I-TASSER predicted spike glycoprotein spike protein monomer model, the PDB file is provided in the supplementary data, and (b) the trimer spike glycoprotein as elucidated by Cryo-EM; PDB:6vxx [19].
The top two proteins structurally close to the spike glycoprotein in the Protein Data Bank (as identified by TM-align) are listed in Table 1. In Table 2 the top five hits of closest Enzyme Commission (EC) numbers and active sites are listed. Our findings showed many molecules, which were structurally close to the spike glycoprotein according to the Enzyme Commission (EC) numbers and active sites. The molecules, as mentioned earlier, included first, the Isoleucyl-tRNA synthetase, which provides the ability to synthesize tRNA. Second, the Crystal structure of the tricorn protease (hydrolase), which provides the ability to hydrolyze the host proteins for viral entry. Third, the Crystal structure of the T. Thermophilus RNA polymerase holoenzyme (transferase), which provides the protein with the ability to synthesize the viral RNA. Fourth, the Crystal structure complex of pyruvate-ferredoxin oxidoreductase from Desulfovibrio africanus and pyruvate (oxidoreductase). Last, the Reovirus core (virus) all might explain the ability of SARS-CoV-2 in getting inside the human host cells.
Table 1.
Proteins structurally close to the spike glycoprotein in the Protein Data Bank (as identified by TM-align).
| Rank | PDB Hit | TM-score | RMSDa | IDENa | Cov |
|---|---|---|---|---|---|
| 1 | 5×58A | 0.827 | 0.41 | 0.751 | 0.828 |
| 2 | 6nzkA | 0.741 | 4.62 | 0.276 | 0.840 |
Protein rankings are based on the structural alignment TM score in the PDB library between the query template and known structures. RMSDa is the root-mean-square deviation among structurally aligned residues of TM-align or it is the measure of the average distance between the atoms (usually the backbone atoms) of superimposed proteins; IDENa is the structurally related region's percentage sequence identity; Cov reflects the alignment range of the TM-alignment and is proportional to the sum by the length of query protein of structurally aligned residues. 5×58A: Prefusion structure of SARS-CoV spike glycoprotein, conformation 1 (viral protein); 6nzkA: Structural basis for human coronavirus attachment to sialic acid receptors (viral protein).
Table 2.
Enzyme Commission (EC) numbers and active sites.
| Rank | CscoreEC | PDB Hit | TM-score | RMSDa | IDENa | Cov | EC Number | Active Site Residues |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.082 | 1ileA | 0.205 | 8.54 | 0.041 | 0.296 | 6.1.1.5 | NA |
| 2 | 0.082 | 1k32A | 0.198 | 9.14 | 0.046 | 0.295 | 3.4.21.- | NA |
| 3 | 0.082 | 3eqIM | 0.201 | 8.65 | 0.046 | 0.288 | 2.7.7.6 | NA |
| 4 | 0.081 | 2pdaA | 0.184 | 10.13 | 0.027 | 0.295 | 1.2.7.1 | NA |
| 5 | 0.081 | 1ej6A | 0.190 | 10.21 | 0.016 | 0.309 | 2.7.7.50 | NA |
The 1ileA is the Isoleucyl-tRNA synthetase (aminoacyl-tRNA synthetase). The 1k32A is the Crystal structure of the tricorn protease (hydrolase). The 3eqlM is the Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with antibiotic myxopyronin (transferase). The 2pdaA is the Crystal structure of the complex between pyruvate-ferredoxin oxidoreductase from Desulfovibrio africanus and pyruvate (oxidoreductase). The 1ej6A is a Reovirus core (virus).
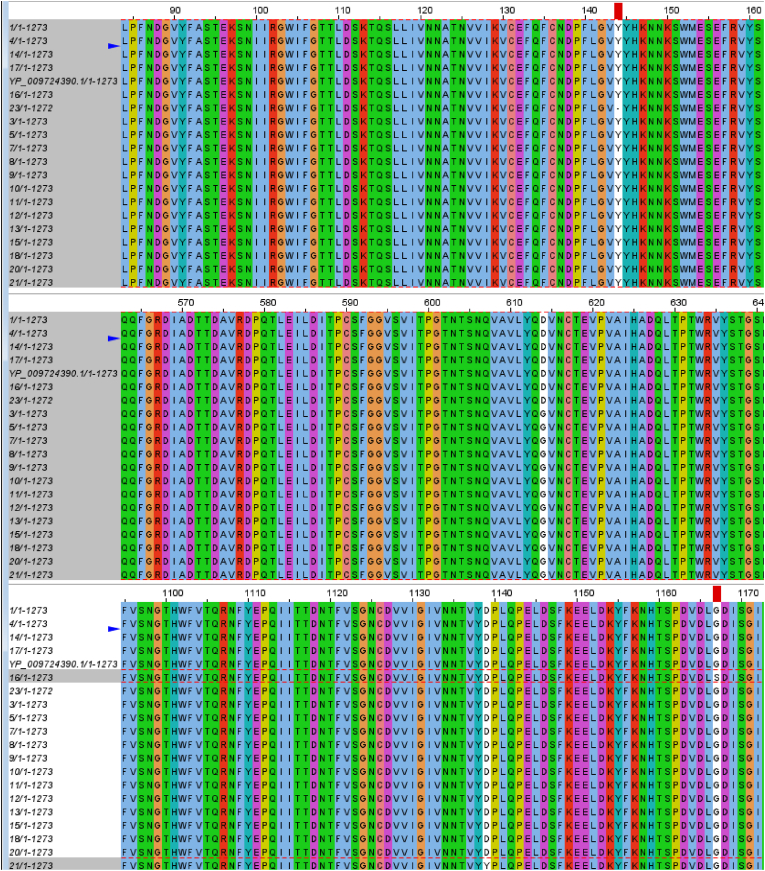
One powerful way of multiple sequence alignment is the Multiple Alignment using Fast Fourier Transform (MAFFT), as shown in Fig. 3 below [13]. Fig. 4 (a) shows the locations of two single amino acid variants, del Y144 & D614G, on spike protein. The del Y144 & D614G have relatively the highest mutation sensitivity scores and (b) shows the mutation sensitivity histograms for all four single amino acid variants on spike protein; Y 144, D 614, D 1139, and G 1167 calculated according to Ref. [17].
Fig. 3.
Multiple Sequence Alignment showing Amino Acid Variant (SAV) viewed by Jalview. The blue arrow and the red box are just cursers; the white columns are the location of the SAV along the full length of the amino acid sequence of spike protein for 20 SARS-CoV-2 samples from Jordan. YP-009724390 is the reference sequence of the spike protein of from Wuhan. The number 1273 is the total number of amino acids in the line. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Single amino acid variant (SAV), and mutation analysis
Our Phyre2 analysis has shown low mutational sensitivity in all variants. The deletion of Y144, D1139, and G1167, in addition to D614G has a low to neutral mutational sensitivity scores indicating a most likely neutral effect on the function of the spike protein. The D614G substitution was previously reported as a dominant mutation in interconnected Europe [16] and later worldwide.
3.4. Percentage of single amino acid variants (SAV) in spike protein
Four amino acid variants (SAV) from 20 samples of SARS-CoV-2 were found in this study. The first variant represented by 5% of samples that showed tyrosine deletion at Y144 located in the SARS-CoV-like_Spike_S1_NTD (N terminal domain). The second variant represented by 62% showed aspartate substitution to glycine at D614G located in the SARS-CoV-2_Spike_S1_RBD (spike recognition binding site). The third variant represented by 5% showed aspartate substitution to tyrosine at D1139Y and forth variant represented by 5% glycine substitution to serine at G1167S the last two located in the Corona_S2 domain. Table 3 shows that D614G has the highest mutation frequency. With a neutral mutation sensitivity, the D614G mutation showed the substitution of aspartate, a bulky amino acid, to glycine, the simplest amino acid.
Table 3.
SAV in spike-protein of SRAS-CoV-2; percentage and mutational sensitivity scores in a Jordanian population.
| SAV in spike-protein of SRAS-CoV-2 | SAV Percentage in a Jordanian population | (Phyre2) Mutational sensitivity Score out of (9 = high mutation sensitivity, 0 = low) | Impact of mutation on S protein |
|---|---|---|---|
| Deletion at Y144 | 5% | 3 | low to neutral |
| D614G | 62% | 5 | neutral |
| D1139Y | 5% | 1 | low |
| G1167S | 5% | 1 | low |
4. Discussion
In this study, we used spike gene sequences from 20 whole-genome sequences of SARS-CoV-2 collected from Jordan. The sequences retrieved from the GISAID database and analyzed at the amino sequence level of the spike glycoprotein versus the reference sequence of the surface glycoprotein [Severe acute respiratory syndrome coronavirus 2; (SARS-CoV-2)] which own the accession number YP_009724390.1 of Wuhan.
From PSIPRED analysis, the D614G mutation showed the substitution of aspartate, a coil amino acid, to glycine, an extracellular amino acid. The meaning of coil amino acid substitution to an extracellular amino acid might be consistent with a recently reported study by Korber et al. (2020), which included that the D614G substitution might change the viral conformational plasticity and hence a potential viral fitness gain [11]. Our PSIPRED results might agree with a recent report published by Henderson et al. (2020) in nature structural & molecular biology, which suggests that the D614G substitution could play a role in two conformational states, ‘up’ and ‘down.’ On the other hand, our mutation sensitivity analysis by Phyre2 for D614G spike protein is consistent with a newly published article, on the 20th of August 2020, in nature scientific reports by Sandra Isabel and her colleagues. The study of Isabel et al. highlighted an inference that the D614G clade might have a most likely neutral effect on the affinity of spike protein with its human ACE2 receptors [20]. However, we have recently reported in Non-coding RNA Research that the 1841A > G substitution at the viral genomic RNA level (D614G at spike protein level) showed many named microRNA sequences in human cells (e.g., hsa-miR-4793-5p to hsa-miR-3620-3p). These named miRNA sequences with an increased target binding score with viral RNA of spike protein from 91% to 92% by utilizing the miRDB database [21]. Our non-coding RNA findings support a new hypothesis of host non-coding RNA based response, which can directly bind and affect the viral RNA replication of the D614G spike protein.
All predicted three-dimensional structures of spike protein monomers of SARS-CoV-2 showed similar structures for all generated structures of the four amino acid variant (SAV). When comparing the predicted structures with the reference sequence of the spike glycoprotein YP_009724390.1 (SARS-CoV-2) versus FASTA sequences of spike glycoproteins from the Jordanian population, no significant change at the three-dimensional structure was noticed. The generated three-dimensional monomer structure of the spike protein of SARS-CoV-2 is consistent with a perfusion conformation structure reported in the literature [4]. The current in silico study is limited with the capabilities of all utilized servers and algorithms; for example, it is highly dependence on the initial templates used for calculations, so if the initial template scoring is low quality then this affect the final output files. At last, with taking the analysis output from all servers applied in this study we draw an inference that what makes the D614G meaningful for the COVID-19 pandemic still unclear and agrees with a previous study [10]; this is due to many reasons. First, a current lack of clinical metadata to link the viral mutations with viral phenotype. Second, lack of fair sample demographic distribution and last, lack of association between viral genomic data with the symptoms degree of severity.
5. Conclusion
Mutations and amino acid variants analysis are essential in understanding the mechanism of binding of spike protein with its receptor to have insights on possibilities to design a peptide or nucleotide-based vaccine for COVID-19. This study predicted the three-dimensional monomer structure of the spike glycoprotein from SARS-CoV-2 of Jordanian specimens. Here, we also reported four amino acid variants. However, the highest mutation frequency in our study, with 62% of samples, showed aspartate substitution to glycine at D614G is consistent with other reports for samples collected in Europe at the same time of our sample collection, in March 2020. In this study, the mutation D614G was the dominant local mutation in Jordan. We expected that the four reported amino acid variants, especially tyrosine deletion at Y144 located in the SARS-CoV-like_Spike_S1_NTD and the aspartate substitution to glycine at D614G located in the SARS-CoV-2_Spike_S1_RBD to have an expliciteffect on the function of the spike protein in the Jordanian population collectively. However, D614G might change the viral conformational plasticity and hence a potential viral fitness gain. These mutations are most likely low to neutral in their effect when looking primarily at the mutation analysis by the Phyre2, were tall red bars represent the highest favorable residues to have a phenotypic effect than shorter blue bars. Still, no tall red bars were noticed at all. It is highly recommended to keep monitoring the mutation rate of SARS-CoV-2 in Jordan on a monthly bases with a higher number of samples to fulfill a statistical power. Some of the low percentages appeared mutations, e.g., 5% might increase if the population size is larger so, in general, one must be cautious about drawing any concrete conclusions about the symptoms severity and viral transmission from genome sequences only. Altered glycosylation at the G614 was not considered in this study, but it could be a motivation in future studies.
Funding
“This research received no external funding.”
Author contributions
Walid Al-Zyoud & Hazem Haddad have contributed equally to Conceptualization, Methodology, Software, Data curation, Writing- Original draft preparation. Visualization, Investigation: Supervision, Software, Validation. Writing Reviewing and Editing. All authors have read and agreed to the published version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors acknowledge BioLab Diagnostic Laboratories (Jordan) & Andersen lab at Scripps Research (USA), who published sequences were retrieved from GISAID, a maintained global database based in Germany.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2020.100896.
Contributor Information
Walid Al-Zyoud, Email: walid.alzyoud@gju.edu.jo.
Hazem Haddad, Email: hazem_haddad1981@just.edu.jo.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020 Mar 12;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y.C., Kuo R.L., Shih S.R. COVID-19: the first documented coronavirus pandemic in history. Biomed. J. 2020;43(4):328–333. doi: 10.1016/j.bj.2020.04.007. Elsevier B.V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020 Feb 22;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. 10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wrapp Daniel, Wang Nianshuang, Corbett Kizzmekia S., Jory A., Goldsmith C.-L.H., Abiona Olubukola, Barney S., Graham J.S.M. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. - PubMed - NCBI. Science. 2020;13;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 Mar 30:1–6. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020 Apr 1;46(4):586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011 May 1;85(9):4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020 Apr 1;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henderson R, Edwards RJ, Mansouri K, Janowska K, Stalls V, Gobeil SMC, et al. Controlling the SARS-CoV-2 Spike Glycoprotein Conformation. [cited 2020 Aug 29]. [DOI] [PMC free article] [PubMed]
- 10.Grubaugh N.D., Hanage W.P., Rasmussen A.L. Making sense of mutation: what D614G means for the COVID-19 pandemic remains unclear. Cell. 2020 Aug 20;182(4):794–795. doi: 10.1016/j.cell.2020.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020 Aug 20;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abio Madeira F., Mi Park Y., Lee J., Buso N., Gur T., Madhusoodanan N. vol. 47. Web Serv issue Publ online; 2019. (The EMBL-EBI Search and Sequence Analysis Tools APIs in 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292(2):195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 15.Moult J., Fidelis K., Kryshtafovych A., Schwede T., Tramontano A. Critical assessment of methods of protein structure prediction (CASP) - round x. Proteins Struct Funct Bioinforma. 2014 Feb;82(SUPPL.2):1–6. doi: 10.1002/prot.24452. [Internet] [cited 2020 Sep 13] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Angyal A, Brown RL, Carrilero L, Green LR, Groves DC, Johnson KJ, et al. Spike Mutation Pipeline Reveals the Emergence of a More Transmissible Form of SARS-CoV-2 on Behalf of the Sheffield COVID-19 Genomics Group#, LaBranche CC2, and Montefiori DC2.
- 17.Yates C.M., Filippis I., Kelley L.A., Sternberg M.J.E. SuSPect: enhanced prediction of single amino acid variant (SAV) phenotype using network features. J. Mol. Biol. 2014 Jul 15;426(14):2692–2701. doi: 10.1016/j.jmb.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015 Jun 30;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 Apr 16;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isabel S., Graña-Miraglia L., Gutierrez J.M., Bundalovic-Torma C., Groves H.E., Isabel M.R. Evolutionary and structural analyses of SARS-CoV-2 D614G spike protein mutation now documented worldwide. Sci. Rep. 2020 Dec 20;10(1):14031. doi: 10.1038/s41598-020-70827-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haddad H., Al-Zyoud Walid. miRNA target prediction might explain the reduced transmission of SARS-CoV-2 in Jordan, Middle East. Non-coding RNA Res. 2020 Aug 1;5(3):135–143. doi: 10.1016/j.ncrna.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.