Abstract
Prostate cancer is the most commonly diagnosed non-cutaneous cancers in North American men. While androgen deprivation has remained as the cornerstone of prostate cancer treatment, resistance ensues leading to lethal disease. Forkhead box A1 (FOXA1) encodes a pioneer factor that induces open chromatin conformation to allow the binding of other transcription factors. Through direct interactions with the Androgen Receptor (AR), FOXA1 helps to shape AR signaling that drives the growth and survival of normal prostate and prostate cancer cells. FOXA1 also possesses an AR-independent role of regulating epithelial-to-mesenchymal transition (EMT). In prostate cancer, mutations converge onto the coding sequence and cis-regulatory elements (CREs) of FOXA1, leading to functional alterations. In addition, FOXA1 activity in prostate cancer can be modulated post-translationally through various mechanisms such as LSD1-mediated protein demethylation. In this review, we describe the latest discoveries related to the function and regulation of FOXA1 in prostate cancer, pointing to their relevance to guide future clinical interventions.
Keywords: FOXA1, pioneer factor, transcription factor, prostate cancer, epigenetics
Introduction
Prostate cancer is the most commonly diagnosed non-cutaneous cancer in North American men (American Cancer Society 2019; Canadian Cancer Society 2019), driven primarily by AR signaling for growth and survival. Given such, targeting AR signaling has been the dominant treatment strategy. Despite advances in AR pathway inhibitors, metastatic disease such as metastatic castration-resistant prostate cancer (mCRPC) and neuroendocrine prostate cancer (NEPC) arises in around 30% of patients (Grossfeld et al. 2003). mCRPC and NEPC remain lethal due to the lack of effective targeted therapies to overcome therapeutic resistance (Watson et al. 2015; Beltran et al. 2016). There is a clear need for novel strategies to manage prostate cancer, especially in the advanced metastatic setting.
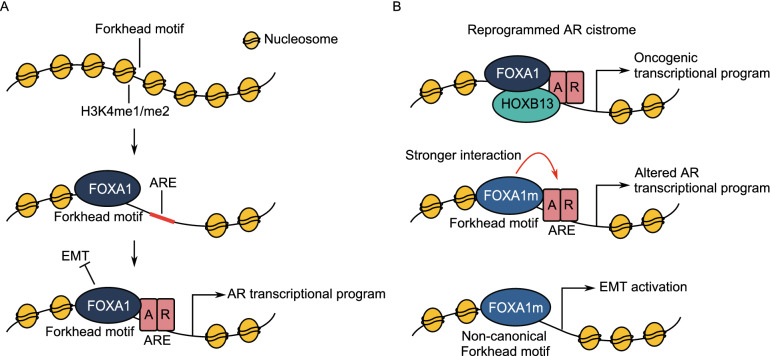
FOXA1 is a driver of prostate cancer onset and progression (Sahu et al. 2011; Gerhardt et al. 2012; Barbieri et al. 2012; Grasso et al. 2012). As a pioneer factor, it induces open chromatin conformations for the subsequent binding of lineage-specific transcription factors such as AR in prostate tissue. FOXA1 recruitment to the chromatin is facilitated by the loss of DNA methylation and the presence of histone methylation, specifically H3K4me1 and H3K4me2 modifications (Fig. 1A) (Lupien et al. 2008; Wang et al. 2009b; Sérandour et al. 2011). By directly interacting with AR to influence its recruitment to discrete genomic regions, FOXA1 regulates transcriptional programs of relevance in both normal prostate tissue and cancer (Gao et al. 2003; Sahu et al. 2013; Pomerantz et al. 2015). The silencing of FOXA1 results in significant alteration in AR binding and gene expression (Wang et al. 2011; Sahu et al. 2011, 2013; Jin et al. 2014). In prostate tumours, FOXA1 is capable of reprogramming AR binding sites and drives oncogenic programs along with transcription factor HOXB13 (Fig. 1B) (Pomerantz et al. 2015, 2020) . In addition, FOXA1 is reported as a suppressor of neuroendocrine differentiation and its loss of expression can promote NEPC progression (Kim et al. 2017; Rotinen et al. 2018). FOXA1 is also reported as the third most frequently mutated gene in prostate cancer (Barbieri et al. 2012). Despite these previous findings, our understanding of the mechanisms and the extent of FOXA1 involvement in prostate cancer remains incomplete. Recent studies reveal that FOXA1 is functionally altered by both coding and noncoding mutations which drives prostate cancer progression (Adams et al. 2019; Parolia et al. 2019; Gao et al. 2019; Xu et al. 2019). In addition, the regulation of FOXA1 through CREs and post-translational modifications is also revealed, opening up a new avenue for FOXA1 targeting (Zhou et al. 2020; Gao et al. 2020). In this review, we summarize the recent research on FOXA1 in prostate cancer and offer a perspective on the clinical value of FOXA1 given these findings.
Figure 1.
Schematic of FOXA1 activity in (A) normal prostate and (B) prostate cancer. (A) FOXA1 preferentially binds to genomic Forkhead motifs marked by H3K4me1/me2 histone modifications. It induces open chromatin conformation and allows for the sequential binding of AR to drive AR transcriptional programs. FOXA1 also inhibits EMT independent of AR. (B) In prostate cancer, FOXA1 can extensively reprogram AR cistrome along with HOXB13. FOXA1 mutations can result in stronger AR binding and significantly alter AR transcriptional programs. FOXA1 mutants can also induce EMT programs to drive cancer metastasis. Abbreviations: ARE, androgen-receptor response element; EMT, epithelial-mesenchymal transition
Coding mutations alter FOXA1 function
FOXA1 is frequently altered by coding mutations
FOXA1 is recurrently mutated in its coding sequence in up to 9% and 13% of primary prostate cancer and mCRPC, respectively (Barbieri et al. 2012; Grasso et al. 2012; Huang et al. 2017; Adams et al. 2019; Parolia et al. 2019). In NEPC, a study of 29 tumour tissues identified single nucleotide variants (SNVs) mapped to FOXA1 in ~25% of the tumours (Beltran et al. 2020). The frequency of FOXA1 mutations has also been recently shown to vary with ethnic background. The analysis into the Chinese Prostate Cancer Genome and Epigenome Atlas (CPGEA) cohort revealed FOXA1 mutation in 41% of localized prostate tumours, a rate significantly higher than Western cohorts (Li et al. 2020). Taken together, FOXA1 is a recurrently mutated gene across various stages of prostate cancer.
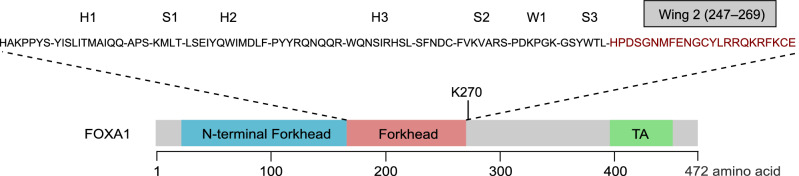
While mutations can occur throughout the FOXA1 coding sequence, >50% of them map to the nucleotides encoding the Forkhead DNA binding domain (Clark et al. 1993; Cancer Genome Atlas Research Network 2015; Adams et al. 2019; Parolia et al. 2019; Gao et al. 2019; Xu et al. 2019; Li et al. 2020). Mutations affecting the Forkhead domain are predominantly missense point mutations and non-truncating insertions and deletions (indels) (Clark et al. 1993; Cancer Genome Atlas Research Network 2015; Adams et al. 2019; Parolia et al. 2019; Gao et al. 2019; Xu et al. 2019). In particular, the majority of mutations in the Forkhead domain cluster at nucleotides encoding residues of the second winged loop (Wing2, residues 247–269), important for DNA binding and nuclear movements (Fig. 2) (Clark et al. 1993; Sekiya et al. 2009; Cancer Genome Atlas Research Network 2015; Adams et al. 2019; Parolia et al. 2019; Li et al. 2020). Wing2 mutations are suggested to originate in the localized stage of prostate cancer and show no enrichment in metastatic diseases (Parolia et al. 2019). Beyond mutations in the Forkhead domain, truncating frameshifts account for 20% of the FOXA1 mutations (Adams et al. 2019). They are restricted to the C-terminal half of the FOXA1 protein downstream of the Forkhead domain and are significantly enriched in metastatic prostate cancers (Adams et al. 2019; Parolia et al. 2019). The C-terminal half of FOXA1 contains an ɑ-helical region recently identified to be critical for histone interactions and chromatin opening (Iwafuchi et al. 2020). FOXA1 coding mutations emerge throughout prostate cancer progression and cluster at hotspot regions that are important for FOXA1 protein function.
Figure 2.
Protein map of FOXA1 functional domains and secondary structures. Residues in red represent Wing2 mutational hotspot. FOXA1 methylation site at lysine 270 (K270) is highlighted. Abbreviation: TA, transactivation domain
Coding mutations of FOXA1 lead to altered transcription factor activity
FOXA1 coding mutations can lead to changes in DNA binding affinity and nuclear movements (Table 1). Mutations affecting the Forkhead Wing2 loop such as M253K, FENG254-7>C (deletion of residues 254 to 257, replaced by C) and F266S decreased DNA binding affinity, resulting in the global reduction of FOXA1 genomic binding (Xu et al. 2019). However, they retain the ability to form base-specific interactions at the Forkhead motifs on DNA sequence (Parolia et al. 2019; Xu et al. 2019). These FOXA1 mutants displayed a loss-of-function phenotype where they failed to rescue FOXA1-mediated gene expression upon wild-type FOXA1 knockdown in LNCaP cells (Xu et al. 2019). In comparison, other mutants such as I176M and R261G in the Forkhead domain displayed increased nuclear movement 5–6 times compared to wild-type FOXA1 which translated to enhanced nuclear activities (Parolia et al. 2019). These mutations also led to 3–6 fold greater transcriptional activation compared to wild-type FOXA1 in luciferase reporter assays. Given the critical function of the Forkhead domain of FOXA1, mutations affecting the region can result in altered DNA binding ability and nuclear movements which lead to functional changes.
Table 1.
Summary of key functionally validated FOXA1 coding mutations
| Type | Location | Mutation | Key functional alterations | Ref |
|---|---|---|---|---|
| Missense | FKHD | I176M | Increased nuclear mobility; higher activation of AR signaling; increased cell growth in androgen-depleted condition | (Parolia et al. 2019) |
| FKHD | R219C, R219S | Decreased transcriptional activity; increased growth of mouse organoid; preference for non-canonical FOXA1 binding motif; promotes EMT | (Adams et al. 2019) | |
| FKHD | D226N, D226G | Increased growth of mouse organoid; mixed results on the effect of AR activity; promotes EMT | (Adams et al. 2019; Gao et al. 2019; Xu et al. 2019) | |
| FKHD Wing2 | H247Q, H247R, H247Y | Increased growth of mouse organoid; mixed results on the effect of AR activity; promotes EMT | (Adams et al. 2019; Gao et al. 2019) | |
| FKHD Wing2 | M253K | Increased growth of mouse organoid; mixed results on the effect of AR activity; reduced chromatin binding; promotes EMT; increased cell growth in androgen-depleted condition | (Adams et al. 2019; Gao et al. 2019; Xu et al. 2019) | |
| FKHD Wing2 | Y259C, Y259S | Increased transcriptional activity; increased growth of mouse organoid | (Adams et al. 2019) | |
| FKHD Wing2 | R261G, R261C | Increased nuclear mobility; higher activation of AR signaling; increased cell growth in androgen-depleted condition; increased growth of mouse organoid | (Adams et al. 2019; Parolia et al. 2019) | |
| FKHD Wing2 | F266L, F266S | Increased growth of mouse organoid; mixed results on the effect of AR activity; reduced chromatin binding; increased cell growth in androgen-depleted condition; promotes EMT | (Adams et al. 2019; Xu et al. 2019) | |
| Indels | FKHD Wing2 | ΔM253–N256 | Increased transcriptional activity; increased growth of mouse organoid | (Adams et al. 2019) |
| FKHD Wing2 | ΔF254/E255 | Increased transcriptional activity; increased growth of mouse organoid | (Adams et al. 2019) | |
| FKHD Wing2 | FENG254-7>C | Reduced chromatin binding; increased cell growth in androgen-depleted condition; promotes EMT | (Xu et al. 2019) | |
| FKHD Wing2 | ΔR265-Q271 | Increased nuclear mobility; higher activation of AR signaling; increased cell growth in androgen-depleted condition | (Parolia et al. 2019) | |
| Frameshift truncations | C-term | P358fs | Higher DNA-binding affinity; increased invasion through activation of WNT pathway | (Parolia et al. 2019) |
| C-term | G275X | Increased transcriptional activity; increased growth of mouse organoid | (Adams et al. 2019) |
FOXA1 mutants can remodel the accessible chromatin landscapes of prostate cancer (Table 1). Through the use of Assay for Transposase-Accessible Chromatin using Sequencing (ATAC-seq), the overexpression of wild-type FOXA1 led to >1000 more accessible chromatin peaks in mouse prostate organoids compared to empty vector control (Adams et al. 2019). In comparison, FOXA1 Forkhead mutants such as ΔF254/E255 and R219S enhanced pioneer factor activity in comparison to wild-type, evident by the ability to induce more accessible sites in total (Adams et al. 2019). The rate of chromatin opening by the mutants was also ~5 times faster than wild-type FOXA1 (Adams et al. 2019). Although ΔF254/E255 and R219S mutants both induced large numbers of accessible sites in the genome, unsupervised clustering analysis of these sites revealed distinct patterns (Adams et al. 2019). ΔF254/E255 mutant showed accessible chromatin peaks at genomic locations similar to wild-type FOXA1. In contrast, R219S mutant induced novel accessible chromatin regions that were enriched for non-canonical Forkhead binding motif (Adams et al. 2019). Other Forkhead mutants such as D226G and M253K led to change in the preference from classic Forkhead binding motifs to non-canonical motifs (Gao et al. 2019). Along with altered ability to induce chromatin accessibility, FOXA1 mutants can induce an unique epigenetic landscape that is distinct from wild-type FOXA1 in prostate cancer.
As a pivotal regulator of AR, FOXA1 mutations can affect FOXA1’s interaction with AR, altering androgen signaling (Table 1). However, both AR promoting and inhibiting functions of FOXA1 have been reported, revealing the complex interplay between FOXA1 and AR. Analysis of large tumour cohorts such as The Cancer Genome Atlas (TCGA) revealed that tumours with FOXA1 mutations have a higher AR transcriptional signature compared to normal tissues and other prostate tumour subtypes (Cancer Genome Atlas Research Network 2015; Parolia et al. 2019). Mouse prostate organoid study further supported the TCGA findings as organoids expressing multiple FOXA1 mutants showed exaggerated AR-positive luminal formation (Adams et al. 2019). Conversely, other studies reported that FOXA1 mutants led to downregulation of AR signaling, suggesting an inhibitory function (Gao et al. 2019; Xu et al. 2019). In LNCaP cells expressing Forkhead mutants such as D226G, H247Y and M253K, FOXA1 and AR binding at enhancers of well-established AR-regulated genes such as NKX3-1 and KLK3 were significantly reduced (Gao et al. 2019). In fact, a global inhibition of AR binding at both FOXA1-dependent and -independent sites was reported (Xu et al. 2019). These mutants may prevent AR binding to chromatin owing to their increased interaction with AR but decreased chromatin binding ability (Fig. 1B) (Gao et al. 2019; Xu et al. 2019). As a result, NKX3-1, KLK3 and other AR-related genes were found significantly downregulated in cells expressing FOXA1 mutants (Gao et al. 2019). FOXA1 mutations resulted in different AR signaling patterns. Both inhibitory and promoting functions were reported and the activity may vary based on the mutations and models used in the study.
Studies on the coding mutations of FOXA1 have revealed distinct activities in terms of regulating AR signaling and open chromatin landscapes (Adams et al. 2019; Parolia et al. 2019; Gao et al. 2019; Xu et al. 2019; Hankey et al. 2020). FOXA1 mutants may contribute to specific stages of prostate cancer progression given their divergent activities due to mutations.
Coding mutations in FOXA1 promote EMT and metastasis
While FOXA1 promotes cell proliferation through AR, it also possesses an AR-independent function of inhibiting EMT and cell invasion (Fig. 1A) (Jin et al. 2013). 22RV1 prostate cancer cells expressing FOXA1 C-terminal truncation mutants showed an accumulation of transcriptionally active β-catenin, indicative of WNT pathway activation (Parolia et al. 2019). Consequently, these cells displayed 2–3 fold increased invasion in vitro and greater ability to metastasize in vivo in zebrafish embryos (Fig. 1B) (Parolia et al. 2019). Mechanistically, EMT activation by C-terminal truncation mutants may be due to the loss of interaction with TLE3 which is a WNT repressor (Parolia et al. 2019). Several missense and non-truncating indel Forkhead mutants also demonstrated similar ability to induce EMT and metastasis (Jin et al. 2013; Adams et al. 2019; Gao et al. 2019; Xu et al. 2019). In particular, RNA sequencing of mouse prostate organoids expressing FOXA1 mutant R219S revealed an enrichment of EMT genes which was not observed with other Forkhead mutants such as D226N and ΔF254/E255 (Adams et al. 2019). R219S is enriched in NEPC compared to prostate adenocarcinoma, a subtype in which EMT is implicated (Adams et al. 2019). EMT was also activated in CWR22-RV1 mCRPC cells expressing D226G, H247Y and M253K FOXA1 Forkhead mutants (Gao et al. 2019). It remains unclear, however, how these FOXA1 mutants induce EMT, whether it is a loss of ability to inhibit EMT or a gain of function of the mutants to directly induce gene expression changes (Xu et al. 2019). Despite some missing knowledge, EMT inhibition may be a novel strategy to treat metastatic prostate cancer with FOXA1 mutations. The ability of FOXA1 mutants to activate EMT may play a role in prostate cancer metastasis and progression in certain types such as NEPC.
Noncoding mutations converge on FOXA1
Mutations converge onto FOXA1 cis-regulatory landscape to drive oncogenic expression
The genome is organized through three-dimensional chromatin looping interactions between CREs (Dixon et al. 2012, 2016; Nora et al. 2012; Szabo et al. 2019). The careful organization of these interactions leads to cell-type specific gene expression programs. In cancer, mutations including SNVs and structural variants (SVs) can alter the activity and interactions between CREs to drive aberrant gene expression, which is seen in the case of FOXA1.
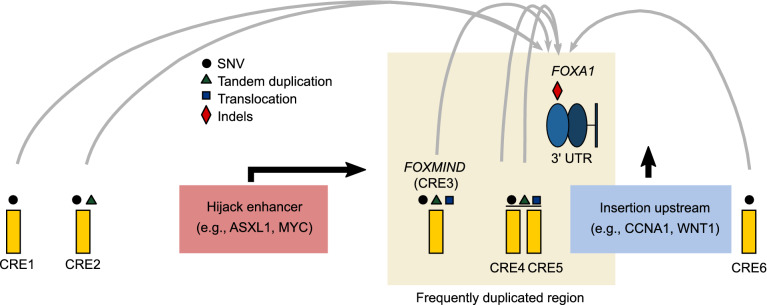
FOXA1 expression is altered by noncoding mutations across prostate cancer stages. Functional studies revealed 6 CREs involved in regulating FOXA1 mRNA expression (Fig. 3) (Parolia et al. 2019; Zhou et al. 2020) with each being targeted by mutations. Consistent with the essentiality of FOXA1, the deletion of each CRE of the plexus resulted in significant reduction in FOXA1 mRNA expression and LNCaP prostate cancer cell growth (Zhou et al. 2020). In primary prostate cancer, 6 SNVs from 5/200 patients (2.5%) were mapped to the 6 CREs. These SNVs were shown to have gain-of-function activity through modulating the binding of AR, FOXA1, HOXB13 and SOX9 in vitro (Zhou et al. 2020). Consistent with the importance of FOXA1 cis-regulatory landscape in disease progression, SVs were found to target loci harbouring the CREs in up to 30% of mCRPC tumours (Parolia et al. 2019). Together with coding mutations, it was estimated that the cumulative frequency of FOXA1 alterations is over 34% in mCRPC (Parolia et al. 2019). The SVs affecting FOXA1 predominantly consist of translocation and tandem duplication and are significantly enriched in mCRPC (Parolia et al. 2019). Two types of translocation events involving the FOXA1 CREs emerge in these tumours. The first involves the repositioning of genes from other chromosomes (i.e., ASXL1, ERG, ETV1, MYC, and SKIL) into the proximity of a FOXA1 CRE termed FOXMIND (Fig. 3) (Quigley et al. 2018; Parolia et al. 2019). The second type of translocation involves the insertion of potent oncogenes, namely WNT1, HOXA1 and CCNA1, upstream of the FOXA1 promoter. Both types of translocation events hijacked the cis-regulatory activity of FOXA1 CREs to significantly upregulate the oncogenic expression of these genes, including FOXA1 in mCRPC tumours (Parolia et al. 2019). Tandem duplications within the FOXA1 locus account for 70% of all SVs identified across >1,400 primary and advanced prostate tumours (Parolia et al. 2019). They predominantly flank the loci harbouring FOXMIND-FOXA1 (Fig. 3) as FOXA1 and FOXMIND are co-duplicated in 89% of the tumours with tandem duplications, driving aberrant FOXA1 and MIPOL1 mRNA expression (Parolia et al. 2019). These efforts highlight that the cis-regulatory plexus of FOXA1 can be recurrently hijacked in prostate tumours to drive aberrant gene expression and disease progression.
Figure 3.
Noncoding mutations converge onto FOXA1. FOXA1 locus is frequently altered by structural rearrangements inclusive of translocations and tandem duplications. Translocations can result in genes hijacking the activity of the FOXMIND enhancer. In addition, translocations can insert oncogenes upstream of FOXA1. Along with FOXMIND, 5 additional functional CREs harbouring SNVs are identified near FOXA1 locus which can affect the expression of FOXA1. The 3′ UTR region of FOXA1 is frequently mutated with indels being the dominant type of mutation. Abbreviations: SNV, single nucleotide variant; 3′ UTR, 3′ untranslated region; CRE, cis-regulatory elements
Noncoding mutations alter the binding of FOXA1 genome-wide
Germline single-nucleotide polymorphisms (SNPs) and somatic SNVs can modulate FOXA1 genomic binding and transcription regulation. Genome-Wide Association Studies (GWAS) identified more than 160 risk loci to be associated with prostate cancer susceptibility (Farashi et al. 2019) with the majority of SNPs map to the noncoding genome (Maurano et al. 2012). These SNPs have the ability to confer disease risk through modulating the binding of transcription factors such as FOXA1 to CREs (Zhang et al. 2014; Zhou et al. 2016). For instance, the rs1859961 variant allele being a G at the 17q24.3 risk locus can significantly decrease FOXA1 binding intensity at the active CRE driving SOX9 oncogenic expression (Zhang et al. 2012). Similar findings were observed at the 8q24 prostate cancer risk locus where rs183373024 can abolish FOXA1 binding (Hazelett et al. 2013). Expanding on this concept, prostate cancer risk SNPs have the propensity to pose allele-specific binding towards AR-FOXA1 complexes (Whitington et al. 2016). Genome-wide analysis of prostate cancer risk SNPs and somatic SNVs demonstrated that they converge and enrich at the binding sites of other master transcription factors besides FOXA1, including AR, HOXB13 and SOX9 (Ahmed et al. 2017; Mazrooei et al. 2019). Deeper analyses revealed that the >270,000 somatic SNVs identified from 200 primary prostate tumours (Fraser et al. 2017; Espiritu et al. 2018) were enriched at tumour-specific and shared binding sites of these master transcription factors, but not at benign-specific binding sites (Mazrooei et al. 2019). While most of these SNVs are likely passengers (Sabarinathan et al. 2016; Mazrooei et al. 2019), a small subset of these SNVs can significantly increase FOXA1 binding at CREs that regulate MYC (Mazrooei et al. 2019) and FOXA1 (Zhou et al. 2020) itself. These SNVs and SNPs add another layer of complexity whereby FOXA1 confers prostate cancer complexity to the regulation of FOXA1 cistrome.
Post-translational modifications alter FOXA1 activity
In addition to transcriptional regulations, FOXA1 activity can also be modulated post-translationally through acetylation, phosphorylation, methylation, and the addition of small ubiquitin-like modifiers (SUMO) (Kohler and Cirillo 2010; Sutinen et al. 2014; Yamaguchi et al. 2017; Gao et al. 2020). FOXA1 is reported to be acetylated by histone acetyltransferase p300 at 11 putative lysine residues, of which 5 of them are clustered within the Forkhead domain. Acetylation can attenuate FOXA1 binding to DNA and reduce its ability to remodel chromatin (Kohler and Cirillo 2010). Similarly, the addition of SUMO to lysine residues of FOXA1 can also result in downregulation of its transcriptional activity and nuclear mobility (Sutinen et al. 2014). Furthermore, phosphorylation by c-Abl is also reported, highlighting that FOXA1 is subjected to various post-translational modifications (Yamaguchi et al. 2017). Recently, methylation mediated by Lysine specific demethylase 1 (LSD1) has been identified as an additional mechanism that regulates FOXA1 activity and holds clinical potential as an avenue to target FOXA1 (Gao et al. 2020). LSD1 is known as a transcriptional repressor that demethylates enhancer-associated H3K4me1/me2 (Shi et al. 2004). In addition to its repressor function, LSD1 is implicated in prostate cancer as a coactivator of AR signaling (Metzger et al. 2005; Wissmann et al. 2007; Cai et al. 2014). Upon LSD1 knockdown with siRNA in LNCaP cells, androgen-stimulated genes were broadly suppressed (Cai et al. 2014). ChIP-seq analysis revealed that the LSD1 genomic binding sites largely associate with FOXA1 and active enhancer marks (Cai et al. 2014). Through immunoprecipitation, FOXA1 was found to physically interact with LSD1 in an AR-independent manner (Cai et al. 2014). Although LSD1 is best characterized as a histone demethylase, it can also demethylate nonhistone proteins (Huang et al. 2007; Wang et al. 2009a). A recent study showed that LSD1 is capable of demethylate K270 residue of FOXA1 (Gao et al. 2020). This residue is located at the carboxyl-end of the Wing2 region and has been shown to be important for the FOXA1 interaction with core histones (Fig. 2) (Iwafuchi et al. 2020). The demethylation of FOXA1 K270 by LSD1 is required for its optimal binding to the chromatin. Upon LSD1 inhibition, a larger fraction of FOXA1 remained methylated at K270 which led to decreased global binding and ability to induce open chromatins. As a result, AR signaling was significantly altered due to impaired FOXA1 pioneer activity (Cai et al. 2014; Gao et al. 2020). The regulation of FOXA1 activity remained unclear for many years which contributed to the difficulty of targeting FOXA1 as a therapeutic target. The recent knowledge that FOXA1 activity can be regulated by post-translational modifiers such as LSD1 opens up a novel avenue to target FOXA1.
From discovery to clinical applications
While recent reports provided new exciting insights on FOXA1, questions regarding their clinical utility remain. Recent studies looking at the implications of FOXA1 across different stages of prostate cancer revealed new functional roles relevant for prognosis, therapeutic response and targeted therapy.
Chemically antagonizing FOXA1 and downstream pathways
Transcription factors such as FOXA1 are known to be difficult to inhibit directly due to their nuclear localization and mechanisms of action (Dang et al. 2017). Modern technologies are expanding our toolkits to inhibit FOXA1 from other angles. As case and point, a drug screen comprising 550 drugs across three breast cancer cell lines identifies CDK1 as a therapeutic target for FOXA1 inhibition (Wang et al. 2018). In the same vein, numerous FDA-approved drug libraries are commercially available for researchers to evaluate new inhibitors for previously undruggable targets. Academic consortia such as Structural Genomic Consortium (Arrowsmith et al. 2012; Scheer et al. 2019; Wu et al. 2019) also offer chemical probes that directly inhibit epigenetic factors implicated in FOXA1 regulation (Lupien et al. 2008; Sérandour et al. 2011; Müller et al. 2018). An example is the new finding that LSD1 regulates FOXA1 activity which implies the use of LSD1 inhibitors may be a viable approach to antagonize FOXA1 activity in prostate cancer (Gao et al. 2020). The LSD1 inhibitors such as GSK2879552 and SP-2509 have shown effectiveness in suppressing prostate tumour cell growth with and without the use of enzalutamide in combination in preclinical animal models (Sehrawat et al. 2018; Gao et al. 2020). In addition, targeting LSD1 presents an advantage as several inhibitors such as IMG-7289 and ORY-1001 are currently in phase 2 clinical trials for essential thrombocythemia, acute myeloid leukemia and other diseases (Fang et al. 2019). Adding LSD1 inhibitors in combination with existing therapies such as enzalutamide may achieve synergistic effect in prostate cancer control (Gao et al. 2020). In addition to epigenetic factors, PARP-2 involved in DNA damage repair pathways may also be a therapeutic target as it is reported as an interactor of FOXA1 that facilitates AR recruitment and transcription (Gui et al. 2019).
Targeting FOXA1-regulated pathways such as EMT may also be a treatment strategy in combination with antiandrogen therapies. FOXA1 downregulation is shown to lead to TGF-β signaling and EMT activation which can be blocked by TGF-β receptor I inhibitor LY2157299 (Song et al. 2019). The use of LY2157299 demonstrated synergistic effects in combination with enzalutamide in effectively suppressing tumour growth and metastasis in mCRPC preclinical mouse models (Song et al. 2019). Similarly, FOXA1 Forkhead coding mutations such as D226G, H247Y and M253K can also result in EMT activation. Consequently, cells expressing these mutant FOXA1 were sensitive towards MET inhibitor crizotinib treatment in mCRPC xenograft models, which targets the c-Met protein known to induce EMT (Gao et al. 2019). Altogether, targeting FOXA1-regulated pathways such as EMT can be another avenue to improve disease control especially for the metastatic cases where EMT is implicated.
Considering the prevalence and differential FOXA1 activity conferred by specific FOXA1 mutations, future studies may consider conducting inhibition-screens with FOXA1 mutant model systems. The recent development of prostate mouse organoids (Adams et al. 2019) that harbor FOXA1 mutants shows promise as a powerful resource towards identifying mutant-specific FOXA1 inhibitors. This is especially important to maximize the therapeutic index of drugging FOXA1 by minimizing toxicity as FOXA1 is also expressed in normal tissues. This ultimately potentiates the idea of precision oncology, selecting patients for tailored therapy based on their FOXA1 mutation status.
Tracking prostate cancer using FOXA1 related mutations
FOXA1 mutations found across various stages of prostate cancer present an opportunity to track disease progression. Their inclusion in targeted sequencing panels from bodily fluids (urine or blood) can serve as a means to track minimal residual disease since normal tissues should not exhibit FOXA1 mutations. A targeted sequencing of 72 prostate cancer driver genes in the plasma cell-free DNA of patients with mCRPC revealed 37 somatic mutations mapped to FOXA1 3′ UTR in 34/290 (12%) of the patients (Fig. 3) (Annala et al. 2018). In fact, FOXA1 harboured the highest 3′ UTR mutation rate among all 72 genes of interest with indels being the dominant type of alteration (Annala et al. 2018). These indels can be found in the early stage of prostate tumours and are not present in other cancer types, such as bladder cancer, indicating prostate cancer specificity (Annala et al. 2018). The study demonstrated that FOXA1 mutations can be readily detected in liquid biopsy and can potentially serve as a prostate cancer-specific biomarker. Recent findings have shown the differential prevalence of FOXA1 mutations across prostate cancer stages, for instance the early emergence of Forkhead Wing2 coding mutations in localized disease and the enrichment of truncation mutations and SVs in metastatic diseases (Adams et al. 2019; Parolia et al. 2019). This further supports the notion that mutation status surrounding FOXA1 can inform disease progression. Based on the varying function of FOXA1 mutants, the prognostic value of different FOXA1 mutations should be further evaluated in future studies. Through identifying inhibitors that antagonize specific FOXA1 mutants, this approach potentiates personalized treatment. With the constant cellular tumour cellular turnover releasing cell-free DNA, liquid biopsy for FOXA1 mutations permits longitudinal tracking for residual disease and early detection for recurrence.
Summary
Recent functional characterization of FOXA1 mutants shed light on their complex modes of action in driving prostate cancer where the function may be divergent depending on the residues affected. Overlaying the prostate cancer epigenome reveals a catalog of CREs critical for FOXA1 mRNA expression that are extensively perturbed through various modes of somatic mutations. In addition, post-translational modifications such as protein demethylation mediated by LSD1 are identified as additional regulations of FOXA1 activity. Deeper understanding of the somatic mutations and regulatory mechanisms may yield novel biomarkers and therapeutic strategies complementary to our existing standard of care with the ultimate goal of improving patient outcomes. With present day high-throughput methodologies and model systems that have been lacking in the past, there is promise in translating the past and recent biological findings regarding FOXA1 into clinical utility.
Acknowledgments
Abbreviations
AR, androgen receptor; ATAC-seq, Assay for Transposase-Accessible Chromatin using Sequencing; ChIP-seq, chromatin immunoprecipitation followed by sequencing; CPGEA, Chinese Prostate Cancer Genome and Epigenome Atlas; CRE, cis-regulatory elements; EMT, epithelial-to-mesenchymal transition; FOXA1, Forkhead box A1; GWAS, Genome-Wide Association Studies; Indels, insertions and deletions; LSD1, lysine specific demethylase 1; mCRPC, metastatic castration-resistant prostate cancer; NEPC, neuroendocrine prostate cancer; SNP, single-nucleotide polymorphism; SNV, single nucleotide variant; SUMO, small ubiquitin-like modifiers; SV, structural variant; TCGA, The Cancer Genome Atlas
Compliance with Ethics Guidelines
Mona Teng, Stanley Zhou, Changmeng Cai, Mathieu Lupien, and Housheng Hansen He declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
Mona Teng and Stanley Zhou have contributed equally to this work.
Contributor Information
Mathieu Lupien, Email: mathieu.lupien@uhnresearch.ca.
Housheng Hansen He, Email: hansenhe@uhnresearch.ca.
References
- Adams EJ, Karthaus WR, Hoover E, Liu D, Gruet A, Zhang Z, Cho H, DiLoreto R, Chhangawala S, Liu Y, et al. FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes. Nature. 2019;571:408–412. doi: 10.1038/s41586-019-1318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M, Sallari RC, Guo H, Moore JH, He HH, Lupien M. Variant Set Enrichment: an R package to identify disease-associated functional genomic regions. BioData Min. 2017;10:9. doi: 10.1186/s13040-017-0129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Cancer Society (2019) Cancer Facts & Figures 2019
- Annala M, Taavitsainen S, Vandekerkhove G, Bacon JVW, Beja K, Chi KN, Nykter M, Wyatt AW. Frequent mutation of the FOXA1 untranslated region in prostate cancer. Commun Biol. 2018;1:122. doi: 10.1038/s42003-018-0128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat J-P, White TA, Stojanov P, Van Allen E, Stransky N, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BVSK, Varambally S, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran H, Romanel A, Conteduca V, Casiraghi N, Sigouros M, Franceschini GM, Orlando F, Fedrizzi T, Ku S-Y, Dann E, et al. Circulating tumor DNA profile recognizes transformation to castration-resistant neuroendocrine prostate cancer. J Clin Invest. 2020;130:1653–1668. doi: 10.1172/JCI131041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C, He HH, Gao S, Chen S, Yu Z, Gao Y, Chen S, Chen MW, Zhang J, Ahmed M, et al. Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell Rep. 2014;9:1618–1627. doi: 10.1016/j.celrep.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canadian Cancer Society (2019) Canadian Cancer Statistics 2019
- Cancer Genome Atlas Research Network The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Dang CV, Reddy EP, Shokat KM, Soucek L. Drugging the “undruggable” cancer targets. Nat Rev Cancer. 2017;17:502–508. doi: 10.1038/nrc.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Gorkin DU, Ren B. Chromatin domains: the unit of chromosome organization. Mol Cell. 2016;62:668–680. doi: 10.1016/j.molcel.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espiritu SMG, Liu LY, Rubanova Y, Bhandari V, Holgersen EM, Szyca LM, Fox NS, Chua MLK, Yamaguchi TN, Heisler LE, et al. The evolutionary landscape of localized prostate cancers drives clinical aggression. Cell. 2018;173:1003–1013.e15. doi: 10.1016/j.cell.2018.03.029. [DOI] [PubMed] [Google Scholar]
- Fang Y, Liao G, Yu B. LSD1/KDM1A inhibitors in clinical trials: advances and prospects. J Hematol Oncol. 2019;12:129. doi: 10.1186/s13045-019-0811-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farashi S, Kryza T, Clements J, Batra J. Post-GWAS in prostate cancer: from genetic association to biological contribution. Nat Rev Cancer. 2019;19:46–59. doi: 10.1038/s41568-018-0087-3. [DOI] [PubMed] [Google Scholar]
- Fraser M, Sabelnykova VY, Yamaguchi TN, Heisler LE, Livingstone J, Huang V, Shiah Y-J, Yousif F, Lin X, Masella AP, et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature. 2017;541:359–364. doi: 10.1038/nature20788. [DOI] [PubMed] [Google Scholar]
- Gao N, Zhang J, Rao MA, Case TC, Mirosevich J, Wang Y, Jin R, Gupta A, Rennie PS, Matusik RJ. The role of hepatocyte nuclear factor-3 alpha (Forkhead Box A1) and androgen receptor in transcriptional regulation of prostatic genes. Mol Endocrinol. 2003;17:1484–1507. doi: 10.1210/me.2003-0020. [DOI] [PubMed] [Google Scholar]
- Gao S, Chen S, Han D, Barrett D, Han W, Ahmed M, Patalano S, Macoska JA, He HH, Cai C. Forkhead domain mutations in FOXA1 drive prostate cancer progression. Cell Res. 2019;29:770–772. doi: 10.1038/s41422-019-0203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Chen S, Han D, Wang Z, Li M, Han W, Besschetnova A, Liu M, Zhou F, Barrett D, et al. Chromatin binding of FOXA1 is promoted by LSD1-mediated demethylation in prostate cancer. Nat Genet. 2020 doi: 10.1038/s41588-020-0681-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt J, Montani M, Wild P, Beer M, Huber F, Hermanns T, Müntener M, Kristiansen G. FOXA1 promotes tumor progression in prostate cancer and represents a novel hallmark of castration-resistant prostate cancer. Am J Pathol. 2012;180:848–861. doi: 10.1016/j.ajpath.2011.10.021. [DOI] [PubMed] [Google Scholar]
- Grasso CS, Wu Y-M, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossfeld GD, Latini DM, Lubeck DP, Mehta SS, Carroll PR. Predicting recurrence after radical prostatectomy for patients with high risk prostate cancer. J Urol. 2003;169:157–163. doi: 10.1016/S0022-5347(05)64058-X. [DOI] [PubMed] [Google Scholar]
- Gui B, Gui F, Takai T, Feng C, Bai X, Fazli L, Dong X, Liu S, Zhang X, Zhang W, et al. Selective targeting of PARP-2 inhibits androgen receptor signaling and prostate cancer growth through disruption of FOXA1 function. Proceedings of the National Academy of Sciences. 2019;116:14573–14582. doi: 10.1073/pnas.1908547116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankey W, Chen Z, Wang Q. Shaping chromatin states in prostate cancer by pioneer transcription factors. Cancer Res. 2020 doi: 10.1158/0008-5472.CAN-19-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelett DJ, Coetzee SG, Coetzee GA. A rare variant, which destroys a FoxA1 site at 8q24, is associated with prostate cancer risk. Cell Cycle. 2013;12:379–380. doi: 10.4161/cc.23201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FW, Mosquera JM, Garofalo A, Oh C, Baco M, Amin-Mansour A, Rabasha B, Bahl S, Mullane SA, Robinson BD, et al. Exome sequencing of African-American prostate cancer reveals loss-of-function ERF mutations. Cancer Discov. 2017;7:973–983. doi: 10.1158/2159-8290.CD-16-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- Iwafuchi M, Cuesta I, Donahue G, Takenaka N, Osipovich AB, Magnuson MA, Roder H, Seeholzer SH, Santisteban P, Zaret KS. Gene network transitions in embryos depend upon interactions between a pioneer transcription factor and core histones. Nat Genet. 2020;52:418–427. doi: 10.1038/s41588-020-0591-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H-J, Zhao JC, Ogden I, Bergan RC, Yu J. Androgen receptor-independent function of FoxA1 in prostate cancer metastasis. Cancer Res. 2013;73:3725–3736. doi: 10.1158/0008-5472.CAN-12-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H-J, Zhao JC, Wu L, Kim J, Yu J. Cooperativity and equilibrium with FOXA1 define the androgen receptor transcriptional program. Nat Commun. 2014;5:3972. doi: 10.1038/ncomms4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Jin H, Zhao JC, Yang YA, Li Y, Yang X, Dong X, Yu J. FOXA1 inhibits prostate cancer neuroendocrine differentiation. Oncogene. 2017;36:4072–4080. doi: 10.1038/onc.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S, Cirillo LA. Stable chromatin binding prevents FoxA acetylation, preserving FoxA chromatin remodeling. J Biol Chem. 2010;285:464–472. doi: 10.1074/jbc.M109.063149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Xu C, Lee HJ, Ren S, Zi X, Zhang Z, Wang H, Yu Y, Yang C, Gao X, et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature. 2020;580:93–99. doi: 10.1038/s41586-020-2135-x. [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazrooei P, Kron KJ, Zhu Y, Zhou S, Grillo G, Mehdi T, Ahmed M, Severson TM, Guilhamon P, Armstrong NS, et al. Cistrome partitioning reveals convergence of somatic mutations and risk variants on master transcription regulators in primary prostate tumors. Cancer Cell. 2019;36:674–689.e6. doi: 10.1016/j.ccell.2019.10.005. [DOI] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AHFM, Günther T, Buettner R, Schüle R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Müller S, Ackloo S, Arrowsmith CH, Bauser M, Baryza JL, Blagg J, Böttcher J, Bountra C, Brown PJ, Bunnage ME, et al. Science forum: donated chemical probes for open science. Elife. 2018;7:e34311. doi: 10.7554/eLife.34311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolia A, Cieslik M, Chu S-C, Xiao L, Ouchi T, Zhang Y, Wang X, Vats P, Cao X, Pitchiaya S, et al. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature. 2019;571:413–418. doi: 10.1038/s41586-019-1347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, Cejas P, Vazquez F, Cook J, Shivdasani RA, et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet. 2015;47:1346–1351. doi: 10.1038/ng.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz MM, Qiu X, Zhu Y, Takeda DY, Pan W, Baca SC, Gusev A, Korthauer KD, Severson TM, Ha G, et al. Prostate cancer reactivates developmental epigenomic programs during metastatic progression. Nat Genet. 2020;52:790–799. doi: 10.1038/s41588-020-0664-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJ, Foye A, Kothari V, Perry MD, Bailey AM, et al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell. 2018;174:758–769.e9. doi: 10.1016/j.cell.2018.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotinen M, You S, Yang J, Coetzee SG, Reis-Sobreiro M, Huang W-C, Huang F, Pan X, Yáñez A, Hazelett DJ, et al. ONECUT2 is a targetable master regulator of lethal prostate cancer that suppresses the androgen axis. Nat Med. 2018;24:1887–1898. doi: 10.1038/s41591-018-0241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabarinathan R, Mularoni L, Deu-Pons J, Gonzalez-Perez A, López-Bigas N. Nucleotide excision repair is impaired by binding of transcription factors to DNA. Nature. 2016;532:264–267. doi: 10.1038/nature17661. [DOI] [PubMed] [Google Scholar]
- Sahu B, Laakso M, Ovaska K, Mirtti T, Lundin J, Rannikko A, Sankila A, Turunen J-P, Lundin M, Konsti J, et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 2011;30:3962–3976. doi: 10.1038/emboj.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu B, Laakso M, Pihlajamaa P, Ovaska K, Sinielnikov I, Hautaniemi S, Jänne OA. FoxA1 specifies unique androgen and glucocorticoid receptor binding events in prostate cancer cells. Cancer Res. 2013;73:1570–1580. doi: 10.1158/0008-5472.CAN-12-2350. [DOI] [PubMed] [Google Scholar]
- Scheer S, Ackloo S, Medina TS, Schapira M, Li F, Ward JA, Lewis AM, Northrop JP, Richardson PL, Kaniskan HÜ, et al. A chemical biology toolbox to study protein methyltransferases and epigenetic signaling. Nat Commun. 2019;10:19. doi: 10.1038/s41467-018-07905-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehrawat A, Gao L, Wang Y, Bankhead A, 3rd, McWeeney SK, King CJ, Schwartzman J, Urrutia J, Bisson WH, Coleman DJ, et al. LSD1 activates a lethal prostate cancer gene network independently of its demethylase function. Proc Natl Acad Sci USA. 2018;115:E4179–E4188. doi: 10.1073/pnas.1719168115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 2009;23:804–809. doi: 10.1101/gad.1775509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sérandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Métivier R, et al. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 2011;21:555–565. doi: 10.1101/gr.111534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Song B, Park S-H, Zhao JC, Fong K-W, Li S, Lee Y, Yang YA, Sridhar S, Lu X, Abdulkadir SA, et al. Targeting FOXA1-mediated repression of TGF-β signaling suppresses castration-resistant prostate cancer progression. J Clin Invest. 2019;129:569–582. doi: 10.1172/JCI122367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutinen P, Rahkama V, Rytinki M, Palvimo JJ. Nuclear mobility and activity of FOXA1 with androgen receptor are regulated by SUMOylation. Mol Endocrinol. 2014;28:1719–1728. doi: 10.1210/me.2014-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Q, Bantignies F, Cavalli G. Principles of genome folding into topologically associating domains. Sci Adv. 2019;5:eaaw1668. doi: 10.1126/sciadv.aaw1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Singh S, Katika M, Lopez-Aviles S, Hurtado A. High throughput chemical screening reveals multiple regulatory proteins on FOXA1 in breast cancer cell lines. International Journal of Molecular Sciences. 2018;19:4123. doi: 10.3390/ijms19124123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15:701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitington T, Gao P, Song W, Ross-Adams H, Lamb AD, Yang Y, Svezia I, Klevebring D, Mills IG, Karlsson R, et al. Gene regulatory mechanisms underpinning prostate cancer susceptibility. Nat Genet. 2016;48:387–397. doi: 10.1038/ng.3523. [DOI] [PubMed] [Google Scholar]
- Wissmann M, Yin N, Müller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Günther T, Buettner R, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- Wu Q, Heidenreich D, Zhou S, Ackloo S, Krämer A, Nakka K, Lima-Fernandes E, Deblois G, Duan S, Vellanki RN, et al. A chemical toolbox for the study of bromodomains and epigenetic signaling. Nat Commun. 2019;10:1915. doi: 10.1038/s41467-019-09672-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Song B, Lu X, Kim J, Hu M, Zhao JC, Yu J. Altered chromatin recruitment by FOXA1 mutations promotes androgen independence and prostate cancer progression. Cell Res. 2019;29:773–775. doi: 10.1038/s41422-019-0204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Shibazaki M, Yamada C, Anzai E, Morii M, Nakayama Y, Kuga T, Hashimoto Y, Tomonaga T, Yamaguchi N. Tyrosine phosphorylation of the pioneer transcription factor FoxA1 promotes activation of estrogen signaling. J Cell Biochem. 2017;118:1453–1461. doi: 10.1002/jcb.25804. [DOI] [PubMed] [Google Scholar]
- Zhang X, Bailey SD, Lupien M. Laying a solid foundation for Manhattan–’setting the functional basis for the post-GWAS era’. Trends Genet. 2014;30:140–149. doi: 10.1016/j.tig.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Cowper-Sal-lari R, Bailey SD, Moore JH, Lupien M. Integrative functional genomics identifies an enhancer looping to the SOX9 gene disrupted by the 17q24.3 prostate cancer risk locus. Genome Research. 2012;22:1437–1446. doi: 10.1101/gr.135665.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Hawley JR, Soares F, Grillo G, Teng M, Madani Tonekaboni SA, Hua JT, Kron KJ, Mazrooei P, Ahmed M, et al. Noncoding mutations target cis-regulatory elements of the FOXA1 plexus in prostate cancer. Nat Commun. 2020;11:441. doi: 10.1038/s41467-020-14318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Treloar AE, Lupien M. Emergence of the Noncoding Cancer Genome: A Target of Genetic and Epigenetic Alterations. Cancer Discov. 2016;6:1215–1229. doi: 10.1158/2159-8290.CD-16-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]