Abstract
Cell migration is an important factor influencing the treatment outcomes of high-grade glioma (World Health Organization grades III–IV). Using immunohistochemical staining, the present study demonstrated that the protein levels of phosphorylated pyruvate dehydrogenase α1 (p-PDHA1) were increased according to the grade of glioma. Moreover, p-PDHA1 mediated tumor necrosis factor-α (TNF-α)-induced cell migration in glioma cells. Phalloidin staining and western blot analysis were used to detect the protein level of p-PDHA1 in U251 glioma cells stimulated by TNF-α at different time points. Phalloidin staining was used to observe the cytoskeletal structure. The effects on the expression of specific migration markers and on the cytoskeletal structure were also detected. Dichloroacetic acid is an inhibitor of PDK. These results indicated that p-PDHA1 served an important role in the migration of glioma cells, and consequently in the development of glioma.
Keywords: glioma, phosphorylated-pyruvate dehydrogenase α1, tumor necrosis factor-α, migration
Introduction
According to the grading system developed by the World Health Organization (WHO), gliomas are classified into grades I–IV (1). Grade I and II gliomas are low-grade gliomas and the prognosis of the patients is relatively good. Grade III and IV gliomas are high-grade gliomas (HGG). The prognosis of the patients with these types of tumors is relatively poor (2). Diffuse astrocytoma is classified as a WHO grade II tumor, whereas anaplastic astrocytoma is a WHO grade III tumor. Glioblastoma is a WHO grade IV tumor (3). In the last 30 years, the 5-year survival rate of gliomas has not significantly improved (4). Based on this evidence, the identification of new treatments for patients with GBM is imperative.
Tumor necrosis factor-α (TNF-α) is a polypeptide cytokine produced by mononuclear macrophages and is considered an important inflammatory factor in the tumor inflammatory microenvironment (5). Tumor necrosis factor receptor 1 (TNFR1) is one of the receptors of TNF-α that mediates the majority of its biological functions, such as antiviral activity (6), apoptosis induction (7) and activation of the NF-κB protein (8,9). The receptor of TNF-α is a type I transmembrane glycoprotein distributed on the surface of the normal cell membrane (10). A previous study has reported that TNFR1 exerts a significant influence on the malignant progression of tumors and that it is associated with the degree of differentiation or the depth of invasion of the tumors (11).
Pyruvate dehydrogenase (PDH) α1 (PDHA1) is a subunit of PDH. PDHA1 has a molecular weight of 43.3 kDa and a genomic DNA full length of 15.92 Kbp which includes 11 exons. It is located on the X chromosome (Xp22.1–22.2) and is also known as PDC E1α (12). In mitochondria, PDH exists in the form of PDH complex (PDHc) and a specific phosphorylation site of PDHc on PDHA1 serves an important role in regulating the activity and energy metabolism of PDHc (13). PDH kinase (PDK)1 primarily acts on PDHA1 in PDHc. The regulation of PDHA1 phosphorylation can increase PDHc activity (14). The normal expression of PDHA1 is a prerequisite for the normal development of the tricarboxylic acid (TCA) cycle and the oxidative phosphorylation in the mitochondria (15). The function of PDHA1 is to transform aerobic glycolysis into oxidative phosphorylation. In the absence of oxygen, pyruvate is reduced to lactate. In the presence of oxygen, pyruvate is oxidized by PDH to acetyl-CoA, which enters the mitochondria and is metabolized further by the TCA cycle. When the phosphorylation of PDHA1 is inhibited, it can inactivate PDHc (16). Furthermore, the Warburg effect is enhanced, and the invasiveness of tumor cells is increased (17). Therefore, the ability of phosphorylated (p)-PDHA1 to mediate TNFα-induced cell migration in glioma cells is worth determining.
Materials and methods
Patients and tissue samples
A total of 80 paraffin embedded samples were obtained from the Affiliated Hospital of Nantong University between January 2013 and January 2017. The present study was approved by the Human Research Ethics Committee of Nantong University, China. The patients were recruited based on: i) Postoperative pathological diagnosis; and ii) Magnetic resonance imaging data. Moreover, all glioma specimens were obtained from patients who had not previously received radiotherapy or chemotherapy. The sections of the glioma samples were analyzed by two professionals from the Department of Pathology, Affiliated Hospital of Nantong University according to the WHO (2007) tumor grading standards (18). The grading system adopted the double-blind method. The follow-up data were collected by telephone visit and concomitant household registration data visit (follow-up time ≤60 months). The follow-up rate was 93.7% (75/80).
Cell lines
The cell lines used were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. Specifically, U251, U87 (glioblastoma of unknown origin), A172, H4 and SHG44 glioma cell lines, and 293T cells were used in the present study. U251, U87, A172, H4 and SHG44 glioma cell lines were cultured in DMEM medium (Gibco; Thermo Fisher Scientific, Inc.). 293T cells were cultured in RPMI 1640 medium (Sigma-Aldrich; Merck KGaA). In total, 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) was added into all culture media. Cell culture was performed at 37°C in a 5% CO2 incubator.
Antibodies
The primary antibodies used in the present study included the following: PDHA1 (1:500; cat. no. 377092 Santa Cruz Biotechnology, Inc.), p-PDHA1 (1:1,000; no. ab115343; Abcam), TNFR1 (1:1,000; cat. no. ab68160; Abcam), MMP2 (1:500; cat. no. 13595; Santa Cruz Biotechnology, Inc.), MMP14 (1:500; cat. no. 373908; Santa Cruz Biotechnology, Inc.), E-cadherin (1:1,000; cat. no. 14472; Cell Signaling Technology, Inc.), GAPDH (1:5,000; cat. no. ab8245; Abcam).
Drug treatment
DCA stock solutions of 1/10 mM (Sigma-Aldrich; Merck KGaA) were diluted in PBS for in vitro experiments. Prior to western blotting experiments, DCA was diluted into 5, 10 and 20 mM concentrations to stimulate U251 cells for 24 h at 37°C in a 5% CO2 incubator. Prior to migration and wound-healing experiments, the concentration of 20 mM DCA was chosen to stimulate U251 cell for 24 and 48 h at 37°C in a 5% CO2 incubator.
Cell protein extraction
Cells at 0, 4, 8, 12 and 24 h were washed three times with PBS at room temperature for 5 min. RIPA cell protein lysate (Thermo Fisher Scientific, Inc.; 400 µl) was added to the cells on ice for 40 min. Samples were then centrifuged at 4°C for 15 min at 12,000 × g.
Cell proliferation assays
The U251 glioma cells were seeded in 96-well plates at a density of 2×106 cells/well for 24 h and subsequently the Cell Counting Kit (CCK)-8 reagent (Dojindo Molecular Technologies, Inc.) was added to each well for 4 h, taking care to avoid light. The absorbance was measured at 450 nm by a microplate reader (Bio-Rad Laboratories, Inc.).
Western blot analysis
Proteins were extracted from the transfected cells using RIPA buffer on ice. Protein extracts were mixed with 2X loading buffer. The protein was quantified using BCA determination method. Proteins (60 µg/lane) were separated by 10% SDS-PAGE. The protein samples were transferred to polyvinylidene fluoride (PVDF) membranes following sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The PVDF membrane was blocked with skimmed milk at room temperature for 2 h, before the aforementioned primary antibodies were added at 4°C overnight. Membranes were then incubation with horseradish peroxidase conjugated goat anti-rabbit secondary antibody (1:5,000; cat. no. ab7090; Abdam) at room temperature for 2 h. Pierce™ Electro-chemiluminescence (Thermo Fisher Scientific, Inc.) was used to detect protein expression. The ImageJ software (v1.50; National Institutes of health) was used to conduct densitometric analysis.
Immunofluorescence staining of cytoskeleton
U251MG cells were cultured in a 6-well plate. After 4 h growth, the cells were collected. The cells were fixed with 4% paraformaldehyde for 1 h at 37°C. Subsequently, they were treated with 0.2% Triton X-100 and blocked with 1% bovine serum albumin (Sigma-Aldrich-Merck KGaA) at room temperature for 30 min. Then cells were immunolabeled with anti-vinculin (1:200; cat. no. 00162950; Sigma-Aldrich; Merck KGaA) at 37°C for 1 h followed by FITC-anti-mouse (1:200; cat. no. F4516; Sigma-Aldrich; Merck KGaA) at room temperature for 2 h. Subsequently, phalloidin-tetramethylrhodamine (1:1,000, Abcam) was added to the cells and placed in a 37°C incubator for 1 h. Finally, the nuclei were stained with 4′,6-diamidino-2-phenylindole at room temperature for 1 h. The cytoskeletal structure was detected by an Olympus fluorescence microscope (magnification, ×1,000).
Wound healing assay
The U251 glioma cells were cultured in a 6-well plate according to different experimental conditions. A line was drawn in the fused cell layer (5×105 cells/well) with a 100 µl micropipette tip. The cells were washed three times with PBS and grown in serum-free DMEM, before being incubated at 37°C for 48 h. Then, the cells were imaged at 0, 24 and 48 h using an inverted Leica phase contrast microscope (magnification, ×200). The wound closure rate was calculated using ImageJ (version 1.6; National Institutes of Health) and relative wound closure was determined.
Cell invasion assay
A total of 200 µl U251 cells (5×105 cells) was added in serum-free DMEM to the upper layer of each chamber and 600 µl DMEM containing 10% fetal bovine serum was added to the lower layer of each chamber. The cells were cultured at 37°C in a 5% CO2 incubator for 24 h. At appropriate time points, the cells in the upper chamber were removed with a cotton swab and the migrated cells were fixed and stained with 4% crystal violet at room temperature for 5 min. The migrated cells were counted using an inverted Leica phase contrast microscope (magnification, ×100).
Immunohistochemical analysis
The tissue slices were put into the oven for 8 h and subsequently into gradient ethanol and xylene. The sections were boiled in citrate solution to retrieve the antigens and washed with PBS three times. The endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide at room temperature for 1 h. The primary antibody (p-PDHA1; 1:1,000; cat. no. ab115343; Abcam), E-cadherin (1:1,000; cat. no. 14472; Cell Signaling Technology, Inc.) was added to the sections at 4°C for overnight incubation. The following day, the sections were washed with PBS and the secondary antibody was added at room temperature for 2 h. The slices were stained with 3,3-diaminobenzidine and hematoxylin at room temperature for 5 min. The sections were differentiated with hydrochloric acid alcohol and dehydrated with gradient alcohol. Finally, the slices were sealed with neutral balsam. The sections were examined by two professional pathologists who did not have previous relevant information regarding the samples. The staining intensity of the sections was classified as follows: 0 (no staining), 1 (weak staining), 2 (moderate staining) and 3 (strong staining). The proportion of cell staining corresponded to the following scores: 0, <1%; 1, 1–20%; 2, 21–50%; 3, 51–75%; and 4, >75%. PBS was used as the negative control and the sections that were stained positive were used as the positive control. Following the non-specific staining of the tumor margin, the necrotic area and adjacent brain tissue area were excluded. The scores of the staining intensity and positive cell proportion were added.
Cell transfection
The small interfering (si)RNA sequences used for PDHA1 knockdown were synthesized by the Shanghai GeneChem Co., Ltd. The sequences were the following: siPDHA1-1, 5′-CAATCAGTGGATCAAGTT-3′, siPDHA1-2, 5′-AATGGAGTTGAAAGCAGAT-3′, siPDHA1-3, 5′-TGGTAGCATCCCGTAATTT-3′, siPDHA1-4, 5′-AGAAATTCTCGCAGAGCTT-3′. The negative control (NC) siRNA sequence (siNC) was the following: 5′-TTCTCCGAACGTGTCACGT-3′. Subsequent experiments were conducted at 48 h post-transfection. The U251 glioma cells were grown at a density of 80% and transfected with 100 nM siRNA targeting specific genes using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific), according to the manufacturer's instructions.
Statistical analysis
The data were analyzed by the SPSS 19.0 software (IBM Corp.). The association between PDHA1 and Ki-67 was evaluated by the Spearman's correlation coefficient. The χ2 test was used to compare the clinicopathological parameters and the levels of the p-PDHA1 protein in 80 patients. The Kaplan-Meier (K-M) by SPSS (v.22.0; IBM Corp.) survival curve was used to analyze the survival rate of the patients. The log rank test was used to compare the survival curve data. The differences between two groups were analyzed using the Student's paired t-test. The differences between multiple groups were analyzed by one-way ANOVA followed by Tukey's post hoc test. Cox proportional analysis was used to analyze the independent effect of p-PDHA1 on the prognosis of patients. All experiments were repeated at least three times. P<0.05 was considered to indicate a statistically significant difference.
Results
p-PDHA1 levels in different grades of human glioma tissues
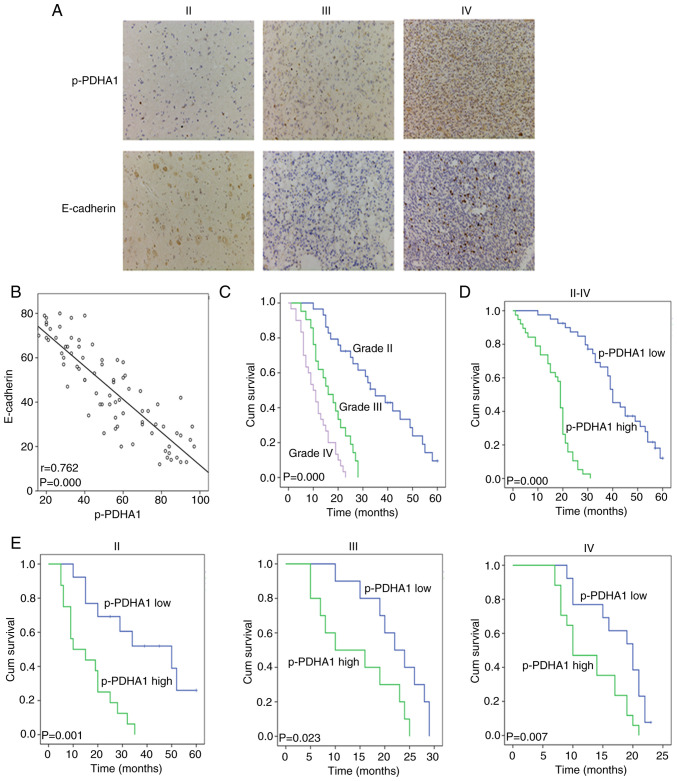
The protein levels of p-PDHA1 and E-cadherin in tissues were detected by immunohistochemistry (Fig. 1A). The staining results of 80 patients with glioma were assessed using the Spearman rank test. The data demonstrated that the protein levels of p-PDHA1 correlated negatively with E-cadherin (Fig. 1B; P<0.001). The clinical data of 80 patients with glioma were analyzed, including 29 patients with grade II glioma, 20 patients with grade III glioma and 31 patients with grade IV glioma. The χ2 test indicated that p-PDHA1 level significantly differed between the different grades of glioma (P=0.019; Table I). The K-M survival curve and log-rank test were used to analyze the survival between different WHO grades and p-PDHA1 level in patients with glioma (Fig. 1C; P=0.000). Subsequently, the association of the survival time between high and low p-PDHA1 level was analyzed using the K-M curve and the log rank test (Fig. 1D; P=0.000). Patients were then divided into high and low p-PDHA1 expression groups. The high expression group was associated with the different grades of glioma (Table I). It was revealed that the survival time of patients with high p-PDHA1 level in glioma was lower than that noted in low p-PDHA1 group. The K-M survival curves of high and low p-PDHA1 level in the different glioma grades indicated that low p-PDHA1 level was associated with longer survival time of patients with glioma in tissues of all grades (Fig. 1E; P<0.05). Multivariate survival analysis demonstrated that p-PDHA1 level was an independent prognostic factor for glioma (P=0.003; Table II).
Figure 1.
p-PDHA1 protein level in glioma tumors of different grade. (A) Immunohistochemical staining was used to detect the level of p-PDHA1 and E-cadherin in glioma tumors of different grade (×400). (B) Scatter plots indicated the correlation between p-PDHA1 and E-cadherin. (C) K-M survival curves of 80 patients with glioma with different WHO grade tumors. (D) K-M survival curve of high and low p-PDHA1 level in 80 patients with glioma. (E) K-M survival curve of 80 patients with glioma with high and low p-PDHA1 level in different WHO grade tumors. p-PDHA1, phosphorylated pyruvate dehydrogenase α1; K-M, Kaplan-Meier; WHO, World Health Organization.
Table I.
Protein level of p-PDHA1 and clinicopathological characteristics in 80 cases of glioma.
| p-PDHA1 expression | |||||
|---|---|---|---|---|---|
| Variables | Total, n | Low | High | χ2-value | P-value |
| Age, years | |||||
| <40 | 38 | 23 | 15 | 0.317 | 0.573 |
| ≥40 | 42 | 29 | 13 | ||
| Sex | |||||
| Female | 43 | 18 | 25 | 0.747 | 0.387 |
| Male | 37 | 20 | 17 | ||
| Tumor location | |||||
| Frontal | 20 | 7 | 13 | 1.312 | 0.726 |
| Parietal | 17 | 9 | 8 | ||
| Occipital | 32 | 14 | 18 | ||
| Temporal | 11 | 5 | 6 | ||
| Tumor size, cm | |||||
| <4 | 35 | 20 | 15 | 0.722 | 0.395 |
| ≥4 | 45 | 31 | 14 | ||
| WHO grade | |||||
| II | 29 | 10 | 19 | 7.885 | 0.019 |
| III | 20 | 6 | 14 | ||
| IV | 31 | 20 | 11 | ||
| Extent of resection | |||||
| Biopsy | 14 | 8 | 6 | 1.564 | 0.458 |
| Total resection | 36 | 15 | 21 | ||
| Subtotal resection | 30 | 17 | 13 | ||
Pearson χ2 test was used for statistical analysis. WHO, World Health Organization; p-PDHA1, phosphorylated pyruvate dehydrogenase α1.
Table II.
Contribution of various potential prognostic factors to survival by Cox regression analysis on 80 glioma specimens.
| Characteristics | Hazard ratio | 95% CI | P-value |
|---|---|---|---|
| Age | 1.089 | 0.636–1.865 | 0.757 |
| Sex | 1.290 | 0.760–2.189 | 0.345 |
| Tumor location | 0.822 | 0.639–1.057 | 0.127 |
| Tumor size | 1.340 | 0.800–2.244 | 0.266 |
| WHO Grade | 13.661 | 7.046–26.488 | 0.000 |
| Extent of resection | 0.944 | 0.688–1.295 | 0.721 |
| p-PDHA1 | 2.432 | 1.344–4.400 | 0.003 |
CI, confidence interval; WHO, World Health Organization; p-PDHA1, phosphorylated pyruvate dehydrogenase α1.
Protein level of p-PDHA1 in glioma cells
Initially, five glioma cell lines were selected as follows: H4, A172, SHG44, U251 and U87. Western blot analysis indicated that the levels of p-PDHA1 were increased in U251 glioma cells compared with total protein (Fig. 2A). Subsequently, TNF-α (50 ng/ml) was used to stimulate U251 glioma cells and the absorbance was detected at 0, 4, 8, 12 and 24 h at 490 nm. CCK-8 analysis indicated that following the stimulation of TNF-α, compared with 0 h, the proliferation of glioma cells was increased (Fig. 2B). Furthermore, U251 glioma cells were treated with TNF-α (50 ng/ml) and the cell protein was collected at 0, 4, 8, 12 and 24 h, in order to assess the highest protein level time point of p-PDHA1. Western blot analysis indicated that p-PDHA1 and TNFR1 levels were increased at 4 h in U251 glioma cells compared with those noted at 0 h (Fig. 2C).
Figure 2.
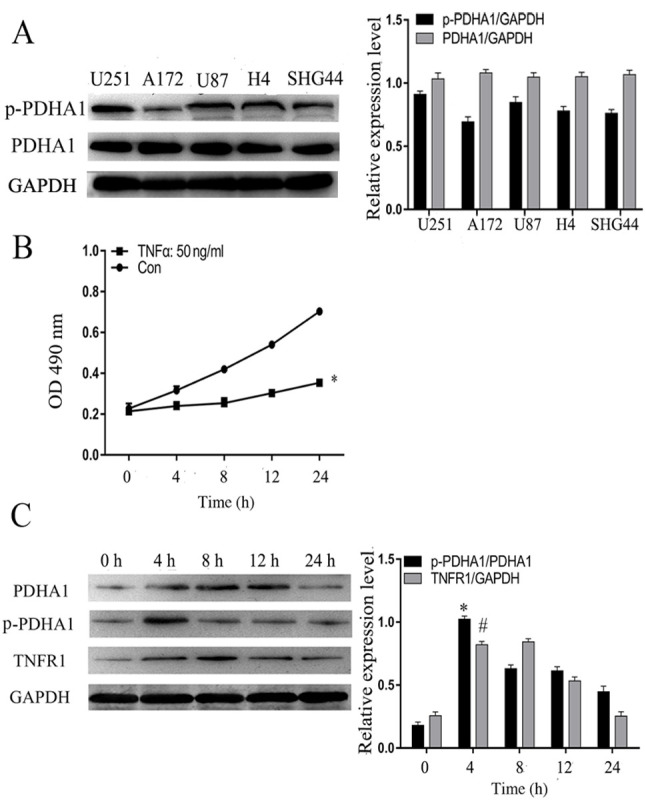
Protein level of p-PDHA1 in glioma cells. (A) Western blotting results of p-PDHA1 level in five glioma cell lines. The bar chart indicated the ratio of p-PDHA1 to PDHA1 levels. (B) Cell Counting Kit-8 analysis of TNF-α (50 ng/ml)-induced U251 glioma cells. *P<0.05 vs. Con. (C) Western blot analysis of TNFR1 and p-PDHA1 levels in U251 glioma cells stimulated by TNF-α (50 ng/ml) at different time points. The bar chart indicates the ratio of p-PDHA1 to PDHA1 expression and the expression levels of TNFR1 compared with those of GAPDH. *P<0.05 and #P<0.05 vs. 0 h. p-, phosphorylated; PDHA1, pyruvate dehydrogenase α1; TNF-α, tumor necrosis factor-α; TNFR1, tumor necrosis factor receptor 1; Con, control.
p-PDHA1 promotes glioma cell migration and invasion following TNF-a stimulation
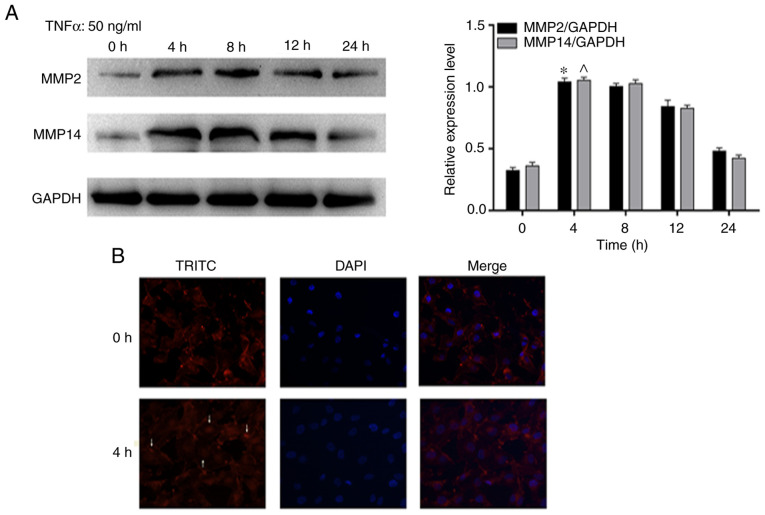
In order to further investigate whether p-PDHA1 exhibited migratory effects on glioma cells, the effects of TNF-α stimulation on specific migratory markers were assessed in U251 glioma cells. The results indicated that the protein levels of MMP2 and MMP14 in glioma cells were significantly increased at 4 and 8 h compared with 0 h. (Fig. 3A). In addition, these results were further verified by immunofluorescence staining of the cytoskeleton. Immunofluorescence microscopy demonstrated that F-actin formed strong and dense stress fibers in U251 glioma cells treated with TNF-α for 4 h (Fig. 3B).
Figure 3.
p-PDHA1 promotes the migration of glioma cells following TNF-α treatment. (A) Western blot analysis of the expression levels of MMP2 and MMP14 following TNF-α stimulation at different time points. The bar chart indicates the expression levels of MMP2 to MMP14 compared with those of GAPDH. *P<0.05; ^P<0.05 vs. 0 h. (B) Immunofluorescence staining of the cytoskeleton indicating the localization of F-actin (red fluorescence, arrow) and of the nuclear region (blue fluorescence) using fluorescence microscopy (magnification, ×1,000). p-, phosphorylated; PDHA1, pyruvate dehydrogenase α1; TNF-α, tumor necrosis factor-α; TRITC, tetramethylrhodamine.
Knockdown of PDHA1 is associated with the migration and invasion capacities of glioma cells
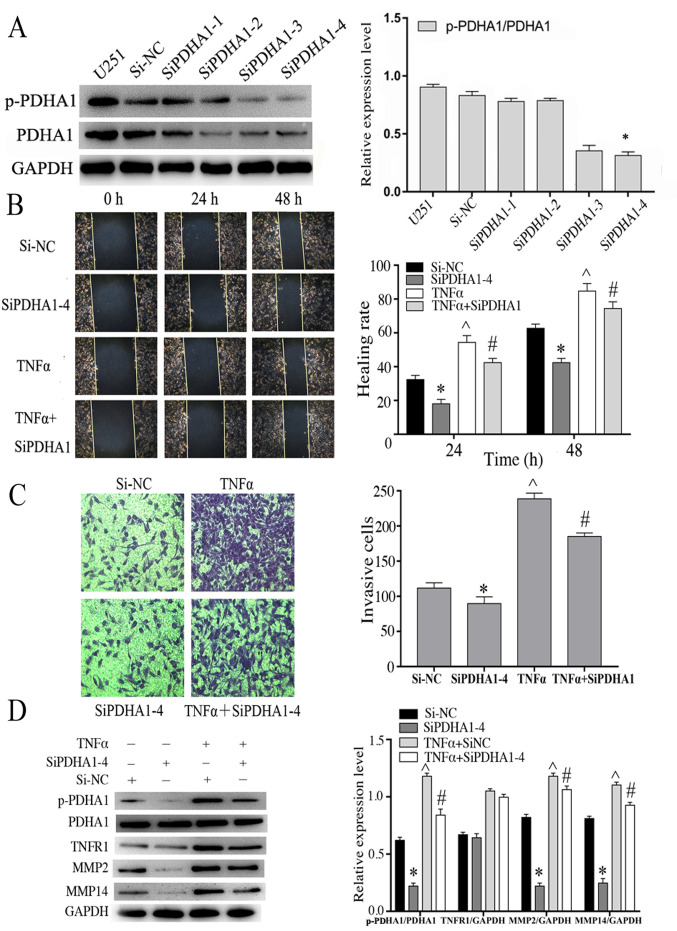
Following stimulation with TNF-α, the expression levels of the migratory markers were increased suggesting that they were also associated with p-PDHA1 levels. U251 cells were transfected with four PDHA1-specific siRNAs and the data demonstrated that siPDHA1-4 exhibited the most significant knockdown efficiency in U251 glioma cells (Fig. 4A). Therefore, siPDHA1-4-transfected U251 cells were selected and grouped according to the experimental design. The stimulation time of TNF-α (50 ng/ml) was 4 h. The association between p-PDHA1 and glioma cell was studied using cell migration and wound-healing assays. The results indicated that knockdown of p-PDHA1 significantly inhibited the migratory and migration efficiency of glioma cells at 24 and 48 h compared with 0 h. TNF-α stimulation did not enhance cell migration in PDHA1-siRNA-transfected cells compared with the TNF-α stimulation group (Fig. 4B and C). Subsequently, western blot analysis was performed, and the data indicated that TNFR1 levels were upregulated following TNF-α stimulation. Knockdown of p-PDHA1 also reduced the expression levels of MMP2 and MMP14. TNF-α stimulation increased the expression levels of MMP2, MMP14 and p-PDHA1/PDHA1 (P<0.05). Whereas it did not increase the expression levels of MMP2, MMP14 and p-PDHA1/PDHA1 (P<0.05) in PDHA1-siRNA transfected cells (Fig. 4D).
Figure 4.
Knockdown of PDHA1 is associated with the migration and invasion capacities of glioma cells. (A) Western blot analysis of the interference efficiency of U251 cells transfected with siPDHA1. The bar chart indicates the ratio of p-PDHA1 to PDHA1 expression. *P<0.05 vs. siNC. (B) Wound healing assays of U251MG cells treated with si-NC, si-PDHA1-4, TNF-α and siPDHA1 + TNF-α. The migration of glioma cells to the wound surface at 0, 24 and 48 h was observed using the inverted Leica phase contrast microscope (magnification, ×200). The bar chart indicates the quantitative results of the wound healing assays *P<0.05, ^P<0.05 vs. siNC; #P<0.05 vs. TNF-α. (C) Transwell invasion assays of U251 glioma cells in the si-NC, siPDHA1-4, TNF-α and siPDHA1 + TNFα groups. The bar chart indicated the quantitative results of the Transwell invasion assays (magnification, ×100). *P<0.05, ^P<0.05 vs. siNC; #P<0.05 vs. TNF-α. (D) Western blot analysis of siPDHA1-transfected and/or TNF-α-stimulated U251 glioma cells. The bar chart indicates the ratio of p-PDHA1 to PDHA1 expression, as well as the expression levels of TNFR1, MMP2 and MMP14 normalized to those of GAPDH. *P<0.05, ^P<0.05 vs. siNC; #P<0.05 vs. TNF-α + siNC. p-PDHA1, phosphorylated pyruvate dehydrogenase α1; PDHA1, pyruvate dehydrogenase α1; TNF-α, tumor necrosis factor-α; TNFR1, tumor necrosis factor receptor 1.
Effects of DCA on the migration and invasion capacities of glioma cells stimulated by TNF-α
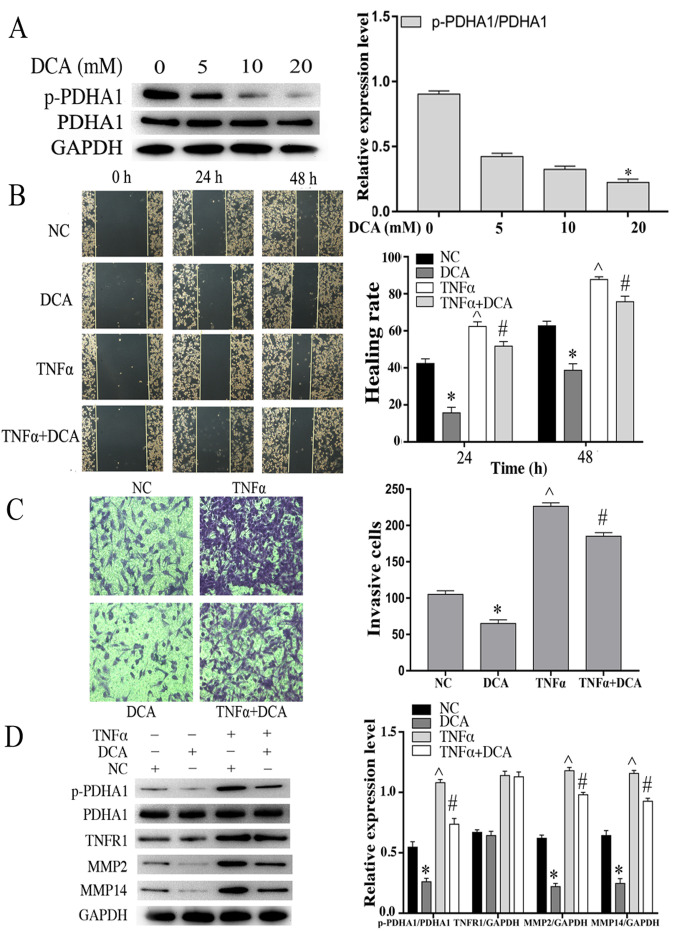
It has been reported that DCA is the inhibitor with preliminary clinical effects on glioblastoma (19). PDK is an important kinase that regulates the activity of the PDHc. Therefore, the protein levels of p-PDHA1 were examined in U251 glioma cells at various concentrations of DCA. The results indicated that p-PDHA1/PDHA1 level (P<0.05) was significantly lower following treatment of the cells with 20 mM DCA compared with that noted in the control cells (Fig. 5A). U251 glioma cells were incubated with 20 mM DCA for 24 h and grouped according to the experimental design. The stimulation time of TNF-α (50 ng/ml) was 4 h. Based on the analysis of the wound healing and transwell invasion assays, DCA-treatment of U251 cells resulted in a significant inhibition in their migratory and invasive efficacy. TNF-α stimulation did not enhance cell migration in the DCA-treated cells compared with the cells treated with TNF-α alone (Fig. 5B and C). Western blot analysis indicated that DCA reduced the expression levels of MMP2 and MMP14. TNF-α stimulation increased the expression levels of MMP2, MMP14 and p-PDHA1/PDHA1 (P<0.05). Whereas it did not increase the expression levels of MMP2, MMP14 and p-PDHA1/PDHA1 (P<0.05) in the DCA-treated glioma cells (Fig. 5D).
Figure 5.
Effects of DCA on the migration and invasion of glioma cells stimulated by TNF-α. (A) Western blot analysis of U251 glioma cells incubated with different concentrations of DCA for 24 h. The bar chart indicates the ratio of p-PDHA1 to PDHA1 expression. *P<0.05 vs. 0 mM. (B) Wound healing assays of NC, DCA, TNF-α and DCA+TNF-α U251 cells. The bar chart is representative of the quantitative results of the wound healing assays. *P<0.05, ^P<0.05 vs. NC; #P<0.05 vs. DCA. (C) Transwell invasion assays of NC, DCA, TNF-α and DCA + TNFα U251 cell groups. The bar chart is indicative of the quantitative results of the Transwell invasion assay. *P<0.05, ^P<0.05 vs. NC; #P<0.05 vs. DCA. (D) Western blot analysis of U251MG cells treated with DCA and/or TNF-α. The bar chart indicated the ratio of p-PDHA1 to PDHA1 expression and the expression levels of TNFR1, MMP2 and MMP14 normalized to those of GAPDH. *P<0.05, ^P<0.05 vs. NC; #P<0.05 vs. DCA. DCA, dichloroacetic acid; TNF-α, tumor necrosis factor-α; p-PDHA1, phosphorylated pyruvate dehydrogenase α1; NC, negative control.
Discussion
Invasion and migration are the main biological characteristics of tumor malignancy. A previous study has reported that the three main characteristics of tumor cell migration are the following: Tumor cells adhere to ECM and degrade it using lyase, cathepsin and autocrine or paracrine cytokines, such as 5-HT, histamine and bradykinin (20). All these processes alter cell morphology and enable the migration movement using pseudopodia (20). This partially explains the poor survival rate of patients with glioma. Migration is the key to invasion; therefore, the control of the migration of glioma cells is particularly important for the effective treatment of this disease (21).
Several inflammatory cytokines have been demonstrated to be involved in the tumor microenvironment, such as interleukin (IL)-1, IL-6, IL-1β, TNF-α and TGF-β, which not only recruit inflammatory cells to the tumor site, but also amplify the inflammatory effect. They also promote the proliferation and metastasis of tumor cells, and accelerate the formation of tumor blood vessels and lymphatic vessels (22–24). Tumor necrosis factor can induce a variety of processes, such as apoptosis, necrosis and cell migration (25–27). There are two types of TNFR receptors: TNFR1 and TNFR2. The majority of the biological effects of TNF-α are produced by TNFR1 (28). In the present study, the role of p-PDHA1 was investigated in the regulation of genes involved in cell migration and TNF-α-induced gene expression in glioma cells.
Normal expression of PDHA1 is a prerequisite for the carboxylic acid cycle and oxidative phosphorylation occurring in the mitochondria. PDHA1 is an important catalytic component of PDHc. Phosphorylation of PDHA1 occurs at three specific serine residues and the single phosphorylation of each site can cause PDHc inactivation. Phosphorylation of site 1 is the most rapid reaction; covalent modification occurs at specific serine residues in PDHA1 (29). Tumor progression promotes the development of hypoxia and induces the expression of hypoxia inducible factors (HIFs). HIFs induce high expression of PDK, which phosphorylates PDHA1 and inactivates PDHc (30). Although aerobic glycolysis has been widely accepted as a metabolic feature of tumors, its mechanism in tumor progression remains unclear. In the present study, the data demonstrated that the protein levels of TNFR1 and p-PDHA1 were altered in glioma cells following the stimulation with TNF-α. The present study examined whether p-PDHA1 was associated with the inflammatory microenvironment, and its biological interaction with the processes of invasion and migration of glioma cells was further explored. Western blotting was used to analyze the activity of glioma cell migration following stimulation by TNF-α. The potential of glioma cell migration was evaluated by immunofluorescence staining of the cytoskeleton. It has been reported that PDK and PDH phosphatase regulate the activity of PDHA1. Phosphorylation of PDHA1 can cause PDHc inactivation. It can enhance the Warburg effect and the invasiveness of tumor cells (17). Therefore, in the present study, the involvement of PDHA1 phosphorylation in glioma cell migration and invasion was further assessed.
Various proteases, including MMPs, are involved in the destruction of normal brain tissue (31). The wound healing and Transwell invasion assays demonstrated that knockdown of the expression of PDHA1 and reduced levels of p-PDHA1 decreased the migration and invasion of glioma cells. TNF-α stimulation promoted glioma cell migration and invasion, whereas this effect was not noted in PDHA1 siRNA-transfected cells. DCA is an inhibitor of PDK, which is an important kinase that regulates the activity of PDHc (19). The present study indicated that DCA treatment significantly inhibited the migratory and invasive efficiency of glioma cells. TNF-α stimulation promoted glioma cell migration and invasion, whereas TNF-α stimulation did not enhance cell migration in the DCA-treated cells.
In conclusion, the present study demonstrated that p-PDHA1 served an important role in cell migration and invasion of glioma cells, suggesting that its level was associated with the development of glioma. Furthermore, the data indicated that TNF-α induced the level of p-PDHA1 in glioma cells. Therefore, p-PDHA1 may represent a key messenger for the regulation of cellular movement and gene expression. A more comprehensive understanding of this pathway may be helpful in establishing new therapeutic strategies for glioma.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant nos. 81572491, 81602201 and 81502169).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
CZ and DW designed the study. ZY and YW performed the experiment and wrote the manuscript. LZ analyzed the clinical data. All authors read, revised and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Nantong University, Jiangsu, China (grant no. 20190313). The present manuscript is one of the results of the project. All patients or their family members provided informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Reference
- 1.Rasmussen BK, Hansen S, Laursen RJ, Kosteljanetz M, Schultz H, Norgard BM, Guldberg R, Gradel KO. Epidemiology of glioma: Clinical characteristics, symptoms, and predictors of glioma patients grade I–IV in the the Danish Neuro-oncology registry. J Neurooncol. 2017;135:571–579. doi: 10.1007/s11060-017-2607-5. [DOI] [PubMed] [Google Scholar]
- 2.Hakyemez B, Erdoğan C, Ercan I, Ergin N, Uysal S, Atahan S. High-grade and low-grade gliomas: Differentiation by using perfusion MR imaging. Clin Radiol. 2005;60:493–502. doi: 10.1016/j.crad.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Brat DJ, Aldape K, Colman H, Holland EC, Louis DN, Jenkins RB, Kleinschmidt-DeMasters BK, Perry A, Reifenberger G, Stupp R, et al. cIMPACT-NOW update 3: Recommended diagnostic criteria for ‘Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV’. Acta Neuropathol. 2018;136:805–810. doi: 10.1007/s00401-018-1913-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anjum K, Shagufta BI, Abbas SQ, Patel S, Khan I, Shah SAA, Akhter N, Hassan SSU. Current status and future therapeutic perspectives of glioblastoma multiforme (GBM) therapy: A review. Biomed Pharmacother. 2017;92:681–689. doi: 10.1016/j.biopha.2017.05.125. [DOI] [PubMed] [Google Scholar]
- 5.Hwang JS, Jung EH, Kwon MY, Han IO. Glioma-secreted soluble factors stimulate microglial activation: The role of interleukin-1β and tumor necrosis factor-α. J Neuroimmunol. 2016;298:165–171. doi: 10.1016/j.jneuroim.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Camelo S, Lafage M, Galelli A, Lafon M. Selective role for the p55 Kd TNF-alpha receptor in immune unresponsiveness induced by an acute viral encephalitis. J Neuroimmunol. 2001;113:95–108. doi: 10.1016/S0165-5728(00)00427-6. [DOI] [PubMed] [Google Scholar]
- 7.Jiang Y, Yu M, Hu X, Han L, Yang K, Ba H, Zhang Z, Yin B, Yang XP, Li Z, Wang J. STAT1 mediates transmembrane TNF-alpha-induced formation of death-inducing signaling complex and apoptotic signaling via TNFR1. Cell Death Differ. 2017;24:660–671. doi: 10.1038/cdd.2016.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ting AT, Bertrand MJM. More to Life than NF-κB in TNFR1 signaling. Trends Immunol. 2016;37:535–545. doi: 10.1016/j.it.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Yin N, Guo A, Zhang Q, Zhang Y, Xu Y, Liu H, Tang B, Lai L. NF-κB signaling and cell-fate decision induced by a fast-dissociating tumor necrosis factor mutant. Biochem Biophys Res Commun. 2017;489:287–292. doi: 10.1016/j.bbrc.2017.05.149. [DOI] [PubMed] [Google Scholar]
- 10.Yang S, Xie C, Chen Y, Wang J, Chen X, Lu Z, June RR, Zheng S. Differential roles of TNFα-TNFR1 and TNFα-TNFR2 in the differentiation and function of CD4+Foxp3+induced Treg cells in vitro and in vivo periphery in autoimmune diseases. Cell Death Dis. 2019;10:27. doi: 10.1038/s41419-018-1266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martínez-Reza I, Díaz L, García-Becerra R. Preclinical and clinical aspects of TNF-α and its receptors TNFR1 and TNFR2 in breast cancer. J Biomed Sci. 2017;24:90. doi: 10.1186/s12929-017-0398-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z, Yu M, Fei B, Fang X, Ma T, Wang D. miR-21-5p targets PDHA1 to regulate glycolysis and cancer progression in gastric cancer. Oncol Rep. 2018;40:2955–2963. doi: 10.3892/or.2018.6695. [DOI] [PubMed] [Google Scholar]
- 13.Ferriero R, Nusco E, De Cegli R, Carissimo A, Manco G, Brunetti-Pierri N. Pyruvate dehydrogenase complex and lactate dehydrogenase are targets for therapy of acute liver failure. J Hepatol. 2018;69:325–335. doi: 10.1016/j.jhep.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stacpoole PW. Therapeutic targeting of the pyruvate dehydrogenase complex/pyruvate dehydrogenase kinase (PDC/PDK) axis in cancer. J Natl Cancer Inst. 2017;109 doi: 10.1093/jnci/djx071. [DOI] [PubMed] [Google Scholar]
- 15.Xu L, Li Y, Zhou L, Dorfman RG, Liu L, Cai R, Jiang C, Tang D, Wang Y, Zou X, et al. SIRT3 elicited an anti-Warburg effect through HIF1α/PDK1/PDHA1 to inhibit cholangiocarcinoma tumorigenesis. Cancer Med. 2019;8:2380–2391. doi: 10.1002/cam4.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu F, Zhang W, You X, Liu Y, Li Y, Wang Z, Wang Y, Zhang X, Ye L. The oncoprotein HBXIP promotes glucose metabolism reprogramming via downregulating SCO2 and PDHA1 in breast cancer. Oncotarget. 2015;29:27199–27213. doi: 10.18632/oncotarget.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu L, Cao J, Zhao J, Li X, Suo Z, Li H. PDHA1 Gene Knockout in Human esophageal squamous cancer cells resulted in greater warburg effect and aggressive features in vitro and in vivo. Onco Targets Ther. 2019;12:9899–9913. doi: 10.2147/OTT.S226851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glotsos D, Spyridonos P, Petalas P, Cavouras D, Ravazoula P, Dadioti PA, Lekka I, Nikiforidis G. Computer-based malignancy grading of astrocytomas employing a support vector machine classifier, the WHO grading system and the regular hematoxylin-eosin diagnostic staining procedure. Anal Quant Cytol Histol. 2004;2:77–83. [PubMed] [Google Scholar]
- 19.Velpula KK, Guda MR, Sahu K, Tuszynski J, Asuthkar S, Bach SE, Lathia JD, Tsung AJ. Metabolic targeting of EGFRvIII/PDK1 axis in temozolomide resistant glioblastoma. Oncotarget. 2017;8:35639–35655. doi: 10.18632/oncotarget.16767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oudin MJ, Weaver VM. Physical and chemical gradients in the tumor microenvironment regulate tumor cell invasion, migration, and metastasis. Cold Spring Harb Symp Quant Biol. 2016;81:189–205. doi: 10.1101/sqb.2016.81.030817. [DOI] [PubMed] [Google Scholar]
- 21.Andreozzi M, Quintavalle C, Benz D, Quagliata L, Matter M, Calabrese D, Tosti N, Ruiz C, Trapani F, Tornillo L, et al. HMGA1 expression in human hepatocellular carcinoma correlates with poor prognosis and promotes tumor growth and migration in in vitro models. Neoplasia. 2016;18:724–731. doi: 10.1016/j.neo.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao X, Huang J, Zhong H, Shen N, Faggioni R, Fung M, Yao Y. Targeting interleukin-6 in inflammatory autoimmune diseases and cancers. Pharmacol Ther. 2014;141:125–139. doi: 10.1016/j.pharmthera.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Castano Z, San Juan BP, Spiegel A, Pant A, DeCristo MJ, Laszewski T, Ubellacker JM, Janssen SR, Dongre A, Reinhardt F, et al. IL-1β inflammatory response driven by primary breast cancer prevents metastasis-initiating cell colonization. Nat Cell Biol. 2018;20:1084–1097. doi: 10.1038/s41556-018-0173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Batlle E, Massague J. Transforming growth factor-β signaling in immunity and cancer. Immunity. 2019;50:924–940. doi: 10.1016/j.immuni.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang BP, Lin CS, Wang CJ, Kao SH. Upregulation of heat shock protein 70 and the differential protein expression induced by tumor necrosis factor-alpha enhances migration and inhibits apoptosis of hepatocellular carcinoma cell HepG2. Int J Med Sci. 2017;14:284–293. doi: 10.7150/ijms.17861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiozaki A, Shimizu H, Ichikawa D, Konishi H, Komatsu S, Kubota T, Fujiwara H, Okamoto K, Iitaka D, Nakashima S, et al. Claudin 1 mediates tumor necrosis factor alpha-induced cell migration in human gastric cancer cells. World J Gastroenterol. 2014;20:17863–17876. doi: 10.3748/wjg.v20.i47.17863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ooppachai C, Limtrakul Dejkriengkraikul P, Yodkeeree S. Dicentrine potentiates TNF-α-induced apoptosis and suppresses invasion of A549 lung adenocarcinoma cells via modulation of NF-κB and AP-1 activation. Molecules. 2019;24:4100. doi: 10.3390/molecules24224100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Horssen R, Ten Hagen TL, Eggermont AM. TNF-alpha in cancer treatment: Molecular insights, antitumor effects, and clinical utility. Oncologist. 2006;11:397–408. doi: 10.1634/theoncologist.11-4-397. [DOI] [PubMed] [Google Scholar]
- 29.Holness MJ, Sugden MC. Regulation of pyruvate dehydrogenase complex activity by reversible phosphorylation. Biochem Soc Trans. 2003;31:1143–1151. doi: 10.1042/bst0311143. [DOI] [PubMed] [Google Scholar]
- 30.Izquierdo-Garcia JL, Viswanath P, Eriksson P, Cai L, Radoul M, Chaumeil MM, Blough M, Luchman HA, Weiss S, Cairncross JG, et al. IDH1 mutation induces reprogramming of pyruvate metabolism. Cancer Res. 2015;75:2999–3009. doi: 10.1158/0008-5472.CAN-15-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou W, Yu X, Sun S, Zhang X, Yang W, Zhang J, Zhang X, Jiang Z. Increased expression of MMP-2 and MMP-9 indicates poor prognosis in glioma recurrence. Biomed Pharmacother. 2019;118:109369. doi: 10.1016/j.biopha.2019.109369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.