Summary
Transcription through noncoding regions of the genome is pervasive. How these transcription events regulate gene expression remains poorly understood. Here, we report that, in S. cerevisiae, the levels of transcription through a noncoding region, IRT2, located upstream in the promoter of the inducer of meiosis, IME1, regulate opposing chromatin and transcription states. At low levels, the act of IRT2 transcription promotes histone exchange, delivering acetylated histone H3 lysine 56 to chromatin locally. The subsequent open chromatin state directs transcription factor recruitment and induces downstream transcription to repress the IME1 promoter and meiotic entry. Conversely, increasing transcription turns IRT2 into a repressor by promoting transcription-coupled chromatin assembly. The two opposing functions of IRT2 transcription shape a regulatory circuit, which ensures a robust cell-type-specific control of IME1 expression and yeast meiosis. Our data illustrate how intergenic transcription levels are key to controlling local chromatin state, gene expression, and cell fate outcomes.
Keywords: IME1, lncRNA, meiosis, Rtt109, yeast, transcription, chromatin, cell fate, H3K56ac, Rme1
Graphical Abstract

Highlights
-
•
Transcription levels of the lncRNA IRT2 regulate opposing local transcription states
-
•
Low levels of IRT2 promote H3K56ac, chromatin disassembly, and TF recruitment
-
•
Increasing IRT2 levels promote chromatin assembly and transcriptional repression
-
•
IRT2 transcription shapes the regulatory circuit for cell-type-specific control of yeast meiosis
Moretto et al. demonstrate that transcription levels of a lncRNA named IRT2 can have opposing effects on local gene transcription, thereby controlling entry into yeast meiosis. Low levels of IRT2 promote H3K56ac, chromatin disassembly, transcription factor recruitment, and, thus, transcriptional activation. Increasing levels promote chromatin assembly and, consequently, transcriptional repression.
Introduction
Transcription in intergenic regions of the genome is widespread. The noncoding RNAs emanating from these transcription events embody a large fraction of the transcriptome (Hon et al., 2017; Iyer et al., 2015; Kung et al., 2013). Particularly, the long noncoding RNAs (lncRNAs) constitute a diverse class of transcripts that control various cellular processes, including cell differentiation and stress, and have been implicated in diseases such as cancer (Guttman et al., 2011; Ponting et al., 2009; Wapinski and Chang, 2011). Despite extensive efforts, the function of the majority of noncoding RNAs remains poorly understood.
Noncoding transcription occurs near protein coding genes in promoter regions or at the 3′ ends of genes where they produce sense or antisense lncRNA transcripts (Neil et al., 2009; Pelechano and Steinmetz, 2013; Tisseur et al., 2011; Xu et al., 2009). Noncoding transcription can locally regulate the expression of coding genes through various mechanisms (Gil and Ulitsky, 2020; Wang and Chang, 2011). A widespread mechanism by which noncoding regions regulate gene expression is through the act of transcription via RNA polymerase II (Pol II) (Kornienko et al., 2013). During noncoding transcription, Pol II recruits chromatin remodelers that modify chromatin locally, thereby regulating the transcription of nearby genes (Ard et al., 2017; Venkatesh and Workman, 2015).
Noncoding transcription events can repress and activate gene transcription. Examples from yeasts have demonstrated that transcription through promoters of protein coding genes exerts gene repression (Ard et al., 2014; Bumgarner et al., 2009; Latos et al., 2012; Martens et al., 2004; Rom et al., 2019; van Werven et al., 2012). At these loci, chromatin regulators such as Facilitator of Chromatin Transcription (FACT), Set2, SET3C, and others mediate repression evoked by lncRNA transcription (Ard and Allshire, 2016; Hainer et al., 2011; Kim et al., 2012; van Werven et al., 2012). Conversely, noncoding transcription can also stimulate opening of chromatin and promote coding gene transcription (Hirota et al., 2008; Takemata et al., 2016). Transcription through enhancers, which produces enhancer RNAs (eRNAs), can contribute to enhancer activity and, thus, promotes gene expression (Li et al., 2016). How some noncoding transcription events promote while others repress gene expression remains unclear.
Two lncRNAs are transcribed in the promoter of the Inducer of Meiosis 1 gene, IME1 (Moretto et al., 2018; van Werven et al., 2012). This master transcription factor (TF) controls the cell fate decision of whether to enter meiosis (Kassir et al., 1988; Nachman et al., 2007). In diploid cells, expression of Ime1 activates the so-called early meiotic genes, thereby driving meiotic entry and the production of four haploid spores (Primig et al., 2000; van Werven and Amon, 2011). The IME1 gene is highly regulated at the level of transcription through its unusually large promoter (about 2.5 kb), at which nutrient and mating-type signals integrate (Tam and van Werven, 2020; van Werven and Amon, 2011). These signals ensure that IME1 transcription is only induced in cells expressing both mating-type loci (MATa and MATα) under starvation conditions.
Mating-type control of IME1 expression is mediated by the transcription of two lncRNAs in the IME1 promoter (Moretto et al., 2018; van Werven et al., 2012). In cells with a single mating type (MATa or MATα), typically haploid cells, transcription of a lncRNA named IME1 regulating transcript 1 (IRT1) represses the IME1 promoter to prevent meiotic entry (van Werven et al., 2012). The act of IRT1 transcription establishes a repressed chromatin state in the IME1 promoter, where TFs important for IME1 activation bind (Kahana et al., 2010; Sagee et al., 1998; Tam and van Werven, 2020; van Werven et al., 2012). In MATa/α diploid cells, IRT1 transcription is reduced, because the a1α2 heterodimer (expressed from opposite mating-type loci) represses the transcriptional activator of IRT1, RME1, enabling IME1 induction and, thus, meiotic entry (Figure 1A) (Mitchell and Herskowitz, 1986; van Werven et al., 2012). Despite the presence of a1α2 in MATa/α diploid cells, Rme1 (and, thus, IRT1) is expressed to moderate levels in various genetic backgrounds (Deutschbauer and Davis, 2005; Gerke et al., 2009). To overcome IRT1 transcription in MATa/α diploid cells, Ime1 activates the transcription of a second lncRNA, named IRT2, located upstream in its own promoter (Moretto et al., 2018). Transcription through IRT2, in turn, interferes with Rme1 recruitment, represses IRT1 transcription, and thereby promotes IME1 expression (Figure 1A).
Figure 1.
IRT2 is required for activation of IRT1 transcription
(A) Scheme of the two lncRNAs, IRT1 and IRT2, expressed in the IME1 promoter. In MATa/α (diploid) cells, RME1, the activator of IRT1, is repressed by a1α2. A feedback regulatory circuit consisting of IRT2, IRT1, and Ime1 facilitates IME1 expression in diploids. In single-mating-type cells (MATa or MATα, haploids), Rme1 is expressed, and IME1 expression is repressed by transcription of IRT1.
(B) IRT1, IRT2 (combined probe), and IME1 expression in MATa/α (FW1509) and MATa (FW1511) cells, as detected by northern blot. Cells were grown in rich medium (YPD) for 24 h and shifted to pre-sporulation medium (pre-SPO) for 16 h before being transferred and sampled in sporulation medium (SPO). SCR1 was used as a loading control. High-contrast blots for IRT1/IRT2 illustrate IRT2 signal in haploids.
(C) IRT1 and IRT2 transcription start site sequencing (TSS-seq), transcription end site sequencing (TES-seq), and RNA-seq data for MATa/α (6 h SPO, top panel) and MATa (4 h SPO, bottom panel). Cells were grown as described in (B). Blue lines indicate different IRT1 RNA isoforms. Red line indicates the IRT2 transcript, and light red line indicates where IRT2 transcription initiates. The y axes are in reads per million (RPMs).
(D) Haploid MATa cells (FW1509) were grown in YPD (24 h) before being transferred to SPO. An IRT2-specific probe is also featured.
(E) Similar to (B), except using MATa Δirt2(−188) (FW1210) and Δirt2(−246) (FW1356) mutants along with WT cells (FW1509).
(F) Rme1 association to the IRT1 promoter as detected by ChIP in mutants described in (E) but also harboring RME1 tagged with V5 epitope (FW4031, FW3132, and FW3140). Cells were grown as in (B). qPCR primer pair was nested over Rme1 binding sites in the IRT1 promoter. Signals were normalized to HMR, which does not bind Rme1. The error bars represent the standard error of the mean (SEM) of n = 5. ∗p < 0.05; ∗∗∗p < 0.005, compared to MATa control on a two-way ANOVA followed by a Fisher’s least significant difference (LSD) test.
See also Figure S1.
Here, we report the surprising finding of a dual function for transcription of the more upstream lncRNA in the IME1 promoter, IRT2. We show that the levels of IRT2 transcription regulate opposing chromatin and transcription states to ensure a robust transition from nutrient to mating-type control of the IME1 promoter. Low levels of IRT2 transcription direct H3 lysine 56 acetylation to chromatin, thereby promoting disassembly of chromatin, Rme1 recruitment, and activation of IRT1 transcription. Remarkably, increasing transcription converts IRT2 into a repressor. The dual function of IRT2 transcription shapes the regulatory circuit that ensures that only cells expressing opposite, but not one of either, mating-type loci (MATa and MATα) enter meiosis in yeast. Our data illustrate how noncoding transcription levels are key to controlling local chromatin state, gene expression, and cell fate outcomes.
Results
IRT2 is required for repression of IME1 in cells with a single mating type
We hypothesized that there is a mechanism to ensure a robust transition from nutrient to mating-type control of yeast meiosis, involving the two lncRNAs expressed in the IME1 promoter. To examine this, we determined IRT1 and IRT2 expression levels and mapped the transcription start sites (TSSs) and polyadenylation sites (transcription end sites; TESs) of both transcripts in cells harboring a single mating type (MATa) and both mating types (MATa/α) in the sporulation-proficient SK1 strain background. As expected, MATa/α diploid cells entering meiosis synchronously induced IME1 expression rapidly and displayed strong induction of IRT2, whereas IRT1 levels remained relatively low (Figures 1A, 1B, and S1A) (van Werven et al., 2012). In these cells (6 h in sporulation medium [SPO]; Figures 1C and S1B), we detected a single IRT2 TES while multiple TSSs spread over ∼215 base pairs (bp) were detected, matching the slight smear observed on the northern blot for the IRT2 transcript (Figure 1B). In haploid (MATa) cells, IRT1 expression was higher than in MATa/α cells, and IME1 expression was repressed (Figures 1A and 1B). The IRT1 TSS mapped to a single region, while three TES regions were mapped: one to the middle of IRT1 and two near the 3′ end (Figures 1C and S1B). Indeed, two distinct IRT1 species were detected by northern blot, which were reduced to one truncated transcript when IRT1 transcription was terminated early (irt1-T) (Figure 1B, lanes 7–10; Figure S1C). Surprisingly, we also detected low levels of IRT2 expression before IRT1 induction in MATa cells (Figure 1B, lane 6). The IRT2 transcript, TSS, and TES were also detectable at low levels in starved MATa cells (SPO 4 h), demonstrating that IRT2 is also expressed in this cell type (Figure S1D). To capture the IRT2 expression window in haploid MATa cells, we sampled during conditions for slow induction of IME1 and IRT1 expression (Figure S1A). Strikingly, we detected IRT2 expression in several time points before IRT1 induction (Figure 1D). Since the IME1 promoter is highly regulated by nutrient signaling, the expression of IRT2 that we detected may reflect changes in nutrient environment, such as glucose availability (van Werven and Amon, 2011).
To test whether IRT2 is required for IRT1 induction, we created deletions in the IRT2 TSS cluster, Δirt2(−188) and Δirt2(−246), while keeping the Rme1 binding sites intact (Figures 1E, S1E, and S1F). Remarkably, in Δirt2(−188) and Δirt2(−246) cells, IRT1 expression decreased; IME1 levels increased; and, as expected, IRT2 expression was not detectable. Both IRT2 mutants also displayed reduced association of Rme1 to the IRT1 promoter (Figure 1F). Thus, in addition to its transcriptional repressor function described in MATa/α cells (Moretto et al., 2018), IRT2 is also required for IRT1 expression and repression of the IME1 gene in cells with a single mating type.
The act of IRT2 transcription is required and sufficient for activating IRT1 expression
We next evaluated whether the act of transcription of IRT2 contributes to IRT1 activation. We integrated a transcriptional terminator between the IRT2 TSS and the Rme1 binding sites to generate the irt2-T allele (Figure 2A). A shorter form of IRT2 was detected in irt2-T cells (IRT2∗; Figure 2B). Remarkably, irt2-T cells showed diminished association of Rme1 with the IRT1 promoter, reduced IRT1 expression and Pol II binding to IRT1, and increased IME1 expression (Figures 2B, 2C, and S2A). A control sequence (irt-I) did not alter IRT1 and IME1 expression (Figure S2B, lanes 2–5 and 12–15). In addition, a transcriptional terminator integrated into IRT1 (irt1-T) showed wild-type (WT)-like Rme1 recruitment and displayed no additional reduction in Rme1 binding when combined with Δirt2(−246), indicating that transcription through IRT1 is not required for inducing IRT1 expression (Figure S2C).
Figure 2.
Transcription of IRT2 is required for induction of IRT1 expression
(A) Scheme of IME1 promoter harboring a transcriptional terminator integrated between the IRT2 TSS and the Rme1 binding site, irt2-T. Primer pairs (p1 and p2) used for ChIP in (C) and (D) are also depicted.
(B) IRT1, IRT2, and IME1 expression in WT and irt2-T MATa cells (FW1509 and FW3596). The asterisk depicts the IRT2 short form caused by early termination in IRT2.
(C) Rme1 association at the IRT1 promoter in WT and irt2-T MATa cells (FW4031 and FW3128) by ChIP. qPCRs were performed using primer pair p1 depicted in (A) Error bars represent SEM; n = 6. ∗∗∗p < 0.005, two-way ANOVA followed by a Fisher’s LSD test.
(D) Pol II association at IRT1 in MATa (control), irt2-T, and Δirt2(−246) (FW8515, FW8512, and FW8510, respectively) cells expressing FLAG-tagged Rpb3 along with a no-tag control (FW1509). qPCRs were performed using the primer pair p2. Error bars represent SEM; n = 4, except for the no-tag condition (n = 3). ∗∗∗p < 0.0005; ∗∗∗∗p < 0.0001, two-way ANOVA followed by a Fisher’s LSD test performed on the whole group of samples, including the one presented in Figure S2E.
(E) Schemes of the ime1-T mutant, which harbors point mutations in the C terminus of IME1 (which impairs Ime1 function), and u6bsΔ, which harbors a deletion of the IRT2 Ume6 binding site.
(F) IRT1 and IRT2 expression in WT and ime1-T MATa cells (FW1509 and FW2189) detected by northern blot. (G) IRT2 transcript start and end sites in u6bsΔ cells determined by TSS-seq and TES-seq during exponential growth in YPD.
(H) Same as (F), except that WT cells (FW1509, lanes 1–11) and u6bsΔ MATa cells (FW2438, lanes 12–22) were used, and cells were shifted to SPO before saturation.
(I) MATa WT cells (FW1509, lanes 1–8), or cells harboring ime1Δ together with the WT IRT2 promoter (FW1555, lanes 9–16) or a lexO1(−10) site (FW7142, lanes 17–24) were grown as in (F), and samples were taken at the indicated time points. Asterisk indicates a longer version of IRT2 originated from lexO1(−10) that has about the same size of one IRT1 isoform.
Additionally, we modulated IRT2 transcription without affecting the IRT2 sequence. We reasoned that, if IRT2 transcription was involved in IRT1 activation, then changing IRT2 expression should affect the IRT1 expression pattern. Therefore, we first abrogated IRT2 activation by introducing point mutations in Ime1 (ime1-T), which impairs Ime1 function (Moretto and van Werven, 2017). Little or no IRT1 expression was detected in the absence of IRT2 transcription (Figures 2E and 2F). Moreover, we constitutively expressed IRT2 by deleting the Ume6 repressor binding site (u6bsΔ) in the IRT2 promoter, which decouples IRT2 expression from IME1 activation (Figure 2E) (Moretto et al., 2018). To confirm that u6bsΔ gives rise to IRT2 transcription, we mapped the transcript (Figure 2G). As expected, the IRT2 start and end sites in u6bsΔ overlapped with positions we identified for IRT2 in WT cells (Figures 1C and S1D). Constitutive levels of IRT2 transcription (u6bsΔ) led to earlier IRT1 transcription and earlier Rme1 recruitment (Figures 2H and S2D–S2G). Furthermore, u6bsΔ rescued the IRT1 expression defect observed when Ime1 function was impaired (ime1-T), but not when IRT2 transcription was terminated earlier (irt2-T) (Figure S2H, lanes 7–9 and 10–12; and S2I, lanes 12–15 and 17–20). We conclude that IRT2 is required for IRT1 activation and that Ime1 activates IRT1 expression via IRT2 transcription.
We next determined whether IRT2 transcription was also sufficient for full activation of IRT1. To control IRT2 transcription, we used a system that can titrate the levels of transcription using a combination of lexO sites together with LexA-ER (estrogen receptor) activator and β-estradiol inducer (Ottoz et al., 2014). We integrated a single lexO site, lexO(−10), upstream of the Ume6 binding site in ime1Δ cells, thus preserving IRT2 repression (Figure 2I). Remarkably, we were able to rescue the IRT1 expression defects of ime1Δ cells with a single lexO(−10) site without the need to activate LexA-ER with β-estradiol (Figure 2I, lanes 17–24). These cells displayed constitutive low levels of IRT2 and marked levels of IRT1 expression across all time points, despite the presence of the Ume6 binding site. A similar result was obtained when using the GAL1-10 promoter (pGAL-IRT2) in cells harboring Gal4-ER (no β-estradiol) (Figure S2J). In the absence of Gal4-ER, we detected neither IRT2 expression nor IRT1 expression in pGAL-IRT2 cells, indicating that Gal4-ER was required for inducing IRT2 transcription and, consequently, IRT1 (Figure S2J). Taken together, low levels of IRT2 transcription are required and sufficient for induction of IRT1 expression in cells with a single mating type.
IRT2 transcription prevents meiosis in cells with a single mating type
Mis-expression of meiotic genes can have detrimental consequences to haploid cells (Lino and Yamamoto, 1985; Wagstaff et al., 1982). Haploid cells harboring a single mating type, but lacking Rme1 or IRT1, undergo a lethal type of meiosis (van Werven et al., 2012). We sought to determine the importance of IRT2-mediated activation of IRT1 in preventing haploid cells from entering meiosis. We found that in irt2 mutants Δirt2(−246) and irt2-T, a large fraction of cells displayed high levels of IME1 expression (more than 30 mRNA copies per cell) (Figures 3A, S3A, and S3B). After a prolonged period of starvation, irt2 mutants Δirt2(−188), Δirt2(−246), and irt2-T also displayed reduced viability, possibly due to entering meiosis (Figure 3B). We also generated diploid cells with a single mating type (MATa/a), mimicking mating-type repression of IME1 expression. Approximately 30% to 40% of MATa/a diploid cells for each irt2 mutant underwent at least one meiotic division (Figure 3C). This was comparable to the rme1 mutant that has impaired IRT1 transcription. Thus, IRT2 is essential for inhibiting meiotic entry in starved cells with a single mating type.
Figure 3.
Transcription of IRT2 prevents meiotic entry in cells with a single mating type
(A) IME1 expression in single cells, as measured by single-molecule RNA fluorescence in situ hybridization (FISH). MATa control, Δirt2(−246), and irt2-T (FW1533, FW3580, and FW3585) cells. The strains used also harbored a flo8 deletion. Formaldehyde-fixed cells were hybridized with probes directed against IME1 and ACT1. Each dot represents the number of IME1 transcripts in a single cell positive for ACT1 expression. Error bars represent SEM; n = 150 cells.
(B) Spot assay of cells on rich medium agar plates in 5-fold serial dilutions after 0 or 14 days in SPO. MATa control, Δirt2(−188), Δirt2(−246), and irt2-T cells (FW1509, FW1210, FW1356, and FW3596).
(C) MATa/a diploid control cells (FW15) and cells harboring rme1Δ, Δirt2(−188), Δirt2(−246), or irt2-T (FW1317, FW3453, FW3402, and FW3629) were grown as described in (A). DAPI masses were counted from cells fixed at 72 h in SPO. Cells harboring >2 DAPI masses were considered to have undergone meiotic divisions (MI + MII). Error bars represent ±SEM; n = 4, except for the control sample (n = 6). ∗∗p < 0.005; ∗∗∗p < 0.0005; and ∗∗∗∗p < 0.0001, on a one-way ANOVA followed by Fisher’s LSD test.
Rtt109 facilitates IRT2 transcription-mediated IRT1 activation
To gain insight into the mechanism by which IRT2 transcription stimulates IRT1 expression, we screened for mutants that displayed decreased IRT1 and increased IME1 expression. We selected a small set of known histone-modifying enzymes as well as several Pol II machinery factors. Gene deletions affecting Pol II transcription fidelity (RPB9 and CTK1), histone acetylation (GCN5 and RTT109), and histone chaperone function (ASF1) were identified (Figure 4A; Table S1). We focused our analyses on two candidate genes: RTT109 and ASF1. Rtt109 is the sole histone acetyltransferase that acetylates histone H3 lysine 56 (H3K56ac) in yeast, whereas Asf1 is involved directly in chromatin assembly and acts as a chaperone for Rtt109-directed H3K56ac (Driscoll et al., 2007; Masumoto et al., 2005; Recht et al., 2006; Schneider et al., 2006; Tsubota et al., 2007). H3K56ac-marked histones are assembled into nucleosomes during DNA replication, where they buffer gene transcription, but they are also present at promoters, where they are incorporated into nucleosomes in a replication-independent manner (Kaplan et al., 2008; Rufiange et al., 2007; Schneider et al., 2006; Voichek et al., 2016; Williams et al., 2008; Xu et al., 2005). Furthermore, nucleosomes harboring H3K56ac mark active transcription and active enhancers in higher eukaryotes (Schneider et al., 2006; Skalska et al., 2015; Värv et al., 2010).
Figure 4.
Rtt109 mediates IRT2-directed activation of IRT1 transcription
(A) Candidate genes involved in IRT2-mediated activation of IRT1. The minus symbol represents lower IRT1 expression compared to MATa WT cells, and the plus symbol represents higher IME1 expression compared to MATa WT cells.
(B) IRT1, IRT2, and IME1 expression in WT MATa (FW1509) and rtt109Δ (FW4077) cells detected by northern blot.
(C) Nascent RNA-seq (Pol II-associated RNA) for the IRT1 and IME1 loci from MATa control (FW4031) and rtt109Δ (FW4075) cells harvested 4 h in SPO. Two replicates are presented. Pol II was purified using the Rpb3-FLAG. The y axes indicate reads per million (RPM).
(D) Nascent RNA-seq read quantifications of selected transcripts (IRT1, IME1, and the controls RME1, ACT1, and SOD1) as obtained in (C). Error bars represent ±SEM; n = 2. ∗∗p < 0.005, parametric unpaired two-tailed Student’s t test.
(E) Pol II ChIP with IRT1 in control MATa and rtt109Δ cells (FW8515 and FW8561). Error bars represent SEM; n ≥ 3. ∗∗p < 0.005, two-way ANOVA followed by a Fisher’s LSD test performed on the whole group of samples, including those presented in Figure 2D.
(F) IRT1 and IRT2 expression in MATa ime1Δ cells with a lexO1(−10) site at IRT2 (FW7142) and in the same cells including a rtt109Δ mutation (FW8555). Cells were grown in YPD (24 h) and shifted to SPO.
(G) IRT1 and IRT2 expression in control MATa (FW1509), u6bsΔ (FW2438), rtt109Δ (FW4077), and u6bsΔ rtt109Δ double-mutant (FW5225) cells.
We found that, in rtt109Δ—and, to a lesser extent, in asf1Δ MATa cells—IRT1 expression was reduced and IME1 expression levels were increased (Figures 4B and S4A). Importantly, Rme1 protein levels were not affected in rtt109Δ cells (Figure S4B). In addition to steady-state RNA measurements, we also determined whether IRT1 and IME1 transcription (nascent RNA sequencing [RNA-seq] and Pol II chromatin immunoprecipitation [ChIP]) were affected in rtt109Δ cells during starvation (Figures 4C–4E, S4C, and S4D). rtt109Δ cells displayed reduced IRT1 transcription, less Pol II binding to IRT1, and increased IME1 transcription. We conclude that Rtt109 regulates IRT1 transcription.
One possible explanation for decreased IRT1 expression in rtt109Δ cells is that IRT2 transcription is affected. Therefore, we de-repressed IRT2 transcription using two different genetic approaches in the rtt109Δ background. First, we measured IRT1 expression in cells when IRT2 transcription was driven from a single lexO(−10) site. Whereas IRT2 was clearly expressed in lexO(−10) rtt109Δ cells, IRT1 expression was still compromised, suggesting that Rtt109 acts in concert with IRT2 transcription in inducing IRT1 (Figures 4F and S4F). Second, we combined the u6bsΔ and rtt109Δ mutants. De-repression of IRT2 transcription in u6bsΔ also did not rescue the IRT1 expression defect in rtt109Δ cells (Figure 4G, lanes 6–10, 11–15, and 16–20; Figure S4E). Furthermore, the IRT2 RNA was clearly detectable in u6bsΔ rtt109Δ and lexO(−10) rtt109Δ cells, suggesting that the IRT2 RNA by itself has little function in IRT2-mediated activation of IRT1 (Figures 4F and 4G, lane 16; Figure S4F). We conclude that Rtt109 likely acts downstream of IRT2 transcription in activating IRT1 transcription.
H3K56ac is directed to chromatin via IRT2 transcription and prevents meiotic entry
To further dissect how Rtt109 contributes to IRT1 activation, we combined the rtt109Δ with the early termination of IRT2 (irt2-T) allele and measured the effect on IRT1 activation. IRT1 expression in the irt2-T rtt109Δ double mutant was affected to a degree comparable to that of the single mutants (Figure 5A, lanes 6–10, 11–15, and 16–20; Figure S5A). Importantly, Rme1 recruitment was also affected to similar levels in the irt2-T rtt109Δ single and double mutants (Figure 5B). This suggests that Rtt109 acts downstream of IRT2 transcription and facilitates Rme1 recruitment and IRT1 activation.
Figure 5.
IRT2 transcription directs histone H3 lysine 56 acetylation to chromatin locally to activate IRT1 transcription
(A) Expression of IRT1, IRT2, and IME1 in MATa WT (FW1509), rtt109Δ (FW4077, lanes), irt2-T (FW3596), and rtt109Δ, irt2-T double-mutant (FW4072) cells.
(B) ChIP of Rme1-V5 at the IRT1 promoter in strains described in (A) but harboring the RME1-V5 allele (FW4031, FW3128, FW4075, and FW4073). Error bars represent ±SEM; n = 4, except for rtt109Δ and irt2-T rtt109Δ (n = 3). ∗∗∗p < 0.0005, two-way ANOVA followed by Fisher’s LSD test.
(C) Histone H3 lysine 56 acetylation (H3K56ac) levels in the IRT1 promoter as measured by ChIP in control (MATa, FW1509) and irt2-T (FW3596) cells. H3K56ac ChIP signals were normalized to histone H3. As control, rtt109Δ and irt2-T rtt109Δ cells (FW4077 and FW4072) were included. Error bars represent ±SEM; n = 5 for control, n = 4 for rtt109Δ, and n = 3 for irt2-T and irt2-T rtt109Δ. ∗p < 0.05; ∗∗p < 0.005, two-way ANOVA followed by Fisher’s LSD test performed on each primer pair individually.
(D) IRT1, IRT2, and IME1 expression in the MATa histone H3 control (FW5102), H3K56A (FW5113), and H3K56R (FW5116) cells.
(E) MATa/a diploid cells harboring rtt109Δ or H3K56R and matching controls (FW15, FW4557, FW7413, and FW7417). DAPI masses were counted from cells fixed at 72 h in SPO. Cells harboring >2 DAPI masses were considered to have undergone meiotic divisions (MI + MII). Error bars represent ±SEM; n = 7 for control and rtt109Δ, and n = 5 for H3 control and H3K56R. ∗∗p < 0.005; ∗∗∗∗p < 0.0001, on an unpaired parametric two-tailed Student t test comparing mutant with respective control.
Rtt109-mediated H3K56ac occurs off chromatin on newly synthesized histones (Driscoll et al., 2007; Han et al., 2007). In addition, H3K56ac can be incorporated in chromatin at promoter and transcribed regions in the genome (Rufiange et al., 2007; Schneider et al., 2006; Värv et al., 2010; Williams et al., 2008). To examine whether there is a link between IRT2 transcription and H3K56ac, we evaluated whether transcription of IRT2 directs H3K56ac to chromatin locally and whether H3K56ac mediates activation of IRT1 transcription. When we measured H3K56ac levels in the IRT2 region, we found that H3K56ac was enriched at the time of IRT2 transcription (0 h and 3 h), but not when IRT2 was repressed during exponential growth (YPD) or when RTT109 was deleted (Figure 5C). Importantly, H3K56ac levels were reduced in irt2-T cells, further supporting that IRT2 transcription is required for H3K56ac deposition (Figure 5C). This suggest that IRT2 transcription facilitates H3K56ac deposition in chromatin.
Although the main substrate of Rtt109 is H3K56, it is also known to acetylate lysine 9 of histone H3 (Adkins et al., 2007). To examine the role of H3K56ac directly, we mutated the H3K56 residue to alanine or arginine (H3K56A and H3K56R) to mimic the absence of H3K56ac in cells. The H3K56R mutant—and, to a lesser extent, H3K56A—displayed reduced IRT1 expression and increased IME1 expression (Figure 5D, lanes 1–5 and 11–15; Figures S5C and S5D). Importantly, IRT1 expression levels in the H3K56R mutant were affected to a degree comparable to that of the rtt109Δ H3K56R double mutant, suggesting that other targets of Rtt109 do not play a major role in IRT2-mediated activation of IRT1 (Figure S5C, lanes 14–16, 10–12, and 6–8, and S5D). H3K56ac was also necessary to prevent meiotic entry in cells with a single mating type. Approximately 20% to 30% of MATa/a diploid cells underwent at least one meiotic division when H3K56ac deposition was impaired (Figure 5E). These data demonstrate that Rtt109-mediated deposition of H3K56ac is critical for the activation of IRT1 and prevention of inappropriate meiotic entry.
IRT2 transcription has a level-dependent effect on local chromatin state
We showed that low levels of IRT2 transcription activate IRT1 expression by directing H3K56ac to chromatin (Figures 2H, 2I, and 4C). How is H3K56ac incorporated into chromatin by IRT2 transcription? Given that Rtt109-mediated H3K56ac occurs off chromatin, perhaps at low levels IRT2 transcription mediates histone exchange to deliver H3K56ac to chromatin (Tsubota et al., 2007). Indeed, it is known that transcription can deliver free histones to nucleosomes in exchange for old ones (Das and Tyler, 2013; Jackson, 1990; Venkatesh and Workman, 2015). To examine whether IRT2 promotes incorporation of new histones, we measured histone H3 exchange rates in the presence or absence of IRT2 transcription. A strain harboring differentially epitope-tagged histone H3, with one copy expressed from the endogenous promoter and the other expressed from a GAL1 inducible promoter, was used for the analysis (Figure 6A) (Schermer et al., 2005). Remarkably, the rate of incorporation of newly synthesized histone H3 significantly increased in the presence of constitutive levels of IRT2 transcription (Figure 6B; compare WT [no IRT2 transcription] to u6bsΔ [IRT2 transcription]). Galactose induction had no effect on IME1 expression under these conditions, which excludes the possibility that the effects were due to changes in IME1 promoter activity (Figure 6C). The histone H3 exchange rates at two control promoters were not affected by IRT2 transcription (Figures 6D and S6A). Importantly, elevated histone H3 exchange rate, as observed in the presence of constitutive levels of IRT2 transcription, correlated with the enrichment of H3K56ac at IRT2. We propose that low levels of IRT2 transcription stimulates histone exchange by a mechanism that is yet to be determined. Consequently, free histone H3, which is typically acetylated at lysine 56, is directed to chromatin. Subsequently, the presence of the H3K56ac mark facilitates chromatin disassembly, recruitment of Rme1, and activation of IRT1 transcription.
Figure 6.
IRT2 transcription has a level-dependent effect on local chromatin and transcription states
(A) Scheme for measuring histone H3 exchange rates.
(B) Histone H3 exchange rate at the IRT1 promoter in the presence or absence of IRT2 transcription. A strain harboring differentially epitope-tagged histone H3, with one copy expressed from the endogenous promoter (Myc-H3) and the other expressed from a GAL1-10 inducible promoter (pGAL-FLAG-H3) were used for the analysis. Constitutive low levels of IRT2 transcription are achieved using the u6bsΔ mutation (FW7880), whereas WT cells (FW7853) display no IRT2 transcription in rich medium (YP). Cells were grown till mid-log in YP raffinose and arrested in G1 with α factor, and FLAG-H3 was induced with galactose. The signals for H3 ChIP (Myc-H3 and FLAG-H3) were normalized to a telomere locus, and ratios for n = 3 (error bars represent ±SEM) are displayed. ∗p < 0.05; ∗∗p < 0.005; and ∗∗∗p < 0.0005, on a two-way ANOVA followed by Fisher’s LSD test. The slopes of the linear regression equations (Y = [0.07105 ⋅ X] + 1.466 for control, and Y = [0.1258 ⋅ X] + 3.325 for u6bsΔ) are significantly different.
(C) Relative expression of IRT2, IRT1, and IME1 in cells and grown as described in (B). qPCR signals were normalized to ACT1. Error bars represent ±SEM; n = 2. ∗p < 0.05; ∗∗p < 0.005, on an unpaired Student’s t test.
(D) Similar to (B), histone exchange at the PHO5 and PGK1 control promoters in strains described in (B). Error bars represent ±SEM; n = 3. The slopes of the linear regression equations for both loci are not significantly different.
(E) Scheme for controlling different levels of IRT2 by LexA-ER (top). Multiple lexA operator (lexO) sequences were integrated in the IRT2 promoter at +96 bp relative to IRT2 start site (lexO(+96)). LexA-ER is activated by β-estradiol. IRT1 and IRT2 levels as quantified by northern blot normalized to SCR1, in cells harboring 1, 2, 3, 4, and 8 lexO(+96) sites in the IRT2 promoter (FW6594, FW6599, FW6607, FW6611, and FW6619) at 1 h in SPO. The MATa LexA-ER control strain (control, FW6560) was included. Cells were treated (+β-estradiol) or not (mock) for 3 h and shifted to SPO plus β-estradiol. The ratio of +β-estradiol/mock is displayed. n = 2 data points and a trend line representing second-degree polynomial fit are shown.
(F) Chromatin structure at the IRT1 promoter in the presence of distinct levels of IRT2 transcription. Control cells (MATa LexA-ER, FW6560) or cells harboring 1 or 4 lexO(+96) sites (FW6594 or FW6611) were treated as described in (E). MNase-digested fragments were subject to qPCRs using primer pairs nested in IRT2. The red arrows indicate the position of the Rme1 binding sites. The signals were normalized over a telomere locus. Error bars represent ±SEM; n = 3. ∗p < 0.05; ∗∗∗p < 0.0005; and ∗∗∗∗p < 0.0001, on a two-way ANOVA followed by Fisher’s LSD test performed on lexO strains compared to control SPO for 3 h.
(G) MATa/α and MATa/a diploid cells (FW1511 and FW15) and MATa/a cells harboring pCUP-IRT2 (FW8923) were shifted to SPO and either treated (+Cu) or not (−Cu) with copper sulfate. DAPI masses were counted from cells fixed at 72 h in SPO. Cells harboring >2 DAPI masses were considered to have undergone meiotic divisions (MI + MII). Error bars represent ±SEM; n = 4 for the controls, and n = 5 for pCUP-IRT2. ∗∗∗∗p < 0.0001, two-way ANOVA followed by Fisher’s LSD test performed on MATa/a strains with or without copper sulfate treatment.
Previously, we showed that transcription of IRT2 is important for repressing IRT1 in MATa/α diploid cells (Moretto et al., 2018). In MATa/α diploid cells, IRT2 levels were much higher than in MATa haploid cells (Figure 1B). This raises the question of how IRT2 transcription levels control opposing regulatory outcomes on IRT1 transcription. In MATa/α diploid cells, IRT2 transcription increases nucleosome assembly around the Rme1 binding sites, which, consequently, interferes with Rme1 recruitment and activation of IRT1 transcription (Moretto et al., 2018). In the wake of transcription, nucleosomes disassemble and re-assemble to maintain the chromatin structure (Venkatesh and Workman, 2015). With this view, increasing IRT2 levels in MATa haploid cells should elevate transcription-coupled chromatin assembly and, thereby, turn IRT2 into a repressor of transcription. To test this, we modulated the level of IRT2 transcription with different degrees in MATa haploid cells. We integrated LexA operator (lexO) sequence repeats near the IRT2 TSS (lexOn(+96), +96 bp from the Ume6 binding site) and measured IRT2 and IRT1 levels together with nucleosome positioning (Figures 6E, 6F, and S6C–S6E). Upon activation by LexA-ER with β-estradiol, IRT2 levels as well as Pol II binding increased with the number of integrated lexOn(+96) repeats (Figures 6E and S6C–S6E). We found that the higher IRT2 transcription was, the greater the repression of IRT1 transcription was. As expected, nucleosome occupancy signal encompassing the Rme1 binding sites was reduced when IRT1 was transcribed (SPO, 3 h) compared to when the locus was repressed (YPD) (Figures 6F and S6F). With increasing levels of IRT2 transcription, the signals of the nucleosome around the Rme1 binding sites increased concomitantly (Figures 6F and S6F). Thus, with increasing IRT2 transcription levels, the rate of chromatin assembly increases as well as the degree of IRT1 transcription repression.
Taken together, these data indicate that low levels of IRT2 transcription direct H3K56ac to chromatin, which, in turn, promotes the recruitment of Rme1 and activation of IRT1 transcription. Conversely, increasing levels of IRT2 transcription set in place a well-positioned nucleosome that likely interferes with Rme1 recruitment and, consequently, represses IRT1 transcription (Moretto et al., 2018).
IRT2 transcription levels regulate the decision to enter meiosis
Our observation that IRT2 transcription displays an opposing effect on chromatin state and IRT1 expression prompted us to examine how increasing IRT2 levels affect the fate of cells. Specifically, we determined the effect of increased IRT2 transcription on meiosis in MATa/a diploids (which behave like MATa haploid cells) by replacing the endogenous promoter with the CUP1 promoter (pCUP-IRT2) (Figure 6G). Under noninducing conditions (−Cu) the CUP1 promoter is not fully repressed, which explains that MATa/a cells did not undergo meiotic divisions (Moretto et al., 2018). Approximately 50% of cells underwent meiosis when we induced IRT2 transcription to high levels (+Cu). These data show that increasing IRT2 transcription levels suppresses IRT1-mediated repression of the IME1 promoter, allowing these cells to enter meiosis. Conversely, low levels of IRT2 are required for activating IRT1 transcription and preventing meiotic entry (Figure 3G).
Mathematic model of how IRT2 transcription levels control meiotic entry
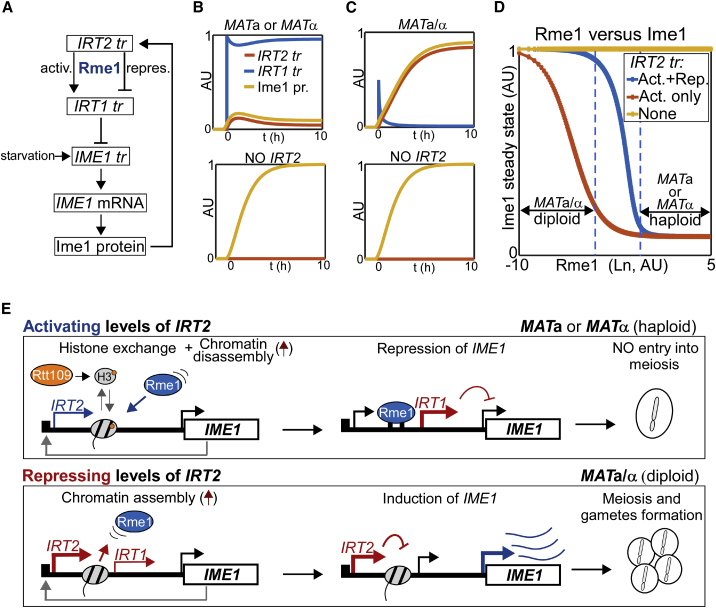
Our data demonstrate that low levels of IRT2 transcription activates IRT1 expression, whereas higher levels of IRT2 transcription repress IRT1 expression in a dose-dependent manner. IRT1 transcription levels are also linked to the levels of Rme1, which vary greatly between cells expressing a single mating type (MATa or MATα) and cells expressing opposite mating types (MATa/α), and between MATa/α cells of different genetic backgrounds (Deutschbauer and Davis, 2005; Gerke et al., 2009; Mitchell and Herskowitz, 1986). To quantitatively dissect how the different signals of IRT2 and Rme1 impinge on Ime1 expression, we developed a mathematical model describing the regulatory circuit consisting of IRT2, Rme1, IRT1, and IME1 (Figures 7A and S7A).
Figure 7.
Modeling of cell-type-specific control of Ime1 expression involving noncoding transcription
(A) Scheme of the mathematical model.
(B and C) Simulation of MATa and MATa/α cells in the presence or absence of IRT2.
(D) Simulation of Ime1 steady-state protein levels over a range of Rme1 concentrations (Ln), in the presence of either WT levels of IRT2 (activating and repressive effects on IRT1), activating levels of IRT2 (activating effect on IRT1 only), or no IRT2.
(E) Model describing how distinct levels of IRT2 control the decision of whether to enter meiosis and form gametes in haploid (MATa or MATα) and diploid (MATa/α) cells.
To test the model, we simulated high and low Rme1 levels representing the single mating type (MATa or MATα, haploid) and diploid (MATa/α) cell type, respectively, which resulted in the repression or activation of Ime1 expression (Figures 7B and 7C). In agreement with our experimental data, the model predicted that, in the absence of IRT2, activation of IME1 transcription is independent of mating-type status. We then assessed the dose-response association between Rme1 and Ime1 (Figures 7D, S7B, and S7C). The analyses revealed a sigmoidal relationship, where over a wide range of high to low Rme1 levels, Ime1 expression was either repressed or activated. The two extremes of the curve, Ime1 expression or repression, represent the ability for diploid (MATa/α) and the inability for haploid (MATa or MATα) cells to enter meiosis. Importantly, the absence of IRT2 transcription or the presence of only activating low levels of IRT2 abrogated the bimodal relationship between Rme1 and Ime1 expression levels (Figures 7D and S7C). The modeling further illustrates the importance of the dual function for IRT2 transcription in controlling the timely expression of Ime1 in a cell-type-specific manner and thereby regulating the decision to enter meiosis in yeast.
Discussion
We showed how transcription levels of an lncRNA have a critical role in regulating gene expression and cell fate outcomes. Specifically, we demonstrated that opposing transcription levels of the lncRNA IRT2 ensure a robust transition from nutrient to mating-type control of IME1 promoter activity. This dual role for IRT2 transcription is an essential component of the regulatory circuit enabling meiosis in yeast cells.
Mechanism of IRT2-mediated activation of transcription
Two lncRNAs, IRT1 and IRT2, are transcribed through different parts of the IME1 promoter to control IME1 expression and the decision to enter meiosis (Moretto et al., 2018; van Werven et al., 2012). Several lines of evidence indicate that IRT2 transcription directly promotes IRT1 transcription in single-mating-type cells. First, partial deletions in IRT2 compromise Rme1 recruitment and activation of IRT1 transcription. Second, insertion of a transcriptional terminator between the Rme1 binding sites and the IRT2 TSS affects IRT1 activation, suggesting that the act of transcription is required. Third, altered IRT2 transcription patterns due to mutations in Ime1 or the Ume6 binding sites upstream of IRT2 affected IRT1 expression in a comparable way. Finally, low levels of IRT2 transcription controlled from a heterologous promoter directly upstream of IRT2 were sufficient to induce IRT1 transcription.
How does IRT2 transcription promote IRT1 activation? We identified Rtt109 as a regulator of IRT1 transcription. Cells with a single mating type can enter meiosis when RTT109 is deleted or when H3K56ac, the main substrate of Rtt109, is directly disrupted. We propose that low levels of IRT2 transcription stimulate H3K56ac incorporation into nucleosomes locally. The H3K56ac mark, in turn, facilitates Rme1 recruitment and activation of IRT1 transcription. Our results are consistent with a model describing that H3K56ac increases nucleosome unwrapping, facilitating TF binding and transcription activation (Bernier et al., 2015; Neumann et al., 2009; Williams et al., 2008).
We show that IRT2 transcription stimulates histone exchange, thereby directing free histone H3K56ac to chromatin. The underlying mechanism, however, remains to be determined. One possibility is that, during Pol II transcription, chaperones facilitate the incorporation of free histones into nucleosomes (Park and Luger, 2008). During DNA replication in yeast, free histones and, thus, H3K56ac are directed to nucleosomes by chromatin assembly factors (Kaplan et al., 2008; Topal et al., 2019). Perhaps these factors also play a role during IRT2 transcription. Another possibility is that, in the wake of transcription, partial disassembly of nucleosomes leads to stochastic exchange of histones (Jamai et al., 2007). In line with both ideas, H3K56ac is enriched in transcribed gene bodies in the set2 deletion mutant—thus, in the absence of H3K36 methylation—suggesting that transcription promotes histone exchange (Venkatesh et al., 2012). It is important to note that free H3K56ac histones are unlikely to promote histone exchange themselves during transcription (Ferrari and Strubin, 2015). Finally, H3K56ac is widespread at promoter proximal nucleosomes where divergent noncoding transcription takes place, which raises the interesting possibility that H3K56ac incorporation via noncoding transcription may be widespread (Topal et al., 2019; Xu et al., 2009).
The transcription-controlled histone exchange and incorporation of acetylated histones into chromatin, as we described here, could be reminiscent of how a class of lncRNAs, called eRNAs, regulate gene expression (Li et al., 2016). Like IRT2 transcription, transcription through enhancers can facilitate recruitment of TFs and correlates with certain histone marks, including histone acetylation. Interestingly, in mammalian cells, H3K56ac is enriched at active enhancers and promoters, perhaps suggesting that noncoding transcription-directed H3K56ac deposition could be conserved (Skalska et al., 2015; Tan et al., 2013).
Model for the dual function of transcription of an lncRNA
Previously, we described the regulatory circuit consisting of IRT2, IRT1, and IME1 (Moretto et al., 2018). We showed that IRT2 transcription interferes with IRT1 activation, which, in turn, leads to the upregulation of IME1 expression and entry into meiosis in MATa/α cells. Here, we demonstrated that the IRT2 effect on gene regulation follows a hormetic pattern. In the absence of IRT2 transcription, no activation of IRT1 transcription will occur. However, relatively low levels of IRT2 will promote IRT1 activation, whereas increasing IRT2 transcription will repress IRT1 (Figure 7E).
How do transcription levels of IRT2 determine whether to activate or repress gene expression? Our data suggest that there is a dynamic interplay between nucleosome assembly, disassembly, and TF concentration. In the wake of transcription, nucleosomes disassemble and re-assemble to maintain chromatin structure (Ard et al., 2017; Venkatesh and Workman, 2015). With this view, as the transcription levels of IRT2 increases, the rate of transcription-coupled nucleosome assembly also increases, eventually leading to repression of IRT1. In activation context, IRT2 transcription directs histone H3K56ac to nucleosomes, which, in turn, facilitates nucleosome disassembly (Kaplan et al., 2008; Rufiange et al., 2007). In principle, one round of transcription can direct histone exchange and, thus, nucleosome disassembly. When the rate of transcription-coupled nucleosome assembly is higher than the rate of nucleosome disassembly, it “tips the scale” toward repression of IRT1 (Figure 7E). As depicted in our mathematical model, the concentration of Rme1 also plays an important role in activation of IRT1 transcription. The higher Rme1 levels are, the earlier a stable association to its binding site will occur and, thus, activate IRT1 transcription. Our model further shows that the dual function of IRT2 and Rme1 concentration in the cell form a circuit to regulate mating-type signaling to Ime1 expression. Taken together, our findings in this study of the IME1 promoter demonstrate that noncoding transcription levels play a determining role on whether repression or activation of gene transcription will occur.
Many examples of gene regulation by lncRNA transcription with different outcomes have been reported (Engreitz et al., 2016; Gil and Ulitsky, 2020; Hirota et al., 2008; Kornienko et al., 2013; Martens et al., 2004; van Werven et al., 2012). Our finding of distinct transcription levels of a noncoding RNA directly opposing chromatin and transcription states illustrates the gene regulatory potential for noncoding transcription events in general. Given that lncRNAs are transcribed across many parts of the genome, from yeast to humans, we propose that the act of noncoding transcription itself through promoters or other regulatory regions may have extensive functions in regulating gene expression (David et al., 2006; Hon et al., 2017; Iyer et al., 2015; Kung et al., 2013).
STAR★methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-V5 tag (mouse) antibody | Thermo Fisher Scientific | R96025; RRID: AB_2556564 |
| Anti-hexokinase (rabbit) antibody | US Biological | H2035; RRID: AB_2629457 |
| Anti-Myc tag (mouse) antibody | Merck Millipore | 05-724; RRID: AB_309938 |
| Anti-FLAG tag (mouse) antibody | Sigma-Aldrich (Merck) | F3165; RRID: AB_259529 |
| Anti-V5 agarose affinity gel | Sigma-Aldrich (Merck) | A7345; RRID: AB_10062721 |
| Anti-Mouse IgG IRDye® 800CW (goat) | LICOR (NE USA) | 925-32210; RRID: AB_2687825 |
| Anti-rabbit IgG IRDye 680RD (donkey) | LICOR (NE USA) | 925-68073; RRID: AB_2716687 |
| Anti-H3 (rabbit) antibody | Abcam | ab1791; RRID: AB_302613) |
| Anti-H3K56ac (rabbit) antibody | Sigma-Aldrich/Millipore (Merck) | 07-677-I; RRID: AB_390167 |
| Chemicals, peptides, and recombinant proteins | ||
| dATP [α-32P] | PerkinElmer | NEG512H500UC |
| Proteinase K | Thermo Fisher Scientific | EO0491 |
| rDNase | Machery-Nagel | 740963 |
| RNA Fragmentation Reagents (Ambion) | Thermo Fisher Scientific | AM8740 |
| Shrimp Alkaline Phosphatase (rSAP) | NEB | M0371L |
| Cap-Clip Acid Pyrophosphatase | CellScript | C-CC15011H |
| T4 RNA ligase 1 (high concentration) | NEB | M0437M |
| SuperScript IV Reverse Transcriptase | Thermo Fisher Scientific | 18090050 |
| RNasin Plus Ribonuclease Inhibitor | Promega | N2115 |
| RNase H | NEB | M0297L |
| RNase cocktail | Thermo Fisher Scientific | AM2286 |
| Dynabeads MyOne Streptavidin C1 | Thermo Fisher Scientific | 65002 |
| Phenol:chloroform: Isoamyl alcohol (125:24:1) | Ambion | AM9520 |
| Random hexamers | ThermoFisher Scientific | N8080127 |
| HighPrep PCR beads | MagBio | AC-60050 |
| Novex 6% TBE gels | Invitrogen | EC62655BOX |
| Costar SpinX column | Corning Incorporrated | 8161 |
| glass pre-filters | Whatman | 1823010 |
| linear acrylamide | Ambion | AM9520 |
| 5-Methylcytosine-dNTPs | Zymo Research | D1030 |
| NEbuffer 2 | NEB | B7002S |
| NAD+ | NEB | B9007S |
| E.coli DNA ligase | NEB | M0205S |
| DNA polymerase I | NEB | M0209S |
| GsuI | ThermoFisher Scientific | ER0461 |
| Halt Protease Inhibitor Cocktail (100X) | ThermoFisher Scientific | 78429 |
| SUPERase•In RNase Inhibitor (20 U/μL) | ThermoFisher Scientific | AM2694 |
| FLAG® Peptide | Millipore | F3290 |
| Critical commercial assays | ||
| Poly(A)Purist MAG kit | Ambion | AM1922 |
| KAPA RNA hyperPrep kit | Roche | KK8540 |
| Qbit RNA high sensitivity assay kit | ThermoFisher Scientific | Q32852 |
| KAPA Hyper Prep Kit | Roche | KK8504 |
| Qubit dsDNA HS assay kit | Invitrogen | Q32851 |
| RNeasy MinElute Cleanup Kits | QIAGEN | 74204 |
| Platinum SYBR mix | ThermoFisher Scientific | 11733046 |
| 2100 Bioanalyzer | Agilent | G2939BA |
| KAPA Hi-Fi hot start ready mix | Roche | KK2601 |
| KAPA single indexed adapters Set A | Roche | KK8701 |
| KAPA single indexed adapters Set B | Roche | KK8702 |
| Prime-It II Random Primer Labeling Kit | Agilent | 300385 |
| IME1 and ACT1 single molecule RNA-FISH probes | Biosearch Technologies | N/A |
| Deposited data | ||
| mRNA-seq, TSS-seq, TES-seq and PolII associated RNA-seq | This paper | GEO: GSE138898 |
| Experimental models: organisms/strains | ||
| S. cerevisiae: Strain background: BY, SK1 see Table S2 | This paper | N/A |
| Oligonucleotides | ||
| Oligonucleotides and primers, see Table S3 | This paper | N/A |
| Recombinant DNA | ||
| Plasmids, see Table S4 | This paper | N/A |
| Software and algorithms | ||
| Cutadapt (version 1.9.1) | (Martin, 2011) | https://cutadapt.readthedocs.io/en/stable/ |
| STAR (version 2.5.2) | (Dobin et al., 2013) | https://github.com/alexdobin/STAR |
| SAMTools (version 1.3.1) | (Li et al., 2009) | http://www.htslib.org/ |
| BEDTools (version 2.26.0) | (Quinlan and Hall, 2010) | https://bedtools.readthedocs.io/en/latest/ |
| BigWig and BigBed | (Kent et al., 2010) | http://hgdownload.cse.ucsc.edu/admin/exe/linux.x86_64/ |
| ImageJ (version 1.48k) | Schneider et al., 2012 | https://imagej.nih.gov/ij/index.html |
| MATLAB 2018a | MATLAB - MathWorks - MATLAB and Simulink | RRID: SCR_001622 |
| StarSearch | Raj Laboratory, University of Pennsylvania | https://rajlab.seas.upenn.edu/StarSearch/launch.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Folkert van Werven (folkert.vanwerven@crick.ac.uk).
Materials availability
Plasmids and strains generated for this study are available upon request to the lead contact.
Data and code availability
The accession number for the RNA sequencing, the TSS and TES sequencing, and the Pol II associated RNA sequencind data reported in this paper is GEO: GSE138898.
The code use for the mathematical modeling is available upon request to the lead contact.
Experimental model and subject details
Yeast strains
Yeast strains used in this paper were derived from the BY and SK1 strain background. Gene or promoter deletions were generated using the one-step deletion protocol as described previously (Longtine et al., 1998). The strain genotypes are listed in Table S2.
Growth and conditions
All experiments were performed at 30°C in a shaker incubator at 300rpm. A protocol for rapid induction of IME1 or IRT1 expression was described previously (van Werven et al., 2012). In short, cells were grown till saturation for 24h in YPD (1.0% (w/v) yeast extract, 2.0% (w/v) peptone, 2.0% (w/v) glucose, and supplemented with uracil (2.5 mg/l) and adenine (1.25 mg/l)), cells were then diluted at OD600 = 0.4 to pre-sporulation medium (1.0% (w/v) yeast extract, 2.0% (w/v) bacto tryptone, 1.0% (w/v) potassium acetate, 50 mM potassium phthalate) grown for about 16 h, subsequently centrifuged, washed with sterile miliQ water, centrifuged again, re-suspended at OD600 = 1.8 in sporulation medium (SPO) (0.3% (w/v) potassium acetate and 0.02% (w/v) raffinose)) and incubated at 30°C. For Figures 1D, 2E, 2H, 4F, and S2I, we used a protocol inducing IRT1 or IME1 expression with slow kinetics. In short, cells were grown till saturation for 24h in YPD then diluted in YPD aiming to reach OD600 = 6 after 16h, several samples were taken from that point till 24h, subsequently cells were washed, and transferred to SPO.
For the lexO/LexA-ER experiments described in Figures 6E, 6F, and S6B–S6E, cells were grown till saturation for 24h in YPD, diluted (OD600 = 0.4) in pre-sporulation medium and grown for another 16h. Cultures were then split and treated with β-estradiol (25nM) or ethanol (mock) for 1h, transferred to SPO (OD600 = 1.8), and incubated up till 3h in the presence of β-estradiol (15 nM) or ethanol.
For the and pGAL-IRT2/GAL4-ER experiment described in Figure S2I, cells were grown till saturation for 24h in YPD, diluted (OD600 = 0.4) in pre-sporulation medium and grown for another 16h. Subsequently samples were taken for the indicated time points.
For measuring histone exchange rates described in Figures 6 and S6, cells were grown till saturation for 24h in YPD then shifted and grown overnight in YP raffinose 2% (YPR) till it reached OD600 = 0.9. Cells were then arrested in G1 with α factor (5 μg/ml) for 2h, subsequently split to YP plus 2% galactose and YPR both containing α factor (5 μg/ml).
Method details
Plasmids and yeasts transformation
The RPB3-FLAG allele was generated through a one-step C-terminal tagging procedure of RPB3 with a FLAG tag cassette which contains three copies of the FLAG epitope (gift Jesper Svejstrup). MATa/a diploid strains were generated by replacing the MATα locus in MATa/α diploids with a plasmid linearized by EcoRI digestion harboring the MATa fragment with URA3 or TRP1 selectable markers (pRS304-MATa, this work; pRS306-MATa)(van Werven et al., 2012). The irt2-T allele harbors the CYC1 terminator sequence, which was integrated in the IRT2 locus using the ‘‘delitto perfetto’’ strategy (Storici et al., 2001). In short, Kluyveromyces lactis URA3 marker was first integrated in the IRT2 locus, subsequently a PCR product containing the CYC1 terminator and homology to the flanking sequences was used to excise the URA3 marker by 5-fluoro-orotic acid counter selection. As a control we also generated an irt2-I allele, which harbors a control insert from pUG6-Myc-Avitag (van Werven and Timmers, 2006). To excise the KanMX marker from irt2-I, we expressed the Cre recombinase from a plasmid (pRS304-GPDpr-CRE-EBD78-CYC1t, which is pTW040 re-cloned into pRS304, gift from Celine Bouchoux) (Terweij et al., 2013). After excision of the KanMX marker, genetic crosses were used to remove the pRS304-GPDpr-CRE-EBD78-CYC1t from irt2-I. Histone H3K56A and H3K56R mutants were generated through site directed mutagenesis using a plasmid carrying histone H3 and H4 genes along with a URA3 marker (HHT1 and HHF1,pDM9 (Sommermeyer et al., 2013), gift from Valerie Borde). Mutant plasmids were transformed into a strain with all four genomic copies of histone H3 and H4 genes deleted and harboring a plasmid wild-type for the histone H3 and H4 genes on TRP1 selectable marker (HHT2 and HHF2, pVB140 (Sommermeyer et al., 2013), gift from Valerie Borde). Genetic crosses were used to generate the histone H3K56A and H3K56R mutants in the absence of the wild-type histone H3 covering plasmid. The plasmids for generating strains with LexA operator (lexO) sequences and LexA fused to the estrogen receptor domain and activation domain (LexA-ER-HA-B112) were described previously (gift from Elçin Ünal) (Chia et al., 2017) (Ottoz et al., 2014). A different number of LexO sites were integrated at IRT2 locus position +96 or −10 bp from the Ume6 binding site. A plasmid expressing LexA-ER-HA-B112 from the GPD1 promoter was linearized with SfiI restriction enzyme and integrated at the TRP1 locus. The strains used for the histone H3 exchange assay described in Figures 6 and S6 were described previously (Schermer et al., 2005). In short, we used a strain with the endogenous copies of histone H3 and H4 deleted and covered by a centromeric plasmid pNOY439 harboring HHT2-Myc (histone Myc-H3) and untagged HHF2 (histone H4) under control of their endogenous promoters. In addition this stain also harbored a plasmid (YIplac211 pGAL1-10 HHF1 FLAG-HHT1) integrated at the URA3 locus with HHT1 FLAG tagged (histone FLAG-H3) and untagged HHF1 (histone H4) under control of the GAL1-10 promoter (Schermer et al., 2005). To generate pGAL-IRT2 strain, the GAL1-10 promoter was integrated 10 bp upstream of the Ume6 binding site. The GAL4-ER expression construct was described previously (Carlile and Amon, 2008). Plasmids are listed in Table S4.
Oligonucleotides Used in This Study
A table of oligonucleotides used in this study is available in Table S3.
Nuclei/DAPI counting
DAPI staining was used to monitor meiotic divisions throughout time courses. Cells were fixed in 80% (v/v) ethanol, pelleted by centrifugation and re-suspended in 100 mM phosphate buffer (pH 7) with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI). Cells were then sonicated for a few seconds and left in the dark at room temperature for at least 5 min. The proportion of cells containing one, two, three or four (meiosis I + II) DAPI masses were counted using a fluorescence microscope.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) experiments were performed as described previously (Moretto et al., 2018). Cells were fixed in 1.0% w/v formaldehyde for 25 min at room temperature and quenched with 100 mM glycine. Cells were lysed in FA lysis buffer (50 mM HEPES–KOH, pH 7.5, 150 mM NaCl, 1mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS and protease cocktail inhibitor used as recommended by the manufacturer (complete mini EDTA-free, Roche)) using beadbeater (BioSpec) and chromatin was sheared by sonication using a Bioruptor (Diagenode, 8 cycles of 30 s on/off). Extracts were incubated for 2 h at room temperature with anti-V5 agarose beads (Sigma) or overnight at 4°C with magnetic Prot A beads (Sigma) coupled with a polyclonal antibody raised against Histone H3 (Ab1791, Abcam) or Histone H3 acetylated lysine 56 (07-677-I, Millipore), washed twice with FA lysis buffer, twice with wash buffer 1 (FA lysis buffer containing 0.5M NaCl), and twice with wash buffer 2 (10 mM Tris–HCl, pH 8.0, 0.25M LiCl, 1 mM EDTA, 0.5% NP-40, 0.5% Na-deoxycholate). Subsequently, reverse cross-linking was done in 1% SDS-TE buffer (100 mM Tris pH 8.0, 10 mM EDTA, 1.0% v/v SDS) at 65°C overnight. After 2 h of proteinase K treatment, samples were purified, and DNA fragments were quantified by real-time PCR using SYBR green mix (Life Technologies) using primers described in Table S3. Signals were normalized over the HMR locus, which showed no binding for Rme1. For Figure 4G, H3K56ac ChIP signals were normalized to histone H3 ChIP. For the histone H3 turnover experiments described in Figures 6 and S6, ChIPs were performed as described above using antibodies against the Myc epitope (clone 9E11, Thermofisher) or against the FLAG epitope (M2 beads, Sigma). Signals were normalized using primers directed against a telomeric region from chromosome VI. For the Rpb3-FLAG ChIPs, we used FLAG-antibody coupled beads (M2 beads, Sigma). Signals were normalized over the HMR locus, which showed no binding for Rpb3.
Micrococcal nuclease (MNase) qPCR
Nucleosome positioning at the IRT2 locus was determined by quantifying the abundance of mononucleosomal DNA using a MNase digestion protocol that was described previously (Rando, 2010). In short, approximately 90 OD600 units of cells were crosslinked for 25 min at 30°C with 1% (v/v) formaldehyde. Reaction was quenched with the addition of glycine to 125 mM. Subsequently, cells were re-suspended in 20 mL of buffer Z (1 M sorbitol, 50 mM Tris-HCl pH 7.4) plus β-mercaptoethanol (10mM) and treated with 250 μg of T100 Zymolyase for 60 min. Next, cells were re-suspended in 1 mL NP buffer (0.5 mM spermidine, 1 mM β-mercaptoethanol (β -ME), 0.075% (w/v) Tergitol solution-type NP-40 detergent (NP-40), 50 mM NaCl, 10 mM Tris-HCl pH 7.4, 5 mM MgCl2, 1 mM CaCl2), vortexed for 10 s, and 100 μL of extract was treated with 0.2 μL of MNase (2mg/ml, NEB) for 30 min at 37°C, the reaction was quenched with 10 mM EDTA, and reverse crosslinked overnight in 1% SDS-TE and 4 units per ml of proteinase K (NEB). Samples were treated with RNase A and purified DNA fragments were separated by gel electrophoresis before gel purification of the mono-nucleosome bands. MNase treated and input samples were quantified by qPCR on a 7500 FAST Real-Time PCR machine (Life Technologies) using Platinum SYBR mix (Thermofisher). The signals were normalized using primers directed against a telomere locus. The scanning primer pairs covering the IRT2 locus and downstream region used for the analysis are available in Table S3.
Northern blotting
Northern blots were performed as described previously (Moretto et al., 2018). In short, total RNA was extracted with Acid Phenol:chloroform:Isoamyl alcohol (125:24:1) and precipitated in ethanol with 0.3 M sodium acetate. RNA samples were denatured in a glyoxal/DMSO mix (1 M deionized glyoxal, 50% v/v DMSO, 10 mM NaPi buffer pH 6.5-6.8) at 70°C for 10 min. Samples were mixed with loading buffer (10% v/v glycerol, 2 mM NaPi buffer pH 6.8, 0.4% w/v bromophenol blue) and separated on an agarose gel (1.1% w/v agarose, 0.01 M NaPi buffer pH 6.8) for 2 h at 80 V. RNAs were then transferred onto nylon membranes overnight by capillary transfer in 0.025 M NaPi buffer pH 6.8. Membranes were blocked for 2–3 h at 42°C in Hybridization buffer (1% w/v SDS, 50% v/v de-ionized formamide, 25% w/v dextran sulfate, 58 g/L NaCl, 200 mg/L herring sperm single strand DNA, 2 g/L BSA, 2 g/L polyvinyl-pyrrolidone, 2 g/L ficoll, 1.7 g/L pyrophosphate, 50 mM Tris pH 7.5) before hybridization. Radioactive probes were synthesized using a Prime-It II Random Primer Labeling Kit (Agilent), a target-specific DNA template and dATP [α-32P] (Perkin-Elmer). To avoid differences in signal intensity due to labeling quality and blotting variations, for each experiment samples were run on a single gel, transferred onto a single membrane, and hybridized with probes in a single hybridization tube. Background normalized quantifications of the Northern blots were done using Imagej software (Schneider et al., 2012). The oligo nucleotide sequences used to generate target-specific DNA template for amplifying the Northern blot probes are displayed in Table S3. Data S1 contains all the raw blots.
RNA-seq
At least 5 μg of total RNA was treated with DNase and purified on column (Macherey-Nagel). At least 500 ng of purified total RNA was used as input for the KAPA mRNA Hyper Prep kit (KK8580, Roche). Libraries were prepared according to manufacturer’s instructions. After bead based clean up, libraries were sequenced on an Illumina HiSeq 2500 to an equivalent of 75 bases single-end reads, at a depth of approximately 20 million reads per library.
TSS-seq and TES-seq
The TSS sequencing approach was adapted and modified from previously published protocols (Adjalley et al., 2016) (Arribere and Gilbert, 2013) (Malabat et al., 2015). At least 5 μg of mRNAs were purified from total RNA using the Poly(A)Purist MAG kit (AM1922, Ambion). poly(A)+ RNA/mRNAs, together with in vitro spike-ins, were fragmented for 3 min at 70°C using a Zinc-based alkaline fragmentation reagent (AM8740, Ambion). RNAs were cleaned up using RNeasy MinElute Cleanup Kits (74204, QIAGEN) to enrich for 200-300 nt fragments. These fragments were dephosphorylated with 30 units of recombinant shrimp alkaline phosphatase (M0371, NEB) for 1 h at 37°C with RNasin Plus, the phosphatase was heat inactivated and the RNA was extracted with Acid Phenol:chloroform: Isoamyl alcohol (125:24:1) and precipitated at −20°C overnight in ethanol with 0.3M sodium acetate and 1 μL linear acrylamide (AM9520, Ambion). RNA was then subjected to a decapping reaction with 2 units of Cap-Clip acid pyrophosphatase (C-CC15011H, Tebu-Bio) and with RNasin Plus. RNAs were then extracted using acid Phenol:chloroform:isoamyl alcohol (125:24:1) and precipitated in ethanol. Some RNA from a SPO (starvation) 0 h time point was set apart without the decapping reaction as a non-decapping control. Subsequently, the RNA was mixed with 10 μM of custom 5′ adaptor (CACTCTrGrArGrCrArArUrArCrC) and the ligation reaction was done using T4 RNA ligase 1 (M0437M, NEB) and with RNasin Plus. The ligation reaction was cleaned up with the RNeasy MinElute Cleanup Kit and RNAs were mixed with 2.5 μM random hexamers (N8080127, ThermoFisher Scientific) and RNasin Plus, denatured at 65°C for 5 min and cooled on ice. Reverse transcription reactions were carried out using SuperScript IV reverse transcriptase (18090010, Invitrogen) at 23°C for 10 min, 50°C for 10 min, 80°C for 10 min and held at 4°C. The RNA templates were degraded by incubating reactions with 5 units of RNase H (M0297, NEB) and 1.0 μL of RNase cocktail enzyme mix (AM2286, Ambion). DNA products were purified using 1.8x volume of HighPrep PCR beads (AC-60050, MagBio). Purified products were subjected to second strand synthesis using 0.3 μM of second strand biotinylated primer (GCAC/iBiodT/GCACTCTGAGCAATACC) and the KAPA Hi-Fi hot start ready mix (KK2601, Roche). The second strand reaction was carried out at 95°C for 3 min, 98°C for 15 s, 50°C for 2 min, 65°C for 15 min and held at 4°C. Double stranded product (dsDNA) was purified with 1.8x volume HighPrep PCR beads and concentration was quantified using the Qubit dsDNA HS assay kit (Q32851, Invitrogen). At least 1 ng of dsDNA was then used as input for the KAPA Hyper Prep Kit (KK8504, Roche) and ligated to KAPA single indexed adapters Set A (KK8701, Roche) or Set B (KK8702, Roche). Samples were processed according to manufacturer’s instructions with one exception: just prior to the library amplification step, samples were bound to MyOne Streptavidin C1 Dynabeads (65001, ThermoFisher Scientific) to capture biotinylated dsDNA. Library amplification was done on the biotinylated dsDNA fraction bound to the beads. Depending on the input amounts, 15 PCR cycles were used to generate libraries. Amplified libraries were quantified by Qubit, and adaptor-dimers were removed by electrophoresing libraries on Novex 6% TBE gels (EC62655BOX, Invitrogen) at 120 V for 1 h, and excising the smear above ∼150 bp. Gel slices containing libraries were shredded by centrifugation at 13000 g for 3 min. Gel shreds were re-suspended in 500 μL crush and soak buffer (500 mM NaCl, 1.0 mM EDTA and 0.05% v/v SDS) and incubated at 65°C for 2 h on a thermomixer (1400 rpm for 15 s, rest for 45 s). Subsequently, the buffer was transferred into a Costar SpinX column (8161, Corning Incorporrated) with two 1 cm glass pre-filters (1823010, Whatman). Columns were centrifuged at 13000 g for 1 min. DNA libraries in the flowthrough were precipitated at −20°C overnight in ethanol with 0.3 M sodium acetate and 1 μL linear acrylamide (AM9520, Ambion). Purified libraries were further quantified and inspected on a Tapestation (Agilent Technologies) and sequenced on an Illumina HiSeq 2500 to an equivalent of 75 bases single-end reads, at a depth of approximately 20 million reads per library.
The TES sequencing approach was adapted and modified from previously published protocols (Lai et al., 2015) (Ng et al., 2005). From the same pool of fragmented mRNAs describe above in the 5′ end sequencing, at least 1 μg was used for 3′ end sequencing. RNA fragments were mixed with 2.5 μM GsuI20TVN primer (/5BiotinTEG/ GAGCTAGTTCTGGAGTTTTTTTTTTTTTTTTTTTTVN), 0.5 mM 5-Methylcytosine-dNTPs (D1030, Zymo Research) and 0.5 μL RNasin Plus. Reaction mixtures were denatured at 65°C for 5 min and held at 50°C without allowing to cool. SuperScript IV, reaction buffer and 0.4 μg of Actinomycin D were added to the hot reaction mixtures and reverse transcription was performed at 50°C for 10 min, 80°C for 10 min and held at 4°C. Samples were cleaned with 1.8x volume HighPrep beads and biotinylated RNA:DNA hybrids were captured on MyOne Streptavidin C1 Dynabeads. After capture, streptavidin beads were washed once with 1x NEbuffer 2 (B7002S, NEB), re-suspended in water and subjected to second strand synthesis. The 50 μL second strand synthesis reaction consisted of 20 μL re-suspended streptavidin beads, 1X NEbuffer 2, 250 μM dNTPs, 26 μM NAD+ (B9007S), 2.5 units RNase H, 10 units E.coli DNA ligase (M0205S), and 15 units DNA polymerase I (M0209S). Second strand synthesis reactions were conducted at 16°C for 2.5 h on a thermomixer (1400 rpm for 15 s, rest for 2 min). After reaction, beads were washed once with 1x binding and washing buffer (5.0 mM Tris-HCl pH 7.5, 0.5 mM EDTA, 1.0 M NaCl) and once with buffer B (10 mM Tris-HCl pH 7.5, 10 mM MgCl2, 0.1 mg/ml BSA). Washed beads were re-suspended in 18 μL buffer B and digested with 10 units of GsuI (ER0461, ThermoFisher Scientific) at 30°C for 1 h on a thermomixer (1400 rpm for 15 s, rest for 2 min). After digestion, the DNA fragments in the supernatant were extracted with Phenol/chloroform and precipitated at −20°C overnight in ethanol with 0.3 M sodium acetate and 1 μL linear acrylamide. The concentration was quantified using the Qubit dsDNA HS assay kit. At least 1 ng of dsDNA was then used as input for the KAPA Hyper Prep Kit (KK8504, Roche) and ligated to KAPA single indexed adapters Set A (KK8701, Roche) or Set B (KK8702, Roche). Samples were processed according to manufacturer’s instructions. Amplified libraries were cleaned and purified by gel extraction using the procedures described in the previous section for TSS sequencing. Purified libraries were further quantified and inspected on a Tapestation (Agilent Technologies) and sequenced on an Illumina HiSeq 2500 to an equivalent of 75 bases single-end reads, at a depth of approximately 20 million reads per library.
Nascent RNA-Seq
The nascent RNA sequencing procedure was adapted as described in (Churchman and Weissman, 2011). Briefly, about one liter of yeast culture was spin down at the harvest time point and pellet snap frozen in liquid nitrogen. Cell pellets were then grinded in fine powder using a cryo-mill instrument with the instrument maintained cooled with liquid nitrogen. All the powder was re-suspended in about 12 to 15 mL of lysis buffer (20 mM HEPES pH 7.4, 110 mM K acetate, 0.5% Triton X-100, 0.1% Tween-20, 10 mM MnCl2, 1X protease inhibitor cocktail HALT, SUPERasin RNase inhibitor 50 U/ml) and lysate subjected to DNase I treatment (1200 U per sample) for 20 min. Lysate was then clarified through two rounds of centrifugation at about 20 000 g for 10 min at 4C. Anti-FLAG immunoprecipitation was performed using 750 μL of anti-FLAG M2 agarose beads per sample and for 2.5 h at 4C on a rotating wheel. Beads were then washed 5 times using 10 mL of cold wash buffer (20 mM HEPES pH 7.4, 110 mM K acetate, 0.5% Triton X-100, 0.1% Tween-20, 10 mM MnCl2, SUPERasin RNase inhibitor 1 U/ml, 1 mM EDTA) for 5 min at 4C. Beads were then cleaned using a Pierce column and two rounds of elution were performed using FLAG peptide (450 μL 10 mg/ml Sigma FLAG peptide) for 30 min at 4C. Elution was then subjected to a phenol:Chloroform extraction and RNA precipitated overnight in ethanol containing 0.3 M Na acetate. RNAs were then resuspended in Nuclease free water, quantified by Qbit RNA high sensitivity assay kit (Q32852, ThermoFisher Scientific) and analyzed by Bioanalyser (Agilent technologies).
At least 500 ng of purified RNA was treated with DNase and purified on column (Macherey-Nagel). Then, about 100 ng of purified total RNA was used as the inputs for the KAPA RNA hyperPrep kit (KK8540, Roche). Libraries were prepared according to manufacturer’s instructions. After bead based clean up, libraries were sequenced on an Illumina HiSeq 4000 to an equivalent of 75 bases single-end reads, at a depth of approximately 50 million reads per library.
Bioinformatic analysis
For RNA-seq data (including nascent RNA-seq), adaptor trimming was performed with cutadapt (version 1.9.1) with parameters “-a AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC–minimum-length=20.” STAR (version 2.5.2) with parameters “–alignIntronMin 3–alignIntronMax 5000” was used to perform the read mapping to the S. cerevisiae SK1 genome assembly from Keeney lab (http://cbio.mskcc.org/public/SK1_MvO/) (Martin, 2011). Alignments with mapping quality of < 10 or soft/hard-clipping were filtered (Dobin et al., 2013) (Martin, 2011). Alignments from forward strand and reverse strand were separated by using “samtools view -b -f 0x10” and “samtools view -b -F 0x10” to split the alignments. The tool “bedtools genomecov” (Quinlan and Hall, 2010) was used to generate the RNA-seq BedGraph tracks across the genome, for both forward and reverse strands. All the reads mapping to the rRNA were trimmed from the analyses prior the normalization. The BedGraph tracks were normalized by the number of usable reads in each library. BedGraph files were converted to bigWig using the tool bedGraphToBigWig from UCSC (Kent et al., 2010).
For the TSS transcript sequencing data, the custom 5′ adaptor sequence specific to the protocol was removed by re-running cutadapt with the parameters “-g CACTCTGAGCAATACC -O 16–minimum-length=20,” and only the reads containing the adaptor sequence were used for further analysis. STAR (version 2.5.2) with parameters “–alignIntronMin 2–alignIntronMax 1” (i.e., not allowing introns) was used to align TSS-seq reads to the SK1 genome assembly (plus three spike-in sequences) (Dobin et al., 2013). The alignments with mapping quality of > = 10 were kept for further analysis. Alignments from forward strand and reverse strand were separated by using “samtools view -b -f 0x10” and “samtools view -b -F 0x10.” The 5′-most nucleotide of aligned reads were extracted to generate the genome-wide tracks of TSSs. The BedGraph tracks were normalized by the number of usable reads in each library.
For the TES transcript sequencing data, Adaptor trimming was performed with cutadapt (version 1.9.1) with parameters “-a AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC–minimum-length=20.”
STAR (version 2.5.2) with parameters “–alignIntronMin 2–alignIntronMax 1” as for the 5′ end sequencing part, and alignments with mapping quality of > = 10 were kept for further analysis. The reads kept were those with soft-clipping at the 3′ end (size of soft-clipping part ≤ 10) and with at least two consecutive non-templated as in the soft-clipping part. Insertions/deletions were also not allowed. Alignments from forward strand and reverse strand were separated by using “samtools view -b -f 0x10” and “samtools view -b -F 0x10.” The 3′-most nucleotide of aligned reads were extracted to generate the genome-wide tracks of TESs. The BedGraph tracks were normalized by the number of usable reads in each library.
Single molecule RNA fluorescent in situ hybridization (FISH)
The single molecule RNA FISH was performed as described previously (Moretto et al., 2018). In short, cells were fixed with formaldehyde overnight, treated with zymolyase and further fixed in 80% ethanol. Subsequently cells were hybridized with fluorophore labeled probes directed to IME1 (AF594) and ACT1 (Cy5) as an internal control. Cells were imaged using a 100x oil objective, NA 1.4, on a Nikon TI-E imaging system (Nikon). DIC, DAPI, AF594 (IME1), Cy5 (ACT1) images were collected every 0.3 micron (20 stacks) using an ORCA-FLASH 4.0 camera (Hamamatsu) and NIS-element software (Nikon). ImageJ software was used to make maximum intensity Z projections of the images (Schneider et al., 2012). StarSearch software (https://rajlab.seas.upenn.edu/StarSearch/launch.html, Raj laboratory, University of Pennsylvania) was used to quantify transcripts in single cells. Comparable thresholds were used to count RNA foci in single cells. Only cells positive for the internal control ACT1 were used for IME1 analysis. At least a total n = 150 cells were counted for each experiment.
Western blotting
Western blots were performed as previously described (Moretto et al., 2018). Protein extracts were prepared using the trichloroacetic acid (TCA) extraction protocol. After SDS-polyacrylamide gel electrophoresis (4%–20% gradient), proteins were transferred onto PVDF membranes. The membranes were then incubated overnight primary antibodies in blocking buffer. Mouse anti-v5 (R96025, Sigma-Aldrich (MO, USA)) was used at a 1:2000 dilution and rabbit anti-hexokinase antibody (H2035, Stratech (Newmarket, UK)) at a 1:8000 dilution. Membranes were then washed in PBST buffer and incubated with IRDye 800CW goat anti-mouse and IRDye 680RD donkey anti-rabbit secondary antibodies (LI-COR (NE, USA)) at a 1:15000 dilution for LI-COR detection. Protein levels were detected on an Odyssey Imager for LI-COR detection.
Spot growth assay
Cells were grown using the protocol for rapid induction of IRT1. After 0 or 14 days in SPO cells were diluted to OD600 = 1 in sterile water, and serial dilutions (5-fold) were spotted onto YPD agar plates. Cells were incubated at 30°C for 2 days before imaging.
Mathematical modeling
To investigate of the complex regulation of IME1 expression by IRT1 and IRT2, we developed a mathematical model. We incorporated new observations described in this manuscript, into the model described previously (Moretto et al., 2018). Specifically, the new mathematical model describes both the activating and repressing properties of IRT2 on IRT1 expression. The IRT2 activating property is incorporated by controlling the IRT1 transcription rate with the indicator function, , which is 1 when IRT2 transcription rate reaches an activating level threshold, . After the activation of IRT1 remains at 1 even when IRT2 transcription rate falls below . In addition, the model includes Rme1 concentration, , which is assumed to be constant during the time-period of Ime1 activation. By changing Rme1 concentrations, , we can simulate both haploid and diploid cell-types, which differ significantly in their Rme1 levels. The model variables are listed below. The parameters have been chosen to reproduce the qualitative behavior of this system.
List of variables (scaled between 0 and 1)
| Starvation signal | |
|---|---|
| IRT1 transcription rate | |
| IRT2 transcription rate | |
| Ime1 transcription rate | |
| Ime1 mRNA concentration | |
| Ime1 protein concentration |
List of parameters used for simulation
| controls the strength of inhibition by | 5 | |
|---|---|---|
| threshold for activating | 0.05 | |
| controls the strength of inhibition by | 10 | |
| degradation rate of | 1 /h | |
| synthesis rate of | 1 /h | |
| degradation rate of | 1 /h | |
| Rme1 concentration | 0-5 | |
| threshold for activating | 0.01 |
Equations
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
The model is simulated with the initial conditions and .
The first term in Equation 5, , describes the IME1 transcription rate.
Equation 1 models the starvation signal as a step input. Upon starvation, IME1 mRNA and in turn Ime1 protein are synthesized. The evolution of is given by Equation 5 and of is given by Equation 6. When reaches the threshold , IRT2 transcription,, turns on. Here we assumed that IRT2 transcription closely follows Ime1 protein concentration (Equation 4). Once reaches the threshold , the indicator function is set at 1 (Equation 2) and IRT1 transcription starts(Equation 3). Note that remains equal to 1, even if falls below , as long as is greater than 0. is driven by the Rme1 concentration, , which is assumed to be constant in the cell through the time period of Ime1 activation. Activation of suppresses to various degrees depending on (Figures 7A and S7A). The mathematical model shows how IRT1 and IRT2 control expression of IME1 and Ime1 protein. Activating IRT2 and no IRT2 transcription are simulated by setting and respectively, for (Figures 7D and S7C). The mathematical model is simulated in MATLAB 2018a using ode45 function. Code is available upon request.
Quantification and statistical analysis
Statistical significance and tests are indicated in the figure legends and were performed using GraphPad Prism 7 and 8. When comparing only two groups of samples, we have used a parametric two-tailed Student’s t test. When assessing the variance of more than two groups we performed an ANOVA analysis (one way or two ways in function of the experimental design), followed by a Fisher LSD test. Fisher’s LSD test confer more power to the test, compared to performing multiple Student’s t tests, in that t tests compute the pooled standard deviations from only the two groups being compared, while the Fisher’s LSD test computes the pooled standard deviations from all the groups while assuming that all population have similar standard deviation.
Acknowledgments
We are grateful to Frank Uhlmann, Jesper Svejstrup, and members of the van Werven lab for advice and critical reading of the manuscript. We thank Elçin Ünal, Philipp Korber, Valerie Borde, and Celine Bouchoux for sharing reagents. N.E.W. is supported by the Cancer Prevention and Research Institute of Texas (RR150058, which was obtained by Andreas Doncic). F.M. and F.J.v.W. are supported by the Francis Crick Institute (FC001203), which receives its core funding from Cancer Research UK (FC001203), the UK Medical Research Council (FC001203), and the Wellcome Trust (FC001203). C.L. and N.M.L. are supported by the Francis Crick Institute (FC010110), which receives its core funding from Cancer Research UK (FC010110), the UK Medical Research Council (FC01011), and the Wellcome Trust (FC010110). M.C. is supported by a fellowship from the Agency for Science, Technology and Research (A∗STAR) of Singapore.
Author contributions
F.M. and F.J.v.W. conceived and designed the study. F.M. and F.J.v.W. designed the experiments. F.M. performed the experiments. F.M. and F.J.v.W. analyzed the data. N.E.W. generated the mathematical model. M.C., C.L., and N.M.L. helped with generating and analyzing sequencing data. F.M. and F.J.v.W. wrote the manuscript, with input from all authors. F.J.v.W. supervised the project.
Declaration of interests
The authors declare no competing interests.
Published: January 19, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108643.
Supplemental information
References
- Adjalley S.H., Chabbert C.D., Klaus B., Pelechano V., Steinmetz L.M. Landscape and Dynamics of Transcription Initiation in the Malaria Parasite Plasmodium falciparum. Cell Rep. 2016;14:2463–2475. doi: 10.1016/j.celrep.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkins M.W., Carson J.J., English C.M., Ramey C.J., Tyler J.K. The histone chaperone anti-silencing function 1 stimulates the acetylation of newly synthesized histone H3 in S-phase. J. Biol. Chem. 2007;282:1334–1340. doi: 10.1074/jbc.M608025200. [DOI] [PubMed] [Google Scholar]
- Ard R., Allshire R.C. Transcription-coupled changes to chromatin underpin gene silencing by transcriptional interference. Nucleic Acids Res. 2016;44:10619–10630. doi: 10.1093/nar/gkw801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ard R., Tong P., Allshire R.C. Long non-coding RNA-mediated transcriptional interference of a permease gene confers drug tolerance in fission yeast. Nat. Commun. 2014;5:5576. doi: 10.1038/ncomms6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ard R., Allshire R.C., Marquardt S. Emerging Properties and Functional Consequences of Noncoding Transcription. Genetics. 2017;207:357–367. doi: 10.1534/genetics.117.300095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J.A., Gilbert W.V. Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome Res. 2013;23:977–987. doi: 10.1101/gr.150342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier M., Luo Y., Nwokelo K.C., Goodwin M., Dreher S.J., Zhang P., Parthun M.R., Fondufe-Mittendorf Y., Ottesen J.J., Poirier M.G. Linker histone H1 and H3K56 acetylation are antagonistic regulators of nucleosome dynamics. Nat. Commun. 2015;6:10152. doi: 10.1038/ncomms10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumgarner S.L., Dowell R.D., Grisafi P., Gifford D.K., Fink G.R. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc. Natl. Acad. Sci. USA. 2009;106:18321–18326. doi: 10.1073/pnas.0909641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile T.M., Amon A. Meiosis I is established through division-specific translational control of a cyclin. Cell. 2008;133:280–291. doi: 10.1016/j.cell.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia M., Tresenrider A., Chen J., Spedale G., Jorgensen V., Unal E., van Werven F.J. Transcription of a 5’ extended mRNA isoform directs dynamic chromatin changes and interference of a downstream promoter. Elife. 2017;6 doi: 10.7554/eLife.27420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C., Tyler J.K. Histone exchange and histone modifications during transcription and aging. Biochim. Biophys. Acta. 2013;1819:332–342. doi: 10.1016/j.bbagrm.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L., Huber W., Granovskaia M., Toedling J., Palm C.J., Bofkin L., Jones T., Davis R.W., Steinmetz L.M. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschbauer A.M., Davis R.W. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat. Genet. 2005;37:1333–1340. doi: 10.1038/ng1674. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll R., Hudson A., Jackson S.P. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz J.M., Haines J.E., Perez E.M., Munson G., Chen J., Kane M., McDonel P.E., Guttman M., Lander E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari P., Strubin M. Uncoupling histone turnover from transcription-associated histone H3 modifications. Nucleic Acids Res. 2015;43:3972–3985. doi: 10.1093/nar/gkv282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke J., Lorenz K., Cohen B. Genetic interactions between transcription factors cause natural variation in yeast. Science. 2009;323:498–501. doi: 10.1126/science.1166426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil N., Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 2020;21:102–117. doi: 10.1038/s41576-019-0184-5. [DOI] [PubMed] [Google Scholar]
- Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G., Young G., Lucas A.B., Ach R., Bruhn L. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer S.J., Pruneski J.A., Mitchell R.D., Monteverde R.M., Martens J.A. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 2011;25:29–40. doi: 10.1101/gad.1975011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Zhou H., Horazdovsky B., Zhang K., Xu R.M., Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- Hirota K., Miyoshi T., Kugou K., Hoffman C.S., Shibata T., Ohta K. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature. 2008;456:130–134. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- Hon C.C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J., Gough J., Denisenko E., Schmeier S., Poulsen T.M., Severin J. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V. In vivo studies on the dynamics of histone-DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry. 1990;29:719–731. doi: 10.1021/bi00455a019. [DOI] [PubMed] [Google Scholar]
- Jamai A., Imoberdorf R.M., Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol. Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- Kahana S., Pnueli L., Kainth P., Sassi H.E., Andrews B., Kassir Y. Functional dissection of IME1 transcription using quantitative promoter-reporter screening. Genetics. 2010;186:829–841. doi: 10.1534/genetics.110.122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan T., Liu C.L., Erkmann J.A., Holik J., Grunstein M., Kaufman P.D., Friedman N., Rando O.J. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 2008;4:e1000270. doi: 10.1371/journal.pgen.1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassir Y., Granot D., Simchen G. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell. 1988;52:853–862. doi: 10.1016/0092-8674(88)90427-8. [DOI] [PubMed] [Google Scholar]
- Kent W.J., Zweig A.S., Barber G., Hinrichs A.S., Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Xu Z., Clauder-Münster S., Steinmetz L.M., Buratowski S. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012;150:1158–1169. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornienko A.E., Guenzl P.M., Barlow D.P., Pauler F.M. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung J.T., Colognori D., Lee J.T. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai D.P., Tan S., Kang Y.N., Wu J., Ooi H.S., Chen J., Shen T.T., Qi Y., Zhang X. Genome-wide profiling of polyadenylation sites reveals a link between selective polyadenylation and cancer metastasis. Hum Mol Genet. 2015;24:3410–3417. doi: 10.1093/hmg/ddv089. [DOI] [PubMed] [Google Scholar]
- Latos P.A., Pauler F.M., Koerner M.V., Şenergin H.B., Hudson Q.J., Stocsits R.R., Allhoff W., Stricker S.H., Klement R.M., Warczok K.E. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 10000 Genomes Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Notani D., Rosenfeld M.G. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat. Rev. Genet. 2016;17:207–223. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- Lino Y., Yamamoto M. Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol. Gen. Genet. 1985;198:416–421. doi: 10.1007/BF00332932. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., 3rd, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Malabat C., Feuerbach F., Ma L., Saveanu C., Jacquier A. Quality control of transcription start site selection by nonsense-mediated-mRNA decay. Elife. 2015;4 doi: 10.7554/eLife.06722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J.A., Laprade L., Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- Martin Marcel. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- Masumoto H., Hawke D., Kobayashi R., Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- Mitchell A.P., Herskowitz I. Activation of meiosis and sporulation by repression of the RME1 product in yeast. Nature. 1986;319:738–742. doi: 10.1038/319738a0. [DOI] [PubMed] [Google Scholar]
- Moretto F., van Werven F.J. Transcription of the mating-type-regulated lncRNA IRT1 is governed by TORC1 and PKA. Curr. Genet. 2017;63:325–329. doi: 10.1007/s00294-016-0639-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretto F., Wood N.E., Kelly G., Doncic A., van Werven F.J. A regulatory circuit of two lncRNAs and a master regulator directs cell fate in yeast. Nat. Commun. 2018;9:780. doi: 10.1038/s41467-018-03213-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman I., Regev A., Ramanathan S. Dissecting timing variability in yeast meiosis. Cell. 2007;131:544–556. doi: 10.1016/j.cell.2007.09.044. [DOI] [PubMed] [Google Scholar]
- Neil H., Malabat C., d’Aubenton-Carafa Y., Xu Z., Steinmetz L.M., Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- Neumann H., Hancock S.M., Buning R., Routh A., Chapman L., Somers J., Owen-Hughes T., van Noort J., Rhodes D., Chin J.W. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol. Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P., Wei C.L., Sung W.K., Chiu K.P., Lipovich L., Ang C.C., Gupta S., Shahab A., Ridwan A. Gene identification signature (GIS) analysis for transcriptome characterization and genome annotation. Nat Methods. 2005;2:105–111. doi: 10.1038/nmeth733. [DOI] [PubMed] [Google Scholar]
- Ottoz D.S., Rudolf F., Stelling J. Inducible, tightly regulated and growth condition-independent transcription factor in Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42:e130. doi: 10.1093/nar/gku616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.J., Luger K. Histone chaperones in nucleosome eviction and histone exchange. Curr. Opin. Struct. Biol. 2008;18:282–289. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V., Steinmetz L.M. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013;14:880–893. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Primig M., Williams R.M., Winzeler E.A., Tevzadze G.G., Conway A.R., Hwang S.Y., Davis R.W., Esposito R.E. The core meiotic transcriptome in budding yeasts. Nat. Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando O.J. Genome-wide mapping of nucleosomes in yeast. Methods Enzymol. 2010;470:105–118. doi: 10.1016/S0076-6879(10)70005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recht J., Tsubota T., Tanny J.C., Diaz R.L., Berger J.M., Zhang X., Garcia B.A., Shabanowitz J., Burlingame A.L., Hunt D.F. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc. Natl. Acad. Sci. USA. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rom A., Melamed L., Gil N., Goldrich M.J., Kadir R., Golan M., Biton I., Perry R.B., Ulitsky I. Regulation of CHD2 expression by the Chaserr long noncoding RNA gene is essential for viability. Nat. Commun. 2019;10:5092. doi: 10.1038/s41467-019-13075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufiange A., Jacques P.E., Bhat W., Robert F., Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Sagee S., Sherman A., Shenhar G., Robzyk K., Ben-Doy N., Simchen G., Kassir Y. Multiple and distinct activation and repression sequences mediate the regulated transcription of IME1, a transcriptional activator of meiosis-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1998;18:1985–1995. doi: 10.1128/mcb.18.4.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermer U.J., Korber P., Hörz W. Histones are incorporated in trans during reassembly of the yeast PHO5 promoter. Mol. Cell. 2005;19:279–285. doi: 10.1016/j.molcel.2005.05.028. [DOI] [PubMed] [Google Scholar]
- Schneider J., Bajwa P., Johnson F.C., Bhaumik S.R., Shilatifard A. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J. Biol. Chem. 2006;281:37270–37274. doi: 10.1074/jbc.C600265200. [DOI] [PubMed] [Google Scholar]
- Skalska L., Stojnic R., Li J., Fischer B., Cerda-Moya G., Sakai H., Tajbakhsh S., Russell S., Adryan B., Bray S.J. Chromatin signatures at Notch-regulated enhancers reveal large-scale changes in H3K56ac upon activation. EMBO J. 2015;34:1889–1904. doi: 10.15252/embj.201489923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommermeyer V., Beneut C., Chaplais E., Serrentino M.E., Borde V. Spp1, a member of the Set1 Complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol Cell. 2013;49:43–54. doi: 10.1016/j.molcel.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Storici F., Lewis L.K., Resnick M.A. In vivo site-directed mutagenesis using oligonucleotides. Nature Biotechnology. 2001;19:773. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- Takemata N., Oda A., Yamada T., Galipon J., Miyoshi T., Suzuki Y., Sugano S., Hoffman C.S., Hirota K., Ohta K. Local potentiation of stress-responsive genes by upstream noncoding transcription. Nucleic Acids Res. 2016;44:5174–5189. doi: 10.1093/nar/gkw142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam J., van Werven F.J. Regulated repression governs the cell fate promoter controlling yeast meiosis. Nat. Commun. 2020;11:2271. doi: 10.1038/s41467-020-16107-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Xue Y., Song C., Grunstein M. Acetylated histone H3K56 interacts with Oct4 to promote mouse embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. USA. 2013;110:11493–11498. doi: 10.1073/pnas.1309914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terweij M., van Welsem T., van Deventer S., Verzijlbergen K.F., Menendez-Benito V., Ontoso D., San-Segundo P., Neefjes J., van Leeuwen F. Recombination-induced tag exchange (RITE) cassette series to monitor protein dynamics in Saccharomyces cerevisiae. G3 (Bethesda) 2013;3:1261–1272. doi: 10.1534/g3.113.006213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisseur M., Kwapisz M., Morillon A. Pervasive transcription - Lessons from yeast. Biochimie. 2011;93:1889–1896. doi: 10.1016/j.biochi.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Topal S., Vasseur P., Radman-Livaja M., Peterson C.L. Distinct transcriptional roles for Histone H3-K56 acetylation during the cell cycle in Yeast. Nat. Commun. 2019;10:4372. doi: 10.1038/s41467-019-12400-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubota T., Berndsen C.E., Erkmann J.A., Smith C.L., Yang L., Freitas M.A., Denu J.M., Kaufman P.D. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven F.J., Amon A. Regulation of entry into gametogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011;366:3521–3531. doi: 10.1098/rstb.2011.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven F.J., Neuert G., Hendrick N., Lardenois A., Buratowski S., van Oudenaarden A., Primig M., Amon A. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell. 2012;150:1170–1181. doi: 10.1016/j.cell.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven F.J., Timmers H.T.M. The use of biotin tagging in Saccharomyces cerevisiae improves the sensitivity of chromatin immunoprecipitation. Nucleic Acids Research. 2006;34 doi: 10.1093/nar/gkl003. 33–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Värv S., Kristjuhan K., Peil K., Lõoke M., Mahlakõiv T., Paapsi K., Kristjuhan A. Acetylation of H3 K56 is required for RNA polymerase II transcript elongation through heterochromatin in yeast. Mol. Cell. Biol. 2010;30:1467–1477. doi: 10.1128/MCB.01151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S., Workman J.L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015;16:178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- Venkatesh S., Smolle M., Li H., Gogol M.M., Saint M., Kumar S., Natarajan K., Workman J.L. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- Voichek Y., Bar-Ziv R., Barkai N. Expression homeostasis during DNA replication. Science. 2016;351:1087–1090. doi: 10.1126/science.aad1162. [DOI] [PubMed] [Google Scholar]
- Wagstaff J.E., Klapholz S., Esposito R.E. Meiosis in haploid yeast. Proc. Natl. Acad. Sci. USA. 1982;79:2986–2990. doi: 10.1073/pnas.79.9.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O., Chang H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. [Google Scholar]
- Williams S.K., Truong D., Tyler J.K. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc. Natl. Acad. Sci. USA. 2008;105:9000–9005. doi: 10.1073/pnas.0800057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Zhang K., Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Münster S., Camblong J., Guffanti E., Stutz F., Huber W., Steinmetz L.M. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA sequencing, the TSS and TES sequencing, and the Pol II associated RNA sequencind data reported in this paper is GEO: GSE138898.
The code use for the mathematical modeling is available upon request to the lead contact.