Abstract
Stress has been shown to disturb the balance of human intestinal microbiota and subsequently causes mental health problems like anxiety and depression. Our previous study showed that ingesting the probiotic strain, Lactobacillus (L.) plantarum P-8, for 12 weeks could alleviate stress and anxiety of stressed adults. The current study was a follow-up work aiming to investigate the functional role of the gut metagenomes in the observed beneficial effects. The fecal metagenomes of the probiotic (n = 43) and placebo (n = 36) receivers were analyzed in depth. The gut microbiomes of the placebo group at weeks 0 and 12 showed a significantly greater Aitchison distance (P < 0.001) compared with the probiotic group. Meanwhile, the Shannon diversity index of the placebo group (P < 0.05) but not the probiotic group decreased significantly at week 12. Additionally, significantly more species-level genome bins (SGBs) of Bifidobacterium adolescentis, Bifidobacterium longum, and Fecalibacterium prausnitzii (P < 0.01) were identified in the fecal metagenomes of the probiotic group, while the abundances of SGBs representing the species Roseburia faecis and Fusicatenibacter saccharivorans decreased significantly (P < 0.05). Furthermore, the 12-week probiotic supplementation enhanced the diversity of neurotransmitter-synthesizing/consuming SGBs and the levels of some predicted microbial neuroactive metabolites (e.g., short-chain fatty acids, gamma-aminobutyric acid, arachidonic acid, and sphingomyelin). Our results showed a potential link between probiotic-induced gut microbiota modulation and stress/anxiety alleviation in stressed adults, supporting that the gut-brain axis was involved in relieving stress-related symptoms. The beneficial effect relied not only on microbial diversity changes but more importantly gut metagenome modulations at the SGB and functional gene levels.
Keywords: Lactobacillus plantarum, Species-Level genome bins, Stress, Neuroactive compounds, Gut-brain axis
Highlights
-
•
Stress and anxiety are related to gut dysbiosis.
-
•
Ingesting Lactobacillus plantarum P-8 (P-8) improved stress/anxiety symptoms.
-
•
Symptoms were relieved by modulating gut microbiota and neuroactive potential.
1. Introduction
Stress is a ubiquitous part of human daily life, it can cause anxiety and even depression in serious cases (Pittenger and Duman, 2008). Depression affects over 300 million people worldwide, accounting for almost 800,000 suicidal deaths per year (Eker, 2018). Recent studies have associated risk of stress and depression with the gut microbiota (Richards et al., 2018; Mohajeri et al., 2018). The gut microbiota can be viewed as a reservoir of collective genomes that harbors at least 100 times as many genes as the human genome (Qin et al., 2010). The co-evolution of the human genome and the gut microbiome resulted in complex bidirectional interactions between the gut, enteric nervous system, and central nervous system (Karl et al., 2018; Cryan and Dinan, 2012). The intestinal microbiota widely participates in the synthesis/release of various hormones and gut-brain axis-related neurotransmitters, which then modulate the brain function and host behavior (Strandwitz, 2018; Mohajeri et al., 2018). Gastrointestinal disturbances are known to influence mental conditions, including stress, anxiety, depression, Parkinson's and Alzheimer’ diseases (Pellegrini et al., 2018).
Psychobiotics are defined as living microorganisms (probiotics) that confer mental health benefits to the host through interactions with commensal gut bacteria when administered in adequate amounts (Dinan et al., 2013). This concept has been further expanded to encompass prebiotics that stimulate the growth of beneficial gut bacteria (Sarkar et al., 2016). Since probiotics modulate the host gut microbial communities and their synthesis/release of certain neuroactive compounds, probiotic-based therapy has thus been proposed as an alternative treatment for some neurological and neurodegenerative diseases (Puebla-Barragan and Reid, 2019; Westfall et al., 2017). Recent clinical studies have shown that some probiotic strains could help relieve stress, anxiety, and depression symptoms (Messaoudi et al., 2011; Takada et al., 2016), particularly, the Bifidobacterium (B.) and Lactobacillus (L.) genera (Zhang et al., 2018). However, most studies have focused on monitoring the clinical effects by administering behavior tests, cognitive assessments and questionnaires to assess relief of symptoms (Allen et al., 2016; Takada et al., 2016). Some studies have attempted to characterize probiotic-induced changes in the taxonomic (16S rRNA-based) profiles of intestinal microbiota (Li et al., 2018; Davis et al., 2016), but failed to elucidate the function of the gut microbiota and its neuroactive potential due to the lack of corresponding reference genomes (Valles-Colomer et al., 2019). Microbial reference genomes are indispensable tools for microbial profiling, annotating metagenomes and deciphering their physiological function (Kyrpides et al., 2014). Recent studies reconstructed metagenome-assembled genomes (MAGs) from metagenomes by binning, which has massively expanded the human gut reference genomes to unveil the role of gut microbiome in human health (Almeida et al., 2019; Nayfach et al., 2019).
High-quality human and animal correlative studies have undoubtedly shown that the gut microbiota plays an influential role in many stress-related conditions (Foster et al., 2017; Rea et al., 2020); however, the causal relationship and extent of contribution of the gut microbiome in stress-related conditions are unclear. Previously, our team performed a 12-week randomized, double-blind, and placebo-controlled human trial, demonstrating the clinical efficacy of the probiotics, L. plantarum P-8, in ameliorating stress-/anxiety-related symptoms in stressed adults (Lew et al., 2019), accompanied by decreased plasma levels of interferon (IFN)-γ, tumor necrosis factor (TNF)-α, and cortisol, as well as enhanced memory and cognitive traits. However, the mechanism behind these effects remained unknown. The current study was a follow-up work aiming to elucidate such mechanism from the perspective of the intestinal microbiome.
2. Materials and methods
2.1. Experimental design
This study was extended from our previous human trial. Each subject received 12-week treatment of daily oral L. plantarum P-8 (2 g; 2x1010 CFU/sachet/day; maltodextrin as excipient) or placebo (only maltodextrin; light yellow powder with identical taste and appearance as the probiotic material; manufactured by Jinhua Yinhe Biological Technology Co. Ltd., China under ISO9001 and HALAL standards). Although 52 probiotic-receivers and 51 placebo-receivers finished the trial, only 43 probiotic-receivers and 36 placebo-receivers donated fecal samples at both weeks 0 and 12 for the current analysis (Fig. S1a; Table S1).
2.2. DNA extraction, metagenomic sequencing and quality control
Metagenomic DNA was extracted from 100 mg stool using the QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany). The DNA quality was examined by agarose gel electrophoresis and a Nanodrop spectrophotometer (260 nm/280 nm ratio). Shotgun metagenomic sequencing was performed using an Illumina HiSeq 2500 instrument. Libraries were constructed by DNA fragments of ~300 bp length; paired-end reads were generated using 150 bp in both forward and reverse directions.
A total of 158 stool samples were shotgun sequenced (n = 43 and 36 for P-8 and placebo groups, sampling at weeks 0 and 12, respectively), generating 1.52 Tbp of high-quality paired-end reads (8.34 ± 1.41 Gbp/sample; range = 4.72–11.96 Gbp) for downstream analysis. The generated metagenomic reads were processed with the KneadData quality control pipeline (http://huttenhower.sph.harvard.edu/kneaddata; v0.7.5), which used Trimmomatic (a flexible trimmer for Illumina sequence data) (Dinan et al., 2013) to filter out low-quality reads of less than 60 nt in length. Human contaminating reads were removed by Bowtie2 (v2.3.5.1) (Langmead and Salzberg, 2012).
2.3. Metagenomic assembly, contig binning, genome dereplication
All samples were singly assembled into scaffolds using MetaSPAdes (Nurk et al., 2017), with standard quality control setting and the options, –k/33,55,77,99,111/–meta, which the average N50 length was 17.19 Kbp (Table S2). The results of metagenomic assembly were evaluated by QUAST (Gurevich et al., 2013), with the parameters, “--min-contig 2000”. Assembled scaffolds >2,000 bp were selected for binning. Afterwards, MAGs were generated for each sample using MaxBin2 (Wu et al., 2016) and MetaBAT2 (Kang et al., 2019), with default options, which yielded 10,545 and 14,534 raw bins from the initial scaffolds, respectively. Then, the bin refinement module of MetaWRAP (Uritskiy et al., 2018) was used to combine the results of the two binners to obtain 6566 bins. Raw reads were mapped back to the corresponding assemblies using BWA-MEM (Liu et al., 2019), and the read depths were calculated using Samtools (Li et al., 2009) and the jgi_summarize_bam_contig_depths function in MetaBAT2.
The completeness and contamination of MAGs were evaluated with CheckM (Parks et al., 2015) using the lineage_wf workflow. The quality of MAGs was classified as high (completeness≥80%, contamination≤5%), medium (completenes≥70%, contamination≤10%), and partial (completeness≥50%, contamination≤5%) (Parks et al., 2017). Genomic comparisons were performed by the dRep tool (Olm et al., 2017) with options -pa 0.95 (primary cluster at 90%) -sa 0.95 (secondary cluster at 95%) to identify groups of essentially identical genomes and select the best genome from each replicate set. Results generated by dRep were used to extract species-level genome bins (SGBs) from the high-quality genomes based on single representative genomes.
2.4. Taxonomic annotation, phylogenetic analysis, and genome comparison
The MAG scaffolds were annotated using the Kraken2 tool (Wood et al., 2019) and NCBI nonredundant Nucleotide Sequence Database (retrieved on 2019.04.21) with default parameters. Putative genes in the scaffolds were predicted using Prodigal (Hyatt et al., 2010). The predicted genes were searched against the UniProt Knowledgebase (UniProtKB, release 2019.04) using the blastp function of DIAMON (Buchfink et al., 2015). Phylogeny analysis was performed using 400 universal PhyloPhlAn markers and visualized using iTOL (Letunic and Bork, 2019).
To evaluate the novelty of SGBs in the current dataset, 589 SGBs were cross-compared with the IGG dataset (https://github.com/snayfach/IGGdb; comprised 16,136 non-redundant representative human gut genomes) and the MAGs dataset (comprised 154,723 non-redundant genomes from gut and other body parts of the global population) (Pasolli et al., 2019).
To identify P-8 in the samples, all metagenomic DNA samples were mapped to the P-8 reference genome using a genome coverage breadth of 40% (Vandeputte et al., 2017). To avoid ambiguity alignment, reads were counted only when the alignment similarity was over 97%.
2.5. Abundance of SGBs
The BBMap tool (https://github.com/BioInfoTools/BBMap) was used to map raw reads to the scaffolds with the parameters “minid = 0.95 idfilter = 0.95”. The coverage of scaffolds was calculated by pileup.sh using sam file as input. The unit, reads per kilobase per million (RPKM), calculated by an in-house script was used to indicate the average content of SGB in each scaffold.
2.6. Identification of neuroactive compounds
A metabolic reconstruction was performed by searching across literature (gut-brain modules, GBMs) (Valles-Colomer et al., 2019) and the metabolite database, MetaCyc (Caspi et al., 2020). For each SGB, open reading frames (ORFs) were predicted using Prodigal. Annotation of metabolic potential and pathways was performed by using the key reactions in the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthologies (KOs) database (as of 2018). Each GBM was assigned to synthesis/release of a neuroactive compound by specific microbes. All pathways were detected by Omixer-RPM (Darzi et al., 2016) and defined in SGBs (parameter: c 0.66).
2.7. Prediction of microbial metabolites
Microbial sequences were used to predict the gut metabolomes. Firstly, one million reads per sample were subsampled using seqtk (https://github.com/lh3/seqtk). The subsampled reads were compared using the blastx function of DIAMON with options --query-cover 90 --id 50. Then, the best hit of each gene was selected for calculation of gene abundances in each sample. Finally, the MelonnPan-predict workflow was employed to convert gene abundances into a predicted metabolomic table (Mallick et al., 2019).
2.8. Data and code availability
Sequencing data and analysis codes are available in NCBI-SRA (BioProject: PRJNA634666) and under https://github.com/TengMa-Cleap/Probiotics-relieve-human-stress-and-anxiety-project.
2.9. Statistical analyses
All statistical analyses were performed using the R software (v.4.0.2). Unless otherwise stated, data were expressed as mean ± SD. To evaluate the structural difference between the microbiota of different groups of samples, non-metric multi-dimensional scaling (NMDS; Bray-Curtis distance) was performed and visualized using the R package vegan (v.2.5-1) and ggpubr, respectively. The stress value reflected the data representation in the NMDS analysis; stress values < 0.05, <0.1, <0.2, and >0.2 were considered excellent, great, good/acceptable, and poor representation after dimensionality reduction, respectively. Wilcoxon test, t-test, and Kruskal-Wallis test were used to evaluate differences in the microbiome and neurometabolome between two or multiple groups. The false discovery rate (FDR) was corrected by using the Benjamini and Hochberg (BH) method with the p.adjust function of the R software with a cut-off confidence level of 0.05. Similarity percentage (SIMPER) analysis was used to determine the SGBs that contributed most to the dissimilarity between sample groups. It was performed by the R software Vegan package. The corr.test function in the ‘psych’ package was used to perform Spearman correlation analysis.
In addition, the clr abundance conversion of the samples were performed using the R package microbiome, and the Aitchison distance was calculated based on the vegan package. To evaluate similarities among groups, analysis of similarities (ANOSIM, permutations = 999) was performed. Procrustes analysis based on the vegan package was used to determine similarity between two multivariate axes, and p-value was generated based on 999 permutations. All graphical presentations were generated under the R and Adobe Illustrator (AI) environment.
3. Results
3.1. Genomic characteristics of gut microbiome in stressed adults
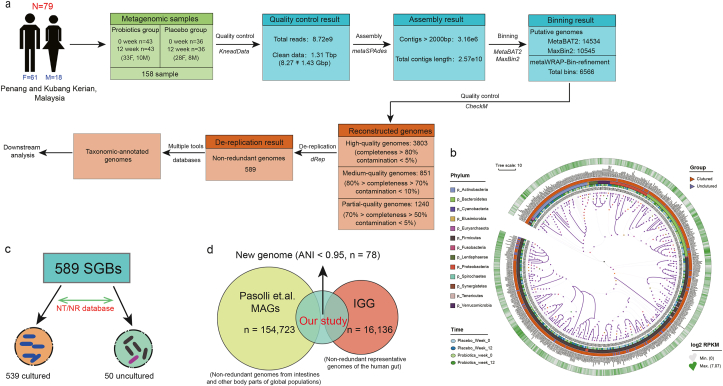
After bin refinement, 3803, 851, and 1240 MAGs were assigned to high-, medium-, and partial-quality MAGs (Table S3 for information of the 3803 high-quality MAGs). The average mappability of 589 SGBs obtained was 78.56 ± 6.46% of metagenomic reads/sample (Fig. 1a), and the high mappability suggested most genomic contents were already known microbial community. The mapped SGBs were distributed across 13 phyla, 23 classes, 30 orders, 49 families, 91 genera, and 341 species. Most SGBs were assigned to the phylum Firmicutes (57.72%), followed by Bacteroidetes (14.60%), Actinobacteria (12.56%), and Proteobacteria (8.15%). Fifty SGBs remained unmapped and uncharacterized at the species level, representing the portion of uncultured species. These uncultured species belonged mainly to three phyla, i.e., Firmicutes (72%), Bacteroidetes (11.67%), and Proteobacteria (4%) (Fig. 1b and c). To estimate the size of the unexplored SGBs, the current dataset was cross-compared with the IGG and MAGs datasets (Pasolli et al., 2019). Seventy-eight SGBs did not match (<95% average nucleotide similarity) any gut microbial genome in the two datasets, suggesting their novelty (Fig. 1d; Table S4).
Fig. 1.
Metagenomic reconstruction pipeline and genomic characteristics. (a) The stepwise pipeline of metagenomic assembly, binning, reconstruction, and de-replication resulted in a total of 589 high-quality species-level genome bins (SGBs). (b) Phylogenetic placement of the 589 high-quality SGBs. The heatmap of the tree shows the log2-fold change of the SGBs in reads per kilobase per million (RPKM) during the trial. (c) Among the 589 SGBs, 539 and 50 were previously cultured and uncultured, respectively. (d) The novelty of the dataset and the 78 SGBs were confirmed by genomic comparison across the IGG database (a collection of 16,136 non-redundant representative human gut genomes) and the Pasolli et al. MAGs database (a dataset collection of 154,723 non-redundant genomes from gut and other body parts of the global population). These 78 SGBs were genomes reconstructed here and without overlapping with existing isolates or metagenomically assembled genomes of the compared human microbiome datasets.
An attempt was made but failed to identify P-8 genome in any of the metagenomic DNA samples by mapping the P-8 reference genome across the metagenomic DNA using a genome coverage breadth of 40%. In general, at least 5-fold sequencing coverage would be required for tracing a specific bacterial strain. Thus, the failure was likely due to the inadequate levels of sequencing amount and genome coverage applied in this work, which were used in similar metagenomic studies (Fig. S1b).
3.2. Probiotic administration modulated gut microbiota composition
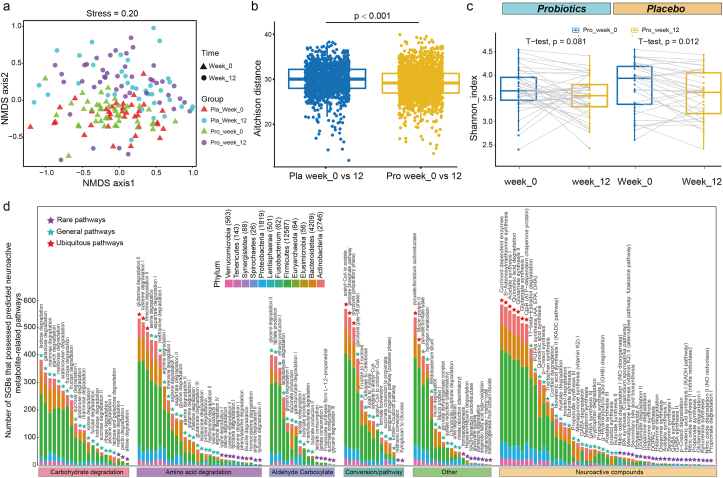
To assess the effect of probiotic consumption on the intestinal microbiome of stressed adults, an NMDS analysis was performed. Symbols representing samples at week 0 for both the placebo and P-8 groups were mainly distributed to the lower quadrants, while symbols representing samples at week 12 located mostly at the upper quadrants (Fig. 2a), suggesting obvious difference between the microbiota structure at weeks 0 and 12. Further analysis by ANOSIM found no significant difference between the probiotic and placebo groups at week 0 (R = 0.003, P = 0.354), but the two groups displayed significant difference at week 12 though with a relatively low R value (R = 0.041, P = 0.028). Interestingly, the placebo-receivers showed a significantly larger difference in the Aitchison distance between weeks 0 and 12 than the probiotic-receivers (P = 0.001; Fig. 2b). Moreover, comparing with week 0, the Shannon diversity index of the placebo-receivers decreased significantly at week 12 (P < 0.05), but such decrease was non-significant for the probiotic-receivers (Fig. 2c). These results together suggested that although both the P-8 and placebo groups exhibited changes in alpha-/beta-diversity after the 12-week-trial, the probiotic-treatment resulted in smaller changes in the gut microbiota diversity and structure.
Fig. 2.
Structure, diversity, and functional features of intestinal microbiota in stressed adult. (a) Non-metric multidimensional scaling (NMDS) analysis (Bray-Curtis similarity index). Symbols representing samples of the placebo (Pla) and probiotic (Pro) groups are shown in different colors. Triangles and circles represent weeks 0 and 12 samples, respectively. (b) Significantly larger Aitchison distance was observed in the gut microbiota (week 0 versus week 12) of the placebo group than the probiotic group. (c) Shannon diversity index of the two groups at two different time points. (d) Distribution of species-level genome bins (SGBs) that possessed predicted neuroactive metabolite-related pathways or gut brain modules (GBMs) across different phyla. The number written next to the phylum name indicates the quantity of SGBs distributed in that phylum. The purple, light blue, and red triangles represent rare, general, and ubiquitous pathways, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The “core microbiota” comprised stable key microorganisms of the human intestine, and it is closely related to human health (Astudillo-García et al., 2017). This study applied the concept of “core microbiota” to identify the “core SGBs” in the current dataset (i.e., SGBs present in >80% of the total sample number). Thirteen core SGBs were identified. No significant difference was observed in the core SGBs, S24A.M011 and S14A.M004 (both belonged to Fecalibacterium prausnitzii), P08.M042 (Agathobaculum butyriciproducens), S37B.M011 (uncultured Blautia sp.), S25A.M021 (Ruminococcus sp. CAG:9-related_41_34), and H2_119.37 (Bacteroides vulgatus), between the probiotic and placebo groups. Differential abundant SGBs were also identified by comparing the abundance of all SGBs in the probiotic group before and after probiotic treatment.
The abundances of six core SGBs and seventeen non-core SGBs changed significantly responding to the probiotic treatment; SIMPER analysis found that these SGBs together accounted for 13.65% of the microbial community variation (Table S5). Meanwhile, the abundances of one core SGB and twelve non-core SGBs changed significantly in the placebo group, together accounting for 4.85% of the microbial community variation (Table S5). At week 12, the probiotic-receivers had significantly more (P < 0.05 in all cases) S33A.M036 (B. adolescentis), S4A.M008 (B. longum), H1_08.M012 (F. prausnitzii), and S34B.M002 (Subdoligranulum sp. 60_17), while significantly less (P < 0.05 in both cases) H2_09.M016 (Roseburia faecis) and S22A.M013 (Fusicatenibacter saccharivorans), contributing to 11.15% of the microbial community variation (Fig. S1c; Table S5). Moreover, ten SGBs (e.g., B. longum, Megamonas funiformis, Subdoligranulum sp. 60_17, Bacteroides sp., and three uncultured species) showed no significant difference between the placebo and probiotic groups at week 0, but they became significantly differential abundant at week 12, accounting for 5.41% of the microbial community variation (Table S5). The results of SIMPER analysis suggested that the microbial community variations between probiotic/placebo treatments or different time points were contributed mainly by overall shifting of the microbial communities but not changes in individual dominating species.
3.3. Probiotic administration modulated gut microbiota-encoded neuroactivity-related pathways
A genome-centric metabolic reconstruction was performed to identify changes in neuroactivity-related pathways of the 589 identified SGBs using the MetaCyc and KEGG databases, focusing mainly on the human intestinal microbial metabolic pathways relating to the sugar fermentation, amino acid utilization, short-chain fatty acids (SCFAs), gut enzyme conversion, and metabolism of human neuroactive compounds. The identified pathways belonged to 12 phyla, distributed mainly to Firmicutes (53.47%), Bacteroidetes (18.79%), and Actinobacteria (12.20%) (Fig. 2d). The three most ubiquitous pathways (prevalence >80% among the SGBs) were corrinoid dependent enzymes (96.69%), acetyl-CoA to acetate (95.93%), and glutamine degradation II (89.98%). Some neuroactive compound pathways were detected in <5% of all SGBs, e.g., propionate degradation I, nitric oxide degradation II (NO reductase), and kynurenine synthesis; these scarce pathways were mainly found in Proteobacteria and Actinobacteria (Table S6).
3.4. Probiotic administration modulated gut microbiota-related neuroactive compounds
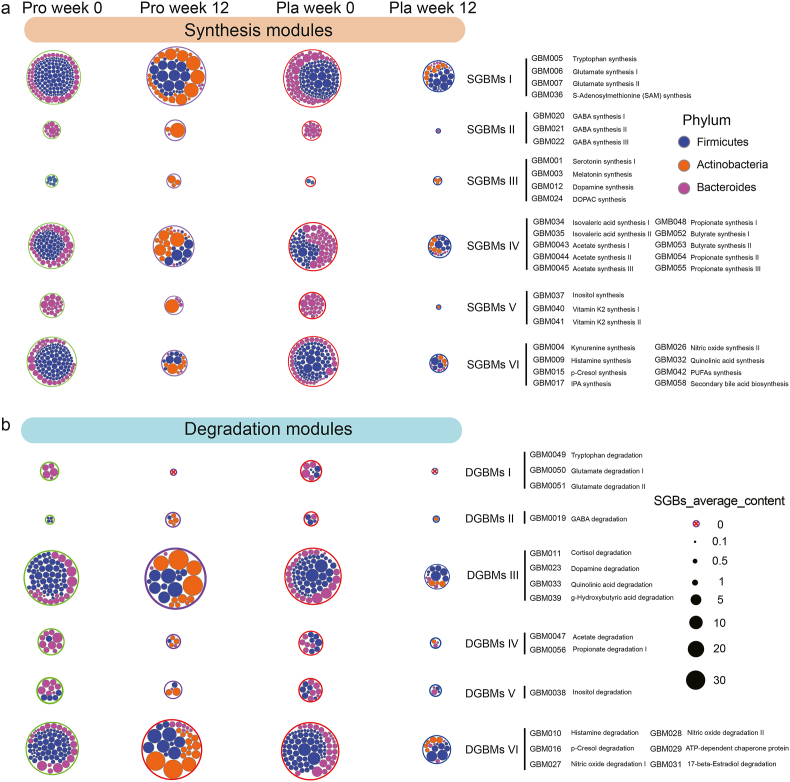
A module-based analysis framework developed by Valles-Colomer et al. (2019) was used to characterize the neuroactive compounds of our samples. Thirty-two and seventeen differential abundant modules were related with neuroactivity synthesis and degradation, respectively (Fig. 3a and b). The synthesis-/degradation-related GBMs could each be classified into six functional subgroups according to their potential of metabolic pathway. Significant differences were found in the total abundance of SGBs encoding synthesis-related GBMs (SGBMs) between the two groups (P < 0.001) only at week 12 but not at week 0. Moreover, the abundances of SGBs encoding SGBMs I (amino acid and derivatives synthesis) and SGBMs IV (SCFAs synthesis) varied greatly between the P-8 and placebo groups at week 12 (P < 0.01; Fig. S1d). Additionally, the abundance of SGBs involved in SGBMs II [neurotransmitter I synthesis; contained three synthetic pathways of gamma-aminobutyric acid (GABA)] was higher in the probiotic group at week 12. On the other hand, significant difference was found in the total abundance of degradation-related GMBs (DGBMs)-possessing SGBs between the two groups at week 12. The abundances of SGBs that encoded DGBMs II (neurotransmitter I degradation) and DGBMs III (neurotransmitter II degradation) were significantly different between weeks 0 and 12 in the probiotic group. The other subgroups of DGBMs, e.g., DGBMs I (amino acid degradation), DGBMs IV (SCFAs degradation), and DGBMs VI (vitamin degradation), only fluctuated mildly after the trial (Table S7).
Fig. 3.
Distribution of gut brain modules (GBMs) at the phylum level. The prevalence of different types of (a) synthesis-related GBMs (SGBMs) and (b) degradation-related GBMs (DGBMs), and their compositions at the phylum level in the probiotic (pro) and placebo (pla) groups are shown. Identified SGBMs/DGBMs were classified into six groups (I to VI) based on their predicted function of metabolite synthesis, respectively, and their associated species-level genome bins (SGBs) were assigned to the corresponding phyla. The size of the outer circle represents the average abundance of the corresponding SGBs.
To profile the key potential neuroactive compounds, GBMs encoded by differential SGBs that showed significant differences between the probiotic and placebo groups were analyzed (Fig. S2a). At week 12, samples of the probiotic-receivers had more diverse SGBs participating in menaquinone synthesis (vitamin K2) I synthesis, GABA and SCFAs metabolism, while the placebo group had more diverse SGBs participating in inositol degradation. Moreover, the probiotic-receivers had more cortisol-degrading SGBs at week 12 than the placebo-receivers. Interestingly, more probiotic-receivers had histamine-synthesizing SGBs at week 12 (8 at week 0 versus 19 at week 12, respectively; Table S8).
3.5. Probiotic administration modulated predicted gut neurometabolome
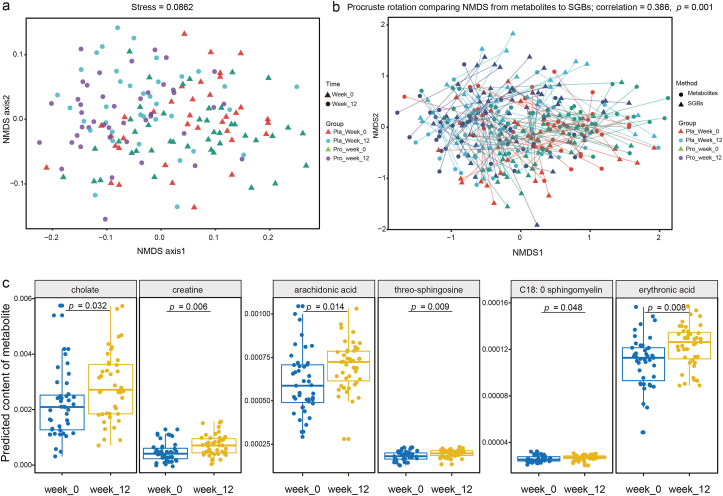
MelonnPan was applied to find correlation between SGBs and potential neurometabolites. Eighty metabolites were predicted; deoxycholic acid, glutamate, and cholate were the three most abundant metabolites. Changes in the subjects’ predicted fecal metabolomes were evaluated by NMDS analysis. Although some overlapping occurred on the NMDS score plot, samples collected at different time points formed clear clustering pattern (stress = 0.0862; Fig. 4a). Procrustes analysis is a statistical technique that displays multi-omics datasets in the low-dimensional space after data dimensionality reduction, and it has been increasingly used in microbiome research to evaluate the similarities and differences between datasets (Chong and Xia, 2017). Procrustes analysis was performed in this study, which found a positive cooperativity between the microbiome and metabolome profiles (correlation = 0.386; P = 0.001; Fig. 4b), suggesting that the metabolite output of the samples was consistent with the SGBs that produced it. Significant changes in the intestinal metabolite profiles occurred after the 12-week-trial for both the P-8 and placebo groups (P < 0.001; comparing between datasets of weeks 0 and 12 of each group), while ANOSIM found no significant difference between the metabolite profiles of the two groups at week 0.
Fig. 4.
Differences between the predicted intestinal metabolomes of the probiotic (pro) and placebo (pla) groups at two time points. (a) Non-metric multidimensional scaling (NMDS, Bray-Curtis similarity index) and (b) Procrustes analyses were performed on the predicted microbiomes and metabolomes of the two groups of subjects at week 0 and week 12, showing a positive cooperativity between microbiomic and metabolomic profiles (correlation = 0.386; P = 0.001). (c) Predicted differential neuroactive metabolites between week 0 and week 12 of the probiotic group. The p value was corrected by the FDR method, and the threshold P < 0.05 indicated significant.
Forty-one and twelve differential abundant metabolites were identified in the P-8 and placebo groups, respectively. These differential abundant metabolites showed significant differences in the predicted levels of abundance between weeks 0 and 12. Some metabolites shared the same trend of change after the clinical trial, e.g., at week 12, both groups had significantly lower predicted abundances of pantothenate, nicotinate, and lithocholate, while significantly increased predicted cytosine level (P < 0.05 in all cases; Table S9). These changes were likely non-specific to the probiotic treatment. In contrast, the average predicted abundances of cholate, arachidonic acid, creatine, threo-sphingosine, erythronic acid, and C18: 0 sphingomyelin were significantly higher for the probiotic-receivers (P < 0.05; Fig. 4c), representing probiotic-specific changes of some neuro-related metabolites.
4. Discussion
The present research observed probiotic-driven changes in the intestinal microbiota structure of moderately stressed subjects after the administration of the P-8 strain. The alleviation effect of probiotics on stress-related symptoms like anxiety and depression through gut microbiota modulation has been reported in previous studies (Westfall et al., 2017; Foster et al., 2017). Interestingly, the Shannon diversity index of the placebo group (P < 0.05) but not the probiotic group decreased significantly at week 12. In another double-blind, placebo-controlled clinical trial of probiotics found that the relief of stress-related symptoms was accompanied by an increase in volunteers' gut microbiota diversity after 8-week intervention (Kato-Kataoka et al., 2016). Generally, a high gut microbiota diversity is assumed to be important for maintaining a healthy physiological state, and unhealthy conditions (gastrointestinal discomforts, Crohn's disease, and some cancers) have been associated with reduced gut microbial diversity; and gut dysbiosis could even partially perturb normal energy metabolism and immunity in athletes (Wosinska et al., 2019). However, Shade (2017) pointed out a number of fallacies that strongly argued against the assumption that a high microbiota diversity is implicitly a good/desirable outcome for emergent properties like stability and productivity.
There are indeed a number of limitations in calculating the microbial diversity using currently available methods, including bias in protocols like extraction methods and inadequate sequencing depth/coverage. Moreover, the Aitchison distance represents differences in microbial structure and composition between two microbiota communities, while the alpha diversity (e.g., Shannon index) reflects the microbial richness and evenness in a sample. Thus, the diversity measures are the outcome of ecological processes but not the ecological process itself. Shade (2017) concluded that a high diversity is not necessarily ‘better’ or ‘healthy’, but “a starting point for further inquiry of ecological mechanisms rather than an ‘answer’ to community outcomes”. Therefore, the divergent responses in the gut microbiota diversity between the placebo and probiotic groups observed in our study could only be interpreted as differential responses towards the respective treatments.
Furthermore, the diversity analysis showed that the placebo had significantly greater within-individual beta diversity (measured by Aitchison distance) compared with the probiotic treatment, suggesting that dynamic microbiome changes not only occurred in the probiotic group but also the placebo group. A recent study provided a new concept called as volatility to describe the dynamic microbiome, which has been characterized relevant to the stress in both animals and humans (Bastiaanssen et al., 2021), therefore suggesting the changes in stress resilience in both placebo and probiotics groups in present study. The placebo subjects received only malodextrin but not the probiotic bacteria, while the probiotic subjects received both materials. Maltodextrin is a sugar-based excipient material used in clinical trials like the current work (Elnaggar et al., 2010), and it is regarded as inert and “generally regarded as safe” by the US Food and Drug Administration. However, several reports have described adverse effect of intake of maltodextrin on the gut environment, worsening chronic inflammatory disorders like inflammatory bowel disease and metabolic syndrome (Arnold and Chassaing, 2019). Laudisi et al. (2019) found that such detrimental effect was independent of mucosa-associated microbiota, as no significant change was observed in the gut mucosal microbiota composition in mice given maltodextrin. On the other hand, Yeo et al. (2010) observed in vitro prebiotic effect of maltodextrin in stimulating the growth of multiple Lactobacilli and Bifidobacteria strains, resulting in significant enhancement of lactic acid production (Yeo and Liong, 2010). Changes in the gut microbiota structure of the placebo group after the intervention period did not seem to be totally random, which was somewhat unexpected. This study was a randomized, double-blind, placebo-controlled trial, which was designed to control the placebo responses, incidental factors, and eliminating biases. The sample processing and metagenomic DNA extraction of samples of both groups for both time points were performed in parallel, so batch effect due to reagents and related technical factors did not exist. Instead, it was more likely that the gut microbiota response of the placebo group was a combined result of directed changes caused by the potential prebiotic effect of maltodextrin, temporal variation, and individual heterogeneity. The logical interpretation for our observations of the volatility of the fecal microbiome structure subject to the interventions was that the probiotic-driven regulation of gut microbiota was not drastic. Thus, to decipher the probiotic-driven gut microbiota response and its role in the beneficial function, it would be necessary to dig deeper to the functional metagenome level to gain understanding of changes in genes coding for metabolic pathways related to physiological stress.
Our results showed that the prevalence of some SGBs significantly increased (e.g., B. adolescentis, B. longum, F. prausnitzii) at week 12, while others decreased significantly (e.g., R. faecis, Fusicatenibacter saccharivorans). Lactobacillus, Bifidobacterium, and Faecalibacterium have been found to correlate positively with brain health and exert regulatory and protective effects on neurological diseases (Teichman et al., 2020). Bifidobacterium adolescentis exerted anxiolytic and antidepressant effect by rebalancing the gut microbiota and lowering inflammatory cytokines (Guo et al., 2019). Intake of B. longum 1714 ameliorated the physiological/psychological responses to acute stressors and improved the visuospatial memory of healthy adults (Allen et al., 2016). Additionally, ingesting F. prausnitzii prevented and reduced depression/anxiety-like behavior in rats (Hao et al., 2019). Our study also found negative correlations among B. adolescentis, B. longum, and F. prausnitzii with the stress/anxiety symptom scores, while R. faecis correlated positively with these scores (Fig. S2b). These results suggested that health maintenance not only relied on a diverse gut microbiota but also specific gut functional species/strains.
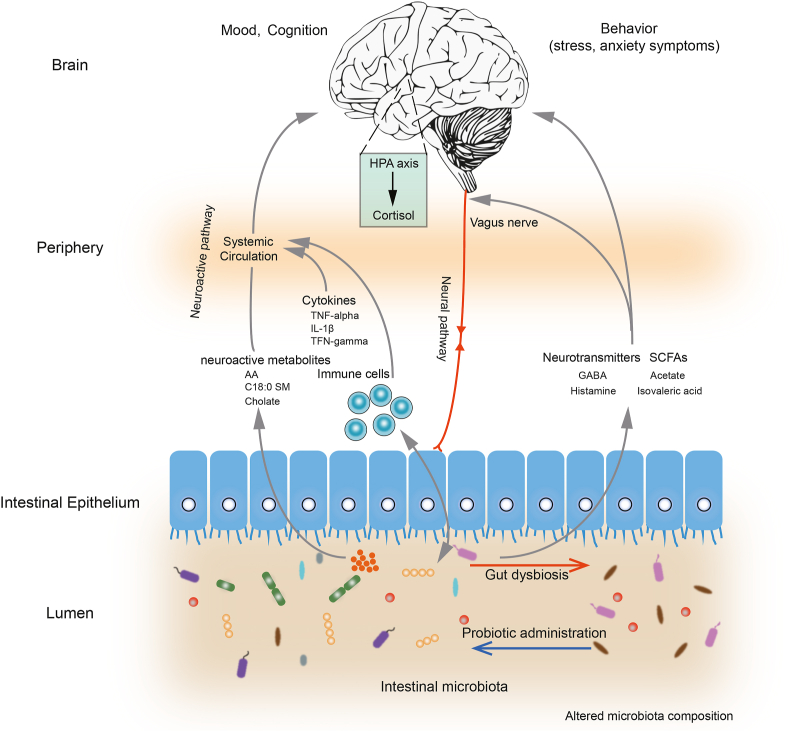
Bastiaanssen et al. point out the gut microbiota responds to stress similarly on the functional level but not on the taxonomical level in different host model (Bastiaanssen et al., 2021). Similarly, Tomizawa et al. found that the gut microbial function involved in Gut-Brain is influenced by the psychotropic consumption (Tomizawa et al., 2020). Collectively, the taxa encoded the GBM seemed to be major players participating in stress resilience, more work is needed to determine the changes of GBM after probiotics consumption. Multiple pathways are involved in the gut-brain-axis communication, which is facilitated by bidirectional interactions between the vagus nerve, the immune system, the endocrine system, and microorganism-originated metabolites/neurotransmitters (e.g., dopamine, serotonin, histamine, SCFAs, GABA, and various neurohormones; Fig. 5) (Teichman et al., 2020). This study investigated probiotic-specific modulation of the neuro-microbiome mediator metabolism by two platforms (i.e., the module-based analytical framework developed by Valles-Colomer et al. (2019) and MelonnPan (Teichman et al., 2020)). The probiotic treatment significantly increased the diversity of SGBs possessing neuroactive potential-related pathways, namely, menaquinone synthesis (vitamin K2) I, SCFAs and GABA metabolism. Vitamin K2 is synthesized by human gut bacteria. It is not only important in blood clotting and anti-inflammation, but also a neuroprotective agent that reduced the severity of relapses in patients with multiple sclerosis (Lasemi et al., 2018) and rescued drosophila from Parkinson's disease-associated mitochondrial defects (Josey et al., 2013). Gut microbe-originated metabolites like SCFAs, especially acetate, butyrate, proprionate, are important neuroimmune endocrine regulators. Our results confirmed that Fecalibacterium was desirable gut SCFAs-producers that correlated with quality of life indicators (Valles-Colomer et al., 2019). Another gut microbe-originated metabolite, GABA, is commonly produced by Bifidobacterium and Lactobacillus (Valles-Colomer et al., 2019). Our data showed that probiotic administration enriched GABA synthesis pathway-possessing B. adolescentis in the gut. Bravo et al. reported that ingesting L. rhamnosus reduced depression/anxiety behavior in mice in a vagus-dependent manner, as vagotomy abolished the anxiolytic/antidepressant effect and the symptom improvement was accompanied by increased GABAergic activity (Bravo et al., 2011). Transmission of nerve signals, e.g., bacterial derived-neurotransmtters like GABA and histamine, via the vagus nerve was the most important neural pathway communicating between gut-brain-axis (Pellegrini et al., 2018). Moreover, a marginal decline was observed in the plasma cortisol level in the probiotic-receivers compared with the placebo-receivers; such decline was accompanied by more intestinal cortisol-degrading SGBs observed in this work. Cortisol is the primary stress hormone that relates with stress response like increasing anxiety/depression. Regular intake of probiotics could lower the level of urinary free cortisol, suggesting some gut bacteria could reduce plasma cortisol (Evrensel and Ceylan, 2015).
Fig. 5.
Schematic diagram illustrating key probiotic-driven pathways that modulated the gut-brain-axis and host response.
Another group of microbial neuroactive metabolites, bile acids and unsaturated fatty acids, also constitutes information flow between the gut-brain-axis (Mohajeri et al., 2018). Our data showed that the predicted levels of cholate, arachidonic acid, and C18:0 sphingomyelin increased significantly after probiotic treatment. Bile acids help regulate the intestinal permeability and the blood-brain barrier, and secondary bile acids like cholate can stimulate 5-hydroxytryptamine biosynthesis (Agus et al., 2018). Ingesting probiotics significantly increased arachidonic acid concentration in rodents; arachidonic acid is vital for brain and optic nerve development, facilitating cognitive processes like memory and learning (Dinan et al., 2015). The immune system also plays an important role in regulating the gut-brain-axis. The P-8-receivers exhibited more obvious reduction in plasma pro-inflammatory cytokines, e.g. IFN-γ, TNF-α, and IL-1ꞵ, compared with the placebo-receivers. Consistently, peripheral administration of pro-inflammatory cytokines induced depression-like behaviors in rodents (Bilbo and Schwarz, 2012), while probiotics might exert immunomodulatory effects to counter stress response by generating T-regulatory cell populations and secreting cytokines (Dinan et al., 2013). Thus, the balance of intestinal microbiota may closely regulate host inflammatory response, activate intestinal and circulating immune pathways, subsequently influencing the mood and brain functions of the host (Pellegrini et al., 2018).
Disclosure of potential conflicts of interest
The authors declare no competing interests.
Funding
This research was supported by the National Natural Science Foundation of China [grant numbers 31720103911, 31972083]; and Inner Mongolia Science & Technology Major Projects [grant number ZDZX2018018].
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the Universiti Sains Malaysia and ClinicalTrials.gov was NCT03268447. Informed consent was obtained from all recruited subjects.
CRediT authorship contribution statement
Teng Ma: Formal analysis, Data curation, Visualization, Writing - original draft. Hao Jin: Formal analysis, Software, Validation. Lai-Yu Kwok: Writing - review & editing, Resources. Zhihong Sun: Writing - review & editing, Resources. Min-Tze Liong: Conceptualization, Methodology. Heping Zhang: Conceptualization, Methodology.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ynstr.2021.100294.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Agus A., Planchais J., Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23(6):716–724. doi: 10.1016/J.CHOM.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Allen A.P., Hutch W., Borre Y.E., Kennedy P.J., Temko A., Boylan G., Murphy E., Cryan J.F., Dinan T.G., Clarke G. Bifidobacterium longum 1714 as a translational psychobiotic: modulation of stress, electrophysiology and neurocognition in healthy volunteers. Transl. Psychiatry. 2016;6(11):e939. doi: 10.1038/TP.2016.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida A., Mitchell A.L., Boland M., Forster S.C., Gloor G.B., Tarkowska A., Lawley T.D., Finn R.D. A new genomic blueprint of the human gut microbiota. Nature. 2019;568(7753):499–504. doi: 10.1038/S41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold A.R., Chassaing B. Maltodextrin, modern stressor of the intestinal environment. Cell. Mol. Gastroenterol. Hepatol. 2019;7(2):475–476. doi: 10.1016/J.JCMGH.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astudillo‐García C., Bell J.J., Webster N.S., Glasl B., Jompa J., Montoya J.M., Taylor M.W. Evaluating the core microbiota in complex communities: a systematic investigation. Environ. Microbiol. 2017;19(4):1450–1462. doi: 10.1111/1462-2920.13647. [DOI] [PubMed] [Google Scholar]
- Bastiaanssen T.F.S., Gururajan A., van de Wouw M., Moloney G.M., Ritz N.L., Long-Smith C.M., Wiley N.C., Murphy A.B., Lyte J.M., Fouhy F., Stanton C., Claesson M.J., Dinan T.G., Cryan J.F. Volatility as a concept to understand the impact of stress on the microbiome. Psychoneuroendocrinology. 2021;124 doi: 10.1016/j.psyneuen.2020.105047. [DOI] [PubMed] [Google Scholar]
- Bilbo S.D., Schwarz J.M. The immune system and developmental programming of brain and behavior. Front. Neuroendocrinol. 2012;33(3):267–286. doi: 10.1016/J.YFRNE.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J.A., Forsythe P., Chew M.V., Escaravage E., Savignac H.M., Dinan T.G., Bienenstock J., Cryan J.F. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. U.S.A. 2011;108(38):16050–16055. doi: 10.1073/PNAS.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12(1):59–60. doi: 10.1038/NMETH.3176. [DOI] [PubMed] [Google Scholar]
- Caspi R., Billington R., Keseler I.M., Kothari A., Krummenacker M., Midford P.E., Ong W.K., Paley S., Subhraveti P., Karp P.D. The MetaCyc database of metabolic pathways and enzymes-a 2019 update. Nucleic Acids Res. 2020;48(D1) doi: 10.1093/NAR/GKZ862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J., Xia J. Computational approaches for integrative analysis of the metabolome and microbiome. Metabolites. 2017;7(4):62. doi: 10.3390/METABO7040062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan J.F., Dinan T.G. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012;13(10):701–712. doi: 10.1038/NRN3346. [DOI] [PubMed] [Google Scholar]
- Darzi Y., Falony G., Vieira-Silva S., Raes J. Towards biome-specific analysis of meta-omics data. ISME J. 2016;10(5):1025–1028. doi: 10.1038/ISMEJ.2015.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D.J., Doerr H.M., Grzelak A.K., Busi S.B., Jasarevic E., Ericsson A.C., Bryda E.C. Lactobacillus plantarum attenuates anxiety-related behavior and protects against stress-induced dysbiosis in adult zebrafish. Sci. Rep. 2016;6(1):33726. doi: 10.1038/SREP33726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinan T.G., Stanton C., Cryan J.F. Psychobiotics: a novel class of psychotropic. Biol. Psychiatr. 2013;74(10):720–726. doi: 10.1016/J.BIOPSYCH.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Dinan T.G., Stilling R.M., Stanton C., Cryan J.F. Collective unconscious: how gut microbes shape human behavior. J. Psychiatr. Res. 2015;63:1–9. doi: 10.1016/J.JPSYCHIRES.2015.02.021. [DOI] [PubMed] [Google Scholar]
- Eker S. Prevalence of depression symptoms in diabetes mellitus. Open access maced. J. Med. Sci. 2018;6(2):340–343. doi: 10.1016/J.JPSYCHIRES.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnaggar Y.S.R., El-Massik M.A., Abdallah O.Y., Ebian A.E.R. Maltodextrin: a novel excipient used in sugar-based orally disintegrating tablets and phase transition process. AAPS PharmSciTech. 2010;11(2):645–651. doi: 10.1208/S12249-010-9423-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrensel A., Ceylan M.E. The gut-brain Axis: the missing link in depression. Clin. Psychopharmacol. Neurosci. Off. Sci. J. korean Coll. Neuropsychopharmacol. 2015;13(3):239–244. doi: 10.9758/CPN.2015.13.3.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J.A., Rinaman L., Cryan J.F. Stress & the gut-brain axis: regulation by the microbiome. Neurobiol. Stress. 2017;7:124–136. doi: 10.1016/J.YNSTR.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Xie J.-P., Deng K., Li X., Yuan Y., Xuan Q., Xie J., He X.-M., Wang Q., Li J.-J., Luo H.-R. Prophylactic effects of Bifidobacterium adolescentis on anxiety and depression-like phenotypes after chronic stress: a role of the gut microbiota-inflammation Axis. Front. Behav. Neurosci. 2019;13:126. doi: 10.3389/FNBEH.2019.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A.A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi: 10.1093/BIOINFORMATICS/BTT086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z., Wang W., Guo R., Liu H. Faecalibacterium prausnitzii (ATCC 27766) has preventive and therapeutic effects on chronic unpredictable mild stress-induced depression-like and anxiety-like behavior in rats. Psychoneuroendocrinology. 2019;104:132–142. doi: 10.1016/J.PSYNEUEN.2019.02.025. [DOI] [PubMed] [Google Scholar]
- Hyatt D., Chen G.-L., LoCascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11(1):119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josey B.J., Inks E.S., Wen X., Chou C.J. Structure-activity relationship study of Vitamin K derivatives yields highly potent neuroprotective agents. J. Med. Chem. 2013;56(3):1007–1022. doi: 10.1021/JM301485D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D., Li F., Kirton E.S., Thomas A., Egan R.S., An H., Wang Z. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019;7(7) doi: 10.7717/PEERJ.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl J.P., Hatch A.M., Arcidiacono S.M., Pearce S.C., Soares J.W. Effects of psychological, environmental and physical stressors on the gut microbiota. Front. Microbiol. 2018;9 doi: 10.3389/FMICB.2018.02013. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato-Kataoka A., Nishida K., Takada M., Suda K., Kawai M., Shimizu K., Kushiro A., Hoshi R., Watanabe O., Igarashi T., Miyazaki K., Kuwano Y., Rokutan K. Fermented milk containing Lactobacillus casei strain Shirota prevents the onset of physical symptoms in medical students under academic examination stress. Benef. Microbes. 2016;7(2):153–156. doi: 10.3920/BM2015.0100. [DOI] [PubMed] [Google Scholar]
- Kyrpides N.C., Hugenholtz P., Eisen J.A., Woyke T., Göker M., Parker C.T. Genomic Encyclopedia of bacteria and archaea: sequencing a myriad of type strains. PLoS Biol. 2014;12(8) doi: 10.1371/JOURNAL.PBIO.1001920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/NMETH.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasemi R., Kundi M., Moghadam N.B., Moshammer H., Hainfellner J.A. Vitamin K2 in multiple sclerosis patients. Wien Klin. Wochenschr. 2018;130(9):307–313. doi: 10.1007/S00508-018-1328-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudisi F., Fusco D., Di Dinallo V., Stolfi C., Grazia A. Di, Marafini I., Colantoni A., Ortenzi A., Alteri C., Guerrieri F., Mavilio M., Ceccherini-Silberstein F., Federici M., MacDonald T.T., Monteleone I., Monteleone G. The Food additive maltodextrin promotes endoplasmic reticulum stress-driven mucus depletion and exacerbates intestinal inflammation. Cell. Mol. Gastroenterol. Hepatol. 2019;7(2):457–473. doi: 10.1016/J.JCMGH.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W259. doi: 10.1093/NAR/GKZ239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew L.-C., Hor Y.-Y., Yusoff N.A.A., Choi S.-B., Yusoff M.S.B., Roslan N.S., Ahmad A., Mohammad J.A.M., Abdullah M.F.I.L., Zakaria N., Wahid N., Sun Z., Kwok L.-Y., Zhang H., Liong M.-T. Probiotic Lactobacillus plantarum P8 alleviated stress and anxiety while enhancing memory and cognition in stressed adults: a randomised, double-blind, placebo-controlled study. Clin. Nutr. 2019;38(5):2053–2064. doi: 10.1016/J.CLNU.2018.09.010. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/BIOINFORMATICS/BTP352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Wang Q., Wang Y., Sun A., Lin Y., Jin Y., Li X. Oral probiotics ameliorate the behavioral deficits induced by chronic mild stress in mice via the gut microbiota-inflammation Axis. Front. Behav. Neurosci. 2018;12:266. doi: 10.3389/FNBEH.2018.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Liu Y., Li J., Guo H., Zang T., Wang Y. deSALT: fast and accurate long transcriptomic read alignment with de Bruijn graph-based index. Genome Biol. 2019;20(1):1–14. doi: 10.1186/S13059-019-1895-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick H., Franzosa E.A., Mclver L.J., Banerjee S., Sirota-Madi A., Kostic A.D., Clish C.B., Vlamakis H., Xavier R.J., Huttenhower C. Predictive metabolomic profiling of microbial communities using amplicon or metagenomic sequences. Nat. Commun. 2019;10(1):3136. doi: 10.1038/S41467-019-10927-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaoudi M., Lalonde R., Violle N., Javelot H., Desor D., Nejdi A., Bisson J.-F., Rougeot C., Pichelin M., Cazaubiel M., Cazaubiel J.-M. Assessment of psychotropic-like properties of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in rats and human subjects. Br. J. Nutr. 2011;105(5):755–764. doi: 10.1017/S0007114510004319. [DOI] [PubMed] [Google Scholar]
- Mohajeri M.H., La Fata G., Steinert R.E., Weber P. Relationship between the gut microbiome and brain function. Nutr. Rev. 2018;76(7):481–496. doi: 10.1093/NUTRIT/NUY009. [DOI] [PubMed] [Google Scholar]
- Nayfach S., Shi Z.J., Seshadri R., Pollard K.S., Kyrpides N.C. New insights from uncultivated genomes of the global human gut microbiome. Nature. 2019;568(7753):505–510. doi: 10.1038/S41586-019-1058-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S., Meleshko D., Korobeynikov A., Pevzner P.A. MetaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27(5):824–834. doi: 10.1101/GR.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olm M.R., Brown C.T., Brooks B., Banfield J.F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017;11(12):2864–2868. doi: 10.1038/ISMEJ.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25(7):1043–1055. doi: 10.1101/GR.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks D.H., Rinke C., Chuvochina M., Chaumeil P.-A., Woodcroft B.J., Evans P.N., Hugenholtz P., Tyson G.W. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2017;2(11):1533–1542. doi: 10.1038/S41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- Pasolli E., Asnicar F., Manara S., Zolfo M., Karcher N., Armanini F., Beghini F., Manghi P., Tett A., Ghensi P., Collado M.C., Rice B.L., DuLong C., Morgan X.C., Golden C.D., Quince C., Huttenhower C., Segata N. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019;176(3):649. doi: 10.1016/J.CELL.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini C., Antonioli L., Colucci R., Blandizzi C., Fornai M. Interplay among gut microbiota, intestinal mucosal barrier and enteric neuro-immune system: a common path to neurodegenerative diseases? Acta Neuropathol. 2018;136(3):345–361. doi: 10.1007/S00401-018-1856-5. [DOI] [PubMed] [Google Scholar]
- Pittenger C., Duman R.S. Stress, depression, and neuroplasticity: a convergence of mechanisms. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2008;33(1):88–109. doi: 10.1038/SJ.NPP.1301574. [DOI] [PubMed] [Google Scholar]
- Puebla-Barragan S., Reid G. Forty-five-year evolution of probiotic therapy. Microb. cell. 2019;6(4):184–196. doi: 10.15698/MIC2019.04.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., Mende D.R., Li J., Xu J., Li Shaochuan, Li D., Cao J., Wang B., Liang H., Zheng H., Xie Y., Tap J., Lepage P., Bertalan M., Batto J.-M., Hansen T., Paslier D. Le, Linneberg A., Nielsen H.B., Pelletier E., Renault P., Sicheritz-Ponten T., Turner K., Zhu H., Yu C., Li Shengting, Jian M., Zhou Y., Li Y., Zhang X., Li Songgang, Qin N., Yang H., Wang Jian, Brunak S., Doré J., Guarner F., Kristiansen K., Pedersen O., Parkhill J., Weissenbach J., Bork P., Ehrlich S.D., Wang Jun. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/NATURE08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea K., Dinan T.G., Cryan J.F. Gut microbiota: a perspective for psychiatrists. Neuropsychobiology. 2020;79(1):50–62. doi: 10.1159/000504495. [DOI] [PubMed] [Google Scholar]
- Richards D., Duffy D., Blackburn B., Earley C., Enrique A., Palacios J., Franklin M., Clarke G., Sollesse S., Connell S. Digital IAPT: the effectiveness & cost-effectiveness of internet-delivered interventions for depression and anxiety disorders in the Improving Access to Psychological Therapies programme: study protocol for a randomised control trial. BMC Psychiatr. 2018;18(1):59. doi: 10.1186/S12888-018-1639-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A., Lehto S.M., Harty S., Dinan T.G., Cryan J.F., Burnet P.W.J. Psychobiotics and the manipulation of bacteria-gut-brain signals. Trends Neurosci. 2016;39(11):763–781. doi: 10.1016/J.TINS.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shade A. Diversity is the question, not the answer. ISME J. 2017;11(1):1–6. doi: 10.1038/ISMEJ.2016.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandwitz P. Neurotransmitter modulation by the gut microbiota. Brain Res. 2018;1693:128–133. doi: 10.1016/J.BRAINRES.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada M., Nishida K., Kataoka-Kato A., Gondo Y., Ishikawa H., Suda K., Kawai M., Hoshi R., Watanabe O., Igarashi T., Kuwano Y., Miyazaki K., Rokutan K. Probiotic Lactobacillus casei strain Shirota relieves stress‐associated symptoms by modulating the gut–brain interaction in human and animal models. Neuro Gastroenterol. Motil. 2016;28(7):1027–1036. doi: 10.1111/NMO.12804. [DOI] [PubMed] [Google Scholar]
- Teichman E.M., O'Riordan K.J., Gahan C.G.M., Dinan T.G., Cryan J.F. When rhythms meet the blues: circadian interactions with the microbiota-gut-brain Axis. Cell Metabol. 2020;31(3):448–471. doi: 10.1016/J.CMET.2020.02.008. [DOI] [PubMed] [Google Scholar]
- Tomizawa Y., Kurokawa S., Ishii D., Miyaho K., Kishimoto T. Effects of psychotropics on the microbiome in patients with depression and anxiety: considerations in a naturalistic clinical setting. Int. J. Neuropsychopharmacol. 2020 doi: 10.1093/ijnp/pyaa070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uritskiy G.V., DiRuggiero J., Taylor J. MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome. 2018;6(1):1–13. doi: 10.1186/S40168-018-0541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valles-Colomer M., Falony G., Darzi Y., Tigchelaar E.F., Wang J., Tito R.Y., Schiweck C., Kurilshikov A., Joossens M., Wijmenga C., Claes S., Oudenhove L. Van, Zhernakova A., Vieira-Silva S., Raes J. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019;4(4):623–632. doi: 10.1038/S41564-018-0337-X. [DOI] [PubMed] [Google Scholar]
- Vandeputte D., Kathagen G., D’hoe K., Vieira-Silva S., Valles-Colomer M., Sabino J., Wang J., Tito R.Y., Commer L. De, Darzi Y., Vermeire S., Falony G., Raes J. Quantitative microbiome profiling links gut community variation to microbial load. Nature. 2017;551(7681):507–511. doi: 10.1038/NATURE24460. [DOI] [PubMed] [Google Scholar]
- Westfall S., Lomis N., Kahouli I., Dia S.Y., Singh S.P., Prakash S. Microbiome, probiotics and neurodegenerative diseases: deciphering the gut brain axis. Cell. Mol. Life Sci. 2017;74(20):3769–3787. doi: 10.1007/S00018-017-2550-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20(1):1–13. doi: 10.1186/S13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosinska L., Cotter P.D., O'Sullivan O., Guinane C. The potential impact of probiotics on the gut microbiome of athletes. Nutrients. 2019;11(10) doi: 10.3390/NU11102270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.W., Simmons B.A., Singer S.W. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2016;32(4):605–607. doi: 10.1093/BIOINFORMATICS/BTV638. [DOI] [PubMed] [Google Scholar]
- Yeo S.-K., Liong M.-T. Effect of prebiotics on viability and growth characteristics of probiotics in soymilk. J. Sci. Food Agric. 2010;90(2):267–275. doi: 10.1002/JSFA.3808. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Lv J., Pan L., Zhang Y. Roles and applications of probiotic Lactobacillus strains. Appl. Microbiol. Biotechnol. 2018;102(19):8135–8143. doi: 10.1007/S00253-018-9217-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data and analysis codes are available in NCBI-SRA (BioProject: PRJNA634666) and under https://github.com/TengMa-Cleap/Probiotics-relieve-human-stress-and-anxiety-project.