Abstract
Background
Approximately half of clinical carbapenem-resistant Enterobacterales (CRE) isolates lack carbapenem-hydrolysing enzymes and develop carbapenem resistance through alternative mechanisms.
Objectives
To elucidate development of carbapenem resistance mechanisms from clonal, recurrent ESBL-positive Enterobacterales (ESBL-E) bacteraemia isolates in a vulnerable patient population.
Methods
This study investigated a cohort of ESBL-E bacteraemia cases in Houston, TX, USA. Oxford Nanopore Technologies long-read and Illumina short-read sequencing data were used for comparative genomic analysis. Serial passaging experiments were performed on a set of clinical ST131 Escherichia coli isolates to recapitulate in vivo observations. Quantitative PCR (qPCR) and qRT–PCR were used to determine copy number and transcript levels of β-lactamase genes, respectively.
Results
Non-carbapenemase-producing CRE (non-CP-CRE) clinical isolates emerged from an ESBL-E background through a concurrence of primarily IS26-mediated amplifications of blaOXA-1 and blaCTX-M-1 group genes coupled with porin inactivation. The discrete, modular translocatable units (TUs) that carried and amplified β-lactamase genes mobilized intracellularly from a chromosomal, IS26-bound transposon and inserted within porin genes, thereby increasing β-lactamase gene copy number and inactivating porins concurrently. The carbapenem resistance phenotype and TU-mediated β-lactamase gene amplification were recapitulated by passaging a clinical ESBL-E isolate in the presence of ertapenem. Clinical non-CP-CRE isolates had stable carbapenem resistance phenotypes in the absence of ertapenem exposure.
Conclusions
These data demonstrate IS26-mediated mechanisms underlying β-lactamase gene amplification with concurrent outer membrane porin disruption driving emergence of clinical non-CP-CRE. Furthermore, these amplifications were stable in the absence of antimicrobial pressure. Long-read sequencing can be utilized to identify unique mobile genetic element mechanisms that drive antimicrobial resistance.
Introduction
Carbapenem-resistant Enterobacterales (CRE) infections are an emerging global health priority and among the most serious antimicrobial resistance (AMR) threats.1 Carbapenem resistance can develop due to acquisition of enzymes that hydrolyse carbapenems, known as carbapenemases, as well as through outer membrane permeability changes and/or drug efflux activity that decrease intracellular carbapenem concentrations.2 Globally, carbapenem-producing Enterobacterales (CPE) remain the most predominant CRE subset group.2 Nevertheless, while most CRE research has focused on characterizing CPE,2,3 recent clinical and molecular epidemiology studies indicate up to 50% of CRE isolates can be non-carbapenemase producers, suggesting that a substantial proportion of CRE isolates develop carbapenem resistance through alternative mechanisms.4,5 Although the contribution of non-carbapenemase-producing CRE (non-CP-CRE) to the overall CRE population can be highly variable by geographical location as well as patient population, many countries report non-CP-CRE as a highly prevalent CRE subset group, suggesting a global impact of these understudied pathogens.6–8
Non-CP-CRE mechanistic studies have been conducted in both basic and clinical research settings.9–14 Clinical non-CP-CRE isolates generally carry ESBL or AmpC-like enzymes with concomitant mutations that alter porin function.9,10 Similarly, laboratory passaging of ESBL-positive Enterobacterales (ESBL-E) strains has indicated that development of carbapenem resistance requires both an ESBL and a decrease in outer membrane porin function.11–14 Additionally, augmented production of β-lactamases without known carbapenemase activity has important effects on carbapenem susceptibility in porin-deficient backgrounds.13,14 Thus, it is generally thought that non-CP-CRE strains arise from precursor strains producing either an ESBL or AmpC enzyme. However, it is likely that transition from ESBL-E to non-CP-CRE primarily occurs during colonization such that having paired ESBL-E and non-CP-CRE clinical isolates is rare.9–17 Thus, there remains a knowledge gap regarding mechanisms by which non-CP-CRE mechanisms evolve in patient populations.
Herein, we sought to address how ESBL-E isolates transition to CRE in the clinical setting by systematically studying a cohort of ESBL-E bacteraemia isolates to assess CRE prevalence and characterize mechanisms of CRE emergence. Through the application of long-read and short-read WGS technologies on serial clinical isolates, as well as through serial passaging experiments, we sought to generate insights into how outer membrane porins become inactivated, as well as characterize β-lactamase amplification mechanisms within a patient cohort.
Materials and methods
Study design and clinical data abstraction
A retrospective review of patients with ESBL-E bacteraemia hospitalized from January 2015 to July 2016 was conducted at MD Anderson Cancer Center (MDACC) in Houston, TX, USA. All patients with one or more episodes of ESBL-E bacteraemia who were 18 years of age or older were eligible for inclusion. Clinical and demographic characteristics were extracted from electronic medical records and recorded using REDCap software (Vanderbilt University, Nashville, TN, USA).18 A waiver of informed consent to collect clinical data from electronic medical records and analyse the isolates was provided by the MDACC IRB (PA15-0799).
Isolate characterization and antimicrobial susceptibility testing
Blood culture isolates are routinely saved at MDACC and stored at −80°C. Carbapenem resistance was defined using CLSI criteria as resistance to either ertapenem or meropenem.19 Recurrent Enterobacterales bacteraemia was defined as identification of the same species in blood culture at any point during the follow-up period following at least one negative blood culture and completion of an antibiotic treatment regimen.
Antibiotic susceptibility testing was performed per routine clinical laboratory practice using an automated system (VITEK 2, bioMérieux, Marcy L’Étoile, France) with additional testing performed as needed using individual antibiotic gradient strips (Etest, bioMérieux). ESBL production was assessed per routine laboratory practice on Escherichia coli, Klebsiella pneumoniae and Klebsiella oxytoca isolates that were resistant to one or more oxyimino-cephalosporins (e.g. cefotaxime, ceftriaxone or ceftazidime) using either the ESBL Etest (bioMérieux) or the Rapid ESBL Screen kit (ROSCO, Taastrup, Denmark). Carbapenemase production was evaluated in the clinical laboratory for any Enterobacterales isolate resistant to one or more of the carbapenems using the Neo-Rapid CARB kit (ROSCO) according to the manufacturer’s instructions.
WGS and normalized coverage depth analysis
All available isolates from recurrent bacteraemia patients underwent WGS via Illumina HiSeq as described previously.20 Normalized coverage depth of short-read mapping to individual genes or genome regions of interest was quantitated relative to the average short-read mapping depth for the PubMLST E. coli housekeeping gene schema using a suite of bioinformatic tools (bwa v0.7.17, minimap2 v2.2.17, SAMtools v1.9, bedtools v2.27.1) to infer approximate copy numbers.21–23 Serial isolates that developed a non-CP-CRE phenotype, isolates used in serial passaging experiments and non-CP-CRE isolates from a previous study20 underwent Oxford Nanopore Technologies (ONT) long-read sequencing. A custom Python script was used to generate hybrid, consensus assemblies using the Flye assembler v2.524 (https://github.com/wshropshire/flye_hybrid_assembly_pipeline). A description of the hybrid assembly pipeline is available in the Supplementary methods, available as Supplementary data at JAC Online. ONT assembly metrics are provided in Table S1.
Clonality analysis
Phylogenetic analysis, in silico MLST and variant calling of serial isolates compared with the index strain genome were used to determine clonality. Roary v3.12.025 was used to perform a core-gene alignment with the aligner tool PRANK.25,26 A maximum-likelihood (ML) phylogenetic tree using RAxML v8.2.1227 was built using the core-gene alignment. In silico MLST was performed using mlst v2.15.1 (https://github.com/tseemann/mlst).28 Recurrent isolates that became non-CP-CRE were checked for clonality using the variant-calling pipeline tool Snippy v4.3.6 (https://github.com/tseemann/snippy) using the consensus assemblies of the index isolates as references.
Genome annotation, AMR gene and mobile genetic element (MGE) identification
Genome annotation was performed using Prokka v1.14.0.29 ABRicate (https://github.com/tseemann/abricate), using the Comprehensive Antibiotic Resistance Database (CARD),30 and PlasmidFinder31 were used to search for AMR determinants, such as carbapenemases, and plasmid signatures, respectively. Annotated Prokka gbk files were used to characterize ORFs of interest. CARD, PlasmidFinder, ISFinder32 and BLAST webtools were utilized during manual inspection of the assemblies. SVAnts v0.133 (https://github.com/EpiBlake/SVAnts) was used to investigate subsets of ONT long reads.
β-lactamase gene copy number, transcript level and impact of β-lactamase expression on susceptibility analysis
Quantitative PCR (qPCR) and qRT–PCR were used to assess DNA copy number and RNA transcript levels, respectively. qPCR and qRT–PCR primers and probe sequences are provided in Table S2. The DNA levels of blaOXA-1 and blaCTX-M-1 group genes were determined relative to the rpsL control gene using the ΔCt method as described previously.14 A full description of the methods used for copy number and gene transcript level quantification can be found in the Supplementary methods. For the purposes of β-lactamase cloning and expression analysis, the ORFs of β-lactamases were amplified from genomic DNA using Q5 polymerase with the primers listed in Table S2. Additional experiment parameters and conditions are provided in the Supplementary methods.
Serial passaging experiments on clinical E. coli ST131 bacteraemia isolates
Two individual passaging experiments of the Patient 4 index strain, i.e. isolate 4A, with antibiotic selection were performed in LB broth under shaking conditions at 37°C. For the first passaging experiment, cells were collected on four consecutive days (strains 4A_1 to 4A_4) while continuing to passage at an ertapenem concentration of 32 mg/L. For the second set of passaging experiments, cells were collected on the first day that the ertapenem MIC reached ≥32 mg/L, serially diluted onto agar plates and two different isolates were studied to assess for heterogeneity (strains 4A_H1 and 4A_H2). Additional serial passaging experiments were conducted using ertapenem-resistant patient strains 4C and 4D in the absence of antibiotic selection, to determine the stability of the amplified translocatable units, with a protocol adapted from previously published methods.34 Additional experimental conditions are included in the Supplementary methods.
Statistical analyses
All statistical analyses were performed using Stata v13.1 (StataCorp LP, College Station, TX, USA). Bivariate comparisons between patients with a single bacteraemia episode and those with recurrent bacteraemia were made with Wilcoxon rank-sum and Fisher’s exact tests, as appropriate, based on covariate distributions. Comparisons of DNA and RNA levels among strains were performed using the Wilcoxon rank-sum test and the Kruskal–Wallis test when two or three strains were being analysed, respectively. MIC comparisons were performed using ANOVA with Dunnett’s test of multiple comparisons. Statistical significance was assigned as a two-sided P value <0.05.
Data availability
Assemblies of index isolates from patients with recurrent bacteraemia as well as long-read and short-read data for recurrent isolates have been deposited in the NCBI BioProject database (PRJNA603908). ONT sequencing data of non-CP-CRE isolates from a previous study20 were deposited in the NCBI BioProject database (PRJNA388450).
Results
ESBL-E bacteraemia cohort
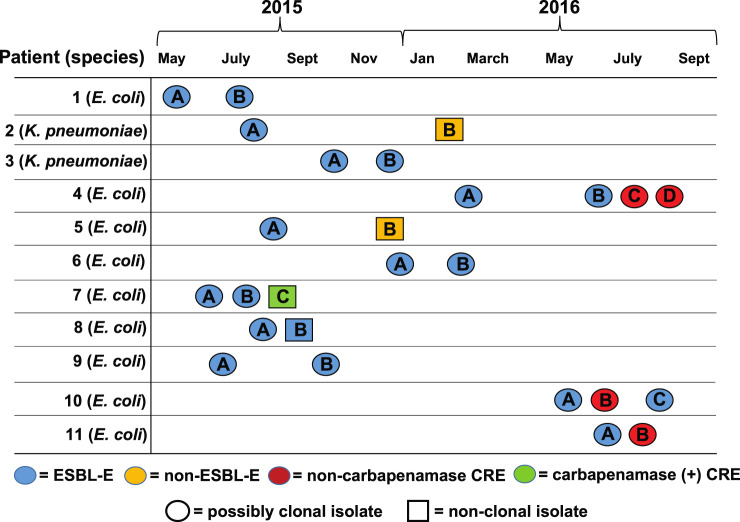
There were 116 patients with carbapenem-susceptible ESBL-E index bacteraemia cases. Clinical and demographic features are presented in Table S3. E. coli was the most common organism isolated (100/116; 86.2%), followed by K. pneumoniae (14/116; 12.1%) and K. oxytoca (2/116; 1.7%). Recurrent bacteraemia was identified in 16/116 (13.8%) patients and primarily occurred either in leukaemia or HSCT patients (14/16 cases, Table S3). Carbapenem-resistant isolates were present in 4/16 (25%) recurrent bacteraemia patients (Figure 1), with all four patients having E. coli serial isolates. Isolates from recurrent bacteraemia patients (n = 26) were available for analysis in 11/16 patients, including all strains from the four patients who had at least one recurrent isolate that developed a carbapenem-resistant phenotype.
Figure 1.
Timeline showing date of serial strain isolation from blood cultures. Patient numbers and species isolated are in the first column. The shape and colour of the isolate labels indicate potential clonality and AMR, respectively. Isolates were considered possibly clonal if they were the same ST and clustered on the phylogenetic tree presented in Figure S1. Patient subgroups (e.g. Patient 1; isolates A and B) refer to the order of isolation.
Identification of recurrent bacteraemia E. coli isolates that developed carbapenem resistance
Short-read sequencing was performed on all 26 isolates (Table S4) from recurrent bacteraemia patients with clonality assessed initially via phylogenetic analysis (Figure S1). The date of infection, antimicrobial susceptibility and clonality status are depicted in Figure 1. We subsequently focused on recurrent bacteraemia isolates that developed carbapenem resistance to identify potential de novo mechanisms of carbapenem resistance. Three of the four carbapenem-resistant serial isolates (those from Patients 4, 10 and 11) had at least one non-CP-CRE recurrent strain. Our short-read WGS data indicated that for each of these three patients, their recurrent E. coli isolates clustered together within the phylogenetic tree, had the same Bayesian hierarchical population structure and belonged to the same ST (Figure S1). We generated long-read (ONT) plus short-read (Illumina) hybrid, consensus assemblies to create highly resolved genomes for the index strains of Patients 4, 10 and 11. We confirmed clonality by measuring pairwise SNP distances using these aforementioned genome assemblies and found less than 20 SNPs for all respective recurrent strains relative to their index strain, suggesting that these three patients had clonal strains that developed carbapenem resistance through a non-carbapenemase mechanism (Table 1). The other set of isolates in which carbapenem resistance developed (Patient 7, strains 7A–C) included both ST405 (7A–B) and ST10 (7C) isolates, with the latter isolate harbouring the class D carbapenemase blaOXA-181, indicating that the patient had been infected with a new E. coli strain rather than the index strain developing carbapenem resistance.
Table 1.
Molecular and genomic characterization of clinical, recurrent bacteraemia Enterobacterales isolates
| Patient | Isolate | ST | Species | Pairwise SNP distance | CAZ MICa | FEP MICa | TZP MICa | ETP MICa | MEM MICa | ompC b , c | ompF c , d | β-lactamase amplification |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 4A | 131 | E. coli | Ref | 16 | >64 | 8 | <0.5 | <0.25 | WT | WT | none |
| 4B | 131 | E. coli | 2 | 16 | 8 | >128 | <0.5 | <0.25 | WT | WT | bla OXA-1 | |
| 4C | 131 | E. coli | 14 | 16 | 64 | >128 | >32 | 4 | disruptione | disruption | bla OXA-1 | |
| 4D | 131 | E. coli | 15 | >64 | >64 | >128 | >32 | 4 | disruptione | disruption | bla OXA-1 | |
| 10 | 10A | 10 | E. coli | Ref | 16 | 2 | 8 | <0.5 | <0.25 | WT | WT | none |
| 10B | 10 | E. coli | 1 | >64 | >64 | >128 | >32 | 8 | disruption | disruption | bla CTX-M-55 | |
| 10C | 10 | E. coli | 0 | >64 | >64 | 16 | <0.5 | <0.25 | WT | WT | bla CTX-M-55 | |
| 11 | 11A | 131 | E. coli | Ref | >64 | >64 | 64 | <0.5 | <0.5 | intactf | disruption |
bla OXA-1, bla CTX-M-15 |
| 11B | 131 | E. coli | 0 | >64 | >64 | >128 | 4 | 1 | disruption | disruption |
bla OXA-1, bla CTX-M-15 |
|
| NA | MB746 | 405 | E. coli | NA | 64 | >64 | >128 | >32 | 4 | disruption | disruption |
bla OXA-1, bla CTX-M-15 |
| NA | MB101 | 37 | K. pneumoniae | NA | >64 | >64 | >128 | >32 | 8 | disruptiong | disruptiong |
bla OXA-1, bla CTX-M-15 |
CAZ, ceftazidime; FEP, cefepime; TZP, piperacillin/tazobactam; ETP, ertapenem; MEM, meropenem; Ref, reference; NA, not available.
All MICs reported in mg/L.
UniProt reference entry names for ompC are A0A192C9D6_ECOLX, OMPC_ECOLI and U9Y7F2_ECOLX for ST131, ST10 and ST405 E. coli isolates, respectively; MB101 porin is ompK36 with UniProt reference entry name A0A0H3H0Y2_KLEPH.
WT indicates WT coding DNA sequence, disruption indicates an INDEL event; all INDEL events are frameshift mutations that leave premature, truncated coding DNA sequences unless otherwise noted.
UniProt reference entry names for ompF are A0A192CJU0_ECOLX, OMPF_ECOLI and S0Z171_ECOLX for ST131, ST10 and ST405 E. coli isolates, respectively; MB101 porin is ompK35 with UniProt reference entry name O87753_KLEPN.
Insertion of MB1860TU_A with variable number of repeating units.
INDEL, Y170_N174delinsKR, creates an in-frame OmpC protein.
Insertion of 1× copy of MB101TPU in ompK36 and nonsense mutation (p.Y36X) in ompK35, respectively.
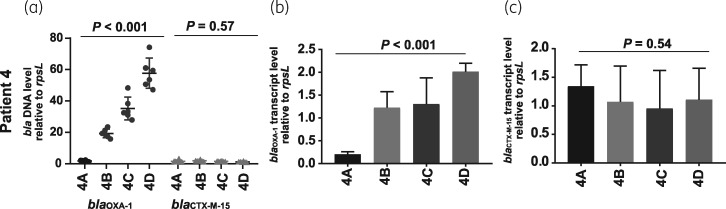
E. coli isolates from Patient 4 (4A–D) and Patient 11 (11A–B) were ST131 whereas Patient 10 isolates (10A–C) were ST10. Each of the serial isolates had increased Illumina short-read and ONT long-read normalized coverage depth for β-lactamase genes indicating amplification (Table S5 and Table S6). Specifically, we identified increased normalized coverage depth of blaOXA-1 in all Patient 4 recurrent isolates, blaCTX-M-55 (a derivative of blaCTX-M-15 that differs by a single A80V amino acid substitution)35 in Patient 10 recurrent isolates and blaOXA-1 and blaCTX-M-15 in both the index and recurrent isolates of Patient 11 (Table 1). We used qPCR to confirm increased normalized coverage depth in our WGS analyses and qRT–PCR to show that the increased normalized coverage depth corresponded to increased transcript levels of the amplified β-lactamase genes for Patient 4 isolates (Figure 2) as well as Patient 10 and Patient 11 isolates (Figure S2). We further characterized outer membrane porin genes with sequencing data given that disruptions of these genes are correlated with carbapenem resistance.2 We consistently identified that non-CP-CRE strains contained disruptions in the ORFs of porin proteins OmpC and OmpF (Table 1). We subsequently focused on the E. coli ST131 serial sets of isolates (i.e. Patient 4 and Patient 11 isolates) as both had non-CP-CRE mechanisms involving blaOXA-1 and blaCTX-M-15 presence, potential amplification of these β-lactamase genes and porin disruption.
Figure 2.
PCR analysis of β-lactamase gene levels and transcript levels for Patient 4 isolates (4A–D). (a) TaqMan qPCR of genomic DNA results collected in triplicate on two separate days (n = 6) for either blaOXA-1 or blaCTX-M-15 relative to the endogenous control gene rpsL. Data shown are individual data points with mean ± SD superimposed. (b) blaOXA-1 transcript level relative to endogenous control gene rpsL. RNA was collected from mid-exponential phase in triplicate on two separate days (n = 6). Data shown are mean ± SD. (c) Similar analysis to (b) except that data are for blaCTX-M-15. P values refer to measurements in the serial isolates relative to the initial isolate using the Kruskal–Wallis test.
TnMB1860 present in both E. coli ST131 Patient 4 and Patient 11 index isolates
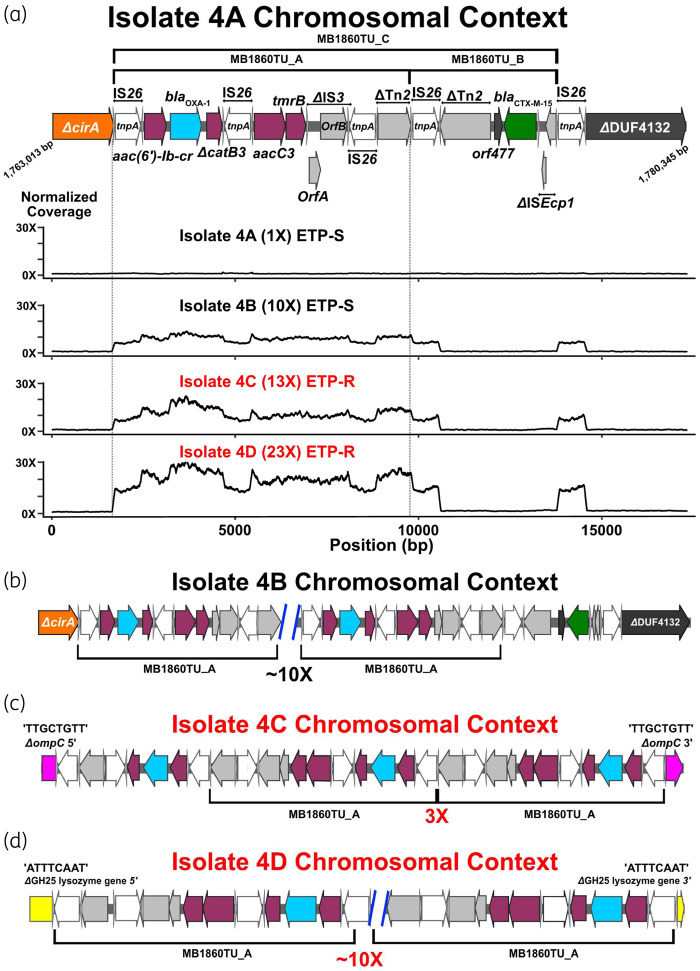
The consensus genome assemblies of index strains for Patient 4 (4A) and Patient 11 (11A) both contained an IS26-mediated translocatable unit (TU), herein designated as MB1860TU_A, which carried blaOXA-1, adjacent and upstream of another TU, designated MB1860TU_B, which carried blaCTX-M-15 (Figure 3a). MB1860TU_C is the combination of both TUs. The entire transposon structure, consisting of overlapping IS26-bounded transposons designated TnMB1860, is shown in Figure 3a.
Figure 3.
Characterization of IS26-flanked transposon, TnMB1860, with transposition of modular TUs in Patient 4 serial isolates (i.e. isolates 4A–D). Terminal left and right inverted repeats (IRL and IRR, respectively) of ISs are specified by grey triangles that bracket complete and incomplete tnpA genes, respectively. ORFs are coloured as follows: non-β-lactamase AMR genes (maroon), blaOXA-1 (blue), blaCTX-M-15 (green), IS26 tnpA (white) and other IS/Tn elements (grey). Font colour for each serial isolate labelled represents ertapenem susceptibility (black) or ertapenem resistance (red). (a) Schematic indicates chromosomal context (GenBank accession #: CP049085) of TnMB1860 locus flanked by directly oriented IS26 transposases found in the 4A isolate. Immediately below the schematic are normalized, short-read coverage depth line graphs for the four Patient 4 serial isolates with MB1860TU_A bracketed by dotted lines. ETP-S, ertapenem susceptible; ETP-R, ertapenem resistant. (b–d) Characterization of MB1860TU_A transposition and ORF disruption events in each of the respective Patient 4 recurrent episode isolates. Black brackets beneath ORFs indicate MB168TU_A. (b) Isolate 4B chromosomal context shows an ∼10× MB1860TU_A amplification event in the original 4A chromosomal locus. (c) Isolate 4C chromosomal context additionally contains transposition and disruption of the ompC porin gene (pink) upstream of the original isolate 4A TnMB1860 locus with three copies of MB1860TU_A. (d) Isolate 4D has previous transposition events found in isolates 4B and 4C as well as another MB1860TU_A transposition, amplification and disruption of a putative glycoside hydrolase gene (i.e. GH25; yellow) downstream of the TnMB1860 locus. The TSDs created by the transposition of the TU are indicated above each respective junction site flanking the insertion and disruption of the ompC and GH25 genes, respectively.
TnMB1860 is located on the chromosome of both index isolate 4A (GenBank accession #: CP049085) and index isolate 11A (GenBank accession #: CP049077). Variable-sized direct repeats called target site duplications (TSDs) flank ISs and are created during the transposition process.36,37 The TnMB1860 transposon on the isolate 11A chromosome has 7 bp TSDs that indicate a transposition within an ORF that putatively is involved in molybdopterin cofactor biosynthesis (Figure S3). TnMB1860 in isolate 4A differs in chromosomal context relative to isolate 11A due to an IS26-mediated intramolecular transposition event that occurred in reverse orientation indicated by 8 bp TSDs at the breakpoint site of a colicin I receptor gene (Figure S4). An alignment of 4A and 11A isolates with two other E. coli ST131 chromosomes available from GenBank, TO217 (GenBank accession #: LS992192.1) and Ecol_AZ146 (GenBank accession #: CP018991.1) indicate similar chromosomal carriage of TnMB1860 with the noted inversion event that has occurred in isolate 4A (Figure S4). This alignment indicates that the strain 11A chromosome has comparable orientation with the aforementioned historical strain chromosomes, in contrast to the strain 4A chromosome, which has undergone an additional IS26-mediated transposition event.
Delineation of MB1860TU_A transposition in ST131 Patient 4 and Patient 11 serial isolates
Patient 11 isolates 11A and 11B had a relatively equal increase in normalized coverage depth for blaOXA-1 and blaCTX-M-15 (Table S5). The consensus assemblies of both 11A and 11B isolates revealed that this increase in normalized coverage depth was due to two copies of blaOXA-1 and blaCTX-M-15 being present on the chromosomal TnMB1860 as well as on IS26-mediated TUs present on a multireplicon, F-type plasmid, p11A_p2 (Figure S3, Figure S4; GenBank accession #: CP049079). The CRE phenotype of strain 11B relative to 11A appears to be driven by additional inactivation of the ompC gene via a frameshift, adenine insertion at position 131 of the coding sequence (CDS) of OmpC (c.131insA), given that the ompF gene is truncated by a deletion of 8 bp in the CDS of OmpF from nucleotide 305 to 312 (c.305_312del) in both strains (Table 1).
In contrast to the Patient 11 isolates, the transposition of the modular TUs that compose TnMB1860 in Patient 4 was present only in a chromosomal context (Figure 3). Within the Patient 4 serial isolates, there was a consistent increase in short-read normalized coverage depth (Figure 3a) of the entire MB1860TU_A structure harbouring blaOXA-1 up to approximately 23-fold in isolate 4D. We first analysed isolate 4B, which had developed resistance to piperacillin/tazobactam but remained carbapenem susceptible (Table 1). In isolate 4B, MB1860TU_A generated an ∼10× tandem array in situ most likely through an IS26-mediated RecA-independent transposition mechanism or homologous recombination (Figure 3b).38,39 The occurrence of the 10× TU amplification at the original TnMB1860 locus was confirmed by aligning the isolate 4B long reads to the reference chromosome of isolate 4A using SVAnts in conjunction with a short-read pileup analysis. All outer membrane porin genes remained intact in isolate 4B. Given that isolate 4B had developed resistance to piperacillin/tazobactam relative to isolate 4A (Table 1), we sought to determine whether overexpression of blaOXA-1 could drive piperacillin/tazobactam resistance. Inducing blaOXA-1 expression through cloning under an arabinose-responsive promoter increased piperacillin/tazobactam MIC 6.8-fold relative to uninduced cells (Figure S5). Nevertheless, blaOXA-1 amplification in the absence of outer membrane porin disruption was not sufficient to drive carbapenem resistance in isolate 4B.
Next, we examined the non-CP-CRE serial isolates 4C and 4D. Interestingly, in both isolates 4C and 4D, a transposition and insertion of MB1860TU_A into ompC was present upstream of the original TnMB1860 chromosomal locus (Figure 3c). We found two individual long reads that were able to span the full length of the MB1860TU_A array (3× copies), which disrupts ompC, for isolate 4C as well as isolate 4D, and confirmed the exact MB1860TU insertion location within ompC (c.504_505ins; Table 1). We were also able to identify 8 bp TSDs (5′-TTGCTGTT-3′) at each end of the ompC insertion sites, which indicates MB1860TU_A replicative transposition. The isolate 4D assembly and individual long reads indicated a second MB1860TU_A transposition and insertion ∼67 kb downstream of TnMB1860 within a GH25 lysozyme gene (Figure 3d). This transposition event could be confirmed by 8 bp TSDs (5′-TTGCTGTT-3′) flanking the inverted repeats of IS26 (Figure 3d). A progressive increase in both DNA and RNA levels for blaOXA-1 but not blaCTX-M-15 in the Patient 4 serial isolates was confirmed using qPCR and qRT–PCR, respectively (Figure 2).
Serial passage of Patient 4 index strain
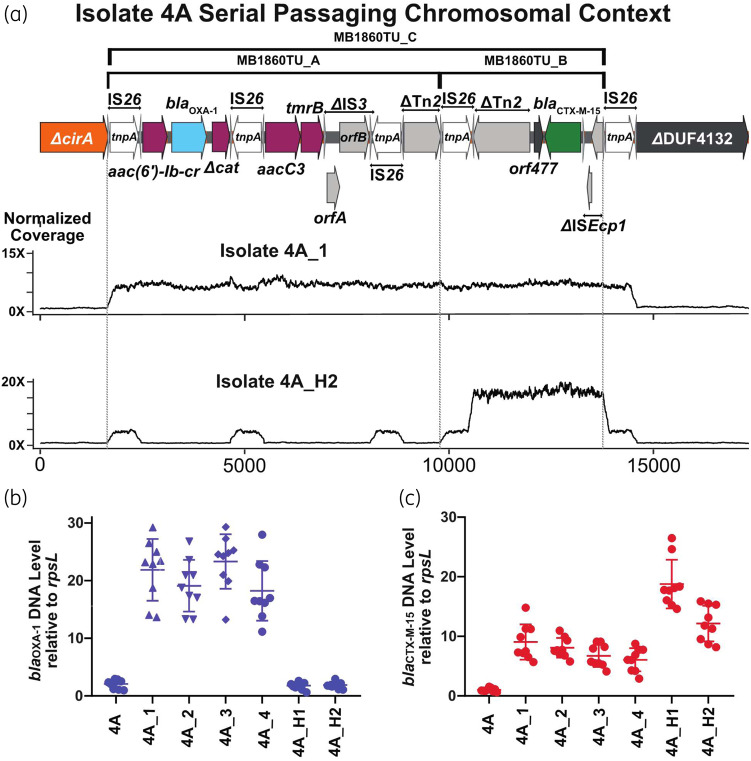
We passaged isolate 4A in increasing concentrations of ertapenem to determine whether antimicrobial exposure was driving the genetic changes observed in our serial clinical isolates. Resistance to ertapenem developed within three passages, corresponding to 3 days. Once ertapenem resistance developed, we collected strains for the next 4 days (4A_1 to 4A_4). ONT and Illumina sequencing on all four isolates indicated that, similar to isolate 4B, there was in situ amplification occurring at the original TnMB1680 chromosomal locus. However, the full-length TU MB1860TU_C (Figure 4a), which harbours both blaOXA-1 and blaCTX-M-15, was the amplifying structure for the passaged isolates in contrast to the clinical Patient 4 isolates where MB1860TU_A was the sole amplifying structure (Table 2). These results were confirmed with qPCR (Figure 4b).
Figure 4.
Identification and characterization of β-lactamase gene amplification following serial passaging of the index isolate 4A under ertapenem exposure. Isolate 4A was grown in the presence of ertapenem with isolates 4A_1 to 4A_4 collected during the first round of passaging and 4A_H1 and 4A_H2 collected during the second round. (a) Schematic of TnMB1860 locus from isolate 4A as detailed in Figure 3. Immediately below the schematic are normalized, short-read coverage depth line graphs for isolates 4A_1 and 4A_H2 aligned to index isolate 4A with the location of MB1860TU_A and MB1860TU_B bracketed by dotted lines. Note amplification of MB1860TU_C (i.e. MB1860TU_A and MB1860TU_C combined) in isolate 4A_1 whereas only MB1860TU_B is amplified in isolate 4A_H2. (b, c) TaqMan qPCR of genomic DNA collected in triplicate on two separate days (n = 6) for either blaOXA-1 (b) or blaCTX-M (c) relative to the endogenous control gene rpsL. Data shown are individual data points with mean ± SD superimposed.
Table 2.
Characterization of index isolate 4A serial passaging experiments
| Sample | ETP MIC (mg/L) | ompC | ompF | bla OXA-1 copy #a | bla CTX-M-15 copy #a |
|---|---|---|---|---|---|
| 4A | <0.5 | WT | WT | 1 | 1 |
| 4A_1 | >32 | c.208T>C; p. Q70X | c.17_18insIS1A | 6 | 7 |
| 4A_2 | >32 | c.208T>C; p. Q70X | WT | 8 | 9 |
| 4A_3 | >32 | c.208T>C; p. Q70X | insIS1Ab | 6 | 7 |
| 4A_4 | >32 | c.208T>C; p. Q70X | insIS1Ab | 6 | 7 |
| 4A_H1 | >32 | IS1A-mediated insertion 96 bp upstream of +1 ompC start site | WT | 1 | 9 |
| 4A_H2 | >32 | IS1A-mediated insertion 96 bp upstream of +1 ompC start site | WT | 1 | 17 |
ETP, ertapenem.
See Materials and methods for normalized coverage depth calculations.
Large IS1A-mediated deletion event (as described in Lee et al.)51 at ompF insertion locus precludes the ability to note correct genomic context. This is due in part to multiple variable-sized deletions detected in the long-read data. This may reflect population heterogeneity or technical difficulties in resolving a single genomic deletion size.
We repeated the experiment and again found ertapenem resistance developed within three passages from the 4A isolate. ONT sequencing of the second round of passaged isolates (4A_H1 and 4A_H2) revealed amplification of MB1860TU_B, which harbours blaCTX-M-15 solely, which we verified using qPCR (Figure 4a and c). Similar to the clinical strains from Patient 4, the serially passaged ertapenem-resistant isolates contained inactivating mutations in ompC, although we did not observe any MB1860TU-mediated interruptions (Table 2). Differential INDELs inactivating the ompF gene were observed in a fraction of the serial isolates, suggesting ompF inactivation may not be necessary for the development of non-CP-CRE, as well as possible population heterogeneity (Table 2).
In order to determine the chromosomal stability of the MB1860TU tandem arrays in the absence of antibiotic selective pressure, both ertapenem-resistant isolates 4C and 4D were passaged for 10 days (∼60 generations) in the absence of ertapenem. Both ertapenem-resistant recurrent strains consistently maintained carbapenem resistance through 60 generations of growth (Figure S6). The 4C and 4D isolates had relative copy number decreases of blaOXA-1 from 33× to 18× (45% decrease) and 53× to 42× (21% decrease) respectively, but the amplifications remained substantially above the index isolate 4A baseline (Figure S6).
Detection of β-lactamase gene amplification and porin disruption by AMR elements in non-serial Enterobacterales strains
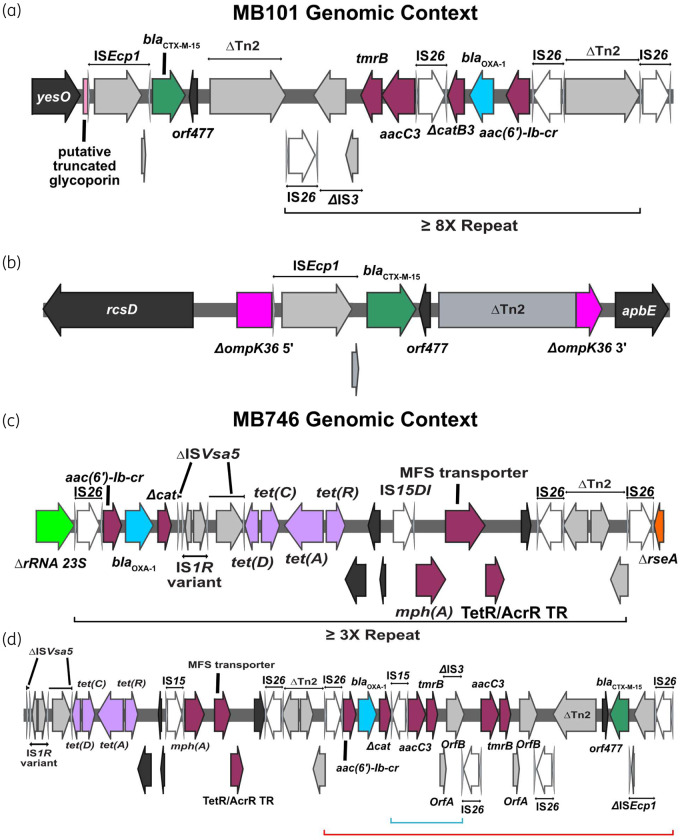
There were two non-CP-CRE strains, MB101 (K. pneumoniae) and MB746 (E. coli), identified in our previous study examining the role of short-read WGS in predicting β-lactam resistance in bacteraemia isolates (Table 1).20 We performed ONT sequencing on these two isolates to determine whether mechanisms of gene amplification and porin disruption were similar to what we observed in the cohort of non-CP-CRE bacteraemia serial isolates. The chromosome of the K. pneumoniae (ST37) MB101 isolate contained amplified IS26-mediated TUs carrying blaOXA-1 nearly identical to MB1860TU_A (Figure 5a). Moreover, OmpK36, the homologue of OmpC in E. coli, was interrupted by a 5390 bp ISEcp1-mediated transposition unit designated as MB101TPU (TPU; used here to delineate from IS26-mediated transposition mechanisms) harbouring blaCTX-M-15 (Figure 5b). The E. coli (ST405) MB746 isolate harboured a chromosomally amplified blaOXA-1 gene, as observed for Patient 4 serial isolates (Figure 5c), as well as a TnMB1860-like element present on an F-type plasmid (Figure 5d). These results suggest the amplification/disruption mechanism may be prevalent in non-CP-CRE clinical isolates within the Houston region.
Figure 5.
Genomic context of MB101 (K. pneumoniae) and MB746 (E. coli) isolates respectively. Terminal left and right inverted repeats (IRL and IRR, respectively) of ISs are specified by grey triangles that bracket complete and incomplete tnpA genes, respectively. ORFs are coloured as follows: AMR genes (maroon), blaOXA-1 (blue), blaCTX-M-15 (green), IS26 tnpA (white) and other IS/Tn elements (grey). Delta (Δ) next to an annotated gene or genetic region indicates a truncation or disruption. (a, b) Chromosomal locations of MB101TU (a) and MB101TPU (b) indicating amplification and transposition of each element, respectively. The black bracket indicates an ∼8× MB101TU repeat. Truncated ompK36 gene (b) is labelled in pink. (c, d) Genomic context of MB746TU and respective modules carrying AMR genes. Black brackets beneath (c) schematic indicate repeating MB746TU. (d) shows FIB plasmid carriage of AMR gene-containing modules. The red bracket indicates an IS26-bounded transposon structure that shares 100% coverage; 99% BLAST similarity with TnMB1860. The blue bracket indicates a small, 2× repeat structure.
Discussion
While recent surveillance studies indicate that the prevalence of non-CP-CRE remains high,4,5 mechanisms contributing to non-CP-CRE emergence within clinical settings are largely unknown. This public health issue is particularly troublesome given the recent finding of similar outcomes for patients with CPE and non-CP-CRE infections.5 Over the past 7 years, there has been an estimated 50% increase in ESBL-E cases in the USA.1 Given that non-CP-CRE is thought to evolve from ESBL-E precursor strains, there are concerns for a subsequent increase in non-CP-CRE infections. The CDC estimated in 2019 that 200 000 cases of ESBL-E infections and 13 000 cases of CRE infections occur in the USA annually.1 Assuming 50% of CRE are non-CP-CRE,4,5 the potential fraction of ESBL-E cases that develop into non-CP-CRE is approximately 3%, which is comparable with what we observed in our cohort. One of the commonly observed ESBL-E organisms detected is the pandemic, extraintestinal pathogenic E. coli ST131 group.40 Therefore, it was concerning that we identified two distinct ST131, non-CP-CRE isolates containing similar TUs harbouring β-lactamase genes and nearly identical TUs were present in previously sequenced ST131 isolates present in GenBank.
Through a combination of comparative genomic methods using clinical isolates and laboratory passaging of an index clinical E. coli ST131 isolate, we demonstrate that IS26-mediated modular amplifications of β-lactamase genes in conjunction with porin inactivation can drive non-CP-CRE emergence. We show that the TUs mediating β-lactamase gene amplification can translocate and disrupt porin genes, thereby simultaneously augmenting β-lactamase production and reducing antimicrobial entry into the periplasmic space. Moreover, our own results from serial passaging of clinical non-CP-CRE isolates without ertapenem exposure suggest that these amplified structures harbouring AMR genes associated with increased ertapenem and meropenem MICs may be fairly stable in the absence of antibiotic selective pressure. These results stand in contrast to the unstable nature of AMR gene amplifications observed following laboratory passaging of clinical isolates,34,41 but are in line with our identification of several non-CP-CRE clinical isolates with β-lactamase gene amplification causing serious infections.
It has long been recognized that increasing gene copy number can expand the range of AMR for a given β-lactamase,42–44 but the genetic complexity of such amplifications has hindered direct observations of mechanisms driving resistance, particularly in clinical isolates.45 While there have been a number of studies that have implicated IS-mediated porin disruptions,46–50 this is, to our knowledge, the first documented IS26-mediated TU disruption of an outer membrane porin with a tandem array of β-lactamase genes. Given the length of the TUs and their ability to generate tandem arrays, it is unlikely that targeted PCR-based strategies or commonly used short-read approaches alone would have been able to identify the correct genomic context underlying this particular non-CP-CRE mechanism. Future studies to determine signalling pathways that regulate and activate TU-mediated transposition, particularly into DNA motifs within outer membrane porins, are warranted. We predict that more widespread application of long-read sequencing technologies will facilitate appreciation of TU- and TPU-mediated transpositions, porin disruptions and gene amplifications on a diverse array of AMR pathogens in the clinical setting.
Supplementary Material
Acknowledgements
We thank the personnel of the clinical microbiology laboratory at MDACC for assistance with collecting isolates. We would also like to thank Jennifer Walker, PhD for her insightful input while drafting the manuscript.
Funding
This work was supported by the Shelby Foundation (R. Lee Clark Fellow Award to S.A.S.). Sequencing was performed at the MDACC DNA sequencing facility, which is supported by the National Cancer Institute (grant number P30-CA016672 via the Bioinformatics Shared Resource).
J.K. is supported by the Cancer Prevention and Research Institute of Texas (RP150596). Other support is provided by the UT Southwestern DocStars award (D.E.G. and J.K.). J.G-.P. is supported by the NIAID (1K01AI143881-01). C.A.A. is supported by NIH/NIAID grants (K24AI121296, R01AI134637, R01AI148342-01, R21AI143229, P01AI152999-01), UTHealth Presidential Award, University of Texas System STARS Award and Texas Medical Center Health Policy Institute Funding Program.
Transparency declarations
No competing interests to declare.
Supplementary data
Supplementary methods, Supplementary references, Tables S1–S6 and Figures S1–S6 are available as Supplementary data at JAC Online.
References
- 1.CDC. Antibiotic Resistance Threats in the United States 2019. 2019. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf
- 2. Logan LK, Weinstein RA.. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 2017; 215: S28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nordmann P, Naas T, Poirel L.. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 2011; 17: 1791–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guh AY, Bulens SN, Mu Y. et al. Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012-2013. JAMA 2015; 314: 1479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Duin D, Arias CA, Komarow L. et al. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study. Lancet Infect Dis 2020; 20: 731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Su CF, Chuang C, Lin YT. et al. Treatment outcome of non-carbapenemase-producing carbapenem-resistant Klebsiella pneumoniae infections: a multicenter study in Taiwan. Eur J Clin Microbiol Infect Dis 2018; 37: 651–9. [DOI] [PubMed] [Google Scholar]
- 7. Hayakawa K, Nakano R, Hase R. et al. Comparison between IMP carbapenemase-producing Enterobacteriaceae and non-carbapenemase-producing Enterobacteriaceae: a multicentre prospective study of the clinical and molecular epidemiology of carbapenem-resistant Enterobacteriaceae. J Antimicrob Chemother 2020; 75: 697–708. [DOI] [PubMed] [Google Scholar]
- 8. Hamzaoui Z, Ocampo-Sosa A, Fernandez Martinez M. et al. Role of association of OmpK35 and OmpK36 alteration and blaESBL and/or blaAmpC genes in conferring carbapenem resistance among non-carbapenemase-producing Klebsiella pneumoniae. Int J Antimicrob Agents 2018; 52: 898–905. [DOI] [PubMed] [Google Scholar]
- 9. Oteo J, Delgado-Iribarren A, Vega D. et al. Emergence of imipenem resistance in clinical Escherichia coli during therapy. Int J Antimicrob Agents 2008; 32: 534–7. [DOI] [PubMed] [Google Scholar]
- 10. Poirel L, Heritier C, Spicq C. et al. In vivo acquisition of high-level resistance to imipenem in Escherichia coli. J Clin Microbiol 2004; 42: 3831–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goessens WH, van der Bij AK, van Boxtel R. et al. Antibiotic trapping by plasmid-encoded CMY-2 β-lactamase combined with reduced outer membrane permeability as a mechanism of carbapenem resistance in Escherichia coli. Antimicrob Agents Chemother 2013; 57: 3941–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tangden T, Adler M, Cars O. et al. Frequent emergence of porin-deficient subpopulations with reduced carbapenem susceptibility in ESBL-producing Escherichia coli during exposure to ertapenem in an in vitro pharmacokinetic model. J Antimicrob Chemother 2013; 68: 1319–26. [DOI] [PubMed] [Google Scholar]
- 13. van Boxtel R, Wattel AA, Arenas J. et al. Acquisition of carbapenem resistance by plasmid-encoded-AmpC-expressing Escherichia coli. Antimicrob Agents Chemother 2017; 61: e01413-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Adler M, Anjum M, Andersson DI. et al. Influence of acquired β-lactamases on the evolution of spontaneous carbapenem resistance in Escherichia coli. J Antimicrob Chemother 2013; 68: 51–9. [DOI] [PubMed] [Google Scholar]
- 15. Kao CY, Chen JW, Liu TL. et al. Comparative genomics of Escherichia coli sequence type 219 clones from the same patient: evolution of the IncI1 blaCMY-carrying plasmid in vivo. Front Microbiol 2018; 9: 1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dupont H, Choinier P, Roche D. et al. Structural alteration of OmpR as a source of ertapenem resistance in a CTX-M-15-producing Escherichia coli O25b:H4 sequence type 131 clinical isolate. Antimicrob Agents Chemother 2017; 61: e00014-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chia JH, Siu LK, Su LH. et al. Emergence of carbapenem-resistant Escherichia coli in Taiwan: resistance due to combined CMY-2 production and porin deficiency. J Chemother 2009; 21: 621–6. [DOI] [PubMed] [Google Scholar]
- 18. Harris PA, Taylor R, Thielke R. et al. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42: 377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CLSI. Performance Standards for Antimicrobial Susceptibility Testing—Twenty-EighthEdition : M100 2018.
- 20. Shelburne SA, Kim J, Munita JM. et al. Whole-genome sequencing accurately identifies resistance to extended-spectrum β-lactams for major Gram-negative bacterial pathogens. Clin Infect Dis 2017; 65: 738–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013; https://arxiv.org/pdf/1303.3997.pdf. [Google Scholar]
- 22. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 2018; 34: 3094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H, Handsaker B, Wysoker A. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kolmogorov M, Yuan J, Lin Y. et al. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 2019; 37: 540–6. [DOI] [PubMed] [Google Scholar]
- 25. Page AJ, Cummins CA, Hunt M. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015; 31: 3691–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Löytynoja A. Phylogeny-Aware Alignment with PRANK. Humana Press, 2014. [DOI] [PubMed] [Google Scholar]
- 27. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30: 1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jolley KA, Maiden MC.. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 2010; 11: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014; 30: 2068–9. [DOI] [PubMed] [Google Scholar]
- 30. Jia B, Raphenya AR, Alcock B. et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 2017; 45: D566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carattoli A, Zankari E, Garcia-Fernandez A. et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 2014; 58: 3895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Siguier P, Perochon J, Lestrade L. et al. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 2006; 34: D32–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hanson B, Johnson J, Leopold S. et al. SVants—a long-read based method for structural variation detection in bacterial genomes. bioRxiv 2019: 822312.
- 34. Adler M, Anjum M, Berg OG. et al. High fitness costs and instability of gene duplications reduce rates of evolution of new genes by duplication-divergence mechanisms. Mol Biol Evol 2014; 31: 1526–35. [DOI] [PubMed] [Google Scholar]
- 35. He D, Chiou J, Zeng Z. et al. Residues distal to the active site contribute to enhanced catalytic activity of variant and hybrid β-lactamases derived from CTX-M-14 and CTX-M-15. Antimicrob Agents Chemother 2015; 59: 5976–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Partridge SR. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol Rev 2011; 35: 820–55. [DOI] [PubMed] [Google Scholar]
- 37. He S, Hickman AB, Varani AM. et al. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio 2015; 6: e00762-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Harmer CJ, Hall RM.. IS26-mediated formation of transposons carrying antibiotic resistance genes. mSphere 2016; 1: e00038-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Harmer CJ, Moran RA, Hall RM.. Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio 2014; 5: e01801-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stoesser N, Sheppard AE, Pankhurst L. et al. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 2016; 7: e02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nicoloff H, Hjort K, Levin BR. et al. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat Microbiol 2019; 4: 504–14. [DOI] [PubMed] [Google Scholar]
- 42. Normark S, Edlund T, Grundstrom T. et al. Escherichia coli K-12 mutants hyperproducing chromosomal β-lactamase by gene repetitions. J Bacteriol 1977; 132: 912–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sandegren L, Andersson DI.. Bacterial gene amplification: implications for the evolution of antibiotic resistance. Nat Rev Microbiol 2009; 7: 578–88. [DOI] [PubMed] [Google Scholar]
- 44. Bradford PA, Urban C, Mariano N. et al. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the foss of an outer membrane protein. Antimicrob Agents Chemother 1997; 41: 563–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Livermore DM, Day M, Cleary P. et al. OXA-1 β-lactamase and non-susceptibility to penicillin/β-lactamase inhibitor combinations among ESBL-producing Escherichia coli. J Antimicrob Chemother 2019; 74: 326–33. [DOI] [PubMed] [Google Scholar]
- 46. Hernández-Allés S, Benedí VJ, Martínez-Martínez L. et al. Development of resistance during antimicrobial therapy caused by insertion sequence interruption of porin genes. Antimicrob Agents Chemother 1999; 43: 937–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Doumith M, Ellington MJ, Livermore DM. et al. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother 2009; 63: 659–67. [DOI] [PubMed] [Google Scholar]
- 48. Vandecraen J, Chandler M, Aertsen A. et al. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit Rev Microbiol 2017; 43: 709–30. [DOI] [PubMed] [Google Scholar]
- 49. Mena A, Plasencia V, Garcia L. et al. Characterization of a large outbreak by CTX-M-1-producing Klebsiella pneumoniae and mechanisms leading to in vivo carbapenem resistance development. J Clin Microbiol 2006; 44: 2831–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zowawi HM, Forde BM, Alfaresi M. et al. Stepwise evolution of pandrug-resistance in Klebsiella pneumoniae. Sci Rep 2015; 5: 15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee H, Doak TG, Popodi E. et al. Insertion sequence-caused large-scale rearrangements in the genome of Escherichia coli. Nucleic Acids Res 2016; 44: 7109–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Assemblies of index isolates from patients with recurrent bacteraemia as well as long-read and short-read data for recurrent isolates have been deposited in the NCBI BioProject database (PRJNA603908). ONT sequencing data of non-CP-CRE isolates from a previous study20 were deposited in the NCBI BioProject database (PRJNA388450).