Abstract
Excessive accumulation of extracellular matrix (ECM) is the hallmark of fibrotic diseases. In the kidney, it is the final common pathway of prevalent diseases, leading to chronic renal failure. While cytokines such as TGF-β play a fundamental role in myofibroblast transformation, recent work has shown that mitochondrial dysfunction and defective fatty acid oxidation (FAO), which compromise the main source of energy for renal tubular epithelial cells, have been proposed to be fundamental contributors to the development and progression of kidney fibrosis. MicroRNAs (miRNAs), which regulate gene expression post-transcriptionally, have been reported to control renal fibrogenesis. To identify miRNAs involved in the metabolic derangement of renal fibrosis, we performed a miRNA array screen in the mouse model of unilateral ureteral obstruction (UUO). MiR-150-5p and miR-495-3p were selected for their link to human pathology, their role in mitochondrial metabolism and their targeting of the fatty acid shuttling enzyme CPT1A. We found a 2- and 4-fold upregulation of miR-150-5p and miR-495-5p, respectively, in both the UUO and the folic acid induced nephropathy (FAN) models, while TGF-β1 upregulated their expressions in the human renal tubular epithelial cell line HKC-8. These miRNAs synergized with TGF-β regarding its pro-fibrotic effect by enhancing the fibrosis-associated markers Acta2, Col1α1 and Fn1. Bioenergetics studies showed a reduction of FAO-associated oxygen consumption rate (OCR) in HKC-8 cells in the presence of both miRNAs. Consistently, expression levels of their mitochondrial-related target genes CPT1A, PGC1α and the mitochondrial transcription factor A (TFAM), were reduced by half in renal epithelial cells exposed to these miRNAs. By contrast, we did not detect changes in mitochondrial mass and transmembrane potential (ΔѰm) or mitochondrial superoxide radical anion production. Our data support that miR-150 and miR-495 may contribute to renal fibrogenesis by aggravating the metabolic failure critically involved in tubular epithelial cells, ultimately leading to fibrosis.
Keywords: MicroRNAS, Kidney fibrosis, Fatty acid oxidation, Extracellular matrix, CPT1A, Mitochondria
Abbreviations: ECM, extracellular matrix; FAO, fatty acid oxidation; miRNAs, microRNAs; FAN, folic acid nephropathy; UUO, unilateral ureteral obstruction; TFAM, mitochondrial transcription factor A; OCR, oxygen consumption rate; CKD, chronic kidney disease; TGF-β, transforming growth factor-β; AKI, acute kidney injury; FBS, fetal bovine serum; ITS, insulin-transferrin-selenium; SPF, specific pathogen free; FDR, false discovery rate; ECAR, extracellular acidification rate; Eto, etomoxir; MMP, mitochondrial membrane potential; ETC, electron transport chain; EMT, epithelial-to-mesenchymal transition
Graphical abstract
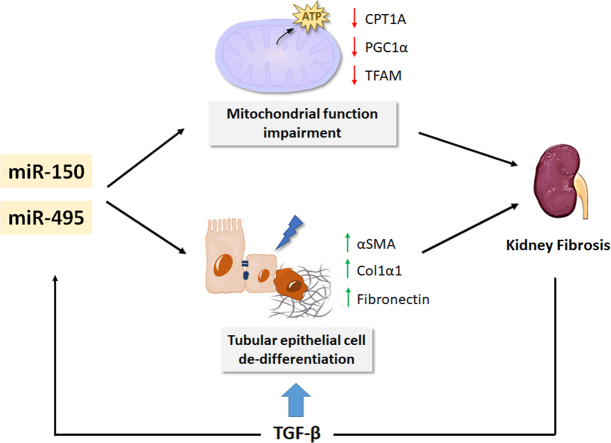
MiR-150-5p and miR-495-3p exert pro-fibrotic effects by synergizing with TGF-β in the fibrotic response and contributing to the mitochondrial impairment associated to renal fibrosis.
Highlights
-
•
MiR-150-5p and miR-495-3p were upregulated both in the UUO and FAN models.
-
•
MiR-150-5p and miR-495-3p synergized with TGF-β profibrotic effects and reduced FAO-associated OCR in renal epithelial cells.
-
•
MiR-150-5p and miR-495-3p did not alter mitochondrial transmembrane potential and superoxide radical anion production.
-
•
MiRNAs 150-5p and 495-3p are contributors to the metabolic impairment leading to renal fibrosis.
1. Introduction
Chronic kidney disease (CKD) is a clinical condition where the reduction of renal function is maintained. It is generally considered to be irreversible and progressive. It represents an important public health problem that can affect 12–14% of the general population [1]. It may be present in about 30–40% of patients with highly prevalent pathologies such as diabetes mellitus and hypertension, where it contributes to dictate evolution and prognosis. Regardless of the disease etiology, progression of CKD leads to tubule-interstitial and glomerular fibrosis [2]. Tubulo-interstitial fibrosis involves the expansion of the space between tubular basement membrane and peritubular capillaries through the deposition of matrix proteins (ECM) (predominantly collagens type I and III and fibronectin) in association with inflammatory cells, tubular cell loss and myofibroblast accumulation [3].
Transforming growth factor-β (TGF-β) is considered a crucial ubiquitous pro-fibrotic cytokine and the master regulator of myofibroblast differentiation in fibrosis [4]. Recent studies have demonstrated that a global defect in fatty acid oxidation (FAO), leading to a compromise in the main source of energy for tubular epithelial cells, plays an important role in the development of kidney fibrosis [5,6]. TGF-β suppresses FAO in a SMAD3-and PGC1A-dependent manner, hence affecting transcriptional regulators of fatty acid uptake and oxidation, including CPT1A [5]. Alterations in FAO finally result in intracellular lipid accumulation and ATP depletion, leading to fibrosis [5,7]. Thus, strategies aimed at restoring the metabolic derangement operating in CKD may provide therapeutic options [8].
MicroRNAs (miRNAs) have emerged as critical regulators of gene expression through their action on posttranscriptional regulation. In the kidney, miRs have been shown to play critical roles in nephrogenesis, homeostasis and disease [9]. MiRNAs are also capable of regulating fibrotic processes and have been termed "fibromiRs" [10]. In addition, due to the lack of an effective therapy to prevent or revert renal fibrogenesis [11], manipulation of miRNA expression by in vivo delivery of miRNA mimics or inhibitors has been proposed as a promising therapeutic strategy for CKD [12]. In addition, circulating miRNAs may constitute diagnostic and even prognostic biomarkers [13]. MiRNA-mediated control of kidney fibrosis mainly occurs throughthe regulation of TGF-β1 signaling in a cell-dependent and context-dependent manner, utilizing the Smad pathway as one of the main targets [14]. In previous work, we have identified several miRNAs that have an impact on renal fibrosis by regulating specific metabolic pathways [15,16]. Over-expression of miR-9-5p prevents tubular epithelial renal cell de-differentiation and reprograms fibrosis-related metabolic derangement [16], while loss of function of miR-33, either genetic or pharmacological, results in increased FAO and prevention of lipid accumulation in tubular epithelial cells. This is associated with increased energy supply, improved mitochondrial bioenergetics and prevention of experimental renal fibrosis [15]. Thus, miRNAs regulating cellular bioenergetics are also capable of regulating renal fibrogenesis. In this study, we have employed a miRNA array-based strategy in the unilateral ureteral obstruction (UUO) chronic kidney injury model to identify miRNAs involved in the renal fibrotic phenotype through the modification of metabolic pathways. By selecting several mitochondrial function related genes (CPT1A, TFAM, PGC1A), we identified both miR-150 and miR-495 as miRNAs capable of reducing FAO-associated oxygen consumption rate (OCR) and promoting TGF-β1-induced fibrogenic transformation in the human renal tubular epithelial cell line HKC-8. Our findings highlight the importance of miRNAs as mitochondrial metabolic regulators involved in renal fibrogenesis.
2. Material and methods
Cell lines and culture conditions. Human renal proximal tubular epithelial cells (HKC-8) were cultured in DMEM/F12 (Dulbecco’s modified Eagle’s medium 1:1 (v/v)) (Corning, New York, NY) supplemented with 15 mM Hepes, 5% (vol/vol) fetal bovine serum (FBS) (HyClone Laboratories, Logan, UT), 1x Insulin-Transferrin-Selenium (ITS) (Gibco, Rockville, MD), 0.5 μg/ml hydrocortisone (Sigma, St. Louis, MO), 50 units/mL penicillin and 50 μg/mL streptomycin (Gibco, Rockville, MD) at 37 °C and 5% CO2. This cell line was kindly provided by Dr. Susztak's lab (Philadelphia, Pennsylvania, USA). Treatments with human recombinant 10 ng/ml TGF-β1 (R&D Systems, Minneapolis, MN) were performed after serum-free starvation of HKC-8 cells for 12 h.
Transfection procedure. Cells were seeded into 60 mm culture dishes to reach a confluence of 70%. They were transfected with 40 nM of either mirVana™ miRNA mimic of miR-150, miR-495 or mirVana™ miRNA mimic negative control (Ambion company, USA) using lipofectamine 2000 (Invitrogen, Carlsbad, CA). Cells were incubated with the lipofectamine-pre-miRNA 37 °C for 6 h. Subsequently, 5 ml fresh medium containing 10% FBS were added to the culture dishes and the cells were maintained in culture for 6 h until used for subsequent experiments. In case of combination with TGF-β1 treatment, it was applied to the cells after miRNAs overexpression according to the previous section.
Immunoblot. Cells were washed in PBS, homogenized and lysed in 100 μL RIPA lysis buffer containing 150 mM NaCl, 0.1% SDS, 1% sodium deoxycholate, 1% NP‐40 and 25 mM Tris–HCl pH 7.6, in the presence of protease (Complete, Roche Diagnostics, Mannheim, Germany) and phosphatase inhibitors (Sigma‐Aldrich, St. Louis, MO) and harvested by scraping. A quarter piece of each kidney sample (obtained after dissection in half both lengthwise and crosswise) was homogenized in 300 μl RIPA buffer with 5 mm stainless steel beads (Qiagen, Valencia, CA) using TissueLyser LT (Qiagen, Valencia, CA) vibrating at 50 Hz for 15 min at 4 °C. Samples were clarified by centrifugation at 10,000 g for 15 min at 4 °C. The pellet was then discarded, and the supernatant was kept as protein lysate. Protein concentrations were determined by the BCA Protein Assay Kit (Thermo Scientific, Rockford, IL) and was measured in Glomax®-Multi Detection system (Promega, Madison, WI). Equal amounts of protein (10–50 μg) from the total extract were separated on 8–10% SDS–polyacrylamide gels and transferred onto nitrocellulose blotting membranes (GE Healthcare, Chicago, IL) at 12 V for 20 min in a semi‐dry Trans‐Blot Turbo system (Bio‐Rad, Hercules, California). Membranes were blocked by incubation for 1 h with 5% non‐fat milk in PBS containing 0.5% Tween‐20 and blotted overnight with the specific antibodies: CPT1A (Ab128568, Abcam; 1:1000; 4 °C overnight), α-SMA (sc-32251, Santa Cruz Biotechnology; 1:1000; 4 °C overnight), GAPDH (MAB374, Millipore; 1:15000; 1 h, room temperature). After incubation with IRDye 800 goat anti‐rabbit and IRDye 600 goat anti-mouse (1:15,000, LI‐COR Biosciences, Lincoln, NE) secondary antibodies, membranes were imaged in triplicates with the Odyssey Infrared Imaging System (LI‐COR Biosciences, Lincoln, NE). Band densitometry was performed using the ImageJ 1.48 software (http://rsb.info.nih.gov/ij) and relative protein expression was determined by normalizing to GAPDH. Fold changes were normalized to values of control condition.
RNA extraction. Total RNA was extracted from HKC-8 cells or mouse kidneys using the miRNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturerʼs instructions. RNA quantity and quality were determined at 260 nm by a Nanodrop-1000 spectrophotometer (Thermo Scientific, Rockford, IL).
Analysis of mRNA expression. Reverse transcription (RT) was carried out with 500 ng of total RNA using the iScript™ cDNA Synthesis kit (Bio‐Rad, Hercules, CA). qRT–PCR was performed with the iQ™SYBR Green Supermix (Bio‐Rad, Hercules, CA), using a 96‐well Bio‐Rad CFX96 RT–PCR System with a C1000 Thermal Cycler (Bio‐Rad, Hercules, CA) according to the manufacturers’ instructions. A Ct value was obtained from each amplification curve using CFX96 analysis software provided by the manufacturer. Relative mRNA expression was determined using the 2−ΔΔCt method [17]. The 18S gene was used for normalization purposes. The primer sequences used for mRNA quantification were: CPT1A (FW: TGCTTTACAGGCGCAAACTG, RV: TGGAATCGTGGATCCCAAA), ACTA (FW: TTCAATGTCCCAGCCATGTA, RV: GAAGGAATAGCCACGCTCAG), COL1A1 (FW: CGGACGACCTGGTGAGAGA, RV: CATTGTGTCCCCTAATGCCTT), FN1 (FW: GTGTTGGGAATGGTCGTGGGGAATG, RV: CCAATGCCACGGCCATAGCAGTAGC), PGC1A (FW: TGCCCTGGATTGTTGACATGA, RV: TTTGTCAGGCTGGGGGTAGG), TFAM (FW: CATCTGTCTTGGCAAGTTGTCC, RV: CCACTCCGCCCTATAAGCATC), 18S (FW: AGCTATCAATCTGTCAATCCTGTC, RV: GCTTAATTTGACTCAACACGGGA). Fold changes were normalized to values of control condition.
Quantification of miRNA expression. Quantification of miRNAs expression was performed using the miRCURY Locked Nucleic Acid (LNA) miRCURY LNA RT Kit (Exiqon, Vedback, Denmark). Following RT, the cDNA template was amplified using microRNA‐specific LNA primers for mature miR150-5p or miR-495-3p (Exiqon, Vedback, Denmark). qRT–PCR was performed in a 96‐well Bio‐Rad CFX96 RealTime PCR System with a C1000 Thermal Cycler using iQ™ SYBR Green Supermix (Bio‐Rad, Hercules, CA) according to the manufacturers’ instructions. A Ct value for triplicate wells was obtained from each amplification curve using the CFX Manager Bio‐Rad software (Bio‐Rad, Hercules, CA). Ct values were normalized to the endogenous control 5S rRNA. Plasma miRNA levels were normalized to miR-103a-5p. Relative miRNA expression was determined using the 2−ΔΔCt method [17]. Fold changes were normalized to values of control condition.
Mouse models of kidney fibrosis. Mice were housed in the Specific Pathogen Free (SPF) animal facility at the CBMSO in accordance with EU regulations for all the procedures. Animals were handled in agreement with the Guide for the Care and Use of Laboratory Animals contained in Directive 2010/63/EU of the European Parliament. Approval was granted by the local ethics review board of the Centro de Biología Molecular “Severo Ochoa” in Madrid, the Ethics committee at CSIC and the Regulatory Unit for Animal Experimental Procedures from the Comunidad de Madrid.
Unilateral ureteral obstruction (UUO): UUO surgery procedure was performed as previously described [18]. Briefly, mice were anesthetized with isoflurane (3–5% for induction and 1–3% for maintenance) and divided into two experimental groups: the UUO group and the sham operation group. In the UUO group, mice were shaved on the left side of the abdomen, a vertical incision was made through the skin with a scalpel and the skin was retracted. A second incision was made through the peritoneum to expose the kidney. The left ureter was ligated twice 15 mm below the renal pelvis with surgical silk and the ureter was then severed between the two ligatures. Then, the ligated kidney was placed gently back into its correct anatomical position and sterile saline was added to replenish loss of fluid. The incisions were sutured and mice were individually caged. The sham operation was performed in a similar manner, but without ureteral ligation. Buprenorphine was used as an analgesic. A first dose was administered 30 min before surgery and then every 12 h for 72 h, at a dose of 0.05 mg/kg subcutaneously. In this model, renal blood flow and glomerular filtration rate become significantly reduced within 24 h and interstitial inflammation (peak at 2–3 days), tubular dilation, tubular atrophy and fibrosis are evident after 7 days. The obstructed kidney reaches maximal dysfunction around 2 weeks after the procedure. Mice were sacrificed by CO2 overdose and control and obstructed kidney and blood samples were harvested after perfusion with PBS at 3, 5, 7, 10 and 15 days after UUO.
Folic acid-induced nephropathy (FAN): In this model, kidney fibrosis was induced by intraperitoneal (i.p) injection with 250 mg folic acid (Sigma‐Aldrich, St. Louis, MO) per kg body weight dissolved in 0.3 M sodium bicarbonate (vehicle) as previously described [19]. Control animals received 0.3 ml of vehicle (i.p). Mice were sacrificed by CO2 overdose and kidneys and blood samples were harvested after perfusion with PBS after 15 days of FA administration.
microRNA profiling. To identify specific miRNAs that regulate the fibrotic outcome, a Custom miRCURY LNA microRNA array (Exiqon, Vedback, Denmark) was selected as the platform for microRNA profiling. This platform consisted of a total of 175 unique assays specific for mouse miRNAs present in the Sanger miRBase v22.1 (http://microrna.sanger.ac.uk). For each microRNA, the prevailing strand was selected for analysis. MiR selection was based on predicted target genes related to renal fibrosis, mitochondrial metabolism, redox processes and circadian rhythm. This analysis was done with the bioinformatics tools for sequence-based miRNA target prediction, microcosm [20] (www.ebi.ac.uk/enright-srv/microcosm), Targetscan 4.1 [21] (www.targetscan.org/vert_40/) Pictar I [22] (www.pictar.mdc-berlin.de), miRWalk [23] (www.mirwalk.umm.uni-heidelberg.de/) and miRanda [24] (www.microrna.org/microrna/). After RNA extraction from 7 day-UUO kidney samples, the miRNA fraction present in 20 ng of total RNA was subjected to reverse transcription using miRCURY LNA RT Kit (Exiqon, Vedback, Denmark) according to the manufacturer's instructions. The resultant cDNAs were analyzed with the miRCURY miRNA PCR Panels in a Roche LightCycler 480 Real-Time PCR system using miRCURY LNA SYBR Green PCR Master Mix (Exiqon, Vedback, Denmark). Each sample was analyzed in duplicate. This procedure was performed in the Genomic Facility of the Fundación Parque Científico de Madrid (Madrid, Spain). The LC480 software was utilized to obtain Cq values (Cq) for each miRNA. The delta Ct (ΔCt) value was calculated by normalizing Ct values to the endogenous housekeeping snRNAU6. The heat map was generated with the real‐time PCR data presented as ΔCt. A hierarchical clustering was built according to ΔCt values. The delta delta Ct (ΔΔCt) value was calculated by subtracting the ΔCt of the reference sample group (control kidney) from the ΔCt of each obstructed kidney sample. Relative quantification (RQ) or fold-change of each miRNA was generated using the 2−ΔΔCt method [17]. Differentially expressed miRNAs were identified by the parametric Limma test. To take the multiple hypotheses testing into account, P‐values were adjusted (adj.) using the Benjamini–Hochberg false discovery rate (FDR) correction [25]. Results were graphed in a volcano plot.
miRNA analysis in blood plasma from CKD patients. The levels of selected miRNAs were analyzed in plasma samples from a cohort of 100 CKD patients, which is detailed below. Peripheral venous blood was collected in EDTA spray-coated vacutainers (BD, Franklin Lakes, NJ). To obtain plasma, blood was centrifuged at 400 g for 20 min, supernatant was transferred into a new tube and centrifuged at 800 g for 20 min. Plasma was collected and stored frozen in aliquots at −80 °C. Plasma miRNA was extracted from blood using miRCURYTM RNA Isolation Kit—Biofluids and UniSp2 Spike-in RNA (Exiqon, Vedbaeck, Denmark). Analyses of miR-150 levels in these samples were performed as described in the quantification of miRNA expression procedure section.
Measurements of oxygen consumption rate. Fatty acid oxidation-associated oxygen consumption rate (OCR) (ligated to oxidative phosphorylation) and extracellular acidification rate (ECAR) (associated with lactate production and glycolysis) were studied using the Seahorse Bioscience metabolic analyzer according to the manufacturer’s instructions [26]. HKC-8 cells were seeded in a p60 plate and microRNA transfection was performed as described above when they reached a confluence of 70% and 48 h later, HKC-8 cells were treated with 10 ng/ml TGF-β1 for 48 h. Then, cells were seeded at 2 × 104 cells per well in a Seahorse Bioscience XFe24 cell culture microplate (Seahorse Bioscience, North Billerica, MA). After cell adherence, growth medium was replaced with substrate-limited medium, Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 0.5 mM Glucose and 1 mM Glutamate. One hour before the assay measurement, cells were incubated with Krebs-Henseleit Buffer (KHB) assay medium supplemented with 0.2% carnitine at 37 °C without CO2. Fifteen minutes before the assay, the CPT1 inhibitor Etomoxir (Eto) (Sigma‐Aldrich, St. Louis, MO) 400 μM was added to the corresponding wells and cells were incubated at 37 °C without CO2. Finally, immediately prior to the assay, BSA or 200 μM Palmitate-BSA FAO Substrate (Agilent Technology, Santa Clara, CA, USA) was added. Immediately, XF Cell Mito Stress Test was performed in a Seahorse XFe24 energy analyzer by adding sequentially several modulators of mitochondrial function. First the ATP synthase inhibitor oligomycin (Sigma‐Aldrich, St. Louis, MO) (1 μM) was used following basal measurement. This decreases electron flow through the ETC, resulting in a reduction in mitochondrial respiration. Next, the uncoupling agent cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) (SigmaAldrich, St. Louis, MO) (3 μM), that collapses the proton gradient and disrupts the mitochondrial membrane potential was added. As a result, electron flow through the ETC is uninhibited, and oxygen consumption by complex IV reaches its maximum. The third and last injection is a mixture of the complex III and I inhibitors antimycin/rotenone (Sigma‐Aldrich, St. Louis, MO) (1 μM). This combination shuts down mitochondrial respiration and enables the calculation of non-mitochondrial respiration driven by processes outside the mitochondria. Substrates/inhibitors were prepared in the same medium in which the experiment was conducted and were injected from the reagent ports automatically at the times indicated. Measurements were registered for 3-min periods of time (over a total period of 2 h) and values were normalized for total protein content. Protein was extracted from wells with 0.1% NP‐40‐PBS solution, and quantified with BCA protein assay (Thermo Scientific, Rockford, IL). Four wells were used for each experimental group. The seahorse XFp Cell Mito Stress Test was used to determine the key parameters of mitochondrial function: basal mitochondrial respiration, ATP-linked respiration, proton leak (non-ATP–linked oxygen consumption), maximal respiration, non-mitochondrial respiration and reserve respiratory capacity, as previously described [27].
Luciferase assay. To characterize the miRNA-150 and miRNA-495 candidate binding sites in the human CPT1A 3′-UTR, a luciferase reporter assay containing the CPT1A 3′-UTR (SwitchGear Genomics, Carlsbad, CA, Product ID S814347) was used. Site-directed mutations in the seed regions of predicted miR-150 and miR-495 sites within the 3′-UTRs were generated by using the Multisite-QuikChange directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol. The primer sequences used were for miR-495 PM1 (FW: GCATCATCCAAGCAGGGTAAACTTTTGTTCTAGAAAAAGAAAAATGTGTTATTCATTGGTGTCCC, RV: GGGACACCAACTTGAATAACACATTTTTCTTTTTCTAGAACAAAAGTTTACCCTGCTTGGATGATGC) and PM3 (FW: TGTCTTAACGCAGCCATGGTTTGAATCTAGAATCTTGGGCTGACCGGTGC, RV: GCACCGGTCAGCCCAAGATTCTAGATTCAAACCATGGCTGCGTTAAGACA) and for miR-150 PM2 (FW: CTCATGCGTGTAATCCCAGCACTTCTAGAGGCCAAGGCGGGCGG, RV: CCGCCCGCCTTGGCCTCTAGAAGTGCTGGGATTACACGCATGAG), All constructs were sequenced before use to confirm their proper structure. HKC-8 cells were seeded in a p24-well plate and were transiently co-transfected with 200 ng pLightSwitch_CPT1A_3′-UTR (intact or mutated constructions) and 4 ng pGL3-Promoter (a firefly luciferase under the control of the SV40 promoter) (Promega Corporation, Madison, WI, USA) reporter plasmids and 40 nM of either mirVana™ miRNA mimic of miR-150, miR-495 or mirVana™ miRNA mimic negative control (Ambion company, USA) using lipofectamine 2000 (Invitrogen, Carlsbad, CA) when they reached a confluence of 70% as described above. Four wells were used for each experimental group. Luciferase assays were performed 24 h later using the Dual-Luciferase reporter system (Promega, Madison, WI). The renilla and firefly luciferase signals were detected using a Glomax multidetection system (Promega Corporation, Madison, WI, USA). The activity of renilla luciferase was normalized by the firefly luciferase activity.
Mitochondrial membrane potential (MMP). Changes in MMP were determined as differences in tetramethylrhodamine methyl ester (TMRM) fluorescence (Invitrogen, Carlsbad, CA, USA). It accumulates in negatively charged polarized mitochondria and fluoresces in orange. When mitochondrial membrane potential collapses in apoptotic or metabolically stressed cells, the TMRM reagent is dispersed throughout the cell cytosol and fluorescence levels drop dramatically. HKC-8 cells were cultured and transfected as described in the cell culture and transfection procedure sections. Next, growth medium was replaced by phenol-red free Hank’s Balanced Salt Solution (HBSS) with 10 mM Hepes. Treatments with oligomycin (5 μM) and FCCP (4 μM) for 5 min were used as positive and negative control conditions. Next, cells were stained with 250 nM TMRM for 30 min at 37 °C. Cells were harvested with trypsin, centrifuged (3000 rpm, 5 min) and the pellet was resuspended in 200 μL HBSS with 1% Bovine Serum Albumin (BSA) and 5 mM ethylenediaminetetraacetic acid (EDTA). Fluorescence intensity was measured by flow cytometry using an emission wavelength of 570 nm for TMRM (FL2) [28] in a BD FacsCantoTM II system (BD Bioscience, San José, CA) and analyzed with the FlowJo 10.2 software (FlowJo, LLC, Ashland, OR). For each experimental condition, at least 20,000 singlets were analyzed in triplicates.
Mitochondrial superoxide radical anion production. Evaluation of superoxide radical anion production was performed by using MitoSOX™ Red mitochondrial superoxide radical anion indicator (Invitrogen, Carlsbad, CA, USA), a highly selective fluorogenic dye for mitochondrial superoxide radical anion in live cells, according to the manufacturer’s instructions. HKC-8 cells were cultured and transfected as described in the cell culture and transfection procedure sections. Next, growth medium was replaced by phenol-red free Hank’s Balanced Salt Solution (HBSS) with 10 mM Hepes. Treatment with antimycin A (150 μM) and carbonyl cyanide m-chlorophenylhydrazone (CCCP) (50 μM) for 5 min were used as positive and negative control conditions. Next, cells were stained with 5 μM MitoSOX™ Red for 30 min at 37 °C. Cells were harvested as described in the mitochondrial membrane potential section. Fluorescence intensity was measured by flow cytometry using an emission wavelength of 580 nm for MitoSOX™ Red (FL2) in a BD FacsCantoTM II system (BD Bioscience, San José, CA) and analyzed with the FlowJo 10.2 software (FlowJo, LLC, Ashland, OR). For each experimental condition, at least 20,000 singlets were analyzed in triplicates.
Mitochondrial labeling. Evaluation of mitochondrial content was performed by using MitoTracker™ green FM (Invitrogen, Carlsbad, CA, USA), which passively diffuses across the plasma membrane and accumulates in active mitochondria regardless of mitochondrial membrane potential, according to the manufacturer’s instructions. HKC-8 cells were cultured and transfected as described in the cell culture and transfection procedure sections. Next, growth medium was replaced by phenol-red free Hank’s Balanced Salt Solution (HBSS) with 10 mM Hepes. Cells were stained with 150 nM with MitoTracker™ green FM for 30 min at 37 °C. For flow cytometry analysis, cells were harvested as described in the mitochondrial membrane potential section. Fluorescence intensity was measured using an emission wavelength of 516 nm for with MitoTracker™ green FM (FL1) in a BD FacsCantoTM II system (BD Bioscience, San José, CA) and analyzed with the FlowJo 10.2 software (FlowJo, LLC, Ashland, OR). For each experimental condition, at least 20,000 singlets were analyzed in triplicates. For fluorescence imaging, nuclei were also stained with DAPI (Sigma, St. Louis, MO) for 5 min at RT. Living cells were visualized by an inverted Zeiss LSM 710 confocal microscope with a Cell observed, a 63X/1.2 Water C-Apochromat Corr UV-VIS-IR M27 objective and analyzed with the Zeiss Zen2010B sp1 software (Zeiss, Oberkochen, Germany).
Mitochondrial copy number determination. Genomic DNA was extracted from HKC-8 cells using the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA) according to the manufacturerʼs instructions. Mitochondrial abundance was determined with the Human Mitochondrial DNA Copy Number Assay Kit (Detroit R&D, Detroit, MI). Relative mtDNA copy number was presented as the mtDNA-to-nuclear DNA ratio.
Human CKD patient samples. A cohort of 100 CKD patients (stage 3–4) from Hospital Principe de Asturias was selected for the analysis of miRNA plasma levels. It included two different subgroups classified according to the evolution of their renal function based on the GFR MDRD indicator [29] over a period of 24 months: 50 patients presented less than 10% of kidney function deterioration while the rest of them had experienced at least a 40% reduction in kidney function or had initiated renal replacement therapy (dialysis). We also quantified the degree of fibrosis in kidney biopsies from a different cohort of 26 patients with graft dysfunction following kidney transplantation from Hospital Ramón y Cajal. All biopsies were evaluated according to the Banff 2007 criteria [30].
Statistical analysis. Data were analyzed using nonparametric tests except where indicated. The difference between two independent groups was examined with Mann-Whitney test, while more than two groups were compared with Kruskall-Wallis test. A P-value of 0.05 or less was considered statistically significant (*/#: P < 0.05, **/##: P < 0.01, ***/###: P < 0.001). Data were analyzed using GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA). Data are reported as mean ± standard error of mean (SEM).
3. Results
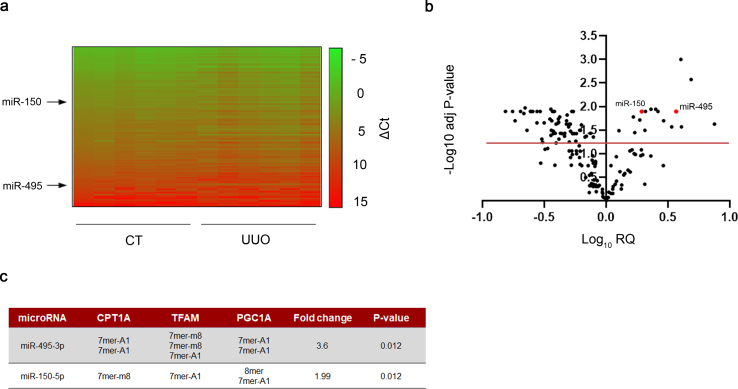
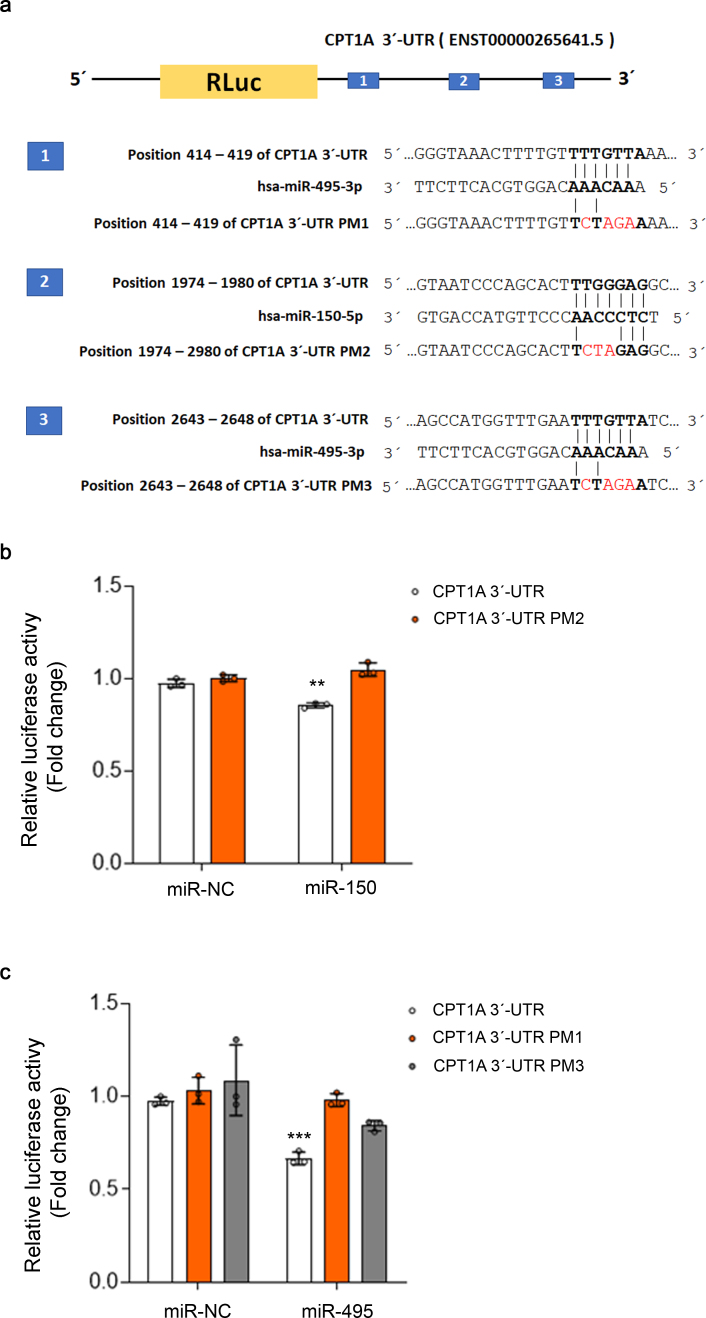
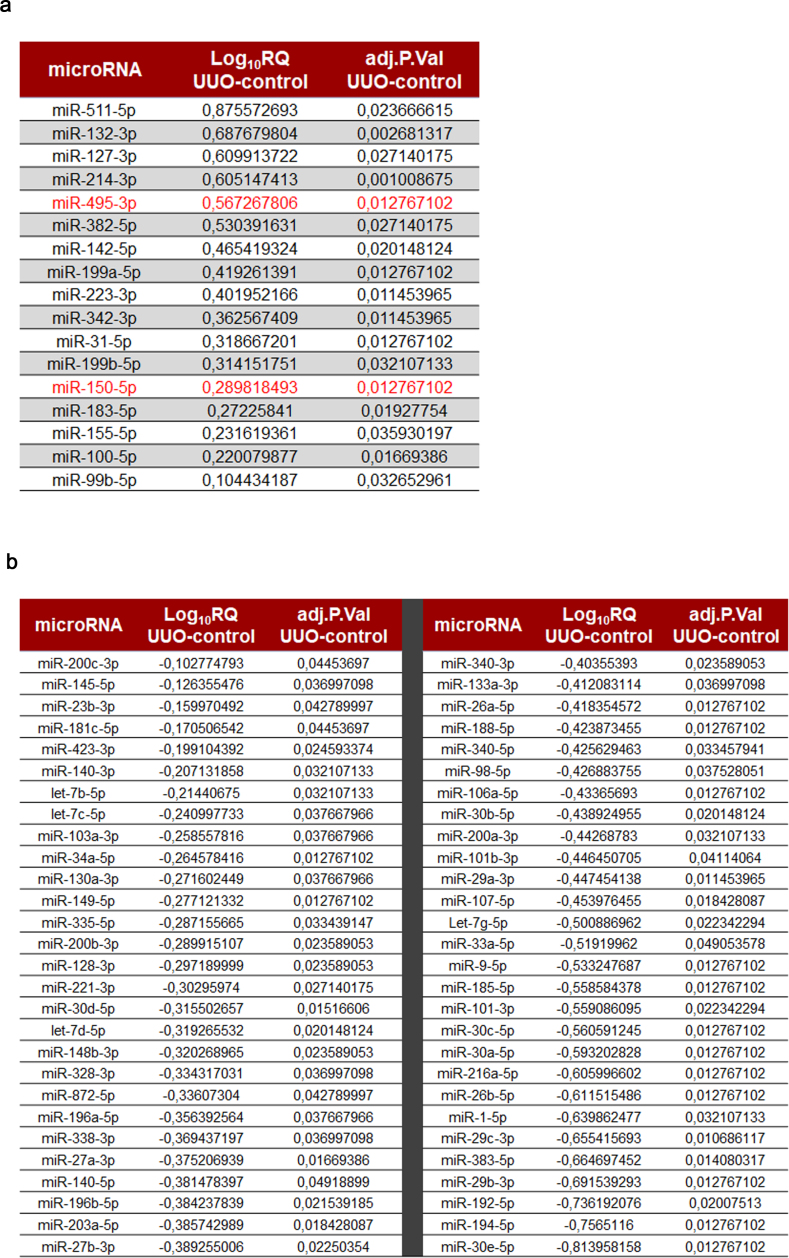
miRNA expression data in UUO-induced fibrotic kidneys. To decipher the contribution of miRNAs to the metabolic regulation of renal fibrogenesis, the 7days UUO model was performed in WT mice. RNA was isolated from control and fibrotic kidney samples and the miRNA expression profile was analyzed in a customized microRNA array. Selection of miRNAs was based on their potential targeting of key enzymes involved in mitochondrial metabolism, redox processes and circadian rhythm after “in silico” analysis (more details in the Methods section). The heat map revealed clearly distinct expression patterns in some miRNAs when comparing fibrotic and control kidney samples (Fig. 1A). The volcano plot analysis showed that 73 of 175 miRNAs were differentially expressed, 17 up‐ and 56 down‐regulated, in the fibrotic kidney samples compared to the control ones (Fig. 1B, Supplementary Table 1). Among them, specific miRNAs that could regulate the fibrotic outcome in the kidney through metabolic pathways were selected. For that purpose, in silico analysis of the 3′UTR (untranslated region) of specific genes involved in metabolic and mitochondrial functions, CPT1A, TFAM, PGC1A was performed with three independent prediction tools (Targetscan, miRWalk and miRanda). Selection criteria also included renal expression level, novelty of targets, degree of knowledge on the metabolic route and biological significance. Link to human pathology, kidney tissue expression level, their role in mitochondrial metabolism and their potential targeting of the fatty acid shuttling enzyme CPT1A encouraged us to focus on three miRNAs, miR-150-5p, miR-495-3p and miR-33-5p, as candidates capable of regulating metabolic alterations associated to renal fibrogenesis (Fig. 1C). Of note, CPT1A 3′-UTR contains one seed site for miR-150 and two seed sites for miR-495. We observed that miR-150 and miR-495 decreased the activity of a luciferase reporter assay containing the CPT1A 3′-UTR by ~15% and 25%, respectively. This downregulation was completely abrogated in the case of point mutations (PMs) in the 3′-UTR of CPT1A for the miR-150 binding site (PM2) and partially absent in both miR-495 binding sites (PM1 and PM3) (Supplementary Fig. 1A and B). These data indicate a direct interaction of CPT1A with miR‐150 and miR-495. MiR-33-5p has been the object of a separate study [15]. MiR-150-5p and miR-495-3p were upregulated in fibrotic kidney samples compared to the control ones, with a fold change of 3.69 (P-value: 0.012) and 1.95 (P-value: 0.012), respectively.
Fig. 1.
miRNA expression data in UUO-induced fibrosis. (A) Heat map showing relative miRNA expression of contralateral and obstructed kidneys of C57/BL6-J mice subjected to the UUO procedure for 7 days (n = 6). The scale bar ranges from green to red (high to low expression) and numbers represent ΔCt values. On the left side are indicated the miRNAs selected for study. (B) Volcano plot analysis of UUO‐modulated miRNAs. Log10 relative quantification (RQ) and negative (−) log10 adjusted (adj.) P‐values are plotted on the x‐ and y‐axis, respectively. Each miRNA is represented by a dot. 73 of 175 miRNAs showed an altered expression in the fibrotic samples (adj. P‐value ≥ 0.05). Selected miRNAs for further analysis are highlighted. (C) miR-150-5p and miR-495-3p were selected on the basis of their power for in silico targeting of the genes CPT1A, TFAM and PGC1A. For each selected microRNA, conserved seed target sites in the 3'UTR of these genes, their fold change and significance (adj. P-value) are indicated.
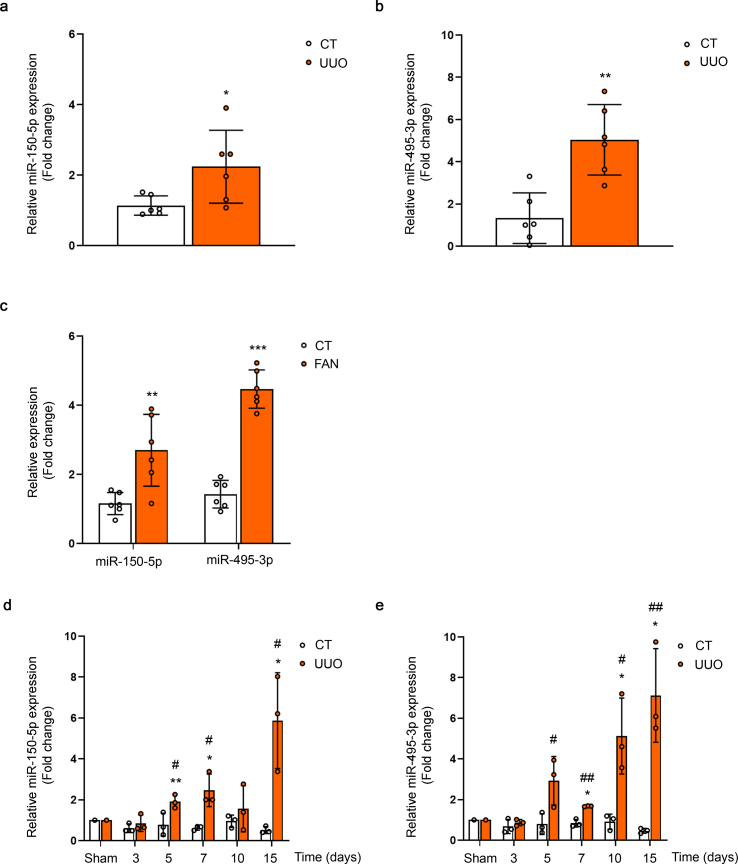
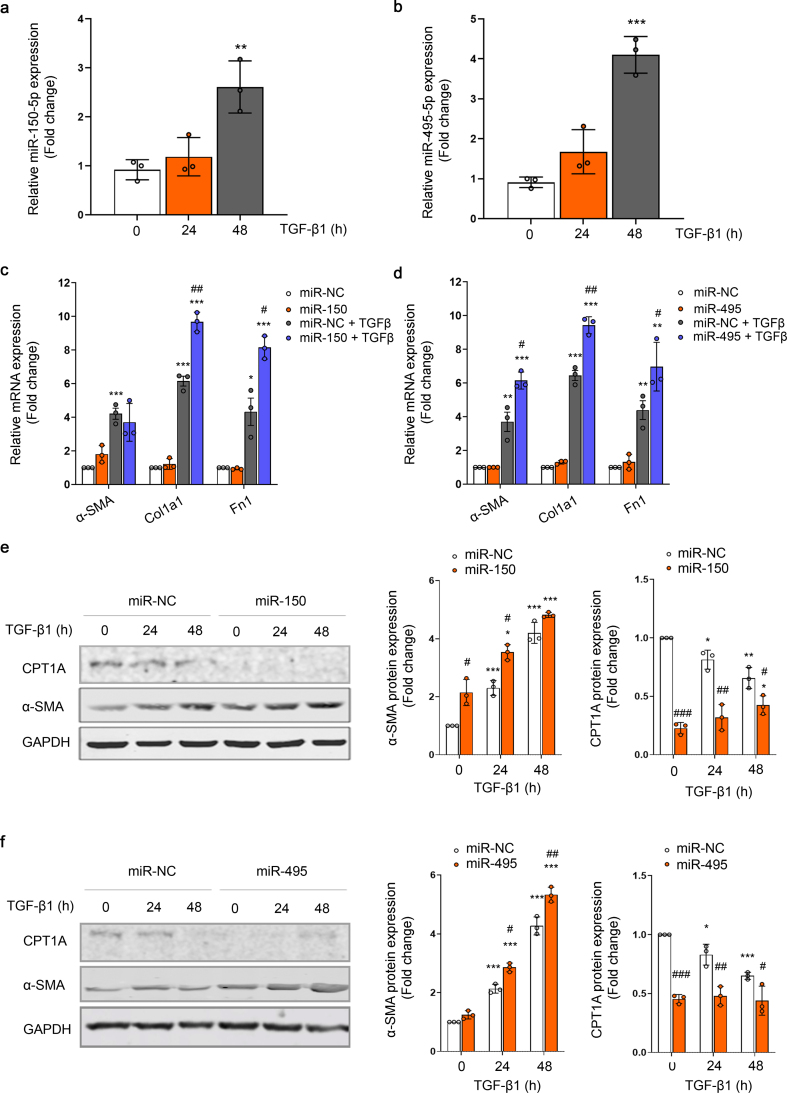
MiR-150 and miR-495 enhance the TGF-β profibrotic response in human renal tubular epithelial cells. Quantitative reverse transcription–polymerase chain reaction (qRT–PCR) was performed to confirm the change of miR‐150‐5p and miR-495-3p expression that had been identified by miRNA expression profiling in the UUO kidney samples (Fig. 2A and B). In addition, kinetics of miR-150-5p and miR-495-3p expression in the kidneys 3, 5, 7, 10 and 15 days after the UUO procedure was analyzed by qRT-PCR. The expression of these miRNAs was significantly enhanced by 2-fold at 5 days after UUO and remained elevated 15 days after UUO (Fig. 2C and D). The expression level of these miRNAs was also evaluated in the folic acid nephropathy (FAN) model. We found a 4- and 2-fold upregulation of miR-495-3p and miR-150-5p, respectively, in fibrotic kidney samples from the FAN model (Fig. 2E). TGF‐β1, one of the master regulators of fibrogenesis, also induced miR‐150 and miR-495 expression in HKC-8 cells more than 2‐fold and 3-fold, respectively, after 48 h, supporting a potential role for these miRNAs in TGF‐β signaling‐related events (Fig. 3A and B). To assess whether miR-150 and miR-495 were involved in the pro-fibrotic transformation of tubular epithelial cells by TGF-β1, the human cell line HKC-8 was transfected with miR-150-5p or miR-495-3p and treated with TGF-β1 for different times. Increasing miR-150-5p and miR-495-3p levels significantly enhanced the TGF-β1-induced mRNA level expression of the fibrosis-associated markers Acta2, Col1α1 and Fn1 (Fig. 3C and D). Similarly, overexpression of miR-150-5p or miR-495-3p strongly reduced CPT1A and enhanced α-SMA protein abundance (Fig. 3E and F). These data suggest that these miRNAs may participate in the pathogenesis of renal fibrosis by promoting TGF-β1-dependent epithelial dedifferentiation of tubular cells, as well as by compromising the expression of a critical enzyme involved in FAO.
Fig. 2.
Expression levels of miR-150-5p and miR-495-5p in kidneys from the UUO and FAN models. (A, B) qRT-PCR analysis of miR-150-5p (A) and miR-495-3p (B) expression in kidneys from mice 7 days after UUO in samples previously analyzed with the microRNA array. (C–E) qRT-PCR analysis of miR-150-5p and miR-495-3p expression after the FAN model (C) and 3, 5, 7, 10 and 15 days after UUO for miR-150-5p (D) and miR-495-3p (E). Bar graphs represent the mean of the fold change expression levels ± s.e.m. from 6 (A, B, E) and 3 (C and D) mice per group. *P < 0.05, **P < 0.01, ***P < 0.001 compared to their corresponding control conditions; #P < 0.05, ##P < 0.01 compared to control kidneys from mice with the same experimental treatment.
Fig. 3.
MiR-150 and miR-495 enhance TGF-β1-dependent pro-fibrotic response in human renal tubular epithelial cells. (A, B) RT–PCR analysis of miR‐150‐5p (A) and miR-495-3p (B) expression in HKC-8 cells treated with 10 ng/ml TGF‐β1 for the indicated times. (C, D) mRNA levels of alpha-smooth muscle actin (α-SMA), alpha 1 type-1 collagen (Col1α1), fibronectin (FN) from cells transfected with miR-NC and mimic miR-150-5p (C) or miR-495-3p (D) were determined by qRT-PCR using Sybr green. Cells were treated with TGF-β1 (10 ng/ml) after miRNA overexpression where indicated (see also methods section). (E, F) Immunoblots depicting CPT1A and α-SMA levels protein levels in cells transfected with miR-NC and mimic miR-150-5p (E) or miR-495-3p (F) for the indicated time points. GAPDH was used for normalization purposes. Bar graphs (right panels) represent the mean of the fold change expression ± s.e.m. from 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 compared to their corresponding control conditions; #P < 0.05, ##P < 0.01, ###P < 0.001 compared to cells treated with miR-NC with the same experimental condition.
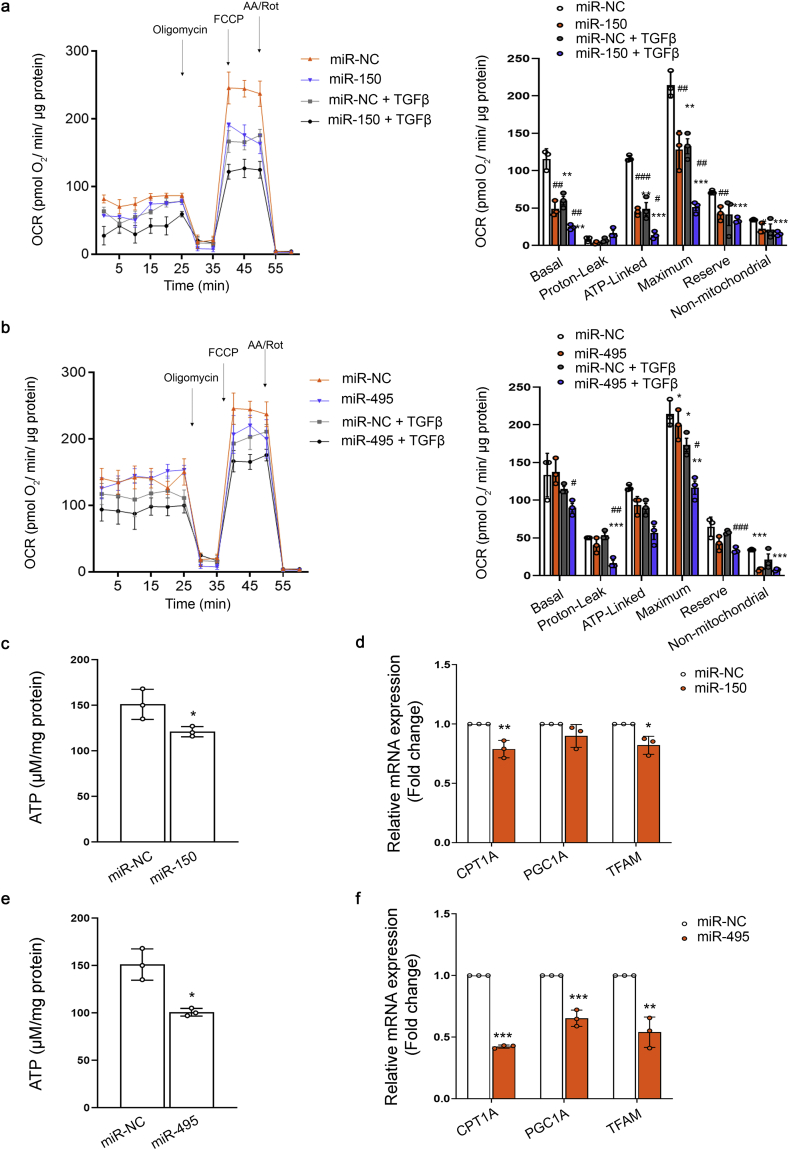
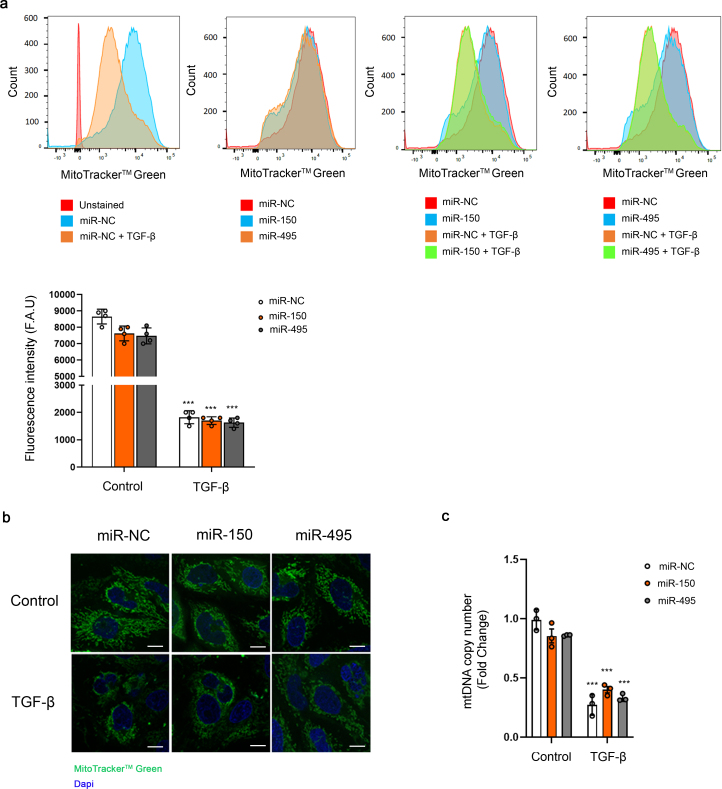
MiR-150 and miR-495 induce mitochondrial bioenergetics changes and reduce the expression of genes involved in crucial mitochondrial function. To gain insight into the metabolic consequences induced by the administration of miR-150-5p or miR-495-3p we studied the bioenergetics profile of human renal tubular epithelial cells. TGF-β1 was used as the model cytokine involved in pro-fibrotic associated changes. To determine the bioenergetics status of these cells, FAO-associated oxygen consumption rate (OCR) and the extracellular acidification rate (ECAR) of HKC-8 cells were measured during sequential treatment with compounds that modulate mitochondrial activity in the presence of palmitate, using a Seahorse XF24 Extracellular Flux Analyzer (more details in the Methods section). After 48h with TGF-β1, HKC-8 cells in the presence of miR-150-5p showed a consistent decrease in basal, OCR coupled to ATP generation, ATP-linked respiration and maximal respiration, which relates to FCCP-sensitive OCR (Fig. 4A). The impairment in the OXPHOS was mirrored by a decrease in ATP content in HKC-8 cells over-expressing miR-150-5p (Fig. 4C). We did not observe that these miRNAs induced variations in ECAR (data not shown). Consistent with this, mRNA expression levels of their mitochondrial-related target genes CPT1A, PGC1α and the mitochondrial transcription factor A (TFAM), were reduced by half in cells treated with miR-150 mimic in comparison to miRNA mimic NC-treated cells (Fig. 4E). Studies using the same experimental approach with miR-495-3p yielded similar results (Fig. 4B, D, F). However, mitochondrial mass, determined by the mtDNA copy number and mitochondrial staining with MitoTrackerTM green FM, was not significantly modulated by miRNA-150 and miRNA-495 both in basal conditions and under TGF-β treatment. A decreasing trend in this parameter was observed basally (Supplementary Fig. 2A, B, C). Overall, these data support that miR-150 and miR-495 promote a pro-fibrotic action in tubular epithelial cells through the impairment of mitochondrial function.
Fig. 4.
MiR-150 and miR-495 enhance TGFβ1-induced FAO repression in HKC-8 cells. (A, B) Oxygen consumption rate (OCR) of HKC-8 cells transfected with 40 nM miR-NC and mimic miR-150-5p (A) or miR-495-3p (B) and exposed to 10 ng/ml TGF-β1 after miRNA overexpression (see also methods section). Bar graphs (right panels) show the rates of OCR associated to basal, proton-leak, ATP-linked, maximum and reserve capacities and non-mitochondrial respiratory statuses. Data are represented after normalization by protein amount. (C, E) ATP levels in HKC-8 cells transfected with miR-NC and mimic miR-150-5p (C) or miR-495-3p (E). (D, F) mRNA levels of CPT1A, PGC1A and TFAM in HKC-8 cells transfected with miR-NC and mimic miR-150-5p (D) or miR-495-3p (F) were determined by qRT-PCR using Sybr green. Bar graphs show the mean ± s.e.m. of 4 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 compared to their corresponding control conditions; #P < 0.05, ##P < 0.01, ###P < 0.001 compared to cells treated with miR-NC with the same experimental condition.
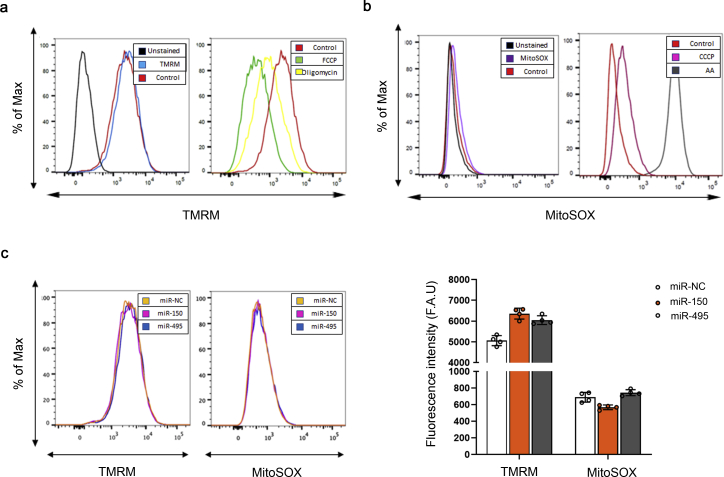
MiR-150 and miR-495 do not alter mitochondrial transmembrane potential (ΔѰm) and mitochondrial superoxide radical anion production. Mitochondrial membrane potential (ΔΨm) generated by proton pumps (Complexes I, III and IV) is an essential component in the process of energy storage during oxidative phosphorylation. Increased ΔѰm can lead to mitochondrial redox perturbation, while its decrease causes a reduction of ATP production, compromising cell viability [254]. Superoxide radical anion (O2•−) is a free radical that can be generated in the mitochondria as a consequence of electron leakage that may occur within several steps of the ETC. Generation of O2•− within the mitochondrial matrix also critically depends on the NADH/NAD+ and CoQH2/CoQ ratios and the local O2 concentration [255]. Its overproduction may lead to the loss of mitochondrial redox homeostasis through different mechanisms, a condition that has been related to many pathological states including AKI and CKD [201]. ΔѰm was examined with the TMRM dye and the superoxide radical anion production with MitoSOX dye. First, we validated these assays by the treatment of cells with FCCP and CCCP as negative controls, for TMRM and MitoSOX respectively, and Oligomycin and Antimycin A as positive controls, for TMRM and MitoSOX respectively, in the human cell line HKC-8. In the case of the TMRM assay, oligomycin treatment induced a 3-fold increase in fluorescence mean of the TMRM compared to the control dye, while FCCP treatment resulted in a 9-fold decrease. Antimycin A and CCCP produced a 35-fold increase and 2-fold decrease, respectively, in the MitoSOX assay (Supplementary Fig. 3A and B). To determine if miR-150 and miR-495 effects on mitochondrial metabolism triggered alterations in these parameters, HKC-8 cells were transfected with a miR-150-5p or miR-495-3p mimic or the corresponding miR-NC. No statistically significant differences in the fluorescence quantification of the TMRM or MitoSOX probes were found between the treatment conditions with the selected miRs and miR-NC (Supplementary Fig. 3C).
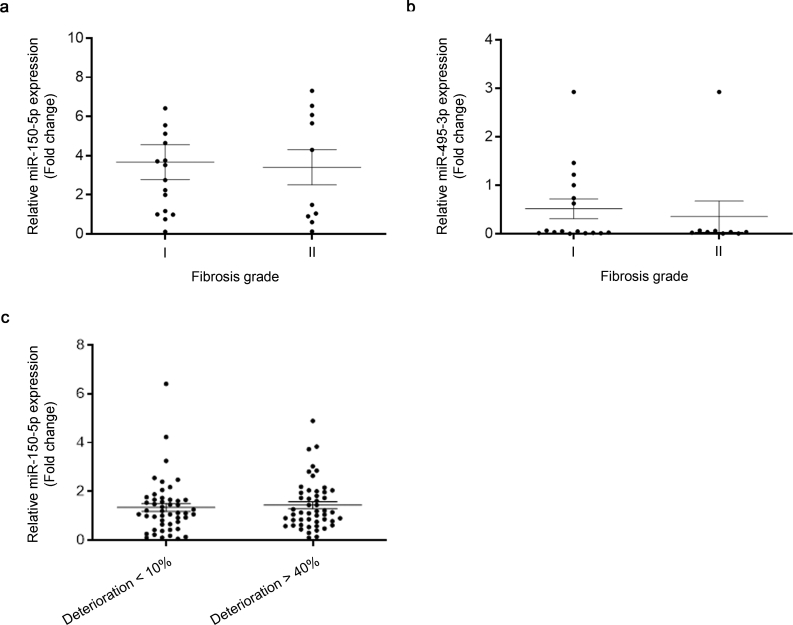
MiR-150 and miR-495 levels are not affected in plasma and kidney samples from CKD patients. We assessed the expression of the selected miRNAs in plasma and kidney biopsies from patients with CKD. Analysis of kidney biopsies from 26 patients (Hospital Ramón y Cajal) did not show correlation between the degree of fibrosis and miR-150-5p and miR-495-3p levels, respectively (Supplementary Fig. 4A and B). Circulating plasma levels of miR-150-5p, the miRNA with the highest kidney expression of the two, were also determined in a cohort of 100 CKD patients (Hospital Príncipe de Asturias). Due to limitations in sample availability, we were unable to determine serum values of miR-495-3p in this cohort. These patients were previously divided in two subgroups based on the evolution of their renal function over a period of 24 months, so that 50 patients presented less than 10% of kidney function deterioration while the rest of them had experienced at least a 40% reduction in kidney function. The qRT-PCR quantification of plasma levels of miR-150-5p did not show a clear trend towards a differential expression in the two subgroups of patients (Supplementary Fig. 4C).
4. Discussion
The discovery of miRNAs as key regulators of pathophysiological processes has fostered research on their use as therapeutic agents in almost every clinical setting, including kidney disease [31]. Renal fibrosis is a highly prevalent outcome of chronic kidney disease (CKD) with independence of the etiology, and its presence is a clear predictor of evolution. Hence, a significant effort has been devoted to identify miRNAs linked to kidney fibrosis [32,33]. In this study we focused on the increasingly important role of metabolic derangement in renal damage and we identified two miRNAs, miR-150 and miR-495, which were previously unknown actors in the scene. Moreover, we found that they exert a profound effect on cellular bioenergetics by synergizing with TGF-β and modifying key metabolic routes related to fatty acid oxidation.
Epithelial injury co-exists with a defect in fatty acid oxidation, the main source of energy for tubular epithelial cells [5]. It leads to their dedifferentiation inducing tubular function impairment, triggering cell cycle arrest and promoting the release of critical pro-fibrotic cytokines that may contribute to activate interstitial myofibroblasts [5,34]. MiR-150 and miR-495 expression was increased in mouse fibrotic kidneys and in human kidney epithelial cells (HKC-8) after treatment with TGF-β, suggesting their involvement in the pro-fibrotic response. Of interest, in silico analysis of the genomic regions coding for miR-150 and miR-495 permits to identify binding sites for transcription factors related to TGF-β signaling and other pathways involved in UUO damage (searches in genecards.org for miR-150 and miR-495). Both miR-150 and miR-495 decreased the expression level of their target genes CPT1A, PGC1α and TFAM, which was reflected in a decreased basal, ATP-linked and maximal OCR and ATP content after treatment with miR-150 and miR-495. This effect was accompanied by an enhanced TGF-β pro-fibrotic response in HKC-8 cells. We found that TGF-β decreased mitochondrial mass and mitochondrial DNA. After treatment with miRNA-150 and miRNA-495 a decreasing trend was observed in mitochondrial content in basal conditions, even though not reaching significance. This points to a mechanism independent from the role of PGC1α or TFAM and most likely related to a metabolic effect linked to CPT1A downregulation as the main variable affected by the presence of the miRNAs. Alterations in mitochondrial bioenergetics are critical in kidney disease [35]. In keeping, we have recently shown that CPT1A is a critical enzyme for the preservation of a healthy tubular compartment, as its overexpression protects from kidney fibrosis [6]. However, PGC1α and TFAM genes are also closely related to mitochondrial function and biogenesis [36]. The impaired expression of these genes is associated with CKD, while its preservation contributes to kidney integrity as shown in several models of damage. Thus, Han et al. found that tubular gain-of-function of PGC1α protected mice against Notch-induced kidney fibrosis and reversed the mitochondrial dysfunction associated with this model [37]. Similarly, re-expression of Tfam in tubule cells prevented Notch-induced metabolic and pro-fibrotic reprogramming [38]. Thus, it is conceivable that the decreased mRNA expression of these mitochondrial genes, induced by miR-150 and miR-495, also contributes to a damaged epithelial phenotype.
The decline we observed in OCR associated with epithelial cell damage is in accordance with previous reports, including our studies with miR-33 and miR-9 [15,16]. Whereas Kang et al. also reported that glucose oxidation was lower in human and mouse models of kidney fibrosis [5], our data show that the microRNA-mediated decrease in OCR does not induce per se variations in ECAR, suggesting that the glycolytic shutdown in fibrotic conditions most likely depends on fibrotic stimuli rather than on impaired oxidative phosphorylation. Several miRNAs have been identified as major modifiers of mitochondrial function, including the archetypal pro-fibrotic miRNA, miR-21 [39]. The suppression of PPAR-α expression by miR-21 has been invoked as a mechanism by which this miRNA exerts its pro-fibrotic action. In addition to inhibiting FAO, the target genes for miR-21 indicated that this miRNA silences a wide range of mitochondrial processes. Other examples include mir-9 [16], miR-33 [15] and miR-30e [40,41]. However, to our knowledge there are few reports addressing the microRNA regulation of CPT1A, PGC1α and TFAM genes in the context of kidney fibrosis. Interestingly, microRNA-214 which promotes epithelial-to-mesenchymal transition (EMT) during renal fibrosis, directly targets TFAM in colon cancer cells [42]. This repression was also reported for miR-590-3p [43]. The miR-29 family, one of the best characterized regulators of ECM production in organ fibrosis, directly targets PGC1α, preserving cardiac homeostasis. Thus, pathological silencing of miR-29 leads to PGC1α upregulation, generating profound alterations in mitochondrial biogenesis that contribute to cardiac disease [44]. While our data are clearly supportive of a role for miR-150 and miR-495 in kidney fibrosis, further in vivo studies would be necessary to confirm this contention.
To evaluate the integrity of mitochondrial electron transport chain (ETC), we focused on two features: ΔѰm and superoxide radical anion production. We observed that miR-150 and miR-495 did not alter these parameters. A stable ΔѰm is required for cell viability and its magnitude varies among cell types [45]. Its maintenance is strictly fine-tuned as an increase in ΔѰm may lead to perturbations in the redox state, while its decrease causes a reduction of ATP production and may trigger apoptosis. In our study, changes in ΔѰm were quantified with the cationic fluorescent dye TMRM, while mitochondrial superoxide radical anion production was measured with the fluorogenic dye MitoSOX Red. The employment of these tools in conditions affecting renal function is not conspicuous in the literature. Nevertheless, in the face of the absence of changes in our study, we did not pursue in depth other strategies to evaluate redox hormesis and hence, we cannot exclude that these miRNAs may alter the nucleophilic tone or the function of the several components of the ETC.
In the clinical setting miR-150 has been suggested as a pro-fibrotic miRNA in lupus nephritis by modulating TGF-β signaling [46,47], while miR-495 was associated with a protective role in diabetic cardiac fibrosis [48]. To find out their potential involvement in CKD, we evaluated their serum and parenchymal levels in two different patient cohorts. Serum levels of miRNAs have been proposed as potential biomarkers for CKD progression [49,50]. Thus, serum circulating miR-21 levels and abundance of miR-29c in urinary exosomes correlate with kidney fibrosis, suggesting their potential role as biomarkers [51,52]. However, miR-150 and miR-495 levels were not different either in plasma or kidney samples from CKD patients. The different microRNA expression patterns among species and cell types may explain this absence of difference in miR-150 and miR-495 levels in fibrotic tissue [53]. While in some cases reported microRNA plasma levels could be reflecting those in the kidney tissue, other mechanisms related to the secretion of miRNAs to the circulatory system or a counterbalance of this secretion by other organs should also be considered [13,54].
Overall, our results support that miR-150 and miR-495 promote renal epithelial cell dedifferentiation most likely through energy deprivation, conveyed by the repression of CPT1A, PGC1α and TFAM mitochondrial function-related genes. They also lend a basis to pursue their role in vivo, as well as their potential usefulness as diagnostic or prognostic biomarkers.
Author contributions
SL conceived and directed research. VM designed, performed and analyzed the majority of experiments. RR assisted with the microRNA array analysis. DRP and LGB performed studies in two different cohorts of CKD patients. All authors helped with the discussion of the results and SL and VM wrote the manuscript.
Declaration of competing interest
The authors have no conflicts of interest.
Acknowledgements
This work was supported by Grants from the Spanish Ministry of Science SAF2015-66107-R and PID2019-104233RB-I00 (SL), PI17/01513 (DRP) co-funded by the European Regional Development Fund, Instituto de Salud Carlos III REDinREN RD12/0021/0009 and RD16/0009/0016 (SL and DRP), FIS PI17/00625 (DRP), Comunidad de Madrid “NOVELREN” B2017/BMD3751 (SL and DRP), a grant-in-aid from the Spanish Society of Nephrology (Fundación Senefro 2017) to SL and Fundación Renal “Iñigo Alvarez de Toledo” (SL) all from Spain. The CBMSO receives institutional support from Fundación “Ramón Areces”. VM was supported by a predoctoral fellowship of the FPI Program (BES-2013-065986) from the Spanish Ministry of Science.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101851.
Contributor Information
Verónica Miguel, Email: vmiguel@cbm.csic.es.
Santiago Lamas, Email: slamas@cbm.csic.es.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. 1.
miR-150 and miR-495 bind to the 3′-UTR of CPT1A. (A) Schematic depicting miR-150 and miR-495 binding sites in the 3′-UTR of CPT1A. Renilla luciferase (RLuc) reporter containing the CPT1A 3′-UTR (ENST00000265641.5; chr11:68,522,351–68,525,111; 2761 bp long) which contains one seed site for miR-150 (numbered as #2) and two seed sites for miR-495 (numbered as #1 and #3). Mutated constructions (in red) contain point mutations (PM) in the 3′-UTR of CPT1A for the miR-150 binding site (PM2) and miR-495 binding sites (PM1 and PM3). (B, C) Relative luciferase activity of CPT1A 3′-UTR WT referred to negative control (miR-NC) in HKC-8 cells co‐transfected with pLightSwitch containing WT or mutated 3′ UTR sequences of human CPT1A and negative control (miR-NC), miRNA mimic of miR-150 (B) or of miR-495 (C). Bar graphs show the mean ± s.e.m. of 3 independent experiments. **P < 0.01, ***P < 0.001 compared to their corresponding control conditions.
Supplementary Fig. 2.
MiR-150-5p and miR-495-3p do not alter mitochondrial mass. (A) Representative flow cytometry histograms showing MitoTracker™ green FM fluorescence intensities in HKC-8 cells transfected with miR-NC, miR-150-5p or miR-495-3p and exposed to TGF-β (10 ng/ml) for 48 h after miRNA overexpression, measured by flow cytometry (upper panels). Bar graph (lower panel) represents the median ± s.e.m. of 4 independent experiments. ***P < 0.001 compared to their corresponding control conditions. (B) Representative images of fluorescence staining with the mitochondrial marker MitoTracker™ green FM and DAPI (nuclei). Scale bar: 20 μm (C) Mitochondrial DNA copy number (mtDNA) was determined in HKC-8 cells transfected with miR-NC, miR-150-5p or miR-495-3p and exposed to TGF-β (10 ng/ml) for 48 h after miRNA overexpression. Bar graphs represent the mean ± s.e.m. of fold changes (n = 3). ***P < 0.01 compared to their corresponding control condition. F.A.U: fluorescence arbitrary units.
Supplementary Fig. 3.
MiR-150-5p and miR-495-3p do not alter mitochondrial transmembrane potential (ΔѰm) or mitochondrial superoxide radical production. (A) Representative flow cytometry histograms showing TMRM fluorescence intensities in HKC-8 cells labeled with the fluorescent dye TMRM (250 nM) exposed to different experimental conditions: unstained, control, FCCP and Oligomycin, measured by flow cytometry. (B) Representative flow cytometry histograms showing mitoSOX fluorescence intensities in HKC-8 cells labeled with the fluorescent dye MitoSOX (5 μM) exposed to different experimental conditions: unstained, control, CCCP and Antimicyn A, measured by flow cytometry. One from three independent experiments is shown in (A) and (B). (C) TMRM and mitoSOX fluorescence histograms in HKC-8 cells transfected with miR-NC, miR-150-5p or miR-495-3p. Bar graph (right panel) represents the mean ± s.e.m. of 4 independent experiments. AA: Antimycin A; F.A.U: fluorescence arbitrary units.
Supplementary Fig. 4.
MiR-150-5p and miR-495-5p levels in kidney samples and plasma from CKD patients. (A) miR150-5p expression levels in 26 human post-transplant renal biopsies with different fibrosis grade were determined by qRT-PCR using Sybr green. (B) miR-495-3p expression levels from the same biopsies as in (A) were determined by qRT-PCR using Sybr green. (C) miR-150-5p circulating plasma levels in a cohort of 100 CKD patients with different kidney function deterioration were determined by qRT-PCR using Sybr green.
Supplementary Table 1.
List of miRNAs included in the array with a differential expression in fibrotic kidney samples. 73 out of 175 analyzed miRNAs were differentially expressed in fibrotic kidneys. From these, 17 miRNAs were upregulated (A) and 56 downregulated (B) in fibrotic kidney samples compared to control ones. In red are those miRNAs selected for further study. Log10 relative quantification (RQ) and negative (−) log10 adjusted (adj.) P‐values
References
- 1.Webster A.C. Chronic kidney disease. Lancet. 2017;389(10075):1238–1252. doi: 10.1016/S0140-6736(16)32064-5. [DOI] [PubMed] [Google Scholar]
- 2.Lopez-Novoa J.M. Etiopathology of chronic tubular, glomerular and renovascular nephropathies: clinical implications. J. Transl. Med. 2011;9:13. doi: 10.1186/1479-5876-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeisberg M., Neilson E.G. Mechanisms of tubulointerstitial fibrosis. J. Am. Soc. Nephrol. 2010;21(11):1819–1834. doi: 10.1681/ASN.2010080793. [DOI] [PubMed] [Google Scholar]
- 4.Huebener P., Schwabe R.F. Regulation of wound healing and organ fibrosis by toll-like receptors. Biochim. Biophys. Acta. 2013;1832(7):1005–1017. doi: 10.1016/j.bbadis.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang H.M. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 2015;21(1):37–46. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miguel V. bioRxiv; 2020. Renal Tubule Cpt1a Overexpression Mitigates Kidney Fibrosis by Restoring Mitochondrial Homeostasis. 2020.02.18.952440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou D., Liu Y. Renal fibrosis in 2015: understanding the mechanisms of kidney fibrosis. Nat. Rev. Nephrol. 2016;12(2):68–70. doi: 10.1038/nrneph.2015.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breyer M.D., Susztak K. The next generation of therapeutics for chronic kidney disease. Nat. Rev. Drug Discov. 2016;15(8):568–588. doi: 10.1038/nrd.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandrasekaran K. Role of microRNAs in kidney homeostasis and disease. Kidney Int. 2012;81(7):617–627. doi: 10.1038/ki.2011.448. [DOI] [PubMed] [Google Scholar]
- 10.Fierro-Fernandez M., Miguel V., Lamas S. Role of redoximiRs in fibrogenesis. Redox Biol. 2016;7:58–67. doi: 10.1016/j.redox.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang-Panesso M., Humphreys B.D. Cellular plasticity in kidney injury and repair. Nat. Rev. Nephrol. 2017;13(1):39–46. doi: 10.1038/nrneph.2016.169. [DOI] [PubMed] [Google Scholar]
- 12.Petrillo F. MicroRNAs in renal diseases: a potential novel therapeutic target. Kidney Dis (Basel) 2017;3(3):111–119. doi: 10.1159/000481730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wonnacott A., Bowen T., Fraser D.J. MicroRNAs as biomarkers in chronic kidney disease. Curr. Opin. Nephrol. Hypertens. 2017;26(6):460–466. doi: 10.1097/MNH.0000000000000356. [DOI] [PubMed] [Google Scholar]
- 14.Meng X.M. TGF-beta/Smad signaling in renal fibrosis. Front. Physiol. 2015;6:82. doi: 10.3389/fphys.2015.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price N.L. Genetic deficiency or pharmacological inhibition of miR-33 protects from kidney fibrosis. JCI Insight. 2019;4(22) doi: 10.1172/jci.insight.131102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fierro-Fernandez M. MiR-9-5p protects from kidney fibrosis by metabolic reprogramming. Faseb. J. 2020;34(1):410–431. doi: 10.1096/fj.201901599RR. [DOI] [PubMed] [Google Scholar]
- 17.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Chevalier R.L., Forbes M.S., Thornhill B.A. Ureteral obstruction as a model of renal interstitial fibrosis and obstructive nephropathy. Kidney Int. 2009;75(11):1145–1152. doi: 10.1038/ki.2009.86. [DOI] [PubMed] [Google Scholar]
- 19.Fink M., Henry M., Tange J.D. Experimental folic acid nephropathy. Pathology. 1987;19(2):143–149. doi: 10.3109/00313028709077125. [DOI] [PubMed] [Google Scholar]
- 20.Griffiths-Jones S. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedman R.C. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krek A. Combinatorial microRNA target predictions. Nat. Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 23.Dweep H., Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat. Methods. 2015;12(8):697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 24.Betel D. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125(1–2):279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 26.van der Windt G.J., Chang C.H., Pearce E.L. Measuring bioenergetics in T cells using a seahorse extracellular Flux analyzer. Curr. Protoc. Im. 2016;113:3 16B 1–3 16B 14. doi: 10.1002/0471142735.im0316bs113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravi S. Defining the effects of storage on platelet bioenergetics: the role of increased proton leak. Biochim. Biophys. Acta. 2015;1852(11):2525–2534. doi: 10.1016/j.bbadis.2015.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chazotte B. Labeling mitochondria with TMRM or TMRE. Cold Spring Harb. Protoc. 2011;2011(7):895–897. doi: 10.1101/pdb.prot5641. [DOI] [PubMed] [Google Scholar]
- 29.Matsushita K. Comparison of risk prediction using the CKD-EPI equation and the MDRD study equation for estimated glomerular filtration rate. J. Am. Med. Assoc. 2012;307(18):1941–1951. doi: 10.1001/jama.2012.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solez K. Banff 07 classification of renal allograft pathology: updates and future directions. Am. J. Transplant. 2008;8(4):753–760. doi: 10.1111/j.1600-6143.2008.02159.x. [DOI] [PubMed] [Google Scholar]
- 31.Badal S.S., Danesh F.R. MicroRNAs and their applications in kidney diseases. Pediatr. Nephrol. 2015;30(5):727–740. doi: 10.1007/s00467-014-2867-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz-Ortega M. Targeting the progression of chronic kidney disease. Nat. Rev. Nephrol. 2020;16(5):269–288. doi: 10.1038/s41581-019-0248-y. [DOI] [PubMed] [Google Scholar]
- 33.Miguel V., Lamas S. Chapter 37 - redox distress in organ fibrosis: the role of noncoding RNAs. In: Sies H., editor. Oxidative Stress. Academic Press; 2020. pp. 779–820. [Google Scholar]
- 34.Chung K.W. Impairment of PPARalpha and the fatty acid oxidation pathway aggravates renal fibrosis during aging. J. Am. Soc. Nephrol. 2018;29(4):1223–1237. doi: 10.1681/ASN.2017070802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duann P., Lin P.H. Mitochondria damage and kidney disease. Adv. Exp. Med. Biol. 2017;982:529–551. doi: 10.1007/978-3-319-55330-6_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jornayvaz F.R., Shulman G.I. Regulation of mitochondrial biogenesis. Essays Biochem. 2010;47:69–84. doi: 10.1042/bse0470069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han S.H. PGC-1alpha protects from notch-induced kidney fibrosis development. J. Am. Soc. Nephrol. 2017;28(11):3312–3322. doi: 10.1681/ASN.2017020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang S. Jagged1/Notch2 controls kidney fibrosis via Tfam-mediated metabolic reprogramming. PLoS Biol. 2018;16(9) doi: 10.1371/journal.pbio.2005233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chau B.N. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci. Transl. Med. 2012;4(121):121ra18. doi: 10.1126/scitranslmed.3003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding E. Plasma microRNAs expression profile in female workers occupationally exposed to mercury. J. Thorac. Dis. 2016;8(5):833–841. doi: 10.21037/jtd.2016.03.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang L. A microRNA-30e/mitochondrial uncoupling protein 2 axis mediates TGF-beta1-induced tubular epithelial cell extracellular matrix production and kidney fibrosis. Kidney Int. 2013;84(2):285–296. doi: 10.1038/ki.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu K. Down-regulation of MicroRNA-214 contributed to the enhanced mitochondrial transcription factor A and inhibited proliferation of colorectal cancer cells. Cell. Physiol. Biochem. 2018;49(2):545–554. doi: 10.1159/000492992. [DOI] [PubMed] [Google Scholar]
- 43.Wu K. Roles of mitochondrial transcription factor A and microRNA5903p in the development of colon cancer. Mol. Med. Rep. 2016;14(6):5475–5480. doi: 10.3892/mmr.2016.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caravia X.M. The microRNA-29/PGC1alpha regulatory axis is critical for metabolic control of cardiac function. PLoS Biol. 2018;16(10) doi: 10.1371/journal.pbio.2006247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zorova L.D. Mitochondrial membrane potential. Anal. Biochem. 2018;552:50–59. doi: 10.1016/j.ab.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou H. miR-150 promotes renal fibrosis in lupus nephritis by downregulating SOCS1. J. Am. Soc. Nephrol. 2013;24(7):1073–1087. doi: 10.1681/ASN.2012080849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luan J. LNA-anti-miR-150 ameliorated kidney injury of lupus nephritis by inhibiting renal fibrosis and macrophage infiltration. Arthritis Res. Ther. 2019;21(1):276. doi: 10.1186/s13075-019-2044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X. MicroRNA-495 inhibits the high glucose-induced inflammation, differentiation and extracellular matrix accumulation of cardiac fibroblasts through downregulation of NOD1. Cell. Mol. Biol. Lett. 2018;23:23. doi: 10.1186/s11658-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25(5):635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 50.Chen X. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22(3):125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 51.Glowacki F. Increased circulating miR-21 levels are associated with kidney fibrosis. PloS One. 2013;8(2) doi: 10.1371/journal.pone.0058014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sole C. miR-29c in urinary exosomes as predictor of early renal fibrosis in lupus nephritis. Nephrol. Dial. Transplant. 2015;30(9):1488–1496. doi: 10.1093/ndt/gfv128. [DOI] [PubMed] [Google Scholar]
- 53.Weber M.J. New human and mouse microRNA genes found by homology search. FEBS J. 2005;272(1):59–73. doi: 10.1111/j.1432-1033.2004.04389.x. [DOI] [PubMed] [Google Scholar]
- 54.Angelini T.G., Emanueli C. MicroRNAs as clinical biomarkers? Front. Genet. 2015;6:240. doi: 10.3389/fgene.2015.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]