Abstract
Splicing factors have recently been shown to be involved in heterochromatin formation, but their role in controlling heterochromatin structure and function remains poorly understood. In this study, we identified a fission yeast homologue of human splicing factor RBM10, which has been linked to TARP syndrome. Overexpression of Rbm10 in fission yeast leads to strong global intron retention. Rbm10 also interacts with splicing factors in a pattern resembling that of human RBM10, suggesting that the function of Rbm10 as a splicing regulator is conserved. Surprisingly, our deep-sequencing data showed that deletion of Rbm10 caused only minor effect on genome-wide gene expression and splicing. However, the mutant displays severe heterochromatin defects. Further analyses indicated that the heterochromatin defects in the mutant did not result from mis-splicing of heterochromatin factors. Our proteomic data revealed that Rbm10 associates with the histone deacetylase Clr6 complex and chromatin remodelers known to be important for heterochromatin silencing. Deletion of Rbm10 results in significant reduction of Clr6 in heterochromatin. Our work together with previous findings further suggests that different splicing subunits may play distinct roles in heterochromatin regulation.
Keywords: Epigenetics, Splicing factor, Schizosaccharomyces pombe, Histone deacetylase, H3K9 methylation
Introduction
In eukaryotic cells, DNA and histones are organized into the highly ordered chromatin structure. Chromatin forms two distinct domains, euchromatin and heterochromatin. Euchromatin is less condensed and associated with actively transcribed genes, whereas heterochromatin is tightly coiled and transcriptionally inactive. Heterochromatin usually consists of highly repetitive sequences, which are commonly found around pericentromeres and telomeres. Heterochromatin plays essential roles in gene expression, chromosome segregation, and genome stability [1–3].
The pattern of histone modifications in heterochromatin contributes to the organization of the chromatin domain. Heterochromatin is characterized by hypoacetylation of histone lysines and methylation of histone H3 on lysine 9 (H3K9me) [1, 3]. Histone deacetylases (HDACs) are responsible for hypoacetylation in heterochromatin, while H3K9me is catalyzed by the histone methyltransferase SUV39, an enzyme conserved from yeasts to humans. H3K9me serves as a binding site for heterochromatin protein 1 (HP1), which recruits additional factors to mediate heterochromatin assembly [1, 3]. Small interfering RNAs (siRNAs) generated by RNA interference pathway have been shown to be important for heterochromatin structure and function in a variety of organisms [1].
The fission yeast Schizosaccharomyces pombe has emerged as an excellent model organism for understanding heterochromatin formation. It harbors three constitutive heterochromatic regions: the pericentromere, telomere, and mating-type region. These regions are enriched with H3K9me, which is recognized by the human HP1 homolog, Swi6. H3K9me in fission yeast heterochromatin is regulated by the multi-subunit CLRC complex, which contains Clr4, the H3K9 methyltransferase, Rik1, Dos1/Raf1, Dos2/Raf2, Cul4, and Lid2, a H3K4 demethylase [4–9]. H3K9 methylation in heterochromatin also requires RNAi. Heterochromatin is transcribed during S phase of the cell cycle [10, 11]. The heterochromatin transcripts are subsequently processed into siRNAs by Ago1, the RDRC complex, and Dicer. The siRNAs are loaded into RITS complex (Ago1, Chp1, and Tas3), which, in turn, guides the CLRC complex to heterochromatin regions [1, 12, 13] to mediate H3K9me. The DNA Pol epsilon subunit, Cdc20, also contributes to the recruitment of CLRC complex during the S phase [14–16].
Like other eukaryotes, histones in fission yeast heterochromatin are hypoacetylated. Fission yeast contains all three subtypes of histone deacetylases (HDACs): Class I, Class II, and Class III. Clr6, a Class I HDAC, is a homolog of mammalian HDAC1 and HDAC2, and deacetylates multiple lysine residues on histone H3 and H4. Clr3 is a Class II HDAC, responsible for H3K14 deacetylation [17–19]. Sir2 is a conserved member of the Sirtuin family of Class III HDACs that uses NAD+ as a cofactor [20, 21]. All of these HDACs contribute to heterochromatin silencing in fission yeast [17, 19]. Our recent results reveal that the histone fold subunit of the DNA Pol epsilon, Dpb4, is important for recruitment of Sir2 during DNA replication [22].
Recently, splicing factors have been linked to heterochromatin formation in fission yeast. However, the mechanisms for how these splicing factors participate in heterochromatin formation remain unclear. A prevailing model proposes that splicing factors, independently from splicing, directly interacts with RNAi components to mediate the recruitments of the CLRC complex to heterochromatic regions [23]. Another study suggests that transcripts derived from pericentromeric repeats contain introns, and splicing factors may mediate the splicing of the intron of the heterochromatin transcripts, which in turn influences siRNA generation [24]. A more recent study by Kallegren et al. suggested that splicing factors may contribute to heterochromatin assembly indirectly by regulating the proper splicing of heterochromatin factors [25].
RBM10 has been recently characterized as a new member of splicing factors, and associated with lung cancer and congenital disorders, such as the TARP syndrome and XLMR [26–29]. RBM10 contains five distinct domains, including two RNA recognition motifs (RRM), two zinc fingers, and one G-patch motif. Splicing involves an ordered, stepwise rearrangement of protein and RNA content in splicesome. An early step in spliceosome assembly is the formation of the prespliceosome (complex A). The prespliceosome rearranges to the precatalytic spliceosome (complex B), which subsequently gives rise to the activated spliceosome (complex B*). Complex B* carries out the first catalytic step of splicing, generating complex C, which in turn carries out the second catalytic step [30]. It has been shown that RBM10 is associated with prespliceosome (complex A) and activated spliceosome (complex B*) in human cells [31–35]. The human RBM10 has been shown to regulate exon skipping by repressing the splicing of introns [26]. In addition to its role in splicing, human RBM10 was also found to be part of histone H2A deubiquitinase complex [36], suggesting a role in linking splicing and chromatin regulation.
In this study, we identified a fission yeast homologue of RBM10, which was named rbm10+. Overexpression of Rbm10 in fission yeast results in strong growth defects and global intron retention. Our study further demonstrated that Rbm10 plays an important role in heterochromatin assembly and suggested a previously unrecognized mechanism underlying splicing factor-mediated heterochromatin silencing.
Results
Human splicing factor RBM10 is conserved in S. pombe
Via homolog search, we identified SPAC57A7.13, an uncharacterized protein, as the homolog of human RBM10 in S. pombe. We hereafter named it Rbm10. The human RBM10 and its S. pombe homolog share an amino acid identity of 26% (Additional file 1: Figure S1). Despite relatively short length of Rbm10 in fission yeast, all five domains are well conserved, namely, two zinc-finger domains, two RRM motifs and a G-patch domain (Fig. 1a). Majority of Zinc-finger domains function as interaction modules that bind DNA, RNA, or proteins [37, 38], whereas RRM motifs and the G-patch domain usually associate with RNA [39]. To determine the cellular distribution of Rbm10, we constructed the GFP-tagged rbm10+ under its native promoter and replaced the endogenous rbm10+. Cells carrying Rbm10-GFP have no detectable defect, indicating that Rbm10-GFP is functional. We found that, similar to human RBM10, Rbm10-GFP is enriched in the nucleus (Fig. 1b).
Fig. 1.
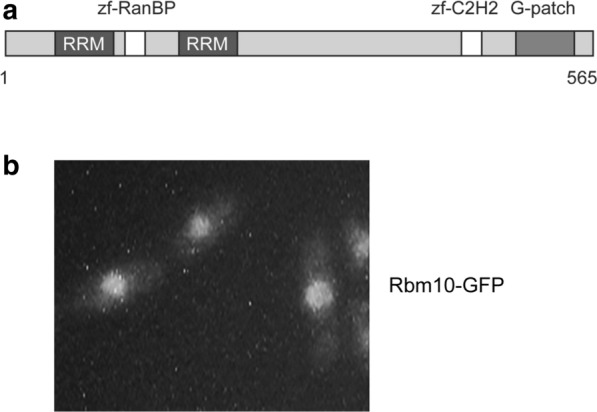
Human RBM10 is conserved in S. pombe. a Schematic diagram showing the domain structure of Rbm10 in S. pombe. b Rbm10 is enriched in the nucleus. Rbm10-GFP was constructed at its native locus under the control of its own promoter
Overexpression of Rbm10 leads to massive intron retention
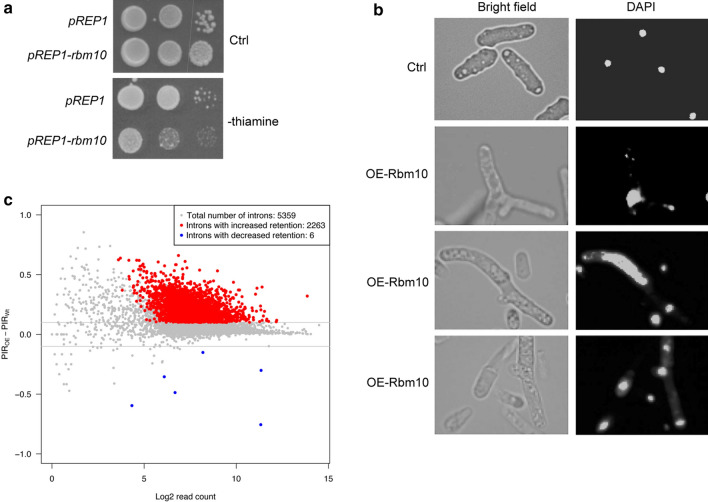
To characterize the function of Rbm10 in fission yeast, we constructed rbm10+ under the strong, inducible nmt1 promoter and overexpress it in wild-type cells. The nmt1 promoter is induced in the absence of thiamine. We found that overexpression of Rbm10 in wild-type results in slow growth (Fig. 2a). In addition to the strong growth defects, cells overexpressing Rbm10 display aberrant cell shapes. They are often massively elongated or branched in to Y-like shapes. The nuclei in these cells also appear to be abnormal. Multiple fragmented nuclei were frequently observed (Fig. 2b). Given that rbm10Δ showed only a mild phenotype as described below, we were surprised to find the observed toxicity resulting from overexpression of Rbm10. To gain insight into the molecular nature of Rbm10 overexpression, we performed unbiased genome-wide RNA-seq using cells overexpressing Rbm10. Cells overexpressing for 20 h were used to minimize secondary effects. We detected massive changes in intron retention from the RNA-seq data: 2263 introns showed increased retention (Fig. 2c and Additional file 2: Table S1), whereas 6 introns have decreased retention (Fig. 2c and Additional file 3: Table S2). These data demonstrated that Rbm10′s function as a splicing regulator is conserved in fission yeast, which can be observed at least under overexpression condition.
Fig. 2.
Overexpression of Rbm10 results in severe growth defects and massive intron retention. a Cells overexpressing Rbm10 display slow growth. Serial dilutions of indicated cells carrying pREP1-rbm10 were plated in the PMG minimal medium without thiamine. Empty pREP1 vector was used as a control. Ctrl, a control plate-containing thiamine. b Cells overexpressing Rbm10 showed abnormal cell shapes and nuclear organization. Left, bright-field microscopic images. Right, DAPI staining. OE, overexpression. c MA plot comparing the intron retention patterns between rbm10-overexpressing cells and WT. Log2 transformed read count for each event was plotted on the X axis, and the PIR difference was on the Y axis. The red and blue dots represented the events with significantly different intron retention patterns. The grey lines indicate 10% PIR difference
Deletion of rbm10+ induces minor changes in gene expression and intron retention
To further understand the role of Rbm10, we constructed a deletion strain, rbm10Δ, in which the complete gene was replaced with a kanamycin selection cassette via homologues recombination. rbm10Δ cells are viable and do not show obvious growth defects, indicating that Rbm10 is not essential for viability.
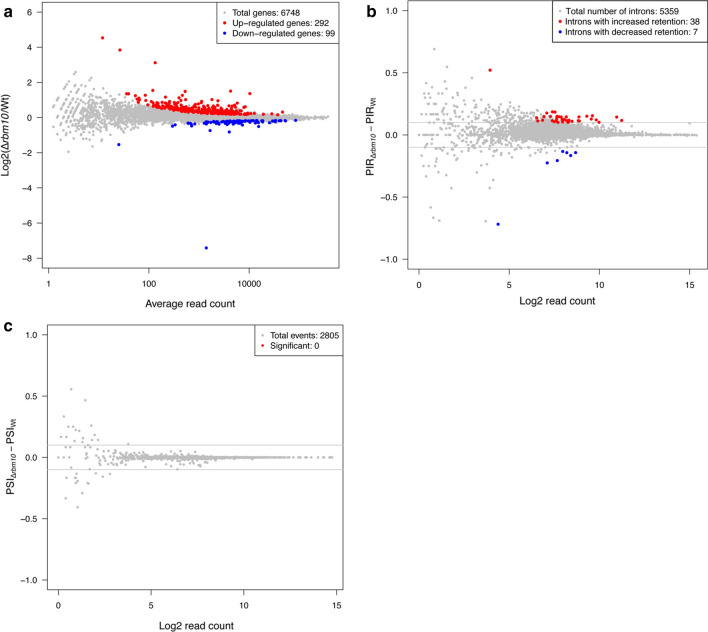
To investigate how Rbm10 affects gene expression and splicing, we performed genome-wide RNA-seq in the rbm10Δ mutant. We reproducibly found that out of 6748 genes, the expressions of 292 genes in rbm10Δ cells were increased compared to wild type (Fig. 3a and Additional file 4: Table S3), whereas the gene expressions of 99 genes were reduced in the mutant (Fig. 3a and Additional file 5: Table S4). However, the fold-changes of these gene expressions were very modest, with only 21 (0.3%) genes whose expression altered more than twofold. Our RNA-seq also allowed us to analyze the effect of rbm10Δ mutant on the intron splicing. Out of 5357 expressed introns, we detected 38 intron retention events in rbm10Δ mutant (Percent Intron Retained difference (ΔPIR) > 10%, Fig. 3b and Additional file 6: Table S5). We also found that 7 intron splicing have been enhanced (ΔPIR < 10%, Fig. 3b and Additional file 7: Table S6). Again, the majority of these changes were affected to a modest level. Previous studies have shown that alternative splicing exists in fission yeast, although not very common [40]. Given that human RBM10 is involved in alternative exon usage [26], we also examined alternative exon usage in rbm10Δ mutant. However, our results show that no case with significant difference between WT and rbm10Δ mutant can be detected (Fig. 3c). Taken together, our results show that deletion of Rbm10 has minor effect on gene expression and splicing.
Fig. 3.
Deletion of Rbm10 has minor effect on gene expression and intron retention. a MA plot comparing the gene expression patterns between rbm10Δ cells and WT. The average read count for each gene was plotted on the X axis, and the log2 transformed fold change was on the Y axis. The red and blue dots represented the genes with significantly different gene expression patterns. b MA plot comparing the intron retention patterns between rbm10Δ cells and WT. Log2 transformed read count for each event was plotted on the X axis, and the PIR difference was on the Y axis. The red and blue dots represented the events with significantly different intron retention patterns. The grey lines indicate 10% PIR difference. c MA plot comparing the exon skipping patterns between rbm10Δ cells and WT. Log2 transformed read count for each event was plotted on the X axis, and the Percent Spliced In difference (ΔPSI) was on the Y axis. The red dots represented the events with significantly different exon skipping patterns. The grey lines indicate 10% PIR (Percent Intron Retained) difference
Wang et al. proposed that human RBM10 promotes exon skipping via binding in the vicinity of splice sides, resulting in delaying their splicing choice; thus, these exons with relatively weaker splice side become more excluded during alternative splicing [26] . However, alternative splicing is a very rare event in fission yeast [40]. It is likely that a delay in splicing resulting from Rbm10 splicing activity does not manifest in alternative splicing, but rather in intron retention. Our data showing a dramatic increase in intron retention in S. pombe cells overexpressing Rbm10 are consistent with this idea.
Deletion of rbm10+ leads to impaired heterochromatin silencing
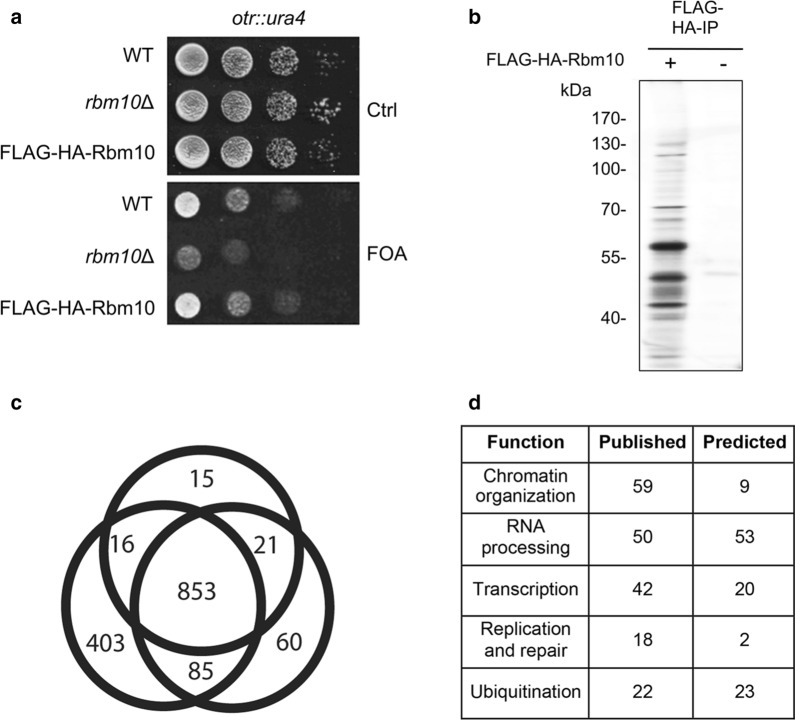
Splicing factors have been implicated in heterochromatin assembly. We next characterized how Rbm10 affects heterochromatin status. Fission yeast harbors three constitutive heterochromatic regions, namely pericentromeres, telomeres, and the mating-type locus. To analyze pericentromeric silencing in the rbm10Δ mutant, a wild-type (WT) strain carrying a ura4+ reporter integrated in the pericentromeric outer repeat (otr) was crossed with the mutant. The resulting rbm10Δ cells with ura4+ reporter at otr region were analyzed by growth assays in the medium without uracil. The rbm10Δ cells grow significantly faster than WT (Fig. 4a), indicating that the pericentromeric silencing in the mutant is disrupted. The strains were further analyzed in the media containing 5-fluoro-orotic acid (FOA), which is toxic to cells expressing uracil. WT cells exhibit strong silencing in otr region, as indicated by their robust growth in the FOA media. The slow growth of the rbm10Δ cells in the FOA media (Fig. 4a) confirmed that the silencing at pericentromeric heterochromatin in rbm10Δ mutant is lost.
Fig. 4.
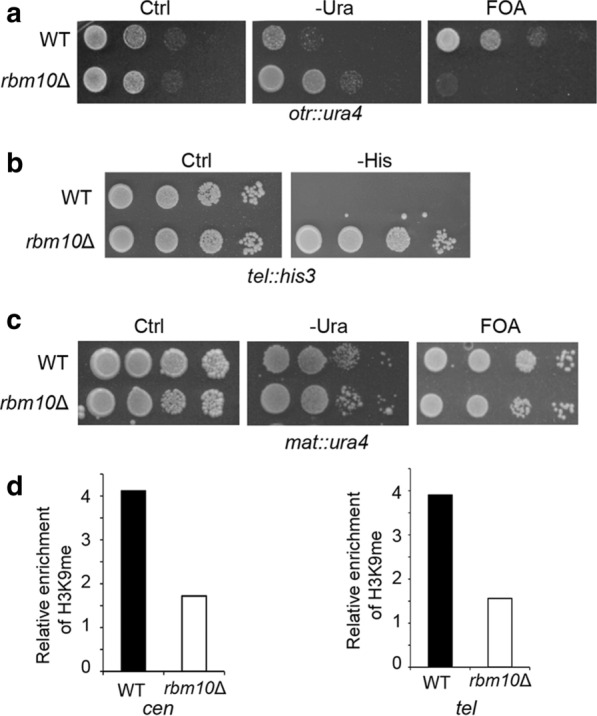
Rbm10 is important for heterochromatin silencing. a Serial dilutions of indicated cells harboring ura4+ at the otr region were plated in the PMG minimal medium without uracil (-Ura) or the EMM medium with the counter-selective FOA. Ctrl, a control plate-containing uracil. WT, wild type. b Serial dilutions of rbm10Δ cells and WT as control carrying his3+ at the telomeric region (tel) were plated in the PMG medium without histidine (-His). c Growth assays of rbm10Δ and WT cells carrying ura4+ at the mating-type region in the PMG medium without uracil (-Ura) or with the FOA media. d Analysis of H3K9 methylation in the otr region by ChIP. ChIP assays were performed using an antibody against H3K9 di-methylation and primers specific for in the otr region. act1+ was used as a control
We also examined the heterochromatin silencing in telomere and mating-type locus in rbm10Δ using his3+ and ura4+ reporters inserted to these regions, respectively. Our growth assays using minimum media without histidine demonstrated that silencing in telomere is strongly lost in rbm10Δ cells (Fig. 4b). However, rbm10Δ cells with ura4+ in mating-type locus did not show major growth deficiency relative to WT in minimum media without uracil and the counter-selective FOA medium (Fig. 4c), indicating that the heterochromatin silencing in this region is not significantly affected by deletion of rbm10+. This different effect between centromere and mating-type heterochromatin in rbm10Δ is likely due to the redundant pathways involved in heterochromatin formation in mating-type locus [41, 42].
We next analyzed H3K9me in centromeres and telomeres using ChIP. We observed that H3K9me level is reduced to approximately 50% of WT at both pericentromeres and telomeres in rbm10Δ (Fig. 4d). The loss of H3K9me at pericentromeres and telomeres is consistent with our growth assays, indicating that heterochromatin silencing is impaired in these regions in rbm10Δ.
It has been shown that mutants of splicing factors, such as Cwf14, can induce mis-splicing of mRNA transcripts from heterochromatin factors, including Ago1, which in turn results in heterochromatin defect [25]. However, our transcriptome profiling from the rbm10Δ mutant showed the minor differences in rbm10Δ in intron retention, indicating that a secondary effect by rbm10Δ via the splicing of heterochromatin factors is unlikely to be the primary cause for heterochromatin defects.
Rbm10 interacts with Clr6 and is important for recruitment of Clr6 to heterochromatin
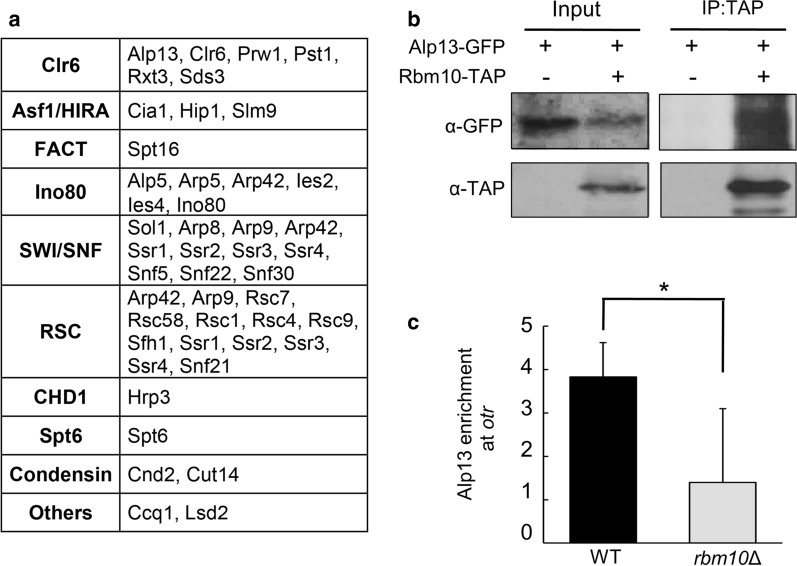
To gain more insight into the function of Rbm10, we determined to identify its interacting proteins using the Tandem Affinity Purification (TAP) method. For this, we constructed a TAP-tag with a combination of FLAG and HA tags at the N-terminus of rbm10+ at its endogenous locus. FLAG-HA-Rbm10 is fully functional, as it does not cause any silencing defects in the pericentromeric otr region (Fig. 5a). However, we observed that the expression level of endogenous FLAG-HA-Rbm10 is extremely low (Data not shown). We thus constructed FLAG-HA-Rbm10 under the nmt1 promoter, and overexpressed FLAG-HA-Rbm10 for TAP-tag purification. We collected cells within 24 h of induction to minimize the potential secondary effects. We performed a two-step affinity purification of FLAG-HA-Rbm10 and subjected the sample to mass spectrometry (MS) analysis (Fig. 5b). From three independent affinity purifications, we obtained 853 interacting proteins (Fig. 5c and Additional file 8: Table S7) found in all replicates but being absent from control purifications. These proteins included factors involved in heterochromatin silencing, chromatin remodeling, and RNA processing, and transcription (Figs. 5d and 6a). GO Term analysis with PANTHER further revealed an overrepresentation of RNA metabolic process (p = 9.75 × 10–5), chromatin organization (p = 2.2 × 10–16), and DNA-dependent transcription (p = 1.06 × 10–2).
Fig. 5.
Mass spectrometry analysis of TAP-tag purified Rbm10. a Cells carrying FLAG-HA tagged Rbm10 under its own promoter expressed from its endogenous locus display no detectable silencing defect. Serial dilutions of indicated cells harboring ura4+ at the otr region were plated in the EMM medium containing FOA. Ctrl, a control plate without FOA. b A silver-stained gel showing the TAP-tag purification of Rbm10 and a control purification from an untagged strain. c Venn diagram showing the overlap of proteins detected by mass spectrometry. d The proteins identified by mass spectrometry were grouped based on their functions, as indicated in the table
Fig. 6.
Rbm10 interacts with Clr6 and chromatin remodeling complexes. a Components of HDAC Clr6 and chromatin remodeling complexes identified by mass spectrometry analysis of Rbm10 were listed. Condensin components and other known heterochromatin factors identified were also shown. b Cell lysates from cells expressing Alp13-GFP and Rbm10-TAP under their own promoter at their native sites were immunoprecipitated with an antibody specific for TAP, and analyzed by immunoblotting using a GFP antibody. Cells expressing Alp13-GFP alone were used as a control. c Analysis of the association of Alp13-GFP in the otr region in the indicated strains by ChIP. ChIP assays were performed using an antibody against GFP. act1+ was used as a control. Error bars indicate SD. (*) p < 0.05
Clr6 HDAC complexes are important for heterochromatin silencing, and fall in two main classes I and II in fission yeast [18]. Five members from the HDAC Clr6 I complex and also Alp13 from the Clr6 II complex can be found to be associated with Rbm10 (Fig. 6a and Additional file 8: Table S7). Particularly, Prw1, a component of the Clr6 complex I, achieves the highest percentage of sequence coverage identified from our MS data.
We note that all the TAP purification experiments were performed using cells expressing FLAG-HA-Rbm10 to a higher level than endogenous Rbm10. To rule out the possibility that the interactions we identified are the artifacts caused by overexpression of Rbm10, we performed co-immunoprecipitation (Co-IP) to validate the interactions of Rbm10-TAP with Alp13-GFP, both of whom are expressed under their own promoter at the endogenous sites.
It has been shown that Alp13 is important for heterochromatin silencing [43]. Rbm10-TAP and Alp13-GFP do not causes any detectable defect (data not shown) and are thus functional. Our co-IP experiments confirmed the interaction of Rbm10 with Alp13 (Fig. 6b). To determine how Rbm10 affects the recruitment of the Clr6 complex to heterochromatin, we analyzed the rbm10Δ mutant carrying Alp13-GFP by ChIP assays. Our results showed that Alp13-GFP in the pericentromeric otr region is significantly reduced in rbm10Δ (Fig. 6c and Additional file 9: Figure S2), whereas deletion of Rbm10 has little effect on association of Alp13-GFP with the rDNA region (Additional file 10: Figure S3). These data indicate that Rbm10 is important for recruitment of the Clr6 complex to pericentromeric heterochromatin.
In addition, we found that Rbm10 is copurified with Asf1/Cia1 and the HIRA histone chaperone complex (Fig. 6a). It has been shown that Asf1 interacts with HIRA to mediate heterochromatin silencing [44, 45]. We also found that Rbm10 associates with FACT, Spt6 and CHD1/Hrp3 chromatin remodelers. These chromatin remodeling factors have also been shown to be important for heterochromatin formation [46–48]. We detected a strong link of Rbm10 with RSC, SWI/SNF, and Ino80 chromatin remodeling complexes. All 13 members of the RSC complex and 11 of 12 members of the SWI/SNF complex were found in the MS results (Fig. 6a and Additional file 8: Table S7). We also found that condensin components in the Rbm10-purified product. Condensin has been implicated in pericentromeric heterochromatin assembly in fission yeast (10, 49, 50). In addition, Rbm10 interacts with Uhp1, an ubiquitinated histone-like protein, and Lsd2, a conserved histone demethylase, both of which are linked to heterochromatin formation [51, 52]. Rbm10 is also copurified together with Ccq1, which is required for silencing in telomere and facultative heterochromatin [53, 54]. Interestingly, Rbm10 associates with transcription factors, including all five members of the PAF complex, and the transcriptional activator Byr3, which was detected with a sequence coverage of 65% and the highest intensity-to-mass ratio in the whole dataset. Heterochromatin in fission yeast can be transcribed during S phase of the cell cycle, and the non-coding transcripts are processed into siRNAs by RNAi machinery [10, 14]. These transcription factors may be involved in regulation of heterochromatin transcription.
Rbm10 interacts with splicing factors
Our MS analysis also detected more than 30 splicing factors interacting with Rbm10 (Table 1). Most of them are from the spliceosome A and B components, such as U1 snRNP, Sf3a and Sf3b, U2AF and NTC complex. This finding is in concordance with interaction data from human RBM10, which has been shown to be part of the spliceosome A and B complex [31–33]. These results demonstrate that Rbm10 interaction with the spliceosome is conserved between human and fission yeast. Our data further indicate that, even though rbm10Δ mutant shows only modest changes in splicing, Rbm10 in fission yeast nevertheless is involved in RNA splicing, consistent with the Rbm10 overexpression data (Fig. 3).
Table 1.
Splicing factors identified by mass spectrometry analysis of purified Rbm10
| ID | Name | Description |
|---|---|---|
| SPAC4D7.13 | Usp104 | U1 snRNP-associated protein Usp104 |
| SPBC4B4.09 | Usp105 | U1 snRNP-associated protein Usp105 |
| SPBC839.10 | Usp107 | U1 snRNP-associated protein Usp107 |
| SPAP8A3.06 | Uaf2 | U2AF small subunit, U2AF-23 |
| SPAC9.03c | Brr2 | U5 snRNP complex subunit Brr2 |
| SPAC644.12 | Cdc5 | Cell division control protein, splicing factor Cdc5 |
| SPBC19C2.01 | Cdc28 | ATP-dependent RNA helicase Prp8 |
| SPBC16A3.18 | Cip1 | RNA-binding protein Cip1 |
| SPBC215.12 | Cwf10 | U5 snRNP GTPase subunit Cwf10 |
| SPCP1E11.07c | Cwf18 | Complexed with Cdc5 protein Cwf18 |
| SPAC3A12.11c | Cwf2 | RNA-binding protein Cwf2 |
| SPBC211.02c | Cwf3 | complexed with Cdc5 protein Cwf3 |
| SPBC31F10.11c | Cwf4 | complexed with Cdc5 protein Cwf4 |
| SPCC550.02c | Cwf5 | RNA-binding protein Cwf5 |
| SPBC354.12 | Gpd3 | Glyceraldehyde 3-phosphate dehydrogenase Gpd3 |
| SPBC29A10.10c | Dbl8 | tRNA-splicing endonuclease positive effector Dbl8 |
| SPAC23H3.02c | Ini1 | RING finger-like protein Ini1 |
| SPBC6B1.07 | Prp1 | U4/U6 x U5 tri-snRNP complex subunit Prp1 |
| SPAC27F1.09c | Prp10 | U2 snRNP-associated protein Sap155 |
| SPCC10H11.01 | Prp11 | ATP-dependent RNA helicase Prp11 |
| SPAPJ698.03c | Prp12 | U2 snRNP-associated protein Sap130 |
| SPAC29A4.08c | Prp19 | Ubiquitin-protein ligase E4 |
| SPBC1861.04c | Prp24 | RNA-binding protein Prp24 (predicted) |
| SPBC119.13c | Prp31 | U4/U6 × U5 tri-snRNP complex subunit Prp31 |
| SPBC16H5.10c | Prp43 | ATP-dependent RNA helicase Prp43 |
| SPBP22H7.07 | Prp5 | WD repeat protein Prp5 |
| SPAC227.12 | Rna4 | U4/U6 × U5 tri-snRNP complex subunit Prp4 family, Rna4 |
| SPAC19G12.07c | Rsd1 | RNA-binding protein Rsd1 (predicted) |
| SPAC22A12.09c | Sap114 | U2 snRNP subunit Sap114 |
| SPBC29A3.07c | Sap14 | U2 snRNP-associated protein SF3B14 Sap14 |
| SPAC22F8.10c | Sap145 | U2 snRNP-associated protein Sap145 |
| SPBC36.09 | Sap61 | U2 snRNP-associated protein sap61 |
| SPAC4F8.12c | Spp42 | U5 snRNP complex subunit Spp42 |
| SPAC4D7.13 | Usp104 | U1 snRNP-associated protein Usp104 |
| SPBC4B4.09 | Usp105 | U1 snRNP-associated protein Usp105 |
| SPBC839.10 | Usp107 | U1 snRNP-associated protein Usp107 |
Discussion
Previous studies have shown that human RBM10 is an RNA splicing protein, responsible for the X-linked TARP syndrome, although the mechanisms by which Rbm10 causes the disease remain unclear [26, 27, 29]. In this study, we identified its homologue Rbm10 in fission yeast. We showed that Rbm10 in fission yeast interacts with a variety of splicing factors, and overexpression of Rbm10 results in strong intron retention. These data suggest that the role of Rbm10 in splicing regulation is conserved. Surprisingly, our genome-wide RNA analysis showed that deletion of rbm10+ in fission yeast results in minor splicing defects. However, heterochromatin formation is severely disrupted in rbm10 mutant. Our data strongly support that abnormality in heterochromatin formation in the mutant arises independently from defects in splicing. We further demonstrated that Rbm10 interacts with heterochromatin factors, including Clr6 complex and chromatin remodelers, suggesting a novel mechanism underlying splicing factor-mediated heterochromatin silencing. Our findings reveal previously unrecognized features of Rbm10 in chromatin regulation, and may help elucidate its roles in human disease.
Role of Rbm10 in heterochromatin formation
As in mammals, heterochromatin in fission yeast is enriched with hypoacetylation of histones and H3K9 methylation. These heterochromatin hallmarks are instrumental for heterochromatin structure and function [1, 3]. In fission yeast, siRNAs generated from heterochromatin transcripts during S phase promotes H3K9me by guiding the CLRC complex containing the H3K9 methyltransferase, Clr4, to heterochromatin [10, 11, 14]. It has been shown that splicing factors are required for RNAi-mediated heterochromatin assembly in fission yeast. However, the mechanism behind the role of splicing factors in heterochromatin assembly remains poorly understood. There is an ongoing discussion in the field as to whether splicing factors participate in heterochromatin formation directly. A study by Bayne et al. has shown that in splicing factor mutants, such as prp10-1, splicing and heterochromatin defects can be uncoupled. In addition, the heterochromatin defects in the prp10-1 mutant cannot rescued by expression of wild-type RNAi genes, ago1+ and hrr1+, which contain introns. They further found that an RNAi component, Cid12, interacts with splicing factors, including Prp10. They proposed that splicing factors interact with the RNAi machinery to directly regulate RNAi-mediated heterochromatin silencing [23]. However, a comprehensive analysis of global splicing changes in these studies was missing, leaving open the possibility that splicing changes of a yet unknown heterochromatin factor might be responsible for the heterochromatin defects. Indeed, the study by Kallgren et al. has shown that splicing factor mutants, such as cwf14, result in the splicing defects in Ago1, which in turn lead to heterochromatin defects [25].
In this study, we performed the first comprehensive analysis on global splicing changes in a splicing mutant showing heterochromatin defects, which could potentially rule out any indirect influences. We show that deletion of rbm10+ has little effect on splicing, but leads to significant loss of silencing and H3K9 methylation in heterochromatin. In addition, Rbm10 interacts with a variety of heterochromatin factors, notably the HDAC Clr6 complex, and chromatin remodeling HIRA, FACT, CHD1, and Spt6 components. Furthermore, deletion of rbm10+ disrupts the association of the Clr6 complex with heterochromatin. These findings strongly suggest that Rbm10 is directly involved in heterochromatin formation. Rbm10-interacting splicing factor Cwf10 has been shown to be associated with pericentromeric transcripts [23]. We propose that Rbm10 binds nascent heterochromatic transcripts, and serves as a platform to recruits the Clr6 complex and chromatin remodelers to heterochromatic region, which in turn promote heterochromatin assembly. Our study, together with previous findings, further suggests that different splicing subunits may play separate roles in regulation of heterochromatin formation.
Role of Rbm10 in regulation of RNA splicing
In addition to its role in heterochromatin formation, our data suggest conserved function of Rbm10 in splicing. We detected that a plethora of Rbm10-interacting partners in fission yeast are from spliceosomal A and B complexes, which recapitulate the known interactions of human RBM10. Furthermore, overexpression of Rbm10 in fission yeast leads to massive intron retention, which can be seen as an equivalent to human RBM10’s function in promoting exon skipping in humans. These results suggest that fission yeast and human Rbm10 have retained similar function in RNA splicing.
However, we only observed the strong effect of Rbm10 on splicing in fission yeast when it is overexpressed. In the deletion mutant strain, only very modest changes in splicing were detected, indicating a mild impact of Rbm10 on splicing at its endogenous expression level. We observed that Rbm10 expression level appears to be extremely low under normal cell growth condition. Given the fact that stoichiometric amounts of splicing factors are needed for splicing, our data indicate that Rbm10 does not serve as a general splicing factor under endogenous expression level. We speculate that higher expression of rbm10+ may be induced under specific environmental conditions, leading to changes of gene expression through altering RNA splicing patterns. Due to its strong toxicity in overexpression, it is likely that rbm10+ expression level is tightly controlled and its dual function might be regulated dose-dependently.
Interactome of Rbm10
We show that Rbm10 interacts with around 850 proteins, with enrichment for splicing factors, HDAC, chromatin remodelers, and transcription factors. These data strongly support the direct role of Rbm10 in heterochromatin formation and splicing. A recent study showed that a splicing factor in Arabidopsis also interacts with HDAC [55]. Both of these processes are involved with various large protein complexes, explaining the high number of interacting partners. Especially, the good correlation between fission yeast and human RBM10 in regard to its interaction with the spliceosome can be seen as a proof-of-principle experiment.
Rbm10 interacts with transcription factors, especially Byr3, which has the highest sequence coverage of the whole dataset. Since heterochromatin formation is also dependent on transcription during S phase, we speculate that this factor may play a role in mediating heterochromatin transcription. Its exact role in heterochromatin regulation still needs further characterization in future studies. Interestingly, we also found that Rbm10 interacts with multiple replication factors (Fig. 5d). Replication factors have also been implicated in heterochromatin and centromere assembly in fission yeast [14, 22, 56–59].
Comparison between fission yeast and human RBM10
The fact that Rbm10 interacts with the spliceosome and its overexpression results in intron retention indicates that the function of Rbm10 as a splicing factor is conserved between human and fission yeast. However, human RBM10 is expressed at substantially higher level than fission yeast Rbm10. This may be due to the fact that human RBM10 regulates exon skipping that generates alternative spliced proteins, but fission yeast Rbm10, on the other hand, is mainly responsible for intron retention, which can lead to premature block of transcription and RNA degradation via nonsense-mediated RNA decay (NMD) pathway [60]. Overexepression and knock-down of human RBM10 lead to massive changes in gene expression [26,27]. Overexepression of human RBM10 also results in nuclear condensation [61]. However, it is still not clear whether the defects are evoked by secondary effects, or result from that fact that human RBM10 has a role in heterochromatin formation and gene silencing. Nevertheless, human RBM10 was found to be part of H2A deubiquitinase complex [36], which has been implicated in heterochromatin regulation [62]. Future work is needed to elucidate whether human RBM10 has a dual function in the regulation of splicing and heterochromatin formation.
Materials and methods
Fission yeast strains and genetic analysis
rbm10Δ, rbm10-GFP, and FLAG-HA-rbm10 strains were constructed via a PCR-based homologous recombination and verified by PCRs. pREP1-FLAG-HA-rbm10 was constructed by inserting their respective cDNAs into NdeI/BamHI or SalI/SmaI sites in the plasmid. Genetic crosses were conducted following standard protocols [63]. Fission yeast strains used in this study are listed in Additional file 11: Table S8. For silencing assays, a series of tenfold dilutions with a starting concentration of 2 × 107 cells/ml were spotted on the designated PMG (Pombe glutamate medium) media or the EMM (Edinburgh Minimal Medium) media with FOA and incubated at 30 °C for 2–3 days.
RNA sequencing
RNA sequencing was performed as described [26]. Briefly, poly (A) RNA was prepared from total RNA by two rounds of oligo (dT)25 Dynabeads (Invitrogen) purification. Following fragmentation at 94 °C for 3.5 min, the RNA was converted to first strand cDNA using random hexamer primer and Superscript II (Invitrogen), followed by second-strand cDNA synthesis with Escherichia coli DNA pol I (Invitrogen) and RNAse H (Invitrogen). The barcoded sequencing library was prepared and sequenced on Illumina HiSeq for 1 × 100 cycles following the standard protocol.
RNA-seq data analysis
RNA-seq reads that pass the Illumina filter were aligned to the S. pombe genome reference sequences (version: ASM294v2) using TopHat with default mapping parameters (version 2.0.8) [64]. The read count for each exon–exon and/or exon–intron junction as well as each gene was produced by custom Perl script. DESeq package was used to identify the differentially expressed genes between rbm10Δ cells and WT [65]. Adjusted p value < 0.05 was used as the threshold for determining significance. Fisher’s exact test was used to compare the splicing pattern of each exon (for exon skipping) or each intron (for intron retention) between rbm10Δ cells and WT. Adjusted p value < 0.05 and |ΔPSI| (or |PIR|) > 10% was used as the threshold for determining significant splicing difference.
ChIP
ChIP was performed by as previously described (66). Briefly, 50 ml of a log-phase yeast culture was cross-linked by adding 37% formaldehyde for 30 min. Cells were collected and sonicated by an Ultrasonic Processor. 1 µl of H3K9me antibody (Abcam ab1220) was used for immunoprecipitation. Immunoprecipitated DNA was purified using a PCR clean up column (Qiagen), and analyzed by PCR using primers listed in Additional file 12: Table S9. The experiments were replicated in two or three independent biological repeats.
RT-PCR
RT-PCR was performed by as previously described with minor modifications [67]. 500 ng starting material of total RNA was used per reaction. First-strand synthesis was performed with Random Primer and Superscript II RT (Invitrogen) at 25 °C for 10 min, then 42 °C for 50 min and 70 °C for 15 min. For the subsequent PCR, 1 µl from the reverse transcriptase reaction was used. Oligos used are listed in Additional file 12: Table S9.
TAP-tag purification and mass spectrometry
Cells carrying pREP1-FLAG-HA-rbm10 was induced for 24 h in minimal media without thiamine. Cell were collected and lysed in lysis buffer with 4 µl/ml Benzonase by bead beating. Protein G beads coated with anti-FLAG-antibody were added to the cell lysate, and incubated for 1 h at 4 °C on rotation. After washing, the sample was eluted using 3 × FLAG peptide (Sigma). The eluate was subsequently incubated with uMACS HA magnetic beads on ice for 30 min with occasionally mixing. The eluate with the beads was loaded on a uMACS column, and washed three times with wash buffer I and twice with wash buffer II. To prepare the sample for MS, proteins on the column were predigested by adding 25 µl 2 M urea in 100 mM Tris–HCl pH 7.5, 1 mM DTT and 150 ng Trypsin for 30 min at RT. The sample was eluted by adding 2 times 50 µl 2 M urea in 100 mM Tris–HCl, pH 7.5, and 5 mM iodoacetamide. The sample was digested overnight at RT. 1 µl trifluoroacetic acid was added to stop the digestion. Mass spectrometry analysis was conducted in the Proteomics platform at Berlin Institute for Medical Systems Biology.
Mass spectrometry data processing and analysis
Raw data were analyzed using the MaxQuant proteomics pipeline (v1.4.0.5) and the built in the Andromeda search engine (Cox, Neuhauser et al. 2011) with the PomBase database, Carbamidomethylation of cysteines was chosen as fixed modification, oxidation of methionine, and acetylation of N-terminus were chosen as variable modifications. The search engine peptide assignments were filtered at 1% FDR. The feature match between runs was left disabled and other parameters were left as default.
Western blot analysis
Western blot was performed by as previously described [68]. Briefly, cell extracts from exponentially growing cells were collected. Extracted proteins were separated on SDS-polyacrylamide gels and blotted onto PVDF membranes. Blots were probed with indicated antibodies.
Co-immunoprecipitation (Co-IP)
Co-immunoprecipitation was conducted as described (59). Cells were collected and resuspended in 100 µl of 1 × lysis buffer with protease inhibitors prior to lysis by bead beating. Lysates were incubated with lgG sepharose (GE Healthcare) at 4 °C for 2 h. After washing with lysis buffer three times, proteins were eluted in SDS loading buffer. Eluates were analyzed by Western blotting using a commercial anti-GFP antibody (Abcam, ab290).
Microscopy
Cells were imaged using the DeltaVision System (Applied Precision, Issaquah, WA). Images were taken as z-stacks of 0.2-µm increments with an oil immersion objective (× 100). Standard DAPI staining and analysis methods for fission yeast nuclei were used.
Data access
All genomic data were available at the GEO database under the accession number GSE93906.
Supplementary Information
Additional file 1: Figure S1. Sequence alignment of fission yeast Rbm10 (YDMD_SCHPO) with Rbm10 orthologs in Callithrix jacchus, human, Macaca mulatta, mouse and rat.
Additional file 2: Table S1. List of genes showing increased intron retention upon Rbm10 overexpression.
Additional file 3: Table S2. List of genes showing decreased intron retention upon Rbm10 overexpression.
Additional file 4: Table S3. List of genes that are up-regulated in Δrbm10 mutant.
Additional file 5: Table S4. List of genes that are down-regulated in Δrbm10 mutant.
Additional file 6: Table S5. List of genes showing increased intron retention in Δrbm10 mutant.
Additional file 7: Table S6. List of genes showing decreased intron retention in Δrbm10 mutant.
Additional file 8: Table S7. List of protein identified as interacting partners of Rbm10.
Additional file 9: Figure S2. Western blot analysis of indicated cells expressing Alp13-GFP using an anti-GFP antibody. Ponceau staining was used as a loading control.
Additional file 10: Figure S3. Analysis of the association of Alp13-GFP with the rDNA region in the indicated strains by ChIP. ChIP assays were performed using an antibody against GFP. act1+ was used as a control.
Additional file 11: Table S8. Strains used in this study.
Additional file 12: Table S9. List of primers used in this study.
Acknowledgements
We thank Qianhua Dong and Jinpu Yang for critical reading of the manuscript. We are grateful to the Japan Yeast Genetic Resource Center for kindly providing strains.
Authors’ contributions
FL and WC designed experiments and wrote the manuscript. MW and HB performed experiments with assistance from QG, and HH. GM and SK performed the mass spectrometry analysis. All authors read and approved the final manuscript.
Funding
This work was supported by National Science Foundation [MCB-1330557 and MCB-1934628 to F.L.].
Ethics approval and consent to participate
Not applied.
Consent for publication
Not applied.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Martina Weigt, Qingsong Gao, and Hyoju Ban have contributed equally to this work as first authors
Contributor Information
Wei Chen, Email: chenw@sustc.edu.cn.
Fei Li, Email: fl43@nyu.edu.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13072-021-00382-y.
References
- 1.Martienssen R, Moazed D. RNAi and heterochromatin assembly. Cold Spring Harb Perspect Biol. 2015;7:a019323. doi: 10.1101/cshperspect.a019323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elgin SC, Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb Perspect Biol. 2013;5:a017780. doi: 10.1101/cshperspect.a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, Jia ST, Jia S. New Insights into the Regulation of Heterochromatin. Trends Genet. 2016;32:284–294. doi: 10.1016/j.tig.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li F, Goto DB, Zaratiegui M, Tang X, Martienssen R, Cande WZ. Two novel proteins, Dos1 and Dos2, interact with Rik1 to regulate heterochromatic RNA interference and histone modification. Curr Biol. 2005;15:1448–1457. doi: 10.1016/j.cub.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 5.Horn PJ, Bastie JN, Peterson CL. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 2005;19:1705–1714. doi: 10.1101/gad.1328005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia S, Kobayashi R, Grewal SI. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol. 2005;7:1007–1013. doi: 10.1038/ncb1300. [DOI] [PubMed] [Google Scholar]
- 7.Thon G, Hansen KR, Altes SP, Sidhu D, Singh G, Verhein-Hansen J, Bonaduce MJ, Klart AJS. The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe. Genetics. 2005;171:1583–1595. doi: 10.1534/genetics.105.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong EJ, Villen J, Gerace EL, Gygi SP, Moazed D. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3–K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2005;2:106–111. doi: 10.4161/rna.2.3.2131. [DOI] [PubMed] [Google Scholar]
- 9.Li F, Huarte M, Zaratiegui M, Vaughn MW, Shi Y, Martienssen R, Cande WZ. Lid2 Is Required for coordinating H3K4 and H3K9 methylation of heterochromatin and euchromatin. Cell. 2008;135:272–283. doi: 10.1016/j.cell.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SIS. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 11.Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 14.Li F, Martienssen R, Cande WZ. Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature. 2011;475:244–248. doi: 10.1038/nature10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaratiegui M, Castel SE, Irvine DV, Kloc A, Ren J, Li F, de Castro E, Marin L, Chang AY, Goto D, et al. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature. 2011;479:135–138. doi: 10.1038/nature10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez M, Li F. DNA replication, RNAi and epigenetic inheritance. Epigenetics. 2012;7:14–19. doi: 10.4161/epi.7.1.18545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bjerling P, Silverstein RA, Thon G, Caudy A, Grewal S, Ekwall K. Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol Cell Biol. 2002;22:2170–2181. doi: 10.1128/MCB.22.7.2170-2181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicolas E, Yamada T, Cam HP, Fitzgerald PC, Kobayashi R, Grewal SI. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol. 2007;14:372–380. doi: 10.1038/nsmb1239. [DOI] [PubMed] [Google Scholar]
- 19.Grewal SI, Bonaduce MJ, Klar AJ. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics. 1998;150:563–576. doi: 10.1093/genetics/150.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shankaranarayana GD, Motamedi MR, Moazed D, Grewal SI. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr Biol. 2003;13:1240–1246. doi: 10.1016/S0960-9822(03)00489-5. [DOI] [PubMed] [Google Scholar]
- 21.Buscaino A, Lejeune E, Audergon P, Hamilton G, Pidoux A, Allshire RC. Distinct roles for Sir2 and RNAi in centromeric heterochromatin nucleation, spreading and maintenance. EMBO J. 2013;32:1250–1264. doi: 10.1038/emboj.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He H, Li Y, Dong Q, Chang AY, Gao F, Chi Z, Su M, Zhang F, Ban H, Martienssen R, et al. Coordinated regulation of heterochromatin inheritance by Dpb3-Dpb4 complex. Proc Natl Acad Sci U S A. 2017;114:12524–12529. doi: 10.1073/pnas.1712961114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bayne EH, Portoso M, Kagansky A, Kos-Braun IC, Urano T, Ekwall K, Alves F, Rappsilber J, Allshire RC. Splicing factors facilitate RNAi-directed silencing in fission yeast. Science. 2008;322:602–606. doi: 10.1126/science.1164029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chinen M, Morita M, Fukumura K, Tani T. Involvement of the spliceosomal U4 small nuclear RNA in heterochromatic gene silencing at fission yeast centromeres. J Biol Chem. 2010;285:5630–5638. doi: 10.1074/jbc.M109.074393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kallgren SP, Andrews S, Tadeo X, Hou H, Moresco JJ, Tu PG, Yates JR, 3rd, Nagy PL, Jia S. The proper splicing of RNAi factors is critical for pericentric heterochromatin assembly in fission yeast. PLoS Genet. 2014;10:e1004334. doi: 10.1371/journal.pgen.1004334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Gogol-Doring A, Hu H, Frohler S, Ma Y, Jens M, Maaskola J, Murakawa Y, Quedenau C, Landthaler M, et al. Integrative analysis revealed the molecular mechanism underlying RBM10-mediated splicing regulation. EMBO Mol Med. 2013;5:1431–1442. doi: 10.1002/emmm.201302663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bechara EG, Sebestyen E, Bernardis I, Eyras E, Valcarcel J. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol Cell. 2013;52:720–733. doi: 10.1016/j.molcel.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 28.Rodor J, FitzPatrick DR, Eyras E, Caceres JF. The RNA-binding landscape of RBM10 and its role in alternative splicing regulation in models of mouse early development. RNA Biol. 2016 doi: 10.1080/15476286.2016.1247148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernandez J, Bechara E, Schlesinger D, Delgado J, Serrano L, Valcarcel J. Tumor suppressor properties of the splicing regulatory factor RBM10. RNA Biol. 2016;13:466–472. doi: 10.1080/15476286.2016.1144004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agafonov DE, Deckert J, Wolf E, Odenwalder P, Bessonov S, Will CL, Urlaub H, Luhrmann R. Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol Cell Biol. 2011;31:2667–2682. doi: 10.1128/MCB.05266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Behzadnia N, Golas MM, Hartmuth K, Sander B, Kastner B, Deckert J, Dube P, Will CL, Urlaub H, Stark H, et al. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 2007;26:1737–1748. doi: 10.1038/sj.emboj.7601631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 35.Hegele A, Kamburov A, Grossmann A, Sourlis C, Wowro S, Weimann M, Will CL, Pena V, Luhrmann R, Stelzl U. Dynamic protein-protein interaction wiring of the human spliceosome. Mol Cell. 2012;45:567–580. doi: 10.1016/j.molcel.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 36.Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, Tempst P, Glass CK, Rosenfeld MG. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell. 2007;27:609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/S0959-440X(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 38.Brown RS. Zinc finger proteins: getting a grip on RNA. Curr Opin Struct Biol. 2005;15:94–98. doi: 10.1016/j.sbi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 39.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332:930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thon G, Bjerling KP, Nielsen IS. Localization and properties of a silencing element near the mat3-M mating-type cassette of Schizosaccharomyces pombe. Genetics. 1999;151:945–963. doi: 10.1093/genetics/151.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 43.Shim YS, Choi Y, Kang K, Cho K, Oh S, Lee J, Grewal SI, Lee D. Hrp3 controls nucleosome positioning to suppress non-coding transcription in eu- and heterochromatin. EMBO J. 2012;31:4375–4387. doi: 10.1038/emboj.2012.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greenall A, Williams ES, Martin KA, Palmer JM, Gray J, Liu C, Whitehall SK. Hip3 interacts with the HIRA proteins Hip1 and Slm9 and is required for transcriptional silencing and accurate chromosome segregation. J Biol Chem. 2006;281:8732–8739. doi: 10.1074/jbc.M512170200. [DOI] [PubMed] [Google Scholar]
- 45.Yamane K, Mizuguchi T, Cui B, Zofall M, Noma K, Grewal SI. Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol Cell. 2011;41:56–66. doi: 10.1016/j.molcel.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lejeune E, Bortfeld M, White SA, Pidoux AL, Ekwall K, Allshire RC, Ladurner AG. The chromatin-remodeling factor FACT contributes to centromeric heterochromatin independently of RNAi. Curr Biol. 2007;17:1219–1224. doi: 10.1016/j.cub.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiely CM, Marguerat S, Garcia JF, Madhani HD, Bahler J, Winston F. Spt6 is required for heterochromatic silencing in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol. 2011;31:4193–4204. doi: 10.1128/MCB.05568-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jae Yoo E, Kyu Jang Y, Ae Lee M, Bjerling P, Bum Kim J, Ekwall K, Hyun Seong R, Dai Park S. Hrp3, a chromodomain helicase/ATPase DNA binding protein, is required for heterochromatin silencing in fission yeast. Biochem Biophys Res Commun. 2002;295:970–974. doi: 10.1016/S0006-291X(02)00797-0. [DOI] [PubMed] [Google Scholar]
- 49.He H, Zhang S, Wang D, Hochwagen A, Li F. Condensin promotes position effects within tandem DNA repeats via the RITS complex. Cell Rep. 2016;14:1018–1024. doi: 10.1016/j.celrep.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J, Li F. Are all repeats created equal? Understanding DNA repeats at an individual level. Curr Genet. 2017;63:57–63. doi: 10.1007/s00294-016-0619-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naresh A, Saini S, Singh J. Identification of Uhp1, a ubiquitinated histone-like protein, as a target/mediator of Rhp6 in mating-type silencing in fission yeast. J Biol Chem. 2003;278:9185–9194. doi: 10.1074/jbc.M212732200. [DOI] [PubMed] [Google Scholar]
- 52.Lan F, Zaratiegui M, Villen J, Vaughn MW, Verdel A, Huarte M, Shi Y, Gygi SP, Moazed D, Martienssen RA. S. pombe LSD1 homologs regulate heterochromatin propagation and euchromatic gene transcription. Mol Cell. 2007;26:89–101. doi: 10.1016/j.molcel.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 53.Wang J, Cohen AL, Letian A, Tadeo X, Moresco JJ, Liu J, Yates JR, 3rd, Qiao F, Jia S. The proper connection between shelterin components is required for telomeric heterochromatin assembly. Genes Dev. 2016;30:827–839. doi: 10.1101/gad.266718.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zofall M, Smith DR, Mizuguchi T, Dhakshnamoorthy J, Grewal SI. Taz1-shelterin promotes facultative heterochromatin assembly at chromosome-internal sites containing late replication origins. Mol Cell. 2016;62:862–874. doi: 10.1016/j.molcel.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daras G, Rigas S, Alatzas A, Samiotaki M, Chatzopoulos D, Tsitsekian D, Papadaki V, Templalexis D, Banilas G, Athanasiadou AM, et al. LEFKOTHEA regulates nuclear and chloroplast mRNA Splicing in plants. Dev Cell. 2019;50(767–779):e767. doi: 10.1016/j.devcel.2019.07.024. [DOI] [PubMed] [Google Scholar]
- 56.He H, Gonzalez M, Zhang F, Li F. DNA replication components as regulators of epigenetic inheritance–lesson from fission yeast centromere. Protein Cell. 2014;5:411–419. doi: 10.1007/s13238-014-0049-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakayama J, Allshire RC, Klar AJS, Grewal SIS. Role for DNA polymerase alpha in epigenetic control of transcriptional silencing in fission yeast. EMBO J. 2001;20:2857–2866. doi: 10.1093/emboj/20.11.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Natsume T, Tsutsui Y, Sutani T, Dunleavy EM, Pidoux AL, Iwasaki H, Shirahige K, Allshire RC, Yamao F. A DNA Polymerase alpha Accessory protein, Mcl 1, is required for propagation of centromere structures in fission yeast. PLoS ONE. 2008;3:14. doi: 10.1371/journal.pone.0002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gonzalez M, He H, Sun S, Li C, Li F. Cell cycle-dependent deposition of CENP-A requires the Dos1/2-Cdc20 complex. Proc Natl Acad Sci USA. 2013;110:606–611. doi: 10.1073/pnas.1214874110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 61.Wang K, Bacon ML, Tessier JJ, Rintala-Maki ND, Tang V, Sutherland LC. RBM10 Modulates apoptosis and influences TNF-alpha gene expression. J Cell Death. 2012;5:1–19. doi: 10.4137/JCD.S9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Braun S, Madhani HD. Shaping the landscape: mechanistic consequences of ubiquitin modification of chromatin. EMBO Rep. 2012;13:619–630. doi: 10.1038/embor.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-L. [DOI] [PubMed] [Google Scholar]
- 64.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dong Q, Yin FX, Gao F, Shen Y, Zhang F, Li Y, He H, Gonzalez M, Yang J, Zhang S, et al. Ccp1 homodimer mediates chromatin integrity by antagonizing CENP-A loading. Mol Cell. 2016;64:79–91. doi: 10.1016/j.molcel.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang J, Sun S, Zhang S, Gonzalez M, Dong Q, Chi Z, Chen YH, Li F. Heterochromatin and RNAi regulate centromeres by protecting CENP-A from ubiquitin-mediated degradation. PLoS Genet. 2018;14:e1007572. doi: 10.1371/journal.pgen.1007572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aristizabal-Corrales D, Yang J, Li F. Cell cycle-regulated transcription of CENP-A by the MBF complex ensures optimal level of CENP-A for centromere formation. Genetics. 2019;211:861–875. doi: 10.1534/genetics.118.301745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Sequence alignment of fission yeast Rbm10 (YDMD_SCHPO) with Rbm10 orthologs in Callithrix jacchus, human, Macaca mulatta, mouse and rat.
Additional file 2: Table S1. List of genes showing increased intron retention upon Rbm10 overexpression.
Additional file 3: Table S2. List of genes showing decreased intron retention upon Rbm10 overexpression.
Additional file 4: Table S3. List of genes that are up-regulated in Δrbm10 mutant.
Additional file 5: Table S4. List of genes that are down-regulated in Δrbm10 mutant.
Additional file 6: Table S5. List of genes showing increased intron retention in Δrbm10 mutant.
Additional file 7: Table S6. List of genes showing decreased intron retention in Δrbm10 mutant.
Additional file 8: Table S7. List of protein identified as interacting partners of Rbm10.
Additional file 9: Figure S2. Western blot analysis of indicated cells expressing Alp13-GFP using an anti-GFP antibody. Ponceau staining was used as a loading control.
Additional file 10: Figure S3. Analysis of the association of Alp13-GFP with the rDNA region in the indicated strains by ChIP. ChIP assays were performed using an antibody against GFP. act1+ was used as a control.
Additional file 11: Table S8. Strains used in this study.
Additional file 12: Table S9. List of primers used in this study.