Abstract
Patients with metastasized malignant melanomas (MM) are regularly treated with immune checkpoint inhibitors (CPI). Within our study, we evaluated the predictive value of tumor infiltrating lymphocyte (TIL) clusters in primary MM and its association to molecular subtypes to predict response to CPI treatment. A cohort of 90 MM patients who received CPI treatment were collected from a single center, as well as a validation cohort of 351 patients from the TCGA database (SKCM) who received standard of care. A deep-convolutional-neural network (U-Net) was trained to detect viable tumor areas on H&E whole-slide-images, following a quantitative detection of TILs with help of a separate additional neural network. The number of TIL clusters was associated with response to CPI in 90 MM patients (AUC = 0.6), even more pronounced within the sub-cohort of BRAF V600E/K-mutated MM patients (AUC = 0.7, n = 32). Interestingly, the TIL clusters in NRAS-mutated as well as wildtype MM (BRAF-wt, NRAS-wt) tumors, did not demonstrate a predictive value of CPI response (AUC = 0.5, n = 25). Moreover, PD-L1 expression had a limited predictive value within our cohort. In parallel, within an independent cohort of MM patients (TCGA, n = 351), the number of TIL clusters was associated with improved survival in BRAF V600E/K mutated MM (p < 0.0001, n = 164) but neither in NRAS-mutated (55.7 months vs. 63.0 months, respectively, p = 0.590, n = 85) nor BRAF/NRAS-wildtype MM patients (52.4 months vs. 47.4 months, respectively, p = 0.581, n = 104). While TILs in MM have been associated with improved survival, we show—for the first time—that TIL clusters are associated with response to immunotherapy in BRAF V600E/K mutated MM.
Subject terms: Cancer, Skin cancer, Tumour biomarkers, Computational biology and bioinformatics, Genetics, Biomarkers, Oncology
Introduction
Immune checkpoint inhibitors (CPI) targeting the programmed death 1/programmed death-ligand 1 (PD-1/PD-L1), as well as the cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4), have demonstrated promising and durable antitumor activity, and revolutionized the treatment of metastasized malignant melanoma (MM) patients1–3. However, a relevant fraction of patients does not benefit from monotherapy or even combined CPI regimens. Despite controversial study results, CPI treatment responses have been shown to correlate with certain quantitative markers, such as PD-L1 expression levels, as well as tumor mutational burden2–7.
Meanwhile, the quantitative assessment of tumor infiltrating lymphocytes (TILs) has been associated with a favorable prognosis in MM8–10. In addition, increased numbers of TILs were linked to response to interferon-alpha treatment in patients with advanced stage MM11. However, to the best of our knowledge, the prognostic value of TIL quantification from H&E images to immune checkpoint inhibition in MM remains elusive.
To explore the predictive value of quantitative assessment of TILs in malignant melanoma as well as regarding treatment response to CPI, we analyzed a cohort of 90 individuals treated with CPI at a single center, real world data and validated our findings within an independent cohort of 351 cases of malignant melanomas from the TCGA database.
Results
Detection of viable tumor areas and tumor infiltrating lymphocyte clusters using digitized H&E whole-slide images
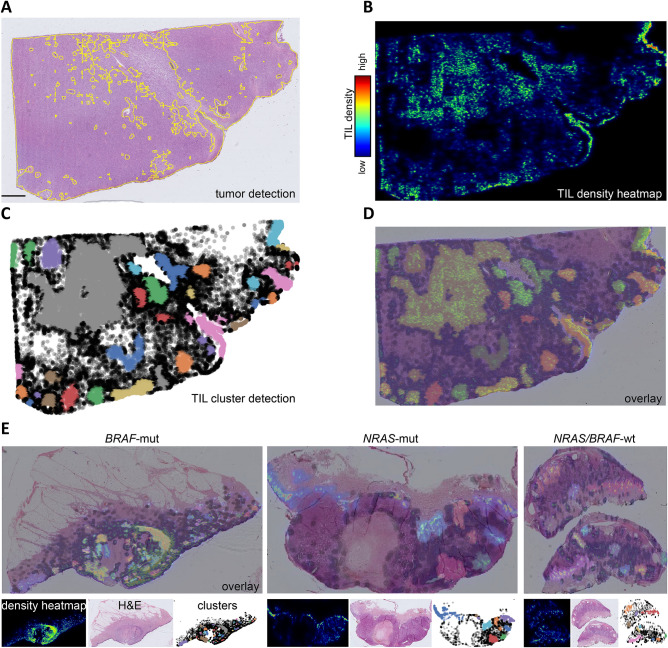
To allow a controllable and efficient detection of viable tumor areas and to avoid a bias of necrotic tumor regions with an accumulation of inflammatory cells, we trained a U-Net deep convolutional neural network for segmentation of tumor regions using digitized whole-slide images (Fig. 1A). Subsequently, we generated a neural network to detect TILs within areas of viable tumor cells (Fig. 1A).
Figure 1.
Detection of viable tumor areas and TIL clusters using regular H&E whole-slide-images of malignant melanomas. Illustration of tumor segmentation using regular H&E stains of cases of malignant melanoma, with visualizations of TIL detection and TIL clustering using single images and combined overlays. (A) H&E stains of one representative case of malignant melanoma with segmentation of viable tumor areas with help of a deep convolutional neural network (U-Net). The yellow line represents the segmentation of tumor area, while necrotic areas are left out. (B) Subsequent TIL detection within viable tumor areas and illustration of TILs using a density heatmap where red indicates high density of TILs and blue indicates low density, according to the legend. (C) Visualization of TIL cluster detection. Black circles indicate individual TILs and colored areas highlight distinct TIL cluster that had been assigned using HDBSCAN12. (D) Overlay of the images from panels A-C with transparent layers combining H&E image, TIL density heatmap and TIL clusters. (E) Panel of BRAF-mut (V600E/K) NRAS-mut and BRAF/NRAS-wildtype samples, where the overlay is shown in the upper panel and the given TIL density heatmap, TIL clustering and H&E images are shown below.
Then, we calculated the distribution of TILs within areas of viable tumor cells by using a clustering algorithm (Fig. 1B–E)12. In addition to a quantitative assessment of TILs, TIL clusters would allow a qualitative assessment of TIL distribution within MM tumors.
Predictive value of tumor infiltrating lymphocyte clusters within BRAF V600E/K mutated melanoma patients receiving CPI
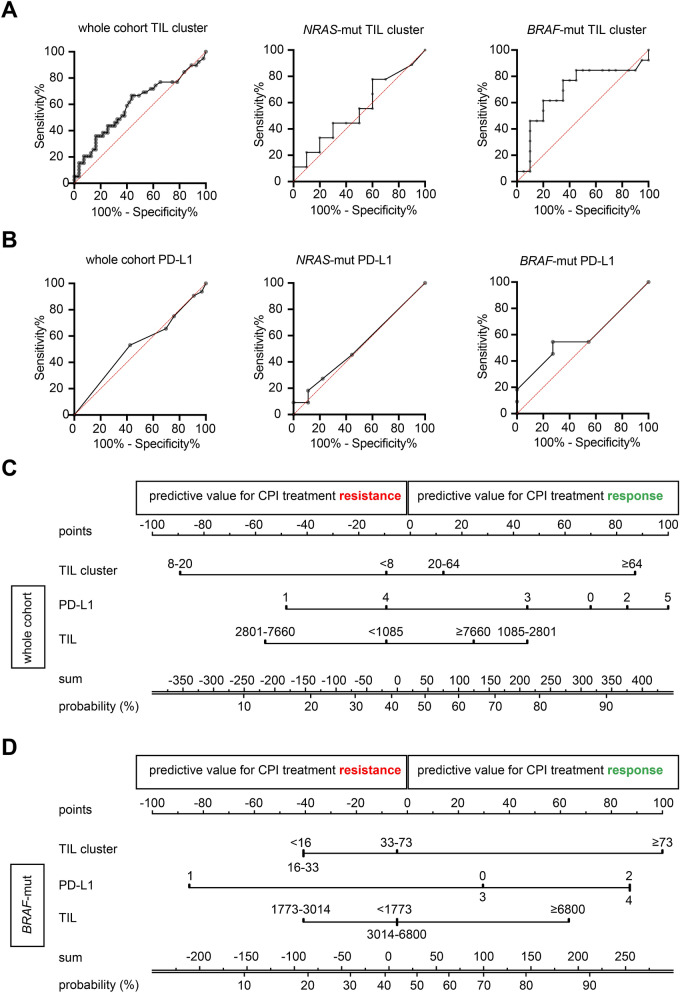
Having built a deep learning-based CNN for tumor segmentation, as well as a neural network for detection of TILs, we applied this method to a cohort of 90 malignant melanoma patients that received immune-checkpoint inhibition (CPI; Table 1) from a single center. Here, the overall predictive relevance to CPI response of TIL clusters was low (AUC = 0.6, n = 90; Fig. 2A). However, in 32 cases of BRAF V600E/K mutated MMs, there was a higher predictive value for TIL clusters (AUC = 0.7, n = 32). Within NRAS-mutated MMs, there was no predictive value of TIL clusters (AUC = 0.5, n = 25).
Table 1.
CPI treatment results.
| Ipilimumab (n = 40) | Pembrolizumab (n = 32) | Nivolumab (n = 11) | Ipilimumab + Nivolumab (n = 7) | |
|---|---|---|---|---|
| Mean applications received ± SD (range) | 3.6 ± 0.7 (1–4) | 7.9 ± 5.8 (1–23) | 8.7 ± 5.2 (3–17) | 3.5 ± 0.8 (2–4) |
| Treatment response | ||||
| Complete response | 2 (5.0%) | 0 | 0 | 0 |
| Partial response | 7 (17.5%) | 11 (34.4%) | 5 (45.5%) | 2 (28.6%) |
| Stable disease | 3 (7.5%) | 5 (15.6%) | 0 | 0 |
| Progress | 28 (70.0%) | 16 (50.0%) | 6 (54.5%) | 4 (57.1%) |
| Not evaluable | 0 | 0 | 0 | 1 (14.3%) |
| Stopped treatment due to side effects | 4 (10.0%) | 1 (3.1%) | 2 (18.2%) | 1 (14.3%) |
Figure 2.
Predictive value of TIL clusters in malignant melanoma subsets. Overview of predictive value of TIL clusters according to molecular subtypes of malignant melanoma using individual ROC curves and a nomogram combining several attributes and their predictive value to predict response to CPI treatment. (A) Area under the curve for TIL clusters for different mutation types of MM tumors (BRAF-mut; BRAF V600E/K). (B) Area under the curve for PD-L1 scoring for different MM mutation subtypes. (C) Nomogram for the whole cohort of MM patients (n = 90). (D) Nomogram for the BRAF-mutated (BRAF-mut; BRAF V600E/K) sub cohort (n = 32) of MM patients. The positive values (points) indicate the prediction towards responses, while negative values reflect the predictive value for therapy resistance.
To compare the predictive value of TIL clusters to CPI treatment, TILs and PD-L1 (Fig. 2B; Table 2), we generated a nomogram for the whole cohort of MM patients (n = 90) as well as BRAF V600E/K mutated MMs (n = 32). Here, TIL clusters showed an improved predictive value compared to TILs in the whole cohort as well as within BRAF V600E/K mutated MMs (Fig. 2C,D). Interestingly, TIL clusters did reveal a predictive value for both response- and resistance to CPI treatment. There was a correlation between the number of TIL clusters and response to CPI treatment within BRAF V600E/K mutated MM (Fig. 2D). Elevated numbers of TIL clusters (above 33) showed a higher probability of response to CPI treatment. Conversely, categorial assessment of PD-L1 showed that scoring of either 2 or 4 were associated with response, while PD-L1 scoring of 1 was linked to resistance to CPI treatment (Fig. 2D). This effect was even pronounced within the whole cohort of MM patients, as scoring of either 1 or 4 was associated with resistance to CPI treatment, compared to response to CPI treatment (scores of 3, 0 and 2; Fig. 2C).
Table 2.
Predictive value of PD-L1 score concerning treatment outcome.
| Disease control (complete/partial response, stable disease) | Treatment non-responders (progress) | p | |
|---|---|---|---|
|
All CPI treatments (n = 78) PD-L1 | |||
| Positive | 13 | 14 | 0.447 |
| Negative | 20 | 31 | |
|
All anti-PD-1 treatments (nivolumab, pembrolizumab, ipilimumab + nivolumab) PD-L1 | |||
| Positive | 8 | 8 | 1.0 |
| Negative | 15 | 15 | |
|
Ipilimumab PD-L1 | |||
| Positive | 5 | 6 | 0.215 |
| Negative | 5 | 16 | |
|
Nivolumab PD-L1 | |||
| Positive | 0 | 1 | 1.0 |
| Negative | 5 | 5 | |
|
Pembrolizumab PD-L1 | |||
| Positive | 7 | 5 | 0.654 |
| Negative | 9 | 9 | |
|
Ipilimumab + Nivolumab PD-L1 | |||
| Positive | 1 | 2 | 1.0 |
| Negative | 1 | 1 | |
Prognostic value of tumor-infiltrating lymphocyte clusters in molecular subtypes of malignant melanomas
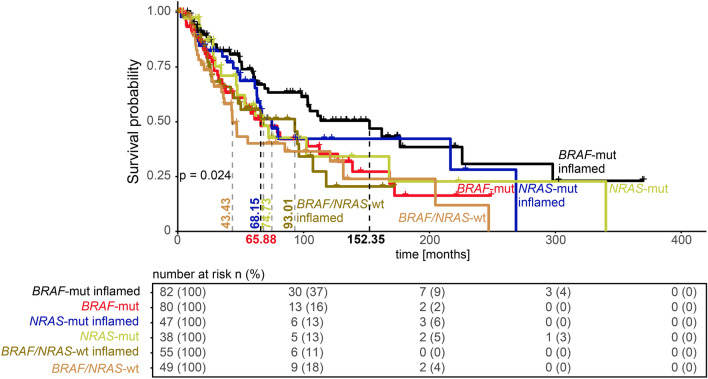
To validate whether TIL cluster counts were associated with a favorable prognosis within molecular subtypes of MM, we analyzed an independent cohort of 351 patients from the TCGA database. Interestingly, within BRAF V600E/K mutated MM tumors, the number of TIL clusters was associated with improved survival (median overall survival rate BRAF V600E/K with low clusters: 48.2 vs. BRAF V600E/K with high clusters: 86.9 months, p < 0.0001, n = 164; Fig. 3) but this effect was not seen in neither NRAS-mutated MM patients, (55.7 months vs. 63.0 months, respectively, p = 0.590, n = 85) nor wildtype MM patients (52.4 months vs. 47.4 months, respectively, p = 0.581, n = 104).
Figure 3.
Prognostic value of TIL clusters within molecular subtypes of malignant melanomas. Kaplan–Meier curve of TCGA MM patients (n = 351) stratified for mutational subtypes (BRAF-mutated, NRAS-mutated, and BRAF/NRAS wildtype). Inflamed tumors are indicated according to their molecular subtype, while inflamed tumors are defined as tumors with TIL clusters above the median value as a cutoff. The median survival time for each molecular subtype and inflammatory status is provided using a vertical line and indicated in letters according to their color code. The risk table is shown below for the given subgroups.
Discussion
There is a clinical need to explore prognostic biomarkers for immune response to immune checkpoint inhibition in solid cancers—especially within MM—where alternative treatment strategies for molecular subtypes are available, but CPI treatment may be associated to long-term remission. Here, BRAF V600-mutated MM may reveal additional treatment options with BRAF and MEK inhibitors13.
Within our study, we highlight the predictive value of quantitative TIL cluster characterization to CPI response within BRAF V600-mutated MMs. Recently, there were two clinical trials performing subgroup analyses concerning response rate, progression-free and overall survival including the BRAF mutation status2,3. In a study by Larkin et al.2, the median overall survival among patients with BRAF-mutated MM was longer in the Nivolumab-plus-Ipilimumab group (more than 60.0 months; 95% CI, 50.7 to not reached), the Nivolumab group (45.5 months; 95% CI, 26.4 to not reached) and the Ipilimumab group (24.6 months; 95% CI, 17.9–31.0) compared to patients without BRAF-mutated MM (Nivolumab-plus-Ipilimumab group: 39.1 months; 95% CI, 27.5 to not reached, Nivolumab group: 34.4 months; 95% CI, 24.1–59.2, Ipilimumab group: 18.5 months; 95% CI, 14.1–22.7). However, the median progression-free survival was only longer among patients with BRAF mutations in the combination treatment group compared to patients without BRAF mutations (16.8 months; 95% CI, 8.3–32.0 versus 11.2 months; 95% CI, 7.0–18.1). On the other hand, Robert et al. 3 reported that response rates of patients receiving either Ipilimumab or Pembrolizumab were similar in patients with tumors with or without BRAF V600E/K mutations.
Given the mounting evidence for TILs as a potential biomarker, our study supports to consider the feature of TIL clusters as a predictive marker for prognosis and response to CPI in MM. Assessing TILs through deep learning showed a predictive value in BRAF V600E/K melanomas that received CPI treatment from a single center. These results emphasize the relevance for future studies to potentially deploy TIL clusters as a biomarker for BRAF V600E/K mutated MM, where either immunotherapy or targeted therapies may be considered as treatment regimens.
Material and methods
Patient and tumor characteristics
90 patients with metastasized MM diagnosed at the University Hospital Cologne receiving first line CPI treatment were included in our study. All patients underwent a clinical follow-up according to the current treatment guidelines for MM. 40 patients were treated with Ipilimumab (44.4%; 3 mg/kg body weight every 3 weeks), 32 patients were treated with Pembrolizumab (35.6%; 2 mg/kg every 3 weeks) and 11 patients were treated with Nivolumab (12.2%, 3 mg/kg body weight every 2 weeks). Other 7 patients (7.8%) received Ipilimumab 3 mg/kg in combination with Nivolumab 1 mg/kg body weight every 3 weeks. Treatment response was categorized as disease control (partial/complete response or stable disease) versus disease progression according to RECIST criteria (Table 1). In general, there were more patients with disease control under Nivolumab and Pembrolizumab versus Ipilimumab (Table 1). Termination of treatment due to side effects was the highest in the Nivolumab (18.2%) and combination treatment group (14.3%; Table 1). There were no significant differences in sex, patient age at initial diagnosis or treatment initiation, tumor depth, localization, or subtype as well as mutation (BRAF, NRAS mutation or wildtype) and PD-L1 status in the different monotherapy groups. The small group of combination therapy (n = 7) contained more male patients harboring melanomas with higher Breslow index. Cases from the TCGA database received standard of care, which did not include CPI treatment at the time of sample processing.
Molecular subtyping of MM samples
Molecular subtyping of single center cases of malignant melanomas was conducted using a targeted panel sequencing approach, covering NRAS (exon 2, 3, 4) and BRAF (exon 11, 15) among other genes as described previously14–16. Activating mutations for BRAF and NRAS were annotated for both TCGA and internal data17–19. Absence of known activating mutations in either BRAF or NRAS was considered as wildtype (BRAF/NRAS-wildtype; BRAF-wt/NRAS-wt), while cases with either BRAF or NRAS activating mutations were considered as BRAF-V600E/K or NRAS-mutated (BRAF-mut/NRAS-mut).
Whole-slide-images and processing
Regular H&E stained slides, following standard protocols, were scanned using a NanoZoomer S360 (Hamamatsu Photonics) whole-slide scanning device at a 40X magnification, as well as slides from the TCGA database being scanned 20X using Leica Aperio slide scanning devices. All digitized slides were evaluated for image quality and included, if more than 90% of the tissue area was in focus. All tumors investigated were primary melanomas excised before beginning of the CPI treatment.
Image segmentation, object detection and clustering
A U-Net was trained to detect vital tumor areas on H&E MM virtual whole slide images20. For this purpose, images were annotated using whole slide images by a trained pathologist. To generate a training dataset that would reflect the heterogeneity that can be observed within MM tumors, 92 image tiles (10,000 × 10,000 pixels, resized to 1000 × 1000 for further training purpose) from a total of 35 cases were used, with an image patch size of 256 pixels for the network to be processed. Training was performed using TCGA cases, as well as cases from a set of internal cases, while all cases were independent to the test set. Training was done on a NVIDIA RTX 6000 using the PyTorch framework and Adam as an optimizer. Several augmentation steps were applied, including grey-scale augmentation with a ten percent probability21. For TIL object detection, we followed a similar approach that has already been published22. To efficiently identify the number of TIL clusters, we used the HDBSCAN algorithm12.
PD-L1 expression on tumor cells
A tumor cell was considered PD-L1 positive if the cell membrane was partially or completely stained, whereas cytoplasmic PD-L1 staining was not considered as a specific immune signal. The tumor proportion score was determined as previously published23.
Statistical analysis
The area under the receiver-operator-curve (ROC curve) was calculated based on the overall ability of the given attributes to discriminate between patients that either responded or progressed under immune-checkpoint inhibition therapy. To visualize the effects of the attributes on the class probabilities (response/progress under therapy), we generated a nomogram using a Naïve-Bayes classifier that was trained on the attributes of treatment results as described previously24. Statistical analysis was performed with Python (version 3.7, https://www.python.org/), R, the R Project (version 4.0.3, https://www.r-project.org/) and the statistical software package IBM SPSS (version 25.0). Statistical testing was carried out by using X2 test, Fisher’s test or Student's t-test. Survival rates were calculated by the Kaplan–Meier method and compared using log-rank. p < 0.05 was considered to be significant.
Ethics approval
The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Ethics Committee of the Medical Faculty of University of Cologne (Registration No. 08-144). Informed consent has been obtained.
Abbreviations
- CNN
Convolutional neural network
- CPI
Immune checkpoint inhibitor
- CTLA-4
Cytotoxic T-lymphocyte-associated antigen 4
- MM
Malignant melanoma
- PD-1/PD-L1
Programmed death 1/programmed death-ligand 1
- TIL
Tumor-infiltrating lymphocyte
Author contributions
S.K. study design, methodical conception, drafting of the manuscript, formal analysis; C.M. formal analysis, data acquisition; manuscript reviewing K.B. methodical conception, formal analysis, manuscript reviewing; K.W.N. data acquisition, manuscript reviewing; S.Kn. data acquisition, manuscript reviewing; R.B. data acquisition, manuscript reviewing; A.Q. study design, data acquisition, manuscript reviewing; D.H. study design, drafting of the manuscript, formal analysis.
Funding
Open Access funding enabled and organized by Projekt DEAL. Funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) - Project-ID 73111208 - SFB 829 (Z4 to D.H. and C.M.). SK, KB, AQ, RB received funding for this work from the European Union Fonds for Regional development (EFRE) and the German State of North Rhine Westphalia (NRW).
Data availability
Data supporting the findings of this manuscript are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sebastian Klein, Email: sebastian.klein@ukmuenster.de.
Doris Helbig, Email: doris.helbig@uk-koeln.de.
References s
- 1.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larkin J, et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med. 2019;381:1535–1546. doi: 10.1056/NEJMoa1910836. [DOI] [PubMed] [Google Scholar]
- 3.Robert C, et al. Pembrolizumab versus ipilimumab in advanced melanoma (KEYNOTE-006): post-hoc 5-year results from an open-label, multicentre, randomised, controlled, phase 3 study. Lancet Oncol. 2019;20:1239–1251. doi: 10.1016/S1470-2045(19)30388-2. [DOI] [PubMed] [Google Scholar]
- 4.Goodman AM, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol. Cancer Ther. 2017;16:2598–2608. doi: 10.1158/1535-7163.Mct-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herbst RS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol. Cancer Ther. 2015;14:847–856. doi: 10.1158/1535-7163.Mct-14-0983. [DOI] [PubMed] [Google Scholar]
- 7.Taube JM, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti–PD-1 therapy. Clin. Cancer Res. 2014;20:5064–5074. doi: 10.1158/1078-0432.Ccr-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azimi F, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J. Clin. Oncol. 2012;30:2678–2683. doi: 10.1200/jco.2011.37.8539. [DOI] [PubMed] [Google Scholar]
- 9.Clemente CG, et al. Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer. 1996;77:1303–1310. doi: 10.1002/(SICI)1097-0142(19960401)77:7<1303::AID-CNCR12>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 10.Saltz J, et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23:181–193.e187. doi: 10.1016/j.celrep.2018.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moschos SJ, et al. Neoadjuvant treatment of regional stage IIIB Melanoma with high-dose interferon Alfa-2b induces objective tumor regression in association with modulation of tumor infiltrating host cellular immune responses. J. Clin. Oncol. 2006;24:3164–3171. doi: 10.1200/jco.2005.05.2498. [DOI] [PubMed] [Google Scholar]
- 12.McInnes L, Healy J, Astels S. hdbscan: hierarchical density based clustering. J. Open Sour. Softw. 2017;2:205. doi: 10.21105/joss.00205. [DOI] [Google Scholar]
- 13.Eroglu Z, Ribas A. Combination therapy with BRAF and MEK inhibitors for melanoma: latest evidence and place in therapy. Ther. Adv. Med. Oncol. 2015;8:48–56. doi: 10.1177/1758834015616934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein S, et al. Integrative analysis of pleomorphic dermal sarcomas reveals fibroblastic differentiation and susceptibility to immunotherapy. Clin. Cancer Res. 2020;26:5638–5645. doi: 10.1158/1078-0432.CCR-20-1899. [DOI] [PubMed] [Google Scholar]
- 15.Ihle MA, et al. Comparison of high resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p.V600E and non-p.V600E BRAF mutations. BMC Cancer. 2014;14:13. doi: 10.1186/1471-2407-14-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helbig D, et al. Oncogene and therapeutic target analyses in atypical fibroxanthomas and pleomorphic dermal sarcomas. Oncotarget. 2016;7:21763–21774. doi: 10.18632/oncotarget.7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoadley KA, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173:291–304.e296. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chakravarty D, et al. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol. 2017 doi: 10.1200/po.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.Cd-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ronneberger, O., Fischer, P. & Brox, T. U-net: convolutional networks for biomedical image segmentation. 2015, pp. 234–241. 10.1007/978-3-319-24574-4_28 (2015).
- 21.Klein S, et al. Deep learning predicts HPV-association in oropharyngeal squamous cell carcinomas and identifies patients with a favorable prognosis using regular H&E stains. Clin. Cancer Res. 2020 doi: 10.1158/1078-0432.CCR-20-3596. [DOI] [PubMed] [Google Scholar]
- 22.Acs B, et al. An open source automated tumor infiltrating lymphocyte algorithm for prognosis in melanoma. Nat. Commun. 2019;10:5440. doi: 10.1038/s41467-019-13043-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheel AH, et al. Harmonized PD-L1 immunohistochemistry for pulmonary squamous-cell and adenocarcinomas. Mod. Pathol. 2016;29:1165–1172. doi: 10.1038/modpathol.2016.117. [DOI] [PubMed] [Google Scholar]
- 24.Možina M, Demšar J, Kattan M, Zupan B. Nomograms for Visualization of Naive Bayesian Classifier. In: Boulicaut JF, Esposito F, Giannotti F, Pedreschi D, editors. Knowledge Discovery in Databases: PKDD 2004. PKDD 2004. Lecture Notes in Computer Science. Berlin : Springer; 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data supporting the findings of this manuscript are available from the corresponding author upon reasonable request.