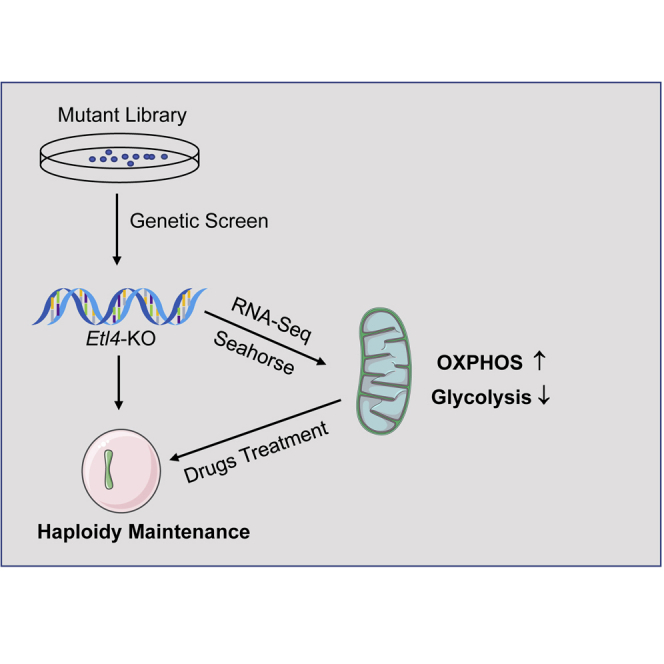
Summary
Mammalian haploid embryonic stem cells (haESCs) hold great promise for functional genetic studies and forward screening. However, all established haploid cells are prone to spontaneous diploidization during long-term culture, rendering application challenging. Here, we report a genome-wide loss-of-function screening that identified gene mutations that could significantly reduce the rate of self-diploidization in haESCs. We further demonstrated that CRISPR/Cas9-mediated Etl4 knockout (KO) stabilizes the haploid state in different haESC lines. More interestingly, Etl4 deficiency increases mitochondrial oxidative phosphorylation (OXPHOS) capacity and decreases glycolysis in haESCs. Mimicking this effect by regulating the energy metabolism with drugs decreased the rate of self-diploidization. Collectively, our study identified Etl4 as a novel haploidy-related factor linked to an energy metabolism transition occurring during self-diploidization of haESCs.
Keywords: haploid embryonic stem cells, Pluripotent stem cell, self-diploidization, genetic screen, Etl4, energy metabolism
Graphical Abstract
Highlights
-
•
A genome-wide genetic screen identifies several haploidy-related factors in haESCs
-
•
Etl4-deficiency stabilizes the haploid state in different haESC lines
-
•
Etl4-deficiency increases mitochondrial OXPHOS and decrease glycolysis in haESCs
-
•
Energy metabolism regulation with drugs decreased the rate of self-diploidization
Established haploid cells are prone to spontaneous diploidization during long-term culture. Here, Huang and Liu et al. found several gene mutations could significantly reduce the rate of self-diploidization in haESCs. Among them, Etl4-deficiency could increase the mitochondrial oxidative phosphorylation capacity and decrease glycolysis in haESCs. Moreover, haploid cells treated with energy metabolism-modifying drugs exhibit a decreased rate of self-diploidization, which suggests that the changes of energy metabolism are associated with self-diploidization.
Introduction
For most metazoans, haploidy is restricted to their gametes, which are terminally differentiated cells and cannot proliferate further. Recently, haploid embryonic stem cells (haESCs) have been generated in various organisms ranging from mouse to human (Elling et al., 2011; Leeb and Wutz, 2011; Li et al., 2012; Sagi et al., 2016; Yang et al., 2012). As they possess only one set of chromosomes, it is quite convenient to generate loss-of-function mutations in haploid cells, and this method holds great promise for forward genetic screening (Cui et al., 2020; Yilmaz et al., 2016).
In spite of their multiple scope of application, haESCs have the major disadvantage to undergo spontaneous diploidization in culture (Elling et al., 2011; Leeb and Wutz, 2011). Until now, several groups focused their research on elucidating the mechanisms of self-diploidization with the aim to prolong haploidy in long-term culture. Recent studies have demonstrated that the mitotic process in haploid cells is different from that in diploid cells, the main differences laying in prolonged G2/M phase (Li et al., 2017) and metaphase (Guo et al., 2017), mitotic slippage (He et al., 2017), or failed cytokinesis (Leng et al., 2017). It was also reported that overexpression of Dnmt3b (He et al., 2018), or deletion of P53 (Olbrich et al., 2017) or P73 (Zhang et al., 2020) in mouse haESCs could decrease self-diploidization. Another study showed that 10-Deacetyl-baccatin-III (DAB), a precursor of Taxol, selectively favors haploidy in mammalian cells (Olbrich et al., 2019). Other works led to contradictory results, as both activation (Takahashi et al., 2014) and inhibition (He et al., 2017) of CDK1, a cyclin involved in diploidy control, could reduce the rate of self-diploidization. Therefore, more efforts are needed to elucidate the mechanisms underlying self-diploidization.
Here, we used a genome-wide mutant library to screen for possible haploidy-maintaining factors. This strategy revealed that the loss of Etl4 function promotes the maintenance of haploidy in different haESC lines. Moreover, RNA-Seq data showed that Etl4-knockout (KO) haESCs exhibit a distinct metabolism state directly correlated with the reduction of self-diploidization. Our study provides a new general strategy for maintaining haploid state during cell cultures via the modulation of cell metabolism.
Results
Genetic Screening to Identify Mutations Stabilizing Haploidy in Mouse haESCs
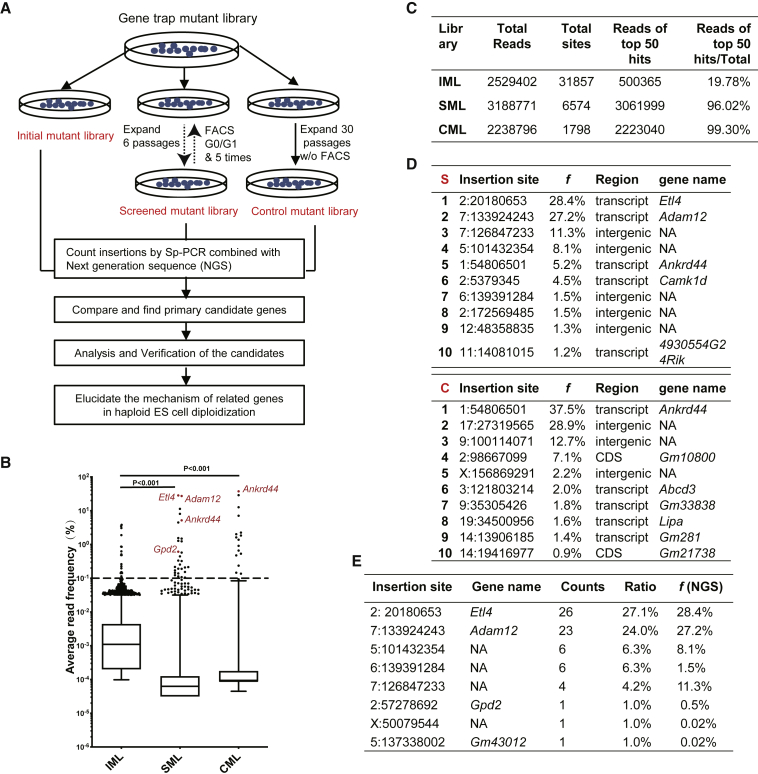
To uncover genes related to the self-diploidization of haESCs, we conducted a high-throughput screening on a previously described genome-wide haploid mutant library that contains ∼70,000 individual AGH-OG-3 (OG3) cell colonies covering 18,841 different genes mutated by the insertion of PiggyBac transposon-based gene trap vector (Liu et al., 2017). The mutant library was first replicated into three copies. One copy was used as Initial Mutant Library (IML). The second copy was used for enrichment in desired mutant haESCs via a multiple-round “cell culture-sorting” strategy. Consecutive cell cultures and five times of fluorescence-activated cell sorting (FACS) for haploid cells resulted in the collection of a Screened Mutation Library (SML). To eliminate possible artifacts due to faster proliferation of some mutants, the third copy was cultured continuously for 30 passages without FACS and served as the Control Mutant Library (CML). SML retained a high haploidy ratio, but this ratio gradually decreased in CML during long-term culture without FACS (Figure S1). Through Splinkerette (Sp)-PCR combined with next generation sequencing (NGS) of the three mutant libraries, self-diploidization–related gene candidates were identified (Figure 1A).
Figure 1.
A Genetic Screen to Identify Factors Stabilizing Haploidy in Mouse haESCs
(A) Schematic representation of the genetic screening procedure.
(B) Box plot of average read frequencies of transposon insertions in IML, SML, and CML. Dots represent the top 1% insertions. The upper vertical line represents the 75%–99% percentile group. Boxes indicate the distribution of 25%–75% of insertions. The lower vertical line represents the 1%–25% percentile group.
(C) The reads for the top 50 hits in SML and CML were increased compared with IML.
(D) Top 10 candidates obtained from the next generation sequencing (NGS) of SML and CML. S and C refer to SML and CML, respectively. Candidate genes obtained from IML, SML, and CML are listed in Table S3.
(E) Validation of the NGS results via Sp-PCR. The ratio represents the percentage of each insertion site to the total in Sp-PCR. The f (NGS) means the percentage of each insertion site to the total in NGS. The primers of Sp-PCR are listed in Table S2.
In comparison with IML, the ratios of the reads obtained for the top 50 hits in SML and CML were higher, confirming the effectivity of the screening strategy (Figures 1B and 1C). Not surprisingly, among the top 10 hits in CML (Figure 1D) were a few genes previously reported to be involved in cell proliferation, such as Ankrd44 (La Ferla et al., 2019) and Lipa (Zhao et al., 2015). Interestingly, Ankrd44 also appeared among the gene selection from SML (Figure 1D), indicating that proliferation-related genes also could be enriched with continuous haploidy sorting and cell passages.
To confirm the NGS results, 96 individual clones were randomly picked up from SML and the transposon-host junctions were amplified by Sp-PCR and submitted to Sanger sequencing. After mapping to the mouse reference genome (GRCm38.p5), 26 insertions were located in Etl4 locus and 23 in Adam12 locus, in keeping with their frequency distribution in the NGS analysis (Figure 1E). Therefore, Etl4 and Adam12 were selected for further study.
Etl4-Deficiency Reduces the Self-Diploidization Rate of Different Haploid ES Cell Lines
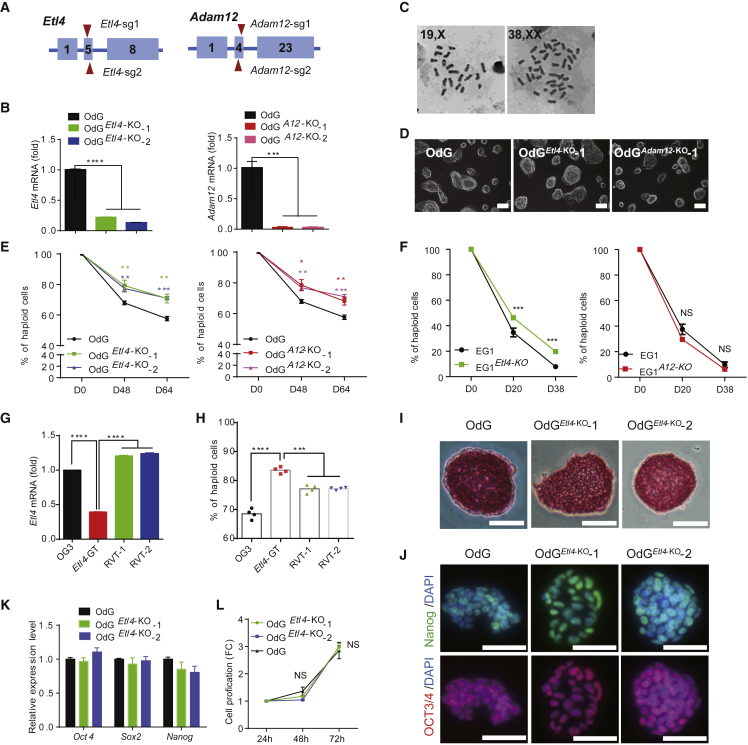
To verify the effects of these two candidates on haploidy maintenance, we used CRISPR/Cas9 technology to delete critical exons of Etl4 and Adam12 in the OdG haESC line (Figure 2A), derived from OG3 (Yang et al., 2012) cells by deletion of the EGFP cassette. Two Etl4-KO (OdGEtl4−KO-1 and OdGEtl4−KO-2) and two Adam12-KO (OdGA12−KO-1 and OdGA12−KO-2) clones were verified by genomic PCR sequencing (Figure S2A) and RT-qPCR (Figure 2B). Standard karyotyping showed that these KO cells contained 20 chromosomes (Figure 2C). Study of their morphology revealed no difference between mutant and wild type (WT) haESCs (Figures 2D and S2B).
Figure 2.
Etl4 Deficiency Facilitates the Maintenance of Mouse haESCs
(A) Schematic diagram of the strategy to knockout Etl4 and Adam12 in OdG haESCs by using CRISPR/Cas9 system. Boxes indicate the exon of the genes and the triangles represent the single guide RNA (sgRNA). The sequences of the sgRNAs are listed in Table S1.
(B) Validation of Etl4 and Adam12 expression level in mutant and WT OdG haESCs by RT-qPCR. The expression level was normalized by Gapdh (n = 3 independent experiments).
(C) Chromosome spreads of Etl4-KO OdG haESCs showing normal haploidy. The right panel depicts the self-diploidized Etl4-KO cells.
(D) Morphology of Etl4 and Adam12-deficient OdG haESCs cultured in M2iL medium. Scale bars, 100 μm.
(E and F) Flow cytometry analysis of the ratio of haploid cells in (E) Etl4-KO, Adam12-KO and (F) two corresponding WT haESC lines during cell passages.
(G) Expressional level of Etl4 in OG3, Etl4-GT, RVT-1, and RVT-2 haESCs tested by RT-qPCR (n = 3 independent experiments).
(H) Percentages of haploid cells in OG3, Etl4-GT, RVT-1, and RVT-2 haESCs after 12 days of culture (n = 3 independent experiments).
(I) Alkaline phosphatase activity in Etl4-KO and WT OdG haESCs. Scale bars, 50 μm.
(J) Immunofluorescence (IF) staining for the indicated pluripotency markers in Etl4-KO and WT OdG haESCs. Scale bars, 50 μm.
(K) Validation of indicated pluripotency-related markers' expression level in Etl4-KO and WT OdG haESCs by RT-qPCR. The expression level was normalized by Gapdh (n = 3 independent experiments).
(L) Cell proliferation rate of Etl4-KO and WT OdG haESCs (n = 3 independent experiments). Data in (B), (E), (F), (G), and (H) are shown as mean ± SEM, ∗p < 0.05 and ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. NS, not significantly different.
Then, we assessed the effects of Etl4 or Adam12 deficiencies on haploidy maintenance. Both Etl4 and Adam12-deficient OdG cell cultures had a higher proportion of haploid cells than the WT during continuous cell passages (Figure 2E). To exclude any effects of the genetic background, we tested AGH-EG-1 (EG1) cells, another haESC line that has a faster self-diploidization rate than OG3 (Yang et al., 2012). Etl4 and Adam12-deficient EG1 haESCs were also generated by CRISPR/Cas9 technology (Figure S2C). After 38 days of culture, approximately 20% of Etl4-deficient EG1 cells remained haploid, whereas the proportion of haploid cells in both WT and Adam12-deficient EG1 cells decreased to less than 10% (Figures 2F and S2D), suggesting that Adam12-deficiency stabilizes haploidy in a cell line–dependent manner. Because the Etl4-deficiency could reduce the rate of self-diploidization, we wondered if rescuing Etl4 expression would accelerate the rate of self-diploidization. We generated two revertant clones, RVT-1 and RVT-2, by reexpressing PiggyBac transposase in the Etl4 gene-trapped OG3 haESC clones (Etl4-GT). RT-qPCR analysis confirmed that the expression of Etl4 in RVT-1 and RVT-2 was restored compared with the Etl4-GT haESCs (Figure 2G). After 12 days of cultures started with 100% of haploid cells, the ratio of haploid cells was lower in revertant than in Etl4-GT haESCs (Figure 2H).
Next, we assessed the possible effect of Etl4-deficiency on basic properties of OdG haESCs. Etl4-deficient haESCs formed alkaline phosphatase (AP)-positive dome-shaped colonies similar to those of the WT cells (Figure 2I), and expressed pluripotency markers-Oct4, Sox2, and Nanog at normal levels (Figures 2J and 2K). In addition, the loss of Etl4 did not affect the proliferation rate of OdG haESCs (Figure 2L). Similarly, Etl4-deficiency had no obvious impact on colony morphology, pluripotency, and cell proliferation of EG1 haESCs (Figures S2E–S2I). Altogether, these data suggest that Etl4 is an obstacle to haploidy maintenance, but does not influence the self-renewing ability of ES cells.
Transcriptome Analysis of Etl4-Deficient haESCs
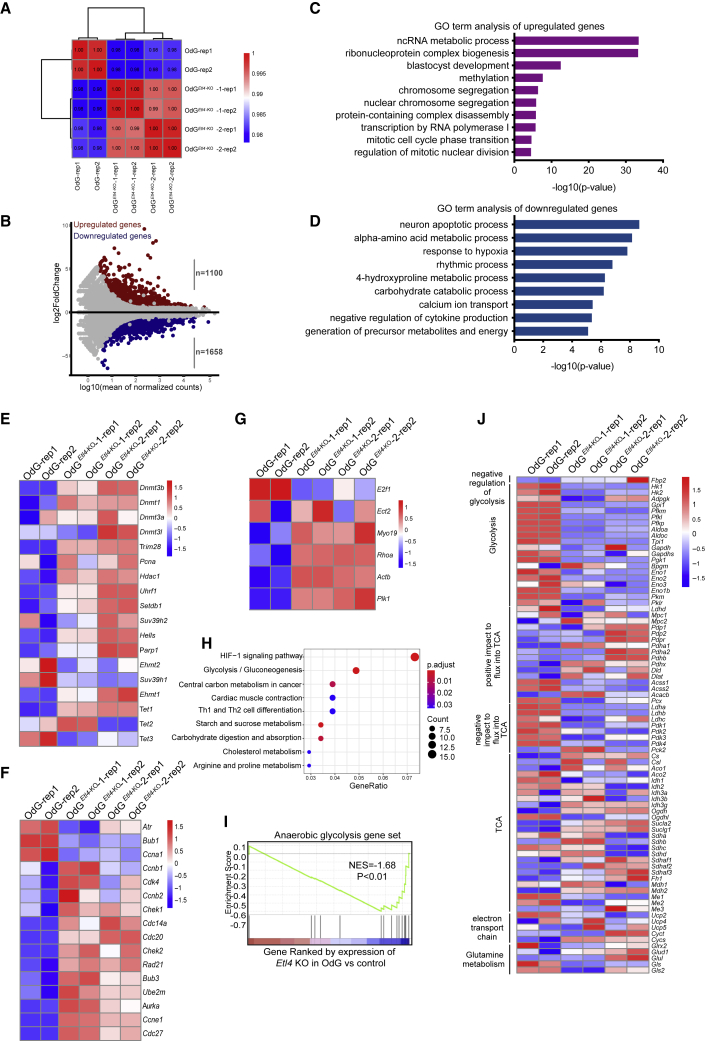
To elucidate the mechanism underlying haploid maintenance in Etl4-deficiency, we analyzed the transcriptomes of Etl4-deficient and WT haESCs. Etl4-deficient OdG cells exhibited a slightly different expression pattern compared with WT haESCs (Figure 3A). MA plots of normalized mean counts versus fold change (log2 scale) showed that 1,100 genes were upregulated and 1,658 genes were downregulated in the Etl4-deficient haESCs (Figure 3B). Gene ontology (GO) analysis indicated that genes related to non-coding RNA metabolic process, methylation, chromosome segregation, and mitotic cell cycle phase transition were upregulated (Figure 3C), whereas some metabolic processes were downregulated in Etl4-deficient haESCs (Figure 3D). Similar results were found in EG1Etl4−KO haESCs (Figures S3A and S3B). A recent study showed that overexpression of DNA methyltransferases 3b (Dnmt3b) effectively improves DNA methylation level and reduces the rate of self-diploidization in androgenetic haESCs (He et al., 2018). Consistently, our RNA-Seq data revealed that Dnmt1 and Dnmt3b were substantially upregulated in Etl4-deficient haESCs (Figures 3E and S3C). This result was confirmed by western blot analysis (Figure S3D). Moreover, the 5-methylcytosine (5-mC) level in Etl4-KO haESCs was higher than in WT haESCs (Figures S3E and S3F), which is consistent with a previous report (He et al., 2018). Several works reported that abnormal cell cycle was the major reason for self-diploidization, and that inducing G2/M phase transition or promoting metaphase/anaphase transition could partially maintain haploid state (He et al., 2017; Takahashi et al., 2014). Similarly, G2/M phase transition-related genes, such as Chek1, Chek2, and Bub3 (Figures 3F and S3G), and the core components of Rho A pathway, which is a central player in the assembly of the contractile ring during cytokinesis, were upregulated in Etl4-deficient haESCs (Figure 3G). Although these results suggested that Etl4-deficiency maintains haploid state through methyltransferases and cell cycle–related genes, to clarify whether other mechanisms operate requires further investigation.
Figure 3.
Transcriptome of Etl4-deficient OdG haESCs
(A) Hierarchical clustering of gene expression profiles from two biologically independent samples based on Pearson correlation coefficient in two Etl4-KO and WT haES clones. Colors from green to red indicate weak to strong correlations.
(B) MA plots showing gene expression changes in Etl4-KO haESCs. Red dots indicate upregulated genes and blue dots indicate downregulated genes.
(C and D) GO analysis of upregulated genes (C) and downregulated genes (D) for biological processes in Etl4-KO OdG haESCs.
(E–G) Heatmap showing the expression of methylation related genes (E), cell-cycle–related genes (F), and Rho A pathway (G) in Etl4-KO and WT OdG haESCs.
(H) Significantly enriched KEGG pathways for genes expressed differentially in Etl4-KO OdG haESCs.
(I) GSEA for anaerobic glycolysis gene set in WT OdG and Etl4-KO haESCs. For the x axis, genes were ranked based on the ratio of Etl4-KO versus WT haESCs.
(J) Heatmap showing changes in RNA expression levels for various enzymes and regulators of central carbon metabolism in Etl4-KO and WT OdG haESCs. The scale represents Z score.
Furthermore, both GO (Figures 3D and S3B) and KEGG enrichment analysis (Figures 3H and S3H) showed a significant enrichment in terms related to amino acid and carbohydrate metabolic processes in Etl4-deficient haESCs. Gene set enrichment analysis (GSEA) also indicated that anaerobic glycolysis gene set was predominantly reduced in Etl4-deficient haESCs (Figures 3I and S3I). Moreover, the expression of various enzymes and regulators participating in central carbon metabolism changed markedly. Generally, glycolytic enzymes and negative regulators of the Tricarboxylic Acid (TCA) cycle tended to be downregulated in Etl4-deficient haESCs, whereas positive regulators of the TCA cycle and TCA cycle itself were upregulated (Figures 3J and S3J). Taken together, these results suggest changes in the metabolic activity of Etl4-deficient haESCs compared with WT haploid cells.
Enhancement of OXPHOS or Suppression of Glycolysis Diminishes the Rate of Self-Diploidization
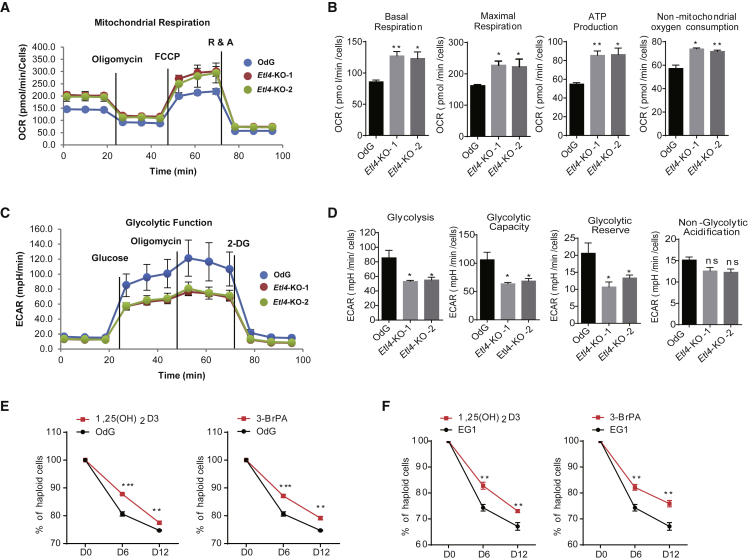
To further explore the metabolic changes in Etl4-deficient haESCs, we measured the respiratory capacity using a Seahorse Extracellular Flux XF24 Analyzer. As indicated by the oxygen consumption rate curves, OdGEtl4−KO-1 and OdGEtl4−KO-2 haESCs showed a significant increment in the basal and maximal mitochondrial respiratory capacities, and in ATP production (Figures 4A and 4B). These results were consistent with the RNA-Seq data indicating a higher expression level of oxidative phosphorylation (OXPHOS)-related genes in Etl4-deficient haESCs (Figure 3J). In addition, we monitored the extracellular acidification rate (ECAR) in Etl4-KO and WT OdG haESCs (Figure 4C). The rate of glycolysis, glycolytic capacity, and glycolytic reserve were decreased in Etl4-deficient haESCs, whereas no differences were observed for nonglycolytic acidification between Etl4-KO and WT cells (Figure 4D). Next, we tested the changes of metabolism in Etl4-GT and RVT haESCs. The restoration of ETL4 level decreased OHPXOS capacity and increased the glycolysis in haESCs (Figures S4A–S4D). Collectively, our results demonstrate that there is a shift from glycolysis to OXPHOS when Etl4 is deficient, with no obvious changes in mitochondria morphology (Figure S4E).
Figure 4.
Reduced Self-Diploidization by Promotion of OXPHOS or Inhibition of Glycolysis
(A) Mitochondrial stress test to detect mitochondrial energy metabolism and respiratory functions in Etl4-KO and WT OdG haESCs.
(B) Quantification of the mitochondrial stress test presented in (A) (n = 3 independent experiments).
(C) The glycolytic stress test to measure glycolytic activities in Etl4-KO and WT OdG haESCs.
(D) Quantification of glycolytic activities in (C) (n = 3 independent experiments).
(E and F) DNA content analysis of haploid cells in OdG (E) and EG1 (F) haESCs treated with 3-BrPA (12.5 μg/mL) and 1,25-(OH)2D3 (5 μM) for 12 days.
Data in (B), (D), (E), and (F) are presented as means ± SEM, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
The energy metabolism is closely linked to many important biological processes, including stem cell self-renewal (Ito and Ito, 2016), cell reprogramming (Cliff and Dalton, 2017), and cell cycle (Icard et al., 2019). Because Etl4-deficient haESCs provoked an increase in OXPHOS level and decrease of glycolysis, we hypothesized that promoting OXPHOS or inhibiting the glycolysis in haESCs would reduce the rate of self-diploidization. To test our conjecture, haploid cells collected by FACS were treated with two classes of energy metabolism–modifying drugs during long-term cultures. Both 1,25-(OH)2D3, an OXPHOS agonist (Ferreira et al., 2015), and 3-Bromopyruvic acid (3-BrPA), a glycolysis inhibitor (Fang et al., 2019), could maintain haploidy to a certain extent in both OdG and EG1 haESCs (Figures 4E and 4F). Furthermore, neither of the two drugs affected the proliferation (Figure S4F) or pluripotency of the haESCs (Figures S4G and S4H). Collectively, here we provide a tractable solution to maintain the haploid state of haESCs through enhancement of OXPHOS or suppression of glycolysis.
Discussion
Here, we report a genome-wide genetic screening and the identification of Etl4-deficiency as a significant reducer of self-diploidization. Etl4 (enhancer trap locus 4, also named Skt) is required for normal development of the intervertebral disc (Semba et al., 2006) and anorectum (Suda et al., 2011). ETL4 protein is cytoplasmic and contains a proline-rich region in its the N terminus and a coiled-coil domain in its middle (Semba et al., 2006). The exact function of ETL4 protein is not clear, but the proline-rich region has been shown to be important for proper folding of cytochrome P450s (Kusano et al., 2001), which catalyzes many metabolic reactions. A previous study also showed that the proline-rich region may play a role in the subcellular localization and enzymatic function (Arreaza and Deutsch, 1999). Thus, here we defined a novel function carried out by Etl4, consisting of the regulation of self-diploidization in haESCs through a metabolic control.
Glycolysis and OXPHOS are two key cellular pathways for energy production. Most cells can switch between these two pathways to adapt to changes in the environment. OXPHOS, taking place in mitochondria, and the different subunits of the OXPHOS complexes are encoded by both mitochondrial (mtDNA) and nuclear DNA (nucDNA) (Quiros et al., 2016). Because haESCs have one-half of the nucDNA, they have a higher ratio of mtDNA to nucDNA compared with diploid cells. In response to the difference in mtDNA/nucDNA ratio, the expression of some OXPHOS-related genes encoded by the nucleus is upregulated in haploid cells (Sagi and Benvenisty, 2017). The relatively higher level of OXPHOS in haESCs may serve the increase in energy demands and be beneficial to haploidy maintenance. Our results clearly show that Etl4-deficiency increases the mitochondrial OXPHOS capacity and decreases glycolysis in haESCs, although the detailed mechanism linking the two phenomena needs to be further investigated.
Previous studies showed that deficient phosphorylation of mitochondrial respiration chain complex I (CI) subunits by Cdk1 results in a loss of G2/M-associated enhancement of mitochondrial activity, and delays G2/M progression (Wang et al., 2014). Loss of mitochondrial function during prolonged mitotic arrest results in the upregulation of glycolysis (Salazar-Roa and Malumbres, 2017). Progression through the cell cycle, including phase transition and structural rearrangements for cell division are energy-demanding (Pederson, 2003). Combined with our finding that both 1,25-(OH)2D3 and 3-BrPA help maintain haploidy in haESCs, it can be proposed that Etl4 deficiency causes cell cycle changes and maintains the haploidy state by increasing mitochondrial metabolism.
Overall, our work identified Etl4 as a novel haploidy-related factor, whose deficiency significantly reduces the rate of self-diploidization in haESCs through increasing OXPHOS or inhibiting glycolysis. Moreover, our study suggests that elucidating the interplay between metabolic changes and haploid state stabilization would provide interesting insights for future hsESCs applications.
Experimental Procedures
Cell Cultures and Drug Treatment
Mouse haESC lines AGH-OG-3 and AGH-EG-1 were kindly provided by Prof. Jinsong Li (Institute of Biochemistry and Cell Biology, Shanghai). Cells were cultured in M2iL mediums, which containing knockout DMEM (10,829-018, Gibco), 15% fetal bovine serum (SH30396.03, HyClone), 1% GlutaMAX (35,050-061, Gibco), 1% nonessential amino acids (11140050, Gibco), 100 μM β-mercaptoethanol (21985-023, Gibco), 100 U Leukemia inhibitory factor (ESG1107, Merck Millipore), 3μM CHIR99021 (S2924, Selleck), and 1μM PD0325901 (S1036, Selleck). The cells were cultured at 37°C with 5% CO2 in a humidified environment. 1,25-(OH)2D3 (Aladdin, C120126) and 3-BrPA (Selleck, S5426) were used at a final concentration of 12.5 μg/mL and 5μM, respectively.
FACS and DNA Content Analysis
For FACS, haESCs were trypsinized and stained with Hoechst33342 (5 μg/mL) (B2261, Sigma) for 30 min at 37°C. After filtering with a 40-μm cell strainer, the haploids in 1n peak were purified with a Beckman Moflo-XDP cell sorter. Diploid (2n) ES cells were used as a control.
For DNA content analysis, haESCs were treated with 0.2 μg/mL Colchicine (D1925, Sigma) for 6 h and then trypsinized into single cells followed by fixation with 75% ethanol at 4°C for more than 2 h. Fixed cells were treated with 25 μg/mL RNase at 37°C for 30 min and then stained with 5 μg/mL propidium iodide (P4170, Sigma) at 4°C for 30 min. Flow cytometry data were recorded by using BD Accuri C6. The percentage of the haploid cells was calculated as hap/(hap + dip), where hap indicates haploid cells, and dip indicates diploid cells.
Data and Code Availability
The accession number for the RNA-Seq data in this paper is GEO: GSE150215.
Author Contributions
Y.H. and G.L. conceived and designed the study; G.Z., Y.S., G.L., and X.W. performed the experiments; X.L. performed bioinformatic analyses; Y.H., G.Z., X.L., Y.S., and G.L. wrote the manuscript.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2016YFA0100103 to Y.H.), CAMS Innovation Fund for Medical Sciences (2016-I2M-3-002 to Y.H. and G.L.), and National Natural Science Foundation of China (31671410, 31970813 to Y.H. and 31501048 to G.L.).
Published: January 12, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.11.016.
Contributor Information
Guang Liu, Email: liuguang@ibms.pumc.edu.cn.
Yue Huang, Email: huangyue@pumc.edu.cn.
Supplemental Information
References
- Arreaza G., Deutsch D.G. Deletion of a proline-rich region and a transmembrane domain in fatty acid amide hydrolase. FEBS Lett. 1999;454:57–60. doi: 10.1016/s0014-5793(99)00774-7. [DOI] [PubMed] [Google Scholar]
- Cliff T.S., Dalton S. Metabolic switching and cell fate decisions: implications for pluripotency, reprogramming and development. Curr. Opin. Genet. Dev. 2017;46:44–49. doi: 10.1016/j.gde.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui T., Li Z., Zhou Q., Li W. Current advances in haploid stem cells. Protein Cell. 2020;11:23–33. doi: 10.1007/s13238-019-0625-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elling U., Taubenschmid J., Wirnsberger G., O'Malley R., Demers S.P., Vanhaelen Q., Shukalyuk A.I., Schmauss G., Schramek D., Schnuetgen F. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell. 2011;9:563–574. doi: 10.1016/j.stem.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Shen Z.Y., Zhan Y.Z., Feng X.C., Chen K.L., Li Y.S., Deng H.J., Pan S.M., Wu D.H., Ding Y. CD36 inhibits beta-catenin/c-myc-mediated glycolysis through ubiquitination of GPC4 to repress colorectal tumorigenesis. Nat. Commun. 2019;10:3981. doi: 10.1038/s41467-019-11662-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira G.B., Vanherwegen A.S., Eelen G., Gutierrez A.C.F., Van Lommel L., Marchal K., Verlinden L., Verstuyf A., Nogueira T., Georgiadou M. Vitamin D3 induces tolerance in human dendritic cells by activation of intracellular metabolic pathways. Cell Rep. 2015;10:711–725. doi: 10.1016/j.celrep.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Guo A., Huang S., Yu J., Wang H., Li H., Pei G., Shen L. Single-cell dynamic analysis of mitosis in haploid embryonic stem cells shows the prolonged metaphase and its association with self-diploidization. Stem Cell Rep. 2017;8:1124–1134. doi: 10.1016/j.stemcr.2017.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Zhang X., Zhang Y., Zheng W., Xiong Z., Hu X., Wang M., Zhang L., Zhao K., Qiao Z. Reduced self-diploidization and improved survival of semi-cloned mice produced from androgenetic haploid embryonic stem cells through overexpression of Dnmt3b. Stem Cell Rep. 2018;10:477–493. doi: 10.1016/j.stemcr.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z.Q., Xia B.L., Wang Y.K., Li J., Feng G.H., Zhang L.L., Li Y.H., Wan H.F., Li T.D., Xu K. Generation of mouse haploid somatic cells by small molecules for genome-wide genetic screening. Cell Rep. 2017;20:2227–2237. doi: 10.1016/j.celrep.2017.07.081. [DOI] [PubMed] [Google Scholar]
- Icard P., Fournel L., Wu Z., Alifano M., Lincet H. Interconnection between metabolism and cell cycle in cancer. Trends Biochem. Sci. 2019;44:490–501. doi: 10.1016/j.tibs.2018.12.007. [DOI] [PubMed] [Google Scholar]
- Ito K., Ito K. Metabolism and the control of cell fate decisions and stem cell renewal. Annu. Rev. Cell Dev. Biol. 2016;32:399–409. doi: 10.1146/annurev-cellbio-111315-125134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano K., Kagawa N., Sakaguchi M., Omura T., Waterman M.R. Importance of a proline-rich sequence in the amino-terminal region for correct folding of mitochondrial and soluble microbial p450s. J. Biochem. 2001;129:271–277. doi: 10.1093/oxfordjournals.jbchem.a002854. [DOI] [PubMed] [Google Scholar]
- La Ferla M., Lessi F., Aretini P., Pellegrini D., Franceschi S., Tantillo E., Menicagli M., Marchetti I., Scopelliti C., Civita P. ANKRD44 gene silencing: a putative role in trastuzumab resistance in her2-like breast cancer. Front. Oncol. 2019;9:547. doi: 10.3389/fonc.2019.00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M., Wutz A. Derivation of haploid embryonic stem cells from mouse embryos. Nature. 2011;479:131–134. doi: 10.1038/nature10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng L., Ouyang Q., Kong X., Gong F., Lu C., Zhao L., Shi Y., Cheng D., Hu L., Lu G., Lin G. Self-diploidization of human haploid parthenogenetic embryos through the Rho pathway regulates endomitosis and failed cytokinesis. Sci. Rep. 2017;7:4242. doi: 10.1038/s41598-017-04602-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Guo A., Xie Z., Tu W., Yu J., Wang H., Zhao J., Zhong C., Kang J., Li J. Stabilization of mouse haploid embryonic stem cells with combined kinase and signal modulation. Sci. Rep. 2017;7:13222. doi: 10.1038/s41598-017-13471-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shuai L., Wan H., Dong M., Wang M., Sang L., Feng C., Luo G.Z., Li T., Li X. Androgenetic haploid embryonic stem cells produce live transgenic mice. Nature. 2012;490:407–411. doi: 10.1038/nature11435. [DOI] [PubMed] [Google Scholar]
- Liu G., Wang X., Liu Y., Zhang M., Cai T., Shen Z., Jia Y., Huang Y. Arrayed mutant haploid embryonic stem cell libraries facilitate phenotype-driven genetic screens. Nucleic Acids Res. 2017;45:e180. doi: 10.1093/nar/gkx857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olbrich T., Mayor-Ruiz C., Vega-Sendino M., Gomez C., Ortega S., Ruiz S., Fernandez-Capetillo O. A p53-dependent response limits the viability of mammalian haploid cells. Proc. Natl. Acad. Sci. U S A. 2017;114:9367–9372. doi: 10.1073/pnas.1705133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olbrich T., Vega-Sendino M., Murga M., de Carcer G., Malumbres M., Ortega S., Ruiz S., Fernandez-Capetillo O. A chemical screen identifies compounds capable of selecting for haploidy in mammalian cells. Cell Rep. 2019;28:597–604 e594. doi: 10.1016/j.celrep.2019.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. Historical review: an energy reservoir for mitosis, and its productive wake. Trends Biochem. Sci. 2003;28:125–129. doi: 10.1016/S0968-0004(03)00030-6. [DOI] [PubMed] [Google Scholar]
- Quiros P.M., Mottis A., Auwerx J. Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 2016;17:213–226. doi: 10.1038/nrm.2016.23. [DOI] [PubMed] [Google Scholar]
- Sagi I., Benvenisty N. Haploidy in humans: an evolutionary and developmental perspective. Dev. Cell. 2017;41:581–589. doi: 10.1016/j.devcel.2017.04.019. [DOI] [PubMed] [Google Scholar]
- Sagi I., Chia G., Golan-Lev T., Peretz M., Weissbein U., Sui L., Sauer M.V., Yanuka O., Egli D., Benvenisty N. Derivation and differentiation of haploid human embryonic stem cells. Nature. 2016;532:107–111. doi: 10.1038/nature17408. [DOI] [PubMed] [Google Scholar]
- Salazar-Roa M., Malumbres M. Fueling the cell division cycle. Trends Cell Biol. 2017;27:69–81. doi: 10.1016/j.tcb.2016.08.009. [DOI] [PubMed] [Google Scholar]
- Semba K., Araki K., Li Z., Matsumoto K., Suzuki M., Nakagata N., Takagi K., Takeya M., Yoshinobu K., Araki M. A novel murine gene, Sickle tail, linked to the Danforth's short tail locus, is required for normal development of the intervertebral disc. Genetics. 2006;172:445–456. doi: 10.1534/genetics.105.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda H., Lee K.J., Semba K., Kyushima F., Ando T., Araki M., Araki K., Inomata Y., Yamamura K. The Skt gene, required for anorectal development, is a candidate for a molecular marker of the cloacal plate. Pediatr. Surg. Int. 2011;27:269–273. doi: 10.1007/s00383-010-2785-0. [DOI] [PubMed] [Google Scholar]
- Takahashi S., Lee J., Kohda T., Matsuzawa A., Kawasumi M., Kanai-Azuma M., Kaneko-Ishino T., Ishino F. Induction of the G2/M transition stabilizes haploid embryonic stem cells. Development. 2014;141:3842–3847. doi: 10.1242/dev.110726. [DOI] [PubMed] [Google Scholar]
- Wang Z., Fan M., Candas D., Zhang T.-Q., Qin L., Eldridge A., Wachsmann-Hogiu S., Ahmed Kazi M., Chromy Brett A., Nantajit D. Cyclin B1/cdk1 coordinates mitochondrial respiration for cell-cycle G2/M progression. Dev. Cell. 2014;29:217–232. doi: 10.1016/j.devcel.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Shi L., Wang B.A., Liang D., Zhong C., Liu W., Nie Y., Liu J., Zhao J., Gao X. Generation of genetically modified mice by oocyte injection of androgenetic haploid embryonic stem cells. Cell. 2012;149:605–617. doi: 10.1016/j.cell.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Yilmaz A., Peretz M., Sagi I., Benvenisty N. Haploid human embryonic stem cells: half the genome, double the value. Cell Stem Cell. 2016;19:569–572. doi: 10.1016/j.stem.2016.10.009. [DOI] [PubMed] [Google Scholar]
- Zhang W., Tian Y., Gao Q., Li X., Li Y., Zhang J., Yao C., Wang Y., Wang H., Zhao Y. Inhibition of apoptosis reduces diploidization of haploid mouse embryonic stem cells during differentiation. Stem Cell Rep. 2020;15:185–197. doi: 10.1016/j.stemcr.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Du H., Ding X., Walls K., Yan C. Activation of mTOR pathway in myeloid-derived suppressor cells stimulates cancer cell proliferation and metastasis in lal(-/-) mice. Oncogene. 2015;34:1938–1948. doi: 10.1038/onc.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-Seq data in this paper is GEO: GSE150215.