Abstract
Objective
Both the gut microbiome and host genetics are known to play significant roles in the pathogenesis of IBD. However, the interaction between these two factors and its implications in the aetiology of IBD remain underexplored. Here, we report on the influence of host genetics on the gut microbiome in IBD.
Design
To evaluate the impact of host genetics on the gut microbiota of patients with IBD, we combined whole exome sequencing of the host genome and whole genome shotgun sequencing of 1464 faecal samples from 525 patients with IBD and 939 population-based controls. We followed a four-step analysis: (1) exome-wide microbial quantitative trait loci (mbQTL) analyses, (2) a targeted approach focusing on IBD-associated genomic regions and protein truncating variants (PTVs, minor allele frequency (MAF) >5%), (3) gene-based burden tests on PTVs with MAF <5% and exome copy number variations (CNVs) with site frequency <1%, (4) joint analysis of both cohorts to identify the interactions between disease and host genetics.
Results
We identified 12 mbQTLs, including variants in the IBD-associated genes IL17REL, MYRF, SEC16A and WDR78. For example, the decrease of the pathway acetyl-coenzyme A biosynthesis, which is involved in short chain fatty acids production, was associated with variants in the gene MYRF (false discovery rate <0.05). Changes in functional pathways involved in the metabolic potential were also observed in participants carrying rare PTVs or CNVs in CYP2D6, GPR151 and CD160 genes. These genes are known for their function in the immune system. Moreover, interaction analyses confirmed previously known IBD disease-specific mbQTLs in TNFSF15.
Conclusion
This study highlights that both common and rare genetic variants affecting the immune system are key factors in shaping the gut microbiota in the context of IBD and pinpoints towards potential mechanisms for disease treatment.
Keywords: inflammatory bowel disease, genetics, intestinal microbiology
Significance of this study.
What is already known about this subject?
Gene–microbiome interactions are important in the pathogenesis of IBD.
Multiple genetic and epidemiological factors have been identified to be associated to changes in gut microbiome homeostasis in both IBD and the general population.
The identified gene–microbiome interactions in IBD contain mostly common genetic variants.
What are the new findings?
Novel associations between common genomic variants located in IBD implicated genes (MYRF, IL17REL, SEC16A and WDR78) or immune-related genes (CABIN1) to the gut microbial features have been identified in both IBD and the general population cohort.
By using high-resolution sequencing data, we were also able to identify rare and deleterious variants in five genes (GPR151, CYP2D6, TPTE2, LEKR1 and CD160) that may also be involved in the regulation of the gut microbiota.
Disease-specific host microbiota interactions were assessed by taking into account potential cofounding factors such as medication use.
How might it impact on clinical practice in the foreseeable future?
Our research revealed the host–microbiota interactions in context of IBD, which helps us to understand the pathology of IBD and potentially move towards new therapeutic targets for IBD.
Introduction
IBD, comprising Crohn’s disease (CD) and UC, is a chronic inflammatory condition of the gut with an increasing incidence in westernised countries.1 Large-scale genome-wide association studies (GWAS) have identified more than 200 genetic loci associated with IBD, including genes implicated in the immune pathways involved in responses to gut microbes.2
Extensive changes in the composition of the gut microbiota have been reported in patients with IBD. Several studies have described similar alteration on the faecal microbiota of patients with IBD, mainly a decreased microbial richness, the depletion of strictly anaerobic commensal species and the expansion of pathobiont.3–5 Despite these observations, the gut microbiota composition of patients with IBD is heterogeneous and mainly influenced by disease behaviour together with the impact of clinical and environmental factors.6 7 As neither genetics nor microbiome studies have revealed the triggering factors for IBD, there is an increasing need to study host–microbial interactions in order to understand the aetiology and progression of the disease.8 9
To date, both mouse models and human studies have shown that IBD-associated genes interact with the intestinal microbiome via regulation of the mucosal physical barrier as well as immune responses. For example, the nucleotide-binding oligomerisation domain (NOD)-like receptor 2 (NOD2) is involved in the bacterial peptidoglycan recognition.10 It has been shown that NOD2 knock-out mice show ineffective recognition and clearance of bacterial pathogens. As a consequence, these mice present increased abundances of pathogenic bacteria from the Bacteroides and Escherichia genera.11–13 Another host–microbiome interaction involves ATG16L1, a gene implicated in autophagy. In patients with CD, ATG16L1-T300A mutation carriers have more pathosymbionts in their gut mucosa.14 Recently, genome-wide host–microbiota association analyses have reported correlations between variants in immune-related genes and microbial features. For example, IL10 has been associated with the abundance of Enterobacteriaceae15 and IL1R2 associated with the overall community composition (beta diversity).16
Host genetics–microbiome association studies have been described in cohorts based on the general population.15 16 These studies tend to miss the genetics signals that are more pronounced in a disease context like IBD. On the other hand, the microbial quantitative trait loci (mbQTL) studies in IBD cohorts available to date have been limited in either sample size or in genomic and microbiome resolution. Also details in phenotypes capturing the heterogeneity present within IBD has been lacking in previous studies.17 18 The discovery of host–microbiota interactions, moreover, has been hampered by the large influence of intrinsic and environmental factors on the gut microbiome and relatively low microbial heritability.19
The aim of this study was to expand current knowledge of host–gut microbiota interactions.20 We combined whole exome sequencing (WES) of the host genome with metagenomics sequencing of faecal samples in a population cohort and in an IBD cohort. In addition to whole-exome-wide analyses, we investigated disease-specific interactions and the influence of rare variants on the gut microbiota in order to identify mechanisms involved in gut homeostasis and disease development.
Methods
Study cohorts
This study included two independent Dutch cohorts: a population-representative cohort (LifeLines-DEEP) from the northern part of the Netherlands and an IBD cohort made up of patients diagnosed in the specialised IBD clinic of the University Medical Center Groningen (Groningen, the Netherlands). The LifeLines-DEEP cohort (M12.113965) was approved by the ethics committee of the University Medical Centre Groningen, with registering at the LifeLines Research Site in Groningen. All individuals were also asked to fill in the questionnaire on GI symptoms. The IBD cohort (IRB-number 2008.338) was approved by University Medical Centre Groningen IRB (online supplementary table 1).
gutjnl-2019-319706supp001.xlsx (12.7MB, xlsx)
WES and data processing
WES was performed on blood samples. Library preparation and sequencing were done at the Broad Institute of MIT and Harvard. On average, 86.06 million high-quality reads were generated per sample and 98.85% of reads were aligned to a human reference genome (hg19). Moreover, 81% of the exonic regions were covered with a read depth >30×. Next, the Genome Analysis Toolkit21 of the Broad Institute was used for variant calling. Variants with a call rate <0.99 or Hardy-Weinberg equilibrium test with p<0.0001 were excluded using PLINK tool (V.1.9). To remove genetic outliers, we combined WES data with genomes of Europeans from publically available 1000 Genome Project (phase 3) data (http://www.internationalgenome.org/), and performed principal component analysis (PCA) analysis based on single nucleotide polymorphisms (SNPs) shared between datasets. Outliers were defined as samples which fall outside of a mean±3 SD interval in both of the first two PCs, and these samples were removed. We also removed sex-mismatching samples based on the inbreeding coefficient (lower than 0.4 for females and higher than 0.7 for males) and related samples with identity-by-descent>0.185.22 GATK germline copy number variant (gCNV)23 was used for copy number variant (CNV) detection. GATK-gCNV uses a Bayesian model to adjust for known bias factors of exome capture and sequencing, such as GC content and mappability, while also controlling for other technical and systematic differences. Raw sequencing files are compressed into read counts over the set of exons defined under Gencode Annotation (V.33). After processing, variant quality and frequency filters (<1% site frequency) are applied to produce the final CNV callset (https://gatkforums.broadinstitute.org/gatk). In summary, 73 164 common variants (minor allele frequency (MAF) >5%), 98 878 rare variants (MAF <5%) and 1046 CNVs (site frequency <1%) from 920 LifeLines-DEEP and 435 individuals with IBD were considered for further analyses.
Metagenomic sequencing and data processing
Metagenomic sequencing was performed for faecal samples, using the Illumina MiSeq platform. Reads belonging to the human genome were removed by mapping the data to the human reference genome (version NCBI37) with kneaddata (V.0.5.1, http://huttenhower.sph.harvard.edu/kneaddata).
Profiling of microbiome taxonomic and functional composition was done using MetaPhlan (V.2.6.0)24 (http://huttenhower.sph.harvard.edu/metaphlan) and HUMAnN2 (V.0.6.1)25 (http://huttenhower.sph.harvard.edu/humann2). For each cohort, taxa present in fewer than 10% of total samples and pathways present in fewer than 25% of samples were excluded from the analyses (online supplementary methods, online supplementary table 2). We then normalised the relative abundances of 242 microbial taxa and 301 pathways present in both cohorts through inverse rank transformation.
gutjnl-2019-319706supp002.pdf (109.3KB, pdf)
Host genetics and gut microbiota differences between cohorts
IBD genetic signature
To assess the similarity of the genetic makeup of our IBD cohort compared with other GWAS studies on IBD, we performed case-control analyses in terms of genetics (population controls vs patients with CD, controls vs patients with UC and controls vs all patients combined) and compared the results with the largest IBD GWAS meta-analysis of populations of European ancestry published to date.2 Logistic regression analysis was used (PLINK V.1.9) adjusting for age, sex and smoking status. P values were adjusted for multiple testing by using the Bonferroni method and an false discovery rate (FDR) <0.05 was considered statistically significant.
IBD-associated gut microbial taxa and pathways
Then, we compared relative abundance of microbial taxa and pathways between the groups. The analyses were performed using Maaslin2 software (https://bitbucket.org/biobakery/maaslin2/src/default/). We selected covariates for our linear models based on factors which have often been used in mbQTL studies to increase comparability to other studies. Furthermore, we added covariates which have shown to have a large impact on the gut microbiome composition.3 15–17 20 26–30 This resulted in the inclusion of the following covariates: age, sex, body mass index, smoking, read depth, medication use (proton pump inhibitors, laxatives and antibiotics) and disease location for the IBD cohort. Bonferroni procedure was used to adjust for multiple testing and an FDR<0.05 was considered statistically significant.
mbQTL analyses
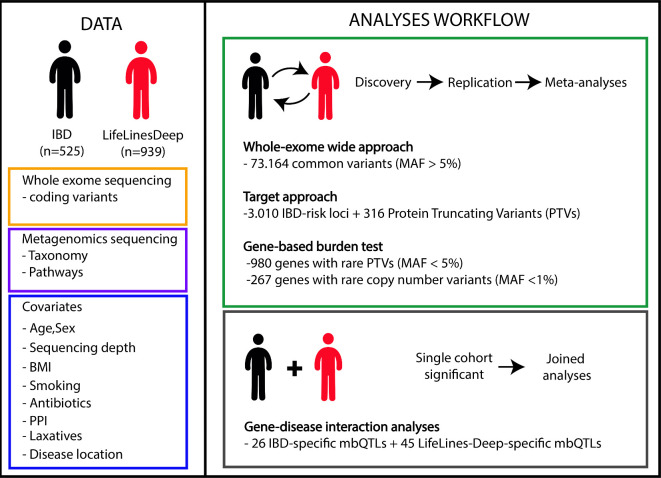
Microbial taxa and functional pathways were treated as quantitative traits. For all analyses, linear regression (where variants were encoded as 0 for homozygote of major allele, 1 for heterozygotes and 2 for homozygote of minor allele, online supplementary methods) was used to adjust for the effect of the confounders mentioned above. The Spearman correlation method was applied to determine the relationship between non-zero microbial data and host genotype in a four-step approach (figure 1).
Figure 1.
Schematic overview of the study. (DATA part) We performed whole exome sequencing of the host genome and whole genome shotgun sequencing of faecal samples of 525 individuals (IBD) and 939 controls (LifeLines-DEEP). Nine covariates (age, sex, body mass index (BMI), smoking status, medication use (antibiotics, proton pump inhibitors (PPIs) or laxatives), disease location (in the IBD cohort) and sequencing read depth) were corrected for relative abundances of 242 taxa and 301 pathways. (ANALYSES WORKFLOW part) A four-step analysis was performed: step 1 includes a meta-analysis (p<6.83 10−7, corresponding to FDR<0.05) in which 73 164 exome-wide common variants with minor allele frequency (MAF) >5% were used for association analyses for microbial traits. Step 2 includes a meta-analysis (p<1.5 10−5, corresponding to FDR<0.05) using a targeted approach that only tested for 3010 variants located in IBD-associated genes known from IBD genome-wide association studies and PTVs with MAF >5%. Step 3 includes a meta-analysis (p<5 10−5, corresponding to FDR<0.05) using a gene-based burden test for 980 genes with rare PTVs (MAF <5%); a meta-analysis (p<1.87 10−4, corresponding to FDR<0.05) using a gene-based test for 267 genes with rare copy number variants (site frequency <1%). Step 4 includes joint analysis combining the two cohorts for disease and genetics interaction analyses. Step 4 focused only on single-cohort-significant microbial quantitative trait loci (mbQTLs) from steps 1 and 2 while adding a disease and a genetic interaction term into the model. All analyses were confined to non-zero values of taxa and pathways. All significance thresholds were set up by Bonferroni correction taking all variants/genes used into account.
Step 1: whole-exome-wide association meta-analyses
Seventy three thousand one hundred and sixty-four common variants (MAF >5%) were correlated with the relative abundances of microbial taxa and metabolic pathways using the same method in the previous study.15 First, we tested associations in the LifeLines-DEEP cohort (discovery stage) and selected signals with p<5 10–5. Second, we replicated these in the IBD cohort and only kept associations with the same allelic direction that passed a replication threshold p<0.05 (replication stage). Third, we performed meta-analyses on these datasets using a weighted-Z-score approach by ‘Metap’ package in R V.3.5.0. The criteria of significance were p values that met a whole-exome-wide threshold of 6.83 10–7, corresponding to exome-wide FDR=0.05 (Bonferroni method, n=73 164 variants). We then repeated this analysis switching the discovery and replication cohorts: using the IBD cohort as discovery and LifeLines-DEEP as replication.
Step 2: meta-analyses of selected variants
We selected two sets of variants for targeted analysis: protein truncating variants (PTVs)31 and variants located in known IBD-associated genes.2 We predicted 316 stop-gain, splice-disrupting and frameshift variants with MAF >5% in this analyses. We selected all genetic variants with an MAF >5% present in genomic loci that have been associated to IBD2 (n=3010). Associations between these variants and microbiome traits were performed following the same procedure described above in step 1. The significance threshold was adjusted according to the number of genetic variants tested: p<0.001 in the discovery cohort, p<0.05 in the replication cohort and a final meta p meeting 1.5 10−5, corresponding to FDR=0.05 (Bonferroni method, n=3309 variants).
Step 3: gene-based burden test meta-analyses
To identify the effect of rare SNPs, we performed gene-based burden tests by using the variant’s score instead of individual genotype in correlation analyses (MetaSKAT packages32 in R V.3.5.0), keeping only PTVs with MAF <5% and calculating per-gene scores.33 The number of genes implicated in this analysis was 980, so the final meta p was 5 10-5, corresponding to gene-wise FDR=0.05, with a discovery p of 0.005 and a replication p of 0.05. To identify the effect of CNVs, we used a strategy similar to the one for rare SNVs and overlapped genes with CNVs. For each gene, a score was assigned based on the number of CNV sites and then used in association tests.33 34 This analysis was conducted for 267 genes with deletions and duplications separately. We chose signals with p<0.05 in each cohort, and the final meta p<1.8710−4, FDR of 0.05 (Bonferroni method, n=267 genes).
Step 4: assessing disease effect in the host–microbiota correlations
Next, we investigated the mbQTLs that were only significant in one of the cohorts in steps 1 and 2. To identify whether the presence and absence of IBD could have an effect on the observed mbQTLs, we performed association analyses combining both cohorts and adding diseases and the interaction between genotype and diseases as covariates (online supplementary methods).35 Significance thresholds at whole-exome-wide level were p<6.83 10−7 (Bonferroni method, n=73 164 variants) for the discovery cohort, p>0.05 for the replication cohort and significant interaction p (IBD ×genotype)<0.0013, corresponding to FDR=0.05 (Bonferroni method, n=38 variants, including 17 IBD-specific and 21 LifeLines-DEEP-specific observed mbQTLs; online supplementary table 3, online supplementary table 4). The criteria for significance in the targeted-level analyses were discovery cohort p<1.5 10−5, replication cohort p>0.05, significant interaction p (IBD ×genotype)<0.0014, corresponding to FDR=0.05 (Bonferroni method, n=36, including 12 IBD-specific and 24 LifeLines-DEEP-specific mbQTLs; online supplementary table 3, online supplementary table 4). To avoid inflated statistics in these analyses, we randomly permutated the disease status across all samples 999 times (online supplementary methods). In addition, taking into account the heterogeneity of patients with IBD, we also considered the clinical IBD subphenotypes and performed a case-control mbQTL analyses in patients with CD and patients with UC separately.
Annotation of genetic variants
To further explore the function of the observed mbQTLs, we examined tissue-specific gene expression (expression quantitative trait loci (eQTLs)) in the GTEx Consortium database36 and used the Enrichr37 and FUMAGWAS38 databases to annotate the biological function and immunological signatures of the genes with a mbQTL effect in the whole-exome-wide analyses.
Results
Cohort description
The two cohorts in this study are derived from the Netherlands. The LifeLines-DEEP cohort comprises 939 individuals (59.74% female, mean age 45.24±13.46) and the IBD cohort comprises 525 patients with IBD (61.33% female, mean age 43.18±14.46), including 291 patients with CD, 202 patients with UC and 32 IBD unclassified (IBDU) patients. Eighteen individuals from LifeLines-DEEP and 17 patients from IBD cohort were removed through genetic PCA analysis. One individual from LifeLines-DEEP and seven patients from IBD were failed in quality control (QC) (online supplementary methods). The presence of an ileoanal pouch or a stoma was an exclusion criterion in the IBD cohort (n=66; online supplementary table 1). Finally, 920 LifeLine-DEEP individuals and 435 patients with IBD (CD=242, UC=161 and IBDU=32) were used for analysis.
Differences on host genetics and gut microbiota between cases and controls
IBD was associated to genomic variants located in previously reported IBD risk loci (FDR<0.05, online supplementary tables 5,6), including genes in human leukocyte antigen (HLA) loci (eg, rs77504727, c.740C>T, p.Arg247His, ORIBD=2.65, PIBD=1.25×10−13, FDRIBD=8.71×10−09, ORCD=2.88, PCD=1.16×10−10, FDRCD=8.12×10−6) and NOD2 (rs2066843, c.1296C>T, ORCD=1.83, PCD=3.35×10−08, FDRCD=0.0023). An increased abundance of the phylum Bacteroidetes was detected in patients with IBD compared with general population controls (FDR=1.30×10−23, online supplementary table 7). In terms of microbial pathways, pathways involved in fermentation of pyruvate to propanoate were decreased in IBD (FDRIBD=3.10×10−6, FDRCD=2.35×10−3, FDRUC=7.14×10−3), while the pathway of fermentation of pyruvate to acetate and lactate was decreased in patients with CD compared with population controls (FDR=1.77×10−11).
Whole-exome-wide analysis reveals mbQTLs in immune-related genes
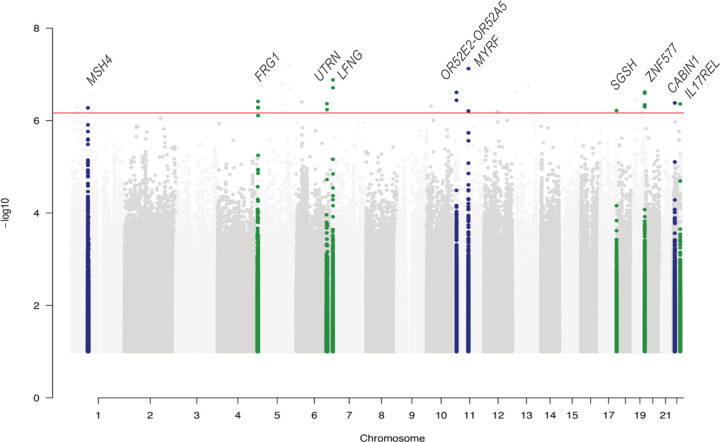
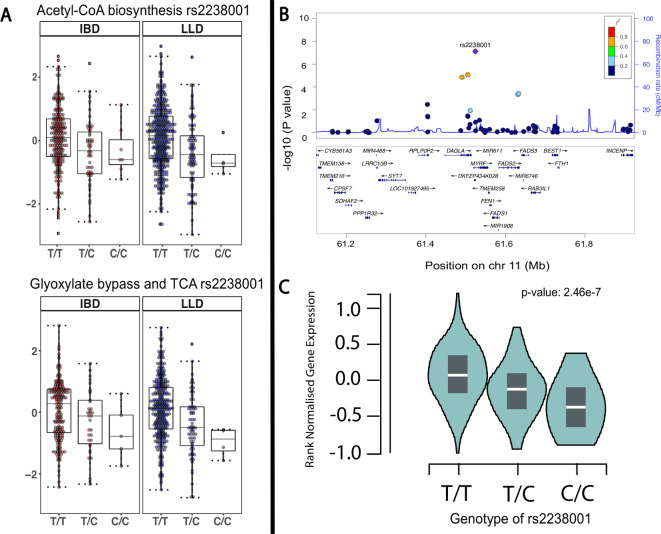
The exome-wide mbQTL analysis (step 1) identified associations between 10 genetic variants and 11 microbial features (FDR<0.05). Four variants were associated to bacterial metabolic pathways involved in degradation of glucarate, the tricarboxylic acid cycle (TCA) cycle, coenzyme A (CoA) biosynthesis and glycogen biosynthesis, while the other six variants were associated with relative abundance of bacteria (figure 2, online supplementary table 8). The most significant associations were found between the minor allele of an intronic SNP (rs2238001, c.46+4245T>C) in the MYRF gene, which is located in an IBD-associated loci,2 and decreased abundance of two microbial pathways involved in carbohydrate metabolism: acetyl-CoA biosynthesis (PWY-5173, meta p=7.50 10−8, FDR=0.0058) and glyoxylate bypass (TCA-GLYOX-BYPASS, meta p=6.16 10−7, FDR=0.048; figure 3A; online supplementary figure 1). In the step 2 analysis, the same SNP was also observed to be associated with another metabolic pathway (GLYCOLYSIS-TCA-GLYOX-BYPASS, meta p=2.73 10−6, FDR=0.02). These pathways are mainly predicted from Escherichia coli. Concordantly, E. coli shows the strongest association among all 242 microbial taxa to MYRF (meta p=6.00 10−3), although it does not meet the statistically significant threshold. Examination of the GTEx database revealed that the rs2238001 has a eQTL effect specific to colon tissue that results in increased expression of MYRF (p=2.50 10−7; figure 3C).
Figure 2.
Whole-exome-wide meta-analysis results from LifeLines-DEEP and IBD cohorts. Seventy three thousand one hundred and sixty-four common variants (minor allele frequency >5%), 242 taxa and 301 pathways (corrected for all covariates) were used in the association analyses. The discovery significance threshold was p<5 10−5 and the replication significance threshold was p<0.05. Manhattan plot displays −log10 p values for all association tests. Green and blue represent taxonomies and pathways, respectively. Red line indicates the whole-exome-wide association significance threshold: meta p<6.83 10−7, corresponding to exome-wide FDR<0.05 (n=73 164 variants, Bonferroni correction).
Figure 3.
Microbial quantitative trait loci and eQTL analyses of MYRF. (A) Spearman correlation between genotype (TT, TC, CC) of rs2238001 in MYRF and the relative abundance of acetyl-coenzyme A (CoA) biosynthesis (IBD cohort, p=1.43 10−3, r=−0.19; LifeLines-DEEP (LLD) cohort, p=1.47 10−5, r=−0.20; meta p=7.50 10−8, FDR<0.05), the glyoxylate bypass and tricarboxylic acid cycle (TCA) MetaCyc pathways (IBD cohort, p=0.0149, r=−0.16; LLD cohort, p=1.04 10−5, r=−0.22; meta p=6.07 10−7, FDR<0.05). (B) The rs2238001 locus zoomed in on the IBD-associated region, including the IBD-associated genes MYRF, FADS2 and FADS3. P values are derived from meta-analyses between variants and the relative abundance of acetyl-CoA biosynthesis. (C) eQTL analysis between rs2238001 and MYRF gene expression in colon tissue from the GTEx database (n=246 tissues, p=2.46 10−7). r, Spearman correlation coefficient.
gutjnl-2019-319706supp003.pdf (142.8KB, pdf)
The minor allele of a synonymous variant in the immune-related gene CABIN1 (rs17854875, c.5745C>T, p.Ala1915Ala) was associated with an increase of D-glucarate degradation (GLUCARDEG-PWY, meta p=4.15 10−7, FDR=0.032). Another SNP located near the gene IL17REL (rs5845912, AC >A) was correlated with a lower abundance of the species Alistipes indistinctus (meta p=4.36 10−7, FDR=0.033). Variants in this gene have been reported to be associated with UC. IL17REL encodes interleukin 17 (IL-17) receptor E-like, a homolog of IL-17 receptor E that is considered to be a part of the IL-17 pathway that initiates a T helper 2–mediated immune response.39
Gene function enrichment analysis of all 10 mbQTLs (table 1) identified enrichment in gene functions related to mature B cell differentiation (GO:0002313, p=0.005, FDR=0.038) and CD4 and CD8 T-cell differentiation pathways (GSE31082, p=2.81 10−6, FDR=0.0103; online supplementary table 9).
Table 1.
Microbial quantitative trait loci associated with microbial taxonomies and pathways identified in a whole-exome-wide approach
| Chr | Pos | Allele | SNP | Gene symbol | Annotation | Microbial taxonomy/pathway | Microbial change* | Meta P value† | Meta FDR | LifeLines-DEEP cohort | IBD cohort | ||
| P value‡ | r§ | P value‡ | r§ | ||||||||||
| 1 | 76 344 011 | A/C | rs1493367 | MSH4 | Splice region variant and intron variant | Superpathway of sulfur amino acid biosynthesis Saccharomyces cerevisiae | – | 5.30×10–7 | 0.041 | 1.36×10–5 | 0.17 | 0.012 | 0.15 |
| 4 | 190 862 155 | T/G | rs4145515 | FRG1 | 5 prime UTR premature start codon gain variant | Genus Actinomyces | – | 5.29×10–7 | 0.041 | 3.53×10–5 | 0.19 | 0.0045 | 0.20 |
| 6 | 144 999 763 | A/C | rs9376837 | UTRN | Intron variant | Species Parabacteroides merdae (strain Parabacteroides merdae) | – | 4.31×10–7 | 0.033 | 4.44×10–6 | −0.18 | 0.031 | −0.15 |
| 7 | 2 566 028 | G/A | rs12700028 | LFNG | Synonymous variant | Family Acidaminococcaceae | – | 1.31×10–7 | 0.01 | 3.89×10–6 | 0.27 | 0.010 | 0.25 |
| 11 | 5 142 270 | T/G | rs10837375 | OR52E2-OR52A5 | Intergenic region | Glycogen biosynthesis I from ADP-D-glucose | – | 2.45×10–7 | 0.019 | 6.63×10–6 | 0.15 | 0.011 | 0.12 |
| 11 | 61 524 507 | T/C | rs2238001 | MYRF | Intron variant | Superpathway of glyoxylate bypass and TCA | Increase in CD | 6.16×10–7 | 0.048 | 1.04×10–5 | −0.22 | 0.015 | −0.16 |
| 11 | 61 524 507 | T/C | rs2238001 | MYRF | Intron variant | Superpathway of acetyl-CoA biosynthesis | Increase in IBD/CD | 7.50×10–8 | 0.0058 | 1.47×10–5 | −0.20 | 0.0014 | −0.19 |
| 17 | 78 188 963 | G/A | rs4889839 | SGSH | Intron variant | Species Ruminococcus sp 5 1 39BFAA (strain GCF 000159975) | Decrease in UC | 6.07×10–7 | 0.047 | 1.25×10–5 | −0.15 | 0.0159 | −0.14 |
| 19 | 52 376 507 | T/C | rs2288868 | ZNF577 | Missense variant | Species Haemophilus parainfluenzae | – | 4.56×10–7 | 0.035 | 7.84×10–6 | 0.32 | 0.015 | 0.24 |
| 22 | 24 564 477 | C/T | rs17854875 | CABIN1 | Synonymous variant | D-glucarate degradation I | Increase in CD | 4.15×10–7 | 0.032 | 1.82×10–5 | 0.27 | 0.0057 | 0.21 |
| 22 | 50 471 620 | A/C | rs5845912 | IL17REL | Intergenic region | Species Alistipes indistinctus (strain GCF 000231275) | – | 4.36×10–7 | 0.033 | 6.18×10–6 | −0.24 | 0.024 | −0.24 |
The whole-exome-wide approach identified 11 significant associations between variants located in 10 genes and microbial taxa and pathways. Seventy three thousand one hundred and sixty-four common variants (minor allele frequency >5%), 242 taxa and 301 pathways (corrected for all covariates) were used in the association analyses. Spearman correlation was performed in the association test in each cohort, followed by a Z-score-based meta-analysis. The discovery significance threshold was p<5 ×10−5, the replication significance threshold was p<0.05 and the final meta threshold was 6.83 ×10−7, corresponding to FDR<0.05.
*Case-control analysis on microbial data. Significant (FDR <0.05) microbial change in IBD are shown (online supplementary table 7).
†Meta p value threshold was decided by the number of total variants (n = 73 164, Bonferroni correction).
‡P values from association analyses in each cohort.
§Correlation coefficients from association analyses in each cohort.
CD, Crohn’s disease; CoA, coenzyme A; TCA, tricarboxylic acid cycle.
Targeted analysis identifies mbQTLs in IBD-associated genes
Two additional IBD-associated genes with mbQTLs were identified in this targeted approach (step 2; table 2; online supplementary table 10). The top significant variant, rs10781497 (c.834G>A, p.Asp278Asp) located in the SEC16A gene, was associated with lower levels of bacterial biosynthesis of thiamin phosphate (THISYN-PWY) and thiazole (PWY-6892) (online supplementary figure 2A), and an SNP in WDR78 (rs74609208, c.2497-18C>A) was associated with higher level of biosynthesis of rhamnose (DTDPRHAMSYN-PWY; online supplementary figure 2B).
Table 2.
Microbial quantitative trait loci associated with microbial taxonomies and pathways identified in a targeted approach
| Chr | Pos | Allele | SNP | Gene symbol | Annotation | Bacterial taxonomy/pathway | Microbial change* | Meta P value† | Meta FDR | LifeLines-DEEP cohort | IBD cohort | ||
| P value‡‡ | r§ | P value‡ | r§ | ||||||||||
| 1 | 67 279 881 | C/A | rs74609208 | WDR78 | Intron variant | dTDP-L-rhamnose biosynthesis I | – | 1.46×10–5 | 0.048 | 0.00035 | 0.12 | 0.014 | 0.12 |
| 9 | 139 371 234 | G/A | rs10781497 |
SEC16A
(INPP5E) |
Synonymous variant | Superpathway of thiamin diphosphate biosynthesis I | – | 1.88×10–6 | 0.0062 | 5.33×10–5 | −0.15 | 0.011 | −0.14 |
| 9 | 139 371 234 | G/A | rs10781497 |
SEC16A
(INPP5E) |
Synonymous variant | Thiazole biosynthesis I Escherichia coli | Increase in CD | 2.44×10–6 | 0.0052 | 6.69×10–5 | −0.15 | 0.012 | −0.14 |
| 11 | 61 524 507 | T/C | rs2238001 | MYRF | Intron variant | Superpathway of acetyl-CoA biosynthesis | Increase in IBD/CD | 7.50×10–8 | 2.5×10–4 | 1.47×10–5 | −0.20 | 0.0014 | −0.19 |
| 11 | 61 524 507 | T/C | rs2238001 | MYRF | Intron variant | Superpathway of glyoxylate bypass and TCA | Increase in CD | 6.16×10–7 | 0.02 | 1.04×10–5 | −0.22 | 0.015 | −0.16 |
| 11 | 61 524 507 | T/C | rs2238001 | MYRF | Intron variant | Superpathway of glycolysis pyruvate dehydrogenase TCA and glyoxylate bypass | Increase in CD | 2.73×10–6 | 0.009 | 2.90×10–5 | −0.21 | 0.024 | −0.15 |
The targeted approach identified six significant associations between variants located in IBD-associated genes and microbial taxa and pathways. Three thousand and ten variants in IBD-associated genomic regions and 316 protein truncating variants and 242 microbial taxa and 301 MetaCyc pathways were used in targeted approach. Spearman correlation was performed in the association test in each cohort, followed by a Z-score-based meta-analysis. The discovery significance threshold was 0.001, the replication significance threshold was 0.05 and the final meta threshold was 1.5 ×10−5 corresponding to FDR<0.05.
*Case-control analysis on microbial data. Significant (FDR<0.05) microbial change in IBD are shown (online supplementary table 7).
†Meta p value threshold decided by the number of total variants (n = 3309, Bonferroni correction).
‡P values from association analyses in each cohort.
§Correlation coefficients from association analyses in each cohort.
CD, Crohn’s disease; TCA, tricarboxylic acid cycle.
gutjnl-2019-319706supp004.pdf (856.2KB, pdf)
Gene-based burden test highlights rare mutation mbQTLs
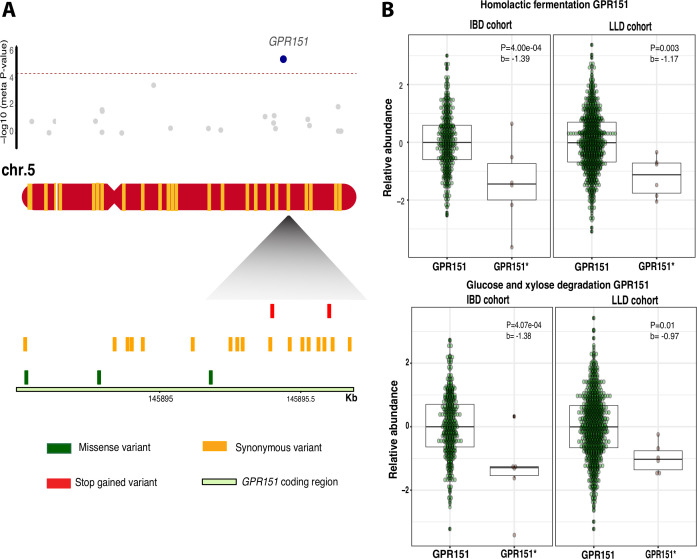
To study the effect of rare variants with predicted protein changing properties and CNVs, we performed gene-based burden tests (step 3). Here, we identified eight associations between four genes and eight microbial pathways (table 3). Two transcriptional stop-gain mutations in the GPR151 gene were significantly associated with lower levels of bacterial carbohydrate metabolism pathways (ANAEROFRUCAT-PWY with meta p=4.78 10−6, FDR=0.0047, GLYCOLYSIS with meta p=5.45 10−6, FDR=0.0053, PWY-5484 with meta p=4.63 10−6, FDR=0.0045, and PWY-6901 with meta p=3.05 10−5, FDR=0.003; figure 4; online supplementary figure 3). In addition, two frameshift variants in the IBD-associated gene CYP2D6 were associated with a decreased level of bacterial biosynthesis of vitamin K (PWY-5838 with meta p=1.45 10−5, FDR=0.014). We also observed that the gene CD160 with exon duplications was significantly associated with decreased abundance of Lachnospiraceae (meta p=1.65 10−4, FDR=0.044, online supplementary table 14).
Table 3.
Rare microbial quantitative trait loci identified by gene-based burden meta-analyses
| Chr | SNP* | Gene symbol | Annotation | Microbial taxonomy/pathway | Microbial change† | Meta P value‡ | Meta FDR | LifeLines-DEEP cohort | IBD cohort | ||
| P value§ | Beta¶ | P value§ | Beta¶ | ||||||||
| 3 | 3:156 570 689 rs200834448 3:156 710 862 | LEKR1 | Stop gain, frameshift variant, splice donor variant and intron variant |
Superpathway of hexitol degradation bacteria | Increase in IBD/CD/UC | 8.42×10–7 | 8.25×10–4 | 1.84×10–5 | 0.61 | 0.016 | 0.54 |
| 5 | rs114285050 rs140458264 | GPR151 | Stop gain, stop gain | Homolactic fermentation | Increase in IBD/CD | 4.78×10–6 | 0.0047 | 0.0032 | −1.17 | 0.00040 | −1.40 |
| 5 | rs114285050 rs140458264 | GPR151 | Stop gain, stop gain | Glycolysis I from glucose 6-phosphate | Increase in IBD/CD | 5.45×10–6 | 0.0053 | 0.0030 | −1.17 | 0.00048 | −1.38 |
| 5 | rs114285050 rs140458264 | GPR151 | Stop gain, stop gain | Glycolysis II from fructose 6-phosphate | Increase in IBD/CD/UC | 4.63×10–6 | 0.0045 | 0.0028 | −1.18 | 0.00044 | −1.38 |
| 5 | rs114285050 rs140458264 | GPR151 | Stop gain, stop gain | Superpathway of glucose and xylose degradation | Increase in IBD/CD | 3.05×10–6 | 0.0030 | 0.015 | −0.97 | 0.00041 | −1.39 |
| 13 | rs139121187 rs150812023 | TPTE2 | Stop gain, splice donor variant and intron variant | Glycolysis IV plant cytosol | – | 4.62×10–6 | 0.0045 | 0.015 | 1.19 | 0.00029 | 2.47 |
| 22 | rs35742686 rs5030655 |
CYP2D6 | Frameshift variant, frameshift variant | Superpathway of menaquinol-8 biosynthesis I | – | 1.45×10–5 | 0.014 | 0.00015 | −0.91 | 0.021 | −0.63 |
| 22 | rs35742686 rs5030655 | CYP2D6 | Frameshift variant, frameshift variant | Superpathway of demethylmenaquinol-8 biosynthesis | – | 1.50×10–5 | 0.015 | 0.00019 | −0.90 | 0.019 | −0.65 |
Eight associations were identified by the gene-based burden test. We collapsed exome-wide protein truncating variants (PTVs) with minor allele frequency <5% into 980 genes. Genetic scores were used instead of single variant dosage in the association analyses in each cohort. Meta-analyses were performed for those associations with discovery p<0.005 and replication p<0.05.
*Rare PTVs located within genes used in the burden test.
†Case-control analysis on microbial data. Significant (FDR<0.05) microbial change in IBD are shown (online supplementary table 7).
‡Meta p value cut-off determined based on the total number of genes (n=980, Bonferroni correction).
§P values from association analyses in each cohort.
¶Effect size in association analyses in each cohort.
CD, Crohn’s disease.
Figure 4.
Associations between gene GPR151 and microbial pathways. (A) Meta p values based on burden test between 30 genes with rare protein truncating variants (PTVs) on chromosome 5 and relative abundance of MetaCyc pathway homolactic fermentation (top). Blue dot represents meta p value of gene GPR151. Lower panel shows the variants found along with the coding region in GPR151. Different colours indicate different variant categories. Red indicates two rare stop-gain mutations, rs114285050 and rs140458264. (B) Box plots for associations between the relative abundance of the homolactic fermentation (meta p=4.78 10−6, FDR<0.05), glucose xylose degradation (meta p=3.05 10−5, FDR<0.05) microbial pathways and GPR151, respectively. b, effect size. GPR151, without rare PTVs. GPR151*, with rare PTVs.
gutjnl-2019-319706supp005.pdf (254.4KB, pdf)
Interaction analyses identifies IBD-specific mbQTLs
Since both the gut microbiota and host genetics are different in patients with IBD compared with the general population, we reanalysed current dataset including an interaction factor between disease and genetics. This analysis identified IBD-specific interactions comprising 18 genetic variants and 19 microbiome features (10 pathways and 9 taxa; FDR<0.05, online supplementary table 12), which were also calibrated by permutation tests to avoid inflated statistics bias (online supplementary methods, online supplementary figure 4). For example, a missense variant
gutjnl-2019-319706supp006.pdf (78KB, pdf)
(rs2076523, c.586T>C, p.Lys196Glu) in the IBD-associated gene BTNL2, which is involved in regulation of T cell proliferation,40 was associated with an increase in Bacteriodes cellulosilyticus in patients with IBD (interaction p=1.31 10−5, interaction FDR=4.98 10−4). We also replicated three previously identified mbQTLs. The well-known association between the LCT gene and Bifidobacterium abundance15 41 42 was confirmed in the population-based cohort (rs748841, GG genotype associated with higher abundance of Bifidobacterium adolescentis, recessive model, p=1.70 10−4, FDR=0.046, online supplementary figure 5, online supplementary table 13), while previously reported genetic variants with mbQTL effect located in the IBD-associated genes TNFSF15 (rs4246905, c.302-63T>C) and HLA-B (rs2074496, c.900C>T, p.Pro300Pro)18 were associated with a glycogen degradation microbial pathway (GLYCOCAT-PWY, interaction p=7.98 10−5, interaction FDR=0.0029) and a strain of Ruminococcaceae bacterium (interaction p=3.32 10−5, interaction FDR=0.0012), respectively.
gutjnl-2019-319706supp007.pdf (490.1KB, pdf)
Finally, we assessed mbQTL effect in patients with CD and UC separately. Two mbQTLs passed the significant threshold in patients with CD (FDR<0.05). For example, rs61732050 (c.1701G>A, p.Ala567Ala, MAFCD=0.052, MAFUC=0.068), located in IBD-associated gene NDST1 and associated with decreased abundance of the family Lachnospiraceae, was only significant in patients with CD (Spearman correlation coefficient=−0.32, p=3.03×10−07, FDR=0.023). The 23 out of 27 IBD-specific mbQTLs identified earlier were nominally significant (p<0.05) in both CD and UC groups (online supplementary table 4), with all 27 showing the same directions of effect.
Discussion
To study the interaction between host genomics and gut microbial features in the context of IBD, we performed a large mbQTL analysis using high-resolution host genomic and gut microbiome data. This identified putative associations between common genomic variants located in IBD (MYRF, IL17REL, SEC16A and WDR78) or immune-related genes (CABIN1) to the abundance of specific microbial taxa and gut microbiome metabolic pathways. The use of WES data also allowed us to identify rare and deleterious variants in five genes (GPR151, CYP2D6, TPTE2 LEKR1 and CD160) that could potentially be involved in the regulation of the gut microbiota. Finally, genetics–disease interaction models revealed disease-specific mbQTL signals.
The patients with IBD in this study showed similarities of their genetic and microbial signatures compared with other studies.2–4 43 For example, NOD2 variants were associated with CD, while the SNPs in HLA loci were associated with both CD and UC. The gut microbiota of patients with IBD was characterised by a decreased abundance of Firmicutes, including Faecalibacterium prausnitzii (FDR=9.69×10−09), and an expansion of Proteobacteria, including E. coli (FDR=0.029), compared with the population controls. These differences were also evident in the predicted microbial pathways, with a decreased abundance of genes involved in short chain fatty acid (SCFA) metabolism.
In whole-exome-wide level analysis, we found that decreased levels of the microbial acetyl-CoA and glyoxylate metabolic pathways correlated with the minor allele (C) of a variant located in the gene MYRF. Acetyl-coA is a precursor in the synthesis of SCFAs, including butyrate and acetate,44 which are important in maintaining gut health.45 Interestingly, the MYRF gene is located in a genomic region that has previously been associated with IBD and other immune-mediated diseases.46 47 This genomic region also contains the FADS1 and FADS2 genes that are involved in the metabolism of polyunsaturated fatty acids,48 and the n-3 polyunsaturated fatty acid has been suggested to have protective effects on IBD.49 Therefore, the current analyses suggest a potential link between inflammation and microbial pathway dysregulation through host genomic variation. Another mbQTL we identified is located in the immune-related gene CABIN1. This gene is involved in negatively regulating T-cell receptor signalling50 and was associated to an increase of D-glucarate degradation pathway. Interestingly, enterobacteria such as E. coli, a potentially pathogenic bacteria known to be enriched in dysbiotic conditions, can use this sugar as a carbon source for growth.51 This implies a potential role between host genetics and a beneficial environment for E. coli to grow. We also found an association between IL17REL, which likely oligomerizes and binds a specific IL17 cytokine, and the bacterium Alistipes. Changes in the abundance of Alistipes have been reported in several conditions, including paediatric CD,52 colorectal cancer53 and obesity.54 Previous studies have reported a negative correlation between the abundance of Alistipes and the lipopolysaccharide (LPS)-induced tumour necrosis factor (TNF) alpha response.55 Therefore, mbQTLs identified at whole-exome-level suggest a potential complex interaction between host genetics, microbial composition and the immune system.
Next, we focused on a subset of selected variants located in genes within IBD-susceptibility regions and predicted protein-disrupting variants that could potentially lead to disease or abnormal phenotype by altering the gut microbiome. Here, we found two mbQTLs located in the IBD-associated genes SEC16A and WDR78. SEC16A is involved in the transitional endoplasmic reticulum and is located within a haplotype block that contains the INPP5E and CARD9 genes.56 The SEC16A-affected pathway biosynthesis of thiamin (vitamin B1, an essential vitamin) is necessary for the proper functioning of the immune system and thiamin is supplied to the host through diet and the gut microbiota.57 WDR78 was associated with L-rhamnose biosynthesis, and L-rhamnose is a precursor of a common enterobacterial antigen. In addition, WDR78, together with genes GPR65 and TNFAIP3, is reported to cooperate in regulation of the macrophage component.58 Therefore, this study reveals a potential link that suggests WDR78 may potentially regulate microbial function through antigen recognition by immune cells.
In contrast to the regular genotyping arrays used in GWAS, WES enables the detection of rare variants with mbQTLs effects. We identified independent rare variants with predicted functional consequences within the G-protein coupled receptor 151, GPR151, that are associated with multiple functional microbial pathways (homolactic fermentation, glucose and xylose degradation). GPR151 is a critical element of antigen recognition and activation of the immune response,59 60 and PTVs in GPR151 have been reported to have a protective effect against obesity and type 2 diabetes in the UK Biobank.61 In addition, lower levels of bacterial carbohydrate degradation lead to lower carbohydrate absorption in the gut by the host, which pinpoints potential mechanisms by which GPR151 variants can protect against metabolic diseases. Limited by the artefacts on capturing exomes using WES, we restricted our analyses on CNV site frequency lower than 1%. The strongest association between genes with CNV and microbiota was CD160, and Lachnospiraceae. CD160 is reported to be highly expressed in small intestine, inducing production of proinflammatory cytokines and antipathogen protein.62 63 Moreover, depletion of gene CD160 has been shown to be associated with increased pathogenic bacteria in mice.64
Finally, we joined the two cohorts to perform genetics–disease interaction analysis, rather than comparing single-cohort-significant mbQTLs separately, to identify disease-specific mbQTLs and to achieve more power. This approach was able to show that genetics potentially exerts a different influence on the microbiome in IBD compared with a healthy situation. The known association between the LCT gene and Bifidobacterium abundance was only present in the population cohort. This could potentially be explained by the fact that Bifidobacterium abundance is decreased in the gut microbiota of CD3 which was observed in this study, and therefore this mbQTL was not present in the IBD cohort. Furthermore, we observed mbQTL effects in known IBD genes18 such as TNFSF15 only in the IBD cohort. When analysing mbQTL effects in patients with CD and UC separately, we could only identify two mbQTLs in patients with CD that reached the significance threshold. This could be due to the limited statistical power resulted by subdividing the IBD group in its two main subtypes.
Heritability studies have shown that part of the microbiome development and composition is under genomic control.41 Studies looking into genome–microbiome interaction have been performed using GWAS technologies in healthy or population-based cohorts.15 16 26 In LifeLines-DEEP cohort, we replicated the association between variants in the LCT gene and abundance of Bifidobacterium, and the association between TIRAP gene (rs560813, T>C, p=0.024) and abundance of genus Holdemania previously reported in Bonder et al,15 which contained partially overlapping samples with the current study. On the level of the general population, the effect of genetic makeup on the variance of microbiome composition is lower compared with the cumulative effect of environmental exposure.20 However, the genetic effects might show more substantial contribution in more specific conditions, such as IBD, which shows more pronounced effects on both genetic and microbial components. Several earlier studies in IBD cohorts have also reported IBD-specific mbQTL variants. We identified variants in the IBD-associated genes TNFSF15 and HLA-B, both genes that have been reported earlier in a study combining mucosal 16s sequencing data and GWAS data.18 The lack of replication of other studies including Lloyd-Price et al 27 could partially be explained by the cohort recruitment, for example, Groningen patients with IBD are over 18 years old with long-term disease problems while half of the patients in Lloyd-Price et al are early onset paediatric cases, which have different IBD genetic makeup and microbial features.65 66 Besides, sample size, datasets, included confounders and analysis strategies might also explain differences in results across studies. In the current study, we performed a large-scale mbQTL analysis of gut microbiome composition and function that combined two high-resolution techniques, WES and shotgun metagenomics, while controlling for major confounders known to influence the gut microbiome. While we are only beginning to dissect the genomic architecture that drives microbiome evolution and composition in health and disease, this study adds considerable insights and provides leads for further functional analyses or targets for therapies in the context of IBD.
This research highlights that both common and rare host genetic variants affecting the immune system are key factors in shaping the gut microbiota taxonomy and function, knowledge which further enhances our understanding of the intricate host–microbiome interaction involved in IBD pathogenesis.
Acknowledgments
The authors thank the LifeLines-DEEP and IBD cohort participants. They thank Kate Mc Intyre for substantive English editing and B.H. Jansen for technical support.
Footnotes
Contributors: Study supervision: RKW and AK. Analysis and drafting: SH, AVV, RG and VC. Data support: CS, MR, RX, MJD, JMF, IW and MET. Critical revision: RKW, AK, AZ, JF, CW, FI, EAF, HMvD, GD, MCV and LB. Shared last authors: AZ, AK and RKW.
Funding: MR is supported by a National Institute of Health Center for Multi- and Trans-Ethnic Mapping of Mendelian and Complex Diseases grant (5U01 HG009080) and by the National Human Genome Research Institute of the National Institutes of Health (NIH) under award R01HG010140. CW is supported by a European Research Council (ERC) Advanced grant (FP/2007-2013/ERC grant 2012-322698), a Netherlands Organization for Scientific Research (NWO) Spinoza prize grant (NWO SPI 92-266) and the Gravitation Netherlands Organ-on-Chip Initiative (024.003.001). JF is supported by grants from NWO (NWO-VIDI 864.13.013) and CardioVasculair Onderzoek Nederland (CVON 2018-27). AZ is supported by an NWO Vidi grant (NWO-VIDI 016.178.056), an ERC Starting Grant (715772), CVON 2018-27 and a Rosalind Franklin Fellowship from the University of Groningen. Copy number variant analyses were supported by NIH MH115957 to MET.
Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were involved in the design, or conduct, or reporting, or dissemination plans of this research. Refer to the Methods section for further details.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available upon reasonable request. The data for the LifeLines DEEP cohort available upon request from the European Genome-Phenome Archive (EGA; https://www.ebi.ac.uk/ega/) at accession number EGAS00001001704. The data for the Groningen IBD cohort can be requested with the accession number EGAS00001002702.
References
- 1. Ng SC, Shi HY, Hamidi N, et al. . Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 2018;390:2769–78. 10.1016/S0140-6736(17)32448-0 [DOI] [PubMed] [Google Scholar]
- 2. de Lange KM, Moutsianas L, Lee JC, et al. . Genome-Wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet 2017;49:256–61. 10.1038/ng.3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vich Vila A, Imhann F, Collij V, et al. . Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med 2018;10. 10.1126/scitranslmed.aap8914. [Epub ahead of print: 19 Dec 2018]. [DOI] [PubMed] [Google Scholar]
- 4. Franzosa EA, Sirota-Madi A, Avila-Pacheco J, et al. . Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol 2019;4:293–305. 10.1038/s41564-018-0306-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schirmer M, Garner A, Vlamakis H, et al. . Microbial genes and pathways in inflammatory bowel disease. Nat Rev Microbiol 2019;17:497–511. 10.1038/s41579-019-0213-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut 2013;62:1505–10. 10.1136/gutjnl-2012-303954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turpin W, Goethel A, Bedrani L, et al. . Determinants of IBD heritability: genes, bugs, and more. Inflamm Bowel Dis 2018;24:1133–48. 10.1093/ibd/izy085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hall AB, Tolonen AC, Xavier RJ. Human genetic variation and the gut microbiome in disease. Nat Rev Genet 2017;18:690–9. 10.1038/nrg.2017.63 [DOI] [PubMed] [Google Scholar]
- 9. Cohen LJ, Cho JH, Gevers D, et al. . Genetic factors and the intestinal microbiome guide development of Microbe-Based therapies for inflammatory bowel diseases. Gastroenterology 2019;156:2174–89. 10.1053/j.gastro.2019.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kobayashi KS, Chamaillard M, Ogura Y, et al. . Nod2-Dependent regulation of innate and adaptive immunity in the intestinal tract. Science 2005;307:731–4. 10.1126/science.1104911 [DOI] [PubMed] [Google Scholar]
- 11. Mondot S, Barreau F, Al Nabhani Z, et al. . Altered gut microbiota composition in immune-impaired Nod2(-/-) mice. Gut 2012;61:634–5. 10.1136/gutjnl-2011-300478 [DOI] [PubMed] [Google Scholar]
- 12. Rehman A, Sina C, Gavrilova O, et al. . Nod2 is essential for temporal development of intestinal microbial communities. Gut 2011;60:1354–62. 10.1136/gut.2010.216259 [DOI] [PubMed] [Google Scholar]
- 13. Butera A, Di Paola M, Pavarini L, et al. . Nod2 deficiency in mice is associated with microbiota variation favouring the expansion of mucosal CD4+ LAP+ regulatory cells. Sci Rep 2018;8:14241. 10.1038/s41598-018-32583-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sadaghian Sadabad M, Regeling A, de Goffau MC, et al. . The ATG16L1-T300A allele impairs clearance of pathosymbionts in the inflamed ileal mucosa of Crohn's disease patients. Gut 2015;64:1546–52. 10.1136/gutjnl-2014-307289 [DOI] [PubMed] [Google Scholar]
- 15. Bonder MJ, Kurilshikov A, Tigchelaar EF, et al. . The effect of host genetics on the gut microbiome. Nat Genet 2016;48:1407–12. 10.1038/ng.3663 [DOI] [PubMed] [Google Scholar]
- 16. Wang J, Thingholm LB, Skiecevičienė J, et al. . Genome-Wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet 2016;48:1396–406. 10.1038/ng.3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aschard H, Laville V, Tchetgen ET, et al. . Genetic effects on the commensal microbiota in inflammatory bowel disease patients. PLoS Genet 2019;15:e1008018. 10.1371/journal.pgen.1008018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Knights D, Silverberg MS, Weersma RK, et al. . Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med 2014;6:107. 10.1186/s13073-014-0107-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kurilshikov A, Wijmenga C, Fu J, et al. . Host genetics and gut microbiome: challenges and perspectives. Trends Immunol 2017;38:633–47. 10.1016/j.it.2017.06.003 [DOI] [PubMed] [Google Scholar]
- 20. Rothschild D, Weissbrod O, Barkan E, et al. . Environment dominates over host genetics in shaping human gut microbiota. Nature 2018;555:210–5. 10.1038/nature25973 [DOI] [PubMed] [Google Scholar]
- 21. McKenna A, Hanna M, Banks E, et al. . The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anderson CA, Pettersson FH, Clarke GM, et al. . Data quality control in genetic case-control association studies. Nat Protoc 2010;5:1564–73. 10.1038/nprot.2010.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Babadi M, Lee S, Smirnov A, et al. . Precise common and rare germline CNV calling with GATK 2018.
- 24. Segata N, Waldron L, Ballarini A, et al. . Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 2012;9:811–4. 10.1038/nmeth.2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Franzosa EA, McIver LJ, Rahnavard G, et al. . Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods 2018;15:962–8. 10.1038/s41592-018-0176-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Turpin W, Espin-Garcia O, Xu W, et al. . Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet 2016;48:1413–7. 10.1038/ng.3693 [DOI] [PubMed] [Google Scholar]
- 27. Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. . Multi-Omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569:655–62. 10.1038/s41586-019-1237-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Falony G, Joossens M, Vieira-Silva S, et al. . Population-Level analysis of gut microbiome variation. Science 2016;352:560–4. 10.1126/science.aad3503 [DOI] [PubMed] [Google Scholar]
- 29. Zhernakova A, Kurilshikov A, Bonder MJ, et al. . Population-Based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016;352:565–9. 10.1126/science.aad3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Imhann F, Vich Vila A, Bonder MJ, et al. . Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2018;67:108–19. 10.1136/gutjnl-2016-312135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rivas MA, Pirinen M, Conrad DF, et al. . Human genomics. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science 2015;348:666–9. 10.1126/science.1261877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee S, Teslovich TM, Boehnke M, et al. . General framework for meta-analysis of rare variants in sequencing association studies. Am J Hum Genet 2013;93:42–53. 10.1016/j.ajhg.2013.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Purcell SM, Moran JL, Fromer M, et al. . A polygenic burden of rare disruptive mutations in schizophrenia. Nature 2014;506:185–90. 10.1038/nature12975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Frenkel S, Bernstein CN, Sargent M, et al. . Genome-Wide analysis identifies rare copy number variations associated with inflammatory bowel disease. PLoS One 2019;14:e0217846. 10.1371/journal.pone.0217846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peters JE, Lyons PA, Lee JC, et al. . Insight into genotype-phenotype associations through eQTL mapping in multiple cell types in health and immune-mediated disease. PLoS Genet 2016;12:e1005908. 10.1371/journal.pgen.1005908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. GTEx Consortium Human genomics. The Genotype-Tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648–60. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuleshov MV, Jones MR, Rouillard AD, et al. . Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90–7. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Watanabe K, Taskesen E, van Bochoven A, et al. . Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017;8:1826. 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Franke A, Balschun T, Sina C, et al. . Genome-Wide association study for ulcerative colitis identifies risk loci at 7q22 and 22q13 (IL17REL). Nat Genet 2010;42:292–4. 10.1038/ng.553 [DOI] [PubMed] [Google Scholar]
- 40. Prescott NJ, Lehne B, Stone K, et al. . Pooled sequencing of 531 genes in inflammatory bowel disease identifies an associated rare variant in BTNL2 and implicates other immune related genes. PLoS Genet 2015;11:e1004955–19. 10.1371/journal.pgen.1004955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Goodrich JK, Davenport ER, Beaumont M, et al. . Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 2016;19:731–43. 10.1016/j.chom.2016.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kolde R, Franzosa EA, Rahnavard G, et al. . Host genetic variation and its microbiome interactions within the human microbiome project. Genome Med 2018;10:6. 10.1186/s13073-018-0515-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu JZ, van Sommeren S, Huang H, et al. . Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979–86. 10.1038/ng.3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Koh A, De Vadder F, Kovatcheva-Datchary P, et al. . From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 2016;165:1332–45. 10.1016/j.cell.2016.05.041 [DOI] [PubMed] [Google Scholar]
- 45. Ríos-Covián D, Ruas-Madiedo P, Margolles A, et al. . Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol 2016;7:185. 10.3389/fmicb.2016.00185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Carethers JM, Jung BH. Genetics and genetic biomarkers in sporadic colorectal cancer. Gastroenterology 2015;149:1177–90. 10.1053/j.gastro.2015.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Costea I, Mack DR, Lemaitre RN, et al. . Interactions between the dietary polyunsaturated fatty acid ratio and genetic factors determine susceptibility to pediatric Crohn's disease. Gastroenterology 2014;146:929–31. 10.1053/j.gastro.2013.12.034 [DOI] [PubMed] [Google Scholar]
- 48. Illig T, Gieger C, Zhai G, et al. . A genome-wide perspective of genetic variation in human metabolism. Nat Genet 2010;42:137–41. 10.1038/ng.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marion-Letellier R, Savoye G, Beck PL, et al. . Polyunsaturated fatty acids in inflammatory bowel diseases: a reappraisal of effects and therapeutic approaches. Inflamm Bowel Dis 2013;19:650–61. 10.1097/MIB.0b013e3182810122 [DOI] [PubMed] [Google Scholar]
- 50. Sun L, Youn HD, Loh C, et al. . Cabin 1, a negative regulator for calcineurin signaling in T lymphocytes. Immunity 1998;8:703–11. 10.1016/S1074-7613(00)80575-0 [DOI] [PubMed] [Google Scholar]
- 51. Gulick AM, Hubbard BK, Gerlt JA, et al. . Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in the active site of D-glucarate dehydratase from Escherichia coli. Biochemistry 2001;40:10054–62. 10.1021/bi010733b [DOI] [PubMed] [Google Scholar]
- 52. Lewis JD, Chen EZ, Baldassano RN, et al. . Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn's disease. Cell Host Microbe 2015;18:489–500. 10.1016/j.chom.2015.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang T, Cai G, Qiu Y, et al. . Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. Isme J 2012;6:320–9. 10.1038/ismej.2011.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ridaura VK, Faith JJ, Rey FE, et al. . Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013;341:1241214. 10.1126/science.1241214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schirmer M, Smeekens SP, Vlamakis H, et al. . Linking the human gut microbiome to inflammatory cytokine production capacity. Cell 2016;167:1125–36. 10.1016/j.cell.2016.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhernakova A, Festen EM, Franke L, et al. . Genetic analysis of innate immunity in Crohn's disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am J Hum Genet 2008;82:1202–10. 10.1016/j.ajhg.2008.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schirmer M, et al. Inflammatory bowel disease gut microbiome. Nat. Microbiol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Peters LA, Perrigoue J, Mortha A, et al. . A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nat Genet 2017;49:1437–49. 10.1038/ng.3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cho H, Kehrl JH. Regulation of immune function by G protein-coupled receptors, trimeric G proteins, and RGS proteins. Prog Mol Biol Transl Sci 2009;86:249–98. 10.1016/S1877-1173(09)86009-2 [DOI] [PubMed] [Google Scholar]
- 60. Gräler MH, Goetzl EJ. Lysophospholipids and their G protein-coupled receptors in inflammation and immunity. Biochim Biophys Acta 2002;1582:168–74. 10.1016/S1388-1981(02)00152-X [DOI] [PubMed] [Google Scholar]
- 61. Emdin CA, Khera AV, Chaffin M, et al. . Analysis of predicted loss-of-function variants in UK Biobank identifies variants protective for disease. Nat Commun 2018;9:1613. 10.1038/s41467-018-03911-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fons P, Chabot S, Cartwright JE, et al. . Soluble HLA-G1 inhibits angiogenesis through an apoptotic pathway and by direct binding to CD160 receptor expressed by endothelial cells. Blood 2006;108:2608–15. 10.1182/blood-2005-12-019919 [DOI] [PubMed] [Google Scholar]
- 63. Cai G, Anumanthan A, Brown JA, et al. . CD160 inhibits activation of human CD4+ T cells through interaction with herpesvirus entry mediator. Nat Immunol 2008;9:176–85. 10.1038/ni1554 [DOI] [PubMed] [Google Scholar]
- 64. Shui J-W, Larange A, Kim G, et al. . HVEM signalling at mucosal barriers provides host defence against pathogenic bacteria. Nature 2012;488:222–5. 10.1038/nature11242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kelsen JR, Dawany N, Moran CJ, et al. . Exome sequencing analysis reveals variants in primary immunodeficiency genes in patients with very early onset inflammatory bowel disease. Gastroenterology 2015;149:1415–24. 10.1053/j.gastro.2015.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. An R, Wilms E, Masclee AAM, et al. . Age-Dependent changes in Gi physiology and microbiota: time to reconsider? Gut 2018;67:2213–22. 10.1136/gutjnl-2017-315542 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2019-319706supp001.xlsx (12.7MB, xlsx)
gutjnl-2019-319706supp002.pdf (109.3KB, pdf)
gutjnl-2019-319706supp003.pdf (142.8KB, pdf)
gutjnl-2019-319706supp004.pdf (856.2KB, pdf)
gutjnl-2019-319706supp005.pdf (254.4KB, pdf)
gutjnl-2019-319706supp006.pdf (78KB, pdf)
gutjnl-2019-319706supp007.pdf (490.1KB, pdf)