Highlights
-
•
CUP occurs in as many as 3–5% of patients when standard diagnostic tests are not able to determine the origin of cancer.
-
•
MI GPSai (Genomic Prevalence Score) is an AI that uses genomic and transcriptomic data to elucidate tumor origin.
-
•
The algorithm was trained on molecular data from 57,489 cases and validated on 19,555 cases.
-
•
MI GPSai predicted the tumor type out of 21 options in the labeled data set with an accuracy of over 94% on 93% of cases.
-
•
When also considering the second highest prediction, the accuracy increases to 97%.
Abstract
Cancer of Unknown Primary (CUP) occurs in 3–5% of patients when standard histological diagnostic tests are unable to determine the origin of metastatic cancer. Typically, a CUP diagnosis is treated empirically and has very poor outcomes, with median overall survival less than one year. Gene expression profiling alone has been used to identify the tissue of origin but struggles with low neoplastic percentage in metastatic sites which is where identification is often most needed. MI GPSai, a Genomic Prevalence Score, uses DNA sequencing and whole transcriptome data coupled with machine learning to aid in the diagnosis of cancer. The algorithm trained on genomic data from 34,352 cases and genomic and transcriptomic data from 23,137 cases and was validated on 19,555 cases. MI GPSai predicted the tumor type in the labeled data set with an accuracy of over 94% on 93% of cases while deliberating amongst 21 possible categories of cancer. When also considering the second highest prediction, the accuracy increases to 97%. Additionally, MI GPSai rendered a prediction for 71.7% of CUP cases. Pathologist evaluation of discrepancies between submitted diagnosis and MI GPSai predictions resulted in change of diagnosis in 41.3% of the time. MI GPSai provides clinically meaningful information in a large proportion of CUP cases and inclusion of MI GPSai in clinical routine could improve diagnostic fidelity. Moreover, all genomic markers essential for therapy selection are assessed in this assay, maximizing the clinical utility for patients within a single test.
Introduction
Carcinoma of Unknown Primary (CUP) represents a clinically challenging heterogeneous group of metastatic malignancies in which a primary tumor remains elusive despite extensive clinical and pathologic evaluation. CUPs comprise approximately 3–5% of cancer diagnoses worldwide [1] and efforts to secure a definitive diagnosis can prolong the diagnostic process and delay treatment initiation. Furthermore, CUP is associated with poor outcome which might be explained by use of suboptimal therapeutic interventions since there is general agreement that CUP tumors retain the biologic properties of the putative primary malignancy [1], [2]. Immunohistochemical (IHC) testing is the gold standard method to diagnose the site of tumor origin, especially in cases of poorly-differentiated or undifferentiated tumors. Meta-analysis of studies assessing the accuracy of IHC in challenging cases reported an accuracy of 60–70% in the characterization of metastatic tumors [3], [4], [5], [6]. Since therapeutic regimens are highly dependent upon diagnosis, this represents an important unmet clinical need. To address these challenges, assays aiming at tissue-of-origin (TOO) identification based on assessment of differential gene expression have been developed and tested clinically. However, integration of such assays into clinical practice is hampered by relatively poor performance characteristics (Table 1) and limited sample availability. Nevertheless, initial clinical studies demonstrate possible benefit of matching treatments to tumor types predicted by the assay [8]. With increasing availability of comprehensive molecular profiling assays, particularly next-generation DNA sequencing, genomic features have been incorporated in CUP treatment strategies [9]. While this approach rarely supports unambiguous identification of the TOO, it does reveal targetable molecular alterations in some patients [9].
Table 1.
Landscape of tissue of origin approaches.
| Assay | Cancer Categories | N Independent Test Set | Accuracy | Cases Called |
|---|---|---|---|---|
| (%) | (%) | |||
| Caris MI GPSai | 21 | 13,661 | 94.7 | 93 |
| 2020 | ||||
| PCAWG | 14 | 1436 | 88 | 100 |
| 2020 [32] | ||||
| MSK IMPACT | 22 | 11,644 | 74.1 | 100 |
| 2019 [10] | ||||
| Cancer Genetics Tissue of Origin | 9 | 27 | 94.1 | 89 |
| 2012 [11] | ||||
| Biotheranostics CancerTYPE ID | 30 | 187 | 83 | 100 |
| 2011 [7] | ||||
| Park SY | 7 | 60 | 75 | 78 |
| 2007 [5] | ||||
| Dennis JL | 7 | 130 | 88 | 100 |
| 2005 [12] | ||||
| Brown RW | 5 | 128 | 66 | 86 |
| 1997 [6] | ||||
| Gamble AR | 14 | 100 | 70 | 100 |
| 1993 [13] |
In the work presented here we pursued a different strategy of TOO identification by utilizing a novel machine-learning approach to build TOO classifiers based on data from a large next-generation DNA sequencing panel in conjunction with data from whole transcriptome sequencing, which are both used broadly for routine molecular tumor profiling. We show that this computational classification system identified TOO at an accuracy significantly exceeding that of other currently available technologies. Moreover, this assay simultaneously determines the presence of genetic abnormalities that guide treatment selection, thus generating substantially increased clinical utility in a single test.
Methods
Next-Generation Sequencing (NGS) - DNA
Genomic DNA isolated from formalin-fixed paraffin-embedded (FFPE) tumor samples was microdissected to enrich tumor purity and subjected to NGS using the NextSeq platform (Illumina, Inc., San Diego, CA). FFPE specimens underwent pathology review to measure percent tumor content and tumor size; a minimum of 20% of tumor content in the area for microdissection was required to enable enrichment and extraction of tumor-specific DNA. Matched normal tissue was not sequenced. A custom-designed SureSelect XT assay was used to enrich 592 or whole exome whole-gene targets (Agilent Technologies, Santa Clara, CA). All variants were detected with > 99% confidence based on allele frequency and probe panel coverage, with an average sequencing depth of coverage of > 500 and an analytic sensitivity of 5%. Prior to molecular testing, tumor enrichment was achieved by harvesting targeted tissue using manual microdissection techniques. Genetic variants identified were interpreted by board-certified molecular geneticists and categorized as ‘pathogenic,’ ‘presumed pathogenic,’ ‘variant of unknown significance,’ ‘presumed benign,’ or ‘benign,’ according to the American College of Medical Genetics and Genomics (ACMG) standards. When assessing mutation frequencies of individual genes, ’pathogenic,’ ‘presumed pathogenic,’ and ‘variants of unknown significance’ were counted as mutations while ‘benign’ and ‘presumed benign’ variants were excluded. Copy number alteration (CNA) was tested by NGS and was determined by comparing the depth of sequencing of genomic loci to a diploid control as well as the known performance of these genomic loci. Calculated gains of 6 copies or greater were considered amplified.
Next-Generation Sequencing (NGS) - RNA
FFPE specimens underwent pathology review to measure percent tumor content and tumor size; a minimum of 20% of tumor content in the area for microdissection was required to enable enrichment and extraction of tumor-specific RNA. Qiagen RNA FFPE tissue extraction kit was used for extraction, and the RNA quality and quantity were determined using the Agilent TapeStation. Biotinylated RNA baits were hybridized to the synthesized and purified cDNA targets and the bait-target complexes were amplified in a post capture PCR reaction. The Illumina NovaSeq 6500 was used to sequence the whole transcriptome from patients to an average of 60 M reads. Raw data was demultiplexed by Illumina Dragen BioIT accelerator, trimmed, counted, PCR-duplicates removed and aligned to human reference genome hg19 by STAR aligner [14]. For transcription counting, transcripts per million molecules was generated using the Salmon expression pipeline [15].
RNA expression
RNA expression, as defined by transcripts per million (TPM) from the Salmon RNA expression pipeline [15] using the Caris Whole Transcriptome Assay, was validated using IHC results from over 5000 human breast adenocarcinoma cases. Protein amounts were measured by FDA-approved antibodies using standard quantitative IHC assays. IHC scores come directly from histopathology review by board-certified pathologists for ER/ESR1 (human estrogen receptor), PR/PGR (human progesterone receptor), AR (human androgen receptor), and HER2/neu/ERBB2 (human Herceptin, receptor tyrosine kinase CD340). 50 IHC 'positive' and 50 IHC 'negative' cases were used to decide the TPM thresholds corresponding to IHC positive and IHC negative for these 4 genes. The thresholds were evaluated on 5197 independent cases and all four markers had a sensitivity > 86% with specificities ranging from 85% to 99%. Full validation results are shown in Supplementary Table S1 and Supplementary Fig. S1.
Additionally, we compared data between the Caris WTS expression assay to the Illumina DASL Expression Microarray and publicly available Affymetrix U133A expression arrays from the expO project (Gene Expression Omnibus accession GSE2109) in a cross-platform comparison method [33]. We selected 10 cases from each dataset from a diagnosed Stage IV uterine carcinoma and 10 cases diagnosed with Stage IV colon adenocarcinoma. We identified 14,473 genes which are common across these three platforms. Although these cases are from different people, we propose that the gene expression profiles from uterine tumors and colon tumors are sufficiently different from each other and sufficiently common within a tumor type that common patterns of over- and under-expression can be seen. To best visualize this, we took the log2 ratio of all 14,473 genes between uterine (numerator) and colon (denominator) cancer and plotted those ratios between panel A {Caris (X axis) and Illumina (Y axis)}, panel B {Illumina (X axis) and Affymetrix (Y axis)}, and panel C {Caris (X axis) and Affymetrix (Y axis)}. Supplementary Fig. S2 shows the ratios plotted against each other with R2 listed in panels A (0.468), B (0.565) and C (0.528). Note that the expression data was averaged across 10 patients. The Pearson's correlation coefficient for each is 0.68, 0.75 and 0.73 respectively.
Compliance statement
This study was conducted in accordance with guidelines of the Declaration of Helsinki, Belmont report, and U.S. Common rule. In keeping with 45 CFR 46.101(b)(4), this study was performed utilizing retrospective, deidentified clinical data. Therefore, this study is considered IRB exempt (WIRB Work Order # 1-1182870-1).
Data statement
Due to the size of the raw data and concerns for patient privacy, raw sequencing data is not available. However, summarized GPS data is available upon request.
Results
Patients
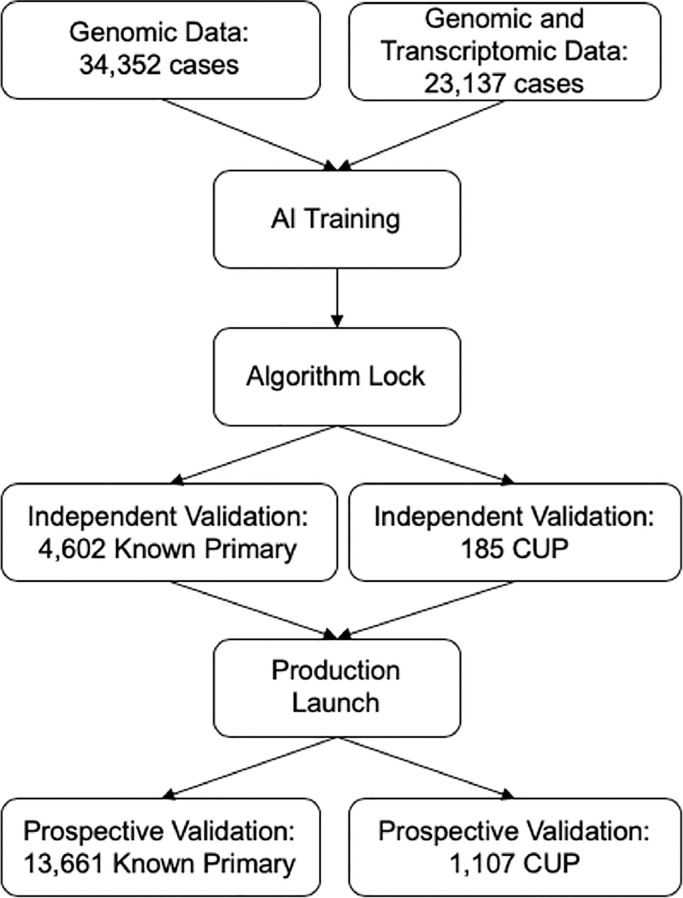
Using the Caris Molecular Database which includes tumors analyzed at the Caris Life Sciences laboratory from 2008 to 2020, we identified 77,044 cases that had next-generation DNA and RNA sequencing results with an available pathology diagnosis including CUP. CUP cases were defined as those assigned a primary tumor site of “Unknown primary site” and for which the “Cancer of Unknown Primary” lineage was selected by the submitting site. The submitted pathological diagnosis was used as the training label. Subsequent independent validation of the classifier was accomplished by including 13,661 cases with a known primary and 1,107 CUP cases that were analyzed prospectively as part of routine tumor profiling (Fig. 1).
Fig. 1.
CONSORT diagram. The DNA and RNA components of MI GPSai were trained using a combined 57,489 patients, which were then validated on 4,602 non-CUP and 185 CUP patients to determine optimal performance settings. Following this evaluation, MI GPSai rendered a prediction on routinely profiled cases resulting in the final prospective validation set and CUP cases.
Artificial intelligence training
Molecular profiles from 57,489 patients were used for initial training of the global tumor classification algorithm designated MI GPSai. This dataset was comprised of 34,352 cases with genomic data and 23,137 with both genomic and transcriptomic data. MI GPSai was generated using an artificial intelligence platform that leverages the Caris Deliberation Analytics (DEAN) framework. DEAN uses biomarker data as feature inputs into an ensemble of over 300 well-established machine learning algorithms, including random forest, support vector machine, logistic regression, K-nearest neighbor, artificial neural network, naïve Bayes, quadratic discriminant analysis, and Gaussian processes models. Multiple feature selection methods were employed to build models along with 5-fold cross validation during training to assess performance. High-performing models deliberate against one another to determine a final result. A set of 115 distinct primary tumor site and histology classes were defined and used to generate subpopulations of patients (Supplementary Table S2). For training the Genomic Profiling Similarity (GPS), all 115 disease types were trained against each other using the training set to generate 6,555 model signatures, where each signature is built to differentiate between a pair of disease types. The signatures were generated using Gradient Boosted Forests. The models were validated using the test cases where each test case was processed individually through all 6,555 signatures, thereby providing a pairwise analysis between every disease type for every case. The results are analyzed in a 115 × 115 matrix where each column and each row is a single disease type and the cell at the intersection is the probability that a case is one disease type or the other. The probabilities for each disease type are summed for each column which results in 115 disease types with their probability sums. These disease types are ranked by their probability sums. The disease types were further grouped into 21 broad cancer categories in order to achieve the highest predictive power for a clinically relevant category that would assist with therapy selection in challenging cases. A higher resolution of tumors such as luminal vs basal breast cancer will be performed in future studies in situations when such distinctions are clinically relevant. A total of 6,559 models were generated and used to determine a final probability (MI GPS Score) for each case belonging to each of the cancer categories.
These MI GPS Scores were then clustered into multidimensional signatures which are empirically evaluated in the Caris Molecular Database to determine the predicted prevalence in each cancer category. The prevalence is the final output of the MI GPSai machine learning platform.
The machine learning approach leveraged molecular features from 592 genes and over 62,000 mRNA transcripts for each of the cancer categories. The top DNA and RNA features that contribute the largest amount of information to predictions made for each of the 21 cancer categories are shown in Supplementary Fig. S3. Key canonical driver mutations are well-represented as the top contributing biomarkers. Examples include IDH1 and EGFR for gliomas, cKIT/PDGFRA in gastrointestinal stromal tumors (GIST), BRAF/NRAS in melanoma, KRAS/CDKN2A in pancreatic cancer, GATA3 and CDH1 in breast cancer, VHL in renal cell carcinoma, BRAF in thyroid, PTEN in endometrial cancer, and FOXL2 in ovarian granulosa cell tumors [16], [17], [18], [19], [20], [21]. Expression of genes relatively specific to tissue lineage are also among the top contributors, e.g., CDX2 in gastroesophageal cancer, KIT in GIST, MITF in melanoma and NKX3–1 in prostate cancer [22], [23], [24], [25]. As only markers that are most useful for differentiating cancer categories are found in these lists, canonical markers such as BRCA1 in Breast Adenocarcinoma are not in the top 10 for the machine learning for Breast as BRCA1 is found in a number of cancer categories. Additional biomarkers that haven't been explicitly associated with the particular cancer types are also included in the algorithm, revealing previously uncovered linkages with biomarkers and pathways that warrants further exploration. Additional details of the machine learning configurations and inputs are described here [26].
Validation of algorithmic disease classification in independent cohorts
Following the lock of the algorithm, predictions made by the MI GPSai platform were first validated in an independent set of 4,602 patients with known cancer category and 185 patients with CUP. MI GPSai provided a top prediction for each case along with a score related to the confidence in the call. When evaluating the MI GPSai top prediction on every case in the cohort irrespective of the score, the top prediction was concordant with the pathologist-assigned disease type in 90.3% of cases. An assessment of the scores in this dataset led us to select 0.835 as a minimum score required to report a result as it was the intersection of accuracy of the top prediction and the call rate (percentage of cases resulted), resulting in 93.3% accuracy on 93.3% of cases with a defined primary and 75.6% of CUP cases (Supplementary Fig. S4). At this threshold, the assay was robust within both primary and metastatic tumors as well as various ranges of tumor purity (Table 2).
Table 2.
Summary of performance in the independent validation cohort at the selected threshold.
| Category | n | Call Rate (%) | Sensitivity (%) |
|---|---|---|---|
| Global | 4602 | 93.3 | 93.3 |
| Primary Specimen | 2544 | 94 | 94.1 |
| Metastatic Specimen | 1969 | 92.2 | 92.5 |
| Percent Tumor >= 20, <= 50 | 2885 | 92.7 | 93.4 |
| Percent Tumor > 50, <= 80 | 1657 | 94.1 | 93.1 |
| Percent Tumor > 80 | 54 | 100 | 100 |
Prospective validation
Subsequently, the assay was used in clinical testing to automatically prospectively evaluate the tumor of each patient profiled at Caris Life Sciences. Pathologists were notified of the MI GPSai score and empirical prevalence tables if the assay returned a MI GPSai Score of >= 0.835 for any cancer category. The tumors of 13,661 non-CUP patients were evaluated by the algorithm as a prospective validation cohort (Table 3). Globally, this cohort exhibited a similar call rate compared to the initial independent validation cohort (93.0% vs 93.3%) and exhibited a higher sensitivity (94.7% vs 93.3%). The sensitivity of the assay remained above 93% in both primary and metastatic tumors regardless of tumor purity (Table 3).
Table 3.
Summary of algorithm performance in the prospective validation cohort.
| Category | n | Above Threshold | Call Rate (%) | Sensitivity in Top 1 (%) | Sensitivity in Top 2 (%) | Sensitivity in Top 3 (%) | Sensitivity in Top 4 (%) | Sensitivity in Top 5 (%) | Rule Outs / Case | Rule Out Accuracy (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Global | 13,661 | 12,699 | 93 | 94.7 | 97.2 | 97.9 | 98.1 | 98.2 | 17.6 | 99.9 |
| Primary Specimen | 7521 | 7087 | 94.2 | 96.1 | 98.2 | 98.7 | 98.8 | 98.9 | 17.8 | 100 |
| Metastatic Specimen | 5942 | 5426 | 91.3 | 93 | 96 | 97 | 97.2 | 97.4 | 17.4 | 99.9 |
| Percent Tumor < 20 | 4 | 3 | 75 | 100 | 100 | 100 | 100 | 100 | 18.7 | 100 |
| Percent Tumor >= 20, <= 50 | 8227 | 7636 | 92.8 | 94.5 | 97 | 97.8 | 97.9 | 98 | 17.4 | 99.9 |
| Percent Tumor > 50, <= 80 | 5189 | 4835 | 93.2 | 95 | 97.7 | 98.2 | 98.4 | 98.5 | 17.9 | 100 |
| Percent Tumor > 80 | 241 | 225 | 93.4 | 96 | 96.4 | 96.4 | 96.4 | 96.9 | 18 | 99.9 |
This prospective dataset also allowed us to evaluate the diagnostic rule-out power (i.e., negative predictive value) of the assay. For all patients, the empirical prevalence tables yielded an average of 17.6 cancer categories that had not been observed per patient (i.e., could be ruled out) for their respective MI GPSai scores. The correct cancer category had a non-zero empirical probability in 98.9% of all cases, and the 1.1% of observations in which the true cancer category was incorrectly ruled out represents less than 0.1% of the total disease types ruled out. Thus, the rule out accuracy exceeds 99.9%.
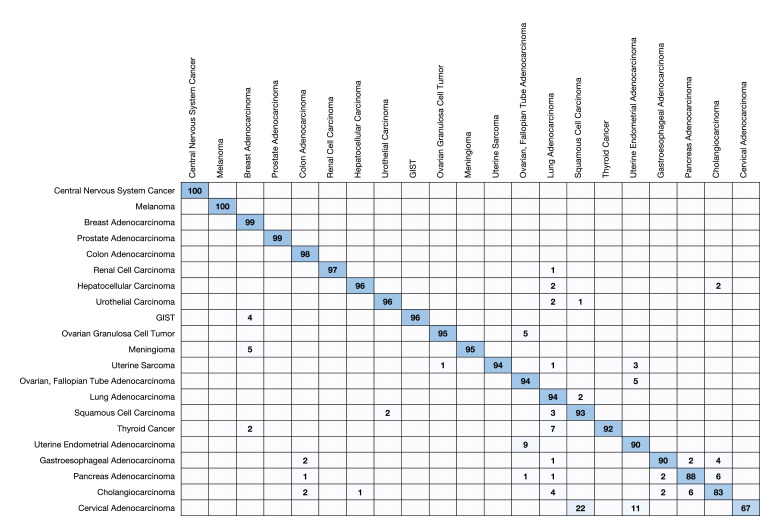
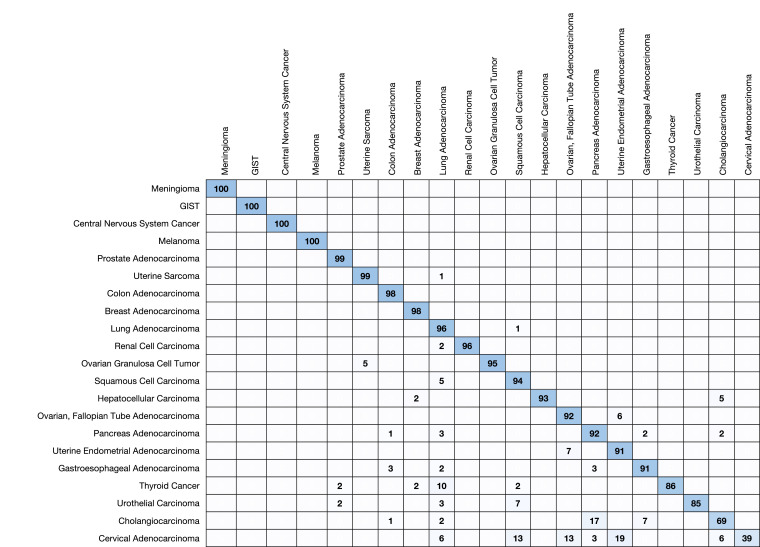
Each of the 21 cancer categories was represented in the prospective validation dataset both with respect to true tumor type and highest prediction (Table 4). Sixteen of the 21 cancer categories had an observed positive predictive value (PPV) of >= 90% and three had a PPV of >= 99%. Strikingly, the minimum rule-out accuracy is 98.0%. Five cancer categories (e.g. central nervous system cancers, GIST, melanoma, meningioma, and prostate) each exhibited > 99% sensitivity while twelve (e.g., breast, colon, gastroesophageal, hepatocellular, lung, two subtypes of ovarian, pancreatic, renal, squamous cell, uterine adenocarcinoma, and uterine sarcoma) more achieved at least 90% sensitivity. We show confusion matrices with respect to prediction and truth for the cancer categories in Fig. 2, Fig. 3, respectively.
Table 4.
Summary of algorithm performance in the prospective validation cohort by cancer category.
| Category | n | Call Rate (%) | Sensitivity (%) | PPV (%) | Rule Out Accuracy (%) |
|---|---|---|---|---|---|
| Breast Adenocarcinoma | 1533 | 98 | 98.4 | 99 | 100 |
| Central Nervous System Cancer | 445 | 99.8 | 99.8 | 100 | 100 |
| Cervical Adenocarcinoma | 60 | 51.7 | 38.7 | 66.7 | 98 |
| Cholangiocarcinoma | 363 | 73.8 | 69.4 | 83 | 99.7 |
| Colon Adenocarcinoma | 2119 | 97 | 98.5 | 98.2 | 100 |
| Gastroesophageal Adenocarcinoma | 613 | 84.5 | 90.9 | 89.5 | 99.9 |
| GIST | 23 | 95.7 | 100 | 95.7 | 100 |
| Hepatocellular Carcinoma | 66 | 84.9 | 92.9 | 96.3 | 99.7 |
| Lung Adenocarcinoma | 2287 | 95 | 96.4 | 93.6 | 100 |
| Melanoma | 373 | 96.5 | 99.7 | 99.7 | 100 |
| Meningioma | 21 | 90.5 | 100 | 95 | 100 |
| Ovarian Granulosa Cell Tumor | 25 | 88 | 95.5 | 95.5 | 100 |
| Ovarian, Fallopian Tube Adenocarcinoma | 1493 | 91.6 | 92.5 | 94.3 | 99.9 |
| Pancreas Adenocarcinoma | 815 | 87.6 | 91.9 | 87.7 | 100 |
| Prostate Adenocarcinoma | 556 | 97.1 | 99.1 | 98.7 | 100 |
| Renal Cell Carcinoma | 176 | 92.6 | 95.7 | 96.9 | 99.8 |
| Squamous Cell Carcinoma | 1193 | 93 | 93.5 | 93.4 | 99.9 |
| Thyroid Cancer | 74 | 85.1 | 85.7 | 91.5 | 99.2 |
| Urothelial Carcinoma | 354 | 90.7 | 85.4 | 96.1 | 99.9 |
| Uterine Endometrial Adenocarcinoma | 989 | 89.4 | 91.4 | 89.7 | 100 |
| Uterine Sarcoma | 83 | 83.1 | 98.6 | 94.4 | 100 |
Fig. 2.
Prediction matrix in the prospective validation set. Each row shows the percentage of the actual disease types observed when a MI GPSai achieves a score > 0.835. The diagonal represents the PPV for the given disease type. Blank cells have values between 0 and 1.
Fig. 3.
Confusion matrix in the prospective validation set. Each column shows observed predictions for each disease type when a MI GPSai achieves a score > 0.835. The diagonal represents the sensitivity for the given disease type. Blank cells have values between 0 and 1.
Analysis of CUP
Of the 1292 CUP cases analyzed by MI GPSai, 71.7% achieved a score exceeding the reportable threshold (Supplementary Fig. S5). Validation of a CUP assay at the individual patient level is fundamentally an impossible task as the “truth” is unknown. As such, comparing the populations generated by MI GPSai for each cancer category in terms of mutation frequencies against the mutation frequencies in populations of known primaries yields insight into the similarities of these populations. The genes with mutation frequencies with a 95% confidence interval which does not overlap with that of any other cancer category along with their frequencies in the populations created by MI GPSai can be seen in Supplementary Table S3. Many of the pathogenic mutation frequencies were similar in the labeled and CUP predicted populations, but not all. In particular, VHL pathogenic mutations were not seen in the 18 CUP cases classified as Renal Cell Carcinoma. This could potentially be due to lower proportions of clear cell carcinoma in CUP [27].
Clinical utility and case examples
For CUP cases, MI GPSai was able to assign a diagnosis in 71.7% of cases. The RNA expression data provide a guide for IHC selection in order to confirm cases. For example, we received an inguinal lymph node biopsy on an 82-year-old man which was sent for molecular profiling. At the time of biopsy, the serum PSA was not elevated, and workup had not identified the primary tumor. Evaluation by the referring pathologist included negative IHC stains with CK7, CK20, PSA, PSAP, CDX2, p40, GATA3, SOX10, and CD45. A cytokeratin stain was positive (AE1/3) and case was diagnosed as carcinoma of unknown primary. Notably, this carcinoma was evaluated appropriately for prostatic lineage with PSA and PSAP IHC, and given the concurrent low serum PSA, prostatic adenocarcinoma was considered ruled out.
MI GPSai predicted that this was prostate adenocarcinoma (MI GPSai score 0.9998) and review of the gene expression data showed high expression of androgen receptor (AR) which guided IHC selection. AR protein was highly expressed which supported the MI GPSai call. Importantly, the molecular profiling also identified pathogenic variants in BRCA2 and PTEN, highlighting the utility of diagnosis and biomarker analysis from the same platform. The patient had a follow-up biopsy of the prostate which confirmed prostatic adenocarcinoma. After discussion with the ordering physician, the diagnosis was changed from CUP to metastatic prostatic adenocarcinoma.
In addition to assigning lineage and identifying biomarker data with CUP cases, MI GPSai also can assist with improving pathologic diagnosis fidelity. We prospectively monitored discrepancies between MI GPSai and the pathologist-assigned diagnoses in 1292 cases. In cases where the pathologist-assigned diagnosis was different than the top MI GPSai prediction and the MI GPSai score for the top prediction exceeded 0.999, an automated email was sent to the pathologist in charge of the case alerting them to this discrepancy. The pathology group was previously educated on the design and performance of MI GPSai and instructed to consider the discrepant cases with their medical judgement. The pathologists were able to review patient clinical history, imaging results if available, order immunohistochemistry, and discuss the case with the referring oncologist and/or pathologist.
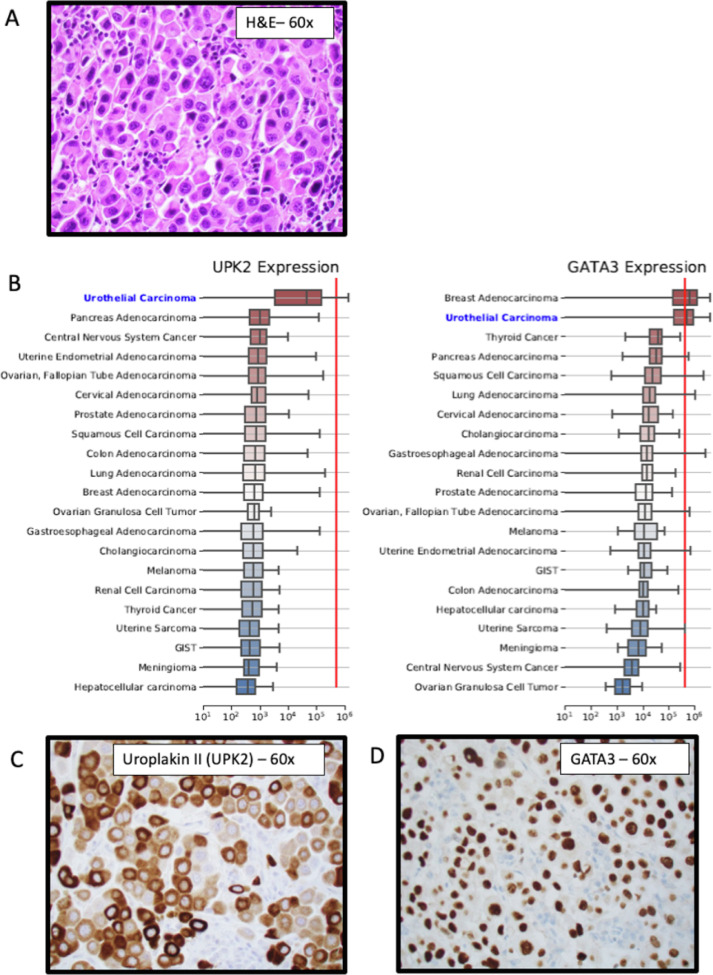
There were 46 cases with a MI GPSai score greater than 0.999 where pathologists were alerted. After review with additional immunohistochemistry and consultation with the referring physician, the diagnosis was changed in 19 cases (41.3%). In 11 cases (23.9%), where the submitted diagnosis was not changed despite MI GPSai predictions, the predicted diagnosis was pancreatic adenocarcinoma, a cancer with limited specific IHC markers for confirmation. All cases did not result in a diagnosis revision for various reasons ranging from a lack of diagnostic IHCs to verify the prediction (such as cholangiocarcinoma vs pancreatic carcinoma) to a lack of response from the oncologist. In one important example, the treatment course was altered. We received a cervical lymph node from a 61-year-old man for molecular profiling (Fig. 4). The referring pathologist assigned a diagnosis of poorly-differentiated squamous cell carcinoma (Fig. 4A). The patient had systemic metastasis and had not responded well to squamous cell carcinoma directed therapy. The MI GPSai predicted diagnosis was urothelial carcinoma (MI GPSai score 0.9999), and additional immunohistochemical workup was pursued based on the results of RNA expression profiling. This additional IHC was positive for Uroplakin II and GATA3 - both relatively specific for urothelial carcinoma and not typically expressed in squamous cell carcinoma (Fig. 4). Importantly, the choice of the PD-L1 clone and scoring system was affected by the lineage of cancer being tested. In this case, the referring pathologist and oncologist asked to change the diagnosis to urothelial carcinoma and run the SP142 PD-L1 antibody according to the label indications for atezolizumab. This PD-L1 score was positive, the patient therapy changed, and the patient response will be tracked in our prospective study. In sum, MI GPSai has significant clinical utility with CUP and diagnostic fidelity.
Fig. 4.
A clinical example showing a representative case in which the pathological diagnosis was reassigned based on MI GPSai predictions using Whole Exome and Whole Transcriptome Sequencing (WES, WTS) data. (A) Molecular profiling was performed using WES and WTS data that was then routed into the MI GPSai pipeline for diagnostic predictions. (B) The whole transcriptome expression data was then used to select for lineage specific gene expression to guide immunohistochemical antibody selection, the current gold-standard for lineage assignment. In the example provided, the mean RNA expression of Uroplakin II and GATA3 of the urothelial carcinoma cases in our database is relatively high (box plots). With the specimen being considered (red line), Uroplakin II and GATA3 RNA expression high. (C) and (D) Immunohistochemical evaluation of the tumor with clinically validated antibodies against Uroplakin II and GATA3 confirmed lineage specific protein expression diagnostic of urothelial carcinoma. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Discussion
Cancer of unknown primary remains a major clinical challenge and outcomes are poor. Molecular predictors of tumor origin can assist in addressing this problem by providing critical information in CUP cases that can inform treatment decisions and potentially improve outcomes. We herein took advantage of a large collection of molecular and pathology data to develop MI GPSai which is, to our knowledge, the first artificial intelligence-derived molecular classifier that utilizes DNA and RNA information to make tumor type predictions across a broad spectrum of diagnostic classes with high accuracy.
Up to this point, development of molecular assays for the identification of cancers of unknown primary has been focused on utilizing RNA profiles which have degraded performance in situations where the tumor is from a site of metastasis or if the tumor percentage is low [7]. Our approach overcomes these limitations since nucleic acid is isolated from microdissected material, thus enriching for tumor cells. The combined analysis of DNA and RNA further reduces susceptibility to the effects of normal cell contamination. As demonstrated in our case examples, availability of mutational and gene expression analysis data further enhances the clinical utility of our approach from a diagnostic and therapeutic perspective.
The accuracy of MI GPSai compares favorably to recent data on the use of DNA NGS panels for tissue of origin identification or guidance of utilization of targeted- and immunotherapies [10], [28]. However, overall accuracy of these approaches is limited. For example, predictions made by a Random Forrest Classifier using results from a 468-gene NGS panel as input, resulted in an overall accuracy of 74.1% [10]. Analysis of circulating tumor DNA data from a commercial 70-gene NGS panel revealed potentially targetable mutations. However, an attempt to identify the underlying TOO was not made [28], possibly due to the limited number of genes analyzed. In contrast, analysis of DNA methylation across the genome might add additional information to above-mentioned assays, as it has been shown to predict a primary tumor in 87% of CUP cases [29].
In addition to its role in understanding CUP, MI GPSai functions also as an outstanding quality control tool when integrated into the pathology laboratory workflow. As part of our prospective evaluation of MI GPSai, pathologists were alerted to discrepancies between submitted diagnosis and MI GPSai prediction, resulting in change in diagnosis in 41.3% of these cases. Considering that the rate of inaccurate diagnosis ranges between 3% and 9% [30], inclusion of MI GPSai in clinical routine could improve diagnostic fidelity overall.
A current limitation of MI GPSai is that it has not yet been trained on certain classes of tumors that need to be evaluated in a traditional pathologic workup. If the differential diagnostic considerations for any particular cancer include non-uterine sarcoma or hematologic malignancy, MI GPSai is not indicated. Limitations to training a model to accurately predict a diagnosis include identification of a sufficient cohort of correctly annotated cases. As data accumulates from routine molecular profiling for predictive biomarker evaluation, additional diagnostic categories will be incorporated into MI GPSai. This highlights our approach that MI GPSai is a diagnostic tool that parses data to be considered by the pathologist in the overall context of the case. Other traditional pieces of information important for rendering a diagnosis such as the clinical, laboratory, and imaging information still maintain salience. Future versions of the tool will be trained on well-characterized cohorts of sarcoma and hematologic malignancy and will also incorporate information from digital pathology image analysis [31] that will further enhance the accuracy of the algorithm.
In summary, MI GPSai displayed robust performance in the diagnostic workup of CUP cases that was consistent across 13,661 cases including both metastatic and low percentage tumors. At the same time, MI GPSai can also play an important role in quality control of anatomical pathology laboratories. Since the MI GPSai analysis uses the results of DNA and RNA profiles obtained as part of routine clinical tumor profiling, both diagnostic and therapeutic information can be returned that optimize patients’ treatment strategy from a single test. This is a substantial improvement over the current standard of multiple tests that require more tissue and increased turnaround time which can delay treatment. Prospective clinical studies are planned to assess the impact of MI GPSai on treatment decisions and outcomes. Other prospective clinical trials are ongoing (CUPISCO; NCT03498521) or completed (GEFCAPI 4; NCT01540058) that evaluate the clinical impact of molecular diagnostic testing on patient outcomes in CUP, but they either do not include an attempt to assign a cancer lineage or have failed to show significant difference between the empiric or cancer-directed therapy. Our approach aims to utilize the context-specific information gained by lineage assignment when considering biomarker-directed therapy.
Funding
This work was supported by Caris Life Sciences.
Disclosures
JA, AH, JX, DM, CN, SA, CJ, MO, WMK and DS are employees of Caris Life Sciences. ABH and JM serve on the scientific advisory board of Caris Life Sciences. E.I. Heath has an unpaid consultant/advisory board relationship with Caris Life Sciences.
CRediT authorship contribution statement
Jim Abraham: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Validation, Visualization, Writing - original draft, Writing - review & editing. Amy B. Heimberger: Writing - review & editing. John Marshall: Writing - review & editing. Elisabeth Heath: Writing - review & editing. Joseph Drabick: Investigation, Methodology. Anthony Helmstetter: Methodology, Software, Visualization, Writing - review & editing. Joanne Xiu: Data curation, Formal analysis, Writing - original draft, Writing - review & editing. Daniel Magee: Formal analysis, Writing - original draft, Writing - review & editing. Phillip Stafford: . Chadi Nabhan: Writing - review & editing. Sourabh Antani: Software, Writing - original draft. Curtis Johnston: Data curation, Investigation, Methodology. Matthew Oberley: Data curation, Formal analysis, Investigation, Methodology, Supervision, Validation, Writing - original draft, Writing - review & editing. Wolfgang Michael Korn: Formal analysis, Investigation, Validation, Writing - original draft, Writing - review & editing. David Spetzler: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank Brady Gilg for his work on the expression pipeline and Erin Morgan for her work on deliberation strategies.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101016.
Appendix. Supplementary materials
References
- 1.Massard C., Loriot Y., Fizazi K. Carcinomas of an unknown primary origin–diagnosis and treatment. Nat. Rev. Clin. Oncol. 2011;8(12):701–710. doi: 10.1038/nrclinonc.2011.158. [DOI] [PubMed] [Google Scholar]
- 2.Varadhachary G.R., Raber M.N. Cancer of unknown primary site. N. Engl. J. Med. 2014;371(8):757–765. doi: 10.1056/NEJMra1303917. [DOI] [PubMed] [Google Scholar]
- 3.DeYoung B.R., Wick M.R. Immunohistologic evaluation of metastatic carcinomas of unknown origin: an algorithmic approach. Semin. Diagn. Pathol. 2000;17(3):184–193. [PubMed] [Google Scholar]
- 4.Anderson G.G., Weiss L.M. Determining tissue of origin for metastatic cancers: meta-analysis and literature review of immunohistochemistry performance. Appl. Immunohistochem. Mol. Morphol. 2010;18(1):3–8. doi: 10.1097/PAI.0b013e3181a75e6d. [DOI] [PubMed] [Google Scholar]
- 5.Park S.Y., Kim B.H., Kim J.H., Lee S., Kang G.H. Panels of immunohistochemical markers help determine primary sites of metastatic adenocarcinoma. Arch. Pathol. Lab. Med. 2007;131(10):1561–1567. doi: 10.5858/2007-131-1561-POIMHD. [DOI] [PubMed] [Google Scholar]
- 6.Brown R.W., Campagna L.B., Dunn J.K., Cagle P.T. Immunohistochemical identification of tumor markers in metastatic adenocarcinoma. A diagnostic adjunct in the determination of primary site. Am. J. Clin. Pathol. 1997;107(1):12–19. doi: 10.1093/ajcp/107.1.12. [DOI] [PubMed] [Google Scholar]
- 7.Erlander M.G., Ma X.J., Kesty N.C., Bao L., Salunga R., Schnabel C.A. Performance and clinical evaluation of the 92-gene real-time PCR assay for tumor classification. J. Mol. Diagn. 2011;13(5):493–503. doi: 10.1016/j.jmoldx.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hainsworth J.D., Rubin M.S., Spigel D.R. Molecular gene expression profiling to predict the tissue of origin and direct site-specific therapy in patients with carcinoma of unknown primary site: a prospective trial of the Sarah Cannon research institute. J. Clin. Oncol. 2013;31(2):217–223. doi: 10.1200/JCO.2012.43.3755. [DOI] [PubMed] [Google Scholar]
- 9.Ross J.S., Wang K., Gay L. Comprehensive genomic profiling of carcinoma of unknown primary site: new routes to targeted therapies [published correction appears in JAMA Oncology. JAMA Oncol. 2015;1(1):40–49. doi: 10.1001/jamaoncol.2014.216. 2019 Aug 1;5(8):1232] [DOI] [PubMed] [Google Scholar]
- 10.Penson A., Camacho N., Zheng Y. Development of genome-derived tumor type prediction to inform clinical cancer care. JAMA Oncol. 2019;6(1):84–91. doi: 10.1001/jamaoncol.2019.3985. [published online ahead of print, 2019 Nov 14] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stancel G.A., Coffey D., Alvarez K. Identification of tissue of origin in body fluid specimens using a gene expression microarray assay. Cancer Cytopathol. 2012;120(1):62–70. doi: 10.1002/cncy.20167. [DOI] [PubMed] [Google Scholar]
- 12.Dennis J.L., Hvidsten T.R., Wit E.C. Markers of adenocarcinoma characteristic of the site of origin: development of a diagnostic algorithm. Clin. Cancer Res. 2005;11(10):3766–3772. doi: 10.1158/1078-0432.CCR-04-2236. [DOI] [PubMed] [Google Scholar]
- 13.Gamble A.R., Bell J.A., Ronan J.E., Pearson D., Ellis I.O. Use of tumour marker immunoreactivity to identify primary site of metastatic cancer. BMJ. 1993;306(6873):295–298. doi: 10.1136/bmj.306.6873.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobin A., Davis C.A., Schlesinger F. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14(4):417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennan C.W., Verhaak R.G., McKenna A. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [published correction appears in Cell. 2014 Apr 24;157(3):753] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah S.P., Köbel M., Senz J. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N. Engl. J. Med. 2009;360(26):2719–2729. doi: 10.1056/NEJMoa0902542. [DOI] [PubMed] [Google Scholar]
- 18.ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium Pan-cancer analysis of whole genomes. Nature. 2020;578(7793):82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez-Vega F., Mina M., Armenia J. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173(2):321–337.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinrich M.C., Corless C.L., Demetri G.D. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J. Clin. Oncol. 2003;21(23):4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan P., Yeoh K.G. Genetics and molecular pathogenesis of gastric adenocarcinoma. Gastroenterology. 2015;149(5):1153–1162. doi: 10.1053/j.gastro.2015.05.059. [DOI] [PubMed] [Google Scholar]
- 23.Miettinen M., Sobin L.H., Sarlomo-Rikala M. Immunohistochemical spectrum of GISTs at different sites and their differential diagnosis with a reference to CD117 (KIT) Mod. Pathol. 2000;13(10):1134–1142. doi: 10.1038/modpathol.3880210. [DOI] [PubMed] [Google Scholar]
- 24.Garraway L.A., Widlund H.R., Rubin M.A. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436(7047):117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 25.Markowski M.C., Bowen C., Gelmann E.P. Inflammatory cytokines induce phosphorylation and ubiquitination of prostate suppressor protein NKX3.1. Cancer Res. 2008;68(17):6896–6901. doi: 10.1158/0008-5472.CAN-08-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abraham J., Spetzler D., Korn W.M., Genomic Profiling Similarity. WO2020146554.
- 27.Greco F.A., Hainsworth J.D. Renal cell carcinoma presenting as carcinoma of unknown primary site: recognition of a treatable patient subset. Clin. Genitourin. Cancer. 2018;16(4):e893–e898. doi: 10.1016/j.clgc.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Kato S., Krishnamurthy N., Banks K.C. Utility of genomic analysis in circulating tumor DNA from patients with carcinoma of unknown primary. Cancer Res. 2017;77(16):4238–4246. doi: 10.1158/0008-5472.CAN-17-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moran S., Martínez-Cardús A., Sayols S. Epigenetic profiling to classify cancer of unknown primary: a multicentre, retrospective analysis. Lancet Oncol. 2016;17(10):1386–1395. doi: 10.1016/S1470-2045(16)30297-2. [DOI] [PubMed] [Google Scholar]
- 30.Peck M., Moffat D., Latham B., Badrick T. Review of diagnostic error in anatomical pathology and the role and value of second opinions in error prevention. J. Clin. Pathol. 2018;71(11):995–1000. doi: 10.1136/jclinpath-2018-205226. [DOI] [PubMed] [Google Scholar]
- 31.Bera K., Schalper K.A., Rimm D.L., Velcheti V., Madabhushi A. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019;16(11):703–715. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiao W., Atwal G., Polak P. A deep learning system accurately classifies primary and metastatic cancers using passenger mutation patterns. Nat. Commun. 2020;11:728. doi: 10.1038/s41467-019-13825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stafford P., Brun M. Three methods for optimization of cross-laboratory and cross-platform microarray expression data. Nucl. Acids Res. 2007;35(10):e72. doi: 10.1093/nar/gkl1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.