Abstract
Osteosarcoma is the most common primary malignancy of the bones, and is associated with a high rate of metastasis and a poor prognosis. A tight association between the tumor microenvironment (TME) and osteosarcoma metastasis has been established. In the present study, the Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) algorithm was applied to calculate the immune and stromal scores of patients with osteosarcoma based on data from The Cancer Genome Atlas database. A metagene approach and deconvolution method were used to reveal distinct TME landscapes in patients with osteosarcoma. Bioinformatics analysis was used to identify differentially expressed genes (DEGs) associated with metastasis and immune infiltration in osteosarcoma, and a risk model was constructed using the DEGs with potential prognostic significance. Subsequently, gene set enrichment and Spearman's correlation analyses were used to delineate the biological processes associated with these prognostic biomarkers. Finally, immunohistochemical (IHC) analysis was performed to evaluate the expression levels of immune infiltrates and prognostic biomarkers in clinical osteosarcoma tissues. The results of the ESTIMATE demonstrated that patients with non-metastatic osteosarcoma presented with higher immune/stromal scores and a more favorable prognosis compared with those with metastatic osteosarcoma. The TME landscapes in patients with osteosarcoma suggested that high levels of tumor-infiltrating immune cells (TIICs) may suppress metastasis. Increased numbers of CD56bright natural killer cells, immature B cells, M1 macrophages and neutrophils, and lower levels of M2 macrophages were observed in the non-metastatic tissues compared with those in the metastatic tissues. A total of 69 DEGs were identified to be associated with metastasis and immune infiltration in osteosarcoma. Of these, GATA3, LPAR5, EVI2B, RIAM and CFH exhibited prognostic potential and were highly expressed in non-metastatic osteosarcoma tissues based on the IHC analysis results. These biomarkers were involved in various immune-related biological processes and were positively associated with multiple TIICs and immune signatures. The risk model constructed using these prognostic biomarkers demonstrated high predictive accuracy for the prognosis of osteosarcoma. In conclusion, the present study proposed a five-biomarker prognostic signature for the prediction of metastasis and immune infiltration in patients with osteosarcoma.
Keywords: osteosarcoma, tumor microenvironment, metastasis, immune infiltration, prognosis
Introduction
Osteosarcoma is a malignancy of the bone that stems from primitive mesenchymal cells and frequently affects the metaphyseal regions of long bones, including the distal femur, proximal tibia and the proximal humerus (1,2). The incidence of osteosarcoma exhibits bimodal distribution, predominantly affecting children and adolescents, with 75% of patients aged <20 years (3). Osteosarcoma is rare, with an annual estimated worldwide incidence of 2–3 cases per 1,000,000 individuals (4). However, it is highly aggressive and primarily metastasizes to the lung; in ~80% of cases, patients exhibit subclinical pulmonary micrometastases at the time of diagnosis (5). The 5-year survival rate of patients with localized osteosarcoma is ~65%, whereas for recurrent and metastatic cases, the long-term survival rate is ~20% (6). Conventionally, treatment strategies of osteosarcoma comprise neoadjuvant chemotherapy, surgical resection and adjuvant chemotherapy (7). However, the survival rates have plateaued in the last three decades, and metastasis remains the principal factor underlying mortality in patients with osteosarcoma (6). Most patients with metastatic osteosarcoma exhibit a poor response to the current standard treatments.
The tumor microenvironment (TME) comprises all factors recruited to the tumor, including non-cellular (microvessels and the extracellular matrix) and cellular (cancer cells, immune cells and stromal cells) components (8,9). During tumor development, cancer cells pathologically affect the TME by inducing various types of stress, including hypoxia, oxidative stress and acidosis (10). These effects trigger aberrant responses from neighboring immune and stromal cells, which promote necrosis and metastasis (11). Considering the importance of tumor-induced immune escape in osteosarcoma recurrence and metastasis, immunotherapy has been proposed as a promising therapeutic option for high-grade osteosarcoma (12). Therapeutic strategies targeting tumor-associated macrophages have been demonstrated to significantly suppress metastasis in cases of advanced osteosarcoma (13,14). In addition, combined therapy using tumor lysate-pulsed dendritic cells (DCs) and an anti-cytotoxic T lymphocyte antigen-4 antibody inhibits the outgrowth of lung metastasis and prolongs patient survival (15). Despite the tremendous amounts of research efforts performed in the past three decades, no effective immunotherapies have been developed for the treatment of osteosarcoma, which has been attributed to the rarity and heterogeneity of osteosarcoma, absence of specific tumor antigens and off-target effects of drugs (16–18). Thus, an improved understanding of the association between metastasis and osteosarcoma TME is urgently required to improve survival outcomes.
Multiple algorithms have been developed for evaluation of the heterogeneity of the TME landscape (19–21). The metagene approach is considered superior to other deconvolution methods such as Tumor Immune Estimation Resource (TIMER) as it is less sensitive to noise caused by sample impurity or sample preparation (19). These two algorithms have been successfully applied to assess immune infiltration and identify clinical signatures in various types of cancer (19–25). However, to the best of our knowledge, the immune profiles of osteosarcoma have not been previously evaluated using these algorithms.
The present study aimed to delineate the distinct profiles of immune infiltration in patients with metastatic and non-metastatic osteosarcoma, and to subsequently identify metastasis- and immune-related genes that may act as potential biomarkers or treatment targets for osteosarcoma. Understanding the aberrant expression of the prognostic biomarkers and the related pathways may facilitate early diagnosis and appropriate therapy for individual patients with osteosarcoma.
Materials and methods
Data preparation and processing
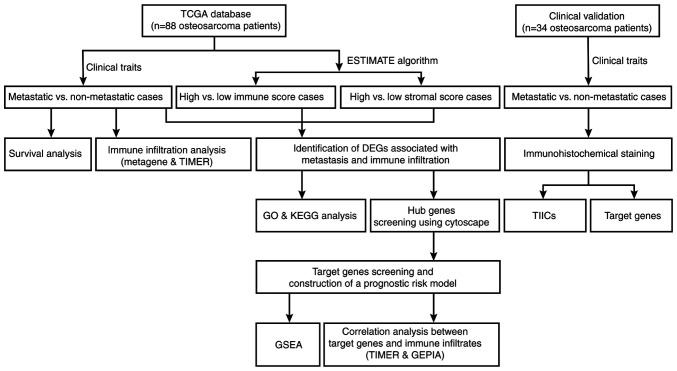
A flowchart of the analysis performed in the present study is outlined in Fig. 1. Level 3 gene transcriptome profiles of patients with osteosarcoma were downloaded from The Cancer Genome Atlas (TCGA; portal.gdc.cancer.gov/) on January 16, 2020. The dataset comprised 88 osteosarcoma cases. Associated clinical data, including sex, age, survival status, metastasis, primary tumor sites, overall survival and event-free survival times were also obtained. The GDCRNATools package version 1.6.0 (bioconductor.org/packages/release/bioc/html/GDCRNATools.html) (26) was used to integrate and normalize the gene expression count matrices in R version 3.6.3 (27). Subsequently, the immune and stromal scores were calculated using the Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) algorithm (28) for predicting the presence of infiltrating immune and stromal cells in tumor tissues. Unpaired Student's t-test was used to compare the differences of immune and stromal scores between metastatic and non-metastatic cases.
Figure 1.
Flowchart of data analysis in present study. TCGA, the Cancer Genome Atlas; TIMER, Tumor IMmune Estimation Resource; ESTIMATE, Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data; DEGs, differentially expressed genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; GSEA, gene set enrichment analysis; GEPIA, Gene Expression Profiling Interactive Analysis; TIICs, tumor-infiltrating immune cells.
Evaluation of immune infiltration in metastatic and non-metastatic osteosarcoma
To comprehensively characterize the cellular composition of the immune infiltrates in osteosarcoma, a metagene approach and the deconvolution method were used independently. The metagene approach relies on a set of genes representing various immune cell types that are not expressed in cancer cell lines or normal tissues. The GSVA package version 1.34.0 (bioconductor.org/packages/release/bioc/html/GSVA.html) (29) was used to calculate the aggregate scores for each tumor-infiltrating immune cells (TIICs) class, with higher scores indicating a higher degree of infiltration. Independently, the processed expression matrices of osteosarcoma cohorts were analyzed using TIMER version 2.0 (cistrome.shinyapps.io/timer/) (20), a web-accessible deconvolution-based resource for systematic analyses and visualization of immune cell abundance. After obtaining the fractions of immune subpopulations in the individual samples, the osteosarcoma cases were classified as metastatic or non-metastatic to establish the differential distribution of TIICs.
Survival analysis
Kaplan-Meier analysis followed by the log-rank test was used to assess the association between metastasis and patient prognosis using the survival (version 3.1–8; cran.r-project.org/web/packages/survival/index.html) (30) and survminer (version 0.4.6; http://CRAN.R-project.org/package=survminer) packages. P<0.05 was considered to indicate a statistically significant difference.
Identification of differentially expressed genes (DEGs)
Next, the limma package version 3.42.0 (bioconductor.org/packages/release/bioc/html/limma.html) (31) was used to identify metastasis-related DEGs in the metastatic group (n=22) vs. the non-metastatic group (n=66) using the following cutoffs: |log2 fold change (FC)|≥log21.5 and P<0.05. The expression pattern of the DEGs was visualized using heatmaps and volcano plots. The 88 osteosarcoma cases were classified into high or low score groups based on the median immune and stromal scores, and the aforementioned approach was used to identify the immune or stromal score-related DEGs. The intersection genes that were differentially expressed (up- or downregulated) between the groups were identified using the VennDiagram package version 1.6.20 (https://CRAN.R-project.org/package=VennDiagram).
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses
To determine the functions and pathways associated with the intersection genes, GO annotation and KEGG pathway enrichment analyses were performed using the clusterProfiler package version 3.14.2 (bioconductor.org/packages/release/bioc/html/clusterProfiler.html) (32). P<0.05 was considered to indicate a statistically significant difference.
Protein-protein interaction (PPI) network construction
The Search Tool for the Retrieval of Interacting Genes (STRING; string.embl.de/) online database was used to establish a PPI network of the intersection genes and evaluate the degree of interactivity at the protein level. The results of the PPI network were visualized with Cytoscape version 3.7.1 (33). A plugin of Cytoscape, cytoHubba (34) was utilized to screen for hub genes using the Maximal Clique Centrality (MCC) algorithm.
Screening for prognostic target genes
Kaplan-Meier survival analysis and univariate Cox regression models were used to identify the prognosis-associated genes amongst the hub genes based on the gene expression values and survival status data. The log-rank test was used for curves without late crossover. The Renyi test was performed to detect differences when survival curves crossed over with the survMisc package version 0.5.5 (https://CRAN.R-project.org/package=survMisc). P<0.05 was considered to indicate a statistically significant difference. The overlapping genes from the two analyses were further screened by the least absolute shrinkage and selection operator (LASSO) Cox regression analysis for variable selection and shrinkage using the glmnet package version 3.0–2 (cran.r-project.org/web/packages/glmnet/index.html) (35). A formula was established to calculate the prognostic risk scores for each patient on the basis of the coefficients derived from the risk model. According to the median value of the risk scores, the patients were grouped into high- and low-risk subsets. Kaplan-Meier survival analysis followed by log-rank test was performed on the two groups, and the receiver operating characteristic (ROC) curves were drawn to evaluate the stability of the model.
Gene set enrichment analysis (GSEA)
The patients with osteosarcoma in TCGA dataset were split into high- and low-expression subgroups according to the expression levels of each target gene (top 50%, high vs. bottom 50%, low). GSEA (software.broadinstitute.org/gsea/index.jsp) was used to determine whether the pre-defined KEGG pathways were enriched (36,37). The enrichment scores, normalized enrichment scores and P-values were calculated. Furthermore, the top five pathways of interest were integrated and visualized in an enrichment plot based on the enrichment scores. P<0.05 and a false discovery rate q<0.05 were considered to indicate a statistically significant difference.
Analysis of correlation between target genes and immune infiltrates
Next, the correlations between TIICs or immune cell markers and the target genes were evaluated using TIMER and Gene Expression Profiling Interactive Analysis (GEPIA; gepia.cancer-pku.cn/) (38), respectively. Spearman correlation analysis was used to determine the correlation coefficients in the two databases. P<0.05 was considered to indicate a statistically significant difference.
Clinical specimens
Ethics approval for the use of clinical material was obtained from the Ethics Committee of the Affiliated Zhujiang Hospital of Southern Medical University (Guangzhou, China; approval no. 2018-GJGBWK-002). Patients were recruited between May 2018 and April 2020. Written informed consent was obtained from all participants or their legal guardians prior to sample collection. A total of 16 metastatic (mean age, 21.56±5.30; range, 14–33 years; 9 male and 7 female patients) and 18 non-metastatic (mean age, 17.56±6.45; range, 10–34 years; 11 male and 7 female patients) osteosarcoma tissues were obtained and stored at −80°C. Osteosarcoma diagnosis was verified by histopathological evaluation. The patient cohort comprised 20 male and 14 female individuals, with a mean age of 19.44±6.19 years. There were eight cases at stage I, 10 cases at stage II and 16 cases at stage III.
Immunohistochemical analysis
All 34 specimens from patients with osteosarcoma were embedded in paraffin. Following dewaxing in xylene and rehydration with graded ethanol series (100, 95, 90, 80 and 70%) at room temperature, the tissue slides were boiled in 0.01 M sodium citrate buffer (pH 6.0; Beijing Zhongshan Golden Bridge Biotech, Co., Ltd.) at 95°C in a water bath for 10 min and immersed in 3% hydrogen peroxide at room temperature for 30 min to block endogenous peroxidase activity. Subsequently, the slides were incubated with 5% bovine serum albumin (Wuhan Servicebio Technology, Co., Ltd.) at room temperature for 30 min and stained with primary antibodies overnight at 4°C. Finally, the slides were incubated with horseradish peroxidase-conjugated secondary antibodies (1:50; cat. no. A0208; Beyotime Institute of Biotechnology) for 60 min at room temperature and stained using a horseradish peroxidase-DAB kit (Tiangen Biotech, Co., Ltd.) according to the manufacturer's protocol. The following primary antibodies were used for immunostaining: CD56bright natural killer (NK) cell-specific antibody CD56 (1:100; cat. no. 14255-1-AP; ProteinTech Group, Inc.), immature B cell-specific antibody CD22 (1:200; cat. no. 21894-1-AP; ProteinTech Group, Inc.), M1 macrophage-specific antibody CD86 (1:100; cat. no. DF6332; Affinity Biosciences), M2 macrophage-specific antibody CD163 (1:100; cat. no. 16646-1-AP; ProteinTech Group, Inc.), neutrophil-specific antibody CD11B (1:200; cat. no. bs-1014R; BIOSS), GATA3 (1:200; cat. no. 10417-1-AP; ProteinTech Group, Inc.), LPAR5 (1:500; cat. no. bs-15366R; BIOSS), EVI2B (1:50; cat. no. 24891-1-AP; ProteinTech Group, Inc.), RIAM (1:100; cat. no. 13500-1-AP; ProteinTech Group, Inc.) and CFH (1:400; cat. no. bs-9525R; BIOSS). Images were captured with a light orthophoto microscope (magnification, ×200). Immunohistochemical staining was evaluated using a semiquantitative scoring method based on the staining intensity and the percentage of positively-stained cells (39). The staining intensity was scored as follows: 0 (no staining), 1 (+), 2 (++) and 3 (+++); the percentage of positive-stained cells was scored as follows: 0 (0-1%), 1 (1-33%), 2 (34-66%) and 3 (67-100%). The total staining scores were defined according to the sum of the intensity and percentage scores: Low expression (0–2); medium expression (3 and 4); and high expression (5 and 6). Statistical significance of the semi-quantified immunohistochemical staining between patients with non-metastatic and metastatic osteosarcoma for each marker was determined by the Mann-Whitney U test.
Statistical analysis
The IHC scores are presented as median and range. Other numerical data are presented as mean ± standard deviation. All statistical analyses were performed using SPSS version 20.0 (IBM Corp.). Prior to analysis, normal distribution and homogeneity of variance of all variables were assumed with Shapiro-Wilk and Levene's tests, respectively. Parametric testing between two groups was performed by unpaired Student's t-test. For non-parametric two-group comparisons, the Mann-Whitney U test was performed. A two-sided P<0.05 was considered to indicate a statistically significant difference.
Results
Differential immune infiltration in metastatic vs. non-metastatic osteosarcoma
TCGA gene expression datasets from 88 patients with osteosarcoma were analyzed. Patient demographics are summarized in Table I. The mean ± SD age of enrolled patients was 15.16±4.86, and 58.0% of these patients were male. Of the 88 osteosarcoma cases, 22 were metastatic. The results of the ESTIMATE analysis revealed that the immune (range, −1,820.49–1,965.19) and stromal (range, −731.86–1,880.48) scores were associated with metastasis. High immune and stromal scores were associated with non-metastatic osteosarcoma, although the associations were not statistically significant (Fig. 2A and B). Kaplan-Meier analysis demonstrated that metastatic osteosarcoma was significantly associated with shorter overall survival (P<0.001) and event-free survival (P<0.001) times compared with non-metastatic osteosarcoma (Fig. 2C and D).
Table I.
Clinical characteristics of the 88 patients with osteosarcoma included in the present study.
| Characteristic | Male | Female | Total |
|---|---|---|---|
| Total, n (%) | 51 (58.0%) | 37 (42.0%) | 88 (100.0%) |
| Age, years | 16.73±5.08 | 13.01±3.59 | 15.16±4.86 |
| Survival status, n (%) | |||
| Alive | 34 (38.7%) | 23 (26.1%) | 57 (64.8%) |
| Dead | 15 (17.0%) | 14 (15.9%) | 29 (32.9%) |
| Unknown | 2 (2.3%) | 0 (0.0%) | 2 (2.3%) |
| Metastasis, n (%) | |||
| Present | 10 (11.4%) | 12 (13.6%) | 22 (25.0%) |
| Absent | 41 (46.6%) | 25 (28.4%) | 66 (75.0%) |
| Primary tumor site, n (%) | |||
| Leg or foot | 47 (53.4%) | 33 (37.5%) | 80 (90.9%) |
| Arm or hand | 2 (2.3%) | 4 (4.5%) | 6 (6.8%) |
| Pelvis | 2 (2.3%) | 0 (0.0%) | 2 (2.3%) |
| Overall survival, years | 4.52±3.22 | 3.65±2.72 | 4.15±3.03 |
| Event-free survival, years | 3.45±2.99 | 3.08±2.80 | 3.30±2.90 |
Continuous variables are presented as the mean ± standard deviation.
Figure 2.
Patients with non-metastatic osteosarcoma present with higher immune/stromal scores and a more favorable prognosis compared with those with metastatic osteosarcoma. Distribution of (A) immune and (B) stomal scores between the metastatic and non-metastatic groups. Kaplan-Meier analyses of patients' (C) OS and (D) EFS between the metastatic and non-metastatic groups. OS, overall survival; EFS, event-free survival.
To further define the patterns of immune infiltration in metastatic vs. non-metastatic osteosarcoma, metagene and TIMER analyses were performed independently. Based on the metagene approach, a set of metagenes for 25 immune cell subpopulations were initially defined (Table SI). Subsequently, the relative expression levels of the metagenes in metastatic vs. non-metastatic osteosarcoma were determined. The results demonstrated that the proportions of CD56bright NK cells, immature B cells and M1 macrophages were significantly higher in the non-metastatic group compared with those in the metastatic group, whereas the proportion of M2 macrophages was significantly lower (Fig. 3A). Higher proportions of activated B cells, activated CD8 T cells, γδ T cells, immature DCs, NK cells, T follicular helper cells, type 1 T helper cells and type 2 T helper cells were observed in the non-metastatic group compared with those in the metastatic group. TIMER analysis revealed a higher proportion of neutrophils in non-metastatic osteosarcoma compared with that in metastatic osteosarcoma (Fig. 3B).
Figure 3.
Distinct patterns of immune infiltrates in patients with metastatic vs. non-metastatic osteosarcoma. Evaluation of immune infiltration in metastatic and non-metastatic osteosarcoma based on (A) the metagene approach and (B) the Tumor Immune Estimation Resource method.
Identification of DEGs associated with metastasis and immune infiltration
A total of 611 metastasis-associated DEGs were identified from the osteosarcoma datasets obtained from TCGA, of which 300 were significantly upregulated and 311 were significantly downregulated in metastatic compared with non-metastatic cases. Due to the close relationship between metastasis and immune infiltration, the cases of osteosarcoma were stratified into high- (n=44) and low- (n=44) score groups based on the median immune and stromal scores. Subsequently, 2,584 DEGs (1,586 upregulated and 998 downregulated) were identified in the high immune score group, and 2,541 DEGs (1,679 upregulated and 862 downregulated) were identified in the high stromal-score group. The expression profiles of DEGs were visualized using heatmaps and volcano plots (Fig. 4A-F). A total of 69 metastasis- and immune-associated DEGs were common to both the metastatic and low immune and stromal score groups (Fig. 4G and H).
Figure 4.
Identification of DEGs associated with metastasis and immune infiltration in osteosarcoma. (A) Heatmap and (D) volcano plot of DEGs between patients with and without metastatic osteosarcoma. (B) Heatmap and (E) volcano plot of DEGs based on immune scores. (C) Heatmap and (F) volcano plot of DEGs based on stromal scores. Venn diagrams of (G) co-upregulated and (H) co-downregulated DEGs amongst the metastatic and low immune/stromal score groups. In the heatmaps, upregulated genes are presented in red, and downregulated genes are presented in blue. DEGs, differentially expressed genes.
Functional enrichment and PPI network analysis
The results of the GO term analysis demonstrated that the intersection DEGs may be associated with several biological processes (BP), including ‘T cell activation’ (GO:0042110), ‘leukocyte differentiation’ (GO:0002521), ‘negative regulation of cytokine production’ (GO:0001818) and ‘regulation of leukocyte activation’ (GO:0002694) (Fig. 5A). The significantly enriched cellular components (CC) included ‘secretory granule membrane’ (GO:0030667), ‘ficolin-1-rich granule’ (GO:0101002), ‘intrinsic component of organelle membrane’ (GO:0031300) and ‘lamellipodium’ (GO:0030027) (Fig. 5B). Among the molecular function (MF) terms, the intersection DEGs were enriched for ‘heparan sulfate proteoglycan binding’ (GO:0043395), ‘cytokine receptor activity’ (GO:0004896), ‘G protein-coupled chemoattractant receptor activity’ (GO:0001637) and ‘chemokine receptor activity’ (GO:0004950) (Fig. 5C). KEGG pathway analysis revealed enrichment for pathways associated with ‘lipid metabolism’ (hsa00565), ‘pantothenate and CoA biosynthesis’ (hsa00770), and ‘glycerophospholipid metabolism’ (hsa00564) (Fig. 5D).
Figure 5.
Functional enrichment analysis of the intersection of genes and the PPI network. Significantly enriched GO terms including (A) biological process, (B) cellular component and (C) molecular function. (D) KEGG enrichment results of the intersection genes. (E) Top 20 hub genes screened using the Maximal Clique Centrality algorithm. The color of a node reflects the connectivity degree; warmer colors indicate a higher degree value. PPI, protein-protein interaction; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
To identify the potential interaction patterns among the transcripts of the 69 intersection DEGs, a PPI network was constructed using the STRING database. Analysis of the PPI network on Cytoscape using the cytoHubba plugin identified the top 20 genes, which were considered the hub genes. These hub genes included PTPRC, LCP2, TLR7, CX3CR1, CX3CL1, NCKAP1L, LPAR5, EVI2B, DOCK8, IL17RA, PLD4, MNDA, GATA3, CLEC7A, RIAM, CD200R1, PIK3CG, JAK3, CFH and KLRB1 (Fig. 5E). In addition, 737 GO terms and 12 KEGG pathways were identified based on additional functional analyses of the hub genes, indicating that the hub genes were primarily enriched in immune-related biological processes, including ‘T cell activation’, ‘leukocyte migration’, ‘cytokine receptor activity’ and ‘chemokine signaling pathway’ (Fig. S1).
Construction of the prognostic risk model
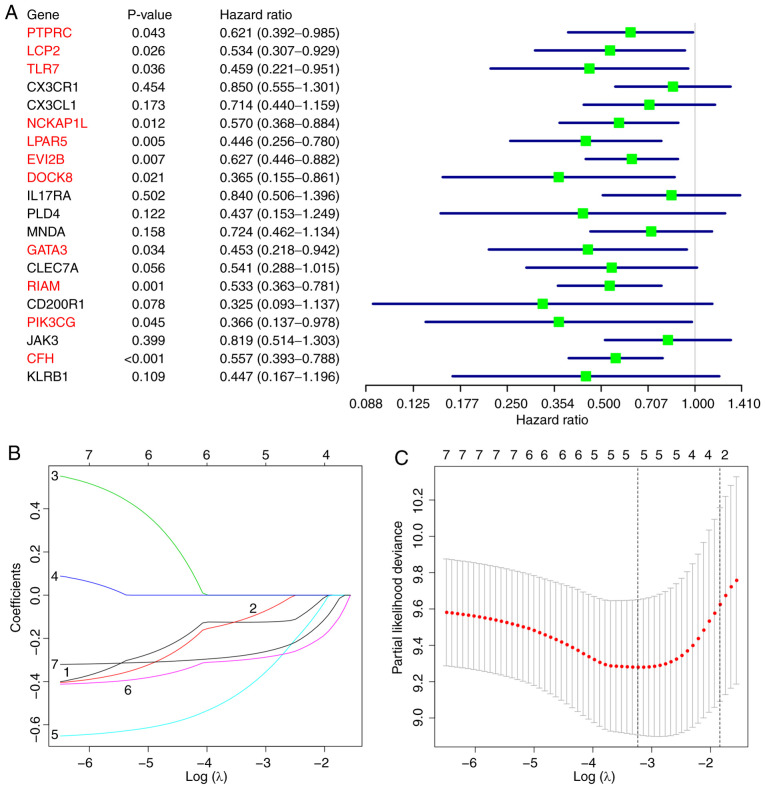
Kaplan-Meier survival analysis revealed that eight of the 20 hub genes were significantly associated with the prognosis of patients with osteosarcoma (Fig. 6). Univariate Cox regression analysis identified 11 hub genes as predictors of favorable prognosis (Fig. 7A). Subsequently, the seven overlapping genes identified by both analytical methods were included in the LASSO Cox regression analysis, which identified five target genes with prognostic potential: GATA3, LPAR5, EVI2B, RIAM and CFH (Fig. 7B and C). The prognostic risk scores for each osteosarcoma case were calculated based on the coefficients and the expression values of the target genes as follows: Risk score = (−0.4107 × expression value of GATA3) + (−0.1253 × expression value of LPAR5) + (−0.0985 × expression value of EVI2B) + (−0.2954 × expression value of RIAM) + (−0.2732 × expression value of CFH).
Figure 6.
Kaplan-Meier survival curves of the hub genes. All hub genes from the protein-protein interaction network were analyzed using Kaplan-Meier analysis, and plots for those with significant differences are presented. (A) GATA3, (B) RIAM, (C) LPAR5, (D) CD200R1, (E) EVI2B, (F) CFH, (G) TLR7 and (H) NCKAP1L. Comparisons between the high and low gene expression groups were performed using the log-rank or Renyi test.
Figure 7.
Construction of a prognostic risk model for osteosarcoma. (A) Univariate Cox regression analysis identified 11 hub genes (highlighted in red) as independent prognostic factors. (B) LASSO coefficient profiles of overlapping hub genes from the Kaplan-Meier survival and univariate Cox regression analyses. (C) Tuning parameter λ selection in the LASSO model. Dotted vertical lines indicate the optimal values using the minimum criteria and the standard error of the minimum criteria. LASSO, least absolute shrinkage and selection operator.
The patients with osteosarcoma in TCGA dataset were stratified into high- and low-risk groups based on the median value of the risk scores. As presented in Fig. 8A and B, a high risk score was associated with shorter survival and a higher incidence of death compared with those in the low risk score group. Heatmap analysis indicated that the expression levels of the five target genes with prognostic potential were downregulated in the high-risk cases compared with those in the low-risk group (Fig. 8C). The results of the survival analysis demonstrated that the 5-year survival rates of patients with high and low risk scores were 38.4 and 88.2%, respectively (Fig. 8D). The ROC curve analysis revealed that the areas under the curve for predicting 1, 3 and 5-year survival were 0.817, 0.849 and 0.876, respectively, indicating that the risk model had a high prognostic capacity (Fig. 8E).
Figure 8.
Robustness of the five-gene signature model. (A) The risk score of each sample in the osteosarcoma cohort from The Cancer Genome Atlas. (B) Survival time and status of each patient in the osteosarcoma cohort. (C) Heatmap of the target genes between the high- and low-risk groups. Red, upregulated genes; blue, downregulated genes. (D) Kaplan-Meier survival curve between the high- and low-risk groups based on the five-gene signature. (E) Receiver operating characteristic analysis of the sensitivity and specificity of the five-gene signature for the prediction of 1-, 3- and 5-year overall survival in patients with osteosarcoma. AUC, area under the curve.
GSEA of the target genes
GSEA was performed for subgroups of patients with osteosarcoma based on the expression levels of each target gene aiming to uncover the significant KEGG pathways enriched in the DEGs between the high- and low-expression subgroups. GATA3 expression was significantly associated with cell adhesion molecules (‘CAMs’), ‘complement and coagulation cascades’, ‘Th1 and Th2 cell differentiation’ and ‘NK cell mediated cytotoxicity’ (Fig. 9A). The LPAR5 high-expression subgroup was enriched for ‘antigen processing and presentation’, ‘BCR signaling pathway’, ‘CAMs’ and ‘TCR signaling pathway’ (Fig. 9B). EVI2B expression was associated with enrichment for ‘antigen processing and presentation’, ‘BCR signaling pathway’, ‘hematopoietic cell lineage’ and ‘CAMs’ (Fig. 9C). The RIAM high-expression subgroup was enriched for ‘FeγR-mediated phagocytosis’, ‘chemokine signaling pathway’, ‘NK cell mediated cytotoxicity’ and ‘TCR signaling pathway’ (Fig. 9D). Finally, CFH was associated with enrichment for the ‘renin-angiotensin system’, ‘TLR signaling pathway’, ‘complement and coagulation cascades’ and ‘apoptosis’ (Fig. 9E). Notably, there was a significant association between all target genes and the ‘NF-κB signaling pathway’.
Figure 9.
GSEA of the target genes. GSEA of (A) GATA3, (B) LPAR5, (C) EVI2B, (D) RIAM and (E) CFH. GSEA, gene set enrichment analysis; Th, T helper cell; NK cell, natural killer cell; CAMs, cell adhesion molecules; BCR, B cell receptor; TCR, T cell receptor; FcγR, Fc γ receptor; TLR, Toll-like receptor.
Analysis of correlations between target genes with TIICs and immune signatures
TIMER analysis was used to evaluate the correlations between the target genes and a range of TIICs in the osteosarcoma TME. The target genes were positively correlated with B cells, CD8 T cells, CD4 T cells, neutrophils and DC infiltration levels (Fig. 10). Similar results were obtained following analysis of the correlations between target genes and crucial immune signatures in the GEPIA database (Table II). Taken together, these results suggested that high levels of the target genes may confer a favorable prognosis for patients with osteosarcoma by modulating immune cells.
Figure 10.
Analysis of the correlation between target genes and infiltration levels of immune cells. Correlation between target genes and infiltration levels of (A) B cells, (B) CD8 T cells, (C) CD4 T cells, (D) neutrophils and (E) dendritic cells based on the Tumor Immune Estimation Resource analysis. *P<0.05, **P<0.01 and ***P<0.001. Cor, Spearman's R value.
Table II.
Correlation analysis between target genes and immune signatures in Gene Expression Profiling Interactive Analysis.
| Spearman's R value | ||||||
|---|---|---|---|---|---|---|
| Immune cells | Markers | GATA3 | LPAR5 | EVI2B | RIAM | CFH |
| B cell | CD19 | 0.32a | 0.51a | 0.54a | 0.51a | 0.36a |
| CD79A | 0.38a | 0.34a | 0.43a | 0.35a | 0.16a | |
| General T cell | CD3D | 0.47a | 0.58a | 0.75a | 0.58a | 0.29a |
| CD3E | 0.49a | 0.57a | 0.74a | 0.56a | 0.31a | |
| CD2 | 0.46a | 0.55a | 0.75a | 0.57a | 0.31a | |
| CD8 T cell | CD8A | 0.50a | 0.55a | 0.66a | 0.52a | 0.30a |
| CD8B | 0.43a | 0.51a | 0.65a | 0.51a | 0.25a | |
| Th1 | TBX21 | 0.46a | 0.50a | 0.69a | 0.56a | 0.28a |
| STAT4 | 0.43a | 0.57a | 0.72a | 0.62a | 0.40a | |
| IFNG | 0.40a | 0.40a | 0.56a | 0.43a | 0.19a | |
| TNF | 0.28a | 0.37a | 0.44a | 0.36a | 0.14a | |
| Th2 | LAMP3 | 0.45a | 0.48a | 0.58a | 0.44a | 0.33a |
| CXCR6 | 0.44a | 0.54a | 0.72a | 0.56a | 0.31a | |
| IL13 | 0.08 | 0.02 | 0.12a | 0.05 | 0.01 | |
| NK cell | KIR2DL1 | 0.28a | 0.23a | 0.38a | 0.34a | 0.15a |
| KIR2DL3 | 0.27a | 0.32a | 0.45a | 0.44a | 0.20a | |
| KIR2DL4 | 0.24a | 0.43a | 0.55a | 0.55a | 0.22a | |
| KIR3DL1 | 0.26a | 0.27a | 0.43a | 0.43a | 0.22a | |
| Neutrophil | CD11B | 0.31a | 0.78a | 0.86a | 0.79a | 0.40a |
| CASP5 | 0.29a | 0.60a | 0.63a | 0.58a | 0.29a | |
| Dendritic cell | HLA-DQB1 | 0.37a | 0.46a | 0.61a | 0.40a | 0.31a |
| HLA-DRA | 0.40a | 0.67a | 0.84a | 0.60a | 0.33a | |
| BDCA-1 | 0.21a | 0.21a | 0.45a | 0.28a | 0.17a | |
| BDCA-4 | 0.11 | 0.11 | 0.16a | 0.29a | 0.39a | |
| CD11c | 0.41a | 0.46a | 0.70a | 0.41a | 0.21a | |
P<0.05. Th, T helper cell; NK cell, natural killer cell.
Immunohistochemical analysis
Immunohistochemical staining was performed in 16 metastatic osteosarcoma tissues and 18 non-metastatic tissues (Fig. 11A). Compared with the metastatic cases, patients with non-metastatic osteosarcoma presented with significantly higher expression levels of the CD56bright NK cell-specific marker CD56, immature B cell-specific marker CD22, M1 macrophage-specific marker CD86 and neutrophil-specific marker CD11B, but lower levels of the M2 macrophage-specific marker CD163 (Fig. 11B). Additionally, these results revealed that the expression levels of all target genes were higher in the non-metastatic tissues compared with those in the metastatic tissues (Table III; Fig. 11).
Figure 11.
Clinical validation of tumor-infiltrating immune cells and target genes in clinical osteosarcoma tissues with and without metastasis. (A) Representative images and (B) statistical analyses of immunohistochemical staining for CD56, CD22, CD86, CD163, CD11B, GATA3, LPAR5, EVI2B, RIAM and CFH. Magnification, ×200. *P<0.05.
Table III.
Quantitative results of immunohistochemical analysis in metastatic and non-metastatic osteosarcoma specimens.
| Expression level | |||||
|---|---|---|---|---|---|
| Marker | Tissue | Low | Medium | High | Median score |
| CD56 | Metastatic | 11 | 3 | 2 | 2 |
| Non-metastatic | 1 | 4 | 13 | 5 | |
| CD22 | Metastatic | 10 | 2 | 4 | 2 |
| Non-metastatic | 2 | 1 | 15 | 5 | |
| CD86 | Metastatic | 12 | 2 | 2 | 2 |
| Non-metastatic | 3 | 2 | 13 | 5 | |
| CD163 | Metastatic | 3 | 1 | 12 | 5 |
| Non-metastatic | 15 | 1 | 2 | 2 | |
| CD11B | Metastatic | 10 | 5 | 1 | 2 |
| Non-metastatic | 2 | 5 | 11 | 5 | |
| GATA3 | Metastatic | 13 | 1 | 2 | 2 |
| Non-metastatic | 1 | 1 | 16 | 5 | |
| LPAR5 | Metastatic | 10 | 3 | 3 | 2 |
| Non-metastatic | 3 | 2 | 13 | 5 | |
| EVI2B | Metastatic | 14 | 2 | 0 | 1 |
| Non-metastatic | 3 | 3 | 12 | 5 | |
| RIAM | Metastatic | 11 | 3 | 2 | 2 |
| Non-metastatic | 3 | 2 | 13 | 5 | |
| CFH | Metastatic | 11 | 4 | 1 | 2 |
| Non-metastatic | 1 | 2 | 15 | 5 | |
Discussion
Osteosarcoma is the most prevalent malignancy of the bone and is characterized by a high propensity for metastasis and a poor patient prognosis (2,5). Treatment outcomes are often poor for patients with recurrent or metastatic osteosarcoma. With the rapid development of molecular biology technology, there has been a growing interest in anticancer immunotherapies, including immune modulators, immune checkpoint inhibitors and genetically modified T cells (40,41). Previous studies have demonstrated that the TME influences the development, recurrence and metastasis of osteosarcoma (42–44). Patients with osteosarcoma lacking immune cell infiltration present with high rates of metastasis and poor clinical outcomes (45). Immune reconstitution has been reported to suppress osteosarcoma recurrence and improve metastatic osteosarcoma survival (46,47). However, the currently available immunotherapy strategies have limited efficacy against metastatic osteosarcoma.
ESTIMATE has been widely used to calculate the proportions of immune and stromal cells in various types of tumor (48,49). In the present study, the results of the ESTIMATE analysis demonstrated that non-metastatic osteosarcomas were associated with higher immune and stromal scores as well as more favorable prognoses compared with those in the metastatic cases. Furthermore, independent analyses using the metagene and TIMER algorithms revealed that the level of favorable TIIC infiltration was lower in patients with metastatic osteosarcoma compared with that in non-metastatic cases, suggesting that impaired immune cell infiltration may promote osteosarcoma progression and metastasis. These results are consistent with previous studies, which have reported that immune infiltrates or immune responses in the local microenvironment serve an important role in the carcinogenesis of osteosarcoma (50–62). The exhaustion of cytotoxic T lymphocytes (CTLs) has been demonstrated to promote osteosarcoma invasion and metastasis, whereas the blockade of programmed cell death-1 (PD-1)/PD-1 ligand 1 interactions efficiently reverse the immunosuppressive effects on CTLs, decreasing the tumor burden of metastatic osteosarcoma (50–52). DCs are involved in the activation of multiple types of adaptive immune cells (53). Preclinical studies have demonstrated the therapeutic potential of DC vaccines in osteosarcoma (54,55). Recently, a role for NK cells in the recruitment of DCs into the TME via C-C motif chemokine ligand 5 and X-C motif chemokine ligand 1 has been reported (56). In osteosarcoma, NK cell-mediated immunotherapy has been associated with favorable clinical outcomes (57,58). In addition, neutrophils have been demonstrated to exert anticancer effects by not only orchestrating the recruitment of other immune cells, but also mediating antibody-dependent cellular cytotoxicity (59,60). According to previous reports, tumor-infiltrating macrophages are classified as antitumor M1-polarized macrophages and pro-tumor M2-polarized macrophages (61,62). Consistent with previous studies, the results of the present study demonstrated high numbers of M1 macrophages and low levels of M2 macrophages in patients with non-metastatic osteosarcoma. However, to the best of our knowledge, no studies are currently available on the function of immature B cells in osteosarcoma.
To determine the molecular mechanisms underlying the changes in the TME, 69 DEGs that were associated with metastasis and immune infiltration in patients with osteosarcoma were identified in the present study. Functional enrichment analyses verified that the DEGs participated in multiple immune-related pathways, including ‘leukocyte differentiation’, ‘regulation of leukocyte activation’ and ‘chemokine receptor activity’. Further analyses of the intersection of DEGs was performed using the MCC algorithm, Kaplan-Meier survival analysis and LASSO Cox regression analysis. Ultimately, five protective biomarkers (GATA3, LPAR5, EVI2B, RIAM and CFH) were used to establish a risk model with a high prognostic capacity for osteosarcoma. The expression levels of the five genes were positively associated with multiple types of TIICs and immune signatures, and negatively associated with the risk scores. Notably, all five genes were associated with the NF-κB signaling pathway, which has been reported to mediate immune escape in osteosarcoma (63).
GATA3 belongs to the zinc-finger transcription factor family and is implicated in the pathogenesis of various diseases, including cancer (64). A reciprocal feedback regulatory loop between GATA3/G9A/MTA3 and ZEB2/G9A/MTA1 has recently been identified; the absence of GATA3 in this axis contributes to breast cancer invasion and metastasis by upregulating the ZEB2 expression levels (65). GATA3 has also been reported to suppress osteosarcoma EMT progression by targeting the transcription factor Slug (66). The present study demonstrated for the first time that GATA3 was involved in the immunomodulation of osteosarcoma. Several immune-related pathways were significantly enriched for GATA3, including ‘complement and coagulation cascades’, ‘Th1 and Th2 cell differentiation’ and ‘NK cell mediated cytotoxicity’, suggesting that GATA3 may modulate immune responses and affect the development of osteosarcoma.
LPAR5, an orphan G protein-coupled receptor, has been reported to encode a subtype of lysophosphatidic acid (LPA) receptors, LPA5 (67). Aberrant LPA receptor function affects the progression of multiple types of cancer including ovarian, bladder, breast and pancreatic cancer (68). However, the role of LPAR5 in osteosarcoma is controversial. It has been reported that LPAR5 may act as a negative regulator of malignant properties in osteosarcoma by inhibiting matrix metalloproteinase-2 activation, thereby suppressing cell migration (69). LPA signaling via LPA5 has also been demonstrated to decrease cell migration and invasion in osteosarcoma and fibrosarcoma cells (70). By contrast, Minami et al (71) have revealed that the inhibition of LPA5 using an antagonist or RNA interference decreased the motility of osteosarcoma MG-63 cells. In the present study, LPAR5 was identified to be a favorable predictor for the clinical outcomes of patients with osteosarcoma and was significantly correlated with the levels of multiple types of TIICs and immune responses.
Although there are limited reports on the biological roles of EVI2B, the gene has been demonstrated to serve an essential role in the maintenance of normal physiological conditions (72). Using bioinformatics analyses based on the osteosarcoma cohorts, EVI2B was revealed to be involved in antigen processing and presentation, B cell receptor signaling pathways and CAMs in the present study. Notably, the transmembrane protein encoded by EVI2B has been reported to be highly expressed in various types of immune cells, including B cells, T cells, monocytes and NK cells (73), and the results of the present study demonstrated a positive correlation between EVI2B and the infiltration levels of multiple TIICs.
RIAM is localized in the cytosol and is recruited to sites of actin dynamics upon activation (74). As a downstream effector of a range of inside-out signaling pathways, RIAM has been implicated in various functions of innate and adaptive immunity. A previous study has demonstrated that RIAM interacts with Rap1, resulting in the activation of αMβ2 integrin, enhancing neutrophil-platelet interactions in the production of neutrophil extracellular pathogen traps and promoting pathogen clearance through complement-mediated phagocytosis and ROS production (75). Additionally, RIAM is essential for the activation of NK cell cytotoxicity (76). In agreement with previous studies, the results of the present study demonstrated that RIAM participated in various immunoregulatory processes in osteosarcoma.
As a major soluble inhibitor of the complement system, CFH can be hijacked by cancer cells and pathogens to escape complement-mediated attack (77). However, other studies have challenged this paradigm, revealing that CFH may serve anticancer roles in specific types of cancer by dampening cancer-related inflammation (78,79). In the present study, CFH was identified for the first time to serve a tumor-suppressive rather than an oncogenic role in osteosarcoma. Notably, a significant association was observed between CFH and various KEGG pathways, including the ‘renin-angiotensin system’, ‘NF-κB signaling pathway’, ‘TLR signaling pathway’, ‘complement and coagulation cascades’ and ‘apoptosis’. These results are in agreement with previous studies reporting that CFH may inhibit excessive tumor angiogenesis and may be actively internalized by apoptotic cells (80,81).
The present study had certain limitations that should be taken into consideration when interpreting the results. First, the number of patients enrolled was relatively small owing to the low incidence of osteosarcoma. Secondly, detailed clinical information, such as chemotherapeutic regimens and tumor stages was lacking, which limited the in-depth subgroup analyses. Finally, further experimental evidence is required to verify the molecular functions of the biomarkers in osteosarcoma, which will be the focus of our future studies.
In conclusion, the results of the present study revealed distinct TME landscapes between metastatic and non-metastatic osteosarcoma patients based on data obtained from TCGA. In addition, five protective biomarkers, including GATA3, LPAR5, EVI2B, RIAM and CFH, were identified, which exhibited high predictive accuracy for the prognosis of osteosarcoma.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was funded by the Science and Technology Program of Guangzhou, China (grant no. 201704020129).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
LL and SZ conceived and designed the study, and substantially revised the manuscript critically for important intellectual content. BY, ZS and GC were responsible for the data collection and bioinformatics analyses. BY and ZS wrote the manuscript. ZZ, JT and GW collected the clinical samples and performed the immunohistochemical tests. BY and ZS confirm the authenticity of all the raw data. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The experimental protocol used in the present study was authorized by the Ethics Committee of the Affiliated Zhujiang Hospital of Southern Medical University (Guangzhou, China; approval no. 2018-GJGBWK-002). Informed consent was obtained from all participants or their legal guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ritter J, Bielack SS. Osteosarcoma. Ann Oncol. 2010;21(Suppl 7):vii320–vii325. doi: 10.1093/annonc/mdq276. [DOI] [PubMed] [Google Scholar]
- 2.Brown HK, Tellez-Gabriel M, Heymann D. Cancer stem cells in osteosarcoma. Cancer Lett. 2017;386:189–195. doi: 10.1016/j.canlet.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Huang L, Huang Z, Lin W, Wang L, Zhu X, Chen X, Yang S, Lv C. Salidroside suppresses the growth and invasion of human osteosarcoma cell lines MG63 and U2OS in vitro by inhibiting the JAK2/STAT3 signaling pathway. Int J Oncol. 2019;54:1969–1980. doi: 10.3892/ijo.2019.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sergi C, Zwerschke W. Osteogenic sarcoma (osteosarcoma) in the elderly: Tumor delineation and predisposing conditions. Exp Gerontol. 2008;43:1039–1043. doi: 10.1016/j.exger.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Jaffe N. Osteosarcoma: Review of the past, impact on the future. The American experience. Cancer Treat Res. 2009;152:239–262. doi: 10.1007/978-1-4419-0284-9_12. [DOI] [PubMed] [Google Scholar]
- 6.Miwa S, Shirai T, Yamamoto N, Hayashi K, Takeuchi A, Igarashi K, Tsuchiya H. Current and emerging targets in immunotherapy for osteosarcoma. J Oncol. 2019;2019:7035045. doi: 10.1155/2019/7035045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reed DR, Hayashi M, Wagner L, Binitie O, Steppan DA, Brohl AS, Shinohara ET, Bridge JA, Loeb DM, Borinstein SC, Isakoff MS. Treatment pathway of bone sarcoma in children, adolescents, and young adults. Cancer. 2017;123:2206–2218. doi: 10.1002/cncr.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reina-Campos M, Moscat J, Diaz-Meco M. Metabolism shapes the tumor microenvironment. Curr Opin Cell Biol. 2017;48:47–53. doi: 10.1016/j.ceb.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meurette O, Mehlen P. Notch signaling in the tumor microenvironment. Cancer Cell. 2018;34:536–548. doi: 10.1016/j.ccell.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Chen JL, Lucas JE, Schroeder T, Mori S, Wu J, Nevins J, Dewhirst M, West M, Chi JT. The genomic analysis of lactic acidosis and acidosis response in human cancers. PLoS Genet. 2008;4:e1000293. doi: 10.1371/journal.pgen.1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roma-Rodrigues C, Mendes R, Baptista PV, Fernandes AR. Targeting tumor microenvironment for cancer therapy. Int J Mol Sci. 2019;20:840. doi: 10.3390/ijms20040840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori K, Rédini F, Gouin F, Cherrier B, Heymann D. Osteosarcoma: Current status of immunotherapy and future trends (Review) Oncol Rep. 2006;15:693–700. [PubMed] [Google Scholar]
- 13.Buddingh EP, Kuijjer ML, Duim RA, Bürger H, Agelopoulos K, Myklebost O, Serra M, Mertens F, Hogendoorn PC, Lankester AC, Cleton-Jansen AM. Tumor-infiltrating macrophages are associated with metastasis suppression in high-grade osteosarcoma: A rationale for treatment with macrophage activating agents. Clin Cancer Res. 2011;17:2110–2119. doi: 10.1158/1078-0432.CCR-10-2047. [DOI] [PubMed] [Google Scholar]
- 14.Ratti C, Botti L, Cancila V, Galvan S, Torselli I, Garofalo C, Manara MC, Bongiovanni L, Valenti CF, Burocchi A, et al. Trabectedin overrides osteosarcoma differentiative block and reprograms the tumor immune environment enabling effective combination with immune checkpoint inhibitors. Clin Cancer Res. 2017;23:5149–5161. doi: 10.1158/1078-0432.CCR-16-3186. [DOI] [PubMed] [Google Scholar]
- 15.Kawano M, Itonaga I, Iwasaki T, Tsumura H. Enhancement of antitumor immunity by combining anti-cytotoxic T lymphocyte antigen-4 antibodies and cryotreated tumor lysate-pulsed dendritic cells in murine osteosarcoma. Oncol Rep. 2013;29:1001–1006. doi: 10.3892/or.2013.2224. [DOI] [PubMed] [Google Scholar]
- 16.Saraf AJ, Fenger JM, Roberts RD. Osteosarcoma: Accelerating progress makes for a hopeful future. Front Oncol. 2018;8:4. doi: 10.3389/fonc.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Li B, Ren Y, Ye Z. T-Cell-based immunotherapy for osteosarcoma: Challenges and opportunities. Front Immunol. 2016;7:353. doi: 10.3389/fimmu.2016.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miwa S, Yamamoto N, Hayashi K, Takeuchi A, Igarashi K, Tsuchiya H. Therapeutic targets for bone and soft-tissue sarcomas. Int J Mol Sci. 2019;20:170. doi: 10.3390/ijms20010170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angelova M, Charoentong P, Hackl H, Fischer ML, Snajder R, Krogsdam AM, Waldner MJ, Bindea G, Mlecnik B, Galon J, Trajanoski Z. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015;16:64. doi: 10.1186/s13059-015-0620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Z, Zhang Y, Xu M, Zheng X, Lin M, Pan J, Ye C, Deng Y, Jiang C, Lin Y, et al. Demethylation and overexpression of CSF2 are involved in immune response, chemotherapy resistance, and poor prognosis in colorectal cancer. Onco Targets Ther. 2019;12:11255–11269. doi: 10.2147/OTT.S216829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan JH, Zhou H, Cooper L, Huang JL, Zhu SB, Zhao XX, Ding H, Pan YL, Rong L. LAYN is a prognostic biomarker and correlated with immune infiltrates in gastric and colon cancers. Front Immunol. 2019;10:6. doi: 10.3389/fimmu.2019.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang S, Liu T, Nan H, Wang Y, Chen H, Zhang X, Zhang Y, Shen B, Qian P, Xu S, et al. Comprehensive analysis of prognostic immune-related genes in the tumor microenvironment of cutaneous melanoma. J Cell Physiol. 2020;235:1025–1035. doi: 10.1002/jcp.29018. [DOI] [PubMed] [Google Scholar]
- 25.Shen Y, Peng X, Shen C. Identification and validation of immune-related lncRNA prognostic signature for breast cancer. Genomics. 2020;112:2640–2646. doi: 10.1016/j.ygeno.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 26.Li R, Qu H, Wang S, Wei J, Zhang L, Ma R, Lu J, Zhu J, Zhong WD, Jia Z. GDCRNATools: An R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC. Bioinformatics. 2018;34:2515–2517. doi: 10.1093/bioinformatics/bty124. [DOI] [PubMed] [Google Scholar]
- 27.R Core Team, corp-author. R Foundation for Statistical Computing; Vienna, Austria: 2012. R: A language and environment for statistical computing. ISBN 3-900051-07-0, URL http://www.R-project.org/ [Google Scholar]
- 28.Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hänzelmann S, Castelo R, Guinney J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Therneau TM, Grambsch PM. Springer; New York, NY: 2000. Modeling Survival Data: Extending the Cox Model. ISBN 0-387-98784-3. [DOI] [Google Scholar]
- 31.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. doi: 10.18637/jss.v033.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 38.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye Z, Zeng Z, Wang D, Lei S, Shen Y, Chen Z. Identification of key genes associated with the progression of intrahepatic cholangiocarcinoma using weighted gene co-expression network analysis. Oncol Lett. 2020;20:483–494. doi: 10.3892/ol.2020.11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsukahara T, Emori M, Murata K, Mizushima E, Shibayama Y, Kubo T, Kanaseki T, Hirohashi Y, Yamashita T, Sato N, Torigoe T. The future of immunotherapy for sarcoma. Expert Opin Biol Ther. 2016;16:1049–1057. doi: 10.1080/14712598.2016.1188075. [DOI] [PubMed] [Google Scholar]
- 41.Wang SD, Li HY, Li BH, Xie T, Zhu T, Sun LL, Ren HY, Ye ZM. The role of CTLA-4 and PD-1 in anti-tumor immune response and their potential efficacy against osteosarcoma. Int Immunopharmacol. 2016;38:81–89. doi: 10.1016/j.intimp.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 42.Heymann MF, Lézot F, Heymann D. The contribution of immune infiltrates and the local microenvironment in the pathogenesis of osteosarcoma. Cell Immunol. 2019;343:103711. doi: 10.1016/j.cellimm.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 43.Ehnman M, Chaabane W, Haglund F, Tsagkozis P. The tumor microenvironment of pediatric sarcoma: Mesenchymal mechanisms regulating cell migration and metastasis. Curr Oncol Rep. 2019;21:90. doi: 10.1007/s11912-019-0839-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng Y, Wang G, Chen R, Hua Y, Cai Z. Mesenchymal stem cells in the osteosarcoma microenvironment: Their biological properties, influence on tumor growth, and therapeutic implications. Stem Cell Res Ther. 2018;9:22. doi: 10.1186/s13287-018-0780-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott MC, Temiz NA, Sarver AE, LaRue RS, Rathe SK, Varshney J, Wolf NK, Moriarity BS, O'Brien TD, Spector LG, et al. Comparative transcriptome analysis quantifies immune cell transcript levels, metastatic progression, and survival in osteosarcoma. Cancer Res. 2018;78:326–337. doi: 10.1158/0008-5472.CAN-17-0576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merchant MS, Melchionda F, Sinha M, Khanna C, Helman L, Mackall CL. Immune reconstitution prevents metastatic recurrence of murine osteosarcoma. Cancer Immunol Immunother. 2007;56:1037–1046. doi: 10.1007/s00262-006-0257-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Merchant MS, Bernstein D, Amoako M, Baird K, Fleisher TA, Morre M, Steinberg SM, Sabatino M, Stroncek DF, Venkatasan AM, et al. Adjuvant immunotherapy to improve outcome in high-risk pediatric sarcomas. Clin Cancer Res. 2016;22:3182–3191. doi: 10.1158/1078-0432.CCR-15-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, Gao Y, Xu Z, Zhang Z, Zheng Y, Qi F. Identification of prognostic genes in adrenocortical carcinoma microenvironment based on bioinformatic methods. Cancer Med. 2020;9:1161–1172. doi: 10.1002/cam4.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang S, Zhang B, Fan W, Zhao Q, Yang L, Xin W, Fu D. Identification of prognostic genes in the acute myeloid leukemia microenvironment. Aging (Albany NY) 2019;11:10557–10580. doi: 10.18632/aging.102477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schell TD, Knowles BB, Tevethia SS. Sequential loss of cytotoxic T lymphocyte responses to simian virus 40 large T antigen epitopes in T antigen transgenic mice developing osteosarcomas. Cancer Res. 2000;60:3002–3012. [PubMed] [Google Scholar]
- 51.Shen JK, Cote GM, Choy E, Yang P, Harmon D, Schwab J, Nielsen GP, Chebib I, Ferrone S, Wang X, et al. Programmed cell death ligand 1 expression in osteosarcoma. Cancer Immunol Res. 2014;2:690–698. doi: 10.1158/2326-6066.CIR-13-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lussier DM, O'Neill L, Nieves LM, McAfee MS, Holechek SA, Collins AW, Dickman P, Jacobsen J, Hingorani P, Blattman JN. Enhanced T-cell immunity to osteosarcoma through antibody blockade of PD-1/PD-L1 interactions. J Immunother. 2015;38:96–106. doi: 10.1097/CJI.0000000000000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steinman RM, Idoyaga J. Features of the dendritic cell lineage. Immunol Rev. 2010;234:5–17. doi: 10.1111/j.0105-2896.2009.00888.x. [DOI] [PubMed] [Google Scholar]
- 54.Yu Z, Qian J, Wu J, Gao J, Zhang M. Allogeneic mRNA-based electrotransfection of autologous dendritic cells and specific antitumor effects against osteosarcoma in rats. Med Oncol. 2012;29:3440–3448. doi: 10.1007/s12032-012-0312-y. [DOI] [PubMed] [Google Scholar]
- 55.Kawano M, Tanaka K, Itonaga I, Iwasaki T, Miyazaki M, Ikeda S, Tsumura H. Dendritic cells combined with anti-GITR antibody produce antitumor effects in osteosarcoma. Oncol Rep. 2015;34:1995–2001. doi: 10.3892/or.2015.4161. [DOI] [PubMed] [Google Scholar]
- 56.Böttcher JP, Bonavita E, Chakravarty P, Blees H, Cabeza-Cabrerizo M, Sammicheli S, Rogers NC, Sahai E, Zelenay S, Reis e Sousa C. NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell. 2018;172:1022–1037.e14. doi: 10.1016/j.cell.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang YH, Connolly J, Shimasaki N, Mimura K, Kono K, Campana D. A chimeric receptor with NKG2D specificity enhances natural killer cell activation and killing of tumor cells. Cancer Res. 2013;73:1777–1786. doi: 10.1158/0008-5472.CAN-12-3558. [DOI] [PubMed] [Google Scholar]
- 58.Kiany S, Huang G, Kleinerman ES. Effect of entinostat on NK cell-mediated cytotoxicity against osteosarcoma cells and osteosarcoma lung metastasis. OncoImmunology. 2017;6:e1333214. doi: 10.1080/2162402X.2017.1333214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beauvillain C, Delneste Y, Scotet M, Peres A, Gascan H, Guermonprez P, Barnaba V, Jeannin P. Neutrophils efficiently cross-prime naive T cells in vivo. Blood. 2007;110:2965–2973. doi: 10.1182/blood-2006-12-063826. [DOI] [PubMed] [Google Scholar]
- 60.Jablonska J, Lang S, Sionov RV, Granot Z. The regulation of pre-metastatic niche formation by neutrophils. Oncotarget. 2017;8:112132–112144. doi: 10.18632/oncotarget.22792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao SJ, Jiang YQ, Xu NW, Li Q, Zhang Q, Wang SY, Li J, Wang YH, Zhang YL, Jiang SH, et al. SPARCL1 suppresses osteosarcoma metastasis and recruits macrophages by activation of canonical WNT/β-catenin signaling through stabilization of the WNT-receptor complex. Oncogene. 2018;37:1049–1061. doi: 10.1038/onc.2017.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Noy R, Pollard JW. Tumor-associated macrophages: From mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li R, Shi Y, Zhao S, Shi T, Zhang G. NF-κB signaling and integrin-β1 inhibition attenuates osteosarcoma metastasis via increased cell apoptosis. Int J Biol Macromol. 2019;123:1035–1043. doi: 10.1016/j.ijbiomac.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 64.Shahi P, Wang CY, Chou J, Hagerling C, Gonzalez Velozo H, Ruderisch A, Yu Y, Lai MD, Werb Z. GATA3 targets semaphorin 3B in mammary epithelial cells to suppress breast cancer progression and metastasis. Oncogene. 2017;36:5567–5575. doi: 10.1038/onc.2017.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Si W, Huang W, Zheng Y, Yang Y, Liu X, Shan L, Zhou X, Wang Y, Su D, Gao J, et al. Dysfunction of the reciprocal feedback loop between GATA3- and ZEB2-nucleated repression programs contributes to breast cancer metastasis. Cancer Cell. 2015;27:822–836. doi: 10.1016/j.ccell.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 66.Ma L, Xue W, Ma X. GATA3 is downregulated in osteosarcoma and facilitates EMT as well as migration through regulation of slug. Onco Targets Ther. 2018;11:7579–7589. doi: 10.2147/OTT.S176534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choi JW, Herr DR, Noguchi K, Yung YC, Lee CW, Mutoh T, Lin ME, Teo ST, Park KE, Mosley AN, Chun J. LPA receptors: Subtypes and biological actions. Annu Rev Pharmacol Toxicol. 2010;50:157–186. doi: 10.1146/annurev.pharmtox.010909.105753. [DOI] [PubMed] [Google Scholar]
- 68.Tsujiuchi T, Araki M, Hirane M, Dong Y, Fukushima N. Lysophosphatidic acid receptors in cancer pathobiology. Histol Histopathol. 2014;29:313–321. doi: 10.14670/HH-29.313. [DOI] [PubMed] [Google Scholar]
- 69.Araki M, Kitayoshi M, Dong Y, Hirane M, Ozaki S, Mori S, Fukushima N, Honoki K, Tsujiuchi T. Inhibitory effects of lysophosphatidic acid receptor-5 on cellular functions of sarcoma cells. Growth Factors. 2014;32:117–122. doi: 10.3109/08977194.2014.911294. [DOI] [PubMed] [Google Scholar]
- 70.Dong Y, Hirane M, Araki M, Fukushima N, Honoki K, Tsujiuchi T. Lysophosphatidic acid receptor-5 negatively regulates cell motile and invasive activities of human sarcoma cell lines. Mol Cell Biochem. 2014;393:17–22. doi: 10.1007/s11010-014-2042-2. [DOI] [PubMed] [Google Scholar]
- 71.Minami K, Ueda N, Ishimoto K, Tsujiuchi T. LPA5-mediated signaling induced by endothelial cells and anticancer drug regulates cellular functions of osteosarcoma cells. Exp Cell Res. 2020;388:111813. doi: 10.1016/j.yexcr.2020.111813. [DOI] [PubMed] [Google Scholar]
- 72.Zjablovskaja P, Kardosova M, Danek P, Angelisova P, Benoukraf T, Wurm AA, Kalina T, Sian S, Balastik M, Delwel R, et al. EVI2B is a C/EBPα target gene required for granulocytic differentiation and functionality of hematopoietic progenitors. Cell Death Differ. 2017;24:705–716. doi: 10.1038/cdd.2017.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matesanz-Isabel J, Sintes J, Llinàs L, de Salort J, Lázaro A, Engel P. New B-cell CD molecules. Immunol Lett. 2011;134:104–112. doi: 10.1016/j.imlet.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 74.Lafuente EM, van Puijenbroek AA, Krause M, Carman CV, Freeman GJ, Berezovskaya A, Constantine E, Springer TA, Gertler FB, Boussiotis VA. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev Cell. 2004;7:585–595. doi: 10.1016/j.devcel.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 75.Patsoukis N, Bardhan K, Weaver JD, Sari D, Torres-Gomez A, Li L, Strauss L, Lafuente EM, Boussiotis VA. The adaptor molecule RIAM integrates signaling events critical for integrin-mediated control of immune function and cancer progression. Sci Signal. 2017;10:eaam8298. doi: 10.1126/scisignal.aam8298. [DOI] [PubMed] [Google Scholar]
- 76.Mace EM, Monkley SJ, Critchley DR, Takei F. A dual role for talin in NK cell cytotoxicity: Activation of LFA-1-mediated cell adhesion and polarization of NK cells. J Immunol. 2009;182:948–956. doi: 10.4049/jimmunol.182.2.948. [DOI] [PubMed] [Google Scholar]
- 77.Meri T, Amdahl H, Lehtinen MJ, Hyvärinen S, McDowell JV, Bhattacharjee A, Meri S, Marconi R, Goldman A, Jokiranta TS. Microbes bind complement inhibitor factor H via a common site. PLoS Pathog. 2013;9:e1003308. doi: 10.1371/annotation/41169409-3260-4295-baf4-a1a4621a8e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bonavita E, Gentile S, Rubino M, Maina V, Papait R, Kunderfranco P, Greco C, Feruglio F, Molgora M, Laface I, et al. PTX3 is an extrinsic oncosuppressor regulating complement-dependent inflammation in cancer. Cell. 2015;160:700–714. doi: 10.1016/j.cell.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 79.Corrales L, Ajona D, Rafail S, Lasarte JJ, Riezu-Boj JI, Lambris JD, Rouzaut A, Pajares MJ, Montuenga LM, Pio R. Anaphylatoxin C5a creates a favorable microenvironment for lung cancer progression. J Immunol. 2012;189:4674–4683. doi: 10.4049/jimmunol.1201654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu J, Hoh J. Loss of complement factor H in plasma increases endothelial cell migration. J Cancer. 2017;8:2184–2190. doi: 10.7150/jca.19452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martin M, Leffler J, Smoląg KI, Mytych J, Björk A, Chaves LD, Alexander JJ, Quigg RJ, Blom AM. Factor H uptake regulates intracellular C3 activation during apoptosis and decreases the inflammatory potential of nucleosomes. Cell Death Differ. 2016;23:903–911. doi: 10.1038/cdd.2015.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.