Abstract
Objective
The aim of study was to characterize Giardia isolates genetically among patients in Chandigarh region, India. For this, nested PCR targeting fragment of the glutamate dehydrogenase (GLUD1 earlier named as GDH) gene was used. Phylogenetic analysis was done by constructing neighbor-joining tree made out of the nucleotide sequences of G. intestinalis isolates obtained in this study and with the known sequences published in GenBank.
Results
Out of 40 samples, GLUD1 gene was amplified in 33 samples (82.5%). The product of GLUD1 gene was successfully sequenced only in 32 samples. In these samples, assemblage B was found in 27 (84.37%) samples whereas 5 (15.6%) samples had assemblage A. Among assemblage B most of them were of BIII. Therefore, genotyping of Giardia would be helpful in conducting epidemiological studies.
Keywords: Giardia, Genotype, GLUD1 (glutamate dehydrogenase), PCR, Sequencing
Introduction
Giardia intestinalis is well known intestinal parasite of humans and mammals. Giardia causes approximately 280 million cases of giardiasis worldwide annually [1]. Most of these cases are associated with lower socioeconomic status. In the year 2004, giardiasis was included in WHO ‘Neglected Diseases Initiative’ because of its high prevalence in communities with low socio-economic status.
Giardiasis is acquired due to ingestion of cysts of Giardia in water or food [2]. Giardia intestinalis is composed of eight major genotypes or assemblages (A–H) [2]. Genotype A and B are common among humans having variable distribution frequency in different geographical locations and these assemblages mainly considered as zoonotic assemblages as they are able to infect both men and animals [3]. These assemblages are further divided into subassemblages on the basis of either digestion by restriction enzyme or sequence analysis. Assemblages A are classified as AI, AII,AIII and AIV. Subassemblages AI and AII were commonly found in humans while AI, AIII and AIV are subassemblages of animals. Zoonotic potential is linked with only subassemblage AI [4]. Assemblage B, catogorised into four sub-assemblages BI, BII, BIII and BIV. As per literature subassemblages BIII and BIV were reported in humans while other two are specific for animals [4, 5]. The BIII sub-assemblage is closer to sub-assemblages BI and BII and therefore has zoonotic potential. The GLUD1 (earlier known as GDH) locus has been utilized for genetic characterization of G. intestinalis isolates in vertebrates [4] hosts and is able to categorize them into sub genotypes/subassemblages. The present work was aimed to determine assemblages and sub-assemblages of Giardia isolates involved in its transmission by using glutamate dehydrogenase (GLUD1) marker.
Main text
Materials and methods
Sample collection
Forty microscopic Giardia positive stool samples were collected from the Routine Laboratory of Department of Medical Parasitology, PGIMER, Chandigarh from August 2019 to December 2019.
DNA extraction
From stool samples, DNA was extracted by using QIAmp Fast DNA Stool Mini Kit (QIAGEN, Germany) as per manufacturer’s instructions with slight modifications. The suspension was initially incubated at 90 °C for 15 min and then for another 30 min at 75 °C. DNA was eluted in 50 µl of AE buffer. DNA concentration was measured by NanoQuant (Infinite® 200 PRO NanoQuant) and stored at − 20 °C until further use.
Polymerase chain reaction amplification
The two-step PCR was employed for the amplification of GLUD1 gene (432 bp) by using previously published primers given by Read et al. [6]. The conditions and primers for both primary and secondary reactions are given as Additional file 1: Table S1. The first set of PCR reaction comprised of 2.0 μL of DNA template, 12.5 μL 2 × Go Taq Green Mix, 1 μL of each primer (10 μM), 1 μL of Bovine Serum Albumin (BSA) whereas in case of secondary PCR, DNA template was replaced by the product of primary reaction. For the negative control, nuclease-free water and for the positive control, DNA of Giardia strain, Portland 1 was used for each PCR reaction. All the precautions were taken to prevent contamination.
PCR product purification and sequencing
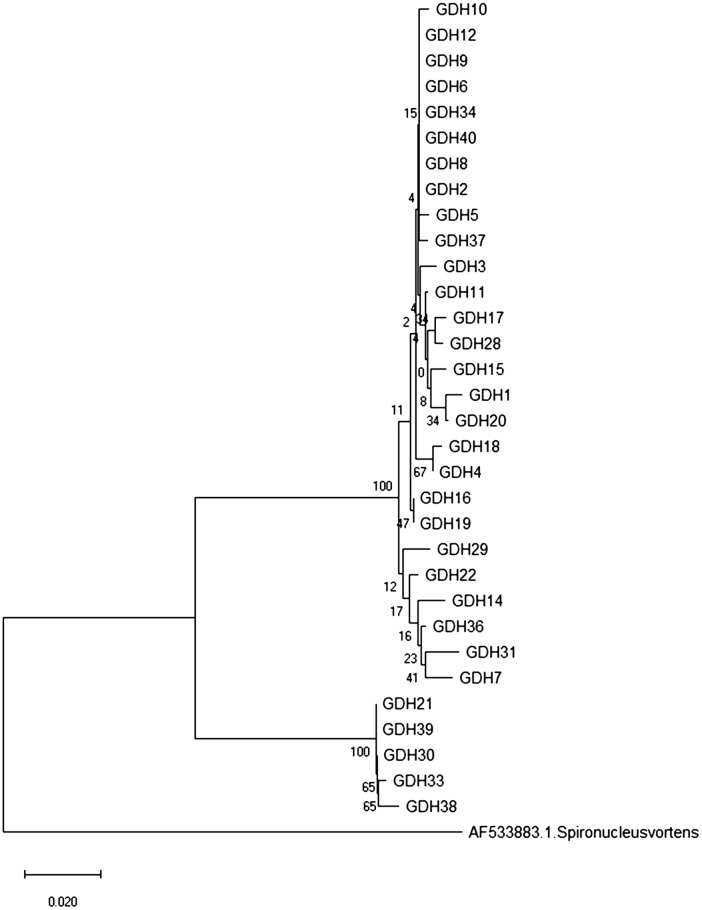
All PCR-positive samples were sequenced using secondary primers. PCR products were sequenced in both forward and reverse directions. By using BLAST, nucleotide similarity of sequenced amplicons was searched in GenBank (http://www.ncbi.nlm.nih.gov/blast). CLUSTAL X was used to determine multiple sequence alignments. Neighbor-joining distance trees were prepared using MEGAX software (https://www.megasoftware.net/) (Fig. 1). Bootstrap values were based on 1000 replicas. All the sequences obtained during the study were submitted to the GenBank (Accession number: MT584168–MT584199).
Fig. 1.
Phylogenetic tree constructed with the neighbor-joining method using nucleotide sequences of GLUD1 gene. The sequence of Spironucleus vortens was used as an out-group
The direct links which are publicly available are as follows:
https://www.ncbi.nlm.nih.gov/nuccore/MT584168
https://www.ncbi.nlm.nih.gov/nuccore/MT584169
https://www.ncbi.nlm.nih.gov/nuccore/MT584170
https://www.ncbi.nlm.nih.gov/nuccore/MT584171
https://www.ncbi.nlm.nih.gov/nuccore/MT584172
https://www.ncbi.nlm.nih.gov/nuccore/MT584173
https://www.ncbi.nlm.nih.gov/nuccore/MT584174
https://www.ncbi.nlm.nih.gov/nuccore/MT584175
https://www.ncbi.nlm.nih.gov/nuccore/MT584176
https://www.ncbi.nlm.nih.gov/nuccore/MT584177
https://www.ncbi.nlm.nih.gov/nuccore/MT584178
https://www.ncbi.nlm.nih.gov/nuccore/MT584179
https://www.ncbi.nlm.nih.gov/nuccore/MT584180
https://www.ncbi.nlm.nih.gov/nuccore/MT584181
https://www.ncbi.nlm.nih.gov/nuccore/MT584182
https://www.ncbi.nlm.nih.gov/nuccore/MT584183
https://www.ncbi.nlm.nih.gov/nuccore/MT584184
https://www.ncbi.nlm.nih.gov/nuccore/MT584185
https://www.ncbi.nlm.nih.gov/nuccore/MT584186
https://www.ncbi.nlm.nih.gov/nuccore/MT584187
https://www.ncbi.nlm.nih.gov/nuccore/MT584188
https://www.ncbi.nlm.nih.gov/nuccore/MT584189
https://www.ncbi.nlm.nih.gov/nuccore/MT584190
https://www.ncbi.nlm.nih.gov/nuccore/MT584191
https://www.ncbi.nlm.nih.gov/nuccore/MT584192
https://www.ncbi.nlm.nih.gov/nuccore/MT584193
https://www.ncbi.nlm.nih.gov/nuccore/MT584194
https://www.ncbi.nlm.nih.gov/nuccore/MT584195
https://www.ncbi.nlm.nih.gov/nuccore/MT584196
https://www.ncbi.nlm.nih.gov/nuccore/MT584197
Results
Out of 40 samples, the GLUD1 gene was amplified in 33 samples (82.5%) (Table1, Fig. 2). The possible reason for this occurrence indicated the less parasitic load in these isolated samples. The product of GLUD1 gene was successfully sequenced only in 32 samples. In these samples, assemblage B was found in 27 (84.37%) among which 18 (66.66%) were sub-genotype BIII and 9 (33.33%) were sub-genotype BIV samples whereas 5 (15.6%) samples had assemblage A. Four of them were AI subgenotype and only1 belonged to sub-genotype AII.
Table 1.
PCR results of samples with their assemblages
| Sample No | PCR | Assemblages |
|---|---|---|
| 1 | +ve | B |
| 2 | +ve | B |
| 3 | +ve | B |
| 4 | +ve | B |
| 5 | +ve | B |
| 6 | +ve | B |
| 7 | +ve | B |
| 8 | +ve | B |
| 9 | +ve | B |
| 10 | +ve | B |
| 11 | +ve | B |
| 12 | +ve | B |
| 13 | +ve | Unable to sequence |
| 14 | +ve | B |
| 15 | +ve | B |
| 16 | +ve | B |
| 17 | +ve | B |
| 18 | +ve | B |
| 19 | +ve | B |
| 20 | +ve | B |
| 21 | +ve | A |
| 22 | +ve | B |
| 23 | −ve | – |
| 24 | −ve | – |
| 25 | −ve | – |
| 26 | −ve | – |
| 27 | −ve | – |
| 28 | +ve | B |
| 29 | +ve | B |
| 30 | +ve | A |
| 31 | +ve | B |
| 32 | −ve | – |
| 33 | +ve | A |
| 34 | +ve | B |
| 35 | −ve | – |
| 36 | +ve | B |
| 37 | +ve | B |
| 38 | +ve | A |
| 39 | +ve | A |
| 40 | +ve | B |
Fig. 2.
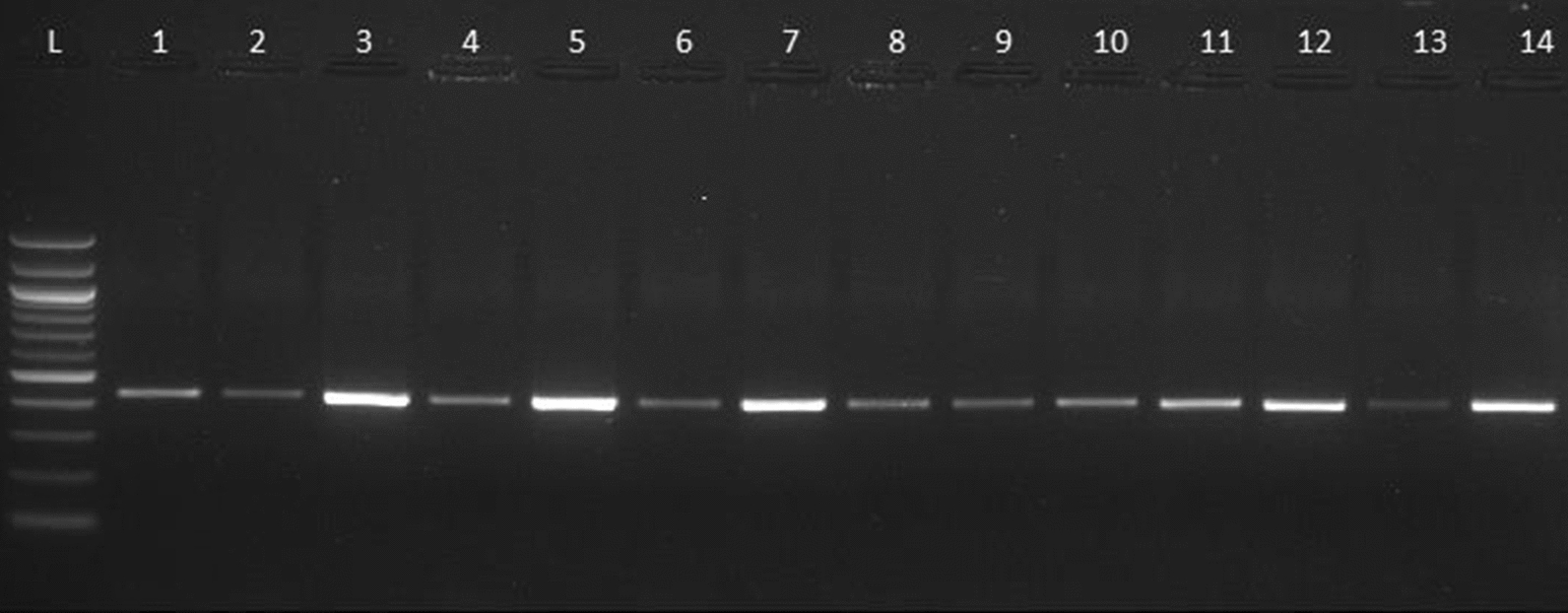
Nested-PCR amplification of Giardia intestinalis GLUD1 gene (432bp) 1.5% agarose gel stained with Ethidium bromide. L, ladder (100bp); lane1–13, Giardia positive samples; lane14, positive control
Discussion
Giardia intestinalis is the most common and frequent intestinal parasitic agent of gastroenteritis mainly in the developing countries [7]. The present study provides data on genetic diversity of Giardia isolates from patients in the Chandigarh region. As per results, assemblage A and B are common among this population, which is in concordance to other studies [8, 9]. In our study, assemblage B was the predominant genotype observed followed by assemblage A. However, similar observations were previously reported and observed worldwide which showed that assemblage B was predominant in comparison to assemblage A [9–12]. There are studies which showed the presence of assemblage B in Rhesus macaques (Macaca mulatta) and potable water resources of Northern India [13, 14]. But other studies have reported the assemblage A as the predominant genotype in other regions [15]. Due to geographical variations, differences were observed in the prevalence of various genotypes and the detection of these variations would be helpful in designing effective therapeutic approaches.
Conclusion
The results showed that PCR sequencing and phylogenetic analysis is an excellent molecular technique for genotyping of Giardia intestinalis. Detection of Giardia intestinalis assemblages and sub-assemblageswould be helpful in conducting epidemiological studies.
Limitation of study
Present study involves only single locus for genotyping and also the sample size is less so it is difficult to interpret zoonotic potential of these isolates. Therefore, multi-locus typing data is required to differentiate between Giardia isolates.
Supplementary Information
Additional file 1: Table S1. Sequence of primers and sgRNA were listed.
Acknowledgements
Authors duly acknowledge Ms Sofia Behad, Senior Research Fellow for providing assistance in alignment of sequences
Abbreviations
- BLAST
Basic Local Alignment Search Tool
- DNA
Deoxyribonucleic Acid
- PCR
Polymerase Chain Reaction
- BSA
Bovine Serum Albumin
Authors’ contributions
ST planned & conducted all the practical work in the laboratory. UK helped in preparing & checking the manuscript. RS helped in designing and drafting the work, provided financial assistance for completing the present work and also helped in analysis of the data of the present work. All authors read and approved the final manuscript.
Funding
None.
Availability of data and materials
All the supporting data related to the present work is available with the authors and the sequences obtained during the study were submitted to the GenBank (Accession number: MT584168–MT584199).
Ethics approval and consent to participate
The study has been approved by Institutional Ethics Committee, PGIMER, Chandigarh vide IEC No- 04/2017-601. Patients were informed of the study objectives and voluntary written consent was sought and obtained before inclusion.
Consent for publication
All the authors have agreed to publish the present research paper.
Competing interests
There are no financial, general and institutional competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13104-020-05419-1.
References
- 1.Einarsson E, Maayeh S, Svärd SG. An up-date on Giardia and giardiasis. Curr Opin Microbiol. 2016;34:47–52. doi: 10.1016/j.mib.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 2.Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. 2011;24(1):110–140. doi: 10.1128/CMR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durigan M, Abreu AG, Zucchi MI, Franco RM, de Souza AP. Genetic diversity of Giardia duodenalis: multilocus genotyping reveals zoonotic potential between clinical and environmental sources in a metropolitan region of Brazil. PLoS ONE. 2014;9(12):e115489. doi: 10.1371/journal.pone.0115489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hooshyar H, Ghafarinasab S, Arbabi M, Delavari M, Rasti S. Genetic variation of Giardia lamblia isolates from food-handlers in Kashan, Central Iran. Iran J Parasitol. 2017;12(1):83. [PMC free article] [PubMed] [Google Scholar]
- 5.Heyworth MF. Giardia duodenalis genetic assemblages and hosts. Parasite. 2016;23:13. doi: 10.1051/parasite/2016013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Read C, Monis P, Thompson R. Discrimination of all genotypes of Giardia duodenalis at the glutamate dehydrogenase locus using PCR-RFLP. Infect Genet Evol. 2004;4(2):125–130. doi: 10.1016/j.meegid.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Thompson RC, Smith A. Zoonotic enteric protozoa. Vet Parasitol. 2011;182:70–78. doi: 10.1016/j.vetpar.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 8.Ajjampur SS, Sankaran P, Kannan A, Sathyakumar K, Sarkar R, Gladstone BP, Kang G. Giardia duodenalis assemblages associated with diarrhea in children in South India identified by PCR-RFLP. Am J Trop Med Hyg. 2009;80(1):16–19. doi: 10.4269/ajtmh.2009.80.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laishram S, Kannan A, Rajendran P, Kang G, Ajjampur SSR. Mixed Giardia duodenalis assemblage infections in children and adults in South India. Epidemiol Infect. 2012;140:2023–2027. doi: 10.1017/S0950268811002767. [DOI] [PubMed] [Google Scholar]
- 10.Minvielle MC, Molina NB, Polverino D, Basualdo JA. First genotyping of Giardia lamblia from human and animal feces in Argentina. S Am Mem Inst Oswaldo Cruz. 2008;103:98–103. doi: 10.1590/S0074-02762008000100015. [DOI] [PubMed] [Google Scholar]
- 11.Tak V, Mirdha BR, Yadav P, Vyas P, Makharia GK, Bhatnagar S. Molecular characterisation of Giardia intestinalis assemblages from human isolates at a tertiary care centre of India. Indian J Med Microbiol. 2014;32(1):19. doi: 10.4103/0255-0857.124290. [DOI] [PubMed] [Google Scholar]
- 12.Chanu NO, Singh TS, Dutta S. Detection and genetic characterization of Giardia intestinalis in children with gastrointestinal symptoms by PCR RFLP in Sikkim, India. J Nat Sci Biol Med. 2018;9(2):193. doi: 10.4103/jnsbm.JNSBM_219_17. [DOI] [Google Scholar]
- 13.Debenham JJ, Tysnes K, Khunger S, Robertson LJ. Occurrence of Giardia, Cryptosporidium, and Entamoeba in wild rhesus macaques (Macaca mulatta) living in urban and semi-rural North-West India. Int J Parasitol Parasites Wildl. 2017;6(1):29–34. doi: 10.1016/j.ijppaw.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Utaaker KS, Joshi H, Kumar A, Chaudhary S, Robertson LJ. Occurrence of Cryptosporidium and Giardia in potable water sources in Chandigarh, Northern India. J Water Supply Res Technol AQUA. 2019;68(6):483–494. doi: 10.2166/aqua.2019.157. [DOI] [Google Scholar]
- 15.Skhal D, Aboualchamat G, Al Mariri A, Al NS. Prevalence of Giardia duodenalis assemblages and sub-assemblages in symptomatic patients from Damascus city and its suburbs. Infect Genet Evol. 2017;47:155–160. doi: 10.1016/j.meegid.2016.11.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Sequence of primers and sgRNA were listed.
Data Availability Statement
All the supporting data related to the present work is available with the authors and the sequences obtained during the study were submitted to the GenBank (Accession number: MT584168–MT584199).