Abstract
Panax notoginseng, a perennial herb of the genus Panax in the family Araliaceae, has played an important role in clinical treatment in China for thousands of years because of its extensive pharmacological effects. Here, we report a high-quality reference genome of P. notoginseng, with a genome size up to 2.66 Gb and a contig N50 of 1.12 Mb, produced with third-generation PacBio sequencing technology. This is the first chromosome-level genome assembly for the genus Panax. Through genome evolution analysis, we explored phylogenetic and whole-genome duplication events and examined their impact on saponin biosynthesis. We performed a detailed transcriptional analysis of P. notoginseng and explored gene-level mechanisms that regulate the formation of characteristic tubercles. Next, we studied the biosynthesis and regulation of saponins at temporal and spatial levels. We combined multi-omics data to identify genes that encode key enzymes in the P. notoginseng terpenoid biosynthetic pathway. Finally, we identified five glycosyltransferase genes whose products catalyzed the formation of different ginsenosides in P. notoginseng. The genetic information obtained in this study provides a resource for further exploration of the growth characteristics, cultivation, breeding, and saponin biosynthesis of P. notoginseng.
Key words: chromosome-level, genome, ginsenoside, P. notoginseng, regulation, transcriptome
This study uses PacBio sequencing and Hi-C-assisted assembly technology to construct a high-quality, chromosome-level reference genome for the famous medicinal plant Panax notoginseng. Furthermore, this study investigates the biosynthesis of saponins in P. notoginseng and characterizes key UGT enzymes.
Introduction
The Chinese medicine Sanchi is prepared from the dried root and rhizome of Panax notoginseng (Burk.) F. H. Chen, a perennial herb that belongs to the Araliaceae ginseng species (Briskin, 2000; Ng, 2006). Generally, Sanchi is collected and washed before P. notoginseng flowers bloom in autumn and is obtained by separating the main root and rhizome after drying (Wang et al., 2016). P. notoginseng has a long history of use in China for eliminating congestion and hemostasis and reducing swelling and pain. The brilliant work of the Ming Dynasty, Compendium of Materia Medica (A.D. 1552–1578), already described P. notoginseng. The medicinal value of P. notoginseng arises from the chemical ingredients it contains. To date, the chemical components isolated from P. notoginseng include mainly saponins, flavones, sugars, volatile oils, and amino acids (Jia et al., 2019). Among these, saponin compounds are the main chemical constituents and are also recognized as the main active ingredients (Xiong et al., 2019). Modern medical research has shown that saponins from P. notoginseng improve myocardial ischemia (Zhang et al., 2017b), protect the liver (Zhong et al., 2019), defend against cardiovascular disease (Chan et al., 2002), lower blood pressure (Pan et al., 2012), and improve arteriosclerosis (Min et al., 2008); they also have antithrombotic (Dang et al., 2015) and anticancer activities. As a rare and valuable medicinal material in China, P. notoginseng is also used in various prescriptions, such as capsules, injections, and powders. It is in widespread use, with total annual output values exceeding 70 billion RMB (Gui et al., 2013; Cui et al., 2014).
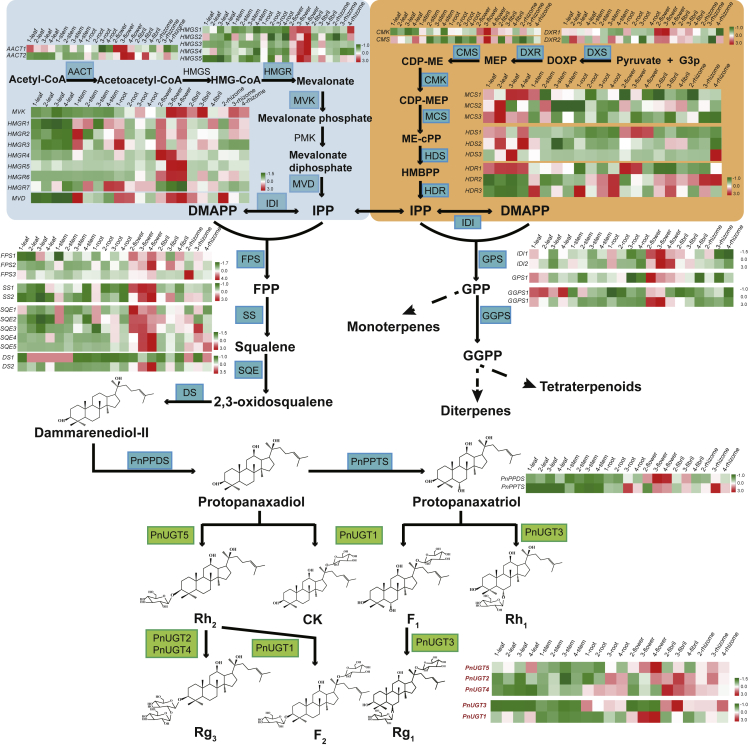
To date, the principal means of obtaining saponins has been to extract and isolate them from the original plants; however, the plant saponin content is low, and this process has a low extraction efficiency and is not environmentally friendly. Therefore, reconstruction of the saponin biosynthetic pathway for heterologous production is an alternative method for obtaining these valuable resources. At present, over 80 tetracyclic triterpenoid saponins have been identified (Xu et al., 2019) from the roots, stems, leaves, flowers, and fruits of P. notoginseng, and these saponins can be divided into protopanaxadiol (PPD) and protopanaxatriol (PPT) based on a hydroxyl substitution at the C-6 position of the molecular structure. The biosynthetic pathway of saponins in P. notoginseng is divided into four main stages. First, the direct precursors isopentenyl allyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) are synthesized by the mevalonate and 2-methyl-D-erythritol-4-phosphate pathways (Deng et al., 2017). Second, isopentenyl transferase and terpene synthases (Niu et al., 2014) catalyze the synthesis of 2,3-oxidosqualene from IPP and DMAPP (Jiang et al., 2017). Third, 2,3-oxidosqualene undergoes cyclization and hydroxylation (Han et al., 2011, 2012) to form the core structures PPD and PPT (Luo et al., 2011; Lu et al., 2018). Finally, the formation of various saponins is catalyzed by a number of glycosyltransferases (GTs) (Yu et al., 2019). The genetic and functional diversity of GTs gives rise to a variety of structurally diverse saponins.
To explore the biosynthetic pathway of ginsenosides, the genome of P. notoginseng has been explored and information mined (Zhang et al., 2017a; Chen et al., 2017). However, because of sequencing technology limitations, existing genomic information generated from second-generation short-read sequencing is insufficient (Shen et al., 2018; Zhao et al., 2019). Here, we present a high-quality P. notoginseng genome obtained using a combination of Illumina, PacBio, and Hi-C (high-throughput chromosome conformation capture) technologies; this is also the first chromosome-level genome of the genus Panax. Using comparative genomics, we explored the evolution and whole-genome duplication (WGD) events of P. notoginseng. We performed detailed transcriptional analysis and explored gene-level regulatory mechanisms that control the formation of characteristic tubercles, the biosynthesis of saponins at temporal and spatial levels, and the regulation of transcription factors. Combined with genomic analysis, we screened a series of UDP-dependent GT (UGT) candidate genes, five of which were identified as having catalytic functions. Our study provides genetic information for further comprehensive analysis of the saponin biosynthetic pathway and the evolution of the ginseng genus, and also describes useful techniques for the breeding of P. notoginseng.
Results
Genome sequencing, assembly, and annotation
According to the K-mer distribution analysis (K = 31), the estimated size of the P. notoginseng genome (2n = 2x = 24 chromosomes) is 2.38 Gb, and the heterozygosity and repeat contents are 0.58% and 69.05%, respectively (Supplemental Figure 1 and Supplemental Table 1). We combined Illumina, PacBio, and Hi-C technologies to sequence and assemble a high-quality, chromosome-level P. notoginseng reference genome. A total of 240.22 Gb of Illumina reads, 284.07 Gb of PacBio long reads, and 340.83 Gb of Hi-C data were generated, resulting in ~325.23× coverage of the P. notoginseng genome (Supplemental Table 2). The final assembled genome was 2.66 Gb in size and consisted of 219 scaffolds, with a scaffold N50 of 216.47 Mb and a contig N50 of 1.12 Mb (Figure 1 and Table 1). The assembled sequence was then anchored onto 12 pseudochromosomes with lengths of 176.58–295.55 Mb. The total length of the pseudochromosomes accounted for 99.89% of the genome sequences, with a scaffold L50 number of 6 (Supplemental Figure 2; Supplemental Table 3). The genome of P. notoginseng had a GC content of 34.45% (Supplemental Table 4).
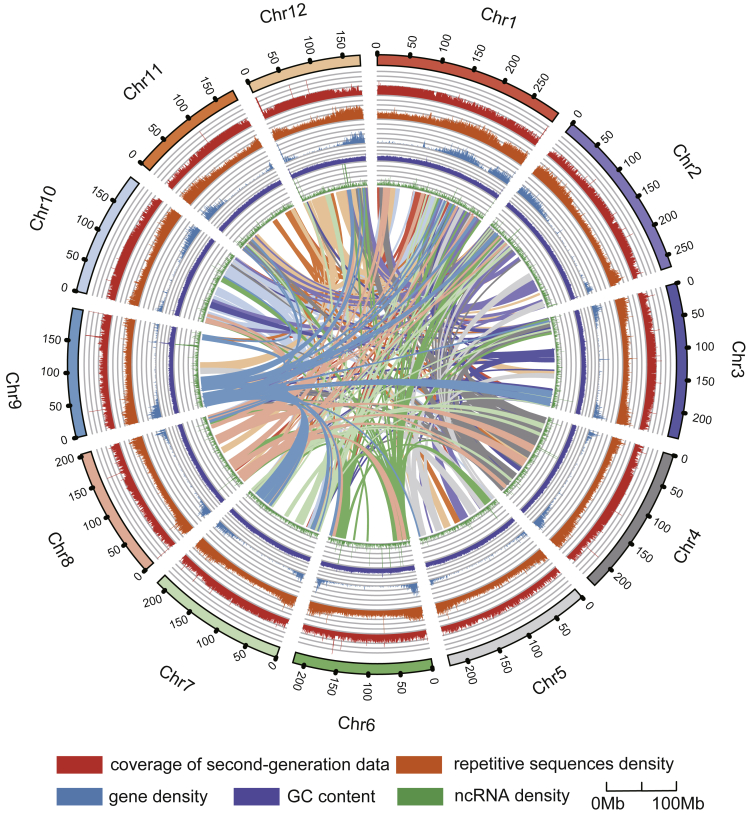
Figure 1.
Genome assembly characterization and chromosome locations of P. notoginseng.
Landscape of the P. notoginseng genome: from outside to inside, chromosome number and length, coverage of second-generation data, density of repetitive sequences, gene density, GC content, noncoding RNA density, and genomic synteny.
Table 1.
Summary of the final genome assembly of P. notoginseng and comparison with published genomes.
| Items | PN201908CCMU | 2017pub-1 | 2017pub-2 |
|---|---|---|---|
| Total length of contigs (Gb) | 2.66 | 1.85 | 2.39 |
| Contig N50 | 1.12 Mb | 13.16 kb | 16 kb |
| Longest contig (bp) | 13.35 Mb | 120.91 kb | 199.81 kb |
| Scaffold N50 | 216.47 Mb | 157.81 kb | 96 kb |
| Longest scaffold (bp) | 295.55 Mb | 1.19 Mb | 834.33 kb |
| GC content (%) | 34.45 | 34.85 | 28.65 |
| Number of genes | 37 606 | 34 369 | 36 790 |
| Average gene length (bp) | 5059.63 | 2705 | 3307.48 |
| BUSCO (%) | 96.6 | N | 82.4 |
| Average CDS length (bp) | 1202.85 | 957 | 942.43 |
| Average exon length (bp) | 231.00 | 251.39 | 211.92 |
| Average exon number per gene | 5.21 | 3.8 | 4.45 |
| Average intron length (bp) | 917.71 | 622.64 | 686.08 |
| Percentage of repeat sequences (%) | 85.85 | 61.31 | 75.94 |
| Percentage of LTR-RTs (%) | 58.88 | 57.41 | 66.72 |
| Reference | This study | (Zhang et al., 2017a) | (Chen et al., 2017) |
To test the coverage of the P. notoginseng genome, the short reads generated from Illumina sequencing were mapped, and 99.82% of these reads could be mapped to the scaffolds with 97.97% overall coverage (Supplemental Table 5). The completeness of the genome assembly was evaluated using BUSCO (Benchmarking Universal Single-Copy Orthologs) (Simao et al., 2015). Based on BUSCO analysis, 96.6% of plant sets were identified as complete (2049 out of 2121 BUSCOs) (Supplemental Table 6). All analyses suggested a high quality of the P. notoginseng genome assembly.
Based on a combination of homology-based and de novo approaches, 85.85% of the assembled P. notoginseng genome (Supplemental Table 7) consisted of repetitive elements; among them, long terminal repeat (LTR) retrotransposons accounted for the largest proportion and made up 58.88% of the genome (Supplemental Table 8; Supplemental Figure 3A and 3B). Compared with the published reference genome version, there were more predictions of repetitive sequences, a phenomenon that also occurs in other highly repetitive genomes (Xia et al., 2017, 2020; Wei et al., 2018; Zhang et al., 2020a). We compared the predicted repeat sequences with the RepBase database and calculated the degree of difference between them, from which LTR retrotransposons broke out at approximately 8% and an unknown outbreak happened earlier at approximately 5% (Supplemental Figure 3C).
An integrated strategy of de novo predictions, homology-based searches, and RNA sequencing was used to predict the protein-coding genes of the P. notoginseng genome. A total of 37 606 genes were annotated, with an average length of 5059.63 bp and an average exon number per gene of 5.21 (Supplemental Table 9; Supplemental Figure 4). The number of genes was similar to the numbers reported in two articles about the P. notoginseng genome published in 2017 (34 369 and 36 790), but other values, such as the average gene length and the average number of exons per gene, have been updated (Supplemental Table 10). Compared with another Araliaceae plant, Panax ginseng C.A. Mey (59 352 genes) (Kim et al., 2018), P. notoginseng has a smaller number of genes, which may be related to the subsequent duplication event of P. ginseng after the divergence of the two plants. Among the annotated P. notoginseng genes, 36 154 (~96.14%) were functionally classified by BLASTing against various functional databases (Supplemental Table 11). We further annotated noncoding RNA genes, obtaining 14 430 microRNA genes, 1513 transfer RNA (tRNA) genes, 314 ribosomal (rRNA) genes, and 272 small nuclear (snRNA) genes (Supplemental Table 12).
Genome evolution and expansion and contraction of gene families
We compared our P. notoginseng assembly with sequenced genomes from seven other plants: P. ginseng, Daucus carota from Apiales, four dicot species (Arabidopsis thaliana, Vitis vinifera, Capsicum annuum L., and Glycyrrhiza uralensis), and a monocot, Oryza sativa. Based on gene family clustering analysis, 30 874 P. notoginseng genes (82.28%) clustered into 15 655 gene families (Supplemental Table 13 and Supplemental Figure 5), which included 7264 gene families shared by all 8 species and 1059 families specific to P. notoginseng (Supplemental Figure 6). Gene ontology (GO) and KEGG enrichment analysis of these P. notoginseng-specific gene families showed that they were mainly involved in a series of biological activities, e.g., mature ribosome assembly, cytosolic part, small-molecule binding, and RNA transport (Supplemental Table 14; Supplemental Figure 7).
We selected 458 single-copy gene families among the 8 species to construct phylogenetic trees. As expected, P. notoginseng clustered with another Araliaceae species, P. ginseng, and these two species were most closely related to the Apiales family (Figure 2A). We estimated that P. notoginseng and P. ginseng diverged from the Apiaceae approximately 62.0 million years ago (mya), and P. notoginseng and P. ginseng diverged around 4.2 mya. These results show that the relationship between P. notoginseng and P. ginseng is very close, consistent with their very similar morphologies and secondary metabolites.
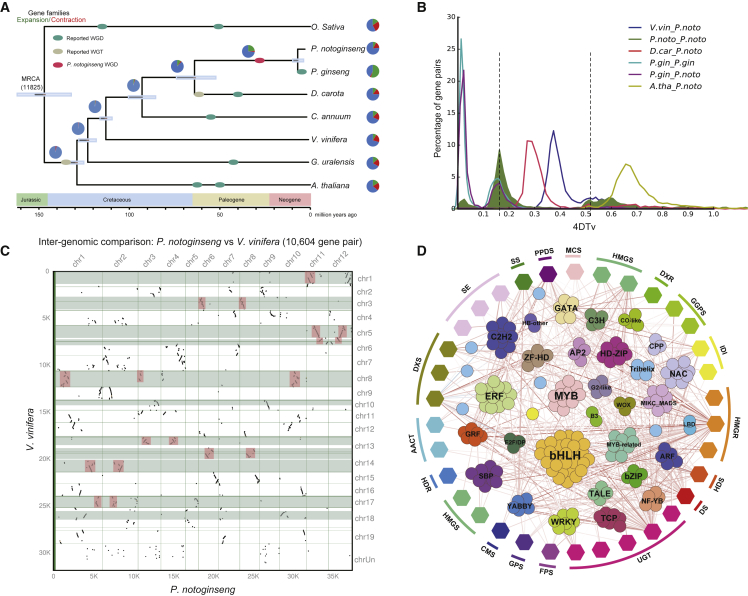
Figure 2.
Genome evolution and transcription factor regulation analysis of P. notoginseng.
(A) Inferred phylogenetic tree with 458 single-copy genes from eight plant species. Gene family expansions are indicated in green, and gene family contractions are indicated in red. The timings of WGD and whole-genome triplications (WGT) are superimposed on the tree. Divergence times are estimated by maximum likelihood (PAML).
(B) Distribution diagram of 4DTv values. The dark green-filled part indicates the 4DTv analysis inside P. notoginseng, and the peaks marked by the dotted line indicate where the two WGD events of P. notoginseng occurred.
(C) Syntenic dot plots show a 2:1 chromosomal relationship between the P. notoginseng genome and the V. vinifera genome. The area in the pink box on each horizontal line represents the collinear block between the two genomes.
(D) Correlation analysis of transcription factors with pathway genes. Pathway genes are represented by hexagons and transcription factors by circles. The line indicates the nature of the correlation: red for a positive correlation and blue for a negative correlation. The darker the color, the higher the correlation.
We compared expanded and contracted gene families in the 8 plant species with their most recent common ancestor. In total, 989 gene families were expanded in P. notoginseng, and 1823 gene families were contracted (Supplemental Figure 8). Compared with P. ginseng (6449), the number of expanded gene families in P. notoginseng was significantly smaller, perhaps because P. ginseng has experienced one more WGD event than P. notoginseng. We performed GO and KEGG enrichment analysis on expanded and contracted gene families in the P. notoginseng genome. The functions of the expanded gene families were mainly enriched in GO terms such as transposition, fatty acid biosynthetic process, respiratory chain, and catalytic activity (Supplemental Figure 9; Supplemental Table 15). The functions of the contracted gene families were mainly enriched in GO terms such as protein phosphorylation, protein modification process, β-glucan biosynthetic process, 1,3-β-D-glucan synthase complex, and purine nucleotide binding (Supplemental Table 16). 1,3-β-D-Glucan is reported to be involved in plant defense against fungi (Lee et al., 2006; Schober et al., 2009), and contraction in associated gene families may be related to the susceptibility of P. notoginseng to fungal pathogens and may explain why it readily develops root rot.
Analysis of WGD and its contribution to terpenoid biosynthesis
To study the WGD events that occurred during the evolution of P. notoginseng, we first analyzed the 4-fold synonymous third-codon transversion rate (4DTv) (Figure 2B) of syntenic gene pairs (Jaillon et al., 2007). There were two peaks in the 4DTv distribution at approximately 0.16 and 0.50 for all syntenic gene pairs in the P. notoginseng genome. The first peak at approximately 0.50 corresponded to the core eudicot γ triplication event, and the second peak at approximately 0.16 revealed that P. notoginseng underwent another WGD event after diverging from V. vinifera and D. carota. By comparing the P. notoginseng genome with the V. vinifera genome, we found that 65% of P. notoginseng gene models were located in syntenic blocks that corresponded to single V. vinifera regions. Meanwhile, 42% of the V. vinifera gene models in syntenic blocks had two orthologous regions, and 22% had one orthologous region (Supplemental Figure 10). The results of a genome collinearity analysis between V. vinifera and P. notoginseng indicated that the WGD event occurred in the P. notoginseng genome and that there was a 1:2 syntenic relationship between P. notoginseng and V. vinifera (Figure 2C and Supplemental Figure 11). Based on the distribution of Ks (Supplemental Figure 12) and 4DTv analysis, we calculated that the WGD event occurred approximately 29.6 mya in the ancestor of P. notoginseng. Compared with P. notoginseng, P. ginseng experienced one additional WGD event (Kim et al., 2018), and this recent event occurred approximately 1.85 mya after divergence from P. notoginseng. The timing of the WGD events was similar to the results of an evolutionary analysis of the P. ginseng genome (28 and 2.2 mya), confirming the accuracy of the present results (Kim et al., 2018).
Through homologous alignment and a Pfam database search, we identified gene families that were potentially involved in terpenoid biosynthesis in the eight species (Supplemental Table 17). The copy numbers of some gene families in the P. notoginseng genome were significantly greater than those in other plant genomes; these included families such as DXS, MCS, HDS, HDR, and SQE. We also observed that the average copy number of most key enzyme genes in P. ginseng was approximately twice that in P. notoginseng (Supplemental Figures 13 and 14). We next performed Ka/Ks analysis of these pathway genes to calculate the duplication times of their gene pairs in the P. notoginseng genome. The gene pair duplication times were concentrated around the time of the WGD event of P. notoginseng (Supplemental Figure 15; Supplemental Table 18), indicating that they may have arisen from the WGD event.
Transcriptome analysis and transcriptional regulation of saponin biosynthesis
To further explore the genetic information in P. notoginseng, we performed detailed transcriptome sequencing of P. notoginseng plants on the basis of the high-quality genome. Samples for transcriptome sequencing were obtained from 1- to 4-year-old P. notoginseng plants that were subdivided into root, stem, leaf, flower, rhizome, fibril, periderm, phloem, and tubercle (Supplemental Figures 16 and 17). Data processing (Supplemental Figures 18 and 19; Supplemental Table 19) and related transcriptome analyses, such as alternative splicing event analysis (Supplemental Figure 20; Supplemental Table 20) (Laloum et al., 2018), new transcription prediction (Dassanayake et al., 2009), single-nucleotide polymorphism analysis (Supplemental Figure 21; Supplemental Table 21) (Sarkar and Maranas, 2020), analysis of gene expression levels (Supplemental Figures 22 and 23), and identification of differentially expressed genes (DEGs) (Supplemental Figure 24; Supplemental Table 22), are detailed in the Supplemental materials.
We further analyzed the regulation of transcription factors in P. notoginseng. A total of 2150 transcription factors from 57 different families (Supplemental Table 23) were identified; we then used correlation analysis to map the gene regulation network (Figure 2D) between terpenoid biosynthetic pathway genes and transcription factors. The transcription factor families that were highly correlated with pathway genes included mainly bHLH (Deng et al., 2020), ERF (Zhang et al., 2020b; Paul et al., 2020), MYB (Li et al., 2020), WRKY (Villano et al., 2020), NAC (Jin et al., 2020), and C2H2 transcription factors (Han et al., 2020), as well as other families that play an important role in plant growth and development, stress resistance, and secondary metabolism.
Based on the expression levels of pathway genes (Supplemental Table 24), we explored the secondary metabolism of saponins in P. notoginseng plants at the temporal and spatial levels. At the temporal level, we compared the expression patterns of 29 genes from the saponin biosynthesis pathway in the same tissues of 1- to 4-year-old plants. In most tissues, highly expressed genes were concentrated in 3- or 4-year-old tissues, but in the stems, highly expressed genes were mainly concentrated in 1- to 2-year-old tissues (Figure 3). In addition, through comparative transcriptome analysis, we identified 7792 DEGs that were highly expressed in 3- to 4-year-old plants but poorly expressed in plants of other ages. At the spatial level, we compared gene expression patterns in different tissues of same-aged plants. Except in 1-year-old plants, most of the pathway genes were specifically expressed in flowers, and a few were highly expressed in rhizomes and roots (Supplemental Figure 25).
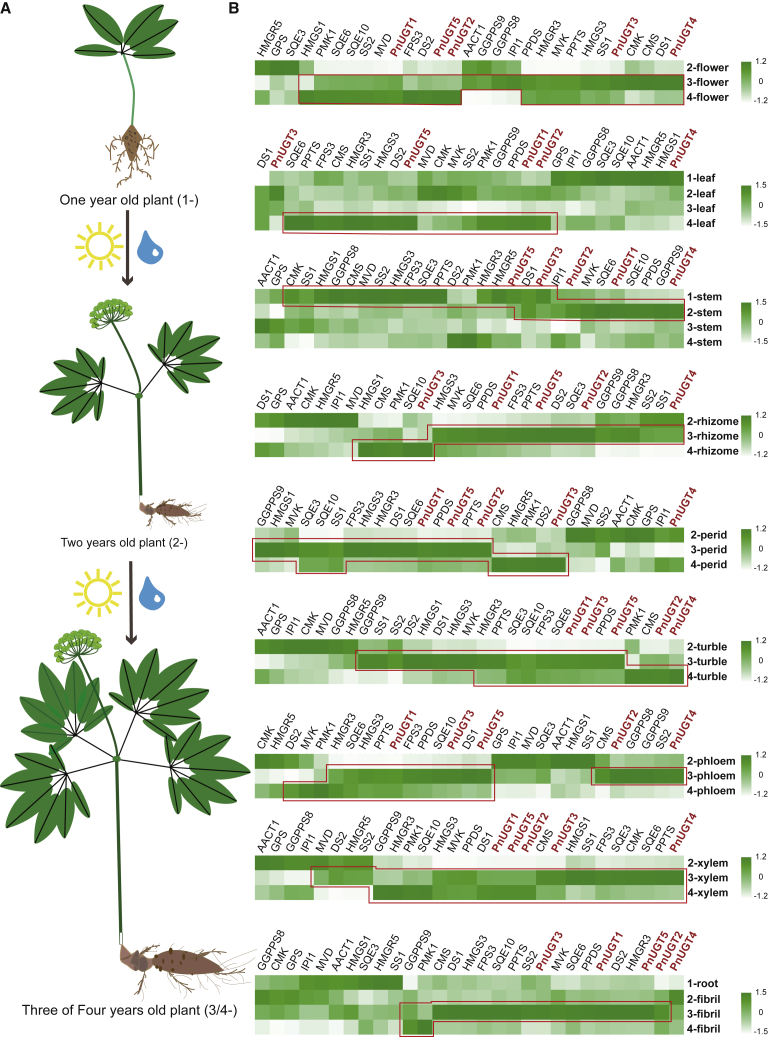
Figure 3.
Temporal expression profile of key enzyme genes in the saponin biosynthesis pathway.
(A) A brief view of the morphological changes in P. notoginseng as the years of growth increase during the cultivation process.
(B) Temporal expression heatmap of terpenoid biosynthetic pathway genes in P. notoginseng. Taking the leaf's heatmap as an example, the Arabic numerals in the label indicate the years of growth; for example, 2-leaf indicates that the sample is the leaf of a 2-year-old P. notoginseng plant. Based on the gene expression levels, the pattern of expression change for any one gene can be observed after the data in each column are standardized. The area marked by the red box indicates high gene expression levels. Each heatmap has its own color scale: the higher the expression, the greener the color.
Analyzing key enzyme genes involved in ginsenoside biosynthesis
The biosynthesis of P. notoginseng saponins is attributed to the activity of a series of key enzyme genes, among which the largest and most diverse gene families are the CYP450s and the UGTs. Phylogenetic analysis of CYP450s showed that more genes were enriched in the CYP71, CYP72, CYP76, CYP716, and CYP94 superfamilies (Supplemental Figure 26; Supplemental Table 25). Most of the genes in these superfamilies are involved in the oxidative stress response (Heitz et al., 2012) and in the biosynthesis of triterpenes (Carelli et al., 2011; Fukushima et al., 2011; Han et al., 2011), sterols, indole alkaloids (Irmler et al., 2000; Collu et al., 2001; Nafisi et al., 2007), geraniol iridoid (Höfer et al., 2013), and so forth.
Most of the saponin compounds in P. notoginseng are triterpene glycosides that contain sugar groups, indicating that UGT genes play a vital role in the modification of these saponins. Phylogenetic analysis of 158 UGT genes showed that most were classified into subfamilies, such as UGT73 (Lim et al., 2002, 2003), UGT71 (Song et al., 2016), UGT94 (Itkin et al., 2016; Ono et al., 2010), UGT91 (Shibuya et al., 2010), UGT85, and UGT74 (Figure 4A; Supplemental Table 26). The UGTs encoded by genes in these subfamilies have been reported to catalyze the glycosylation of flavonoids, isoflavones (Modolo et al., 2007), diterpenes, triterpenes, benzoate, lignans (Grubb et al., 2014; Tanaka et al., 2014; Dai et al., 2015), and other compounds.
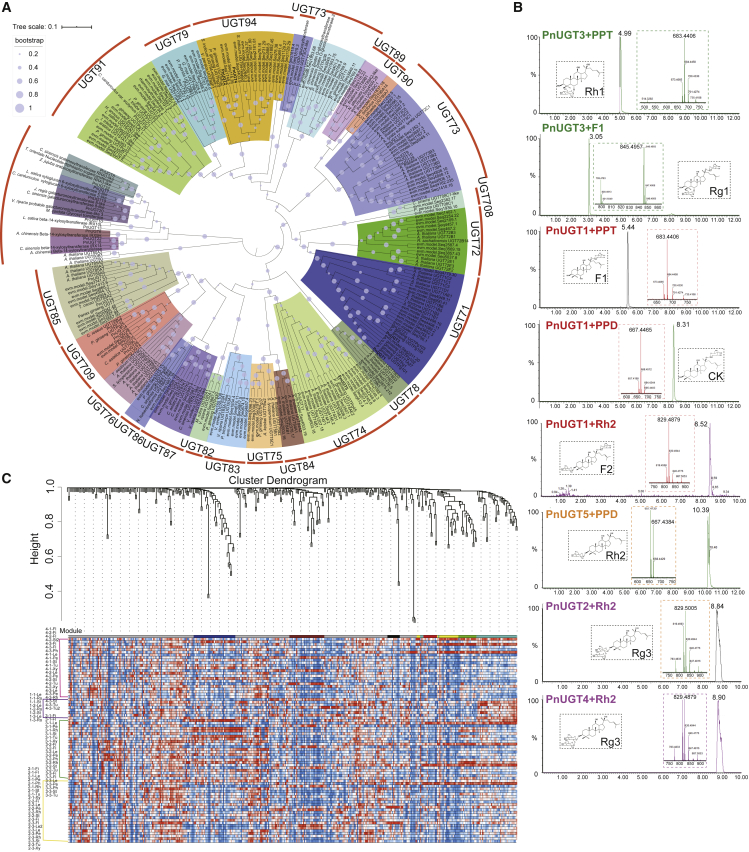
Figure 4.
Screening for candidate UGT genes and functional verification of five UGT genes.
(A) Phylogenetic analysis of UGT genes. The UGT gene families clustered into one clade are represented by different colors. The bootstrap value associated with each branch is represented by a light-purple circle: the larger the radius, the greater the bootstrap value.
(B) UPLC/Q-TOF analysis of five functional UGT genes. In catalytic reactions, PnUGT3 uses PPT and F1 as substrates, PnUGT1 uses PPD, PPT, and Rh2 as substrates, PnUGT5 uses PPD as a substrate, and PnUGT2 and PnUGT4 use Rh2 as a substrate to generate corresponding ginsenoside compounds. The chemical structures and characteristic mass spectrum peaks of products from each reaction are displayed in the dashed box of each track.
(C) WGCNA analysis of UGT genes.
We used UGTs involved in terpene biosynthesis as queries (Wang et al., 2015; Wei et al., 2015; Yan et al., 2014) to search for homologous UGT candidate genes in the P. notoginseng genome and designed primers for cloning (Supplemental Table 27). We ultimately cloned the full lengths of 32 UGT genes (Supplemental Figure 27) and named them PnUGT1–PnUGT32. Then, by expressing their proteins in Escherichia coli, we determined that five of them (PnUGT1–5) had catalytic functions in the biosynthesis of ginsenosides. We used an E. coli-expressed empty vector as the negative control (Supplemental Figure 28). Using PPT and F1 (Monoglycoside; PPT-C20-glucosyl) as substrates, the crude enzyme of gene PnUGT3 could add a glucosyl group at the C6 position to produce Rh1 (Monoglycoside; PPT-C6-glucosyl) and Rg1 (Diglycoside; PPT-C6-glucosyl, C20-glycosyl), respectively (Figure 4B and Supplemental Figure 29). Its functions are therefore consistent with the functions of UGTPg1 and UGTPg101 from P. ginseng (Wei et al., 2015; Yan et al., 2014), but this is a new gene cloned for the first time in P. notoginseng. Using PPD and PPT as substrates, the crude enzyme of gene PnUGT1 could add a glucosyl group at the C20 position to produce CK (Monoglycoside; PPD-C20-glucosyl) and F1 (Monoglycoside; PPT-C20-glucosyl), consistent with the functions of UGTPg100 and UGTPg101 from P. ginseng (Wei et al., 2015). In addition, PnUGT1 could catalyze the production of ginsenoside F2 (Diglycoside; PPD-C3-glucosyl, C20-glycosyl) from Rh2 (Monoglycoside; PPD-C3-glucosyl) (Figure 4B and Supplemental Figure 30), which is the first reported new function in P. notoginseng. The crude enzyme of gene PnUGT5 could catalyze the production of Rh2 from PPD, and crude enzymes of genes PnUGT2 and PnUGT4 could then extend the sugar chain and generate Rg3 (Monoglycoside; PPD-C3-glucosyl-glucosyl) from Rh2 (Figure 4B and Supplemental Figure 31), consistent with the functions of UGTPg45 and UGTPg29 from P. ginseng (Wang et al., 2015). In addition, the last four genes have also been experimentally shown to perform catalytic functions in Saccharomyces cerevisiae (Wang et al., 2020).
Besides the more common ginsenoside compounds mentioned above, there are many unique saponins in P. notoginseng, such as notoginsenoside R1, notoginsenoside R2, notoginsenoside R4, and notoginsenoside Fc, which have better water solubility and good pharmacological activities (Supplemental Figure 32). To screen out more UGT genes, we conducted weighted gene co-expression network analysis (WGCNA) and expression profile consistency analysis. Through WGCNA, we constructed a correlation network between all genes annotated as UGT in the P. notoginseng genome and identified 7 gene modules with strong correlation, including 29 pathway genes and 139 UGT genes (Figure 4C and Supplemental Figure 33). Among them, PnUGT2 was included in the blue module, and PnUGT1 and PnUGT5 were included in the green module. We further analyzed the annotation information and GO enrichment of these candidate UGT genes and found that most were enriched in GO terms such as GO:0008152 (metabolic process) or GO:0071555 (cell wall organization) and had different transferase activities (Supplemental Table 28). We then compared the expression patterns of genes in the terpene biosynthesis pathway and identified UGT genes with similar expression patterns. By comparing the expression levels in each transcriptome sample, the expression patterns of key enzyme genes could be divided into three categories (Figure 5): most were highly expressed in flowers, some were highly expressed in roots (each part), and a small number were most highly expressed in leaves (Supplemental Figure 34). A total of 35 UGT genes that were highly expressed and clustered with the pathway genes were screened from the correlation evolution tree (Supplemental Figure 35; Supplemental Table 29). Combining the results of the two analyses above, we identified candidate UGT genes that may be involved in the notoginsenoside biosynthetic pathway, although the specific functions of the encoded enzymes have yet to be experimentally verified.
Figure 5.
Overview of the saponin biosynthetic pathway in P. notoginseng and expression profiles of key enzyme genes.
The genes in the green box are the UGT genes identified in this study.
Discussion
P. notoginseng, one of the most widely used Chinese medicinal plants from the family Araliaceae, is renowned in China and worldwide for its good efficacy. The main active ingredients in P. notoginseng are saponins, including higher contents of ginsenoside Rg1, ginsenoside Rb1, and notoginsenoside R1 (Su et al., 2016; Duan et al., 2017; Zhang et al., 2018, 2019), and other active compounds, such as ginsenoside Rd, ginsenoside Rg3, ginsenoside Re (Xie et al., 2020), notoginsenoside R2, and notoginsenoside Fc (Liu et al., 2018). The biosynthesis of saponins in P. notoginseng has attracted extensive attention from researchers, and some key enzyme genes, such as HMGR, AACT, SS, PMK, MVK, IDI, and CYP450, have been identified. However, the complete biosynthetic pathways of unique notoginsenosides have not yet been resolved, and further research and exploration are needed.
Gene mining of high-quality genomic and transcriptomic data can provide resources for further exploration of plant growth and secondary metabolism mechanisms (Tu et al., 2020). As early as 2017, two P. notoginseng reference genomes were published (Zhang et al., 2017a; Chen et al., 2017); however, the quality of these genomes was insufficient because of the limited sequencing capacity at that time. We therefore performed whole-genome sequencing of P. notoginseng from Genuine Producing Areas based on third-generation PacBio sequencing technology and used Hi-C technology to construct a high-quality, chromosome-level genome. The assembled genome was 2.66 Gb in size, with a scaffold N50 of 216.47 Mb and a contig N50 of 1.12 Mb. In addition to the depth or accuracy of gene sequencing, this reference genome was greatly improved compared with previous genomes and was resolved to the chromosome level, which can more intuitively reveal the gene distribution and overall genomic landscape.
In addition to P. notoginseng, other plants belonging to Araliaceae are used as medicines, including the well-known plants P. ginseng, Panax quinquefolius L., and Panax zingiberensis C.Y. Wu et K.M. Feng. Based on chemotaxonomy, plants of Panax L. can be divided into two groups. The chemical composition of the first group mainly comprises dammarane-type tetracyclic triterpenes, and there are obvious similarities in plant morphology, including a short and erect rhizome and a carrot-like fleshy root. In terms of geographical distribution, plants in this group show a characteristic narrow and intermittent distribution, which has been observed in an ancient group of Panax plants. Representative plants include P. notoginseng, P. ginseng, P. quinquefolius, and others. The saponins of the second group are mainly oleanane-type pentacyclic triterpenes, and their plant morphology includes a long and creeping rhizome and an undeveloped fleshy root. They are distributed over a wide and continuous geographical area and may represent an evolutionary group of Panax plants. Representative plants from this group include P. zingiberensis C.Y. Wu et K.M. Feng, P. stipuleanatus H.T. Tsai et K.M. Feng, Panax japonicus (T. Nees) C.A. Mey. and Panax japonicus C.A. Mey. var. major (Burk.) C.Y. Wu et K.M. Feng, and others. Based on cytotaxonomy analysis, we found that Panax plants had different ploidy types. For example, P. notoginseng and P. japonicus are diploid, and P. ginseng and P. quinquefolius are tetraploid, further indicating that P. notoginseng is in a relatively primitive evolutionary position among Panax plants. By comparing genomes, we found that after diverging from carrots, an independent WGD event occurred in P. notoginseng. We then studied the distribution of Ka/Ks values of key enzyme gene pairs in the saponin biosynthesis pathway and found that the WGD event may have contributed to the generation of these gene pairs, directing the metabolic flux toward the production of saponins. Based on the locations of coding genes on the chromosomes, we also found two sets of gene cluster duplication. Notably, upstream HDR, SS, and SE genes and downstream CYP450 and UGT genes that are known to be involved in ginsenoside biosynthesis are close to each other in the P. notoginseng genome (Supplemental Figure 36). The gene cluster also contains some UGT and transcription factor genes identified in this study, which are likely to participate in the biosynthesis and regulation of saponins. Compared with P. notoginseng, P. ginseng experienced one additional WGD event, which was manifested in the larger genome size, more expanded gene families, and multiple copies of key enzyme genes. In summary, we analyzed and explored the genetic information of P. notoginseng, one of the more primitive Panax plants, laying a solid foundation for subsequent evolutionary research on the genus Panax.
In addition, we also established a detailed transcript database of P. notoginseng through sequencing and analysis of different tissues from 1- to 4-year-old plants. Through comparative transcriptome analysis, we explored the molecular regulation mechanism of tubercles, a characteristic phenotype of P. notoginseng. The associated DEGs were mainly involved in the biosynthesis of plant hormones such as strigolactone, cytokinin, and auxin. The synergistic effects of these phytohormones result in the production of a tubercle phenotype, and further study of the functions of related DEGs will more fully reveal the molecular mechanisms of tubercle formation.
We next explored the saponin biosynthesis pathway in P. notoginseng plants at temporal and spatial levels. We compared the expression patterns of saponin biosynthesis genes in the same tissues of 1- to 4-year-old plants and found that most genes in tissues other than stems were highly expressed in 3- or 4-year-old plants. This indicates that as plant age increases, saponin biosynthesis gene expression levels also increase, as does the content of accumulated saponins. The quality of P. notoginseng harvested after more than 3 years of growth is therefore optimal, but because of diseases, insect pests, and continuous cropping obstacles, most materials circulated in the market are 3-year-old P. notoginseng. At the spatial level, most pathway genes were specifically expressed in flowers, and a few were highly expressed in rhizomes and roots, including the postmodification UGT enzyme genes PnUGT2, PnUGT3, and PnUGT4. These results indicate that saponin compounds or their precursors may be synthesized in the flowers first and then transferred to the roots or further modified in the roots, consistent with a previous report.
The ultimate step in the saponin biosynthesis pathway is glycosylation catalyzed by UGTs. This is the most critical step in determining the structure and pharmacological effects of the compounds, and we therefore focused on identifying candidate UGT genes. First, we conducted a systematic evolutionary analysis of all P. notoginseng genes that contained the conserved GT domain. As expected, we obtained five UGT genes that catalyzed the glycosylation of ginsenosides. Second, we performed WGCNA analysis on all genes annotated as UGTs and key enzyme genes of the P. notoginseng saponin biosynthetic pathway and screened out seven modules of highly correlated genes. Among these seven modules, two (module blue and module green) contained genes with identified functions, indicating that the genes enriched in these modules were likely to participate in the biosynthesis of saponins. Third, we conducted a consistency analysis of expression profiles and identified 20 UGT genes with high expression levels and expression patterns consistent with those of pathway genes. Combining the results of the two analyses above, we identified candidate UGT genes to lay a foundation for further comprehensive analysis of the complete notoginsenoside biosynthesis pathway.
In summary, we constructed a high-quality, chromosome-level P. notoginseng reference genome as a comprehensive genetic inventory for evolutionary phylogenomic studies of Panax plants. Using detailed transcriptome data, we explored the molecular mechanism of tubercle formation, investigated the biosynthesis pathway of saponins, and provided many promising candidate genes to fully reveal the biosynthetic pathway of notoginsenosides in P. notoginseng.
Materials and Methods
Plant materials, DNA extraction, and library construction
Individual plants of P. notoginseng (Burk.) F. H. Chen were collected in August 2019 from Wenshan County, Yunnan Province, China (26°49′55″N, 100°3′20″E, 2630 m above sea level). Fresh and healthy leaves were harvested, immediately frozen in liquid nitrogen, and preserved at −80°C. High-quality genomic DNA was extracted from the P. notoginseng leaves using the modified phenol-chloroform isoamyl alcohol extraction method. The quality and quantity of the isolated DNA were assessed using a NanoPhotometer (Implen, CA, USA) and a Qubit 2.0 Fluorometer (Life Technologies, CA, USA). Illumina (350 bp), PacBio, and Hi-C libraries were constructed following the operation guide for each technology.
Genome sequencing, assembly, and quality assessment
For PacBio libraries, the whole genome was sequenced on the PacBio Sequel II System based on single-molecule real-time sequencing technology, and 284.07 Gb (~106.79×) of data were obtained. The Illumina library was sequenced on the Illumina HiSeq X Ten platform following standard Illumina protocols. After filtering out adapter sequences and low-quality and duplicated reads, we obtained 231.06 Gb (~86.86×) of clean data. The subreads obtained from PacBio libraries were assembled into contigs using Canu (v1.8), and the consensus genome was polished by referring to the Illumina reads with BWA (v0.7.9a) and Pilon (v1.22). For Hi-C libraries, Illumina HiSeq X Ten was used for sequencing with PE150, and a total of 340.83 Gb (~128.13×) of data were retained. Finally, based on Hi-C technology using BWA-mem and LACHESIS, the final genome was 2.66 Gb in size, and the contig and scaffold N50 were 1.12 and 216.47 Mb, respectively. We used BUSCO (v3.0.1, default parameters), Illumina reads, and transcriptome mapping to the P. notoginseng genome with BWA-mem to confirm the high quality of the assembled genome.
Genome annotation
We used homology-based, de novo, and transcriptome-based predictions to predict the protein-coding genes in the P. notoginseng genome. The gene sets predicted by various strategies were integrated into a non-redundant and more complete gene set using EVidenceModeler. Gene functional annotation was performed mainly by searching against various functional databases, such as Swiss-Prot, NT (Nucleotide Sequence Database), NR (Non-Redundant Protein Sequence Database), Pfam, eggNOG (Evolutionary Genealogy of Genes: Non-supervised Orthologous Groups), and GO. Repetitive sequences were annotated using an ab initio prediction method and a homolog-based approach. We detected noncoding RNA by comparison with known noncoding RNA libraries and Rfam, and we also predicted rRNAs, snRNAs, microRNAs, and so on.
Analysis of genomic evolution and WGD events
We used the OrthoMCL package (1.4) to identify and cluster gene families (clusters) from P. notoginseng and seven other plant species: P. ginseng, D. carota, V. vinifera, C. annuum L., G. uralensis, A. thaliana, and O. sativa. After gene family clustering, we aligned all 458 single-copy gene protein sequences using MUSCLE and constructed a phylogenetic tree using PhyML. Based on the gene family cluster analysis and after filtering gene families with abnormal gene numbers in individual species, we used the CAFÉ program to identify the expansion and contraction of gene families in each species. To explore the evolution of the P. notoginseng genome, we calculated the 4DTv of syntenic blocks and the distribution of synonymous substitutions per synonymous site (Ks) to identify WGD events.
Integrated genomic and transcriptomic analysis
One- to four-year-old P. notoginseng plants were collected from Wenshan County, Yunnan Province, China. There were three biological replicates for each sample, and samples were taken at least five meters apart. After harvesting, we subdivided the plants into different tissue parts, including the root (xylem), stem, leaf, flower, rhizome, fibril, periderm, phloem, and tubercle. All samples were transported on dry ice, washed with ultrapure water three times, immediately frozen in liquid nitrogen, and stored at −80°C before RNA extraction. Total RNA was extracted from each tissue using a modified cetyltrimethylammonium bromide method. The RNA purity was checked using a kaiaoK5500 spectrophotometer (Kaiao, Beijing, China), and the RNA integrity and concentration were assessed using the RNA Nano 6000 Assay Kit for the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). cDNA libraries were constructed using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs, USA) following the manufacturer's recommendations. After cluster generation, the libraries were sequenced on an Illumina NovaSeq S2 platform, and 150 bp paired-end reads were generated.
Genes encoding key enzymes thought to be involved in the saponin biosynthetic pathway were annotated by BLAST (2.2.28). Their predicted proteins were aligned with the Pfam database using HMMER (3.1b1), and their expression levels in different tissues were obtained from transcriptome data. We used MeV software (4.9.0) to create a heatmap of gene expression and analyze gene expression patterns. In addition, we identified transcription factor genes in the P. notoginseng genome by comparison with the PlantTFDB database. We used an R script to calculate the Pearson correlation coefficients between transcription factors and genes in batches and Cytoscape software to draft the correlation map.
Screening and functional verification of candidate UGT genes
Multiple sequence alignments were generated using DNAMAN to visualize the conserved motifs. For phylogenetic tree analysis, the amino acid sequences of UGTs from other species were downloaded from the National Center for Biotechnology Information (NCBI) database and aligned using ClustalW. Then, a neighbor-joining tree was built using MEGA X software (Kumar et al., 2016) with 1000 bootstrap iterations. P. notoginseng cDNA was prepared using the PrimeScript 1st Strand cDNA Synthesis Kit (Takara, Dalian, China). After designing primers, we cloned a total of 32 UGT genes, and the PCR products were ligated into the N-terminal MBP fusion expression vector HIS-MBP-pET28a (HIS, histidine; MBP, maltose-binding protein) (Li et al., 2018) according to the protocol of the Seamless Cloning Kit (Beyotime, Shanghai, China). We transformed the successfully sequenced positive strains into E. coli BL21 (DE3) (Transgen Biotech, Beijing, China) and maintained the cultures in Luria-Bertani liquid medium with kanamycin (50 μg/mL) at 37°C in a shaking incubator until the optical density at 600 nm reached 0.6–0.8. Then, 1 M isopropyl β-D-thiogalactopyranoside was added to a final concentration of 50 μM, and cultures were maintained at 16°C and 120 rpm for 16 h to allow expression of recombinant proteins. pET28a-transformed E. coli BL21 (DE3) cells were treated in parallel as a control. The recombinant cells were harvested by centrifugation at 10 000 g and 4°C, then resuspended in 100 mM phosphate buffer (pH 8.0) that contained 1 mM phenylmethanesulfonylfluoride and sonicated in an ice-water bath for 10 min (lysed for 5 s, paused for 5 s). The sample lysates were centrifuged for 20 min at 12 000 g and 4°C to separate crude enzymes from cell debris. A UGT activity assay was performed in a total volume of 100 μl that contained 100 mM crude enzyme buffer (pH 8.0), 1 mM UDP-glucose, and 0.1 mM acceptor substrate for 2 h in a 35°C water bath and was terminated by the addition of 200 μl methanol. Precipitated proteins were removed by centrifugation (10 000 g for 10 min) and filtered through 0.22 μm filters before injection. Glycosylated products were detected using ultra-high-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (UPLC/Q-TOF-MS, Waters, Milford, MA) using a Waters ACQUITY UPLC HSS T3 analytical column (2.1 × 100 mm, 1.8 μm). Data analysis was performed using MassLynx software (version 4.1). Standards of saponin compounds and UDP-glucose were purchased from Yuanye Bio-Technology (Shanghai, China). To screen additional candidate UGT genes, we also conducted WGCNA using R and expression profile consistency analysis.
Data availability
The data supporting the findings of this work are available within the paper and its Supplemental Information files. The nucleotide sequencing data for UGT genes identified in this study have been deposited at NCBI GenBank under accession numbers MT551198 to MT551202. The genome sequence data of P. notoginseng have been deposited under NCBI BioProject number PRJNA658419https://www.Ncbi.nlm.nih.gov/bioproject/SUB7934826. In addition, the whole-genome sequence data reported in this paper have been deposited in the Genome Warehouse in the National Genomics Data Center (National Genomics Data Center and Partners, 2020), Beijing Institute of Genomics (China National Center for Bioinformation), Chinese Academy of Sciences, under accession number GWHAOSA00000000 and are publicly accessible at https://bigd.big.ac.cn/gsa.
Funding
We acknowledge support from the National Natural Science Foundation of China (nos. 81891010, 81891013), the Key Project at central government level: The ability establishment of sustainable use for valuable Chinese medicine resources (no. 2060302-1806-03), the High-level Teachers in Beijing Municipal Universities in the Period of 13th Five-year Plan (no. CIT&TCD20170324), and the National Program for Special Support of Eminent Professionals.
Author contributions
W.G., L.H., and Z.J. conceived and initiated the study. Z.J., T.L., and W.Y. performed the genome sequencing and bioinformatics analyses. X.C. provided the original plant material of P. notoginseng. Z.J. performed most of the experiments with the assistance of Y.Z., T.H., B.M., Y. Lu, J.G., J.Z., Y. Liu, N.L., X.W., and Y.S., and Y.T. provided assistance with chemical experiments. Z.J. wrote the manuscript and Y.Z., T.L., L.T., W.G., and L.H. revised the manuscript. All authors read and approved the final manuscript.
Acknowledgments
The authors declare no competing interests.
Published: September 20, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental Information is available at Plant Communications Online.
Contributor Information
Luqi Huang, Email: huangluqi01@126.com.
Wei Gao, Email: weigao@ccmu.edu.cn.
Supplemental information
References
- Briskin D. Medicinal plants and phytomedicines. Linking plant biochemistry and physiology to human health. Plant Physiol. 2000;124:507–514. doi: 10.1104/pp.124.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli M., Biazzi E., Panara F., Tava A., Scaramelli L., Porceddu A., Graham N., Odoardi M., Piano E., Arcioni S. Medicago truncatula CYP716A12 is a multifunctional oxidase involved in the biosynthesis of hemolytic saponins. Plant Cell. 2011;23:3070–3081. doi: 10.1105/tpc.111.087312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P., Thomas G., Tomlinson B. Protective effects of trilinolein extracted from Panax notoginseng against cardiovascular disease. Acta Pharmacol. Sin. 2002;23:1157–1162. [PubMed] [Google Scholar]
- Chen W., Kui L., Zhang G., Zhu S., Zhang J., Wang X., Yang M., Huang H., Liu Y., Wang Y. Whole-genome sequencing and analysis of the Chinese herbal plant Panax notoginseng. Mol. Plant. 2017;10:899–902. doi: 10.1016/j.molp.2017.02.010. [DOI] [PubMed] [Google Scholar]
- Collu G., Unver N., Peltenburg-Looman M.G.A., Heijden R.v.d., Verpoorte R., Memelink J. Geraniol 10-hydroxylase, a cytochrome P450 enzyme involved in terpenoid indole alkaloid biosynthesis. Fed. Eur. Biochem. Soc. 2001;508:215–220. doi: 10.1016/s0014-5793(01)03045-9. [DOI] [PubMed] [Google Scholar]
- Cui X., Huang L., Guo L., Liu D. Chinese Sanqi industry status and development contermeasures. Chin. J. Chin. Mater. Med. 2014;39:553–557. [PubMed] [Google Scholar]
- Dai L., Liu C., Zhu Y., Zhang J., Men Y., Zeng Y., Sun Y. Functional characterization of cucurbitadienol synthase and triterpene glycosyltransferase involved in biosynthesis of mogrosides from Siraitia grosvenorii. Plant Cell Physiol. 2015;56:1172–1182. doi: 10.1093/pcp/pcv043. [DOI] [PubMed] [Google Scholar]
- Dang X., Miao J., Chen A.-q., Li P., Chen L., Liang J.-r., Xie R.-m., Zhao Y. The antithrombotic effect of RSNK in blood-stasis model rats. J. Ethnopharmacol. 2015;173:266–272. doi: 10.1016/j.jep.2015.06.030. [DOI] [PubMed] [Google Scholar]
- Dassanayake M., Haas J.S., Bohnert H.J., Cheeseman J.M. Shedding light on an extremophile lifestyle through transcriptomics. New Phytol. 2009;183:764–775. doi: 10.1111/j.1469-8137.2009.02913.x. [DOI] [PubMed] [Google Scholar]
- Deng B., Zhang P., Ge F., Liu D.-Q., Chen C.-Y. Enhancement of triterpenoid saponins biosynthesis in Panax notoginseng cells by co-overexpressions of 3-hydroxy-3-methylglutaryl CoA reductase and squalene synthase genes. Biochem. Eng. J. 2017;122:38–46. [Google Scholar]
- Deng C., Wang J., Lu C., Li Y., Kong D., Hong Y., Huang H., Dai S. CcMYB6-1 and CcbHLH1, two novel transcription factors synergistically involved in regulating anthocyanin biosynthesis in cornflower. Plant Physiol. Biochem. 2020;151:271–283. doi: 10.1016/j.plaphy.2020.03.024. [DOI] [PubMed] [Google Scholar]
- Duan L., Xiong X., Hu J., Liu Y., Li J., Wang J. Panax notoginseng saponins for treating coronary artery disease: a functional and mechanistic overview. Front. Pharmacol. 2017;8 doi: 10.3389/fphar.2017.00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima E.O., Seki H., Ohyama K., Ono E., Umemoto N., Mizutani M., Saito K., Muranaka T. CYP716A subfamily members are multifunctional oxidases in triterpenoid biosynthesis. Plant Cell Physiol. 2011;52:2050–2061. doi: 10.1093/pcp/pcr146. [DOI] [PubMed] [Google Scholar]
- Grubb C.D., Zipp B.J., Kopycki J., Schubert M., Quint M., Lim E.K., Bowles D.J., Pedras M.S., Abel S. Comparative analysis of Arabidopsis UGT74 glucosyltransferases reveals a special role of UGT74C1 in glucosinolate biosynthesis. Plant J. 2014;79:92–105. doi: 10.1111/tpj.12541. [DOI] [PubMed] [Google Scholar]
- Gui Q., Yang Y., Ying S., Zhang M. Xueshuatong improves cerebral blood perfusion in elderly patients with lacunar infarction. Neural Regen. Res. 2013;8:792–801. doi: 10.3969/j.issn.1673-5374.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G., Lu C., Guo J., Qiao Z., Sui N., Qiu N., Wang B. C2H2 zinc finger proteins: master regulators of abiotic stress responses in plants. Front. Plant Sci. 2020;11:115. doi: 10.3389/fpls.2020.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.Y., Hwang H.S., Choi S.W., Kim H.J., Choi Y.E. Cytochrome P450 CYP716A53v2 catalyzes the formation of protopanaxatriol from protopanaxadiol during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2012;53:1535–1545. doi: 10.1093/pcp/pcs106. [DOI] [PubMed] [Google Scholar]
- Han J.Y., Kim H.J., Kwon Y.S., Choi Y.E. The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from dammarenediol-II during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2011;52:2062–2073. doi: 10.1093/pcp/pcr150. [DOI] [PubMed] [Google Scholar]
- Heitz T., Widemann E., Lugan R., Miesch L., Ullmann P., Desaubry L., Holder E., Grausem B., Kandel S., Miesch M. Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. 2012;287:6296–6306. doi: 10.1074/jbc.M111.316364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfer R., Dong L., André F., Ginglinger J.-F., Lugan R., Gavira C., Grec S., Lang G., Memelink J., Krol S.V.D. Geraniol hydroxylase and hydroxygeraniol oxidase activities of the CYP76 family of cytochrome P450 enzymes and potential for engineering the early steps of the (seco)iridoid pathway. Metab. Eng. 2013;20:221–232. doi: 10.1016/j.ymben.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Irmler S., Schroder G., St-Pierre B., Crouch N., Hotze M., Schmidt J., Stract D., Matern U., Schroder J. Indole alkaloid biosynthesis in Catharanthus roseus: new enzyme activities and identification of cytochrome P450 CYP72A1 as secologanin synthase. Plant J. 2000;24:797–804. doi: 10.1046/j.1365-313x.2000.00922.x. [DOI] [PubMed] [Google Scholar]
- Itkin M., Davidovich-Rikanati R., Cohen S., Portnoy V., Doron-Faigenboim A., Oren E., Freilich S., Tzuri G., Baranes N., Shen S. The biosynthetic pathway of the nonsugar, high-intensity sweetener mogroside V from Siraitia grosvenorii. Proc. Natl. Acad. Sci. U S A. 2016;113:E7619–E7628. doi: 10.1073/pnas.1604828113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O., Aury J.M., Noel B., Policriti A., Clepet C., Casagrande A., Choisne N., Aubourg S., Vitulo N., Jubin C. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Jia L., Zuo T., Zhang C., Li W., Wang H., Hu Y., Wang X., Qian Y., Yang W., Yu H. Simultaneous profiling and holistic comparison of the metabolomes among the flower buds of Panax ginseng, Panax quinquefolius, and Panax notoginseng by UHPLC/IM-QTOF-HDMSE-based metabolomics analysis. Molecules. 2019;24:2188. doi: 10.3390/molecules24112188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Rong Q., Chen Y., Yuan Q., Shen Y., Guo J., Yang Y., Zha L., Wu H., Huang L. Molecular cloning and functional analysis of squalene synthase (SS) in Panax notoginseng. Int. J. Biol. Macromol. 2017;95:658–666. doi: 10.1016/j.ijbiomac.2016.11.070. [DOI] [PubMed] [Google Scholar]
- Jin J.F., Wang Z.Q., He Q.Y., Wang J.Y., Li P.F., Xu J.M., Zheng S.J., Fan W., Yang J.L. Genome-wide identification and expression analysis of the NAC transcription factor family in tomato (Solanum lycopersicum) during aluminum stress. BMC Genomics. 2020;21:288. doi: 10.1186/s12864-020-6689-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.H., Jayakodi M., Lee S.C., Choi B.S., Jang W., Lee J., Kim H.H., Waminal N.E., Lakshmanan M., van Nguyen B. Genome and evolution of the shade-requiring medicinal herb Panax ginseng. Plant Biotechnol. J. 2018;16:1904–1917. doi: 10.1111/pbi.12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloum T., Martin G., Duque P. Alternative splicing control of abiotic stress responses. Trends Plant Sci. 2018;23:140–150. doi: 10.1016/j.tplants.2017.09.019. [DOI] [PubMed] [Google Scholar]
- Lee T., Grinshpun S.A., Kim K.Y., Iossifova Y., Adhikari A., Reponen T. Relationship between indoor and outdoor airborne fungal spores, pollen, and (1→3)-β-D-glucan in homes without visible mold growth. Aerobiologia. 2006;22:227–235. [Google Scholar]
- Li S.F., Allen P.J., Napoli R.S., Browne R.G., Pham H., Parish R.W. MYB-bHLH-TTG1 regulates Arabidopsis seed coat biosynthesis pathways directly and indirectly via multiple tiers of transcription factors. Plant Cell Physiol. 2020;61:1005–1018. doi: 10.1093/pcp/pcaa027. [DOI] [PubMed] [Google Scholar]
- Li Y., Lin H.X., Wang J., Yang J., Lai C.J., Wang X., Ma B.W., Tang J.F., Li Y., Li X.L. Glucosyltransferase capable of catalyzing the last step in neoandrographolide biosynthesis. Org. Lett. 2018;20:5999–6002. doi: 10.1021/acs.orglett.8b02146. [DOI] [PubMed] [Google Scholar]
- Lim E.K., Baldauf S., Li Y., Elias L., Worrall D., Spencer S.P., Jackson R.G., Taguchi G., Ross J., Bowles D.J. Evolution of substrate recognition across a multigene family of glycosyltransferases in Arabidopsis. Glycobiology. 2003;13:139–145. doi: 10.1093/glycob/cwg017. [DOI] [PubMed] [Google Scholar]
- Lim E.K., Doucet C.J., Li Y., Elias L., Worrall D., Spencer S.P., Ross J., Bowles D.J. The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J. Biol. Chem. 2002;277:586–592. doi: 10.1074/jbc.M109287200. [DOI] [PubMed] [Google Scholar]
- Liu J., Jiang C., Ma X., Wang J. Notoginsenoside Fc attenuates high glucose-induced vascular endothelial cell injury via upregulation of PPAR-gamma in diabetic Sprague-Dawley rats. Vascul Pharmacol. 2018;109:27–35. doi: 10.1016/j.vph.2018.05.009. [DOI] [PubMed] [Google Scholar]
- Lu J., Li J., Wang S., Yao L., Liang W., Wang J., Gao W. Advances in ginsenoside biosynthesis and metabolic regulation. Biotechnol. Appl. Biochem. 2018;65:514–522. doi: 10.1002/bab.1649. [DOI] [PubMed] [Google Scholar]
- Luo H., Sun C., Sun Y., Wu Q., Li Y., Song J., Niu Y., Cheng X., Xu H., Li C. Analysis of the transcriptome of Panax notoginseng root uncovers putative triterpene saponin-biosynthetic genes and genetic markers. BMC Genomics. 2011;12:S5. doi: 10.1186/1471-2164-12-S5-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S., Jung S., Cho K., Kim D. Antihyperlipidemic effects of red ginseng, Crataegii fructus and their main constituents ginsenoside Rg3 and ursolic acid in mice. Biomol. Ther. 2008;16:364–369. [Google Scholar]
- Modolo L.V., Blount J.W., Achnine L., Naoumkina M.A., Wang X., Dixon R.A. A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula. Plant Mol. Biol. 2007;64:499–518. doi: 10.1007/s11103-007-9167-6. [DOI] [PubMed] [Google Scholar]
- Nafisi M., Goregaoker S., Botanga C.J., Glawischnig E., Olsen C.E., Halkier B.A., Glazebrook J. Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell. 2007;19:2039–2052. doi: 10.1105/tpc.107.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Genomics Data Center Members and Partners Database resources of the national genomics data center in 2020. Nucleic Acids Res. 2020;48:D24–D33. doi: 10.1093/nar/gkz913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T.B. Pharmacological activity of sanchi ginseng (Panax notoginseng) J. Pharm. Pharmacol. 2006;58:1007–1019. doi: 10.1211/jpp.58.8.0001. [DOI] [PubMed] [Google Scholar]
- Niu Y., Luo H., Sun C., Yang T.J., Dong L., Huang L., Chen S. Expression profiling of the triterpene saponin biosynthesis genes FPS, SS, SE, and DS in the medicinal plant Panax notoginseng. Gene. 2014;533:295–303. doi: 10.1016/j.gene.2013.09.045. [DOI] [PubMed] [Google Scholar]
- Ono E., Ruike M., Iwashita T., Nomoto K., Fukui Y. Co-pigmentation and flavonoid glycosyltransferases in blue Veronica persica flowers. Phytochemistry. 2010;71:726–735. doi: 10.1016/j.phytochem.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Pan C., Hou Y., An X., Singh Gurbakhshish, Chen M., Yang Z., Pu J., Li J. Panax notoginseng and its components decreased hypertension via stimulation of endothelial-dependent vessel dilatation. Vasc. Pharmacol. 2012;56:150–158. doi: 10.1016/j.vph.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Paul P., Singh S.K., Patra B., Liu X., Pattanaik S., Yuan L. Mutually regulated AP2/ERF gene clusters modulate biosynthesis of specialized metabolites in plants. Plant Physiol. 2020;182:840–856. doi: 10.1104/pp.19.00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D., Maranas C.D. SNPeffect: identifying functional roles of SNPs using metabolic networks. Plant J. 2020;103:512–531. doi: 10.1111/tpj.14746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober M.S., Burton R.A., Shirley N.J., Jacobs A.K., Fincher G.B. Analysis of the (1,3)-beta-D-glucan synthase gene family of barley. Phytochemistry. 2009;70:713–720. doi: 10.1016/j.phytochem.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Shen Q., Zhang L., Liao Z., Wang S., Yan T., Shi P., Liu M., Fu X., Pan Q., Wang Y. The genome of Artemisia annua provides insight into the evolution of Asteraceae family and artemisinin biosynthesis. Mol. Plant. 2018;11:776–788. doi: 10.1016/j.molp.2018.03.015. [DOI] [PubMed] [Google Scholar]
- Shibuya M., Nishimura K., Yasuyama N., Ebizuka Y. Identification and characterization of glycosyltransferases involved in the biosynthesis of soyasaponin I in Glycine max. FEBS Lett. 2010;584:2258–2264. doi: 10.1016/j.febslet.2010.03.037. [DOI] [PubMed] [Google Scholar]
- Simao F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Song C., Zhao S., Hong X., Liu J., Schulenburg K., Schwab W. A UDP-glucosyltransferase functions in both acylphloroglucinol glucoside and anthocyanin biosynthesis in strawberry (Fragaria × ananassa) Plant J. 2016;85:730–742. doi: 10.1111/tpj.13140. [DOI] [PubMed] [Google Scholar]
- Su P., Du S., Li H., Li Z., Xin W., Zhang W. Notoginsenoside R1 inhibits oxidized low-density lipoprotein induced inflammatory cytokines production in human endothelial EA.hy926 cells. Eur. J. Pharmacol. 2016;770:9–15. doi: 10.1016/j.ejphar.2015.11.040. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Hayashi K.-i., Natsume M., Kamiya Y., Sakakibara H., Kawaide H., Kasahara H. UGT74D1 catalyzes the glucosylation of 2-oxindole-3-acetic acid in the auxin metabolic pathway in Arabidopsis. Plant Cell Physiol. 2014;55:218–228. doi: 10.1093/pcp/pct173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L., Su P., Zhang Z., Gao L., Wang J., Hu T., Zhou J., Zhang Y., Zhao Y., Liu Y. Genome of Tripterygium wilfordii and identification of cytochrome P450 involved in triptolide biosynthesis. Nat. Commun. 2020;11:971. doi: 10.1038/s41467-020-14776-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villano C., Esposito S., D'Amelia V., Garramone R., Alioto D., Zoina A., Aversano R., Carputo D. WRKY genes family study reveals tissue-specific and stress-responsive TFs in wild potato species. Sci. Rep. 2020;10:7196. doi: 10.1038/s41598-020-63823-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Wang J., Shi Y., Li R., Fan F., Huang Y., Li W., Chen N., Huang L., Dai Z. Elucidation of the complete biosynthetic pathway of the main triterpene glycosylation products of Panax notoginseng using a synthetic biology platform. Metab. Eng. 2020;61:131–140. doi: 10.1016/j.ymben.2020.05.007. [DOI] [PubMed] [Google Scholar]
- Wang P., Wei Y., Fan Y., Liu Q., Wei W., Yang C., Zhang L., Zhao G., Yue J., Yan X. Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metab. Eng. 2015;29:97–105. doi: 10.1016/j.ymben.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Wang T., Guo R., Zhou G., Zhou X., Kou Z., Sui F., Li C., Tang L., Wang Z. Traditional uses, botany, phytochemistry, pharmacology and toxicology of Panax notoginseng (Burk.) F.H. Chen: a review. J. Ethnopharmacol. 2016;188:234–258. doi: 10.1016/j.jep.2016.05.005. [DOI] [PubMed] [Google Scholar]
- Wei C., Yang H., Wang S., Zhao J., Liu C., Gao L., Xia E., Lu Y., Tai Y., She G. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. U S A. 2018;115:E4151–E4158. doi: 10.1073/pnas.1719622115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Wang P., Wei Y., Liu Q., Yang C., Zhao G., Yue J., Yan X., Zhou Z. Characterization of Panax ginseng UDP-glycosyltransferases catalyzing protopanaxatriol and biosyntheses of bioactive ginsenosides F1 and Rh1 in metabolically engineered yeasts. Mol. Plant. 2015;8:1412–1424. doi: 10.1016/j.molp.2015.05.010. [DOI] [PubMed] [Google Scholar]
- Xia E., Tong W., Hou Y., An Y., Chen L., Wu Q., Liu Y., Yu J., Li F., Li R. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into genome evolution and adaptation of tea plants. Mol. Plant. 2020;13:1013–1026. doi: 10.1016/j.molp.2020.04.010. [DOI] [PubMed] [Google Scholar]
- Xia E.H., Zhang H.B., Sheng J., Li K., Zhang Q.J., Kim C., Zhang Y., Liu Y., Zhu T., Li W. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant. 2017;10:866–877. doi: 10.1016/j.molp.2017.04.002. [DOI] [PubMed] [Google Scholar]
- Xie W., Zhou P., Qu M., Dai Z., Zhang X., Zhang C., Dong X., Sun G., Sun X. Ginsenoside Re attenuates high glucose-induced RF/6A injury via regulating PI3K/AKT inhibited HIF-1alpha/VEGF signaling pathway. Front. Pharmacol. 2020;11:695. doi: 10.3389/fphar.2020.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Chen L., Man J., Hu Y., Cui X. Chemical and bioactive comparison of Panax notoginseng root and rhizome in raw and steamed forms. J. Ginseng Res. 2019;43:385–393. doi: 10.1016/j.jgr.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Wang W., Wang B., Zhang T., Cui X., Pu Y., Li N. Analytical methods and biological activities of Panax notoginseng saponins: recent trends. J. Ethnopharmacol. 2019;236:443–465. doi: 10.1016/j.jep.2019.02.035. [DOI] [PubMed] [Google Scholar]
- Yan X., Fan Y., Wei W., Wang P., Liu Q., Wei Y., Zhang L., Zhao G., Yue J., Zhou Z. Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Res. 2014;24:770–773. doi: 10.1038/cr.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Chen Y., Shi J., Wang R., Yang Y., Yang L., Zhao S., Wang Z. Biosynthesis of rare 20(R)-protopanaxadiol/protopanaxatriol type ginsenosides through Escherichia coli engineered with uridine diphosphate glycosyltransferase genes. J. Ginseng Res. 2019;43:116–124. doi: 10.1016/j.jgr.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Zhang J., Zhang C., Zhang X., Ye J., Kuang S., Sun G., Sun X. Notoginsenoside R1 protects against diabetic cardiomyopathy through activating estrogen receptor alpha and its downstream signaling. Front. Pharmacol. 2018;9:1227. doi: 10.3389/fphar.2018.01227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Zhang X., Zhang C., Shen Q., Sun G., Sun X. Notoginsenoside R1 protects db/db mice against diabetic nephropathy via upregulation of Nrf2-mediated HO-1 expression. Molecules. 2019;24:247. doi: 10.3390/molecules24020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Li W., Xia E.-h., Zhang Q.-j., Liu Y., Zhang Y., Tong Y., Zhao Y., Niu Y.-c., Xu J.-h. The medicinal herb Panax notoginseng genome provides insights into ginsenoside biosynthesis and genome evolution. Mol. Plant. 2017;10:903–907. doi: 10.1016/j.molp.2017.02.011. [DOI] [PubMed] [Google Scholar]
- Zhang E.Y., Gao B., Shi H.L., Huang L.F., Yang L., Wu X.J., Wang Z.T. 20(S)-Protopanaxadiol enhances angiogenesis via HIF-1alpha-mediated VEGF secretion by activating p70S6 kinase and benefits wound healing in genetically diabetic mice. Exp. Mol. Med. 2017;49:e387. doi: 10.1038/emm.2017.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.J., Li W., Li K., Nan H., Shi C., Zhang Y., Dai Z.Y., Lin Y.L., Yang X.L., Tong Y. The chromosome-level reference genome of tea tree unveils recent bursts of non-autonomous LTR retrotransposons to drive genome size evolution. Mol. Plant. 2020;13:935–938. doi: 10.1016/j.molp.2020.04.009. [DOI] [PubMed] [Google Scholar]
- Zhang S., Zhu C., Lyu Y., Chen Y., Zhang Z., Lai Z., Lin Y. Genome-wide identification, molecular evolution, and expression analysis provide new insights into the APETALA2/ethylene responsive factor (AP2/ERF) superfamily in Dimocarpus longan Lour. BMC Genomics. 2020;21:62. doi: 10.1186/s12864-020-6469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Yang J., Cui M.Y., Liu J., Fang Y., Yan M., Qiu W., Shang H., Xu Z., Yidiresi R. The reference genome sequence of Scutellaria baicalensis provides insights into the evolution of wogonin biosynthesis. Mol. Plant. 2019;12:935–950. doi: 10.1016/j.molp.2019.04.002. [DOI] [PubMed] [Google Scholar]
- Zhong H., Wu H., Bai H., Wang M., Wen J., Gong J., Miao M., Yuan F. Panax notoginseng saponins promote liver regeneration through activation of the PI3K/AKT/mTOR cell proliferation pathway and upregulation of the AKT/Bad cell survival pathway in mice. BMC Comp. Altern. Med. 2019;19:122. doi: 10.1186/s12906-019-2536-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this work are available within the paper and its Supplemental Information files. The nucleotide sequencing data for UGT genes identified in this study have been deposited at NCBI GenBank under accession numbers MT551198 to MT551202. The genome sequence data of P. notoginseng have been deposited under NCBI BioProject number PRJNA658419https://www.Ncbi.nlm.nih.gov/bioproject/SUB7934826. In addition, the whole-genome sequence data reported in this paper have been deposited in the Genome Warehouse in the National Genomics Data Center (National Genomics Data Center and Partners, 2020), Beijing Institute of Genomics (China National Center for Bioinformation), Chinese Academy of Sciences, under accession number GWHAOSA00000000 and are publicly accessible at https://bigd.big.ac.cn/gsa.