Sir,
Klebsiella pneumoniae ranks at the top of the global priority list of pathogens regarding demand for new treatment options.1 Colistin represents a last-resort antibiotic for treatment of MDR K. pneumoniae infections.2 Colistin resistance in K. pneumoniae is attributed to the accumulation of chromosomal mutations, mainly in pmrAB, phoPQ, ccrAB and mgrB.3 In order to maintain the efficacy of such last-resort antibiotics, we need to design rational treatment strategies that consider resistance development associated with clinical antibiotic concentration–time profiles. In vitro experiments in the context of pharmacokinetic (PK) profiles represent a relevant approach to characterize the evolution of resistance. However, experimental evolution studies for colistin have mainly focused on approaches such as serial passaging that do not consider PK. The role of the experimental model and in particular the impact of clinical drug concentrations on evolution of resistance is poorly understood.
We aimed to study the impact of experimental evolution approaches for the development of resistance to colistin in K. pneumoniae. We performed experiments with a continuous culture chemostat replicating clinical PK profiles of colistin, a static clinical colistin concentration and standard serial passaging experiments, comparing the phenotypic and the genotypic changes.
Two colistin-susceptible clinical isolates of K. pneumoniae (KML9749 and KML9884) obtained from Leiden University Medical Centre (the Netherlands) were included in this study. DNA extraction followed by WGS using 2 × 150 bp paired-end reads on an Illumina NovaSeq 6000 platform revealed that the strains KML9749 (ST45, KL62 and O2v1) and KML9884 (ST3248, KL102 and O2v2) were genetically unrelated (Table S1, available as Supplementary data at JAC Online), while both encoded common Klebsiella-related virulence factors (Table S2). Both strains carried an identical FIIk/FIBk plasmid encoding multidrug resistance, whereas neither of them carried chromosomal mutations, acquired mcr-1 to mcr-10 genes or IS elements inserted in chromosomal genes previously associated with colistin resistance.3
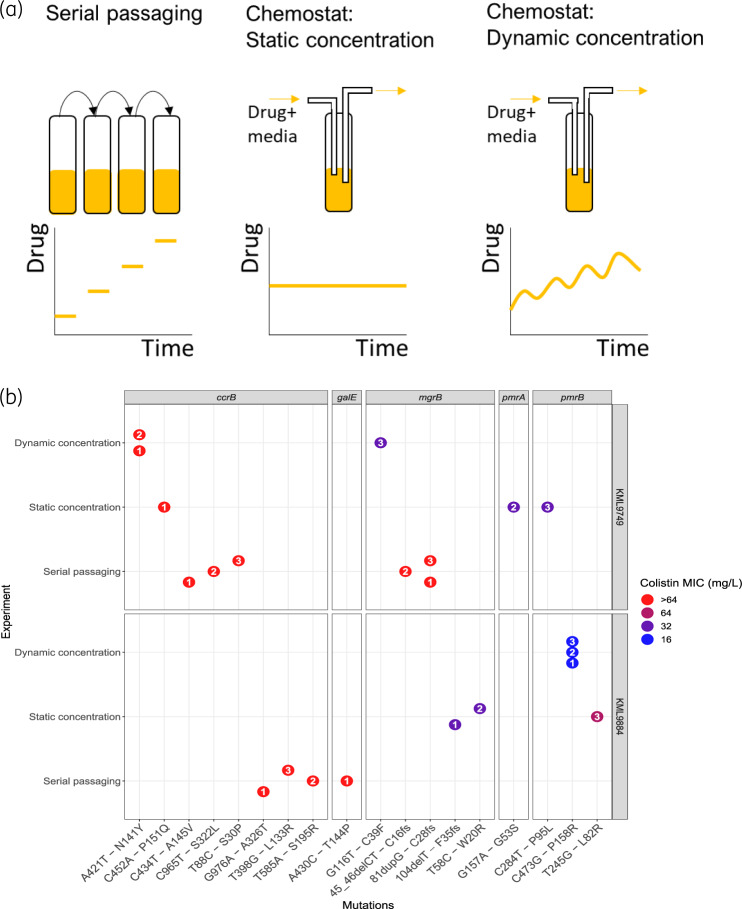
Both strains were separately evolved into colistin-resistant mutants in three different in vitro experiments (Figure 1a), using: (i) daily serial passaging in colistin; (ii) chemostat time–kill experiments with a simulated time-varying PK profile towards a steady-state concentration of 2.75 mg/L;4 and (iii) chemostat time–kill experiments with a static concentration of colistin (2.75 mg/L). In short, the serial passaging experiments were performed in 10 mL of CAMHB (BD, New Jersey, USA) at 37°C under constant agitation. The same culture conditions were used for the 100 mL chemostat time–kill experiments. The chemostat is an open experimental setup with in- and outflow of culture medium. The flow rate of the medium was set to 0.103 mL/min to achieve an elimination half-life of approximately 11 h, corresponding to a typical individual with a creatinine clearance of 70 mL/min.4 Antibiotic-free CAMHB was gradually supplemented with colistin (17.5 μg/h) to achieve a slowly increasing concentration profile clinically observed due to the in vivo conversion of colistimethate sodium (CMS) to colistin. The simulated PK profile corresponded to a standard dosing regimen of colistin (3 × 109 IU CMS q8h).5 Static concentration conditions were obtained by constantly supplementing the initial colistin-containing CAMHB with fresh medium with the same colistin concentration. All experiments were performed in triplicate for a duration of 48 h, with an initial bacterial density of approximately 106 cfu/mL, resulting in 18 end-of-experiment mutants that were analysed by WGS in comparison with their parental strains (Figure 1b, Table S3). Sequence reads for the whole-genome-sequenced isolates from this study are available from the NCBI Sequence Read Archive (SRA) under the bioproject with accession code: PRJNA596368. The analysis pipeline is available from GitHub (https://github.com/vanhasseltlab/KlebsiellaColistinWGS). MICs determined by broth microdilution (MICRONAUT MIC-Strip Colistin, Merlin Diagnostika, Bornheim, Germany) revealed that all mutants showed an increased MIC of colistin compared with their parental WT (MICWT 0.5 mg/L).
Figure 1.
(a) Overview of experimental in vitro models for evolution of antibiotic resistance. (b) SNPs identified in mutants evolved from two parental clinical isolates of Klebsiella pneumoniae, KML9884 and KML9749, in three different in vitro experiments in which the bacteria were treated with colistin over 48 h. Mutated genes are shown as panels with the specific nucleotide substitution followed by the amino acid substitution on the x-axis. Deletions, duplications and frameshift mutations are denoted del, dup and fs, respectively. Colour indicates colistin MIC values (mg/L) and the enclosed numbers represents replicate identity. The MICs of both parental strains were ≤1 mg/L. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Serial passaging evolution experiments in colistin up to 32 mg/L resulted in a considerable increase in MIC for all mutants (>64 mg/L). WGS showed that all of the KML9749 mutants harboured mutations in various nucleotide positions of the ccrB gene (T88C, C434T or C965T) and frameshift mutations in the mgrB gene, while KML9884 mutants harboured ccrB mutations (T398G, T585A or G976A), with one of them also carrying a mutation in galE (A430C) (Figure 1b, Table S3).
For the chemostat in vitro PK experiments, all KML9749 replicates acquired mutations in ccrB (A421T), accompanied in one of them by an mgrB gene mutation (G116T), conferring MICs of >64 and 32 mg/L, respectively. In vitro PK simulations also uniquely promoted mutations in pmrB (C473G) for KML9884 with an MIC of 16 mg/L (Figure 1b, Table S3).
The chemostat experiments with a static colistin concentration did not show consistent mutations within or between the strains. All KML9749 replicates harboured different colistin resistance-associated mutations in pmrA (G157A), pmrB (C284T) or ccrB (C452A), with MICs ranging between 32 and >64 mg/L. However, two of the KML9884 mutants which evolved under these conditions harboured point (T58C) or frameshift (104delT) mutations in the mgrB gene, showing an MIC increase to 32 mg/L, while the remaining one harboured a mutation in pmrB (T245G), conferring a colistin MIC of 64 mg/L (Figure 1b, Table S3).
All three in vitro experimental evolution models resulted in the emergence of mutants encoding exclusively colistin resistance-related mutations.3 Our results suggest that serial passaging leads to convergent evolution of both strains tested, with mutations emerging in ccrB and mgrB genes. In contrast, chemostat time–kill experiments, reflecting a clinically relevant pharmacokinetic profile or a static plasma concentration of colistin while maintaining availability of nutrients at a constant level, resulted in mutants encoding diverse colistin resistance mutations within and/or between the strains tested. These findings indicate that the evolutionary trajectories leading to colistin resistance are dependent on the in vitro experimental system used, potentially due to the different selection pressure exerted on the bacterial cells by these experimental conditions.6 As a consequence, the choice of experimental system should be considered very carefully when studying evolutionary trajectories, in particular when aiming to identify clinically relevant colistin resistance mutations.
In contrast to the in vitro PK chemostat time–kill experiments that constrained the evolution within strains to either the ccrB (KML9749) or pmrB (KML9884) gene, the static experiment resulted in divergent evolution within and between strains tested, with mutations in diverse colistin resistance-related genes. In accordance with previous studies,7 we observed intra- and interstrain diversity of mutations in K. pneumoniae after exposure to colistin, highlighting the effect of the genetic background of the strain on the de novo emergence of resistance.
In conclusion, we have shown that evolutionary trajectories leading to antibiotic resistance depend not only on the genetic background of the strain, but also on the in vitro experimental system used to study these trajectories.
Supplementary Material
Funding
This work was supported by funding from ZonMW Enabling Technology Hotels (project number 435004015) and the Stichting Elise Mathilde Fonds/Leiden University Fund.
Transparency declarations
None to declare.
Supplementary data
Tables S1 to S3 are available as Supplementary data at JAC Online.
References
- 1.WHO. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. 2017. https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf.
- 2. Petrosillo N, Giannella M, Lewis R. et al. Treatment of carbapenem-resistant Klebsiella pneumoniae: the state of the art. Expert Rev Anti Infect Ther 2013; 11: 159–77. [DOI] [PubMed] [Google Scholar]
- 3. Poirel L, Jayol A, Nordmann P.. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 2017; 30: 557–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Karaiskos I, Friberg LE, Pontikis K. et al. Colistin population pharmacokinetics after application of a loading dose of 9 MU colistin methanesulfonate in critically ill patients. Antimicrob Agents Chemother 2015; 59: 7240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Food and Drug Administration. Coly-Mycin M Parenteral (colistimethate for injection, USP) NDA 50-108/S-026 https://www.accessdata.fda.gov/drugsatfda_docs/label/2009/050108s026lbl.pdf.
- 6. Gresham D, Dunham MJ.. The enduring utility of continuous culturing in experimental evolution. Genomics 2014; 104: 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim SJ, Ko KS.. Diverse genetic alterations responsible for post-exposure colistin resistance in populations of the same strain of Klebsiella pneumoniae. Int J Antimicrob Agents 2018; 52: 425–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.