Abstract
The dental follicle is an ectomesenchymal tissue surrounding developing tooth germ that contains osteoblastic-lineage-committed stem/progenitor cells. MicroRNAs (miRNAs) are small non-coding RNAs that regulate gene expression during stem cell growth, proliferation, and differentiation. The aim of this study was to investigate the key regulators of miRNA during osteogenic differentiation in human dental follicle cells (hDFC). We analyzed miRNA expression profiles in hDFC during osteoblastic differentiation. Expression of miR-204 was decreased in hDFC during osteogenic induction on microarray analysis. Real-time and RT-PCR analysis also showed that the expression of miR-204 was decreased in all three hDFC during osteogenic differentiation. To investigate whether miR-204 has an effect on osteogenic differentiation, miR-204 was predicted to target alkaline phosphatase (ALP), secreted protein acidic and rich in cysteine (SPARC), and Runx2 in the in the 3'-UTRs by in silico analysis. When miR-204 was transfected into hDFC, the activity of ALP and protein levels of SPARC and Runx2 were decreased. mRNA levels of ALP, SPARC and Runx2 were also decreased by miR-204 transfection. Our data suggest that miR-204 negatively regulates the osteogenic differentiation of hDFC by targeting the bone-specific transcription factor Runx2, the mineralization maker ALP and the bone extracellular matrix protein SPARC.
Keywords: Micro RNA, Human dental follicle, MicroRNA 204, Osteogenic differentiation
Introduction
MicroRNAs (miRNAs) are a class of small non-coding RNAs that regulate protein translation or mRNA stability by binding to the 3′-untranslated region (3′-UTR) of the target gene in a sequence-specific manner.1,2 They post-transcriptionally regulate several physiological processes, including development, proliferation, differentiation and oncogenesis.3, 4, 5 An increasing number of research groups are now focusing on the involvement of miRNAs in osteogenic differentiation and bone development.6, 7, 8, 9 Consequently, several miRNAs have been reported to influence the differentiation of mesenchymal stem cells (MSC) or osteoblasts.10 However, specific miRNAs and their target genes in the regulation of MSC differentiation have not been well characterized and remain poorly understood.
The dental follicle, an ectomesenchymal tissue that surrounds the developing tooth germ, contains stem cells and lineage-committed progenitor cells or osteoblast/cementoblast precursor cells.11, 12, 13 Human dental follicle cells (hDFC) also have the capacity to commit to multiple cell types, not only to cells of osteoblastic lineage,11 but also to cells of adipogenic12 and neurogenic lineages.14 We have previously reported the gene expression profiles of hDFC during osteogenic differentiation and the potential regulators of this differentiation in hDFC.15
In this study, we analyzed miRNA expression profiles in hDFC during osteoblastic differentiation, then investigated the miRNAs and target genes regulating differentiation and/or mineralization.
Materials and methods
Cell cultures
In this study, hDFC were obtained using a previously reported method.11 Briefly, normal human impacted third molars were surgically removed and collected from patients aged bwtween 14 and 18 years, after obtaining informed consent. Dental follicle tissues were minced with sterilized scalpels, then digested in a solution of 0.1 U/ml collagenase type I and 1 U/ml dispase (Roche, Mannheim, Germany) for 1 h at 37 °C. Attached hDFC were cultured in 100-mm dishes using MSC growth medium (GM; consisting of MSC basal medium supplemented with fetal bovine serum, l-glutamine and penicillin/streptomycin; Lonza, Walkersville, Germany) in a humidified incubator (CO2 incubator MCO-175M; Sanyo, Osaka, Japan) in the presence of 5% CO2 in air at 37 °C. Experiments using hDFC were performed in accordance with the guidelines established by the Ethics Committee of Nihon University School of Dentistry at Matsudo (Recognition number: 10–036).
Osteogenic differentiation
For induction of osteogenic differentiation, hDFC from the 5th to 6th passage were seeded at 3.1 × 103 cells/cm2 in GM. After 24 h (day 0), media were replaced with MSC osteogenic induction medium (OIM) consisting of osteogenic basal medium, osteogenic SingleQuots containing l-glutamine, penicillin/streptomycin, dexamethasone, ascorbate and β-glycerophosphate (Lonza). Medium was replaced every 3 days.
Total RNA isolation
Total RNA from hDFC was isolated using miRNeasy Mini Kits (QIAGEN, Venlo, NLD) according to the instructions from the manufacturer. RNA was stored at −80 °C until use.
Microarray analysis
In order to identify miRNAs associated with osteogenic differentiation, expression of miRNAs was measured in hDFC cultured with OIM or GM on culture day 7 using miRNA microarray analysis. miRNA microarray analysis was performed using Agilent human miRNA Rel. 12.0 arrays (Agilent, Santa Clara, CA, USA) in accordance with Agilent protocols. To identify mRNAs associated with osteogenic differentiation, expression of mRNAs was measured in hDFC cultured with OIM or GM on culture days 3 and 10 using DNA microarray analysis. DNA microarray analysis was performed using the GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA, USA), in accordance with Affymetrix protocols. Data of miRNA microarray and DNA microarray was performed using total RNA isolated from one hDFC sample (female, 14 years old). Gene expression analysis of miRNA and DNA microarrays was performed using GeneSpring GX software (Agilent). Changes in gene expression were determined by hDFC cultured OIM compared to that GM. The regulated genes were showed a greater than twofold difference in intensity between GM culture and OIM culture.
Signaling pathway analysis
Each dataset for regulated miRNAs and mRNAs that were differentially expressed in hDFC cultured OIM compared to that in GM were uploaded into the Ingenuity Pathway Knowledge Base (IPA; IngenuityR Systems, www.ingenuity.com, QIAGEN). Target genes of uploaded miRNAs ware experimentally validated in previous reports using IPA, and were predicted using the miRNA target prediction programs TargetScan, TarBase and miRecords. Targets were then prioritized and filtered as “Experimentally Observed”, “Predicted High Confidence”, or “Predicted Moderate Confidence” using the IPA miRNA Target Filter. The prioritized and filtered miRNA targets were imported into GeneSpring GX software, then paired with genes that were differentially expressed in hDFC during osteogenic differentiation, as assessed by DNA microarray analysis. The regulated miRNAs and these target mRNAs regulated by osteogenic induction were uploaded into the IPA again, and analyzed for gene ontology and biological interactions by IPA core analysis.
miRNA transfection
For transfection of miRNA, HiPerFect Transfection Reagent (QIAGEN) was used in this study, in accordance with the manufacturer's protocol. Briefly, hDFC were seeded at 3.1 × 103 cells/cm2 in GM. After 24 h (day 0), media were replaced with OIM, then the transfection mixture was added. The transfection mixture contained 0.6% HiPerFect Transfection Reagent, and 5 nM miR-204-5p mimic (QIAGEN) or 5 nM AllStars Negative Control siRNA (QIAGEN) in MSC basal medium. Transfection was performed on day 0 and day 3.
Real-time polymerase chain reaction (PCR)
miRNA: cDNA was synthesized using a miScript Reverse Transcription Kit (QIAGEN). Real-time PCR was performed with Primer Assay (QIAGEN) and a miScript SYBR® Green PCR Kit (QIAGEN). PCR mixture was subjected to amplification using a DNA Engine Opticon 1 (Bio-Rad, Hercules, CA, USA), with preheating at 95 °C for 15 min, followed by 40 cycles at 94 °C for 15 s, 55 °C for 30 s and 70 °C for 30s. Amplicons were directly detected by measuring the increase in fluorescence caused by binding of SYBR Green using a DNA Engine Opticon 1 (Bio-Rad). Gene expression levels were calculated using the ΔΔcT method with normalization against RNU6. Each assay was normalized against RNU6 mRNA levels. The mRNA: cDNA was synthesized using a GeneAmp RNA PCR kit (Applied Biosystems, Foster City, CA). Real-time PCR was performed using a DyNAmo SYBR Green qPCR kit (Thermo Fisher Scientific, Waltham, MA, USA). PCR mixture, containing 20 pmol of forward and reverse primers and 2 μl of cDNA, was subjected to amplification with a DNA Engine Opticon 1 (BioRad), with preheating at 95 °C for 10 min, followed by 40 cycles of 94 °C for 15 s, 55 °C for 30 s and 72 °C for 30s. Amplicons were directly detected by measuring the increase in fluorescence caused by binding of SYBR Green using a DNA Engine Opticon 1 (Bio-Rad). Gene expression levels were calculated using the ΔΔcT method with normalization against GAPDH. Each assay was normalized against GAPDH mRNA levels. Primer sequences used for real-time PCR analysis are shown in Table 1.
Table 1.
Primer sequences used for real-time PCR.
| Gene | Primers | bp |
|---|---|---|
| ALP | F: 5′-gtcatcatgttcctgggaga-3′ | 123 |
| R: 5′-gaaggggaacttgtccatct-3′ | ||
| Runx2 | F: 5′-cccagaactgagaaactcaa-3′ | 125 |
| R: 5′-atcacagatggtccctaatg-3′ | ||
| SPARC | F:5′-tgtgatgaagggagagtagg-3′ | 203 |
| R: 5′-actaatcatgcatggctctc-3′ | ||
| GAPDH | F: 5′-atcaccatcttccaggag-3′ | 315 |
| R: 5′-atcgactgtggtcatgag-3′ |
Alkaline phosphate assay
Alkaline phosphatase (ALP) activity was assayed using a StemTAG™ Alkaline Phosphatase Activity Assay Kit (Cell Biolabs, San Diego, CA, USA) according to the instructions from the manufacturer. ALP activity is given as units indicating the release of 1 μmol of p-nitrophenol per min at 37 °C.
Western blot analysis
Whole-cell lysates for Western blotting were extracted with a Qproteome Mammalian Protein Prep kit (QIAGEN). The Bradford method was used for measurement of protein concentrations. Aliquots of cell lysates containing 5 μg of soluble protein were loaded onto a 7.5% TGX gel (Bio-Rad) and electrophoresed. Proteins were electrophoretically transferred to a nitrocellulose membrane (Bio-Rad), which was subsequently treated with antibodies against secreted protein acidic and rich in cysteine (SPARC: rabbit polyclonal, dilution 1:1000; Santa Cruz Biotechnology, Dallas, TX, USA), Runx2 (rabbit polyclonal, dilution 1:1000; Santa Cruz Biotechnology) and β-actin (rabbit monoclonal, dilution 1:1000; Cell Signaling Technology, Beverly, MA, USA). HRP-conjugated secondary antibodies (anti-rabbit; Cell Signaling Technology; dilution 1:2000) were added, followed by incubation at room temperature. Signals were developed using ECL™ Prime Western Blotting Detection Reagent (GE Healthcare, Buckinghamshire, UK) and subsequent densitometric analysis was performed using ImageQuant LAS400mini (GE Healthcare).
Results
miRNA expression profiles in hDFC-induced osteogenic differentiation
We performed microarray analysis to determine the miRNAs regulated in hDFC during osteogenic differentiation. Among the 960 miRNAs tested, 307 were expressed in hDFC cultured with OIM and/or GM for 7 days. A total of 68 miRNAs were defined as regulated miRNAs that displayed a greater than two-fold change in expression in hDFC cultured with OIM, as compared to hDFC cultured with GM. These 68 miRNAs comprised 33 up-regulated miRNAs and 35 down-regulated miRNAs (Fig. 1A and B; Table 2).
Figure 1.
Microarray analysis of hDFC during osteogenic differentiation. miRNA expression profiles of hDFC were compared between culture Day 7 GM and Day 7 OIM (A) Among 960 miRNAs tested, 307 were expressed in hDFC (B) 68 miRNAs (up: 33 miRNA; down: 35 miRNA) showed a greater than two-fold differentiation in OIM cultures when compared with GM cultures (C) Network of miRNAs – mRNAs, as analyzed by IPA.
Table 2.
68 miRNAs (up: 33 miRNA; 35 miRNA) showed a greater than two-fold differentiation in OIM cultures, as compared to GM cultures.
| Fold Change up-regulated genes | ||
|---|---|---|
| MicroRNA Name | Accession No | Fold Change |
| hsa-miR-582-5p | MIMAT0003247 | 186.74 |
| hsa-let-7f-1* | MIMAT0004486 | 131.43 |
| hsv-miR-H6-3p | MIMAT0008404 | 129.72 |
| hsa-miR-1539 | MIMAT0007401 | 129.65 |
| hsa-miR-664 | MIMAT0005949 | 95.21 |
| hsa-miR-892b | MIMAT0004918 | 86.81 |
| hsa-miR-30c-2 | MIMAT0004550 | 62.65 |
| hsa-miR-889 | MIMAT0004921 | 57.02 |
| hsa-miR-129-3p | MIMAT0004605 | 56.09 |
| hsa-miR-132* | MIMAT0004594 | 48.50 |
| hsa-miR-634 | MIMAT0003304 | 48.14 |
| hsa-miR-598 | MIMAT0003266 | 47.12 |
| hsa-miR-563 | MIMAT0003227 | 45.04 |
| hsa-miR-129* | MIMAT0004548 | 44.24 |
| hsa-miR-15b* | MIMAT0004586 | 38.90 |
| hsa-miR-490-5p | MIMAT0004764 | 30.55 |
| hsa-miR-15a* | MIMAT0004488 | 30.28 |
| hsa-miR-602 | MIMAT0003270 | 30.28 |
| hsa-miR-550 | MIMAT0004800 | 25.65 |
| hsa-miR-493 | MIMAT0003161 | 24.89 |
| hsa-let-7b* | MIMAT0004482 | 6.52 |
| hsa-miR-138 | MIMAT0000430 | 4.52 |
| hsa-miR-483-3p | MIMAT0002173 | 3.57 |
| hsa-miR-431 | MIMAT0001625 | 3.17 |
| hsa-miR-33b* | MIMAT0004811 | 2.88 |
| hsa-miR- 425* | MIMAT000 1343 | 2.72 |
| hsa-miR-933 | MIMAT0004976 | 2.62 |
| hsa-miR- 33a | MIMAT0000091 | 2.39 |
| hsa-miR-100* | MIMAT0004512 | 2.26 |
| hsa-miR-370 | MIMAT0000722 | 2.09 |
| hsa-miR-487a | MIMAT0002178 | 2.07 |
| hsa-miR-551b | MIMAT0003233 | 2.06 |
| hsa-miR-30a* | MIMAT0000088 | 2.04 |
| hsa-miR-150* | MIMAT0004610 | −1122.43 |
| hcmv-miR-UL70-3p | MIMAT0003343 | −437.43 |
| hsa-miR-371-5p | MIMAT0004687 | −179.83 |
| hsa-miR-1224-5p | MIMAT0005458 | −100.76 |
| hsa-miR-629* | MIMAT0003298 | −93.75 |
| hsa-miR-663 | MIMAT0003326 | −76.00 |
| hsa-miR-630 | MIMAT0003299 | −71.15 |
| hsa-miR-424* | MIMAT0004749 | −69.67 |
| hsa-miR-204 | MIMAT0000265 | −52.35 |
| hsa-miR-373* | MIMAT0000725 | −45.45 |
| hsa-miR-193b | MIMAT0004767 | −36.16 |
| hsa-miR-671-5p | MIMAT0003880 | −32.73 |
| kshv-miR-K12-10b | MIMAT0002180 | −31.01 |
| hsa-miR-188-5p | MIMAT0000457 | −29.84 |
| hsa-miR-7-1* | MIMAT0004553 | −28.58 |
| hsa-miR-423-3p | MIMAT0001340 | −14.24 |
| hshv-miR-K12-3 | MIMAT0002193 | −11.18 |
| hsa-miR-335 | MIMAT0000765 | −8.35 |
| hsa-miR-1915 | MIMAT0007892 | −7.79 |
| hsa-miR-874 | MIMAT0004911 | −5.32 |
| hsa-miR-1181 | MIMAT0005826 | −4.00 |
| hsa-miR-1225-5p | MIMAT0005572 | −3.57 |
| hsa-miR-1207-5p | MIMAT0005871 | −3.32 |
| hsa-miR-424 | MIMAT0001341 | −3.00 |
| hsa-miR-135a* | MIMAT0004595 | −2.65 |
| hsa-miR-455-5p | MIMAT0003150 | −2.65 |
| hsa-miR-503 | MIMAT0002874 | −2.56 |
| hsa-miR-31* | MIMAT0004504 | −2.44 |
| hsa-miR-125a-3p | MIMAT0004602 | −2.36 |
| hsa-miR-296-5p | MIMAT0000690 | −2.30 |
| hsa-miR-382 | MIMAT0000737 | −2.27 |
| hsa-miR-455-3p | MIMAT0004784 | −2.25 |
| hsa-miR-638 | MIMAT0003308 | −2.21 |
| hsa-miR-210 | MIMAT0000267 | −2.08 |
| hsa-miR-34a* | MIMAT0004557 | −2.08 |
Target genes of regulated miRNAs
The 68 regulated miRNAs were uploaded into the IPA database, and target genes were predicted (TargetScan, TarBase, miRecords and the Ingenuity® Knowledge Base). The miRNA targets were then prioritized as experimentally validated and predicted using the IPA miRNA Target Filter. Among the targets of the 33 up-regulated miRNA by osteogenic induction, 68 genes were experimentally validated as targets, and 2856 and 8067 genes were predicted as targets with high and moderate confidence, respectively. On the other hand, among the targets for the 35 down-regulated miRNAs by osteogenic induction, 277 genes were experimentally validated as targets, and 7491 genes and 12,806 genes were predicted as targets with high and moderate confidence, respectively (Table 3). Next, miRNA-mRNA pairings were investigated for the biological context of mRNAs. A total of 1798 regulated genes (913 up-regulated genes and 885 down-regulated genes), which were defined as regulated mRNAs displaying a greater than two-fold change in expression in hDFC cultured with OIM, as compared to hDFC cultured with GM using oligonucleotide microarrays, were uploaded into IPA and then paired to miRNA target genes using the PA miRNA Target Filter.
Table 3.
Number of target genes for miRNAs.
| Uploaded to IPA | Number of target Genes |
|||
|---|---|---|---|---|
| Total | Experimentally observed | Predicted |
||
| High Confidence | Moderate Confidence | |||
| Up-regulated 33 microRNA | 10091 (1528) | 68 (11) | 2856 (504) | 8067 (1013) |
| Down-regulated 35 microRNA | 20574 (3233) | 277 (53) | 7491 (1258) | 12806 (1922) |
The regulated target genes for up-regulated miRNAs in hDFC during osteogenic induction included 14 of 68 experimentally validated genes, and those predicted as targets with high and moderate confidence were 502 genes among 2856 genes and 1083 genes among 8067 genes, respectively. In contrast, regulated target genes for down-regulated miRNAs in hDFC during osteogenic induction included 57 of 277 experimentally validated genes, and those predicted as targets with high and moderate confidence were 1258 genes among 7491 genes and 1922 genes among 12,806 genes, respectively.
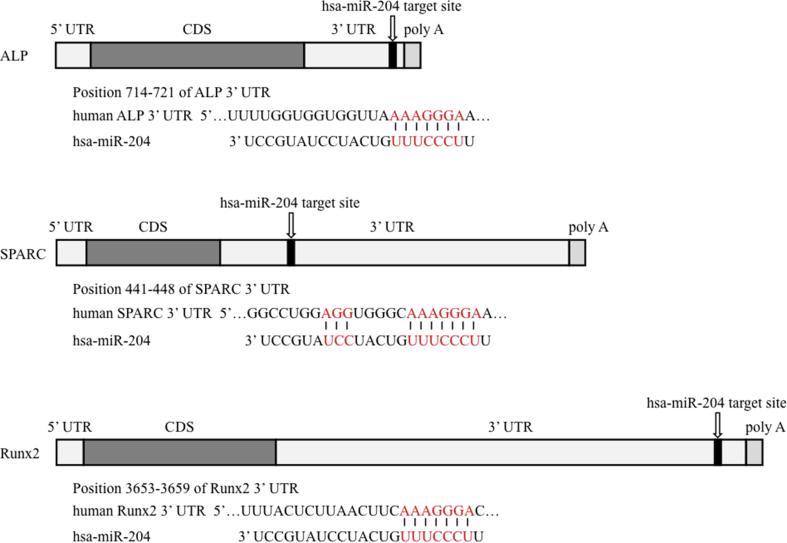
The 68 regulated miRNAs and the 71 (14 + 57) experimentally validated target genes were subjected to pathway analysis using IPA to determine their biological interactions. We focused on miR-204, which was ranked 9 among the down-regulated genes during induced osteoblastic differentiation (Fig. 1B; Table 2), and it appeared to target ALP, SPARC (also known as osteonectin) and Runx2, and to be associated with osteoblast differentiation (Fig. 1C). The putative binding sites of miR-204 are the 3′-UTRs of ALP, SPARC and Runx2 mRNAs. These seed regions are evolutionarily well-conserved among vertebrates (Fig. 2).
Figure 2.
Putative miR-204 binding sites in ALP, SPARC and Runx2 (Prediction scores from Target Scan database; ALP: −0.45, SPARC: −0.21, and Runx2: −0.10).
Expression of miR-204 in hDFC during osteogenic induction
Expression of miR-204 was markedly decreased in hDFC during osteogenic induction on microarray analysis. Because miRNA microarray technology was only used for preliminary screening, miR-204 expression during osteogenic differentiation was confirmed in three hDFC samples isolated from three patients. Real-time RT-PCR analysis showed that expression of miR-204 was significantly lower in all three hDFC samples cultured with OIM, as compared to those cultured with GM on culture days 4, 7 and 14 (Fig. 3).
Figure 3.
Gene expression of miR-204 in hDFC isolated from three donors at the indicated times during osteogenic differentiation. miR-204 gene expression was examined by real-time PCR. Values represent means ± SD of the results from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.005, as compared with GM culture at the indicated time points.
miR-204 regulates expression of ALP, SPARK and Runx2
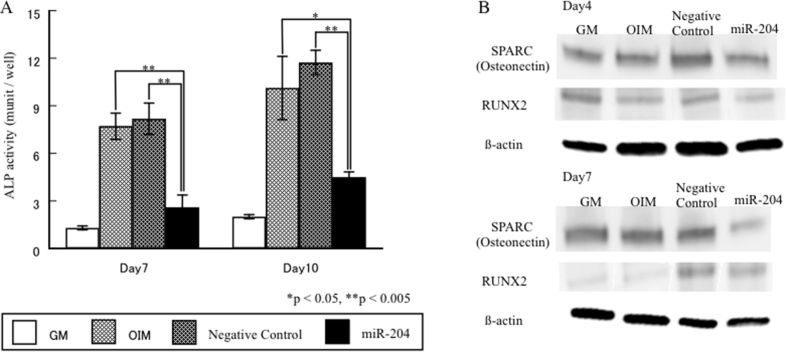
According to bioinformatic analyses to predict target genes for miR-204 using a combination of miRNA databases and signaling pathway analysis, ALP, SPARC and Runx2 were found to be potential target genes for miR-204. To investigate whether miR-204 has an effect on osteoblast differentiation, miR-204 was transfected into hDFC on culture days 0 and 3. ALP activity was measured in hDFC cultured with OIM on days 7 and 10 after transfection with miR-204, as ALP activity in hDFC cultured in OIM increased significantly from culture day 7. ALP activity was suppressed in hDFC transfected with miR-204, but was not suppressed in hDFC transfected with negative control (Fig. 4A). Protein levels of Runx2 and SPARC were examined in hDFC transfected with miR-204 by Western blot analysis. Protein levels of Runx2 were up-regulated in hDFC during osteogenic induction on culture day 4. Protein levels of Runx2 were decreased in hDFC transfected with mir-204 on day 4 (Fig. 4B). In contrast, protein levels of SRARC were clearly detected in hDFC by osteogenic induction on culture day 7. Protein levels of SPARC were decreased in hDFC transfected with miR-204 on day 7, but protein levels of SPARC and Runx2 in hDFC were unaffected after transfection with negative control (Fig. 4B).
Figure 4.
Effects of miR-204 in osteogenic differentiation of hDFC. Cells were cultured with GM, OIM + Negative control and OIM + miR-204 for the indicated time periods (A) ALP activity in hDFC during osteogenic differentiation. Values represent means ± SD of the results from three independent experiments. *p < 0.005, as compared to Negative control at the indicated time points (B) Western blot was performed to confirm that miR-204 decreased SPARC and Runx2 protein levels.
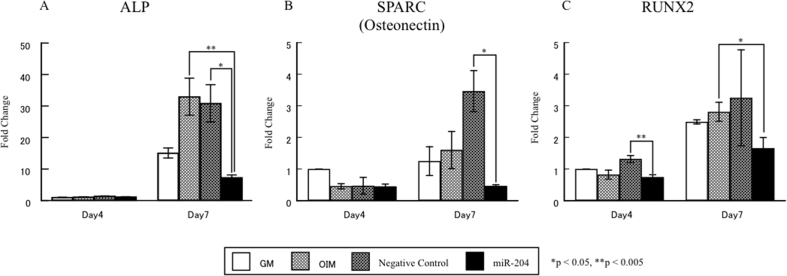
Next, mRNA levels of ALP, SPARC and Runx2 were examined in hDFC transfected with miR-204. In hDFC transfected with miR-204 on culture day 7, mRNA levels of ALP and SPARC were also decreased (Fig. 5A and B). Runx2 was decreased in hDFC transfected with miR-204 on culture days 4 and 7 (Fig. 5C).
Figure 5.
Effects of miR-204 in osteogenic differentiation of hDFC. Cells were cultured with GM, OIM + Negative control and OIM + miR-204 for the indicated time periods. Gene expression was examined using real-time PCR. Values represent means ± SD of the results from three independent experiments. *p < 0.05, **p < 0.05, as compared to OIM and Negative control at the indicated time points.
Discussion
This study shows that miR-204 negatively regulates the osteogenic differentiation of hDFC by targeting the bone-specific transcription factor Runx2, the mineralization marker ALP and the bone extracellular matrix protein SPARC. The processes of osteogenic differentiation and/or mineralization are conducted in a tissue-specific manner via numerous miRNAs. Recently, several miRNAs that are down-regulated during osteoblastic differentiation have been reported to inhibit translation of numerous osteogenic mRNAs, including transcription factors, signal transduction factors and corresponding molecules, which are required for osteoblast formation.16 We used miRNA microarrays to profile miRNA expression in hDFC cultured with OIM, as compared to that with GM on culture day 7. Among these differentially expressed miRNAs, miR-204 was markedly down-regulated in hDFC with OIM culture, and was predicted to target ALP, SPARC and Runx2 in the 3′-UTRs by in silico analysis. We confirmed miR-204 expression in hDFC isolated from three patients using real time-PCR. Expression of miR-204 was significantly decreased in three hDFC cultured with OIM when compared to culture with GM, although expression levels of miR-204 in hDFC varied between patients.
The activity and expression of ALP, as an early marker of osteogenic differentiation,17 were also decreased in hDFC after transfection with miR-204. ALP activity is reportedly reduced in BMP-2-treated C2C12, osteoblastic differentiating MC3T3-E1 and hMSC cultured with OIM after miR-204 transfection,18,19 suggesting that inhibition of ALP activity by miR-204 is Runx2-dependent. A recent study indicated that miR-204 also directly targets ALP, the 3′-UTR segment of which contains a miR-204 seed sequence.19 Co-transfection with miR-204 and pMIR-REPORT vector to generate the ALP 3′-UTR was demonstrated to significantly reduce luciferase activity. We have previously reported that ALP activity increases in hDFC cultured with OIM from culture day 7.15 ALP activity was measured in hDFC cultured with OIM on days 7 and 10 after transfection with miR-204, and ALP activity was suppressed in hDFC transfected with miR-204.
SPARC, also known as osteonectin, is a glycoprotein expressed in a variety of mammalian tissues, and is the most abundant non-collagen extracellular matrix protein in bone.20 In the skeleton, SPARC is critical for normal bone remodeling and the maintenance of bone mass. SPARC-null mice display increased marrow adiposity, develop profound low-turnover osteopenia in the trabecular compartment, and have cortical bone with decreased mechanical properties and matrix quality.18 Transfection of hDFC with miR-204 mimic decreased SPARC expression. We therefore believe that down-regulation of miR-204 in hDFC by osteogenic induction may correlate with increased SPARC protein during matrix maturation and mineralization during osteogenic differentiation.
Runx2 is a key regulator in osteogenesis.21 Expression of miR-204 has been shown to be down-regulated in BMP2-treated C2C12 cells, and the seed sequences for miR-204 are located downstream of a putative conserved polyadenylation site that could reduce the length of the Runx2 3′-UTR and eliminate suppression of Runx2.19 Another study reported that miR-204 is up-regulated in mesenchymal progenitor cells and bone marrow stromal cells (BMSC), as miR-204 inhibits Runx2 protein levels.22 In our study, protein levels and gene expression of Runx2 were also decreased in hDFC by transfection with miR-204 mimic. These data suggest that miR-204 acts as an important endogenous negative regulator of Runx2, which inhibits osteogenesis and promotes adipogenesis of mesenchymal progenitor cells. Expression of miR-204 has also been reported to be up-regulated in osterix (Osx)−/− calvaria and down-regulated in MC3T3 cells over-expressing Osx.19 These results suggest that over-expression of Osx at the onset of preosteoblast differentiation may positively regulate (either directly or indirectly) transcription of miR-204. This suggests a tight correlation between Osx and miRNAs in bone formation, and in maintaining appropriate levels of Runx2 proteins for optimal differentiation and function of osteoblasts.
Numerous recent studies have shown dynamic changes in expression of miRNAs during osteoblast differentiation. Some of these miRNAs are able to redirect mesenchymal stem cells into an adipogenic cell fate with concomitant up-regulation of key lineage-specific transcription factors. Those studies suggest that a process with multiple miRNAs controls mesenchymal lineage progression by selectively blocking differentiation of osteoblasts to control skeletal development. During early MSC differentiation, several miRNAs, including Runx2-targeting miRNAs, are down-regulated, enabling progression of osteogenic differentiation.16,23,24 Among these miRNAs, expression of miR-15b, miR-23a, miR-30b, miR-30c and miR-34c decreased in hDFC cultured with OIM, as compared to that with GM, on culture day 7, and these miRNAs were included in the strongly regulated miRNA group, while expressions of mir-133a, miR-205, miR-217, miR-338-3p and miR-433 were not detected in hDFC. Each mRNA generally has multiple miRNA binding sites. Expression of Runx2 may be up-regulated in hDFC by miRNAs, including miR-204, as well as miR-15b, miR-23a, miR-30b, miR-30c and miR-34c, during osteogenic differentiation.
In conclusion, we defined a characteristic miRNA signature in hDFC during osteogenic differentiation and mineralization. Moreover, we demonstrated the involvement of miR-204 in the regulation of the bone-specific transcription factor Runx2 and the mineralization marker ALP. Our results warrant further investigation in order to improve our knowledge regarding the implications of consistently modulated miRNAs in osteogenic differentiation.
Declaration of Competing Interest
The authors declare no conflicts of interest associated with this manuscript.
Acknowledgements
We would like to thank Ms. Asayo Imaoka for her invaluable assistance with the oligonucleotide microarray technology. This study was supported by Grants-in-Aid for Scientific Research (C) (23592947 and 26463020) and a Grant–in Aid for Research Activity Start-up (25893263) from the Japan Society for the Promotion of Science.
References
- 1.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 2.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Gao J., Yang T., Han J. MicroRNA expression during osteogenic differentiation of human multipotent mesenchymal stromal cells from bone marrow. J Cell Biochem. 2011;112:1844–1856. doi: 10.1002/jcb.23106. [DOI] [PubMed] [Google Scholar]
- 4.Png K.J., Halberg N., Yoshida M., Tavazoie S.F. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2011;481:190–194. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 5.Ebert M.S., Sharp P.A. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie Q., Wang Z., Bi X. Effects of miR-31 on the osteogenesis of human mesenchymal stem cells. Biochem Biophys Res Commun. 2014;446:98–104. doi: 10.1016/j.bbrc.2014.02.058. [DOI] [PubMed] [Google Scholar]
- 7.Chen S., Yang L., Qe Jie. MicroRNA-125b suppresses the proliferation and osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. Mol Med Rep. 2014;9:1820–1826. doi: 10.3892/mmr.2014.2024. [DOI] [PubMed] [Google Scholar]
- 8.Hupkes M., Sotoca A.M., Hendriks J.M., van Zoelen E.J., Dechering K.J. MicroRNA miR-378 promotes BMP2-induced osteogenic differentiation of mesenchymal progenitor cells. BMC Mol Biol. 2014;27:15. doi: 10.1186/1471-2199-15-1. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papaioannou G., Mirzamohammadi F., Kobayashi T. MicroRNAs involved in bone formation. Cell Mol Life Sci. 2014;71:4747–4761. doi: 10.1007/s00018-014-1700-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordeladze J.O., Reseland J.E., Duroux-Richard I., Apparailly F., Jorgensen C. From stem cells to bone: phenotype acquisition, stabilization, and tissue engineering in animal models. ILAR J. 2009;51:42–61. doi: 10.1093/ilar.51.1.42. [DOI] [PubMed] [Google Scholar]
- 11.Morsczeck C., Götz W., Schierholz J. Isolation of precursor cells (PCs) from human dental follicle of wisdom teeth. Matrix Biol. 2005;24:155–165. doi: 10.1016/j.matbio.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Yao S., Pan F., Prpic V., Wise G.E. Differentiation of stem cells in the dental follicle. J Dent Res. 2008;87:767–771. doi: 10.1177/154405910808700801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yagyuu T., Ikeda E., Ohgushi H. Hard tissue-forming potential of stem/progenitor cells in human dental follicle and dental papilla. Arch Oral Biol. 2009;55:68–76. doi: 10.1016/j.archoralbio.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Völlner F., Ernst W., Driemel O., Morsczeck C. A two-step strategy for neuronal differentiation in vitro of human dental follicle cells. Differentiation. 2009;77:433–441. doi: 10.1016/j.diff.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Aonuma H., Ogura N., Takahashi K. Characteristics and osteogenic differentiation of stem/progenitor cells in the human dental follicle analyzed by gene expression profiling. Cell Tissue Res. 2012;350:317–331. doi: 10.1007/s00441-012-1477-6. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y., Xie R.L., Croce C.M. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A. 2011;108:9863–9868. doi: 10.1073/pnas.1018493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsunaga S. Histological and histochemical investigations of constant direct current stimulated intramedullary callus. Nihon Seikeigeka Gakkai Zasshi. 1986;60:1293–1303. [PubMed] [Google Scholar]
- 18.Mansergh F.C., Wells T., Elford C. Osteopenia in Sparc (osteonectin)-deficient mice: characterization of phenotypic determinants of femoral strength and changes in gene expression. Physiol Genom. 2007;32:64–73. doi: 10.1152/physiolgenomics.00151.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Q., Liu W., Sinha K.M., Yasuda H., de Crombrugghe B. Identification and characterization of microRNAs controlled by the osteoblast-specific transcription factor Osterix. PLoS One. 2013;8 doi: 10.1371/journal.pone.0058104. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Termine J.D. Non-collagen proteins in bone. Ciba Found Symp. 1988;136:178–202. doi: 10.1002/9780470513637.ch12. [DOI] [PubMed] [Google Scholar]
- 21.Ducy P., Zhang R., Geoffroy V., Ridall A.L., Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 22.Huang J., Zhao L., Xing L., Chen D. MicroRNA-204 regulates Runx 2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–456. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassan M.Q., Gordon J.A., Beloti M.M. A network connecting Runx2, SATB2, and the miR-23ã27ã24-2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci U S A. 2010;107:19879–19884. doi: 10.1073/pnas.1007698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vimalraj S., Partridge N.C., Selvamurugan N. A positive role of microRNA-15b on regulation of osteoblast differentiation. J Cell Physiol. 2014;229:1236–1244. doi: 10.1002/jcp.24557. [DOI] [PMC free article] [PubMed] [Google Scholar]