Abstract
Background
The etiology of prostate cancer (PCa) is multiple and complex. Among the causes recently cited are chronic infections engendered by microorganisms that often go unnoticed. A typical illustration of such a case is infection due to mollicutes bacteria. Generally known by their lurking nature, urogenital mollicutes are the most incriminated in PCa. This study was thus carried out in an attempt to establish the presence of these mollicutes by PCR in biopsies of confirmed PCa patients and to evaluate their prevalence.
Methods
A total of 105 Formalin-Fixed Paraffin-Embedded prostate tissues collected from 50 patients suffering from PCa and 55 with benign prostate hyperplasia were subjected to PCR amplification targeting species-specific genes of 5 urogenital mollicutes species, Mycoplasma genitalium, M. hominis, M. fermentans, Ureaplasma parvum, and U. urealyticum. PCR products were then sequenced to confirm species identification. Results significance was statistically assessed using Chi-square and Odds ratio tests.
Results
PCR amplification showed no positive results for M. genitalium, M. hominis, and M. fermentans in all tested patients. Strikingly, Ureaplasma spp. were detected among 30% (15/50) of PCa patients. Nucleotide sequencing further confirmed the identified ureaplasma species, which were distributed as follows: 7 individuals with only U. parvum, 5 with only U. urealyticum, and 3 co-infection cases. Association of the two ureaplasma species with PCa cases proved statistically significant (P < 0.05), and found to represent a risk factor. Of note, Ureaplasma spp. were mostly identified in patients aged 60 and above with prostatic specific antigen (PSA) level > 4 ng/ml and an invasive malignant prostate tumor (Gleason score 8–10).
Conclusions
This study uncovered a significant association of Ureaplasma spp. with PCa arguing in favour of their potential involvement in this condition. Yet, this finding, though statistically supported, warrants a thorough investigation at a much larger scale.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13027-021-00344-9.
Keywords: Prostate cancer, Urogenital mollicutes, PCR, Sequencing
Background
Cancer is a major public health issue, considered as one of the leading causes of death worldwide. According to latest records of the International Agency for Research on Cancer, World Health Organization (WHO), the Prostate Cancer (PCa) ranked fourth among the most common cancers worldwide with 1.28 million cases [1]. The main established risk factors for PCa are increasing age, ethnicity, and family history [2], yet compelling evidence suggests a role for environmental factors and infections [3]. In fact, it was found that chronic inflammation associated with infections could tip the balance towards abnormal cellular growth and proliferation. The mechanisms governing the inflammation process that triggers cancer development are beginning to be investigated. In prostate cancer for example, inflammatory reaction was found to be orchestrated by a multiprotein complex known as inflammasomes. Once activated with an infection and/or another stimulus (cell injury, hormonal variation, dietary or environmental factors …), these inflammatory regulators induce a cascade of proinflammatory signaling molecules capable of modulating different cellular activities. This often results in cell damage and enhances abnormal cell proliferation [4]. Thus, inflammation has been considered as an important carcinogenesis promoting condition for some organs, including prostate [5, 6]. Different types of cancer such as those of colon, stomach, liver, and prostate have been linked to persistent inflammation due to chronic infections [7–9]. Additionnaly, according to the American Cancer Society, about 15 to 20% of cancers worldwide are directly related to infectious agents [10]. For oncoviruses for example, the causal relationship with cancer has been well documented since the early 1980s. Since then, viruses are admitted as bona-fide causes of many human cancer types [11, 12]. Apart from viruses, the link between bacterial infections and the development of malignancy was also well explored. It was suggested that several prokaryotes are able to exhibit tumorigenic properties [13, 14]. For instance, many convincing proofs have attributed Streptococcus bovis to colon cancer [15], Salmonella thyphii to gallbladder cancer [16], Chlamydia pneumoniae to lung cancer [17], Bartonella species to vascular cancer [18], and Helicobacter pylori to gastric cancer [19, 20]. Yet, while the latter association has arguably been established [21], it is still unclear whether these infections are themselves directly involved in tumorogenesis or represent a promoting factor.
Regarding mycoplasmas, the smallest known free-living bacteria, their oncogenic potential and role in cancer development have been investigated as early as the 1960s [22]. Actually, the association between malignancy and human mycoplasmas was initially suggested for the human respiratory pathogen Mycoplasma pneumoniae, which was among the first species suspected to be associated with human leukemia [23]. Since then, many other studies suggesting a possible role for mycoplasmas in malignancy have been reported, but no evidence of causation has been brought forward [24]. However, the potential to cause cancer of other mycoplasma species, such as M. fermentans, M. penetrans, and M. hyorhinis has been demonstrated in vitro [25–27]. Additionally, it was also shown that mycoplasma infection is able to transform embryonic cells in mouse by promoting the expression of some oncogenes like H-ras and c-myc. Prolonged infection with mycoplasma was found to lead to prominent chromosomal alteration (rearrangement, translocation, altered pattern of DNA …), to morphological changes, and to uncontrolled cell growth [28]. For M. hominis, M. genitalium, and U. urealyticum, a potential involvment in PCa has been suggested since these species were detected by PCR in patients suffering from this type of cancer [29–33]. These findings are in line with the ability of these mycoplasma species to establish chronic infections in individuals with prostate cancer as evoked since the mid-twentieth century [34, 35].
A rising incidence of PCa was observed in Asian patients from China, India, and Malaysia [36, 37]. It’s worth mentioning that this type of cancer was ranked second most commonly diagnosed cancer and six leading cause of cancer-related death in men worldwide. Likewise, the situation in the Kingdom of Saudi Arabia (KSA) regarding PCa incidence was considered critical as the statistics were getting worse. Indeed, according to the Saudi Cancer Registry and referring to local literature, an upward trend in PCa incidence, from 2.7% in 1994 to 6.3% in 2008, was reported [38, 39]. Although alarming in the KSA, this serious disease is neither studied enough nor the related data being updated.
Here, we investigated the possible involvment of urogenital mollicutes in PCa in a cohort of Saudi patients from Aseer region. We sought to demonstrate the presence of M. genitalium, M. hominis, M. fermentans, U. urealyticum, and U. parvum by PCR amplification targeting species-specific genes.
Methods
Patients
A total of 105 Saudi patients with inflamed, swelling, and painful prostate gland, referred to Aseer Central Hospital (Abha, southwestern KSA) during the years 2010 to 2017, were enrolled in this work. All patients were presenting suspicious and abnormal digital rectal examination (DRE) findings and/or elevated PSA (Prostate Specific Antigen) level (≥ 4 ng/ml). Patients age ranged from 46 to 104 years. Data about Gleason score and PSA were recorded.
Prostate specimens preparation
Prostate specimens were obtained from patients either by transrectal ultrasound (TRUS)-guided biopsy or by surgery (transurethral resection of the prostate: TURP). Both procedures were carried out by specilized physicians in the Central Hospital of Aseer Region (Abha town, KSA).
TRUS biopsy was performed using an endfiring 7.5-MHz biplane transrectal probe attached to an ultrasonography device (bk 3000 ultrasound systems, Denmark). No local anesthetic injection was used as most of the patients tolerated the procedure very well. However, all of them received a general pain killer (75 mg intramuscular pethidine (demerol) injection pre-procedural statin). Biopsy was performed half an hour after pethidine injection. To prevent patients from post-interventional complications, three oral doses of Ciprofloxacillin (500 mg) were administrated to them before, during, and after the biopsy procedure.
TURP was undertaken by urological surgeons. The procedure was carried out under general anaesthesia using bipolar devices. Gentamicin antibiotic (160 mg) was administrated intravenously to patients 1 h before the TURP surgical procedure.
Histopathological examination
All patients specimens were placed in biocassettes and were dehydrated in histokinette with 10% formalin solution for 24 h. Then, they were embedded in paraffin blocks, sectioned with a microtome into slices of 4 μm thick and stained with hematoxylin-eosin (H & E). Differentiation between the benign and malignant prostatic specimens, as well as the aggressiveness classification of the cancerous tissues according to Gleason score [40] were done by experienced Saudi pathologists using light microscopy. Overall, 50 patients were diagnosed with prostate cancer (Cohort 1). Based on anatomopathological examination and Gleason score attribution, tumors were defined as aggessive (poorly differentiated, Gleason score 8–10) in 38 patients and as non-aggessive (moderately differentiated, Gleason score 6–7) in 12 patients. The other 55 patients were diagnosed with Benign Prostate Hyperplasia (BPH) and used as negative controls (Cohort 2).
Formalin-Fixed Paraffin-Embedded (FFPE) prostate tissues were forwarded to the laboratory of mycoplasmas, Institut Pasteur, Tunis, for further manipulations.
DNA isolation from prostate specimens and from mycoplasma cultures
Total genomic DNA was extracted from sections of FFPE prostate tissue using the QIAamp® DNA FFPE Tissue Kit (Qiagen, Germany) according to the supplier’s instructions. For each biopsy, up to eight sections of 5 to 10 μm thick were cut using a microtome and placed in a sterile 2-ml microcentrifuge tube. Sections of FFPE prostate tissue were subjected to paraffin dissolution using xylene (Honeywell, France), followed by homogenization and enzymatic digestion with proteinase K and lysis buffer. Samples were incubated overnight at 56 °C and lysates were purified using the QIAamp MinElute columns to obtain high quality DNA for PCR.
Using the classical phenol/chloroform DNA extraction method [41], genomic DNA was also extracted from the urogenital mollicutes species, M. hominis PG21 (ATCC 23114), M. genitalium G37 (ATCC 33530), M. fermentans PG18 (ATCC 19989), U. urealyticum (ATCC 27618), and U. parvum (ATCC 27813), in order to be used as controls in the PCR reactions. These mollicutes species were cultivated on appropriate SP4 broth medium, prepared according to the previously described formulation [42]. Antigen of each species was collected and washed by a series of centrifugations at 16000 rpm at 4 °C for 30 min using PBS buffer.
Quantity and quality of DNA were assessed by Nanodrop absorbance measurement and by electrophoresis on 1% agarose gel stained with CYBR safe. The obtained purified DNAs were stored at − 20°С until needed.
Standard PCR assays for screening of urogenital mollicutes DNA in the FFPE samples
For each mollicutes species, PCR assays were designed to amlify 16S rRNA gene along with a specific gene fragment; p120’ gene (coding for DUF885 family protein) for M. hominis, mgpA gene (coding for Adhesin P1 protein) for M. genitalium, the gene conding for the Insertion Sequence like-element (IS1630I) for M. fermentans, and ureB gene (coding for beta subunit of the urease enzyme) for Ureaplasma spp. An internal control specific to the human mitochondrial cytochrome c oxidase subunit III gene (named MT-CO3) was also included. Details about these target genes and their respective primer sequences are presented in Table 1. PCRs were performed in a total volume of 50 μl in a thermal cycler (Applied biosystems, USA model 2720). All PCR mixtures contained 10X Taq buffer, 1 mM MgCl2, 0.2 mM dNTP (Sigma, Germany), 1 pmol of each primer, 1.5 U of HotStarTaq DNA polymerase (Qiagen, Germany), and 1–2 μl (approximately 150 ng) of genomic DNA. All samples were subjected to a first denaturation step at 95°С for 10 min and then to 35 cycles at 94°С for 1 min (denaturation), 55°С for 1 min (annealing), and 72°С for 1 min (elongation). The cycling was ended by an extension step at 72 °C for 10 min. Ten microliters of each amplicon were electrophoresed on a 2% agarose gel stained with CYBR safe and visualized under UV light.
Table 1.
Characteristics of primers used for detection of mollicutes investigated in this study
| Mycoplasma species | Primer designation | Target gene | Gene locus | Product | Amplicon size (bp) | Sequence (5′-3′) | References |
|---|---|---|---|---|---|---|---|
| M. hominis |
Mhp120’F Mhp120’R |
p120’ | MHO_RS02125 | DUF885 family protein | 190 |
GTTTTAGAACGTAAAATTCTA CATCAGCAGGATTGTTTAAAGCT |
This studya |
|
Mh16SF Mh16SR |
rrnA16S | MHO_RS01540 | 16S ribosomal RNA | 164 |
GGTAATGGCCCACCAAGACTAT GAAGACCTTCATCGTGCACGCTG |
This studya | |
| M. genitalium |
MgPa335F MgPa432R |
mgpA | MG_RS01075 | Adhesin protein | 78 |
GAGAAATACCTTGATGGTCAGCAA GTTAATATCATATAAAGCTCTACCGTTGTTATC |
[43] |
|
Mg16SF MG16SR |
rrnA16S | MG_RS00775 | 16S ribosomal RNA | 195 |
TAGCTAATACCGCATAAGAACTTT CTCCCGTAGGAGTATGGGCCGTGT |
This studya | |
| M. fermentans |
RW 005 RW 004 |
IS1630I | MBIO_RS03870 | IS30-like element | 205 |
GGTTATTCGATTTCTAAATCGCCT GGACTATTGTCTAAACAATTTCCC |
[44] |
|
Mf16SF Mf16SR |
rrnA16S | MBIO_RS4090 | 16S ribosomal RNA | 186 |
TTGACGGTACCTTATTAGAAA TCTACACAACTCTAGCCTG |
This studya | |
| U. urealyticum |
UUureF UUureR |
ureB | UUR8_RS02385 | Urease subunit beta | 100 |
GATCACATTTCCACTTATTTGAAACA AAACGACGTCCATAAGCAACTTTA |
[45] |
|
Uu16SF Uu16SR |
rrnA16S | UUR8_RS00640 | 16S ribosomal RNA | 181 |
TGCCTGGGTAGTACATTCGCAAGAA GACAACCATGCACCACCTGTC |
This studya | |
| U. parvum |
UPureF UPureF |
ureB | UPA3_RS02275 | Urease subunit beta | 99 |
GATCACATTTTCACTTGTTTG AAGTG AACGTCGTCCATAAGCAACTTTG |
[45] |
|
Up16SF Up16SF |
rrnA16S | UPA3_RS00595 | 16S ribosomal RNA | 183 |
CTTTTATATGGGAAGAAACGCT GCATCTAGATTTAATACC |
This studya | |
| Human DNA |
HmitoDNAF HmitoDNAR |
MT-CO3 | YP_003024032.1 | Cytochrome c oxidase subunit III | 208 |
GGATAATCCTATTTATTACCTCAGAAG AGACTATGGTGAGCTCAGGTGATTGAT |
This studya |
aPrimers for each gene were designed based on sequences available in GenBank
Sequencing and bioinformatics analysis
PCR products were cleaned-up using the ExoSAP protocol. For this purpose, exonuclease I (Biolabs, England) and shrimp alkaline phosphatase (Biolabs, England) enzymes were used to remove leftover primers and remaining dNTPs, respectively. Sequencing was performed using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, USA), according to the manufacturer’s instructions. Sequencing products were run in Applied Biosystems® 3130 Genetic Analyzer (USA) available at the sequencing facility of Institut Pasteur, Tunis. For each PCR amplicon, both strands were sequenced. The derived nucleotide sequences were aligned with reference sequences downloaded from GenBank using the BioEdit Sequence Alignment Editor, version 7.2.5.
Statistics
Statistical analyses were performed using Excel tools (Microsoft, USA) to assess the prevalence of mollicutes species among the studied population. The chi-square test was computed to evaluate the potential relationship between the urogenital mollicutes infections and PCa. P value < 0.05 was considered statistically significant. Odds ratio with a 95% confidence interval was also calculated to estimate the role of mollicutes as a risk factor in PCa disease. Odds ratio values > 1 indicated that mollicutes infections might be a risk factor for the PCa. All these tests were performed using the Statistical Package for Social Science (SPSS) 20.0 software (IBM Corp. Armonk, NY, USA).
Results
The mean age of Saudi men included in this study was 76.08 years for those suffering from PCa and 72.18 years for those diagnosed with BPH. Within both groups, patients aged between 70 and 79 years were the most frequent (46 and 40%, respectively). In both groups, the least common patients were those aged under 60, particularly in the PCa category with only 2 cases (4%). Regarding PSA records, levels were > 4 ng/ml in 92% (48/50) of patients with PCa and 70% (38/55) of patients displaying BPH. Interestingly, high PSA levels exceeding 20 ng/ml were recorded for a large number of PCa patients (62%), whereas only 13% amongst BPH cases showed such high levels. After anatomopathology examination, Gleason score attribution was only applied to patients suffering from PCa. Patients with aggressive PCa were found to be nearly three times more frequent than those with non-aggressive PCa (76% versus 24%, respectively). The characteristics of the study population are summarized in Table 2.
Table 2.
Characteristics of the studied population
| PCa (n = 50) | BPH (n = 55) | |||
|---|---|---|---|---|
| Age (years) | ||||
| < 60 | 2 | (4%) | 5 | (9%) |
| 60–69 | 9 | (18%) | 17 | (31%) |
| 70–79 | 23 | (46%) | 22 | (40%) |
| > 79 | 16 | (32%) | 11 | (20%) |
| PSA level (ng/ml) | ||||
| 0–4 | 4 | (8%) | 17 | (31%) |
| 4.1–10 | 7 | (14%) | 17 | (31%) |
| 10.1–20 | 8 | (16%) | 14 | (25%) |
| > 20 | 31 | (62%) | 7 | (13%) |
| Gleason score | ||||
| 6–7 | 12 | (24%) | – | – |
| 8–10 | 38 | (76%) | – | – |
PCa Prostate Cancer, BPH Benign Prostate Hyperplasia, PSA Prostate Specific Antigen
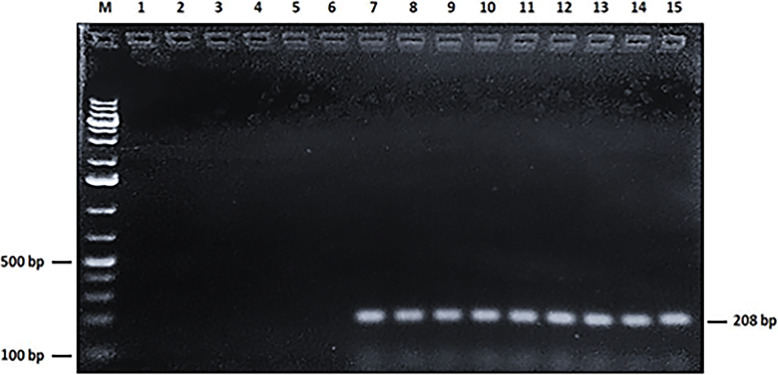
Electrophoresis and Nanodrop quantification have shown that genomic DNA extracted from all mollicutes reference strains as well as DNA collected from all FFPE human samples were of high quality and quantity (DNA concentration ranged from 100 to 1750 ng/μl). This has been confirmed by the efficient amplification of a 208-bp fragment of the human mitochondrial DNA. Indeed, all the 105 samples (100%) produced this targeted fragment at its expected size (Fig. 1).
Fig. 1.
PCR amplification of the 208-bp fragment of human mitochondrial DNA (internal positive control). Lane M: 1Kb + DNA ladder. Lane 1–5: Specificity of the human mitochondrial DNA primers with genomic DNA from M. genitalium, M. hominis, M. fermentans, U. parvum, and U. urealyticum, respectively. Lane 6: No DNA (negative control). Lane 7: Human DNA (positive control). Lane 8–15: DNA from prostate biopsies
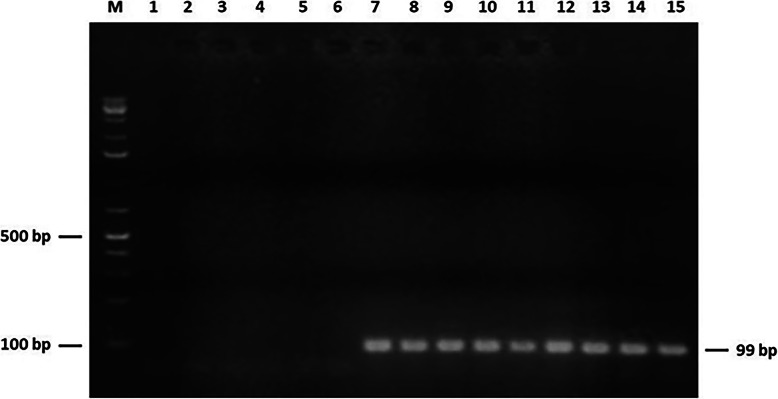
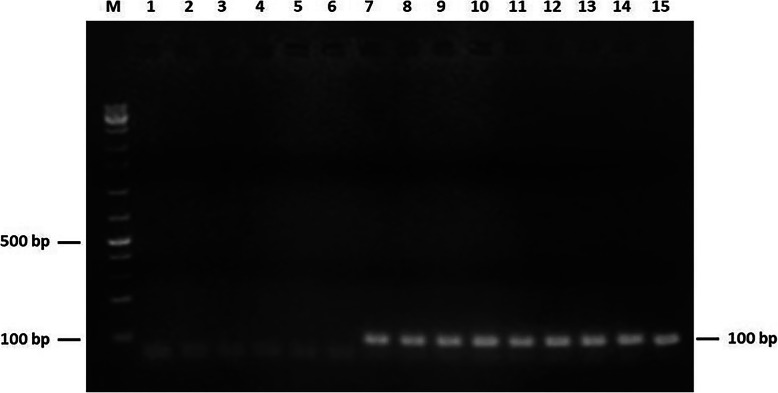
None of the 105 prostate samples was found to contain DNA of mycoplasma species. Indeed, all PCR amplifications targeting 16S rRNA and specific genes of M. genitalium, M. hominis, and M. fermentans were negative (Data not shown). Interestingly, PCR-based detection of ureaplasma species was positive in 18 cases. The biotyping of Ureaplasma spp. revealed U. parvum and U. urealyticum in ten (20%) and eigth (16%) samples, respectively. The 99-bp and 100-bp PCR products corresponding to the amplification of U. parvum and U. urealyticum specific urease coding genes are shown in Figs. 2 and 3, respectively. This result was further confirmed by positive amplification of the targeted segments within 16S rRNA gene in Ureaplasma spp. (Figures S1 and S2). Both forward and reverse strands of these PCR amplicons were sequenced. Alignement of the derived nucleotide sequences with corresponding Ureaplasma spp. reference sequences showed a 100% identity and thus confirmed species identification. Of note, these ureaplasma-positive PCR were only observed in prostate samples of patients diagnosed with PCa. Chi-square test was computed for each ureaplasma species to assess the significance of its potential association with PCa. PCR results relative to these two species in both cohorts group patients were taken into account. Both calculated P values proved statistically significant (P = 0.0005 for U. parvum and P = 0.002 for U. urealyticum). The Odds Ratio values were found to be infinite, suggesting that infection with each of ureaplasma species could constitute a risk factor in PCa disease. Results of mollicutes detection in archival tissues are summed up in Table 3.
Fig. 2.
PCR detection of the 99-bp fragment of Ureaplasma parvum ureB gene. Lane M: 1 kb + DNA ladder. Lanes 1–5: Specificity of primers of U. parvum ureB gene with DNA from human mitochondrion, M. hominis, M. fermentans, M. genitalium, and U. urealyticum. Lane 6: No DNA (negative control). Lane 7: DNA from U. parvum type strain ATCC 27845 (positive control). Lanes 8–15: DNA from prostate biopsies of patients (PCa cohort)
Fig. 3.
PCR amplification of the 100-bp fragment of Ureaplasma urealyticum ureB gene. Lane M: 1 kb + DNA ladder. Lanes 1–5: Specificity of primers of U. urealyticum ureB gene with DNA from human mitochondrion, M. hominis, M. fermentans, M. genitalium, and U. parvum. Lane 6: No DNA (negative control). Lane 7: DNA from U. urealyticum type strain ATCC 27618 (positive control). Lanes 8–15: DNA from prostate biopsies of patients (PCa cohort)
Table 3.
Prevalence of urogenital Mollicutes identified in biopsies of prostate with malignant or benign cancer
| Target organism | Overall prevalence | P value | |||
|---|---|---|---|---|---|
| Malignant tumor tissues | Benign tumor tissues | ||||
| n = 50 | % | n = 55 | % | ||
| Ureaplasma parvum | 10 | 20 | 0 | 0 | 0.0005 |
| Ureaplasma urealyticum | 8 | 16 | 0 | 0 | 0.002 |
| Mycoplasma hominis | 0 | 0 | 0 | 0 | NA |
| Mycoplasma genitalium | 0 | 0 | 0 | 0 | NA |
| Mycoplasma fermentans | 0 | 0 | 0 | 0 | NA |
NA Not applicable
The 18 Ureaplasma spp. positive PCR were detected in 15 patients with PCa. These positive cases were distributed as follows; 7 patients were found to be infected with only U. parvum, 5 with only U. urealyticum, and 3 were co-infected with both species. Based on their Gleason score, tumors of most of the PCa patients infected with U. urealyticum are categorized as aggressive (7 out 8 patients; 87.5%). For those infected with U. parvum, the rate of aggressive tumors was also high (7 out of 10 patients; 70%).
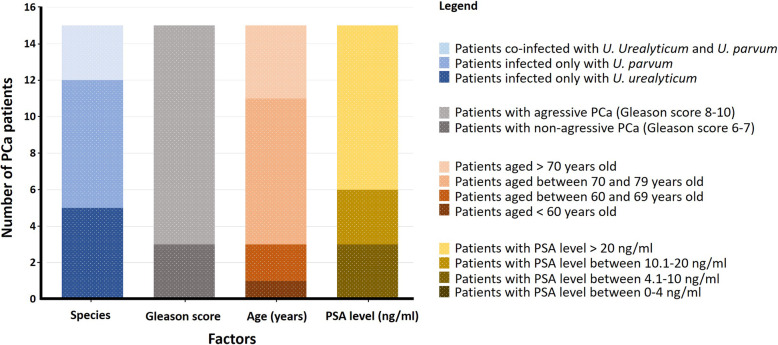
Considering both ureaplasma species and taken into account the three cases of co-infection, statistics showed that among the 15 patients in which Ureaplasma spp. were detected, 12 were diagnosed with an aggressive PCa (Gleason score between 8 and 10), which represents 80% (12/15). These 15 patients were found to belong to different age intervals. Aside from one PCa patient, all the others (14/15; 93.33%) are aged more than 60 years. Moreover, the majority of these patients (9/15; 60%) showed high PSA levels, exceeding 20 ng/ml. The caracterization based on different parameters (Species, Gleason score, age, and PSA level) of the group of patients diagnosed with PCa and found to harbor Ureaplasma spp. DNA is presented in Fig. 4.
Fig. 4.
Distribution of the 15 patients diagnosed with PCa and found to be infected with Ureaplasma species (Y-axis) on the basis of various factors (X-axis)
Discussion
Because of its wide-ranging impact on the human health and social life, cancer has prompted scientific research to delve into its causes and risk factors. Millions of new cases and deaths are recorded worlwide every year [46]. Several studies on determinants of cancer’s origins have led to the suspicion that chronic infections are involved in its establishment or progression [47, 48]. In the begining, this assumption was always contreversial given that cancer is usually assumed to have a genetic background. Over the last few decades, the biology and mechanisms governing the cancer-infectious agents relationship are beginning to be unraveled. One of these mechanisms is the infection-related inflammation. For example, the activation of the nuclear factor kappaB (NF-κB) inflammatory pathway, involved in the maintenance of the cell cycle stability, was associated to cancer. This activation could inhibit apoptosis and promote cell transformation. This was reported for the bacterium Helicobacter pylori in the case of gastric cancer [49] as well as for many oncoviruses like hepatitis virus [50].
While the role of viruses in carcinogenesis has long been established, the possible involvement of bacteria is beginning to gain more and more acceptance [51–57]. With regard to prostate cancer, it was suggested that sexually transmitted infections could be a risk factor for its developement [58]. Recent advances in techniques of molecular microbiology have allowed the identification of many microorganisms in prostate cancer tissues, such as the sexually transmitted bacteria Chlamydia trachomatis and Trichomonas vaginalis, the prostatitis-associated Escherichia coli and Pseudomonas, human papilloma virus, and the pro-inflammatory bacteria Propionibacterium acnes [58]. However, as yet, no association between PCa and infection with any pathogen could firmly be established [59, 60]. Very recently, a link has been evoked between prostate cancer and the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, causing the current 2020 pandemic COVID-19. A gene named transmembrane serine protease 2 (TMPRSS2) for which alteration was historically associated with prostate tumorigenesis was recently confirmed to be involved in facilitating the SARS-CoV-2 entry into host cells. Yet, it is premature to speculate about any involvement of SARS-CoV-2 in PCa [61, 62].
In this study, we have searched for the presence of a panoply of urogenital mollicutes species (M. hominis, M. genitalium, M. fermentans, U. urealyticum, and U. parvum) in prostate tissues from KSA patients with PCa or BPH, using PCR coupled to nucleotide sequencing. In KSA, no studies have been undertaken so far dealing with the etiopathogenesis of PCa. The only available data are about the incidence and mortality rates related to this cancer [63]. Epidemiologic data relating to the time period 2001–2008 have reported a steady increase in PCa incidence and mortality rates [39], however, since then, the situation has not been updated. This further highlights the importance of PCa disease in KSA and prompts the investigation of a potential involvement of urogenital mollicutes in the progression of this cancer.
PCR results showed no amplification of M. genitalium, M. fermentans, or M. hominis DNAs from PCa tissues of Saudi patients. Such a result could be subject to variation, since in Iran the same study confirmed the absence of M. genitalium in PCa [30], while in Australia and Japan, M. genitalium was identified with rates of 8% [31] and 40% [29], respectively. M. hominis was detected at a rate of 15% in Russian men suffering from PCa [33], but not in Japanease patients [29]. Regarding M. fermentans species, it was suspected in the prostate carcinogenesis in Turkey, but no precise results about its prevalence were communicated [32].
A total of 15 Saudi patients tested were found to harbor Ureaplasma spp. DNA. All of them were diagnosed with PCa. Twelve patients were infected with only one Urealplasma species while the 3 others were co-infected with both Ureaplasma species simultaneously. U. urealyticum was detected in 8 among the 50 PCa patients (16%). This rate is the highest among the reported cases. Indeed, only one case of positive U. urealyticum PCR was reported in each of the Australian and the Iranian studies already mentioned above [30, 31]. In this study, U. parvum was identified at a rate of 20% (10/50). This is the first study that reports the detection of this species in patients with PCa. Furthermore, the coinfection with U. urealyticum and U. parvum of the same patient with PCa is a new finding reported for the first time by our study. However, this result remains to be consolidated. Apart from some new clues unveiled by our study, we found some similarities with other studies in terms of species identification and rates. This could explain the role played by several factors such as hereditary susceptibility, environmental factors, or even the methods used to identify Mycoplasma species and their genes [64]. The fact that we have detected Ureaplasma spp. only in patients suffering from PCa lends further support to the hypothesis suggesting their involvement in the etiology of this type of cancer.
In the case of our study, though the results are statistically supported, one can not formally conclude that the presence of Ureaplasma spp. is the primary cause of prostate carcinogenesis. However, according to the Gleason score values (8–10), the presence of Ureaplasma spp. could have contributed to the worsening of cancer progression. Indeed, patients with PCa harbouring ureaplasmas displayed invasive malignant tumours of the prostate. These findings are recorded in patients over 60 years old and with PSA values > 4 ng/ml. Like viruses, mycoplasmas are notoriously known for their ability to cause low-grade chronic inflammation and silent infection, a hallmark thought to gradually compromise the immune system of their hosts, thereby promoting cancer transformation. Nevertheless, their detection in tumour microenvironment does not imply that they are causally involved in carcinogenesis. Actually, many factors like the high prevalence of the infectious agent in the general population, the extended latency, the asymptomatic infections, and the immune system status of patients could make difficult establishing a causality link [9]. Also, one can not ignore the idea that detection of mycoplasma (or any other microorganism) in cancerous tissues could be a consequence of the compromised immune response in tested patients since cancer is known for its association with a wide range of immunological disorders. Indeed, it was suggested that viruses could be activated as consequence of carcinogenesis rather than being a cause of it, and that their carcinogenic potential could be related to the dramatically compromised immune system [9].
To sum up, the role of microbes in cancer is still a subject of debate and it is obvious that all these results further lead us to provide more evidence of the role that mollicutes can play in prostate cancer. Appropriate animal models and in vitro cell cultures could be very useful to achieve this goal.
Conclusions
To our knowledge, this study is the first to be conducted in KSA aiming to characterize the PCa from an etiological standpoint. The finding that only patients with PCa were significantly associated with Ureaplasma spp. infection is in favor of their potential involvement in prostate cancer or, most likely, points to their contribution in worsening its progression. These data are worth to be considered by the Saudi health policy makers during decision-making processes regarding the control and follow-up of PCa epidemiology in KSA.
Supplementary Information
Additional file 1: Figure S1. PCR amplification of Ureaplasma parvum 16S rRNA gene. Lane M: 1 kb + DNA ladder. Lanes 1–5: Specificity of primers of U. parvum 16S rRNA gene with DNA from human mitochondrion, M. hominis, M. fermentans, M. genitalium, and U. urealyticum. Lane 6: No DNA (negative control). Lane 7: DNA from U. parvum type strain ATCC 27845 (positive control). Lanes 8–15: DNA from prostate biopsies of patients (PCa cohort).
Additional file 2: Figure S2. PCR detection of Ureaplasma urealyticum 16S rRNA gene. Lane M: 1 kb + DNA ladder. Lanes 1–5: Specificity of primers of U. urealyticum 16S rRNA gene with DNA from human mitochondrion, M. hominis, M. fermentans, M. genitalium, and U. parvum. Lane 6: No DNA (negartive control). Lane 7: DNA from U. urealyticum type strain ATCC 27618 (positive control). Lanes 8–15: DNA from prostate biopsies of patients (PCa cohort).
Acknowledgements
The authors thank the Deanship of Scientific Research at King Khalid University, Kingdom of Saudi Arabia) for financial support of this work.
Abbreviations
- BPH
Benign Prostate Hyperplasia
- DRE
Digital Rectal Examination
- ExoSAP
Exonuclease Shrimp Alkaline Phosphatase
- FFPE
Formalin-Fixed Paraffin-Embedded
- KSA
Kingdom of Saudi Arabia
- PCa
Prostate Cancer
- PSA
Prostate Specific Antigen
- TRUS
TransRectal UltaSound
- TURP
TransUrethral Resection of the Prostate
- WHO
World Health Organization
Authors’ contributions
Conceived and designed the experiments: BBAM, EY, MHAS, and OMSAW. Performed the experiments: EY and NS. Analyzed the data: EY and BBAM. Contributed reagents/materials/analysis tools: OMSAW, MHAS, BBAM, NS, MSSAF, HH, AHAM, BM, and EY. Wrote the paper: EY and BBAM. All authors read and approved the final manuscript.
Funding
This study was supported by the Deanship of Scientific Research at King Khalid University (Abha town, Kingdom of Saudi Arabia) through General Research Project [grant number G.R.P1/39/39].
Availability of data and materials
All data generated and analysed during this study are included in this published article and its supplementary information files. Materials are available upon reasonable request.
Ethics approval and consent to participate
This study is about a collaborative project between King Khalid University in KSA and Institut Pasteur de Tunis in Tunisia. This study has been peer-approved by the Internal Review Board Committee of Aseer Central Hospital, Academic Affairs for Training and Research (Abha, Saudi Arabia) (26-12-2017) and by the Biomedical Ethical Committee of Institut Pasteur de Tunis (Tunis, Tunisia) (2019/5/I/LR16IPT).
Consent for publication
Before the collection of biopsy, all patients were informed about the procedures they would undergo and their possible complications. Medical charts were retrieved from the medical records of patients.
Competing interests
All the authors declare that there are no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO. 2018. https://www.who.int/news-room/fact-sheets/detail/cancer. Accessed 30 Oct 2020.
- 2.Gann PH. Risk factors for prostate cancer. Rev Urol. 2002;4(Suppl 5):S3–S10. [PMC free article] [PubMed] [Google Scholar]
- 3.Schmid HP, Engeler DS, Pummer K, Schmitz-Dräger BJ. Prevention of prostate cancer: more questions than data. Recent Results in Cancer Res. 2014;174:101–107. doi: 10.1007/978-3-540-37696-5_10. [DOI] [PubMed] [Google Scholar]
- 4.Kolb R, Liu G-H, Janowski AM, Sutterwala FS, Zhang W. Inflammasomes in cancer: a double-edged sword. Protein Cell. 2014;5(1):12–20. doi: 10.1007/s13238-013-0001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Marzo AM, Platz EA, Sutcliffe S, Xu J, Gronberg H, Drake CG, Nagai Y, Isaacs WB, Nelson WG. Inflammation in prostate carcinogenesis. Nat Rev Cancer. 2007;7:256–269. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karan D, Dubey S. From inflammation to prostate cancer: the role of inflammasomes. Adv Urol. 2016;31(5):1–5. doi: 10.1155/2016/3140372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson WG, De Marzo AM, DeWeese TL, et al. The role of inflammation in the pathogenesis of prostate cancer. J Urol. 2004;172(5):S6–S12. doi: 10.1097/01.ju.0000142058.99614.ff. [DOI] [PubMed] [Google Scholar]
- 9.Kuper H, Adami HO, Trichopoulos D. Infections as a major preventable cause of human cancer. J Intern Med. 2000;248(3):171–183. doi: 10.1046/j.1365-2796.2000.00742.x. [DOI] [PubMed] [Google Scholar]
- 10.Can infections cause cancer? https://www.cancer.org/cancer/cancer-causes/infectious-agents/infections-that-can-lead-to-cancer/intro.html. Accessed 22 Oct 2020.
- 11.zur Hausen H. Viruses in human cancers. Science. 1991;254(5035):1167–1173. doi: 10.1126/science.1659743. [DOI] [PubMed] [Google Scholar]
- 12.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 13.Vogelmann R, Amieva MR. The role of bacterial pathogens in cancer. Curr Opin Microbio. 2007;10:76–81. doi: 10.1016/j.mib.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Mager D. Bacteria and cancer: cause, coincidence or cure? A review. J Transl Med. 2006;4:14. doi: 10.1186/1479-5876-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellmerich S, Scholler M, Duranton B, Gosse F, Galluser M, Klein JP, Raul F. Promotion of intestinal carcinogenesis by Streptococcus bovis. Carcinogenesis. 2000;21:753–756. doi: 10.1093/carcin/21.4.753. [DOI] [PubMed] [Google Scholar]
- 16.Lazcano-Ponce EC, Miquel JF, Munoz N, Herrero R, Ferrecio C, Wistuba II, Alonso de Ruiz P, Aristi UG, Nervi F. Epidemiology and molecular pathology of gallbladder cancer. CA Cancer J Clin. 2001;51:349–364. doi: 10.3322/canjclin.51.6.349. [DOI] [PubMed] [Google Scholar]
- 17.Littman AJ, Jackson LA, Vaughan TL. Chlamydia pneumoniae and lung cancer: epidemiologic evidence. Cancer Epidemiol Biomark Prev. 2005;14:773–778. doi: 10.1158/1055-9965.EPI-04-0599. [DOI] [PubMed] [Google Scholar]
- 18.Dehio C. Bartonella-host-cell interactions and vascular tumour formation. Nat Rev Microbiol. 2005;3:621–631. doi: 10.1038/nrmicro1209. [DOI] [PubMed] [Google Scholar]
- 19.zur Hausen H. The search for infectious causes of human cancers: where and why. Virolo. 2009;392:1–10. doi: 10.1016/j.virol.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Touati E. When bacteria become mutagenic and carcinogenic: lessons from H. pylori. Mutat Res. 2010;703:66–70. doi: 10.1016/j.mrgentox.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 21.Marshall BJ, Windsor HM. The relation of Helicobacter pylori to gastric adenocarcinoma and lymphoma: pathophysiology, epidemiology, screening, clinical presentation, treatment, and prevention. Med Clin North Am. 2005;89:313–344. doi: 10.1016/j.mcna.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Fallon RJ, Grist NR, Inman DR, Lemcke RM, Negroni G, Woods DA. Further studies of agents isolated from tissue cultures inoculated with human leukaemic bone-marrow. Br Med J. 1965;2(5458):388–391. doi: 10.1136/bmj.2.5458.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayflick L, Koprowski H. Direct agar isolation of mycoplasmas from human leukaemic bone marrow. Nature. 1965;205:713–714. doi: 10.1038/205713b0. [DOI] [PubMed] [Google Scholar]
- 24.Cimolai N. Do mycoplasmas cause human cancer? Can J Microbiol. 2001;47:691–697. doi: 10.1139/w01-053. [DOI] [PubMed] [Google Scholar]
- 25.Zhang S, Tsai S, Wu TT, Li B, Shih JWK, Lo SC. Mycoplasma fermentans infection promotes immortalization of human peripheral blood mononuclear cells in culture. Blood. 2004;104:4252–4259. doi: 10.1182/blood-2004-04-1245. [DOI] [PubMed] [Google Scholar]
- 26.Tsai S, Wear DJ, Shih JW, Lo SC. Mycoplasmas and oncogenesis: persistent infection and multistage malignant transformation. Proc Natl Acad Sci U S A. 1995;92:10197–10201. doi: 10.1073/pnas.92.22.10197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidhauser C, Dudler R, Schmidt T. A mycoplasmal protein influences tumour cell invasiveness and contact inhibition in vitro. J Cell Sci. 1990;95:499–506. doi: 10.1242/jcs.95.3.499. [DOI] [PubMed] [Google Scholar]
- 28.Zhang B, Shih JW, Wear DJ, Tsai S, Lo SC. High-level expression of H-ras and c-myc oncogenes in mycoplasma-mediated malignant cell transformation. Proc Soc Exp Biol Med. 1997;214:359–366. doi: 10.3181/00379727-214-44104. [DOI] [PubMed] [Google Scholar]
- 29.Miyake M, Ohnishi K, Hori S, Nakano A, Nakano R, Yano H, et al. Mycoplasma genitalium infection and chronic inflammation in human prostate cancer: detection using prostatectomy and needle biopsy specimens. Cells. 2019;8:212. doi: 10.3390/cells8030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eslami G, Goudarzi H, Baseri N, Ghalavand Z, Taherpour A, Zhaam H, et al. The prevalence of Ureaplasma urealyticum and Mycoplasma genitalium in patients with prostate cancer in Shohada Hospital in Tehran, Iran. Novel Biomed. 2015;3(2):73–78. [Google Scholar]
- 31.Yow MA, Tabrizi SN, Severi G, Bolton DM, Pedersen J, Longano A, et al. Detection of infectious organisms in archival prostate cancer tissues. BMC Cancer. 2014;14:579. doi: 10.1186/1471-2407-14-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erturhan SM, Bayrak O, Pehlivan S, Ozgul H, Seckiner I, Sever T, et al. Can mycoplasma contribute to formation of prostate cancer? Int Urol Nephrol. 2013;45(1):33–38. doi: 10.1007/s11255-012-0299-5. [DOI] [PubMed] [Google Scholar]
- 33.Barykova YA, Logunov DY, Shmarov MM, Vinarov AZ, Fiev DN, Vinarova NA, et al. Association of Mycoplasma hominis infection with prostate cancer. Oncotarget. 2011;2(4):289–297. doi: 10.18632/oncotarget.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heshmat MY, Kovi J, Herson J, Jones GW, Jackson MA. Epidemiologic association between gonorrhea and prostatic carcinoma. Uro. 1975;6:457–460. doi: 10.1016/0090-4295(75)90627-5. [DOI] [PubMed] [Google Scholar]
- 35.Steele R, Lees REM, Kraus AS, Chandar R. Sexual factors in the epidemiology of cancer of the prostate. J Chronic Dis. 1971;24(1):29–37. doi: 10.1016/0021-9681(71)90056-7. [DOI] [PubMed] [Google Scholar]
- 36.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun M. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 37.Ito K. Prostate cancer in Asian men. Nat Rev Urol. 2014;11(4):197–212. doi: 10.1038/nrurol.2014.42. [DOI] [PubMed] [Google Scholar]
- 38.Mosli HA, Abdel-Meguid TA, Al-Maghrabi JA, Kamal WK, Saadah HA, Farsi HM. The clinicopathologic patterns of prostatic diseases and prostate cancer in Saudi patients. Saudi Med J. 2009;30(11):1439–1443. [PubMed] [Google Scholar]
- 39.Alghamidi IG, Hussain II, Alghamdi MS, El-Sheemy MA. The incidence rate of prostate cancer in Saudi Arabia: an observational descriptive epidemiological analysis of data from the Saudi Cancer Registry 2001-2008. Hematol Oncol Stem Cell Ther. 2014;7(1):18–26. doi: 10.1016/j.hemonc.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Gleason DF. Histologic grading of prostate cancer: a perspective. Hum Pathol. 1992;23:273–279. doi: 10.1016/0046-8177(92)90108-F. [DOI] [PubMed] [Google Scholar]
- 41.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 42.Tully JG, Whitcomb RF, Clark HF, Williamson DL. Pathogenic mycoplasmas: cultivation and vertebrate pathogenicity of a new spiroplasma. Science. 1977;195:892–894. doi: 10.1126/science.841314. [DOI] [PubMed] [Google Scholar]
- 43.Jensen JS, Björnelius E, Dohn B, Lidbrink P. Use of TaqMan 5′ nuclease real-time PCR for quantitative detection of Mycoplasma genitalium DNA in males with and without urethritis who were attendees at a sexually transmitted disease clinic. J Clin Microbiol. 2004;42:683–692. doi: 10.1128/JCM.42.2.683-692.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang RYH, HU WS, Dawson MS, et al. Selective detection of Mycoplasma fermentans by polymerase chain reaction and by using a nucleotide sequence within the insertion sequence-like element. J Clin Microbiol. 1992;1(30):245–248. doi: 10.1128/JCM.30.1.245-248.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mallard K, Schopfer K, Bodmer T. Development of real-time PCR for the differential detection and quantification of Ureaplasma urealyticum and Ureaplasma parvum. J Microbiol Methods. 2005;60:13–19. doi: 10.1016/j.mimet.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 46.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics. 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 47.De Flora S, La Maestra S. Epidemiology of cancers of infectious origin and prevention strategies. J prev med hyg. 2015;56:E15–E20. [PMC free article] [PubMed] [Google Scholar]
- 48.De Flora S, Bonanni P. The prevention of infection-associated cancers. Carcinogenesis. 2011;32(6):787–795. doi: 10.1093/carcin/bgr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takayama S, Takahashi H, Matsuo Y, Okada Y, Manabe T. Effects of Helicobacter pylori infection on human pancreatic cancer cell line. Hepatogastroenterology. 2007;54(80):2387–2391. [PubMed] [Google Scholar]
- 50.Guan YS, He Q, Wang MQ, Li P. Nuclear factor kappa B and hepatitis viruses. Expert Opin Ther Targets. 2008;12(3):265–280. doi: 10.1517/14728222.12.3.265. [DOI] [PubMed] [Google Scholar]
- 51.Koyi H, Branden E, Gnarpe J, Gnarpe H, Steen B. An association between chronic infection with Chlamydia pneumoniae and lung cancer. A prospective 2-year study. APMIS. 2001;109:572–580. doi: 10.1034/j.1600-0463.2001.d01-177.x. [DOI] [PubMed] [Google Scholar]
- 52.Klein RS, Catalano MT, Edberg SC, Casey JI, Steigbigel NH. Streptococcus bovis septicemia and carcinoma of the colon. Ann Intern Med. 1979;91:560–562. doi: 10.7326/0003-4819-91-4-560. [DOI] [PubMed] [Google Scholar]
- 53.Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, Sibley RK. Helicobacter pylori infection and the risk of gastric carcinoma. New Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 54.Nath G, Singh YK, Kumar K, Gulati AK, Shukla VK, Khanna AK, et al. Association of carcinoma of the gallbladder with typhoid carriage in a typhoid endemic area using nested PCR. J Infect Dev Ctries. 2008;2(4):302–307. doi: 10.3855/jidc.226. [DOI] [PubMed] [Google Scholar]
- 55.Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, et al. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30(Suppl 5):F55–F70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 56.Yang P, Markowitz GJ, Wang X-F. The hepatitis B virus-associated tumor microenvironment in hepatocellular carcinoma. Natl Sci Rev. 2014;1(3):396–412. doi: 10.1093/nsr/nwu038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Coghill AE, Hildesheim A. Epstein-Barr virus antibodies and the risk of associated malignancies: review of the literature. Am J Epidemiol. 2014;180(7):687–695. doi: 10.1093/aje/kwu176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sutcliffe S. Sexually transmitted infections and risk of prostate cancer: review of historical and emerging hypotheses. Future Oncol. 2010;6(8):1289–1311. doi: 10.2217/fon.10.95. [DOI] [PubMed] [Google Scholar]
- 59.Huang W-Y, Hayes R, Pfeiffer R, Viscidi RP, Lee FK, Wang YF, et al. Sexually transmissible infections and prostate cancer risk. Cancer Epidemiol Biomark Prev. 2008;17(9):2374–2381. doi: 10.1158/1055-9965.EPI-08-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sutcliffe S, Giovannucci E, Gaydos CA, Viscidi RP, Jenkins FJ, Zenilman JM, et al. Plasma antibodies against Chlamydia trachomatis, human papillomavirus, and human herpesvirus type 8 in relation to prostate cancer: a prospective study. Cancer Epidemiol Biomark Prev. 2007;16(8):1573–1580. doi: 10.1158/1055-9965.EPI-07-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mollica V, Rizzo A, Massari F. The pivotal role of TMPRSS2 in coronavirus disease 2019 and prostate cancer. Future Oncol. 2020. 10.2217/fon-2020-0571. [DOI] [PMC free article] [PubMed]
- 62.Bhowmick NA, Oft J, Dorff T, Pal S, Agarwal N, Figlin RA, et al. COVID-19 and androgen-targeted therapy for prostate cancer patients. Endocr Relat Cancer. 2020;27(9):R281–R292. doi: 10.1530/ERC-20-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mosli HA. Prostate cancer in Saudi Arabia in 2002. Saudi Med J. 2003;24(6):573–581. [PubMed] [Google Scholar]
- 64.Sfanos KS, Isaacs JT. The “infectious” nature of human prostate cancer: a cautionary note. Oncotarget. 2011;2(4):281–283. doi: 10.18632/oncotarget.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. PCR amplification of Ureaplasma parvum 16S rRNA gene. Lane M: 1 kb + DNA ladder. Lanes 1–5: Specificity of primers of U. parvum 16S rRNA gene with DNA from human mitochondrion, M. hominis, M. fermentans, M. genitalium, and U. urealyticum. Lane 6: No DNA (negative control). Lane 7: DNA from U. parvum type strain ATCC 27845 (positive control). Lanes 8–15: DNA from prostate biopsies of patients (PCa cohort).
Additional file 2: Figure S2. PCR detection of Ureaplasma urealyticum 16S rRNA gene. Lane M: 1 kb + DNA ladder. Lanes 1–5: Specificity of primers of U. urealyticum 16S rRNA gene with DNA from human mitochondrion, M. hominis, M. fermentans, M. genitalium, and U. parvum. Lane 6: No DNA (negartive control). Lane 7: DNA from U. urealyticum type strain ATCC 27618 (positive control). Lanes 8–15: DNA from prostate biopsies of patients (PCa cohort).
Data Availability Statement
All data generated and analysed during this study are included in this published article and its supplementary information files. Materials are available upon reasonable request.