Abstract
The ongoing SARS-CoV-2 pandemic has led to the focused application of resources and scientific expertise toward the goal of developing investigational vaccines to prevent COVID-19. The highly collaborative global efforts by private industry, governments and non-governmental organizations have resulted in a number of SARS-CoV-2 vaccine candidates moving to Phase III trials in a period of only months since the start of the pandemic. In this review, we provide an overview of the preclinical and clinical data on SARS-CoV-2 vaccines that are currently in Phase III clinical trials and in few cases authorized for emergency use. We further discuss relevant vaccine platforms and provide a discussion of SARS-CoV-2 antigens that may be targeted to increase the breadth and durability of vaccine responses.
Keywords: SARS-CoV-2 vaccine, Pre-fusion S protein, ORF3a, Vaccine platform, Phase III vaccine trials, COVID-19
Graphical abstract
1. Introduction
The current international efforts toward quelling the ongoing SARS-CoV-2 pandemic are unprecedented in pace, resource allocation and scientific focus. In less than one year since the first cases of COVID-19 were identified, vaccines showing early signs of promise are already in the later phases of clinical testing and in some cases are approved for emergency use. The scientific development process has been guided largely by insights gained from platforms and viral targets that have already been used in successful anti-viral vaccines for humans [1]. Additionally, data from pre-clinical and limited clinical trials of vaccine candidates that were developed following the severe acute respiratory syndrome (SARS) outbreak in 2003 and the Middle East respiratory syndrome (MERS) outbreak in 2012, along with research from recent years characterizing new/next-generation vaccine platforms and adjuvants have all contributed to the rapid development of numerous SARS-CoV-2 vaccine candidates [[2], [3], [4]].
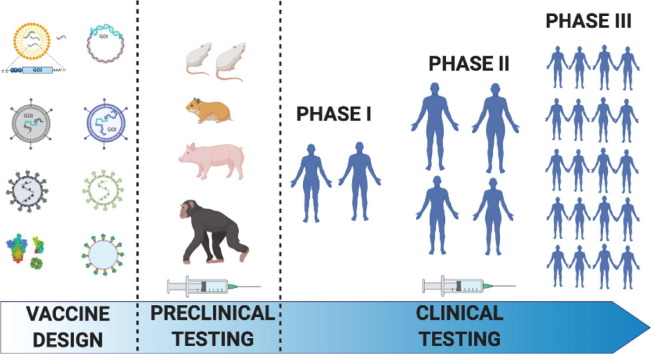
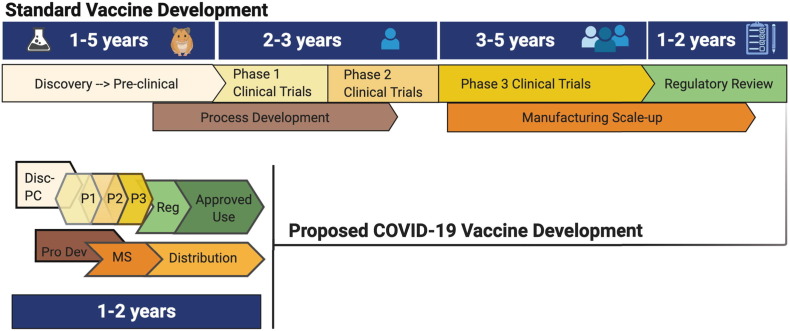
The process of vaccine research and development is typically an investment of 10–15 years from discovery to licensure, prior to manufacturing and distribution. Here, the aspirational timeline is 12–18 months (Fig. 1 ), with the help of newly created programs like HHS’ Operation Warp Speed (OWS), NIH's Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV) partnership, NIH's Rapid Acceleration of Diagnostics (RADx) initiative, and the WHO's Solidarity Vaccine Trial. All of these programs are designed to fund and coordinate the accelerated development, manufacturing and global distribution of SARS-CoV-2 pandemic countermeasures. Apart from infrastructure, there are a number of novel vaccine technologies that are accelerating the time from formulation to preclinical testing and to formal clinical trials (as reviewed by K. Ainslie, this issue).
Fig. 1.
An outline of the standard timeline for vaccine development and the expedition for SARS-CoV-2 vaccines. Standard vaccine development can take up to 15 years before a successful candidate makes it to market. Due to the expense and time of running clinical trials that meet regulatory benchmarks for approval, the length of time can vary significantly at each stage of the process. Compression of the normal timeline is being attempted in order to deliver a vaccine for SARS-CoV-2 within ~18 months of this novel pathogen being identified. Leveraging existing infrastructure for new, but as yet unproven, vaccine platforms like mRNA- and DNA-based vaccines, as well as ongoing work with related CoVs, like SARS1 and MERS, has enabled a shortened time from the preclinical data acquisition stage to almost immediate entry into Phase I clinical trials for some of the leading candidates. Additionally, conducting overlapping Phase I, II and III Clinical Trials, while scaling up the manufacturing of multiple candidates will provide support for rapid distribution once emergency approval is granted.
With the accelerated timeline for clinical testing of SARS-CoV-2 vaccines, a primary concern is ensuring their safety. In the absence of animal models or other assays that can reliably predict adverse events, SARS-CoV-2 vaccine safety is largely being assessed in human trials. An important goal for Phase III clinical trials has thus been to ensure that study cohorts are large and diverse enough so as not to miss serious safety concerns for a given vaccine candidate. At this time, few adverse events have been reported from SARS-CoV-2 vaccine clinical trials and the safety of vaccines which have been most extensively tested is promising.
In addition to safety, the clear requirement for any worthwhile vaccine is effectiveness. Given the urgent need for some level of population immunity against SARS-CoV-2, the early recommendations by the FDA set a relatively low bar for efficacy that should be observed before considering SARS-CoV-2 vaccines for wide distribution. The FDA has recommended that vaccines should show at least 50% efficacy relative to placebo, where efficacy is defined by a primary endpoint of: 1) reduction in COVID-19 cases, 2) reduction in COVID-19 severity (from severe to mild disease), or 3) reduction in SARS-CoV-2 infections [5]. Each of these endpoints has unique benefits and caveats. Determining COVID-19 cases is the most efficient and straightforward option; one caveat to this approach however is that it does not necessarily select for a vaccine that reduces mortality associated with COVID-19 since reduction in mild cases alone will pass for efficacy. Reducing disease severity is a more meaningful endpoint yet this requires substantially larger cohorts in order to achieve the statistical power to separate severe from mild cases. Finally, reduction in infections is an extremely ambitious endpoint that requires regular testing of all subjects in order to detect subclinical infections. This latter endpoint is ideal since it will necessarily limit the spread of SARS-CoV-2 and will provide the most protection for populations at greatest risk of death from this virus. Fortunately, the first data from Phase III studies suggests that SARS-CoV-2 vaccines will likely greatly outperform the 50% efficacy bar, defined at least by the number of PCR confirmed COVID-19 cases among study participants [6,7].
Given the unparalleled pace of COVID-19 vaccine development, this landscape is extremely dynamic. According to the draft landscape of COVID-19 candidate vaccines released by World Health Organization, as of November 12, 2020 there were already 48 vaccine candidates in clinical evaluation and more than 164 in various stages of preclinical evaluation. Of these, 11 candidates are currently in Phase III trials [8]. Six vaccines which have not completed Phase III trials are approved for limited or early use in Russia and China [9]. Two of the vaccines (mRNA-1273 and BNT162b2 developed by Moderna/NIAID and Pfizer/BioNTech/Fosun respectively) have been authorized for emergency use by the US Food and Drug Administration under an Emergency Use Authorization (EUA). Front-line healthcare workers and long-term nursing home residents are being prioritized for vaccination during the first phase of vaccine rollout. While both of these vaccines are mRNA-based and have been reported to confer 95% efficacy (Moderna – 94.5% (N ≈ 30,000), Pfizer – 95% (N ≈43,000)), the vaccines differ with regard to dosage, storage temperature and dosing interval for their prime/boost regimen. With more vaccines likely to be approved in the near future, it is important to note that none of the clinical studies reported to date were designed to evaluate the interaction between two different candidates. Therefore, exercising an abundance of caution, vaccine health experts suggest avoiding switching between different vaccine formulations [[10], [11], [12], [13]]. If the current advanced vaccine candidates are successful, this effort will provide a lasting infrastructure for vaccine production on novel platforms and rapid global vaccine manufacturing and distribution to facilitate the response to future pandemics.
In this review, we discuss SARS-CoV-2 vaccine targets and platforms and we have reviewed aspects of the pre-clinical and clinical data on vaccine candidates that are currently in/or have completed Phase III clinical trials (for which data are available).
2. The virus and vaccine targets
SARS-CoV-2 is an enveloped, single-stranded, and positive-sense RNA virus that belongs to a group of viral pathogens called betacoronaviruses, one of four genera within the family Coronaviridae [14,15]. Despite gaps in our knowledge about modes of possible transmission, exposure to respiratory droplets carrying infectious SARS-CoV-2 virions is thought to be the primary route of transmission [16,17]. There is still debate about the efficiency of airborne transmission via aerosols [18,19], though studies have shown that the virus is viable in aerosols for at least up to 3 h [20] and can be a source of infection when combined with factors like poor ventilation, lack of social distancing and prolonged exposure to infected people in indoor settings [19,21,22]. Other modes of transmission such as fecal-oral or contact with surfaces harboring infectious particles [20] are still being explored. Though viral RNA and in rare cases infectious virus have been detected in stool samples of COVID-19 patients [[23], [24], [25]], there are currently no reports of fecal-oral transmission of SARS-CoV-2. After productive infection, the virus has an incubation period of approximately 5–6 days [16,26,27] and there is a wide spectrum of disease severity ranging from asymptomatic (17–20% of all infections [28]) to severe COVID-19 that can cause death. In the absence of any intervention, there is efficient viral transmission in the population as evidenced by basic reproductive numbers (R0) of 2–3 [[29], [30], [31]]. Asymptomatic patients can transmit virus yet studies have shown that their transmission potential is reduced compared to transmission by pre-symptomatic and symptomatic individuals [28,32,33].
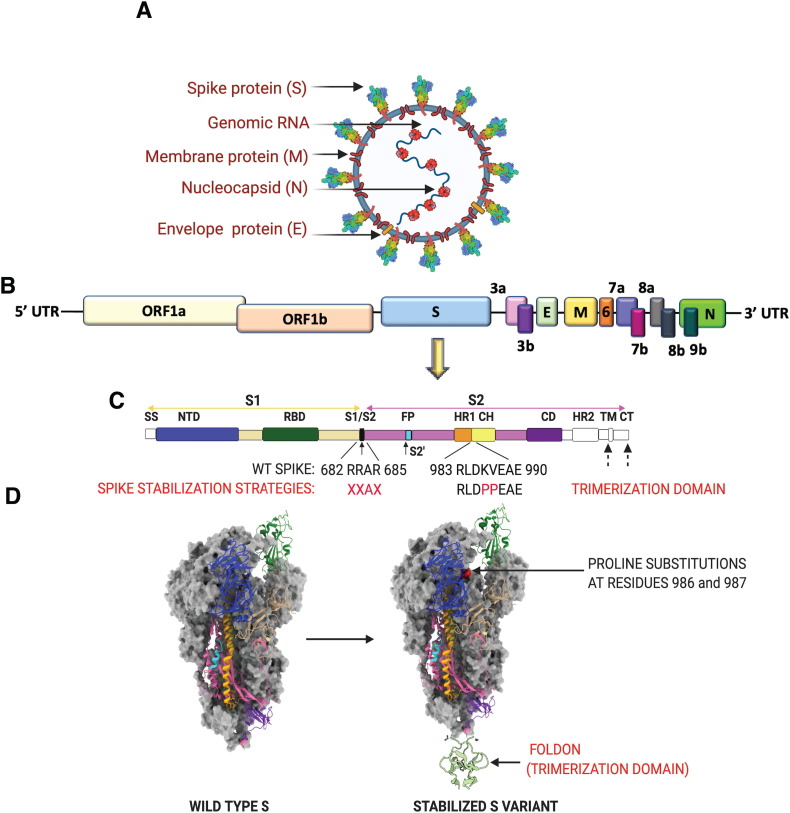
The genome of SARS-CoV-2 is approximately 30 kb with its 15 non-structural proteins located at the 5′-terminus and 12 structural and accessory proteins encoded on the 3′-terminus [34]. There are four major structural proteins, of which, three, the spike (S), the membrane (M) and the envelope (E) are expressed on the surface of the viral particle. The fourth major structural protein, nucleocapsid (N), is found within the virion associated with the viral RNA [35]. The functions of the eight SARS-CoV-2 accessory proteins encoded by open read frame (ORF) 3a, 3b, 6, 7a, 7b, 8a, 8b or 9b have not been fully characterized, but prior work with SARS-CoV-1 suggests that they are not required for replication (Fig. 2 a, b). In SARS-CoV-1, select accessory proteins have been shown to influence the pathogenicity and virulence of the virus by modulating the cytokine response, Type I interferon signaling pathways, and cellular apoptosis [36,37]. As our understanding of SARS-CoV-2 evolves – specifically concerning the nomenclature of orthologous accessory genes and overlapping genes discovered within ORFs [38] – we anticipate that new genes and proteins may yet be discovered. Of note, accessory proteins encoded by open reading frame (ORF) 3a [39] and ORF7a [40] were found on SARS-CoV-1 viral particles, but have yet to be validated for SARS-CoV-2.
Fig. 2.
SARS-CoV-2 virion, genome and strategies for stabilizing the spike protein. (A) SARS-CoV-2 virion with structural proteins (spike (S), membrane (M), nucleocapsid (N) and envelope (E)) and genome depicted. (B) Organization of the SARS-CoV-2 genome. (C) SARS-CoV-2 S protein organization. The S1 subunit (tan) consists of a 5′ signal sequence (SS) followed by the N-terminal domain (NTD) and the receptor binding domain (RBD). Arrows denote the two protease cleavage sites: the polybasic furin site between S1/ S2 and the S2′ site. Cleavage at these two sites in the S protein exposes the hydrophobic fusion peptide (FP) and triggers the fusion process. The other domains of the S2 subunit are the heptad repeat 1 (HR1), CH-central helix, CD-connector domain, heptad repeat 2 (HR2), transmembrane domain (TM) and cytoplasmic tail (CT). Domains that have no corresponding residues in the cryo-EM structures shown in (D) are colored in white. Multiple protein engineering strategies have been adopted to stabilize the pre-fusion conformation. Most of the full-length vaccine candidates have adopted one or all of the following strategies: 1) Introduction of two stabilizing proline mutations at residues 986 and 987 in the loop between HR1 and CH, 2) Removal of the polybasic cleavage site between S1 and S2, and 3) Stabilizing the trimeric spike by addition of a trimerization motif to maintain integrity of conformational epitopes. (D) Prefusion structure of the SARS-CoV-2 spike protein determined by cryo-EM (PDB ID: 6VSB). The spike is a homotrimeric protein. Two of the monomers are colored in gray, whereas the various structural domains have been mapped on one of the monomers in the identical color as shown in (C).
In this section, we have focused on describing the four structural proteins which are the major targets of the adaptive immune response during SARS-CoV-2 infection. In addition, we discuss the accessory proteins ORF3a and ORF7a as there is evidence that these proteins are expressed on infected cell membranes (unlike the structural proteins), potentially making them targets to explore in the context of SARS-CoV-2 vaccines, in addition to the spike protein.
2.1. Spike (S) protein
The surface glycoprotein S is responsible for binding to the major host receptor for SARS-CoV-2, angiotensin converting enzyme 2 (ACE2), and for mediating fusion between the viral and host cell membranes. This makes S protein the principal target of neutralizing antibody responses [[41], [42], [43]] which are so far the best described correlate of protection against infection in humans [[44], [45], [46], [47], [48], [49], [50], [51], [52]], and in animal challenge studies [45,[53], [54], [55], [56], [57]]. The SARS-CoV-2S is a class I homotrimeric transmembrane protein that is expressed on the surface of the viral particle and exists in a prefusion metastable conformation. A monomer is composed of two subunits, S1 and S2, that have distinct functions responsible for viral entry. The S1 subunit contains the receptor binding domain (RBD) and allows recognition and binding to ACE2, while the S2 subunit contains the fusion peptide that mediates the fusion of the host and viral membrane (Fig. 2c) [58,59].The binding of the S1 subunit to ACE2 and its subsequent shedding [60] combined with the cleavage at the S1/S2 and S2’ sites by host proteases like furin, transmembrane serine protease 2 (TMPRSS2) or cathepsins [[61], [62], [63]] trigger drastic conformational changes that lead to the more stable post-fusion structure [64,65]. This complex process drives the fusion of virus and host cell membranes, which allows the release of the ribonucleoprotein complex containing the viral genome into the cytoplasm of the host cell.
Studies have now shown that IgG antibody titers against S and also RBD correlate well with virus-neutralizing titers in vitro [66,67]. Given the metastable nature of the prefusion state of S and its importance in eliciting antibody-mediated neutralization, a number of strategies have been adopted to stabilize S-based vaccine candidates in the proper prefusion conformation. These include first the introduction of two consecutive proline mutations in the loop between the first heptad repeat and the central helix [58,59]. These “2P” mutations prevent conformational rearrangements, presumably by increasing the activation barrier for fusion. “2P” mutations have been used as a generalized strategy across betacoronaviruses for preventing spontaneous transition to the post-fusion S conformation [68,69]. A second stabilizing strategy is removal of the polybasic cleavage site between S1 and S2. Highly pathogenic viruses such as SARS-CoV-2 can have enhanced virulence due to the presence of a polybasic cleavage site in the membrane protein that mediates viral fusion. This site increases protease susceptibility and thus the efficiency of fusion and infection. Removal of this site impedes S cleavage, preventing transition to the post-fusion conformation [65,70,71]. Yet another strategy for stabilizing the trimeric spike is the addition of trimerization motifs such as the bacteriophage fibritin foldon or a yeast-derived “GCN4” domain [58,69,72,73]. Addition of these relatively small domains (~10 kDa) that spontaneously trimerize promotes the formation of a functional trimer, stabilizing the integrity of conformational S epitopes (Fig. 2d).
Aside from mediating virus neutralization, specific, proinflammatory, anti-S responses have been associated with severe COVID-19. Hospitalized COVID-19 patients produce high levels of afucosylated IgG1 and IgG3 anti-S IgGs [74]; these antibody forms have high affinity for activating FcγRs and mediate inflammatory cellular functions [75]. Afucosylated anti-S IgG1 in SARS-CoV-2 immune complexes trigger inflammatory cytokine production by human monocytes, including IL-6 and TNF, which are associated with severe COVID-19 [76]. Subsequent studies showed that, among severe COVID-19 patients, higher levels of afucosylated anti-S IgG1 are found in serum of those with acute respiratory distress syndrome (ARDS) [77]. It is important to note that all the studies describing proinflammatory IgGs have been observed only in patients with severe disease outcomes following infection and data support increased afucosylation during severe viral disease [74,78]. Taken together, with data from COVID-19 vaccine clinical trials, we do not anticipate the generation of such proinflammatory IgGs post vaccination that can cause antibody dependent enhancement (ADE) or vaccine-associated enhanced respiratory disease (VAERD).
2.2. Membrane and envelope proteins
The membrane (M) and envelope (E) proteins, along with the S protein, are the main structural components forming the shell of the viral particle. While the S is the major determinant of viral entry, the role of M is in viral assembly and budding [79] and the E is a putative ion channel involved in viral replication [[80], [81], [82]]. The M protein is the most abundant viral protein [83] and, in combination with either the N [84,85] or E [[86], [87], [88]], can drive the budding of virus-like particles. E protein is largely expressed in the endoplasmic reticulum and Golgi-complex of the host cell and is not highly incorporated in viral particles [[89], [90], [91], [92]]. Antibody [93] and T cell [94] responses against the M protein have been reported in people infected with SARS-CoV-2. However, the role of M-based immunity in protection is not yet known. The immune response to E has not yet been well characterized and there is currently little available data that highlights its immunogenicity and/or role in immunity.
2.3. Nucleocapsid protein
The N protein is a multi-functional protein that can be divided into an N-terminal RNA binding domain and a C-terminal oligomerization region that are connected by a disordered linker region [95]. The N is involved in many aspects of viral replication including RNA packaging, replication and pathogenesis [96]. Via the RNA binding region, N binds to the viral genome and packages it into a long and helical ribonucleoprotein complex which protects the viral RNA from degradation. The nucleocapsid-RNA interaction in the context of genomic and subgenomic (sg) viral RNA is intimately involved in viral transcription and translation [97,98]. Multiple interactions between N and host cellular pathways that may contribute to viral pathogenesis have been described. These include disruption of the host translation machinery, cell cycle, inhibition of Type I interferon signaling, induction of apoptosis and modulation of the transforming growth factor beta pathway [[83], [84], [85], [86], [87],99].
The N of SARS-CoV-1 and -2 is highly immunogenic and is the target of antibody [[100], [101], [102], [103], [104], [105]] and T cell-mediated responses [[106], [107], [108], [109], [110], [111], [112]]. Of note, some SARS-CoV-1 studies demonstrated an enhanced respiratory disease (ERD) after SARS-CoV-1 challenge in mice [[113], [114], [115], [116]] and in ferrets [[117], [118], [119]], where the pathogenesis of ERD was thought to be driven by a Th2 response [113] against the N protein [114,116]. ERD in challenged mice was not reproduced following passive transfer of immune sera that was enriched for N-specific antibodies suggesting that these antibodies may not have been responsible for the ERD phenotype [114]. The role of N antibodies in protection may be limited since it is not thought to be expressed on the surface of viral particles or infected cells. However, the role of N antibody responses in protection or disease warrants further investigation. A recent study suggested that individuals who succumbed to COVID-19 had a stronger N-specific antibody response than patients with severe disease who ultimately recovered [120]. By contrast, the contribution of T cell-mediated immunity against N may be supported by studies from SARS-CoV-1, SARS-CoV-2 and MERS-CoV [121,122]. Indeed, a growing body of literature suggests that N is a major target of CD4+ and CD8+ T cells [94,123] and these responses may correlate with the resolution of COVID-19 in some patients [124].
2.4. ORF3a
Much of what is known about the protein encoded by ORF3a is based on studies performed with SARS-CoV-1. The ORF3a of SARS-CoV-1, also known as ORF3 [125], X1 [126], ORF3a [127] and U274 [128], is located at the 3′ end of the genome residing between the S and the E genes and encodes for the largest of the accessory proteins. It is 274 amino acids long and has an extracellular N-terminal ectodomain, three transmembrane domains and a C-terminal cytosolic domain. Putatively characterized as an ion channel [129] or viroporin, ORF3a expression has been detected in the rough endoplasmic reticulum [130], the Golgi complex [127] and on the plasma membrane [127,130,131] of infected cells. Of note, it is also found as extracellular membrane-associated structures secreted by virus-infected cells [132]. The SARS-CoV-2 ORF3a protein is ~72% identical to the ORF3a of SARS-CoV-1 and current literature indicates it largely shares structural features with other betacoronavirus ORF3a proteins [133,134]. Additional studies suggest a possible role in pathogenesis and virulence [[135], [136], [137], [138], [139], [140]].
While ORF3a may be expressed on viral particles at some level, it is not generally associated with viral entry suggesting that anti-ORF3a antibodies will not neutralize virus by preventing binding or fusion. Yet data supporting its immunogenicity and expression on infected cells suggests that it may be an important target for the many antibody and T cell mediated mechanisms other than virus neutralization that can contribute to broad and durable antiviral immunity.
2.5. ORF7a
Based on its SARS-CoV-1 orthologue, the ORF7a encodes for an accessory type I transmembrane protein that is 122 amino acid long [141,142]. The SARS-CoV-2 orthologue (YP_009724395.1) is 121 amino acids long and is ~86% identical to the ORF7a of SARS-CoV-1 (NP_828857.1). Deletional mutant studies demonstrate that the ORF7a protein is not required for replication in vivo and in vitro [[142], [143], [144]]. It is, however, involved in the induction of caspase-dependent apoptosis [142,145,146]. Expression of ORF7a can be found intracellular in the Golgi complex and on the surface of the plasma membrane of infected cells.
The role of ORF7a in virulence and immunity is not entirely known. The finding that ORF7a is on the surface of infected cells makes it a likely target of the antibody response and recent work by several groups has demonstrated antibody responses against SARS-CoV-2 ORF7a [147,148]. Others have also found ORF7a to be targeted by T cell responses [94,149]. The ORF7a region underwent positive selection during the SARS-CoV-1 zoonotic outbreak earlier in the century [150] and more recently, deletional variants in the SARS-CoV-2 pandemic have been found in circulation [151,152]. Whether or not these mutations occurred due to immunological pressure or natural selection remains to be determined. Nonetheless, ORF7a is immunogenic and is recognized by both arms of the adaptive immune system – whether or not this response contributes to protection warrants further investigation.
3. Vaccine platforms
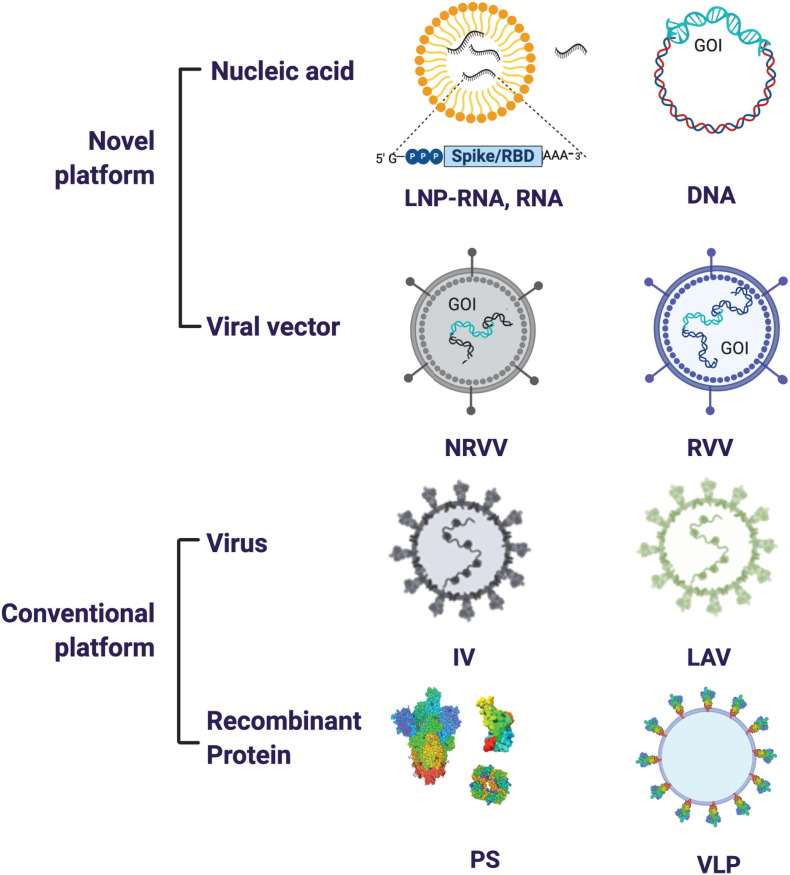
A wide array of vaccine platforms, each with its own advantages and disadvantages, is represented among the experimental vaccines being trialed to prevent COVID-19. Each of these platforms is represented among SARS-CoV-2 vaccine candidates that are in pre-clinical and/or clinical stages of development (Fig. 3 ). The vaccine platforms can be broadly classified as follows (platforms already in use in humans for non-SARS-CoV-vaccines are denoted by *):
Fig. 3.
SARS-CoV-2 vaccine platforms. The major vaccine platforms that are being used in current SARS-CoV-2 vaccine candidates. A combination of conventional and novel vaccine platforms are being tested. Conventional vaccine platforms that have been licensed for human use are the inactivated virus (IV), live attenuated virus (LAV), and recombinant protein-based (Protein Subunit (PS)). Novel vaccine platforms include nucleic acid based (DNA and RNA encoding the gene of interest (GOI)) and viral vector-based (non-replicating viral vector (NRVV) and replicating viral vector (RVV).
3.1. Virus
3.1.1. * Whole inactivated virus (IV)
Inactivated virus (IV) vaccines are being developed based on SARS-CoV-2 isolates cultured from hospitalized COVID-19 patients. The virus is passaged in Vero cells- a WHO certified cell line for vaccine production and is subsequently chemically inactivated using β-propiolactone [[153], [154], [155]]. This platform has been licensed for use in humans for a number of infectious diseases including influenza, polio and Hepatitis A [156,157]. A potential significant benefit of this platform is that it exposes the vaccinated individual to multiple viral proteins rather than one target. However, the mode of inactivation can affect the structural integrity of the antigens and may skew the T-cell response toward a Th2 profile [158,159]. A caveat to this platform is that production requires BSL3-level facilities and expertise which can be prohibitive [160]. There are three inactivated SARS-CoV-2 virus vaccine candidates which are being evaluated in Phase III trials and will be discussed below.
3.1.2. * Live attenuated virus (LAV)
The LAV platform most closely mimics natural viral infection. Here, infectious virus may be attenuated using one of multiple strategies. These include serial passaging, using codon deoptimized genomes that code for the wildtype virus, but that are not optimally translated in the hosts [158,161,162] . More recently, engineering of rational mutations and/or deletions into genes to reduce virulence such as the envelope (E) protein of SARS-CoV-1 and MERS-CoV have been reported [163,164]. Because LAV vaccines cause a low-level of infection, they often elicit robust and enduring humoral and cellular immune responses without requiring repeated vaccinations or adjuvants [165,166]. Additionally, for respiratory viruses, they can be administered intranasally and elicit a strong mucosal immune response [167]. Numerous LAV vaccines are licensed for use in humans, including the yellow fever vaccine which is widely considered to be among the most effective vaccines ever developed [168]. Other examples are LAV vaccines used to prevent influenza viruses, dengue viruses, and measles, mumps, and rubella viruses (MMR vaccine) [156,161]. Disadvantages of the LAV platform include the requirement for extensive quality control and characterization to ensure that the vaccines do not revert back to a less attenuated form and they require cold storage (2–8 °C) [169]. Further, these vaccines cannot be used in the immunocompromised and elderly [[170], [171], [172]]. There are currently three LAV SARS-CoV-2 vaccine candidates in preclinical stages of evaluation.
3.1.3. Viral vector (VV)
VV-based platforms are attenuated or non-replicating viruses that encode an immunogen of choice from an unrelated pathogen [173,174]. As with traditional LAV vaccines, attenuated VV-based vaccines can infect cells and elicit both T cell and antibody responses [165,175]. Antigen expression can be controlled by the strength of the promoter under which the gene coding for the vaccine antigen is cloned, potentially enabling greater potency than traditional vaccines as it is not limited by the input dose as are other vaccine types [176]. Also, this platform (and LAV vaccines) can be adapted to intranasal or oral administration, which can be extremely efficient and well-tolerated routes for vaccination [177,178]. A limitation of this platform can be the presence of pre-existing immunity against the vector which can suppress immune responses to the vaccine antigen and limit effectiveness of these vaccines [179,180]. A workaround for this is the use of vectors that are less immunogenic or that are derived from animal viruses like the chimpanzee adenovirus (ChAd) [175,[181], [182], [183], [184]]. As with LAV vaccines, VV-based vaccines are not suitable for use in immunocompromised populations and they can be reactogenic (causing “expected” adverse reactions after vaccination) in healthy people [185,186]. Finally, there are not yet any licensed VV-based vaccines for use in humans, though Ebola virus and Chikungunya virus vaccines using this platform are in advanced phases of clinical trials; early safety and immunogenicity data for these vaccines are promising [[187], [188], [189], [190], [191]]. For SARS-CoV-2 VV-based vaccines, the S protein (and stabilized variants) are the most common antigens being trialed.
Based on the property of the viral vector, this category of vaccine platform can be further classified, as follows:
-
a)
Non-replicating viral vector (NRVV)
Here, the viral vectors themselves are replication incompetent and their function is to simply deliver the transgene cargo. Four out of ten of the VV-based SARS-CoV-2 vaccines that are in Phase III trials are based on NRVV platforms and all use adenoviral vectors [48,50,51,192]. Other vectors that are in Phase I and preclinical evaluation are gorilla adenovirus, Sendai virus vector, adeno-associated virus vector, modified vaccinia virus Ankara, parainfluenza virus 5, deactivated rabies virus and influenza A virus H1N1 vectors [8,[193], [194], [195], [196]]
-
b)
Replicating viral vector (RVV)
Multiple SARS-CoV-2 vaccines are also being developed with attenuated viral vectors that can replicate at the site of immunization and can thus elicit stronger immune responses using relatively small vaccine doses. There are two SARS-CoV-2 candidates in Phase I/II clinical trials that use measles (NCT04497298) or vesicular stomatitis virus (VSV) (NCT04569786) both developed by Merck Sharp & Dohme [198]. Additionally, there are multiple candidates in preclinical evaluation based on yellow fever, measles virus, horsepox vector, attenuated influenza virus, Newcastle disease virus (NDV) and avian paramyxovirus (APMV) vectors [8,[199], [200], [201]].
3.2. Nucleic acid
Nucleic acid-based platforms utilize delivery of either plasmid DNA or RNA that codes for an immunogen of interest. Following cellular uptake, they are either transcribed and translated (DNA) or directly translated (RNA) to protein antigens that can elicit both humoral and T-cell mediated immune responses. These vaccines can be extremely useful in a pandemic setting as they are scalable and can be rapidly synthesized without handling infectious virus. There are currently no licensed human vaccines based on these platforms, but a number are in clinical evaluation for influenza virus [[202], [203], [204]], Ebola virus [205], Zika virus [206] and MERS [207].
3.2.1. DNA
These vaccines are designed by cloning of a eukaryotic protein expression cassette consisting of the gene of interest downstream of a strong promoter into a bacterial plasmid that can be propagated in organisms like Escherichia coli for mass production. Upon immunization and cellular uptake at immunization sites, the plasmid must traverse the nuclear membrane for transcription, followed by expression of the immunogen in the cell cytoplasm. Because of the complex cellular requirements for protein production, these vaccines may not be as immunogenic as RNA or protein-based vectors and may require specialized delivery techniques like electroporation [176,208]. There are currently four DNA vaccines under clinical evaluation [8]. Pre-clinical data from one candidate DNA vaccine (INO-4800) developed by Inovio demonstrated immunogenicity in mouse and guinea pig models [209]. In a rhesus macaque model, vaccination with two doses of a DNA-encoding S protein protected against SARS-CoV-2 challenge 3 months after vaccination [210].
3.2.2. RNA
RNA platforms are either self-amplifying (sa) alphavirus-derived RNA (saRNA) or non-replicating mRNA (nrmRNA). Current SARS-CoV-2 RNA vaccines encode the stabilized S protein and are encapsulated in lipid nanoparticles for efficient delivery. These comprise some top SARS-CoV-2 candidates in clinical and pre-clinical evaluation, as discussed below [[211], [212], [213]]. Compared to the DNA vaccine platform, RNAs are more immunogenic as they can be translated into the protein of interest without requiring nuclear entry for transcription (they are translated directly in the cytosol following uptake into cells at the injection site) [214]. RNA vaccines are thought to result in production of natively folded and processed proteins, leading to effective antigen presentation and B cell activation. RNA vaccines do not require electroporation as DNA vaccines can [215,216]. Potentially serious considerations around RNA vaccines are related to their stability, a requirement for any SARS-CoV-2 vaccine that will be widely administered [217] . Further, large scale safety data in humans are lacking for nucleic acid platforms [218]. Moderna, the company developing the SARS-CoV-2 nrmRNA vaccine currently in Phase III trials has previously developed two nrmRNA vaccines against Zika virus [219,220]. Both of these Zika virus vaccines are in Phase I clinical trials following promising data from preclinical animal studies, but no data has yet been published from either trial.
3.3. * Recombinant protein subunit (PS)
Immunogenic viral proteins expressed in bacterial, insect or mammalian cells generally make effective and safe vaccines and can be administered with an adjuvant to boost the immune response [221,222]. Another strategy to enhance immunogenicity of subunit protein vaccines is multimeric antigen display and presentation by conjugation to carrier nanoparticles, a platform that is being explored for SARS-CoV-2 vaccine design [223,224]. Although relative to nucleic acid-based or viral vector-based vaccines, protein subunit vaccines are expensive and laborious to produce, there are already several licensed recombinant protein-based vaccines including for hepatitis B and influenza viruses [156,[225], [226], [227]]. There are currently 11 protein subunit SARS-CoV-2 vaccines in Phase I/II clinical trials and one in advanced Phase III trial [8,228].
3.4. * Virus-like particles (VLP)
A special category of recombinant protein-based platform, VLPs are made from viral proteins that self-assemble into supramolecular structures that are meant to mimic native virions in geometry and size [229,230]. Because of this, they are often highly immunogenic and elicit strong innate and adaptive immune responses (both humoral and cell-mediated) [231]. Licensed vaccines against human papillomavirus and hepatitis B viruses use the VLP platform and are widely used in humans [232]. A plant derived VLP vaccine generated by Medicago that is adjuvanted with one of the two different adjuvants (GSK's pandemic adjuvant technology and Dynavax's CpG 1018) is currently in Phase I clinical trials [233].
4. SARS-CoV-2 vaccine candidates in phase III clinical trials
Here, we provide a summary of SARS-CoV-2 vaccine candidates currently in Phase III clinical trials. We have summarized aspects of the preclinical and clinical data, including available information on the vaccines themselves as well as selected information related to safety, immunogenicity and efficacy.
Early-phase clinical studies evaluate safety and immunogenicity of vaccine candidates. With respect to immunogenicity, antibody responses are typically studied by measuring serum titers (usually by ELISA) to S or RBD; in this context, some studies specify the antibody isotype being characterized (particularly IgG). Neutralizing antibody titers are generally measured in in vitro assays using a replicating SARS-CoV-2 virus or non-replicating pseudovirus.
In addition to measuring antibodies, SARS-CoV-2 vaccine clinical trials have also carried out preliminary characterization of the cell mediated immune response. Typically, in a subset of randomly selected volunteers, the frequency of T cells (CD3 + CD4+ and CD3 + CD8+) and cytokine secretion was measured at different time-points, with or without stimulation using overlapping peptide pools from the S protein to evaluate Th1 polarization, which is considered to be an important marker for vaccine selection. Vaccine studies that use whole virus-based vaccine platforms may evaluate antibody and T cell responses against non-S proteins of SARS-CoV-2, but most studies focus on S protein as this is the sole antigen in nearly all vaccines being tested in Phase III studies.
4.1. Developer: Beijing Institute of Biological Products/Sinopharm
4.1.1. Vaccine: BBIBP-CorV
Platform: inactivated virus
Vaccine target: whole virus
A SARS-CoV-2 isolate (19nCoV-CDC-Tan-HB02 (HB02)) from a hospitalized COVID-19 patient was used to develop this vaccine. The HB02 isolate was passaged seven times in Vero cells to generate the vaccine stock which was then inactivated with β-propionolactone. Electron microscopy and western blot analysis of the inactivated virus were performed to characterize the integrity of the viral particles and their surface antigens following inactivation [153].
4.1.2. Preclinical data
Immunogenicity of BBIBP-CorV was first assessed in BALB/c mice. Animals were immunized intraperitoneally with 2, 4, or 8 μg of the vaccine which was adjuvanted with aluminum hydroxide (alum). Several immunization regimens were tested: a) one vaccination at day 0 (D0), b) three prime-boost regimens with 7 (D0/D7), 14 (D0/D14), or 21 (D0/D21) day intervals between the prime and boost, and c) a three-injection schedule at days 0, 7 and 14. For the prime only regimen, 100% seroconversion was achieved across all dose groups at day 7 post-vaccination. The nAb titers increased until day 21 in the low and middle dose group but did not change in the high dose group after day 14. In the two-dose immunization group, where different time points between prime and boost regimen were tested, the D0/D21 regimen elicited the highest nAb level on day 7 post-boost. Immunogenicity of the two-dose regimen was significantly enhanced over the one-dose schedule. Immunogenicity of the three-dose (D0/D7/D14) regimen was highest among all regimens, at all dose levels. Immunogenicity was also measured in additional animal models: rabbits, guinea pigs, rats, and cynomolgus monkeys. In all models, 100% of animals had produced nAbs on day 21 with higher titers elicited by three immunizations compared to one [153].
Immunogenicity and protection were then evaluated in rhesus macaques. All macaques were immunized twice on D0 and D14 with either placebo (saline), or with a “low” dose (2 μg/dose) or “high” dose (8 μg/dose) of BBIBP-CorV. The Geometric mean titers (GMTs) of nAb in the low-dose and high-dose groups reached 215 and 256, respectively at D24, the day of challenge. Animals were challenged with 106 TCID50 of SARS-CoV-2. All macaques in the placebo group showed sustained high viral load up to 7 days after virus challenge from both throat and anal swab samples. In contrast, the viral load in the throat swabs of the low dose group peaked at 5 days and decreased by day 7 post-infection to a level that was significantly lower than that of the placebo group. Three out of four macaques in the low-dose group had no detectable viral load at 7 days post-infection. In the high dose group, all four macaques had undetectable viral load in their throat swabs. Moreover, no viral load was detected in the anal swabs of 2 out of 4 macaques. At 7 days post-infection, lung tissues from euthanized animals were examined for pathology and viral load. No macaques in the low-dose or high-dose groups had detectable lung viral loads and lung histology was largely normal. In comparison, the placebo group showed severe interstitial pneumonia and high lung viral loads [153].
4.1.3. Clinical data
Phase I
Placebo controlled: yes
Participants: healthy adults, 18–59 years of age (n = 96) or ≥ 60 year of age (n = 96)
Immunization Schedule: prime-boost (days 0 and 28), multiple dosages tested, intramuscular administration
A prime-boost regimen was tested in which doses were 2, 4, or 8 μg BBIBP-CorV adjuvanted with aluminum hydroxide (n = 24 per dose group). The placebo group received saline plus adjuvant. For vaccine recipients aged 18–59 years, 79% to 96% seroconverted, defined by nAb production, by day 14 (with a dose-dependent increase in nAb titers). 100% of vaccine recipients produced nAbs by day 28. The vaccine was somewhat less immunogenic in the older cohorts and seroconversion was delayed. Seroconversion rates in those ≥60 year of age were 4% in the 2 μg group and 46% for the 4 μg and 8 μg groups on day 14 post-boost but increased to 91% to 96% in the three dose groups by day 28. NAb GMTs in the 2 μg cohorts were significantly lower than the 8 μg cohorts, irrespective of age. In 6 randomly selected participants from the 4 μg dose group, sera collected on day 42 had neutralizing activity against ten different natural SARS-CoV-2 isolates, four of which contained the well-described D614G spike mutation [234].
All adverse reactions (ARs) were mild or moderate in severity with 29% of vaccine recipients reporting at least one AR within the first 7 days of vaccination. No serious adverse events (AEs) were reported within the 28 days after vaccination [47].
Phase II:
Placebo controlled: yes
Participants: healthy adults, 18–59 years of age (n = 448)
Immunization Schedule: prime only or prime-boost (variable interval), intramuscular administration
Participants received one of four different BBIBP-CorV immunization schedules: a) a prime-boost regimen of 4 μg per dose where the boost was given on day 14, 21 or 28, or b) a single vaccination with 8 μg total protein. Mean nAb titers in the single vaccination cohort were significantly lower than those elicited in any of the two-dose immunization schedules. For the different prime-boost regimens, the participants boosted on either day 21 or 28 produced significantly greater nAb titers than those boosted on day 14 group. These results suggest that the interval between prime and boost impacted nAb titers, in turn, potentially impacting vaccine efficacy.
Self-limiting AEs of mild to moderate severity were reported with no report of severe AEs. 23% of vaccine recipients reported at least one AR within the first 7 days after either vaccination. The reactogenicity profile was similar to the Phase I study where pain at the site of injection was the most common local AE, with significantly higher number in the vaccine group as compared to the placebo. Most common systemic adverse reaction in the vaccine recipient group was fever reported in 2% of the vaccine group [47].
Phase III
Placebo controlled: yes
Participants: healthy adults, 18 years and above (n = 15,000)
Immunization Schedule: prime-boost vaccination. Intramuscular administration
This Phase III trial of BBIBP-CorV is currently underway in Abu Dhabi where 5000 participants are receiving placebo, 5000 participants are receiving BBIBB-CorV and another 5000 participants are receiving another inactivated vaccine also manufactured by Sinopharm (described below). The primary endpoint is protective efficacy of the vaccine 14 days after two doses of immunization and secondary endpoint being protection from severe cases of SARS-CoV-2 pneumonia and deaths accompanied by COVID-19 (ChiCTR2000034780).
4.2. Developer: Wuhan Institute of Biological Products/Sinopharm
4.2.1. Vaccine: no name announced
Platform: inactivated virus
Vaccine target: whole virus
This vaccine is based on a SARS-CoV-2 isolate (WIV04 strain) from a patient in the Jinyintan Hospital in Wuhan and was propagated in Vero cells. Cell supernatant containing virus was inactivated with β-propiolactone, followed by clarification by ultracentrifugation and a second β-propiolactone treatment. The inactivated virus was then adsorbed to alum [154].
4.2.2. Preclinical data
No data available
4.2.3. Clinical data
Phase I/II
Placebo controlled: yes
Participants: healthy adults 18–59 years of age (n = 96 for Phase I and n = 224 for Phase II).
Immunization schedule: three immunizations (Phase I) or prime-boost (Phase II), multiple doses tested, intramuscular administration.
In the Phase I trial, participants were assigned to one of three vaccine dose groups of 2.5, 5, or 10 μg per dose. Three vaccinations of each dose (or placebo) were given on days 0, 28, and 56. In the Phase II trial, participants received a prime-boost consisting of 5 μg per dose (or placebo). The boost was administered on day 14 or 21. The placebo consisted of alum adjuvant only.
In the Phase I study, 100% of participants in the low and high dose groups seroconverted whereas 96% in the medium dose group seroconverted 14 days after the third dose. In the Phase II study, 97.6% seroconverted 14 days post boost regardless of the interval between prime and boost vaccinations. The nAb GMT was 2-fold higher in the group who received injections on days 0/21 as compared to the group who received injections on days 0/14.
Because prior studies have reported that using alum as an adjuvant can bias the vaccine response toward a Th2 phenotype which can lead to VAERD [235,236], an attempt was made to assess whether alum adjuvantation of the Wuhan Institute of Biological Products/Sinopharm vaccine had any such effects. Blood lymphocyte subsets (NK, CD4+ T, CD8+ T and B cells) and serum cytokines were measured in the peripheral blood of participants in Phase I trials at day 0 and 14 days after each injection. An extensive panel of serum cytokines (IL-1, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12P70, IL-17A, IL-17F, IFN-γ, TNF-α, TNF-β) was measured. There were no notable changes between the vaccine or alum-only groups over time, with respect to lymphocyte subsets or the measured cytokines (including the Th2 related cytokines IL-4, IL-5 and IL-10) [154]. T-cell mediated immune responses after antigen stimulation were not measured.
The inactivated vaccine was safe in all dose groups and vaccination schedules. All adverse reactions were mild (grade 1 or 2), transient, and self-limiting, and did not require any treatment [154].
Phase III
Placebo controlled: yes
Participants: healthy adults, ≥18 years of age (n = thousands between multiple trials)
Immunization schedule: intramuscular administration.
There are currently three international clinical trials of the vaccine underway. In Peru, 6000 healthy adults ≥18 years of age are being enrolled (NCT04612972). In the United Arab Emirates.
(Abu Dhabi) 15,000 healthy adults aged ≥18 years are being enrolled (ChiCTR2000034780) where participants will either receive BBIBP-CorV or this vaccine or placebo (n = 5000 each group). In both of these studies, the primary outcome measured is “protective effect against COVID-19, after 14 days following the full course of vaccination”. A third study is being conducted in Morocco, consisting of 600 healthy adults aged ≥18 years old (ChiCTR2000039000). In Morocco, the primary goal is “to evaluate the 4-fold increase rate, GMT and GMI of anti-SARS-CoV-2 neutralizing antibody 28 days after full course of immunization”.
4.3. Developer: Sinovac
4.3.1. Vaccine: CoronaVac (previously known as PiCoVacc)
Platform: inactivated virus
Antigen: whole virus
This vaccine is based on a SARS-CoV-2 isolate (CN2) from a hospitalized COVID-19 patient which was then adapted for efficient growth in Vero cells. Genetic stability of the strain was ascertained by passaging 10 times in culture followed by whole genome sequencing. The virus was inactivated using β-propiolactone. Following inactivation, the structural and antigenic integrity of viral particles was assessed by cryo–electron microscopy which showed intact, crown-like spikes of the prefusion S protein decorating the viral surface [155].
4.3.2. Preclinical data
Immunogenicity was tested in BALB/c mice immunized at days 0 and 7 with various doses of CoronaVac mixed with alum adjuvant (0, 1.5, 3, or 6 μg per dose). No inflammation or other vaccine associated adverse effects were observed in the vaccinated animals. Anti-S, RBD, and N antibody responses were evaluated by ELISA. Rapid elicitation of S-specific and RBD-specific IgG titers occurred, peaking at week six post-vaccination. 50% of the anti-S IgG were directed against RBD, establishing it's immunodominance. The vaccine did not induce a robust anti-N response, with ~30-fold lower titers at week 6 as compared to RBD and S. When benchmarked against sera from convalescent COVID-19 patients, immunized mice had 10-fold higher anti-S and RBD titers at week 6 across all dose groups. NAbs showed a dose dependent increase in titers which peaked at week 6. Sera from immunized mice exhibited broad nAb titers against a panel of 9 viral strains which are circulating globally. CoronaVac had a similar immunogenicity profile in Wistar rats [155].
The immunogenicity and protective efficacy of CoronaVac was also evaluated in rhesus macaques. Animals were immunized three times intramuscularly with a “medium” dose (3 μg per dose) or “high” dose (6 μg per dose) of alum-adjuvanted CoronaVac on days 0, 7, and 14 (n = 4). In both the dose groups, S-specific IgG and nAb titers were similar to those from recovered COVID-19 patients by week 3 post-vaccination. At week 3, immunized animals were challenged with 106 TCID50 of SARS-CoV-2. After challenge, all control macaques (which received either adjuvant alone or saline) had high copies of viral genomic RNA in the airways and lung tissue, along with severe interstitial pneumonia. Viral loads were significantly lower in all vaccinated animals with no detectable virus in pharynx, crissum, or lungs on day 7 post-challenge [155].
4.3.3. Clinical data
Phase I
Placebo controlled: yes
Participants: healthy adults,18–59 years of age (n = 144)
Immunization schedule: prime-boost (variable intervals), multiple doses tested, intramuscular administration
Participants received primary vaccination followed by a boost vaccination on either day 14 or 28 post-prime. Doses were either 3 or 6 μg protein, adjuvanted in alum. Seroconversion rates (measured by serum nAb activity) in those boosted on day 14 were 46% (3μg dose) or 50% (6μg dose). In those boosted on day 28, seroconversion was observed in 83% (3μg dose) or 79% (6μg dose). RBD-specific IgG was detected in 100% of participants only in the 6 μg dose in those boosted at day 14, whereas 100% participants in both dose groups seroconverted if they were boosted on day 28. Serum inflammatory factors (IL-1, IL-6 and TNF-α) were measured in the blood and urine samples collected 7d after each dose using sandwich ELISA, and no significant difference was observed between the vaccine and placebo groups. T cell responses were evaluated in samples by measuring IFN-γ collected at different time-points after dose 1 of the vaccine or placebo. The average IFN-γ spot forming cells (SFCs) per 105 cells was highest for the 3 μg group in both the vaccination cohorts (d0-d14 and d0-d28) in comparison to the 6 μg and placebo groups.
Low reactogenicity was observed with most of the reported AEs being mild and resolving after day two post-vaccination. One case of an acute hypersensitivity reaction (urticaria) was reported in a 6 μg dose group after primary vaccination; this event was considered to be vaccine associated and graded as a severe AE. The participant remained in the study and did not report a similar reaction after the booster dose. Overall, there was no significant difference in AEs among study groups [237].
Phase II
Placebo controlled: yes
Participants: healthy adults aged 18–59 years old (n = 600)
Immunization schedule: prime-boost (same as the Phase I study - variable intervals, variable doses), intramuscular administration.
This was a larger study performed with identical vaccine regimens and doses as in the Phase I trial. Seroconversion rates (measured by serum nAb activity) in those boosted on day 14 were 92% (3μg dose) or 98% (6μg dose). In participants boosted on day 28, seroconversion was observed in 97% (3μg dose) or 100% (6μg dose). When benchmarked against convalescent sera from recovered COVID-19 patients, nAb titers after the second dose in sera of vaccine recipients was lower in all participants. When data from Phase I and Phase II studies were combined, the correlation coefficient between the nAb titer and RBD-specific IgG titer 28 days post vaccination was 0.85. Higher nAb and binding IgG titers in the Phase II trial (compared to Phase I) was due to a different vaccine production process for the Phase II study which inadvertently increased the content of spike protein in the vaccine used for Phase II studies by almost two-fold. T cell responses were determined by measuring IFN-γ in an ELISpot assay wherein PBMCs were cultured with overlapping peptides from the S protein or controls peptides [237].
The reactogenicity profile was similar to the Phase I with most AEs being mild in severity and no reported serious adverse events in the 28 days post-boost vaccination. Considering the combined safety and immunogenicity data, the 3μg dose of CoronaVac was suggested for Phase III trials [237].
Phase III trial
Placebo controlled: yes
Participants: multiple trials
Immunization schedule: prime-boost regimen 14 days apart, intramuscular administration.
CoronaVac has obtained an emergency approval for use in China, and three Phase III clinical trials there are ongoing. In Brazil, ~13,000 health care workers, aged 18 years and older are being enrolled (ClinicalTrials.gov NCT04456595). In Indonesia, enrollment consists of ~1620 healthy adults aged 18–59 years (ClinicalTrials.gov NCT04508075). Finally, in Turkey, a total of ~13,000 adults aged 18–59 years are being stratified into two separate cohorts, 1300 healthcare workers and 11,150 “people at normal risk” (ClinicalTrials.gov NCT04582344). The primary outcome of protection rate against PCR-confirmed COVID-19 starting 2 weeks after the booster dose is being followed in all current Phase III trials. In the Brazil cohort, a primary outcome of frequency of ARs in 7 days post vaccination is also being measured. Initial results released by researchers in Brazil reported the vaccine to be about 50.4% effective at preventing both severe and mild disease in healthcare workers. These results differ from results seen in the earlier trials for this vaccine that took place in Turkey (91.25% effective) and Indonesia (65.3% effective), but are still encouraging in that no one who received the vaccine needed to be hospitalized [286].
4.4. Developer: CanSino Biological inc./Beijing institute of biotechnology
4.4.1. Vaccine: Ad5-nCoV
Platform: a non-replicating, adenovirus type 5 (Ad5)-vector
Vaccine target: SARS-CoV-2 S protein designed to be stabilized in the pre-fusion conformation
4.4.2. Preclinical data
No data available
4.4.3. Clinical data
Phase I:
Placebo controlled: no
Participants: healthy adults, 18–29 years of age (n = 31); 30–39 years of age (n = 42); 40–49 years of age (n = 18); 50–60 years of age (n = 17)
Immunization schedule: prime only or prime-boost, multiple doses tested, intramuscular administration.
Safety was assessed in 108 participants with a median age of 37 and no participants >60 years old. Three dose regimens were tested. A single dose regimen consisting of “low” (5 × 1010 particles) or “medium” (1 × 1011 particles) doses, and a third prime-boost regimen with a “high” (1·5 × 1011 particles) vaccine dose were trialed. Approximately 87% of the participants reported at least one AE within 7 days of vaccination. Overall, >95% of the participants seroconverted to RBD at day 28 post vaccination as measured by ELISA, with those in the high dose group exhibiting the highest titer of nAb responses against live SARS-CoV-2 (GMT 12.7). The greatest number of participants reported grade 3 AEs in the high dose group (17%) as compared to the low and medium dose groups (6% for each). The higher immunogenicity in the high dose group was judged to be offset by higher reactogenicity. Low and medium doses were thus selected for continued testing in a Phase II efficacy trial [52]. Antigen-specific T cell responses were quantified in d14 and d28 post-vaccination samples by measuring IFN-γ using an ELISpot assay by stimulating fresh PBMCs with overlapping S protein peptide pools for 12-24 h before detection. Vaccine induced CD4+ and CD8+ T cell responses were also determined by measuring IFN-γ, IL-2, and TNF-α by ICS after a 6 h S protein peptide pool stimulation. The T cell response peaked at d14 post-vaccination, with a slight decrease by d28 across all dosage groups. The proportion of positive responders across all the dosage groups was estimated to range between 83 and 97% with a dose-dependent increase. Also, the frequency of polyfunctional phenotypes observed in the memory CD4+ T cell subset was higher than those from CD8+ T cells.
Phase II:
Placebo controlled: yes
Participants: healthy adults: ages 18–44 (n = 309); ages 45–54 (n = 134) ≥55 (n = 65).
Immunization schedule: single administration, multiple doses tested, intramuscular administration.
Efficacy was evaluated in 508 participants, of whom 13% were > 55. In this study, a single injection of either “low” dose 5 × 1010 particles or “high” dose 1 × 1011 particles (equal to the “medium” dose in Phase I) was administered. The rate of seroconversion 28 days post vaccination was 49% with a nAb GMT of 18.3 in the low-dose group, and 59% with a nAb GMT of 19.5 against wild-type SARS-CoV-2 in the high-dose group. Approximately 90% of participants in both dosage groups also showed SARS-CoV-2 spike glycoprotein-specific IFNγ-ELISpot responses at day 28 with median values of 10–11 spot-forming cells per 105 peripheral blood mononuclear cells (PBMCs), corresponding to 10-fold increases over baseline pre-vaccination levels. Sex and age of the participants did not impact the T cell responses induced by vaccination. However, age and pre-existing Ad-5 immunity were inversely correlated with the degree of antigen specific immune responses, particularly seroconversion, raising the concern that a single dose of the vaccine may not be adequate in the elderly or in people with high levels of pre-existing Ad5 immunity. The low dose was selected for Phase III trial.
Both doses were reactogenic with most common AEs reported within 14 days of immunization being fever, fatigue and pain at the site of injection in >50% of individuals. 9% of AEs reported by participants receiving high dose of 1 × 1011 viral particles had severe (grade 3) adverse reactions, which was significantly higher than the participants receiving the lower dose. Encouragingly, the grade 3 AEs resolved without any medical intervention within 3–4 days of vaccination. None of the study participants reported any serious AEs within 28 days
Phase III:
Placebo-controlled: yes
Participants: adults >18 years of age (n≥ 40,000)
Immunization Schedule: single administration, intramuscular administration
This trial of a single administration of 5 × 1010 particles in an estimated 40,000 participants is currently underway. The primary outcomes are preventing virologically confirmed COVID-19 (day 28–12 months post vaccination) and the incidence of severe adverse events (clinicaltrials.gov NCT04526990).
4.5. Developer: Janssen pharmaceutical
4.5.1. Vaccine: Ad26.COV2·S
Platform: a non-replicating, adenovirus type 26 (Ad26)-vectored platform
Vaccine target: SARS-CoV-2 S protein designed to be stabilized in the pre-fusion conformation (Ad26.COV2·S)
The Ad26.COV2·S vaccine candidate is an S variant that includes an intact transmembrane domain with a cytoplasmic tail and two stabilizing proline mutations to retain the pre-fusion S conformation, as well as mutations that remove the polybasic cleavage site (originally termed Ad26S.PP and subsequently named Ad26.COV2·S).
4.5.2. Preclinical data
Vaccine target: in pre-clinical studies, vectors encoding multiple variants of the full-length SARS-CoV-2 S protein were tested.
Route of administration: intramuscular or intranasal in preclinical tests.
Immunogenicity and vaccine efficacy were tested in two pre-clinical animal challenge models: 1) nonhuman primates (NHPs) in which SARS-CoV-2 infection causes mild to moderate disease and 2) hamsters, which develop a severe disease characterized by pneumonia, weight loss and mortality. Multiple variants of the full-length S protein in the Ad-26 vector were tested and Ad26.COV2·S was the most efficacious and is the focus of this review. In the hamster model, a single shot of either 109 or 1010 particles was immunogenic, triggering seroconversion by week 4 with neutralizing antibodies measured in a pseudovirus neutralization assay. Upon challenge, hamsters were significantly protected from pneumonia, weight loss and mortality relative to unvaccinated animals. ELISA and nAb titers at week 4 inversely correlated with weight loss, indicating that vaccine-elicited antibodies were a correlate of protection. Similarly in rhesus macaques, one dose of 1011 Ad26.COV2·S particles protected animals from disease when challenged at week 6 post vaccination. Protection in NHP was judged by an absence of subgenomic viral RNA (sgRNA) in the bronchoalveolar lavage (BAL) collected from all the animals and only one out of 6 animals having a positive nasopharyngeal swab by PCR. As in the hamster studies, NAb titers at week 4 were inversely correlated with peak sgRNA levels in BAL. Antigen-specific cellular immune responses as measured by ELISPOT and intracellular cytokine staining (ICS) did not correlate with protection [53,238].
4.5.3. Clinical trial data
Phase I/IIa:
Placebo controlled: yes
Participants: healthy adults, 18–55 years of age (2 cohorts: 1a n = 377 or 1b n = 25) or > 65 years of age (cohort 3 n = 394)
Immunization schedule: prime only or prime-boost, multiple doses tested, intramuscular administration
Interim data on safety, reactogenicity and immunogenicity has been published. Participants received either 5 × 1010 (low dose) or 1 × 1011 (high dose) Ad26 particles or placebo. In this ongoing trial, a subset of participants is scheduled to receive a second vaccine dose equivalent to the first. This boost will occur 8 weeks after the primary vaccination. Immunogenicity data is available for all participants 18–55 years old, but only for 15 participants >65 years of age. Seroconversion rates measured by S binding antibody titers day 29 post vaccination was 99% for cohort 1a participants for both dose levels with no significant difference in GMTs. Seroconversion rate for cohort 3 was 100% with GMTs of 507 and 248 for the low and the high doses respectively. Neutralizing antibody titers were also assessed at day 29 in a subset of participants (n = 50 per dose group for those <55 years old and n = 6 per dose group for >65 year old). In both dosing regimens, 92% of participants <55 years old developed nAbs, with nAb GMTs between 214 and 243, while for the >65-year subset, 100% or 83% developed nAbs at the low, or high dose, respectively. In addition to a higher rate of nAb generation, the lower dose elicited higher nAb levels in subjects >65 years of age, with GMTs of 196 in the low dose and 127 in the high dose group. T cell responses were measured in cohorts 1a and 3 at d14 post immunization by ICS following stimulation by two S protein peptide pools of 15mers overlapping by 11 amino acids. A Th1 (IFN-γ/IL-2) skewed CD4+ T cell response was observed in 80% and 83% of the assayed participants from cohorts 1a and 3 respectively. No/low Th2 responses (IL-4, IL-5, IL-13 and CD40L) were observed. Robust CD8+ T cell responses (IFN-γ/IL-2) were observed in both cohort 1a and 3. Though some reactogenicity was observed across groups, higher reactogenicity was observed in subjects who were 18–55 and who were given the higher vaccine dose, with 72% of 18–55 year old and 46% >65 year old reporting AEs [50].
Phase III:
Placebo controlled: yes
Participants: adults, ≥18 years of age (n = up to 60,000)
Immunization Schedule: one dose, intramuscular administration
This trial of a single administration of the low dose (5 × 1010 particles) vaccine is currently underway. The primary trial outcome is PCR-confirmed moderate to severe COVID-19 occurring ≥15 days post-vaccination in individuals who were seronegative at baseline (clinicaltrials.gov NCT04505722).
4.6. Developer: Oxford University and AstraZeneca
4.6.1. Vaccine: AZD1222 (formerly ChAdOx1 nCoV-19)
Platform: replication-deficient simian adenovirus vector ChAdOx1
Vaccine target: full-length, codon-optimized S protein
AZD1222 vaccine candidate has full-length codon-optimized S with a tissue plasminogen activator leader sequence cloned in the replication-deficient simian adenovirus vector ChAdOx1.
4.6.2. Preclinical data
Immunogenicity studies of AZD1222 were performed in mice and in two large animal models- 1) pigs and 2) rhesus macaques, and vaccine efficacy was reported from a virus challenge study in rhesus macaques. The vaccine consists of the replication-deficient simian adenovirus vector ChAdOx1, containing the full-length codon-optimized S protein, with a tissue plasminogen activator leader sequence. The choice of the viral vector was based on preclinical studies showing that a single dose of ChAdOx1 encoding the spike protein of MERS-CoV protected non-human primates from disease after challenge. Additional Phase I clinical data demonstrated safety of the ChAdOx1 MERS-CoV vaccine. The immunogenicity of AZD1222 was tested in a prime only or a prime-boost regimen (28 days apart) by using 108 infectious units (IU) for the mouse studies and with 109 IU, a dose that is being tested in humans, in the pig studies. Anti-S IgG titers in serum were determined by ELISA and neutralization assays. One dose of the vaccine was immunogenic in two strains of mice tested (BALB/c and CD1) and there was a small but significant increase in titers following the boost in BALB/c mice only. No benefit of a second vaccine dose was observed in the outbred CD1 mice. Both binding and neutralizing IgG titers were significantly enhanced after a homologous boost in pigs as measured on day 42 post-vaccination. SARS-CoV-2 S-specific T-cell responses were also assessed in both mice and pigs, and revealed no statistically significant difference between the prime-only and prime-boost vaccination regimens in either strain of mouse, but showed an increase in IFN-γ + T cells in pigs after the prime-boost compared to prime-only on day 42 post-vaccination [239].
In a challenge study, six rhesus macaques per group were vaccinated intramuscularly with 2.5 × 1010 AZD1222 virus particles in either a prime-only or a prime–boost regimen. Spike-specific antibodies were detected 14 days after vaccination and IgG titers were significantly boosted after the second immunization. Virus-specific neutralizing antibodies were also significantly increased after secondary immunization and were detectable in all vaccinated animals before challenge. There was no correlation between induction of SARS-CoV-2 virus-neutralizing titer and neutralizing titer against the ChAdOx1 vector. Post-challenge, viral RNA and subgenomic RNA were detected in BAL fluid from only two of the vaccinated animals on 3 d.p.i., and viral load was significantly lower as compared to the vector-only vaccinated control group. Viral RNA was detected in nose swabs from all animals and no difference was found on any day between vaccinated and control animals. Lung tissue collected from vaccinated monkeys and examined for pulmonary pathology at day 7 after challenge, showed no evidence of viral pneumonia, immune-enhanced inflammatory disease or detectable SARS-CoV-2 antigen by immunohistochemistry. In contrast, 50% of control animals developed some degree of viral interstitial pneumonia, and five out of six control animals had viral antigen in type-I and II pneumocytes, as well as in alveolar macrophages. The clinical scores of vaccinated animals were lower than those of control animals, suggesting absence of immune-enhanced disease with vaccination [240].
4.6.3. Clinical trial data
Phase I/II (COV001)
Placebo controlled: yes. A licensed meningococcal vaccine was used.
Participants: healthy adults, 18–55 years of age (n = 1077)
Immunization schedule: prime only or prime-boost, intramuscular administration
Study participants in the vaccine group received a single dose of AZD1222 containing 5 × 1010 viral particles. Ten participants received an additional booster vaccination with AZD1222 that was administered 28 days after the first dose.
IgG titers against SARS-CoV-2 S peaked by day 28 and remained elevated at day 56 in participants in both dosing regimens, but levels were higher in the ten participants who received a booster dose. Neutralizing titers were measured by three different wild-type virus neutralizing antibody assays and a pseudovirus neutralization assay. Pseudoviral titers were measured in 35 participants who received 1 dose of the vaccine and 100% achieved neutralizing titers with a median titer of 218 (IQR 122–395) at day 28. The results from live and pseudovirus assays were highly correlated. Interferon-γ ELISpot responses against SARS-CoV-2 spike peptides peaked at 856 spot-forming cells per million PBMCs (IQR 493–1802; n = 43) at day 14, declining to 424 (221–799; n = 43) by day 56 after vaccination. Freshly isolated PBMCs were stimulated with peptide pools (10 μg/ml) comprising a total of 253 synthetic peptides (15mers overlapping by 10 peptides) spanning the entire vaccine insert, including the vector leader sequence (tPA). T cell responses were observed in both the prime only and prime-boost groups. The vaccine induced a Th1 response determined by CD4+ cytokine secretion (IFN-γ and TNF-α). A polyfunctional, cytotoxic CD8+ T cell response was also observed upon vaccination [241] . Pre-existing antibodies to the viral vector in a subset of participants did not correlate with the antibody responses induced against SARS-CoV-2 S. The reactogenicity profile in the AZD1222 group was higher than in the control meningococcal vaccine group. No serious AEs were reported in either vaccine group [51].
Though the study participants were initially planned to receive a single dose of AZD1222, the protocol was subsequently modified to a two-dose prime-boost regimen for the Phase II cohorts due to generation of a more robust immune response in the 10 participants who received a boost in the Phase I trial.
Another placebo controlled Phase I/II trial is underway in South Africa (COV005) in 2130 adults aged 18–65 who are either healthy or HIV positive (n = 100) on anti-retroviral therapy for at least three months and have a viral load of <1000 copies/ml. Participants are receiving 2 doses of 5–7.5 × 1010 vp in a prime – boost regimen 28 days apart (clinicaltrials.gov NCT04536051).
Phase II/III
Placebo controlled: yes
Participants: adults ≥18 years of age (n = 12,390 for UK (COV002), n = 40,000 for US, n = 10,300 for Brazil (COV003))
Immunization Schedule: The study protocol for doses and regimen are variable, intramuscular administration.
Interim analysis of efficacy and safety has been reported using pooled data from four trials- COV001 (Phase I/II), COV002 (Phase II/III), COV003 (Phase III) and COV005 (Phase I/II) [242]. Data from all four trials were used for assessing safety, whereas efficacy analysis was limited to data from a subset of participants of COV002 and COV003. The vaccine was safe and number of recorded AEs in participants were similar in the placebo and the vaccine groups. There were 3 reported cases of transverse myelitis, two in the vaccine group and one in the placebo group. Only one of them was judged to be vaccine related and had caused a temporary pause in the study.
Though efficacy was evaluated by pooling data from the COV002 and COV003 trials, there were differences in vaccination regimen and doses within cohorts of the COV002 trial and also between the two trials. For the UK COV002 study, participants aged 18–55 were recruited first followed by phased recruitments of older cohorts (56–69 years and > 70 years). Firstly, due to an error in the method used to quantify the number of viral particles in the vaccine during the early stages of the trial, a subset of participants in the 18–55 age cohort (n = 1367) received half (2.2 × 1010 viral particles) of the intended standard dose (SD) (5 × 1010 viral particles) for the prime. Secondly, the study was initially intended to be a single dose efficacy study but was later amended to be a two dose regimen based on Phase I results. So, there was considerable lag between the prime and boost (>12 weeks, median gap of 84 days) especially for the participants in the 18–55 age cohort, many of whom received a half dose (LD) for their prime. For the other participants of COV0002 the median interval between the prime and boost was 69 days. For the COV003 study, all participants received or are receiving two shots of the vaccine at a dose of 3.5–6.5 × 1010 viral particles with administration up to 12 weeks apart (median interval of 36 days). The total efficacy when both the low dose followed by standard dose (LD/SD) and 2 standard doses (SD/SD) participants were combined was 70.4% (95.8% CI 54.8–80.6, whereas for SD/SD participants it was 62.1% (95% CI 41.0–75.7) and for LD/SD it was 90.0% (95% CI 67.4–97.0). There was a non-significant increase in efficacy when the interval between prime and boost was >6 weeks (65.4%) as opposed to a gap of <6 weeks (53.4%). A subset of participants was also screened for asymptomatic infection, and the LD/SD group had an efficacy of 58.9% (95% CI 1.0%- 82.9%) versus 3.8% (95% CI -72.4% to 46.3%) for the SD/SD group. Though the vaccine has similar immunogenicity and better reactogenicity profile in older adults (>55 years old) [243], not enough cases were accrued to assess efficacy in this age bracket due to later recruitment. Anti-S protein T cell responses measured by IFN-γ ELISpot assay peaked at 14d after a single dose of AZD1222 (18–55 years: median 1187 spot-forming cells [SFCs] per million PBMCs [IQR 841–2428], n = 24; 56–69 years: 797 SFCs [383–1817], n = 29; and ≥ 70 years: 977 SFCs [458–1914], n = 48) and did not increase significantly after the booster dose.
Phase III studies of participants receiving two immunizations with AZD1222 or placebo are underway in US (clinicaltrials.gov NCT04516746). The primary study endpoints are: 1) PCR-confirmed COVID-19 cases ≥15 days post-boost vaccination, 2) safety and tolerability of the vaccination protocol, 3) reactogenicity.
AZD1222 has been authorized by the UK Medicines and Healthcare products Regulatory Agency (MHRA) for emergency supply for active immunization of adults 18 years or older with a recommendation of administration of two doses with an interval of between four and 12 weeks [244].
4.7. Developer: Gamaleya research institute
4.7.1. Vaccine: Sputnik V
Platform: two non-replicating viral vectors, adenovirus type 5 (rAd5) and adenovirus type 26 (rAd26)
Vaccine target: SARS-CoV-2 full-length glycoprotein S
This is non-replicating viral vectored vaccine with two different adenoviral vectors (recombinant Ad26 [rAd26] and recombinant Ad5 [rAd5]), both carrying the gene for SARS-CoV-2 spike glycoprotein in a prime-boost regimen rationalized to overcome boosting of viral vector specific antibodies.
Preclinical: No published data
Phase I/II:
Placebo controlled: no
Participants: healthy adults,18–60 years of age (n = 76)
Immunization schedule: prime only or prime-boost, intramuscular administration
Study participants received either a frozen vaccine formulation or a lyophilized vaccine formulation. In Phase I, a group of participants received one dose of either rAd26-S or rAd5-S and were assessed for safety over 28 days. In Phase II, a separate subset of participants received one dose of rAd26-S on day 0 and one dose of rAd5-S on day 21. For the prime-only regimen in Phase I, 100% of participants seroconverted to anti-RBD antibodies in the 21 days after vaccination. NAb titers were 61% and 77% for rAd26-S and rAd5-S vaccinated groups, respectively (these numbers were obtained by the authors after pooling data from the lyophilized and the frozen groups, since n was small in each group and there was no significant difference in response between the two vaccine formulations). For the Phase II participants, the seroconversion rate was also 100% when measured by RBD binding antibodies. 100% of participants in the prime-dose regimen of the Phase II study developed nAb titers. There was no significant correlation between baseline nAb titer against the viral vector and the titer of RBD-specific IgGs elicited after vaccination. Also, vaccination with one recombinant vector did not boost titers for the other vector, demonstrating lack of cross-reactivity between the two distinct viral vectors. Antigen-specific CD4+ and CD8+ T cell responses were observed following PBMC stimulation (samples collected after primary immunization) as measured by flow cytometry and IFN-γ secretion in 100% of the volunteers, particularly by d28. Significant cell proliferation was also observed in response to S protein stimulation. While IFN-γ is a marker of Th1-biased cellular responses, Phase III clinical trials will be further supplemented with more focus on Th1 and Th2 polarization. Data from the small number of participants showed that frozen and lyophilized vaccine formulations were equally immunogenic.
The vaccine was well tolerated during both phases of the trial with most of the AEs reported being transient, self-limiting and mild in severity. No serious AEs were reported.
Sputnik V received a highly controversial approval from Russian regulatory authorities, limiting rollout of the vaccine in the general population without a larger Phase III study. According to Gamaleya, the vaccine was used in high risk “red zones” of Russian hospitals. To date, the release says 10,000 people have received the vaccine under this authorization and the efficacy rate was more than 90%, but no data has been published to support these claims [245].
Phase III
Placebo controlled: yes
Participants: healthy adults 18–111 years of age (stratified as 18–30, 31–40, 41–50, 50–60 and 60+) (n = 40,000)
Immunization schedule: prime-boost, intramuscular
A Phase III trial in Russia is underway in which participants are randomized between two groups to receive either Sputnik V combined vector vaccine, in a prime-boost regimen on days 1 (component I rAd26-S) and 21 (component II rAd5-S) or two placebo doses. The primary outcome is the incidence of PCR-confirmed COVID-19 within 6 months of the first vaccine dose. The Gamaleya National Center of Epidemiology and Microbiology in Moscow and the Russian Direct Investment Fund released data from an interim analysis reporting 91.4% vaccine efficacy from 22,714 participants, though data from more participants are required to ascertain the final vaccine efficacy [246].
4.8. Developer: Moderna/NIAID
4.8.1. Vaccine: mRNA-1273
Platform: mRNA encoding a stabilized S is encapsulated in lipid nanoparticles.
Vaccine target: SARS-CoV-2 full-length S protein, designed with two proline mutations (S-2P) to help stabilize the prefusion conformation.
Route of administration: intramuscular.
4.8.2. Preclinical data
Immunogenicity and vaccine efficacy were tested in two pre-clinical animal challenge models: 1) mouse and 2) NHP (rhesus macaques). In mice, a prime/boost regimen was used with 3 different vaccine doses of 0.01, 0.1, or 1 μg mRNA. A dose-dependent induction of S-reactive antibodies was observed after each dose. Serum nAb activity, as assessed in a pseusdovirus neutralization assay, was observed only in the 1 μg/highest dose group. The T cell profile following mRNA vaccination was also assessed and compared to the response after vaccination with the S-2P protein immunogen. The protein immunogens were delivered with one of two adjuvants - Sigma Adjuvant System (SAS) or alum. Both the mRNA and protein platforms of S-2P elicited robust Th1 biased CD4+ T cell response when adjuvanted with SAS, but not with alum. Protective efficacy was assessed in BALB/cJ mice by challenging with 105 PFU of mouse-adapted SARS-CoV-2, which are replication competent in both the upper and lower respiratory tracts. Challenge occurred 5-week post-boost and animals receiving the 1 μg prime/boost vaccine regimen of mRNA-1273 showed complete protection from viral replication in lungs, and 6/7 mice had undetectable virus in the upper respiratory tract washes. Overall, protection, as measured by infectious virus in lungs after challenge, was dependent on vaccine dose. Mice challenged 13-weeks after prime/boost vaccination were similarly protected, demonstrating some durability of the response. Further, a single dose of 1 or 10 μg of mRNA-1273 was also completely protective in mice challenged 7 weeks after vaccination [247].
In the NHP model, a prime/boost regimen was also used with either 10 or 100 μg of mRNA-1273. mRNA-1273 induced robust S-specific nAb responses in NHPs at both doses in a neutralization assay using a SARS-CoV-2-luciferase virus strain, with nAb GMTs of 501 and 3481, in low and high dose groups, respectively. This was 12 to 84-fold higher than nAb titers from convalescent-phase serum from COVID-19 patients. Modest Th1 biased CD4+ T cell responses were elicited, but little or no CD8+ T cell response was observed. Four weeks after vaccination, animals were challenged with 7.6 × 105 plaque-forming units (PFU) (approximately equivalent to 106 TCID50) of SARS-CoV-2. At two days post-challenge, no detectable virus was present in the BAL fluid of 7/8 animals in both vaccinated groups, and in nasal swabs of any of the eight animals in the 100 μg dose group. Interestingly, whereas there was an observed dose-dependent increase in BAL antibody levels after vaccination, serum antibody levels in vaccinated animals remained stable. Taken together these data may indicate that vaccine elicited nAbs in the lung were protective and sterilizing and could be used as a correlate of protection [248]. Based on these encouraging preclinical data, Phase I trials were conducted in humans to assess vaccine safety and immunogenicity.
4.8.3. Clinical trial data
Phase I:
Placebo controlled: no
Participants: healthy adults, 18–55 years of age (n = 45), or ≥ 56 years of age (n = 40).
Immunization schedule: prime-boost, intramuscular
Healthy adults aged 18–55 years received a prime/boost regimen of 25 μg, 100 μg, or 250 μg mRNA per dose. All participants seroconverted by day 15 as measured by ELISA, and median antibody levels in participants, after one shot of either 100 μg or 250 μg mRNA-1273, was similar to the median magnitude observed in convalescent serum specimens. NAb titers were measured in recipients of the 25μg and 100μg doses, and dose-dependent increases in nAb titer were observed at day 43 post vaccination. Reduction of SARS-CoV-2 infectivity by 80% or more (PRNT80) was detected in all participants, with geometric mean PRNT80 responses of 339.7 in the 25 μg group and 654.3 in the 100 μg group. Neutralizing activity of these sera were superior compared with three convalescent specimens from COVID-19 patients. Binding to the vaccine antigen S-2P and neutralization titers were positively correlated. T cell responses against the S protein were measured by ICS. PBMCs were stimulated for 6 h with two peptide pools (S1 and S2) comprising of 15-mer peptides overlapping by 10 amino acids. S1 pool covered the N-terminus of SARS-CoV-2 S protein up to the furin cleavage site, while the C-terminus of the SARS-CoV-2 S protein up to the furin cleavage site was represented by the S2 pool. CD4+ T cell immune profiles were Th1 biased (TNF-α > IL-2 > IFN-γ) in both the 25 μg and 100 μg dose groups, similar to what was observed in preclinical NHP studies. Low levels of CD8+ T cell responses to the S2 peptide pool could be detected only in the 100 μg dose group after the second vaccination.
At least one AE was reported by all study participants, but only one participant in the 25 μg dose group was withdrawn because of vaccine associated transient urticaria and did not receive a second dose. Most of the AEs were mild to moderate while some severe AEs were associated with the 250μg dose group and were reported after the second vaccination [213].
The Phase I trial was then expanded to include 40 participants who were stratified further into two subgroups of people either 56–70 or > 71 years of age. In contrast to the younger adults, these participants received only the two lower doses of 25μg or 100μg mRNA-1273 because the highest dose was associated with increased toxicity in the 18–55 year-old. The vaccine was immunogenic in this cohort in both groups, with neutralizing activity detected in all participants after the boost. NAb titers were similar to those observed in the 18–55 year-old cohort. The cellular responses were also comparable to the 18–55 year-old cohort. Thus, in this small cohort, mRNA-1273 was safe and immunogenic in adults of all ages [49] The durability of the vaccine elicited humoral response has been studied in a subset of 34 participants who received two 100μg doses of the vaccine 28 days apart spread across all three age groups (18–55, 56–70 and > 71) [249] Levels of RBD-binding antibody titers as measured by ELISA and neutralization titers as determined by both live-virus plaque-reduction neutralization and pseudovirus neutralization assay remained elevated in all participants 90 days after the second dose (119 days after the first dose) demonstrating the induction of a robust and durable antibody response.
Phase III:
Placebo controlled: yes
Participants: adults >18 years of age with no known history of SARS-CoV-2 infection (n = 30,420)
Immunization schedule: 100 μg mRNA-1273 or placebo prime-boost, intramuscular
The trial of a prime/boost regimen with two 100 μg doses of mRNA-1273 has been completed. The primary outcome measures were the number of participants with a first instance of symptomatic COVID-19 14 days after receiving the second vaccine dose and the number of participants with solicited and unsolicited AEs or ARs with a time frame of up to 2 years after the second dose (ClinicalTrials.gov NCT04470427). Initial data from the Phase III trial has been reported. The vaccine was safe with a reactogenicity profile similar to that reported in the Phase I trials. The number of serious AEs reported were similar across the vaccine and the placebo arms. There was no evidence of vaccine-associated enhanced respiratory disease. mRNA-1273 met the statistical criteria pre-specified in the study protocol for efficacy, with an estimated vaccine efficacy of 94.1% (95% CI, 89.3 to 96.8%) based on 196 COVID-19 cases diagnosed for primary analysis, of which 185 cases of COVID-19 were observed in the placebo group versus 11 cases observed in the mRNA-1273 group. 30 severe cases (as defined in the study protocol) were reported with all in the placebo group and none in the mRNA-1273 vaccinated group, showing 100% efficacy in preventing severe COVID-19. Age, sex, race, ethnicity and presence of risk factors for severe COVID-19 did not have any impact on vaccine efficacy [250]. mRNA-1273 was authorized for distribution and use under an EUA on December 18, 2020 by the FDA in people 18 years or older [251].
4.9. Developer:Pfizer/BioNTech/Fosun
4.9.1. Vaccine: BNT162b2
Platform: lipid nanoparticle (LNP)-encapsulated, nucleoside-modified mRNA
Vaccine target: SARS-CoV-2 S protein designed to be stabilized in the pre-fusion conformation with a native furin cleavage site (BNT162b2)
Route of administration: intramuscular
BioNTech currently has 4 different candidate vaccines under evaluation based on different combinations of RNA vaccine platforms and antigen. Out of those, two lipid nanoparticle (LNP)-formulated nucleoside-modified mRNA (modRNA) candidates- BNT162b1 [252], encoding trimerized RBD and BNT162b2, encoding the full length prefusion, stabilized, S with a native furin cleavage site were advanced for Phase I clinical trials. BNT162b2 was chosen to advance to international Phase II/III clinical trials based on milder systemic reactogenicity profile and similar immunogenicity to BNT162b1 in Phase I studies. Here, we review data related to BNT162b2.
4.9.2. Preclinical data
Immunogenicity of the vaccine candidate was assessed in BALB/c mice. Animals were immunized once with any one of three different doses (0.2, 1, or 5 μg) of BNT162b2. The vaccine was immunogenic with a rapid dose-dependent development of robust serum IgG titers. On day 28 post immunization, the 50% pseudovirus neutralization GMTs were 26, 176, and 296 respectively for the 0.2, 1, and 5 μg dose groups. Th1 biased T cell responses were detected in recall assays performed 12 and 28 days after vaccination. As compared to control mice that received no antigen, mice that received 5μg dose of BNT162b2 had an increased frequency of plasma cells, class switched B cells, and overall germinal center activity at 12 days post-vaccination [253].
In a non-human primate model of infection, protective efficacy of the vaccine was tested in male 2–4 year-old rhesus macaques. The animals were immunized twice with 30 or 100 μg of BNT162b2, 21 days apart. Antibody concentrations were assessed by Luminex microsphere assay 7 days after the second dose. The geometric mean concentrations (GMCs) in macaques were almost 50-fold higher than the GMC of a panel of 38 SARS-CoV-2 convalescent human sera obtained at a time when the participants were asymptomatic. Peak neutralization GMT of 962 (14 days after booster dose of 30 μg immunogen) or 1689 (7 days after booster dose of 100 μg) were observed as compared to a GMT of 94 for the convalescent serum panel. GMTs of 285 and 310 for the 30μg and 100 μg doses 56 days after the first vaccination were observed attesting to the short-term durability of the response. Consistent with data from mice, Th1-biased T cell responses were observed. Animals that were immunized with two doses of 100 μg of BNT162b2 were challenged 55 days after the second dose with 1.05 × 106 PFU of SARS-CoV-2. There was no detectable viral RNA in BAL fluid at any time point sampled after challenge and no detectable viral RNA in nasal swabs three days and at subsequent time points after challenge. Thus, the vaccine protected animals from SARS-CoV-2 infection [253].
4.9.3. Clinical trial data
Phase I:
Placebo controlled: yes
Participants: healthy adults, 18–55 years of age (n = 45); age 65–85 (n = 45)
Immunization schedule: prime-boost, multiple doses tested, intramuscular administration
Safety and immunogenicity of BNT162b2 were assessed at three doses of 10 μg, 20 μg, or 30 μg (n = 15 per dose group, 12 received the vaccine and 3 received placebo) in Phase I clinical trials. Healthy participants received two 0.5 ml injections 21 days apart at any of the three dose levels. The immunogenicity was measured by neutralization assay and RBD–binding or S1-binding IgG by direct Luminex immunoassays on serum samples collected at days 0, 7, 21, 28 and 35 after the first vaccination. Convalescent sera collected from 38 patients, most of whom had moderate disease and were not hospitalized, were used for benchmarking the response. Antigen-binding IgG and virus-neutralizing titers exhibited a dose-response and were boosted further by the second dose in both age groups. The anti-S1 IgG response and the 50% neutralizing GMTs elicited by the 30 μg dose were measured on day 35. These were 13 and 1.7 times the GMT of the convalescent serum panel, respectively, among participants 18 to 55 years of age. In those 65 to 85 years of age, the anti-S1 IgG response and the 50% neutralizing GMTs were 9.5 and 2.2 times the GMT of the convalescent serum panel, respectively. The vaccine was less immunogenic in the older cohort both for binding IgG and neutralization titers across dose levels on day 28. As reported in a preprint, antigen-specific CD8+ and Th1-type (IFN-γ+) CD4+ T cell responses were observed in most participants who received two 30 μg doses of BNT162b2. A poly-epitopic CD4+ T cell response directed against both N- and C- terminal portions of the S protein was observed. The magnitude of S-specific CD4+ and CD8+ T cell responses correlated with S1-binding IgG, indicating a convergent development of the humoral and cellular adaptive immunity upon vaccination. In 3 participants, the CD8+ T cell response was also evaluated using pMHC-tetramers. The CD8+ T cells showed an early-differentiated effector-memory phenotype (CCR7loCD45RAlo) and markers associated with cognate activation (CD38, HLA-DR and PD-1), with single specificities reaching 0.01–3% of circulating CD8+ T cells [254].
Most of the AEs reported were mild to moderate in severity, showed dose-dependence and increased in incidence after the second injection. Though a small fraction of adults in the younger cohort developed severe systemic events, none of the older participants reported any severe symptom. None of the participants reported grade four local or systemic AEs [255].
Phase II/III:
Placebo controlled: yes
Participants: healthy people and participants with but not limited to chronic, stable human immunodeficiency virus (HIV), Hepatitis C virus (HCV), or Hepatitis B virus (HBV) infections stratified into three age groups:12–15, 16–55 and > 55 years of age (n = 43,448).
Immunization schedule: prime-boost, intramuscular administration
Based on the safety data from Phase I studies, two injections of BNT162b2 at the 30 μg dose 21 days apart was advanced to Phase II/III international trials. Numerous primary endpoints for this study are listed including PCR-confirmed COVID-19 occurring from 7 days after the second injection in participants with and without evidence of infection before vaccination, incidence of AEs and local and systemic reactions (ClinicalTrials.gov NCT04368728). Pfizer released data from Phase III trial meeting all primary efficacy endpoints. Analysis found that the vaccine had a favorable safety profile and reactogenicity was similar to Phase I trials with similar number of serious adverse events in the placebo and the vaccine groups (0.6% and 0.5%, respectively). The vaccine showed 95% (95% CI, 90.3% - 97.6%) efficacy against COVID-19 at least 7 days after the second dose. Over 36,523 participants who were seronegative at baseline there were 170 confirmed COVID-19 cases of which 162 were in the placebo group and 8 in the vaccine group. The efficacy was not impacted when subgroup analyses were done based on age, sex, race, ethnicity, obesity, and presence of a coexisting condition. A total of 10 cases of severe COVID-19 after the first dose of vaccine administration was registered, of which there was one case in the vaccine group as opposed to 9 cases in the placebo group. Though adolescents aged 12–15 years were included in the trial, efficacy and immunogenicity data from this cohort has not been reported in this study [256]. BNT162b2 was authorized for use under an EUA by the FDA on December 11, 2020 for active immunization to prevent COVID-19 in individuals 16 years of age and older [257].
4.10. Developer: Novavax
4.10.1. Vaccine: NVX-CoV2373
Platform: recombinant protein nanoparticle plus Matrix-M1 adjuvant
Vaccine target: Trimeric SARS-CoV-2 S protein designed to be stabilized in the pre-fusion conformation
Route of administration: intramuscular
NVAX-CoV2373 is a protein subunit vaccine based on a full-length S, including the transmembrane and the cytoplasmic tail. To stabilize the prefusion conformation, the polybasic cleavage site was removed, and proline substitutions were introduced, as discussed above. The S protein was expressed and purified from insect cells in the presence of detergent which led to some higher order nanoparticle formation, as determined using electron microscopy. Imaging revealed that the antigen exists as mixture of free trimers as well as higher order multi-trimer rosettes [258].
4.10.2. Preclinical data
Immunogenicity was assessed in BALB/c mice that were immunized with a range (0.01, 0.1, 1 and 10 μg) of NVX-CoV2373 adjuvanted with Matrix-M. Mice received a single dose or a prime-boost regimen spaced 14-days apart. In the single vaccination group, 0.1-10 μg NVX-CoV2373 elicited anti-S IgG titers but nAb responses were only detected in the 10μg group. Animals immunized with the prime/boost regimen had significantly higher levels of anti-S IgG and nAb titers as compared to the prime only group across all doses. NAb responses were not significantly different between the 1 μg and 10 μg groups that received the prime-boost regimen. Importantly, use of Matrix-M adjuvant with NVX-CoV2373 lead to significantly higher anti-S IgG titers across all dose groups when compared to mice immunized with 10 μg NVX-CoV2373 alone [259].
Challenge studies were performed in mice that were immunized with a similar regimen as described above. Mice were made permissive to wild-type SARS-CoV-2 virus infection by intranasal infection of adenovirus expressing hACE2 (Ad/hACE2) four days before challenge. Following challenge, weight was monitored daily, and pulmonary histology and viral load were studied. A dose dependent decrease of virus titer in the animals immunized with vaccine and adjuvant was observed in the single vaccination regimen. In the prime-boost groups, mice immunized with all doses except the lowest dose had very low or undetectable lung titers after challenge. All animals that were immunized with adjuvanted vaccine were protected from weight loss compared to placebo or vaccine-only groups. Histopathology showed that NVX-CoV2373 in presence of Matrix-M adjuvant reduced lung inflammation after challenge. Addition of Matrix-M adjuvant was also associated with enhanced germinal center formation and T cell responses [259].
The immunogenicity of the vaccine was next evaluated in adult baboons with a dose range (1, 5 and 25 μg) of NVX-CoV2373 with 50 μg Matrix-M adjuvant in a prime boost regimen with a 21-days interval. The role of adjuvant was assessed by immunizing an additional group of baboons with 25 μg of NVX-CoV2373 only. Robust anti-S nAb titers were observed following the prime-boost with NVX-CoV2373/Matrix-M across all doses. Animals that received the vaccine without adjuvant did not show appreciable antibody responses. Animals receiving either of the two highest doses of vaccine/adjuvant showed the best T cell responses which were 5-fold higher than the vaccine only groups [259].
The protective efficacy of NVX-CoV2373/Matrix-M was determined in cynomolgus macaques that were immunized with the prime-boost regimen of 2.5, 5, or 25 μg NVX-CoV2373 and 25 or 50 μg Matrix-M adjuvant in the two highest vaccine dose groups. Vaccinated animals that developed significant nAb titers relative to convalescent sera were challenged with SARS-CoV-2. There was no detectable viral RNA in nasal swabs of vaccinated animals and virtually no detectable RNA in the BAL fluid. Placebo control animals had moderate-to-severe lung inflammation whereas little or no inflammation was observed in the lungs of macaques immunized with NVX-CoV2373/Matrix-M [260].
4.10.3. Clinical data
Phase I
Placebo controlled: yes
Participants: 131 healthy adults, ages 18–59
Immunization schedule: multiple doses +/− adjuvant tested in a prime-boost schedule (day 0 and 21). Intramuscular administration.
For the Phase I trial study participants were divided into multiple groups receiving 5 or 25μg NVX-CoV2373 with or without Matrix-M in a prime only or prime-boost schedule.
Anti-spike IgG were detected in all adjuvanted (5 or 25μg) groups after the first vaccination with 10-fold higher levels in those receiving adjuvant as compared to the unadjuvanted group. A single vaccination with adjuvant (both 5 or 25μg dose groups) resulted in IgG titers that were similar to those from COVID-19 patients who had subclinical infections. No significant differences were observed in the antibody responses between a prime-boost regimen with 5 μg or 25 μg NVX-CoV2373 plus Matrix-M. T-cell responses measured by ICS following a 6 h stimulation with rSARS-CoV-2 in 16 randomly selected participants from select groups showed that adjuvanted regimens induced antigen-specific polyfunctional CD4+ T-cell responses at d28 which were skewed to a Th1 phenotype (IFN-γ, TNF-α, IL-2).
Reactogenicity was largely absent or mild and self-limited. Mild unsolicited AEs were reported that were similarly distributed across the groups receiving adjuvanted and unadjuvanted vaccine. No severe AEs were reported [228].
Phase IIa/b
Placebo controlled: yes
Participants: Cohort 1: healthy adults, 18–84 years of age (n = up to 4164); Cohort 2: medically stable HIV-positive (HIV+) adults, 18–64 years of age (n = 240)
Immunization schedule: prime-boost schedule (day 0 and 21), intramuscular administration.
This trial is underway in South Africa. One cohort of healthy adults and one cohort of medically stable HIV+ adults are being immunized with two doses of 5 μg NVX-CoV2373 + 50 μg Matrix-M1 adjuvant. There are multiple primary endpoints for this study including the number of PCR-confirmed COVID-19 cases and AEs in both groups (ClinicalTrials.gov NCT04533399).
Phase III
Placebo controlled: yes
Participants: healthy adults 18–84 years of age (n = 15,000)
Immunization schedule: prime-boost, intramuscular administration.
This Phase III trial underway in the United Kingdom is targeting enrollment of at least 25% of participants who are ≥65 years of age. In addition, other groups that are most affected by COVID-19 are being prioritized for enrollment, including racial and ethnic minorities. The trial is designed to study the efficacy, immunogenicity and safety of two doses of 5 μg NVX-CoV2373 + 50 μg Matrix-M1 adjuvant compared with placebo. An additional study arms are receiving the NVX-CoV2373+ Matrix-M1 vaccine along with the seasonal influenza virus vaccine or the seasonal influenza virus vaccine alone. The primary outcome is the number of participants with first occurrence of PCR-confirmed COVID-19 with onset from Day 28 through the length of the study (ClinicalTrials.gov NCT04583995).
Another Phase III clinical trial of NVX-CoV2373/Matrix-M1 with 30,000 participants is ongoing in the United States and Mexico (ClinicalTrials.gov NCT04611802).
5. Key takeaways
The critical need to address the COVID-19 pandemic has spurred the development of multiple promising vaccine candidates which can now be harnessed to address the global vaccine burden. The successes that have already been reported related to vaccine efficacy, as outlined above, further validate the foundation laid by decades of scientific research in immunology and vaccine development. While we have assessed the pros and cons of the different vaccine candidates, comparing them in a head-to-head manner is generally not possible due to dissimilar study parameters and absence of defined correlates of immunity to predict effectiveness.
With this in mind, some general principles emerge from the review of ongoing late-stage vaccine candidates. All of the advanced SARS-CoV-2 vaccine candidates have reported high efficacy, typically >90% from early-to-late-stage Phase III trials. Based on the results from Phase I/II trials, most studies are evaluating a prime-boost regimen and in some cases with two different dosages. Interestingly, the three studies (Ad5-nCoV, Ad26.COV2·S and AZD1222) that are continuing to evaluate a single immunization in Phase III trials use a non-replicating adenovirus vector platform.
In terms of dosage, typically, higher doses of each vaccine candidate were evaluated in Phase I/II studies relative to Phase III. The inactivated virus candidates are being tested at doses ranging between 2.5 and 10 μg. Viral vector-based vaccine candidates are being tested at doses ranging between 5 × 1010–1.5 × 1011 viral particles. Of the two mRNA-based vaccines, BNT162b2 was tested at a lower dose (10-30 μg) in comparison to mRNA-1273 (25-250 μg) in Phase I/II trials. Subsequently, based on follow-up studies in Phase III trials, the emergency authorized doses for mRNA-1273 and BNT162b2 will contain 100 μg and 30 μg respectively. The only protein-based vaccine candidate, NVX-CoV2373, reviewed here was tested at 5 μg and 25 μg in Phase I trials.
While a dose-dependent increase in the immunogenicity profile was consistently observed, the higher doses were also associated with an increased incidence of AEs and thus often discontinued in Phase III trials. The early phases of testing demonstrate the need for evaluation of multiple dosages to balance the trade-off between dose sparing and immunogenicity. Another feature that is being observed consistently across all the different vaccine platforms from Phase I/II trials is that the optimal window for a booster immunization is generally between 21 and 28 days, consistent with previous knowledge in human vaccinology. A second immunization at shorter intervals (<21 days) often did not result in a significant increase in antigen-specific antibody titers [47,154,237]. All of the vaccine candidates induced high-levels of seroconversion in healthy adults, independent of assay variability, and in the vast majority cases, the early neutralization titers correlated well with the binding antibody titers.
6. Discussion
The early data from clinical trials of SARS-CoV-2 vaccine candidates are very encouraging. While these reports give hope that multiple vaccines will soon be available to prevent infections in many people, one significant issue with the vast majority of ongoing Phase III studies is absence of diversity in study cohorts and little or no representation of populations with underlying conditions that confer risk for severe COVID-19. While the vaccines have thus far been reasonably safe and well tolerated, safety profiles in populations that are at highest risk – those other than healthy adults – remain unknown.
Another caveat in the vaccine development pipeline is that commonly used measures of immunogenicity are often extrapolated to make predictions about efficacy. Though these readouts give some important information about the vaccine candidate, their relevance for predicting vaccine efficacy in humans is speculative. Furthermore, determinants of protection are likely to vary between groups of people based on age and other health considerations [261]. In addition to safety and efficacy, the durability of vaccine responses is an extremely important consideration for vaccine selection, yet it is one that will not be a factor in the selection of the first SARS-CoV-2 vaccines for use in humans.
The most important next step is to define these determinants of immunity and the viral targets of protective immune responses against SARS-CoV-2. Based on current published literature, a robust neutralizing antibody response against the S protein is ideal for providing sterilizing immunity. Thus, a majority of the initial analyses of immune correlates of protection have been focused on the S protein. However, our understanding of host factors and viral targets that provide immunity against SARS-CoV-2 infection or disease remains incomplete. These are also likely to vary among people based on factors such as age, sex, and comorbidities.
While the first wave of vaccines in development is mostly focused on eliciting neutralizing antibodies, a longer-term goal for SARS-CoV-2 vaccine design is to elicit broad immunity against distinct virus variants and strains. Early reports have found little antigenic variation among SARS-CoV-2 isolates and sequencing shows a rate of genetic variation in SARS-CoV-2 that is 2 to 6-fold lower than that observed in influenza viruses [262]. This is somewhat encouraging, though we expect that the rate of antigenic drift in SARS-CoV-2 will increase over time, under the selective pressure of neutralizing antibodies in the population.The end of 2020 presented us with just this scenario, as reports have come out describing new variants that have independently emerged out of the UK (B.1.1.7), South Africa (501Y.V2 also known as B.1.351) and Brazil (B.1.1.28) [[280], [281], [282]]. These have caught the attention of scientists due to multiple mutations that are carried together in the SARS-CoV-2 spike protein, and the fact that they each have been linked to a rapid increase in new cases spreading across their respective countries. The South African variant is also of particular concern due to additional mutations that are carried at key residues in the RBD. Preliminary studies are suggesting that convalescent serum from individuals infected with other strains of SARS-CoV2, as well as serum from vaccinated individuals, may be less effective at neutralizing the 501Y.V2 strain [[283], [284], [285]]. Therefore, development of broad vaccines against SARS viruses or more broadly against coronaviruses will be important. This will almost certainly require immune responses that can recruit specific cellular effector functions through broadly reactive, potentially non-neutralizing antibody responses and that engage T cell responses against conserved domains of coronavirus proteins [[263], [264], [265], [266], [267]]. A consideration here is that, unlike viruses that bud directly from the cell membranes which express a high level of viral proteins on the surface of infected cells, coronavirus particles are released from infected cells by exocytosis. Because of this, relatively little viral protein may be present on the surface of infected cells. Thus, targeting infected cells by recruiting broadly reactive antibody responses will likely require an understanding of which viral proteins are expressed at higher levels on infected cells; these may not be the same as the proteins that are expressed on the surface of virions themselves. As discussed in Section 2, potential candidates to study for this purpose are ORF3A and ORF7A as they may be expressed on the plasma membrane of SARS-CoV-2-infected cells. Overall, one clear strategy for improving the breadth of the immune response is to incorporate other proteins in addition to the S into vaccines. These should be proteins, or subunits of proteins, that are relatively conserved among SARS viruses, and potentially other types of coronaviruses. Much can be learned on this topic from research on influenza virus vaccine design, largely over the last 15 years, where great strides have been made toward the development of broadly protective vaccines [[268], [269], [270], [271], [272], [273], [274], [275], [276], [277]].
Finally, SARS-CoV-2 vaccination can stem the pandemic only if enough of the population is vaccinated to achieve herd immunity. The threshold for herd immunity through vaccination is a function of R0 of the pathogen. For SARS-CoV-2, this is estimated to be 50%–67% for a population with no preexisting immunity and equal susceptibility [278]. The accuracy of this prediction is contingent not only on the ability to properly estimate the reproductive number, but also on the ability of the vaccine to prevent continued transmission. Thus, even with an extremely effective vaccine, uptake will need to be high, and likely greater than 67%, in order to achieve reasonable population immunity. On this topic, data from the Vaccine Confidence Project (VCP), which tracks the beliefs and attitudes of people around the world about vaccine safety and effectiveness, shows that we need to dramatically improve education about how vaccines work and how they have been used to control infectious diseases over the past ~100 years. Recent research from the VCP has shown that 83% of individuals in the United States are worried about the safety of vaccines that are currently being expedited for use [279]. This is not altogether surprising given the maelstrom of political rhetoric and misinformation that exists around SARS-CoV-2 in the media, yet it suggests that a challenge is on the horizon - to promote broad and expedient compliance with uptake of the new SARS-CoV-2 vaccines. Nonetheless, the combined forces that have come together to stop the SARS-CoV-2 pandemic are certain to result in the approval of multiple highly effective vaccines that will save lives. This is but one profound demonstration of the impact that basic science research can have on human health and should serve as a reminder that the combined efforts of physicians and scientists are required to drive advancements in medicine..
Declaration of Competing Interest
The authors have no competing interests to declare.
Acknowledgments
We thank JoEllen Barnett for helpful comments and suggestions. Support was received from Stanford University, the Chan Zuckerberg Biohub, Fast Grants, CEND COVID Catalyst Fund, the National Institute Of Allergy And Infectious Diseases of the National Institutes of Health under Award Numbers U19AI111825, U54CA260517 and R01AI139119. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Ahmed R., Burton D.R. Viral vaccines: past successes and future challenges. Curr. Opin. Virol. 2013;3:307–308. doi: 10.1016/j.coviro.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Padron-Regalado E. Vaccines for SARS-CoV-2: lessons from other coronavirus strains. Infect. Dis. Ther. 2020:1–20. doi: 10.1007/s40121-020-00300-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Riel D., de Wit E. Next-generation vaccine platforms for COVID-19. Nat. Mater. 2020;19:810–812. doi: 10.1038/s41563-020-0746-0. [DOI] [PubMed] [Google Scholar]
- 4.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 5.U.S.D.o.H.a.H.S.F.a.D.A.C.f.B.E.a . In: Research, Development and Licensure of Vaccines to Prevent COVID-19 Guidance for Industry. FDA, editor. 2020. [Google Scholar]
- 6.P.a. BioNTech Pfizer and Biontech Conclude Phase 3 Study of COVID-19 Vaccine Candidate, Meeting all Primary Efficacy Endpoints. 2020. https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-conclude-phase-3-study-covid-19-vaccine
- 7.Moderna Moderna's COVID-19 Vaccine Candidate Meets its Primary Efficacy Endpoint in the First Interim Analysis of the Phase 3 COVE Study. 2020. https://investors.modernatx.com/news-releases/news-release-details/modernas-covid-19-vaccine-candidate-meets-its-primary-efficacy?utm_source=All+Contacts&utm_campaign=d501d4d76b-EMAIL_CAMPAIGN_2018_08_21_06_59_COPY_01&utm_medium=email&utm_term=0_ec5e481dc7-d501d4d76b-182537029
- 8.W.H. Organization . 2020. DRAFT Landscape of COVID-19 Candidate Vaccines. [Google Scholar]
- 9.New York Times. 2020
- 10.Pfizer B. In: Vaccines and Related Biological Products Advisory Committee Meeting December 10, 2020. FDA, editor. 2020. https://www.fda.gov/media/144245/download. [Google Scholar]
- 11.ModernaTX . In: Vaccines and Related Biological Products Advisory Committee Meeting December 17, 2020. FDA, editor. 2020. https://www.fda.gov/media/144434/download. [Google Scholar]
- 12.CDC Pfizer-BioNTech COVID-19 Vaccine. 2020. https://www.cdc.gov/vaccines/covid-19/info-by-product/pfizer/index.html
- 13.CDC Moderna COVID-19 Vaccine. 2020. https://www.cdc.gov/vaccines/covid-19/info-by-product/moderna/index.html
- 14.Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta Mol. basis Dis. 2020;1866 doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cevik M., Kuppalli K., Kindrachuk J., Peiris M. Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ. 2020;371:m3862. doi: 10.1136/bmj.m3862. [DOI] [PubMed] [Google Scholar]
- 17.M. The Lancet Respiratory COVID-19 transmission-up in the air. Lancet Respir. Med. 2020;8:1159. doi: 10.1016/S2213-2600(20)30514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klompas M., Baker M.A., Rhee C. Airborne transmission of SARS-CoV-2: theoretical considerations and available evidence. JAMA. 2020;324:441–442. doi: 10.1001/jama.2020.12458. [DOI] [PubMed] [Google Scholar]
- 19.Prather K.A., Wang C.C., Schooley R.T. Reducing transmission of SARS-CoV-2. Science. 2020;368:1422–1424. doi: 10.1126/science.abc6197. [DOI] [PubMed] [Google Scholar]
- 20.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith J.O., de Wit E., Munster V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang M., Wei J., Yuan J., Guo J., Zhang Y., Hang J., Qu Y., Qian H., Zhuang Y., Chen X., Peng X., Shi T., Wang J., Wu J., Song T., He J., Li Y., Zhong N. Probable evidence of fecal aerosol transmission of SARS-CoV-2 in a high-rise building. Ann. Intern. Med. 2020;173:974–980. doi: 10.7326/M20-0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.C.f.D.C.a. Prevention . 2020. Scientific Brief: SARS-CoV-2 and Potential Airborne Transmission. [Google Scholar]
- 23.Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., Yang Y., Liu B., Wang W., Wei C., Yang J., Ye G., Cheng Z. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020;92:833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- 24.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K., V.C.I.T. Washington State -nCo First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao F., Sun J., Xu Y., Li F., Huang X., Li H., Zhao J., Huang J., Zhao J. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020;26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., Azman A.S., Reich N.G., Lessler J. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buitrago-Garcia D., Egli-Gany D., Counotte M.J., Hossmann S., Imeri H., Ipekci A.M., Salanti G., Low N. Occurrence and transmission potential of asymptomatic and presymptomatic SARS-CoV-2 infections: a living systematic review and meta-analysis. PLoS Med. 2020;17 doi: 10.1371/journal.pmed.1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kissler S.M., Tedijanto C., Goldstein E., Grad Y.H., Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science. 2020;368:860–868. doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruan L., Wen M., Zeng Q., Chen C., Huang S., Yang S., Yang J., Wang J., Hu Y., Ding S., Zhang Y., Zhang H., Feng Y., Jin K., Zhuge Q. New measures for the coronavirus disease 2019 response: a lesson from the Wenzhou experience. Clin. Infect. Dis. 2020;71:866–869. doi: 10.1093/cid/ciaa386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou T., Liu Q., Yang Z., Liao J., Yang K., Bai W., Lu X., Zhang W. Preliminary prediction of the basic reproduction number of the Wuhan novel coronavirus 2019-nCoV. J. Evid. Based Med. 2020;13:3–7. doi: 10.1111/jebm.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyerowitz E.A., Richterman A., Bogoch I.I., Low N., Cevik M. Towards an accurate and systematic characterisation of persistently asymptomatic infection with SARS-CoV-2. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30837-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu X., Nergiz A.I., Maraolo A.E., Bogoch I.I., Low N., Cevik M. Defining the role of asymptomatic and pre-symptomatic SARS-CoV-2 transmission – a living systematic review. medRxiv. 2020 doi: 10.1101/2020.09.01.20135194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Kain A. Mac, Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., Garcia-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu D.X., Fung T.S., Chong K.K., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narayanan K., Huang C., Makino S. SARS coronavirus accessory proteins. Virus Res. 2008;133:113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson C.W., Ardern Z., Goldberg T.L., Meng C., Kuo C.H., Ludwig C., Kolokotronis S.O., Wei X. Dynamically evolving novel overlapping gene as a factor in the SARS-CoV-2 pandemic. Elife. 2020;9 doi: 10.7554/eLife.59633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ito N., Mossel E.C., Narayanan K., Popov V.L., Huang C., Inoue T., Peters C.J., Makino S. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 2005;79:3182–3186. doi: 10.1128/JVI.79.5.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang C., Ito N., Tseng C.T., Makino S. Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. J. Virol. 2006;80:7287–7294. doi: 10.1128/JVI.00414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., Acton O.J., Jaconi S., Guarino B., Minola A., Zatta F., Sprugasci N., Bassi J., Peter A., De Marco A., Nix J.C., Mele F., Jovic S., Rodriguez B.F., Gupta S.V., Jin F., Piumatti G., Presti G. Lo, Pellanda A.F., Biggiogero M., Tarkowski M., Pizzuto M.S., Cameroni E., Havenar-Daughton C., Smithey M., Hong D., Lepori V., Albanese E., Ceschi A., Bernasconi E., Elzi L., Ferrari P., Garzoni C., Riva A., Snell G., Sallusto F., Fink K., Virgin H.W., Lanzavecchia A., Corti D., Veesler D. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042. doi: 10.1016/j.cell.2020.09.037. (e1021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J.B., Chen R.E., Havenar-Daughton C., Snell G., Telenti A., Virgin H.W., Lanzavecchia A., Diamond M.S., Fink K., Veesler D., Corti D. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 43.Tortorici M.A., Veesler D. Structural insights into coronavirus entry. Adv. Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., Bentlage A.E.H., van Haaren M.M., Guerra D., Burger J.A., Schermer E.E., Verheul K.D., van der Velde N., van der Kooi A., van Schooten J., van Breemen M.J., Bijl T.P.L., Sliepen K., Aartse A., Derking R., Bontjer I., Kootstra N.A., Wiersinga W.J., Vidarsson G., Haagmans B.L., Ward A.B., de Bree G.J., Sanders R.W., van Gils M.J. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., Guo X.V., Cerutti G., Bimela J., Gorman J., Zhou T., Chen Z., Yuen K.Y., Kwong P.D., Sodroski J.G., Yin M.T., Sheng Z., Huang Y., Shapiro L., Ho D.D. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 46.Addetia A., Crawford K.H.D., Dingens A., Zhu H., Roychoudhury P., Huang M.L., Jerome K.R., Bloom J.D., Greninger A.L. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with a high attack rate. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xia S., Zhang Y., Wang Y., Wang H., Yang Y., Gao G.F., Tan W., Wu G., Xu M., Lou Z., Huang W., Xu W., Huang B., Wang H., Wang W., Zhang W., Li N., Xie Z., Ding L., You W., Zhao Y., Yang X., Liu Y., Wang Q., Huang L., Yang Y., Xu G., Luo B., Wang W., Liu P., Guo W., Yang X. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2021;21(1):39–51. doi: 10.1016/S1473-3099(20)30831-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Logunov D.Y., Dolzhikova I.V., Zubkova O.V., Tukhvatullin A.I., Shcheblyakov D.V., Dzharullaeva A.S., Grousova D.M., Erokhova A.S., Kovyrshina A.V., Botikov A.G., Izhaeva F.M., Popova O., Ozharovskaya T.A., Esmagambetov I.B., Favorskaya I.A., Zrelkin D.I., Voronina D.V., Shcherbinin D.N., Semikhin A.S., Simakova Y.V., Tokarskaya E.A., Lubenets N.L., Egorova D.A., Shmarov M.M., Nikitenko N.A., Morozova L.F., Smolyarchuk E.A., Kryukov E.V., Babira V.F., Borisevich S.V., Naroditsky B.S., Gintsburg A.L. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet. 2020;396:887–897. doi: 10.1016/S0140-6736(20)31866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A.B., Flach B., Lin B.C., Doria-Rose N.A., O’Dell S., Schmidt S.D., Corbett K.S., Swanson P.A., 2nd, Padilla M., Neuzil K.M., Bennett H., Leav B., Makowski M., Albert J., Cross K., Edara V.V., Floyd K., Suthar M.S., Martinez D.R., Baric R., Buchanan W., Luke C.J., Phadke V.K., Rostad C.A., Ledgerwood J.E., Graham B.S., Beigel J.H. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 2020;383(25):2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sadoff J., Le Gars M., Shukarev G., Heerwegh D., Truyers C., de Groot A.M., Stoop J., Tete S., Van Damme W., Leroux-Roels I., Berghmans P.-J., Kimmel M., Van Damme P., De Hoon J., Smith W., Stephenson K., Barouch D., De Rosa S., Cohen K., McElrath J., Cormier E., Scheper G., Hendriks J., Struyf F., Douoguih M., Van Hoof J., Schuitemaker H. Safety and immunogenicity of the Ad26.COV2.S COVID-19 vaccine candidate: interim results of a phase 1/2a, double-blind, randomized, placebo-controlled trial. medRxiv. 2020 doi: 10.1101/2020.09.23.20199604. [DOI] [Google Scholar]
- 51.Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E.A., Dold C., Faust S.N., Finn A., Flaxman A.L., Hallis B., Heath P., Jenkin D., Lazarus R., Makinson R., Minassian A.M., Pollock K.M., Ramasamy M., Robinson H., Snape M., Tarrant R., Voysey M., Green C., Douglas A.D., Hill A.V.S., Lambe T., Gilbert S.C., Pollard A.J., Oxford C.V.T.G. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu F.C., Li Y.H., Guan X.H., Hou L.H., Wang W.J., Li J.X., Wu S.P., Wang B.S., Wang Z., Wang L., Jia S.Y., Jiang H.D., Wang L., Jiang T., Hu Y., Gou J.B., Xu S.B., Xu J.J., Wang X.W., Wang W., Chen W. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395:1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., McMahan K., Tostanoski L.H., He X., Martinez D.R., Rutten L., Bos R., van Manen D., Vellinga J., Custers J., Langedijk J.P., Kwaks T., Bakkers M.J.G., Zuijdgeest D., Huber S.K. Rosendahl, Atyeo C., Fischinger S., Burke J.S., Feldman J., Hauser B.M., Caradonna T.M., Bondzie E.A., Dagotto G., Gebre M.S., Hoffman E., Jacob-Dolan C., Kirilova M., Li Z., Lin Z., Mahrokhian S.H., Maxfield L.F., Nampanya F., Nityanandam R., Nkolola J.P., Patel S., Ventura J.D., Verrington K., Wan H., Pessaint L., Van Ry A., Blade K., Strasbaugh A., Cabus M., Brown R., Cook A., Zouantchangadou S., Teow E., Andersen H., Lewis M.G., Cai Y., Chen B., Schmidt A.G., Reeves R.K., Baric R.S., Lauffenburger D.A., Alter G., Stoffels P., Mammen M., Van Hoof J., Schuitemaker H., Barouch D.H. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., Zatta F., De Marco A., Guarino B., Bianchi S., Lauron E.J., Tucker H., Zhou J., Peter A., Havenar-Daughton C., Wojcechowskyj J.A., Case J.B., Chen R.E., Kaiser H., Montiel-Ruiz M., Meury M., Czudnochowski N., Spreafico R., Dillen J., Ng C., Sprugasci N., Culap K., Benigni F., Abdelnabi R., Foo S.C., Schmid M.A., Cameroni E., Riva A., Gabrieli A., Galli M., Pizzuto M.S., Neyts J., Diamond M.S., Virgin H.W., Snell G., Corti D., Fink K., Veesler D. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A., Martinez D.R., Loos C., Atyeo C., Fischinger S., Burke J.S., Slein M.D., Chen Y., Zuiani A., Lelis F.J.N., Travers M., Habibi S., Pessaint L., Van Ry A., Blade K., Brown R., Cook A., Finneyfrock B., Dodson A., Teow E., Velasco J., Zahn R., Wegmann F., Bondzie E.A., Dagotto G., Gebre M.S., He X., Jacob-Dolan C., Kirilova M., Kordana N., Lin Z., Maxfield L.F., Nampanya F., Nityanandam R., Ventura J.D., Wan H., Cai Y., Chen B., Schmidt A.G., Wesemann D.R., Baric R.S., Alter G., Andersen H., Lewis M.G., Barouch D.H. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schafer A., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Martinez D.R., Williamson L.E., Chen E.C., Jones T., Day S., Myers L., Hassan A.O., Kafai N.M., Winkler E.S., Fox J.M., Shrihari S., Mueller B.K., Meiler J., Chandrashekar A., Mercado N.B., Steinhardt J.J., Ren K., Loo Y.M., Kallewaard N.L., McCune B.T., Keeler S.P., Holtzman M.J., Barouch D.H., Gralinski L.E., Baric R.S., Thackray L.B., Diamond M.S., Carnahan R.H., Crowe J.E., Jr. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., Gong Y., Xiao H., Fan Z., Tan S., Wu G., Tan W., Lu X., Fan C., Wang Q., Liu Y., Zhang C., Qi J., Gao G.F., Gao F., Liu L. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. (e286) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walls A.C., Tortorici M.A., Snijder J., Xiong X., Bosch B.J., Rey F.A., Veesler D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 2017;114:11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bestle D., Heindl M.R., Limburg H., Van T. Van Lam, Pilgram O., Moulton H., Stein D.A., Hardes K., Eickmann M., Dolnik O., Rohde C., Klenk H.D., Garten W., Steinmetzer T., Bottcher-Friebertshauser E. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Allianc. 2020;3 doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heald-Sargent T., Gallagher T. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4:557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Jr., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walls A.C., Xiong X., Park Y.J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., Zambon M., Rey F.A., Corti D., Veesler D. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039. doi: 10.1016/j.cell.2018.12.028. (e1015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., Hagglof T., Oliveira T.Y., Viant C., Hurley A., Hoffmann H.H., Millard K.G., Kost R.G., Cipolla M., Gordon K., Bianchini F., Chen S.T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A.W., Waltari E., Pak J.E., Huey-Tubman K.E., Koranda N., Hoffman P.R., West A.P., Jr., Rice C.M., Hatziioannou T., Bjorkman P.J., Bieniasz P.D., Caskey M., Nussenzweig M.C. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krammer F., Simon V. Serology assays to manage COVID-19. Science. 2020;368:1060–1061. doi: 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- 68.Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., Kong W.P., Andres E.L., Kettenbach A.N., Denison M.R., Chappell J.D., Graham B.S., Ward A.B., McLellan J.S. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kirchdoerfer R.N., Wang N., Pallesen J., Wrapp D., Turner H.L., Cottrell C.A., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8:15701. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walls A.C., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J.M., DiMaio F., Rey F.A., Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tortorici M.A., Walls A.C., Lang Y., Wang C., Li Z., Koerhuis D., Boons G.J., Bosch B.J., Rey F.A., de Groot R.J., Veesler D. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019;26:481–489. doi: 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walls A., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J., DiMaio F., Rey F.A., Veesler D. Crucial steps in the structure determination of a coronavirus spike glycoprotein using cryo-electron microscopy. Protein Sci. 2017;26:113–121. doi: 10.1002/pro.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tao Y., Strelkov S.V., Mesyanzhinov V.V., Rossmann M.G. Structure of bacteriophage T4 fibritin: a segmented coiled coil and the role of the C-terminal domain. Structure. 1997;5:789–798. doi: 10.1016/s0969-2126(97)00233-5. [DOI] [PubMed] [Google Scholar]
- 74.Chakraborty S., Gonzalez J., Edwards K., Mallajosyula V., Buzzanco A.S., Sherwood R., Buffone C., Kathale N., Providenza S., Xie M.M., Andrews J.R., Blish C.A., Singh U., Dugan H., Wilson P.C., Pham T.D., Boyd S.D., Nadeau K.C., Pinsky B.A., Zhang S., Memoli M.J., Taubenberger J.K., Morales T., Schapiro J.M., Tan G.S., Jagannathan P., Wang T.T. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat. Immunol. 2021;22:67–73. doi: 10.1038/s41590-020-00828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang T.T. IgG Fc glycosylation in human immunity. Curr. Top. Microbiol. Immunol. 2019;423:63–75. doi: 10.1007/82_2019_152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Del Valle D.M., Kim-Schulze S., Huang H.H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., Marron T.U., Xie H., Patel M., Tuballes K., Van Oekelen O., Rahman A., Kovatch P., Aberg J.A., Schadt E., Jagannath S., Mazumdar M., Charney A.W., Firpo-Betancourt A., Mendu D.R., Jhang J., Reich D., Sigel K., Cordon-Cardo C., Feldmann M., Parekh S., Merad M., Gnjatic S. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Larsen M.D., de Graaf E.L., Sonneveld M.E., Plomp H.R., Nouta J., Hoepel W., Chen H.J., Linty F., Visser R., Brinkhaus M., Sustic T., de Taeye S.W., Bentlage A.E.H., Toivonen S., Koeleman C.A.M., Sainio S., Kootstra N.A., Brouwer P.J.M., Geyer C.E., Derksen N.I.L., Wolbink G., de Winther M., Sanders R.W., van Gils M.J., de Bruin S., Vlaar A.P.J., U.M.C.C.-B.S.G. Amsterdam, Rispens T., den Dunnen J., Zaaijer H.L., Wuhrer M., van der Schoot C. Ellen, Vidarsson G. Afucosylated IgG characterizes enveloped viral responses and correlates with COVID-19 severity. Science. 2020 doi: 10.1126/science.abc8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang T.T., Sewatanon J., Memoli M.J., Wrammert J., Bournazos S., Bhaumik S.K., Pinsky B.A., Chokephaibulkit K., Onlamoon N., Pattanapanyasat K., Taubenberger J.K., Ahmed R., Ravetch J.V. IgG antibodies to dengue enhanced for FcgammaRIIIA binding determine disease severity. Science. 2017;355:395–398. doi: 10.1126/science.aai8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Castano-Rodriguez C., Honrubia J.M., Gutierrez-Alvarez J., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeno J.M., Regla-Nava J.A., Fernandez-Delgado R., Verdia-Baguena C., Queralt-Martin M., Kochan G., Perlman S., Aguilella V.M., Sola I., Enjuanes L. Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. mBio. 2018;9 doi: 10.1128/mBio.02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Verdia-Baguena C., Nieto-Torres J.L., Alcaraz A., DeDiego M.L., Torres J., Aguilella V.M., Enjuanes L. Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology. 2012;432:485–494. doi: 10.1016/j.virol.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nieto-Torres J.L., DeDiego M.L., Verdia-Baguena C., Jimenez-Guardeno J.M., Regla-Nava J.A., Fernandez-Delgado R., Castano-Rodriguez C., Alcaraz A., Torres J., Aguilella V.M., Enjuanes L. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004077. e1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang Y., Yang Z.Y., Kong W.P., Nabel G.J. Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: implications for assembly and vaccine production. J. Virol. 2004;78:12557–12565. doi: 10.1128/JVI.78.22.12557-12565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hatakeyama S., Matsuoka Y., Ueshiba H., Komatsu N., Itoh K., Shichijo S., Kanai T., Fukushi M., Ishida I., Kirikae T., Sasazuki T., Miyoshi-Akiyama T. Dissection and identification of regions required to form pseudoparticles by the interaction between the nucleocapsid (N) and membrane (M) proteins of SARS coronavirus. Virology. 2008;380:99–108. doi: 10.1016/j.virol.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bai B., Hu Q., Hu H., Zhou P., Shi Z., Meng J., Lu B., Huang Y., Mao P., Wang H. Virus-like particles of SARS-like coronavirus formed by membrane proteins from different origins demonstrate stimulating activity in human dendritic cells. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hsieh P.K., Chang S.C., Huang C.C., Lee T.T., Hsiao C.W., Kou Y.H., Chen I.Y., Chang C.K., Huang T.H., Chang M.F. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005;79:13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ho Y., Lin P.H., Liu C.Y., Lee S.P., Chao Y.C. Assembly of human severe acute respiratory syndrome coronavirus-like particles. Biochem. Biophys. Res. Commun. 2004;318:833–838. doi: 10.1016/j.bbrc.2004.04.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yuan Q., Liao Y., Torres J., Tam J.P., Liu D.X. Biochemical evidence for the presence of mixed membrane topologies of the severe acute respiratory syndrome coronavirus envelope protein expressed in mammalian cells. FEBS Lett. 2006;580:3192–3200. doi: 10.1016/j.febslet.2006.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Raamsman M.J., Locker J.K., de Hooge A., de Vries A.A., Griffiths G., Vennema H., Rottier P.J. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 2000;74:2333–2342. doi: 10.1128/jvi.74.5.2333-2342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nieto-Torres J.L., Dediego M.L., Alvarez E., Jimenez-Guardeno J.M., Regla-Nava J.A., Llorente M., Kremer L., Shuo S., Enjuanes L. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415:69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lim K.P., Liu D.X. The missing link in coronavirus assembly. Retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins. J. Biol. Chem. 2001;276:17515–17523. doi: 10.1074/jbc.M009731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C., Yip C.C., Cai J.P., Chan J.M., Chik T.S., Lau D.P., Choi C.Y., Chen L.L., Chan W.M., Chan K.H., Ip J.D., Ng A.C., Poon R.W., Luo C.T., Cheng V.C., Chan J.F., Hung I.F., Chen Z., Chen H., Yuen K.Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. (e1415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chang C.K., Sue S.C., Yu T.H., Hsieh C.M., Tsai C.K., Chiang Y.C., Lee S.J., Hsiao H.H., Wu W.J., Chang W.L., Lin C.H., Huang T.H. Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 2006;13:59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chang C.K., Hou M.H., Chang C.F., Hsiao C.D., Huang T.H. The SARS coronavirus nucleocapsid protein–forms and functions. Antivir. Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Baric R.S., Nelson G.W., Fleming J.O., Deans R.J., Keck J.G., Casteel N., Stohlman S.A. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J. Virol. 1988;62:4280–4287. doi: 10.1128/jvi.62.11.4280-4287.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Narayanan K., Kim K.H., Makino S. Characterization of N protein self-association in coronavirus ribonucleoprotein complexes. Virus Res. 2003;98:131–140. doi: 10.1016/j.virusres.2003.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee H.K., Lee B.H., Seok S.H., Baek M.W., Lee H.Y., Kim D.J., Na Y.R., Noh K.J., Park S.H., Kumar D.N., Kariwa H., Nakauchi M., Heo S.J., Park J.H. Production of specific antibodies against SARS-coronavirus nucleocapsid protein without cross reactivity with human coronaviruses 229E and OC43. J. Vet. Sci. 2010;11:165–167. doi: 10.4142/jvs.2010.11.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Leung D.T., Tam F.C., Ma C.H., Chan P.K., Cheung J.L., Niu H., Tam J.S., Lim P.L. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J. Infect. Dis. 2004;190:379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gudbjartsson D.F., Norddahl G.L., Melsted P., Gunnarsdottir K., Holm H., Eythorsson E., Arnthorsson A.O., Helgason D., Bjarnadottir K., Ingvarsson R.F., Thorsteinsdottir B., Kristjansdottir S., Birgisdottir K., Kristinsdottir A.M., Sigurdsson M.I., Arnadottir G.A., Ivarsdottir E.V., Andresdottir M., Jonsson F., Agustsdottir A.B., Berglund J., Eiriksdottir B., Fridriksdottir R., Gardarsdottir E.E., Gottfredsson M., Gretarsdottir O.S., Gudmundsdottir S., Gudmundsson K.R., Gunnarsdottir T.R., Gylfason A., Helgason A., Jensson B.O., Jonasdottir A., Jonsson H., Kristjansson T., Kristinsson K.G., Magnusdottir D.N., Magnusson O.T., Olafsdottir L.B., Rognvaldsson S., le Roux L., Sigmundsdottir G., Sigurdsson A., Sveinbjornsson G., Sveinsdottir K.E., Sveinsdottir M., Thorarensen E.A., Thorbjornsson B., Thordardottir M., Saemundsdottir J., Kristjansson S.H., Josefsdottir K.S., Masson G., Georgsson G., Kristjansson M., Moller A., Palsson R., Gudnason T., Thorsteinsdottir U., Jonsdottir I., Sulem P., Stefansson K. Humoral Immune Response to SARS-CoV-2 in Iceland. N. Engl. J. Med. 2020;383:1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F., Wang D.Q., Hu Y., Ren J.H., Tang N., Xu Y.Y., Yu L.H., Mo Z., Gong F., Zhang X.L., Tian W.G., Hu L., Zhang X.X., Xiang J.L., Du H.X., Liu H.W., Lang C.H., Luo X.H., Wu S.B., Cui X.P., Zhou Z., Zhu M.M., Wang J., Xue C.J., Li X.F., Wang L., Li Z.J., Wang K., Niu C.C., Yang Q.J., Tang X.J., Zhang Y., Liu X.M., Li J.J., Zhang D.C., Zhang F., Liu P., Yuan J., Li Q., Hu J.L., Chen J., Huang A.L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 104.Burbelo P.D., Riedo F.X., Morishima C., Rawlings S., Smith D., Das S., Strich J.R., Chertow D.S., Davey R.T., Cohen J.I. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J. Infect. Dis. 2020;222:206–213. doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Flinck H., Rauhio A., Luukinen B., Lehtimaki T., Haapala A.M., Seiskari T., Aittoniemi J. Comparison of 2 fully automated tests detecting antibodies against nucleocapsid N and spike S1/S2 proteins in COVID-19. Diagn. Microbiol. Infect. Dis. 2021;99(1) doi: 10.1016/j.diagmicrobio.2020.115197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., Sagar F. Daul, Lago M. Salvat, Decker A., Luxenburger H., Binder B., Bettinger D., Sogukpinar O., Rieg S., Panning M., Huzly D., Schwemmle M., Kochs G., Waller C.F., Nieters A., Duerschmied D., Emmerich F., Mei H.E., Schulz A.R., Llewellyn-Lacey S., Price D.A., Boettler T., Bengsch B., Thimme R., Hofmann M., Neumann-Haefelin C. Characterization of pre-existing and induced SARS-CoV-2-specific CD8(+) T cells. Nat. Med. 2020;27(1):78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- 107.Habel J.R., Nguyen T.H.O., van de Sandt C.E., Juno J.A., Chaurasia P., Wragg K., Koutsakos M., Hensen L., Jia X., Chua B., Zhang W., Tan H.X., Flanagan K.L., Doolan D.L., Torresi J., Chen W., Wakim L.M., Cheng A.C., Doherty P.C., Petersen J., Rossjohn J., Wheatley A.K., Kent S.J., Rowntree L.C., Kedzierska K. Suboptimal SARS-CoV-2-specific CD8(+) T cell response associated with the prominent HLA-A*02:01 phenotype. Proc. Natl. Acad. Sci. U. S. A. 2020;117:24384–24391. doi: 10.1073/pnas.2015486117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tsao Y.P., Lin J.Y., Jan J.T., Leng C.H., Chu C.C., Yang Y.C., Chen S.L. HLA-A*0201 T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus nucleocapsid and spike proteins. Biochem. Biophys. Res. Commun. 2006;344:63–71. doi: 10.1016/j.bbrc.2006.03.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao J., Huang Q., Wang W., Zhang Y., Lv P., Gao X.M. Identification and characterization of dominant helper T-cell epitopes in the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:6079–6088. doi: 10.1128/JVI.02568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Peng H., Yang L.T., Wang L.Y., Li J., Huang J., Lu Z.Q., Koup R.A., Bailer R.T., Wu C.Y. Long-lived memory T lymphocyte responses against SARS coronavirus nucleocapsid protein in SARS-recovered patients. Virology. 2006;351:466–475. doi: 10.1016/j.virol.2006.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cheung Y.K., Cheng S.C., Sin F.W., Chan K.T., Xie Y. Investigation of immunogenic T-cell epitopes in SARS virus nucleocapsid protein and their role in the prevention and treatment of SARS infection. Hong Kong Med. J. 2008;14(Suppl. 4):27–30. [PubMed] [Google Scholar]
- 112.Vlasova A.N., Zhang X., Hasoksuz M., Nagesha H.S., Haynes L.M., Fang Y., Lu S., Saif L.J. Two-way antigenic cross-reactivity between severe acute respiratory syndrome coronavirus (SARS-CoV) and group 1 animal CoVs is mediated through an antigenic site in the N-terminal region of the SARS-CoV nucleoprotein. J. Virol. 2007;81:13365–13377. doi: 10.1128/JVI.01169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tseng C.T., Sbrana E., Iwata-Yoshikawa N., Newman P.C., Garron T., Atmar R.L., Peters C.J., Couch R.B. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Deming D., Sheahan T., Heise M., Yount B., Davis N., Sims A., Suthar M., Harkema J., Whitmore A., Pickles R., West A., Donaldson E., Curtis K., Johnston R., Baric R. Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLoS Med. 2006;3 doi: 10.1371/journal.pmed.0030525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bolles M., Deming D., Long K., Agnihothram S., Whitmore A., Ferris M., Funkhouser W., Gralinski L., Totura A., Heise M., Baric R.S. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 2011;85:12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yasui F., Kai C., Kitabatake M., Inoue S., Yoneda M., Yokochi S., Kase R., Sekiguchi S., Morita K., Hishima T., Suzuki H., Karamatsu K., Yasutomi Y., Shida H., Kidokoro M., Mizuno K., Matsushima K., Kohara M. Prior immunization with severe acute respiratory syndrome (SARS)-associated coronavirus (SARS-CoV) nucleocapsid protein causes severe pneumonia in mice infected with SARS-CoV. J. Immunol. 2008;181:6337–6348. doi: 10.4049/jimmunol.181.9.6337. [DOI] [PubMed] [Google Scholar]
- 117.Agrawal A.S., Tao X., Algaissi A., Garron T., Narayanan K., Peng B.H., Couch R.B., Tseng C.T. Immunization with inactivated Middle East respiratory syndrome coronavirus vaccine leads to lung immunopathology on challenge with live virus. Hum. Vacc. Immunother. 2016;12:2351–2356. doi: 10.1080/21645515.2016.1177688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Weingartl H., Czub M., Czub S., Neufeld J., Marszal P., Gren J., Smith G., Jones S., Proulx R., Deschambault Y., Grudeski E., Andonov A., He R., Li Y., Copps J., Grolla A., Dick D., Berry J., Ganske S., Manning L., Cao J. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J. Virol. 2004;78:12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Czub M., Weingartl H., Czub S., He R., Cao J. Evaluation of modified vaccinia virus Ankara based recombinant SARS vaccine in ferrets. Vaccine. 2005;23:2273–2279. doi: 10.1016/j.vaccine.2005.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Atyeo C., Fischinger S., Zohar T., Slein M.D., Burke J., Loos C., McCulloch D.J., Newman K.L., Wolf C., Yu J., Shuey K., Feldman J., Hauser B.M., Caradonna T., Schmidt A.G., Suscovich T.J., Linde C., Cai Y., Barouch D., Ryan E.T., Charles R.C., Lauffenburger D., Chu H., Alter G. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. 2020;53:524–532. doi: 10.1016/j.immuni.2020.07.020. (e524) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., Chia W.N., Chen M.I., Wang L.F., Ooi E.E., Kalimuddin S., Tambyah P.A., Low J.G., Tan Y.J., Bertoletti A. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 122.Zhao J., Alshukairi A.N., Baharoon S.A., Ahmed W.A., Bokhari A.A., Nehdi A.M., Layqah L.A., Alghamdi M.G., Al Gethamy M.M., Dada A.M., Khalid I., Boujelal M., Al Johani S.M., Vogel L., Subbarao K., Mangalam A., Wu C., Ten Eyck P., Perlman S., Zhao J. Recovery from the Middle East respiratory syndrome is associated with antibody and T-cell responses. Sci. Immunol. 2017;2 doi: 10.1126/sciimmunol.aan5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M., Haagmans B.L., de Swart R.L., Sette A., de Vries R.D. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E., Jia X., Nicholson S., Catton M., Cowie B., Tong S.Y.C., Lewin S.R., Kedzierska K. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 126.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 127.Padhan K., Tanwar C., Hussain A., Hui P.Y., Lee M.Y., Cheung C.Y., Peiris J.S.M., Jameel S. Severe acute respiratory syndrome coronavirus Orf3a protein interacts with caveolin. J. Gen. Virol. 2007;88:3067–3077. doi: 10.1099/vir.0.82856-0. [DOI] [PubMed] [Google Scholar]
- 128.Tan Y.J., Teng E., Shen S., Tan T.H., Goh P.Y., Fielding B.C., Ooi E.E., Tan H.C., Lim S.G., Hong W. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 2004;78:6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lu W., Zheng B.J., Xu K., Schwarz W., Du L., Wong C.K., Chen J., Duan S., Deubel V., Sun B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yuan X., Li J., Shan Y., Yang Z., Zhao Z., Chen B., Yao Z., Dong B., Wang S., Chen J., Cong Y. Subcellular localization and membrane association of SARS-CoV 3a protein. Virus Res. 2005;109:191–202. doi: 10.1016/j.virusres.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yu C.J., Chen Y.C., Hsiao C.H., Kuo T.C., Chang S.C., Lu C.Y., Wei W.C., Lee C.H., Huang L.M., Chang M.F., Ho H.N., Lee F.J. Identification of a novel protein 3a from severe acute respiratory syndrome coronavirus. FEBS Lett. 2004;565:111–116. doi: 10.1016/j.febslet.2004.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huang C., Narayanan K., Ito N., Peters C.J., Makino S. Severe acute respiratory syndrome coronavirus 3a protein is released in membranous structures from 3a protein-expressing cells and infected cells. J. Virol. 2006;80:210–217. doi: 10.1128/JVI.80.1.210-217.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Issa E., Merhi G., Panossian B., Salloum T., Tokajian S. SARS-CoV-2 and ORF3a: nonsynonymous mutations, functional domains, and viral pathogenesis. mSystems. 2020;5 doi: 10.1128/mSystems.00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hassan S.S., Choudhury P.P., Basu P., Jana S.S. Molecular conservation and differential mutation on ORF3a gene in Indian SARS-CoV2 genomes. Genomics. 2020;112:3226–3237. doi: 10.1016/j.ygeno.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yue Y., Nabar N.R., Shi C.S., Kamenyeva O., Xiao X., Hwang I.Y., Wang M., Kehrl J.H. SARS-coronavirus open Reading frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018;9:904. doi: 10.1038/s41419-018-0917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Freundt E.C., Yu L., Goldsmith C.S., Welsh S., Cheng A., Yount B., Liu W., Frieman M.B., Buchholz U.J., Screaton G.R., Lippincott-Schwartz J., Zaki S.R., Xu X.N., Baric R.S., Subbarao K., Lenardo M.J. The open reading frame 3a protein of severe acute respiratory syndrome-associated coronavirus promotes membrane rearrangement and cell death. J. Virol. 2010;84:1097–1109. doi: 10.1128/JVI.01662-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Law P.T.W., Wong C.H., Au T.C.C., Chuck C.P., Kong S.K., Chan P.K.S., To K.F., Lo A.W.I., Chan J.Y.W., Suen Y.K., Chan H.Y.E., Fung K.P., Waye M.M.Y., Sung J.J.Y., Lo Y.M.D., Tsui S.K.W. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol. 2005;86:1921–1930. doi: 10.1099/vir.0.80813-0. [DOI] [PubMed] [Google Scholar]
- 138.Padhan K., Minakshi R., Towheed M.A.B., Jameel S. Severe acute respiratory syndrome coronavirus 3a protein activates the mitochondrial death pathway through p38 MAP kinase activation. J. Gen. Virol. 2008;89:1960–1969. doi: 10.1099/vir.0.83665-0. [DOI] [PubMed] [Google Scholar]
- 139.Siu K.L., Yuen K.S., Castano-Rodriguez C., Ye Z.W., Yeung M.L., Fung S.Y., Yuan S., Chan C.P., Yuen K.Y., Enjuanes L., Jin D.Y. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chan W.S., Wu C., Chow S.C., Cheung T., To K.F., Leung W.K., Chan P.K., Lee K.C., Ng H.K., Au D.M., Lo A.W. Coronaviral hypothetical and structural proteins were found in the intestinal surface enterocytes and pneumocytes of severe acute respiratory syndrome (SARS) Mod. Pathol. 2005;18:1432–1439. doi: 10.1038/modpathol.3800439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nelson C.A., Pekosz A., Lee C.A., Diamond M.S., Fremont D.H. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure. 2005;13:75–85. doi: 10.1016/j.str.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schaecher S.R., Touchette E., Schriewer J., Buller R.M., Pekosz A. Severe acute respiratory syndrome coronavirus gene 7 products contribute to virus-induced apoptosis. J. Virol. 2007;81:11054–11068. doi: 10.1128/JVI.01266-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Frieman M.B., Yount B., Sims A.C., Deming D.J., Morrison T.E., Sparks J., Denison M., Heise M., Baric R.S. SARS coronavirus accessory ORFs encode luxury functions. Adv. Exp. Med. Biol. 2006;581:149–152. doi: 10.1007/978-0-387-33012-9_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yount B., Roberts R.S., Sims A.C., Deming D., Frieman M.B., Sparks J., Denison M.R., Davis N., Baric R.S. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 2005;79:14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tan Y.J., Fielding B.C., Goh P.Y., Shen S., Tan T.H., Lim S.G., Hong W. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J. Virol. 2004;78:14043–14047. doi: 10.1128/JVI.78.24.14043-14047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tan Y.X., Tan T.H., Lee M.J., Tham P.Y., Gunalan V., Druce J., Birch C., Catton M., Fu N.Y., Yu V.C., Tan Y.J. Induction of apoptosis by the severe acute respiratory syndrome coronavirus 7a protein is dependent on its interaction with the Bcl-XL protein. J. Virol. 2007;81:6346–6355. doi: 10.1128/JVI.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hachim A., Kavian N., Cohen C.A., Chin A.W.H., Chu D.K.W., Mok C.K.P., Tsang O.T.Y., Yeung Y.C., Perera R., Poon L.L.M., Peiris J.S.M., Valkenburg S.A. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 2020;21:1293–1301. doi: 10.1038/s41590-020-0773-7. [DOI] [PubMed] [Google Scholar]
- 148.Wang H., Wu X., Zhang X., Hou X., Liang T., Wang D., Teng F., Dai J., Duan H., Guo S., Li Y., Yu X. SARS-CoV-2 proteome microarray for mapping COVID-19 antibody interactions at amino acid resolution. ACS Cent. Sci. 2020;6:2238–2249. doi: 10.1021/acscentsci.0c00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang F., Gan R., Zhen Z., Hu X., Li X., Zhou F., Liu Y., Chen C., Xie S., Zhang B., Wu X., Huang Z. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Sig. Transduct. Target Ther. 2020;5:156. doi: 10.1038/s41392-020-00263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tang X., Li G., Vasilakis N., Zhang Y., Shi Z., Zhong Y., Wang L.F., Zhang S. Differential stepwise evolution of SARS coronavirus functional proteins in different host species. BMC Evol. Biol. 2009;9:52. doi: 10.1186/1471-2148-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Holland L.A., Kaelin E.A., Maqsood R., Estifanos B., Wu L.I., Varsani A., Halden R.U., Hogue B.G., Scotch M., Lim E.S. An 81-nucleotide deletion in SARS-CoV-2 ORF7a identified from sentinel surveillance in Arizona (January to March 2020) J. Virol. 2020;94 doi: 10.1128/JVI.00711-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Islam M.R., Hoque M.N., Rahman M.S., Alam A., Akther M., Puspo J.A., Akter S., Sultana M., Crandall K.A., Hossain M.A. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci. Rep. 2020;10:14004. doi: 10.1038/s41598-020-70812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang H., Zhang Y., Huang B., Deng W., Quan Y., Wang W., Xu W., Zhao Y., Li N., Zhang J., Liang H., Bao L., Xu Y., Ding L., Zhou W., Gao H., Liu J., Niu P., Zhao L., Zhen W., Fu H., Yu S., Zhang Z., Xu G., Li C., Lou Z., Xu M., Qin C., Wu G., Gao G.F., Tan W., Yang X. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell. 2020;182:713–721. doi: 10.1016/j.cell.2020.06.008. (e719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Xia S., Duan K., Zhang Y., Zhao D., Zhang H., Xie Z., Li X., Peng C., Zhang Y., Zhang W., Yang Y., Chen W., Gao X., You W., Wang X., Wang Z., Shi Z., Wang Y., Yang X., Zhang L., Huang L., Wang Q., Lu J., Yang Y., Guo J., Zhou W., Wan X., Wu C., Wang W., Huang S., Du J., Meng Z., Pan A., Yuan Z., Shen S., Guo W., Yang X. Effect of an inactivated vaccine against SARS-CoV-2 on safety and immunogenicity outcomes: interim analysis of 2 randomized clinical trials. JAMA. 2020;324:951–960. doi: 10.1001/jama.2020.15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., Gao H., Ge X., Kan B., Hu Y., Liu J., Cai F., Jiang D., Yin Y., Qin C., Li J., Gong X., Lou X., Shi W., Wu D., Zhang H., Zhu L., Deng W., Li Y., Lu J., Li C., Wang X., Yin W., Zhang Y., Qin C. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.U.S.F.D. Administration . 2020. Vaccines Licensed for Use in the United States. [Google Scholar]
- 157.Schultz-Cherry S., Jones J.C. Influenza vaccines: the good, the bad, and the eggs. Adv. Virus Res. 2010;77:63–84. doi: 10.1016/B978-0-12-385034-8.00003-X. [DOI] [PubMed] [Google Scholar]
- 158.Tregoning J.S., Brown E.S., Cheeseman H.M., Flight K.E., Higham S.L., Lemm N.M., Pierce B.F., Stirling D.C., Wang Z., Pollock K.M. Vaccines for COVID-19. Clin. Exp. Immunol. 2020;202:162–192. doi: 10.1111/cei.13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Moghaddam A., Olszewska W., Wang B., Tregoning J.S., Helson R., Sattentau Q.J., Openshaw P.J. A potential molecular mechanism for hypersensitivity caused by formalin-inactivated vaccines. Nat. Med. 2006;12:905–907. doi: 10.1038/nm1456. [DOI] [PubMed] [Google Scholar]
- 160.Funk C.D., Laferriere C., Ardakani A. A snapshot of the global race for vaccines targeting SARS-CoV-2 and the COVID-19 pandemic. Front. Pharmacol. 2020;11:937. doi: 10.3389/fphar.2020.00937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Minor P.D. Live attenuated vaccines: historical successes and current challenges. Virology. 2015;479-480:379–392. doi: 10.1016/j.virol.2015.03.032. [DOI] [PubMed] [Google Scholar]
- 162.Plotkin S. History of vaccination. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12283–12287. doi: 10.1073/pnas.1400472111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Regla-Nava J.A., Nieto-Torres J.L., Jimenez-Guardeno J.M., Fernandez-Delgado R., Fett C., Castano-Rodriguez C., Perlman S., Enjuanes L., DeDiego M.L. Severe acute respiratory syndrome coronaviruses with mutations in the E protein are attenuated and promising vaccine candidates. J. Virol. 2015;89:3870–3887. doi: 10.1128/JVI.03566-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Almazan F., DeDiego M.L., Sola I., Zuniga S., Nieto-Torres J.L., Marquez-Jurado S., Andres G., Enjuanes L. Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. mBio. 2013;4 doi: 10.1128/mBio.00650-13. (e00650-00613) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pulendran B., Ahmed R. Immunological mechanisms of vaccination. Nat. Immunol. 2011;12:509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yong C.Y., Ong H.K., Yeap S.K., Ho K.L., Tan W.S. Recent advances in the vaccine development against Middle East respiratory syndrome-coronavirus. Front. Microbiol. 2019;10:1781. doi: 10.3389/fmicb.2019.01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Belshe R., Lee M.S., Walker R.E., Stoddard J., Mendelman P.M. Safety, immunogenicity and efficacy of intranasal, live attenuated influenza vaccine. Exp. Rev. Vacc. 2004;3:643–654. doi: 10.1586/14760584.3.6.643. [DOI] [PubMed] [Google Scholar]
- 168.Barrett A.D.T. Yellow fever live attenuated vaccine: a very successful live attenuated vaccine but still we have problems controlling the disease. Vaccine. 2017;35:5951–5955. doi: 10.1016/j.vaccine.2017.03.032. [DOI] [PubMed] [Google Scholar]
- 169.Jang Y.H., Seong B.L. Principles underlying rational design of live attenuated influenza vaccines. Clin. Exp. Vacc. Res. 2012;1:35–49. doi: 10.7774/cevr.2012.1.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sarmiento J.D., Villada F., Orrego J.C., Franco J.L., Trujillo-Vargas C.M. Adverse events following immunization in patients with primary immunodeficiencies. Vaccine. 2016;34:1611–1616. doi: 10.1016/j.vaccine.2016.01.047. [DOI] [PubMed] [Google Scholar]
- 171.Neven B., Perot P., Bruneau J., Pasquet M., Ramirez M., Diana J.S., Luzi S., Corre-Catelin N., Chardot C., Moshous D., Leclerc Mercier S., Mahlaoui N., Aladjidi N., Le Bail B., Lecuit M., Bodemer C., Molina T.J., Blanche S., Eloit M. Cutaneous and visceral chronic granulomatous disease triggered by a rubella virus vaccine strain in children with primary Immunodeficiencies. Clin. Infect. Dis. 2017;64:83–86. doi: 10.1093/cid/ciw675. [DOI] [PubMed] [Google Scholar]
- 172.Amanna I.J. Balancing the efficacy and safety of vaccines in the elderly. Open Longev Sci. 2012;6:64–72. doi: 10.2174/1876326X01206010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Choi Y., Chang J. Viral vectors for vaccine applications. Clin. Exp. Vacc. Res. 2013;2:97–105. doi: 10.7774/cevr.2013.2.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ura T., Okuda K., Shimada M. Developments in viral vector-based vaccines. Vacc. (Basel) 2014;2:624–641. doi: 10.3390/vaccines2030624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Coughlan L. Factors which contribute to the immunogenicity of non-replicating adenoviral vectored vaccines. Front. Immunol. 2020;11:909. doi: 10.3389/fimmu.2020.00909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Rauch S., Jasny E., Schmidt K.E., Petsch B. New vaccine technologies to combat outbreak situations. Front. Immunol. 2018;9:1963. doi: 10.3389/fimmu.2018.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zheng Z., Diaz-Arevalo D., Guan H., Zeng M. Noninvasive vaccination against infectious diseases. Hum. Vacc. Immunother. 2018;14:1717–1733. doi: 10.1080/21645515.2018.1461296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang S., Liu H., Zhang X., Qian F. Intranasal and oral vaccination with protein-based antigens: advantages, challenges and formulation strategies. Protein Cell. 2015;6:480–503. doi: 10.1007/s13238-015-0164-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Buchbinder S.P., Mehrotra D.V., Duerr A., Fitzgerald D.W., Mogg R., Li D., Gilbert P.B., Lama J.R., Marmor M., Del Rio C., McElrath M.J., Casimiro D.R., Gottesdiener K.M., Chodakewitz J.A., Corey L., Robertson M.N. T. Step study protocol, efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the step study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ledgerwood J.E., Costner P., Desai N., Holman L., Enama M.E., Yamshchikov G., Mulangu S., Hu Z., Andrews C.A., Sheets R.A., Koup R.A., Roederer M., Bailer R., Mascola J.R., Pau M.G., Sullivan N.J., Goudsmit J., Nabel G.J., Graham B.S., Team V.R.C.S. A replication defective recombinant Ad5 vaccine expressing Ebola virus GP is safe and immunogenic in healthy adults. Vaccine. 2010;29:304–313. doi: 10.1016/j.vaccine.2010.10.037. [DOI] [PubMed] [Google Scholar]
- 181.Ahi Y.S., Bangari D.S., Mittal S.K. Adenoviral vector immunity: its implications and circumvention strategies. Curr. Gene Ther. 2011;11:307–320. doi: 10.2174/156652311796150372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fausther-Bovendo H., Kobinger G.P. Pre-existing immunity against ad vectors: humoral, cellular, and innate response, what’s important? Hum. Vacc. Immunother. 2014;10:2875–2884. doi: 10.4161/hv.29594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.van Doremalen N., Haddock E., Feldmann F., Meade-White K., Bushmaker T., Fischer R.J., Okumura A., Hanley P.W., Saturday G., Edwards N.J., Clark M.H.A., Lambe T., Gilbert S.C., Munster V.J. A single dose of ChAdOx1 MERS provides protective immunity in rhesus macaques. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aba8399. (eaba8399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hassan A.O., Kafai N.M., Dmitriev I.P., Fox J.M., Smith B.K., Harvey I.B., Chen R.E., Winkler E.S., Wessel A.W., Case J.B., Kashentseva E., McCune B.T., Bailey A.L., Zhao H., VanBlargan L.A., Dai Y.N., Ma M., Adams L.J., Shrihari S., Danis J.E., Gralinski L.E., Hou Y.J., Schafer A., Kim A.S., Keeler S.P., Weiskopf D., Baric R.S., Holtzman M.J., Fremont D.H., Curiel D.T., Diamond M.S. A single-dose intranasal ChAd vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell. 2020;183:169–184. doi: 10.1016/j.cell.2020.08.026. (e113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sakurai A., Ogawa T., Matsumoto J., Kihira T., Fukushima S., Miyata I., Shimizu H., Itamura S., Ouchi K., Hamada A., Tani K., Okabe N., Yamaguchi T. Regulatory aspects of quality and safety for live recombinant viral vaccines against infectious diseases in Japan. Vaccine. 2019;37:6573–6579. doi: 10.1016/j.vaccine.2019.08.031. [DOI] [PubMed] [Google Scholar]
- 186.Eckart R.E., Love S.S., Atwood J.E., Arness M.K., Cassimatis D.C., Campbell C.L., Boyd S.Y., Murphy J.G., Swerdlow D.L., Collins L.C., Riddle J.R., Tornberg D.N., Grabenstein J.D., Engler R.J., T. Department of Defense Smallpox Vaccination Clinical Evaluation Incidence and follow-up of inflammatory cardiac complications after smallpox vaccination. J. Am. Coll. Cardiol. 2004;44:201–205. doi: 10.1016/j.jacc.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 187.Ledgerwood J.E., DeZure A.D., Stanley D.A., Coates E.E., Novik L., Enama M.E., Berkowitz N.M., Hu Z., Joshi G., Ploquin A., Sitar S., Gordon I.J., Plummer S.A., Holman L.A., Hendel C.S., Yamshchikov G., Roman F., Nicosia A., Colloca S., Cortese R., Bailer R.T., Schwartz R.M., Roederer M., Mascola J.R., Koup R.A., Sullivan N.J., Graham B.S., Team V.R.C.S. Chimpanzee Adenovirus Vector Ebola Vaccine. N. Engl. J. Med. 2017;376:928–938. doi: 10.1056/NEJMoa1410863. [DOI] [PubMed] [Google Scholar]
- 188.Wang Y., Li J., Hu Y., Liang Q., Wei M., Zhu F. Ebola vaccines in clinical trial: the promising candidates. Hum. Vacc. Immunother. 2017;13:153–168. doi: 10.1080/21645515.2016.1225637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ewer K., Rampling T., Venkatraman N., Bowyer G., Wright D., Lambe T., Imoukhuede E.B., Payne R., Fehling S.K., Strecker T., Biedenkopf N., Krahling V., Tully C.M., Edwards N.J., Bentley E.M., Samuel D., Labbe G., Jin J., Gibani M., Minhinnick A., Wilkie M., Poulton I., Lella N., Roberts R., Hartnell F., Bliss C., Sierra-Davidson K., Powlson J., Berrie E., Tedder R., Roman F., De Ryck I., Nicosia A., Sullivan N.J., Stanley D.A., Mbaya O.T., Ledgerwood J.E., Schwartz R.M., Siani L., Colloca S., Folgori A., Di Marco S., Cortese R., Wright E., Becker S., Graham B.S., Koup R.A., Levine M.M., Volkmann A., Chaplin P., Pollard A.J., Draper S.J., Ballou W.R., Lawrie A., Gilbert S.C., Hill A.V. A monovalent chimpanzee adenovirus ebola vaccine boosted with MVA. N. Engl. J. Med. 2016;374:1635–1646. doi: 10.1056/NEJMoa1411627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ramsauer K., Schwameis M., Firbas C., Mullner M., Putnak R.J., Thomas S.J., Despres P., Tauber E., Jilma B., Tangy F. Immunogenicity, safety, and tolerability of a recombinant measles-virus-based chikungunya vaccine: a randomised, double-blind, placebo-controlled, active-comparator, first-in-man trial. Lancet Infect. Dis. 2015;15:519–527. doi: 10.1016/S1473-3099(15)70043-5. [DOI] [PubMed] [Google Scholar]
- 191.Wressnigg N., Hochreiter R., Zoihsl O., Fritzer A., Bezay N., Klingler A., Lingnau K., Schneider M., Lundberg U., Meinke A., Larcher-Senn J., Corbic-Ramljak I., Eder-Lingelbach S., Dubischar K., Bender W. Single-shot live-attenuated chikungunya vaccine in healthy adults: a phase 1, randomised controlled trial. Lancet Infect. Dis. 2020;20:1193–1203. doi: 10.1016/S1473-3099(20)30238-3. [DOI] [PubMed] [Google Scholar]
- 192.Zhu F.C., Guan X.H., Li Y.H., Huang J.Y., Jiang T., Hou L.H., Li J.X., Yang B.F., Wang L., Wang W.J., Wu S.P., Wang Z., Wu X.H., Xu J.J., Zhang Z., Jia S.Y., Wang B.S., Hu Y., Liu J.J., Zhang J., Qian X.A., Li Q., Pan H.X., Jiang H.D., Deng P., Gou J.B., Wang X.W., Wang X.H., Chen W. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chiuppesi F., Salazar M.D.A., Contreras H., Nguyen V., Martinez J., Park S., Nguyen J., Kha M., Iniguez A., Zhou Q., Kaltcheva T., Levytskyy R., Ebelt N., Kang T., Wu X., Rogers T., Manuel E., Shostak Y., Diamond D., Wussow F. Development of a multi-antigenic SARS-CoV-2 vaccine using a synthetic poxvirus platform. Nat. Commun. 2020;11(1):6121. doi: 10.1038/s41467-020-19819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Capone S., Raggioli A., Gentile M., Battella S., Lahm A., Sommella A., Contino A.M., Urbanowicz R.A., Scala R., Barra F., Leuzzi A., Lilli E., Miselli G., Noto A., Ferraiuolo M., Talotta F., Tsoleridis T., Castilletti C., Matusali G., Colavita F., Lapa D., Meschi S., Capobianchi M., Soriani M., Folgori A., Ball J.K., Colloca S., Vitelli A. Immunogenicity of a new gorilla adenovirus vaccine candidate for COVID-19. bioRxiv. 2020 doi: 10.1101/2020.10.22.349951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hodoniczky J., Zheng Y.Z., James D.C. Control of recombinant monoclonal antibody effector functions by Fc N-glycan remodeling in vitro. Biotechnol. Prog. 2005;21:1644–1652. doi: 10.1021/bp050228w. [DOI] [PubMed] [Google Scholar]
- 196.Routhu S.G. Nanda Kishore, Cheedarla Narayanaiah, Ayalnesh Shiferaw, Sahoo Sheikh Abdul Rahman Anusmita, Shi Pei-Yong, Vineet D., Menachery Katharine Floyd, Fischinger Stephanie, Atyeo Caroline, Alter Galit, Suthar Mehul S., Amara Rama Rao. Modified vaccinia ankara based SARS-CoV-2 vaccine expressing full-length spike induces strong neutralizing antibody response. bioRxiv. 2020 doi: 10.1101/2020.06.27.175166. [DOI] [Google Scholar]
- 198.Jon C., Merck One of big pharma’s biggest players, reveals its COVID-19 vaccine and therapy plans. Science. 2020 doi: 10.1126/science.abd0121. [DOI] [Google Scholar]
- 199.Yahalom-Ronen Y., Tamir H., Melamed S., Politi B., Shifman O., Achdout H., Vitner E.B., Israeli O., Milrot E., Stein D., Cohen-Gihon I., Lazar S., Gutman H., Glinert I., Cherry L., Vagima Y., Lazar S., Weiss S., Ben-Shmuel A., Avraham R., Puni R., Lupu E., David E. Bar, Sittner A., Erez N., Zichel R., Mamroud E., Mazor O., Levy H., Laskar O., Yitzhaki S., Shapira S.C., Zvi A., Beth-Din A., Paran N., Israely T. A single dose of recombinant VSV-ΔG-spike vaccine provides protection against SARS-CoV-2 challenge. bioRxiv. 2020 doi: 10.1101/2020.06.18.160655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hörner C., Schürmann C., Auste A., Ebenig A., Muraleedharan S., Herrmann M., Schnierle B., Baric R.S., Mühlebach M.D. A highly immunogenic and effective measles virus-based Th1-biased COVID-19 vaccine. Proc. Natl. Acad. Sci. U. S. A. 2020;117(51):32657–32666. doi: 10.1073/pnas.2014468117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sun W., McCroskery S., Liu W.C., Leist S.R., Liu Y., Albrecht R.A., Slamanig S., Oliva J., Amanat F., Schafer A., Dinnon K.H., 3rd, Innis B.L., Garcia-Sastre A., Krammer F., Baric R.S., Palese P. A newcastle disease virus (NDV) expressing a membrane-anchored spike as a cost-effective inactivated SARS-CoV-2 vaccine. Vacc. (Basel) 2020;8 doi: 10.3390/vaccines8040771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Smith L.R., Wloch M.K., Ye M., Reyes L.R., Boutsaboualoy S., Dunne C.E., Chaplin J.A., Rusalov D., Rolland A.P., Fisher C.L., Al-Ibrahim M.S., Kabongo M.L., Steigbigel R., Belshe R.B., Kitt E.R., Chu A.H., Moss R.B. Phase 1 clinical trials of the safety and immunogenicity of adjuvanted plasmid DNA vaccines encoding influenza a virus H5 hemagglutinin. Vaccine. 2010;28:2565–2572. doi: 10.1016/j.vaccine.2010.01.029. [DOI] [PubMed] [Google Scholar]
- 203.Ledgerwood J.E., Wei C.J., Hu Z., Gordon I.J., Enama M.E., Hendel C.S., McTamney P.M., Pearce M.B., Yassine H.M., Boyington J.C., Bailer R., Tumpey T.M., Koup R.A., Mascola J.R., Nabel G.J., Graham B.S., Team V.R.C.S. DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. Lancet Infect. Dis. 2011;11:916–924. doi: 10.1016/S1473-3099(11)70240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Jones S., Evans K., McElwaine-Johnn H., Sharpe M., Oxford J., Lambkin-Williams R., Mant T., Nolan A., Zambon M., Ellis J., Beadle J., Loudon P.T. DNA vaccination protects against an influenza challenge in a double-blind randomised placebo-controlled phase 1b clinical trial. Vaccine. 2009;27:2506–2512. doi: 10.1016/j.vaccine.2009.02.061. [DOI] [PubMed] [Google Scholar]
- 205.Tebas P., Kraynyak K.A., Patel A., Maslow J.N., Morrow M.P., Sylvester A.J., Knoblock D., Gillespie E., Amante D., Racine T., McMullan T., Jeong M., Roberts C.C., Park Y.K., Boyer J., Broderick K.E., Kobinger G.P., Bagarazzi M., Weiner D.B., Sardesai N.Y., White S.M. Intradermal SynCon(R) ebola GP DNA vaccine is temperature stable and safely demonstrates cellular and humoral immunogenicity advantages in healthy volunteers. J. Infect. Dis. 2019;220:400–410. doi: 10.1093/infdis/jiz132. [DOI] [PubMed] [Google Scholar]
- 206.Modjarrad K., Lin L., George S.L., Stephenson K.E., Eckels K.H., De La Barrera R.A., Jarman R.G., Sondergaard E., Tennant J., Ansel J.L., Mills K., Koren M., Robb M.L., Barrett J., Thompson J., Kosel A.E., Dawson P., Hale A., Tan C.S., Walsh S.R., Meyer K.E., Brien J., Crowell T.A., Blazevic A., Mosby K., Larocca R.A., Abbink P., Boyd M., Bricault C.A., Seaman M.S., Basil A., Walsh M., Tonwe V., Hoft D.F., Thomas S.J., Barouch D.H., Michael N.L. Preliminary aggregate safety and immunogenicity results from three trials of a purified inactivated Zika virus vaccine candidate: phase 1, randomised, double-blind, placebo-controlled clinical trials. Lancet. 2018;391:563–571. doi: 10.1016/S0140-6736(17)33106-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Modjarrad K., Roberts C.C., Mills K.T., Castellano A.R., Paolino K., Muthumani K., Reuschel E.L., Robb M.L., Racine T., Oh M.D., Lamarre C., Zaidi F.I., Boyer J., Kudchodkar S.B., Jeong M., Darden J.M., Park Y.K., Scott P.T., Remigio C., Parikh A.P., Wise M.C., Patel A., Duperret E.K., Kim K.Y., Choi H., White S., Bagarazzi M., May J.M., Kane D., Lee H., Kobinger G., Michael N.L., Weiner D.B., Thomas S.J., Maslow J.N. Safety and immunogenicity of an anti-Middle East respiratory syndrome coronavirus DNA vaccine: a phase 1, open-label, single-arm, dose-escalation trial. Lancet Infect. Dis. 2019;19:1013–1022. doi: 10.1016/S1473-3099(19)30266-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Xu Z., Patel A., Tursi N.J., Zhu X., Muthumani K., Kulp D.W., Weiner D.B. Harnessing recent advances in synthetic DNA and electroporation technologies for rapid vaccine development against COVID-19 and other emerging infectious diseases. Front. Med. Technol. 2020;2 doi: 10.3389/fmedt.2020.571030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., Gary E.N., Walker S.N., Schultheis K., Purwar M., Xu Z., Walters J., Bhojnagarwala P., Yang M., Chokkalingam N., Pezzoli P., Parzych E., Reuschel E.L., Doan A., Tursi N., Vasquez M., Choi J., Tello-Ruiz E., Maricic I., Bah M.A., Wu Y., Amante D., Park D.H., Dia Y., Ali A.R., Zaidi F.I., Generotti A., Kim K.Y., Herring T.A., Reeder S., Andrade V.M., Buttigieg K., Zhao G., Wu J.M., Li D., Bao L., Liu J., Deng W., Qin C., Brown A.S., Khoshnejad M., Wang N., Chu J., Wrapp D., McLellan J.S., Muthumani K., Wang B., Carroll M.W., Kim J.J., Boyer J., Kulp D.W., Humeau L., Weiner D.B., Broderick K.E. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Patel A., Walters J., Reuschel E.L., Schultheis K., Parzych E., Gary E.N., Maricic I., Purwar M., Eblimit Z., Walker S.N., Guimet D., Bhojnagarwala P., Doan A., Xu Z., Elwood D., Reeder S.M., Pessaint L., Kim K.Y., Cook A., Chokkalingam N., Finneyfrock B., Tello-Ruiz E., Dodson A., Choi J., Generotti A., Harrison J., Tursi N.J., Andrade V.M., Dia Y., Zaidi F.I., Andersen H., Lewis M.G., Muthumani K., Kim J.J., Kulp D.W., Humeau L.M., Ramos S., Smith T.R.F., Weiner D.B., Broderick K.E. Intradermal-delivered DNA vaccine provides anamnestic protection in a rhesus macaque SARS-CoV-2 challenge model. bioRxiv. 2020 doi: 10.1101/2020.07.28.225649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Abbasi J. COVID-19 and mRNA vaccines-first large test for a new approach. JAMA. 2020;324:1125–1127. doi: 10.1001/jama.2020.16866. [DOI] [PubMed] [Google Scholar]
- 212.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R., Zhou J., Bouton C.R., Rogers P., Polra K., Lin P.J.C., Barbosa C., Tam Y.K., Barclay W.S., Shattock R.J. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 2020;11:3523. doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O'Dell S., Schmidt S.D., Swanson P.A., 2nd, Padilla M., Mascola J.R., Neuzil K.M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J.E., Graham B.S., Beigel J.H. An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Zhang C., Maruggi G., Shan H., Li J. Advances in mRNA Vaccines for infectious diseases. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Maruggi G., Zhang C., Li J., Ulmer J.B., Yu D. mRNA as a transformative technology for vaccine development to control infectious diseases. Mol. Ther. 2019;27:757–772. doi: 10.1016/j.ymthe.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Jocelyn K. Temperature concerns could slow the rollout of new coronavirus vaccines. Science. 2020 doi: 10.1126/science.abf7422. [DOI] [Google Scholar]
- 218.Jackson N.A.C., Kester K.E., Casimiro D., Gurunathan S., DeRosa F. The promise of mRNA vaccines: a biotech and industrial perspective. NPJ Vacc. 2020;5:11. doi: 10.1038/s41541-020-0159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R., Wagner W., Granados A., Greenhouse J., Walker M., Willis E., Yu J.S., McGee C.E., Sempowski G.D., Mui B.L., Tam Y.K., Huang Y.J., Vanlandingham D., Holmes V.M., Balachandran H., Sahu S., Lifton M., Higgs S., Hensley S.E., Madden T.D., Hope M.J., Kariko K., Santra S., Graham B.S., Lewis M.G., Pierson T.C., Haynes B.F., Weissman D. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C., Ciaramella G., Diamond M.S. Modified mRNA vaccines protect against zika virus infection. Cell. 2017;169:176. doi: 10.1016/j.cell.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 221.Legastelois I., Buffin S., Peubez I., Mignon C., Sodoyer R., Werle B. Non-conventional expression systems for the production of vaccine proteins and immunotherapeutic molecules. Hum. Vacc. Immunother. 2017;13:947–961. doi: 10.1080/21645515.2016.1260795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Nascimento I.P., Leite L.C. Recombinant vaccines and the development of new vaccine strategies. Braz. J. Med. Biol. Res. 2012;45:1102–1111. doi: 10.1590/S0100-879X2012007500142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Walls A.C., Fiala B., Schafer A., Wrenn S., Pham M.N., Murphy M., Tse L.V., Shehata L., O'Connor M.A., Chen C., Navarro M.J., Miranda M.C., Pettie D., Ravichandran R., Kraft J.C., Ogohara C., Palser A., Chalk S., Lee E.C., Guerriero K., Kepl E., Chow C.M., Sydeman C., Hodge E.A., Brown B., Fuller J.T., Dinnon K.H., 3rd, Gralinski L.E., Leist S.R., Gully K.L., Lewis T.B., Guttman M., Chu H.Y., Lee K.K., Fuller D.H., Baric R.S., Kellam P., Carter L., Pepper M., Sheahan T.P., Veesler D., King N.P. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell. 2020;183:1367–1382. doi: 10.1016/j.cell.2020.10.043. (e1317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Powell A.E., Zhang K., Sanyal M., Tang S., Weidenbacher P.A., Li S., Pham T.D., Pak J.E., Chiu W., Kim P.S. A single immunization with spike-functionalized ferritin vaccines elicits neutralizing antibody responses against SARS-CoV-2 in mice. bioRxiv. 2020 doi: 10.1101/2020.08.28.272518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Manini I., Domnich A., Amicizia D., Rossi S., Pozzi T., Gasparini R., Panatto D., Montomoli E. Flucelvax (Optaflu) for seasonal influenza. Exp. Rev. Vacc. 2015;14:789–804. doi: 10.1586/14760584.2015.1039520. [DOI] [PubMed] [Google Scholar]
- 226.Cox M.M., Patriarca P.A., Treanor J. FluBlok, a recombinant hemagglutinin influenza vaccine. Influenza Other Respir. Viruses. 2008;2:211–219. doi: 10.1111/j.1750-2659.2008.00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Mast E.E., Margolis H.S., Fiore A.E., Brink E.W., Goldstein S.T., Wang S.A., Moyer L.A., Bell B.P., Alter M.J., P. Advisory Committee on Immunization A comprehensive immunization strategy to eliminate transmission of hepatitis B virus infection in the United States: recommendations of the Advisory Committee on Immunization Practices (ACIP) part 1: immunization of infants, children, and adolescents. MMWR Recomm. Rep. 2005;54:1–31. [PubMed] [Google Scholar]
- 228.Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H., Smith G., Patel N., Frieman M.B., Haupt R.E., Logue J., McGrath M., Weston S., Piedra P.A., Desai C., Callahan K., Lewis M., Price-Abbott P., Formica N., Shinde V., Fries L., Lickliter J.D., Griffin P., Wilkinson B., Glenn G.M. Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020;383(24):2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Frietze K.M., Peabody D.S., Chackerian B. Engineering virus-like particles as vaccine platforms. Curr. Opin. Virol. 2016;18:44–49. doi: 10.1016/j.coviro.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Syomin B.V., Ilyin Y.V. Virus-like particles as an instrument of vaccine production. Mol. Biol. 2019;53:323–334. doi: 10.1134/S0026893319030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Rodriguez-Limas W.A., Sekar K., Tyo K.E. Virus-like particles: the future of microbial factories and cell-free systems as platforms for vaccine development. Curr. Opin. Biotechnol. 2013;24:1089–1093. doi: 10.1016/j.copbio.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Roldao A., Mellado M.C., Castilho L.R., Carrondo M.J., Alves P.M. Virus-like particles in vaccine development. Exp. Rev. Vacc. 2010;9:1149–1176. doi: 10.1586/erv.10.115. [DOI] [PubMed] [Google Scholar]
- 233.Ward B.J., Gobeil P., Séguin A., Atkins J., Boulay I., Charbonneau P.-Y., Couture M., D’Aoust M.-A., Dhaliwall J., Finkle C., Hager K., Mahmood A., Makarkov A., Cheng M., Pillet S., Schimke P., St-Martin S., Trépanier S., Landry N. Phase 1 trial of a candidate recombinant virus-like particle vaccine for Covid-19 disease produced in plants. medRxiv. 2020 doi: 10.1101/2020.11.04.20226282. [DOI] [Google Scholar]
- 234.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield C.-G.G., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. (e819) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Fulginiti V.A., Eller J.J., Downie A.W., Kempe C.H. Altered reactivity to measles virus. Atypical measles in children previously immunized with inactivated measles virus vaccines. JAMA. 1967;202:1075–1080. doi: 10.1001/jama.202.12.1075. [DOI] [PubMed] [Google Scholar]
- 236.Kim H.W., Canchola J.G., Brandt C.D., Pyles G., Chanock R.M., Jensen K., Parrott R.H. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 237.Zhang G.Z.Y., Pan H., Li C., Hu Y., Chu K., Han W., Chen Z., Tang R., Yin W., Chen X., Hu X.L.Y., Jiang C., Li J., Yang M., Song Y., Wang X., Gao Q., Zhu F. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18–59 years: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Tostanoski L.H., Wegmann F., Martinot A.J., Loos C., McMahan K., Mercado N.B., Yu J., Chan C.N., Bondoc S., Starke C.E., Nekorchuk M., Busman-Sahay K., Piedra-Mora C., Wrijil L.M., Ducat S., Custers J., Atyeo C., Fischinger S., Burke J.S., Feldman J., Hauser B.M., Caradonna T.M., Bondzie E.A., Dagotto G., Gebre M.S., Jacob-Dolan C., Lin Z., Mahrokhian S.H., Nampanya F., Nityanandam R., Pessaint L., Porto M., Ali V., Benetiene D., Tevi K., Andersen H., Lewis M.G., Schmidt A.G., Lauffenburger D.A., Alter G., Estes J.D., Schuitemaker H., Zahn R., Barouch D.H. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat. Med. 2020;26:1694–1700. doi: 10.1038/s41591-020-1070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Graham S.P., McLean R.K., Spencer A.J., Belij-Rammerstorfer S., Wright D., Ulaszewska M., Edwards J.C., Hayes J.W.P., Martini V., Thakur N., Conceicao C., Dietrich I., Shelton H., Waters R., Ludi A., Wilsden G., Browning C., Bialy D., Bhat S., Stevenson-Leggett P., Hollinghurst P., Gilbride C., Pulido D., Moffat K., Sharpe H., Allen E., Mioulet V., Chiu C., Newman J., Asfor A.S., Burman A., Crossley S., Huo J., Owens R.J., Carroll M., Hammond J.A., Tchilian E., Bailey D., Charleston B., Gilbert S.C., Tuthill T.J., Lambe T. Evaluation of the immunogenicity of prime-boost vaccination with the replication-deficient viral vectored COVID-19 vaccine candidate ChAdOx1 nCoV-19. NPJ Vacc. 2020;5:69. doi: 10.1038/s41541-020-00221-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V.A., Bushmaker T., Flaxman A., Ulaszewska M., Feldmann F., Allen E.R., Sharpe H., Schulz J., Holbrook M., Okumura A., Meade-White K., Perez-Perez L., Edwards N.J., Wright D., Bissett C., Gilbride C., Williamson B.N., Rosenke R., Long D., Ishwarbhai A., Kailath R., Rose L., Morris S., Powers C., Lovaglio J., Hanley P.W., Scott D., Saturday G., de Wit E., Gilbert S.C., Munster V.J. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586:578–582. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Ewer K.J., Barrett J.R., Belij-Rammerstorfer S., Sharpe H., Makinson R., Morter R., Flaxman A., Wright D., Bellamy D., Bittaye M., Dold C., Provine N.M., Aboagye J., Fowler J., Silk S.E., Alderson J., Aley P.K., Angus B., Berrie E., Bibi S., Cicconi P., Clutterbuck E.A., Chelysheva I., Folegatti P.M., Fuskova M., Green C.M., Jenkin D., Kerridge S., Lawrie A., Minassian A.M., Moore M., Mujadidi Y., Plested E., Poulton I., Ramasamy M.N., Robinson H., Song R., Snape M.D., Tarrant R., Voysey M., Watson M.E.E., Douglas A.D., Hill A.V.S., Gilbert S.C., Pollard A.J., Lambe T., Oxford C.V.T.G. T cell and antibody responses induced by a single dose of ChAdOx1 nCoV-19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nat. Med. 2020 doi: 10.1038/s41591-020-01194-5. [DOI] [PubMed] [Google Scholar]
- 242.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., Bibi S., Briner C., Cicconi P., Collins A.M., Colin-Jones R., Cutland C.L., Darton T.C., Dheda K., Duncan C.J.A., Emary K.R.W., Ewer K.J., Fairlie L., Faust S.N., Feng S., Ferreira D.M., Finn A., Goodman A.L., Green C.M., Green C.A., Heath P.T., Hill C., Hill H., Hirsch I., Hodgson S.H.C., Izu A., Jackson S., Jenkin D., Joe C.C.D., Kerridge S., Koen A., Kwatra G., Lazarus R., Lawrie A.M., Lelliott A., Libri V., Lillie P.J., Mallory R., Mendes A.V.A., Milan E.P., Minassian A.M., McGregor A., Morrison H., Mujadidi Y.F., Nana A., O’Reilly P.J., Padayachee S.D., Pittella A., Plested E., Pollock K.M., Ramasamy M.N., Rhead S., Schwarzbold A.V., Singh N., Smith A., Song R., Snape M.D., Sprinz E., Sutherland R.K., Tarrant R., Thomson E.C., Torok M.E., Toshner M., Turner D.P.J., Vekemans J., Villafana T.L., Watson M.E.E., Williams C.J., Douglas A.D., Hill A.V.S., Lambe T., Gilbert S.C., Pollard A.J., Oxford C.V.T.G. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2020;397(10269):99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ramasamy M.N., Minassian A.M., Ewer K.J., Flaxman A.L., Folegatti P.M., Owens D.R., Voysey M., Aley P.K., Angus B., Babbage G., Belij-Rammerstorfer S., Berry L., Bibi S., Bittaye M., Cathie K., Chappell H., Charlton S., Cicconi P., Clutterbuck E.A., Colin-Jones R., Dold C., Emary K.R.W., Fedosyuk S., Fuskova M., Gbesemete D., Green C., Hallis B., Hou M.M., Jenkin D., Joe C.C.D., Kelly E.J., Kerridge S., Lawrie A.M., Lelliott A., Lwin M.N., Makinson R., Marchevsky N.G., Mujadidi Y., Munro A.P.S., Pacurar M., Plested E., Rand J., Rawlinson T., Rhead S., Robinson H., Ritchie A.J., Ross-Russell A.L., Saich S., Singh N., Smith C.C., Snape M.D., Song R., Tarrant R., Themistocleous Y., Thomas K.M., Villafana T.L., Warren S.C., Watson M.E.E., Douglas A.D., Hill A.V.S., Lambe T., Gilbert S.C., Faust S.N., Pollard A.J., Oxford C.V.T.G. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial. Lancet. 2021;396:1979–1993. doi: 10.1016/S0140-6736(20)32466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.AstraZeneca . 2020. AstraZeneca’s COVID-19 Vaccine Authorised for Emergency Supply in the UK. [Google Scholar]
- 245.Jon C. Russia’s claim of a successful COVID-19 vaccine doesn’t pass the ‘smell test,’ critics say. Science. 2020 doi: 10.1126/science.abf6791. [DOI] [Google Scholar]
- 246.S. V . 2020. The Sputnik V Vaccine’s Efficacy is Confirmed at 91.4% based on Data Analysis of the Final Control Point of Clinical Trials. [Google Scholar]
- 247.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schafer A., Ziwawo C.T., DiPiazza A.T., Dinnon K.H., Elbashir S.M., Shaw C.A., Woods A., Fritch E.J., Martinez D.R., Bock K.W., Minai M., Nagata B.M., Hutchinson G.B., Wu K., Henry C., Bahl K., Garcia-Dominguez D., Ma L., Renzi I., Kong W.P., Schmidt S.D., Wang L., Zhang Y., Phung E., Chang L.A., Loomis R.J., Altaras N.E., Narayanan E., Metkar M., Presnyak V., Liu C., Louder M.K., Shi W., Leung K., Yang E.S., West A., Gully K.L., Stevens L.J., Wang N., Wrapp D., Doria-Rose N.A., Stewart-Jones G., Bennett H., Alvarado G.S., Nason M.C., Ruckwardt T.J., McLellan J.S., Denison M.R., Chappell J.D., Moore I.N., Morabito K.M., Mascola J.R., Baric R.S., Carfi A., Graham B.S. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O'Connell S., Bock K.W., Minai M., Nagata B.M., Andersen H., Martinez D.R., Noe A.T., Douek N., Donaldson M.M., Nji N.N., Alvarado G.S., Edwards D.K., Flebbe D.R., Lamb E., Doria-Rose N.A., Lin B.C., Louder M.K., O'Dell S., Schmidt S.D., Phung E., Chang L.A., Yap C., Todd J.M., Pessaint L., Van Ry A., Browne S., Greenhouse J., Putman-Taylor T., Strasbaugh A., Campbell T.A., Cook A., Dodson A., Steingrebe K., Shi W., Zhang Y., Abiona O.M., Wang L., Pegu A., Yang E.S., Leung K., Zhou T., Teng I.T., Widge A., Gordon I., Novik L., Gillespie R.A., Loomis R.J., Moliva J.I., Stewart-Jones G., Himansu S., Kong W.P., Nason M.C., Morabito K.M., Ruckwardt T.J., Ledgerwood J.E., Gaudinski M.R., Kwong P.D., Mascola J.R., Carfi A., Lewis M.G., Baric R.S., McDermott A., Moore I.N., Sullivan N.J., Roederer M., Seder R.A., Graham B.S. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Widge A.T., Rouphael N.G., Jackson L.A., Anderson E.J., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A.B., Flach B., Lin B.C., Doria-Rose N.A., O’Dell S., Schmidt S.D., Neuzil K.M., Bennett H., Leav B., Makowski M., Albert J., Cross K., Edara V.V., Floyd K., Suthar M.S., Buchanan W., Luke C.J., Ledgerwood J.E., Mascola J.R., Graham B.S., Beigel J.H. Durability of responses after SARS-CoV-2 mRNA-1273 vaccination. N. Engl. J. Med. 2020;384(1):80–82. doi: 10.1056/NEJMc2032195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., McGettigan J., Kehtan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B.S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T., C.S. Group Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Moderna U.S. 2020. CDC Advisory Committee on Immunization Practices Recommends Vaccination with Moderna’s COVID-19 Vaccine for Persons 18 Years and Older. [Google Scholar]
- 252.Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.Y., Tureci O., Tompkins K.R., Walsh E.E., Frenck R., Falsey A.R., Dormitzer P.R., Gruber W.C., Sahin U., Jansen K.U. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 253.Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., Kranz L.M., Walzer K.C., Hein S., Güler A., Loschko J., Maddur M.S., Tompkins K., Cole J., Lui B.G., Ziegenhals T., Plaschke A., Eisel D., Dany S.C., Fesser S., Erbar S., Bates F., Schneider D., Jesionek B., Sänger B., Wallisch A.-K., Feuchter Y., Junginger H., Krumm S.A., Heinen A.P., Adams-Quack P., Schlereth J., Kröner C., Hall-Ursone S., Brasky K., Griffor M.C., Han S., Lees J.A., Mashalidis E.H., Sahasrabudhe P.V., Tan C.Y., Pavliakova D., Singh G., Fontes-Garfias C., Pride M., Scully I.L., Ciolino T., Obregon J., Gazi M., Carrion R., Alfson K.J., Kalina W.V., Kaushal D., Shi P.-Y., Klamp T., Rosenbaum C., Kuhn A.N., Türeci Ö., Dormitzer P.R., Jansen K.U., Sahin U. A prefusion SARS-CoV-2 spike RNA vaccine is highly immunogenic and prevents lung infection in non-human primates. bioRxiv. 2020 doi: 10.1101/2020.09.08.280818. [DOI] [Google Scholar]
- 254.Sahin U., et al. BNT162b2 induces SARS-CoV-2-neutralising antibodies and T cells in humans. Med RXiv. 2020 doi: 10.1101/2020.12.09.20245175. [DOI] [Google Scholar]
- 255.Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.Y., Tureci O., Tompkins K.R., Lyke K.E., Raabe V., Dormitzer P.R., Jansen K.U., Sahin U., Gruber W.C. Safety and immunogenicity of two RNA-based covid-19 vaccine candidates. N. Engl. J. Med. 2020;383(25):2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Jr., Hammitt L.L., Tureci O., Nell H., Schaefer A., Unal S., Tresnan D.B., Mather S., Dormitzer P.R., Sahin U., Jansen K.U., Gruber W.C., C.C.T. Group Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Pfizer U.S. 2020. CDC Committee of Independent Health Experts Recommends Vaccination with Pfizer and Biontech Covid-19 Vaccine for Persons Ages 16 Years and Older. [Google Scholar]
- 258.Bangaru S., Ozorowski G., Turner H.L., Antanasijevic A., Huang D., Wang X., Torres J.L., Diedrich J.K., Tian J.H., Portnoff A.D., Patel N., Massare M.J., Yates J.R., 3rd, Nemazee D., Paulson J.C., Glenn G., Smith G., Ward A.B. Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate. Science. 2020;370(6520):1089–1094. doi: 10.1126/science.abe1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Tian J.-H., Patel N., Haupt R., Zhou H., Weston S., Hammond H., Lague J., Portnoff A.D., Norton J., Guebre-Xabier M., Zhou B., Jacobson K., Maciejewski S., Khatoon R., Wisniewska M., Moffitt W., Kluepfel-Stahl S., Ekechukwu B., Papin J., Boddapati S., Wong C.J., Piedra P.A., Frieman M.B., Massare M.J., Fries L., Bengtsson K. Lövgren, Stertman L., Ellingsworth L., Glenn G., Smith G. SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 elicits immunogenicity in baboons and protection in mice. bioRxiv. 2020 doi: 10.1101/2020.06.29.178509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Guebre-Xabier M., Patel N., Tian J.H., Zhou B., Maciejewski S., Lam K., Portnoff A.D., Massare M.J., Frieman M.B., Piedra P.A., Ellingsworth L., Glenn G., Smith G. NVX-CoV2373 vaccine protects cynomolgus macaque upper and lower airways against SARS-CoV-2 challenge. Vaccine. 2020;38:7892–7896. doi: 10.1016/j.vaccine.2020.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Weisberg S.P., Connors T.J., Zhu Y., Baldwin M.R., Lin W.H., Wontakal S., Szabo P.A., Wells S.B., Dogra P., Gray J., Idzikowski E., Stelitano D., Bovier F.T., Davis-Porada J., Matsumoto R., Poon M.M.L., Chait M., Mathieu C., Horvat B., Decimo D., Hudson K.E., Zotti F.D., Bitan Z.C., La Carpia F., Ferrara S.A., Mace E., Milner J., Moscona A., Hod E., Porotto M., Farber D.L. Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat. Immunol. 2020;22(1):25–31. doi: 10.1038/s41590-020-00826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Day T., Gandon S., Lion S., Otto S.P. On the evolutionary epidemiology of SARS-CoV-2. Curr. Biol. 2020;30:R849–R857. doi: 10.1016/j.cub.2020.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Chen Z., John Wherry E. T cell responses in patients with COVID-19. Nat. Rev. Immunol. 2020;20:529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Chakraborty S., Gonzalez J., Edwards K., Mallajosyula V., Buzzanco A.S., Sherwood R., Buffone C., Kathale N., Providenza S., Xie M.M., Andrews J.R., Blish C.A., Singh U., Dugan H., Wilson P.C., Pham T.D., Boyd S.D., Nadeau K.C., Pinsky B.A., Zhang S., Memoli M.J., Taubenberger J.K., Morales T., Schapiro J.M., Tan G.S., Jagannathan P., Wang T.T. Proinflammatory IgG fc structures in patients with severe COVID-19. Nat. Immunol. 2020;22(1):67–73. doi: 10.1038/s41590-020-00828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.DiLillo D.J., Palese P., Wilson P.C., Ravetch J.V. Broadly neutralizing anti-influenza antibodies require fc receptor engagement for in vivo protection. J. Clin. Invest. 2016;126:605–610. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.DiLillo D.J., Tan G.S., Palese P., Ravetch J.V. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat. Med. 2014;20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Henry Dunand C.J., Leon P.E., Huang M., Choi A., Chromikova V., Ho I.Y., Tan G.S., Cruz J., Hirsh A., Zheng N.Y., Mullarkey C.E., Ennis F.A., Terajima M., Treanor J.J., Topham D.J., Subbarao K., Palese P., Krammer F., Wilson P.C. Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe. 2016;19:800–813. doi: 10.1016/j.chom.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Thulin N.K., Wang T.T. The role of Fc gamma receptors in broad protection against influenza viruses. Vacc. (Basel) 2018;6 doi: 10.3390/vaccines6030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Pica N., Hai R., Krammer F., Wang T.T., Maamary J., Eggink D., Tan G.S., Krause J.C., Moran T., Stein C.R., Banach D., Wrammert J., Belshe R.B., Garcia-Sastre A., Palese P. Hemagglutinin stalk antibodies elicited by the 2009 pandemic influenza virus as a mechanism for the extinction of seasonal H1N1 viruses. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2573–2578. doi: 10.1073/pnas.1200039109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ekiert D.C., Bhabha G., Elsliger M.A., Friesen R.H., Jongeneelen M., Throsby M., Goudsmit J., Wilson I.A. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Sui J., Hwang W.C., Perez S., Wei G., Aird D., Chen L.M., Santelli E., Stec B., Cadwell G., Ali M., Wan H., Murakami A., Yammanuru A., Han T., Cox N.J., Bankston L.A., Donis R.O., Liddington R.C., Marasco W.A. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Sutton T.C., Chakraborty S., Mallajosyula V.V.A., Lamirande E.W., Ganti K., Bock K.W., Moore I.N., Varadarajan R., Subbarao K. Protective efficacy of influenza group 2 hemagglutinin stem-fragment immunogen vaccines. NPJ Vacc. 2017;2:35. doi: 10.1038/s41541-017-0036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Maamary J., Wang T.T., Tan G.S., Palese P., Ravetch J.V. Increasing the breadth and potency of response to the seasonal influenza virus vaccine by immune complex immunization. Proc. Natl. Acad. Sci. U. S. A. 2017;114(38):10172–10177. doi: 10.1073/pnas.1707950114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Leon P.E., He W., Mullarkey C.E., Bailey M.J., Miller M.S., Krammer F., Palese P., Tan G.S. Optimal activation of Fc-mediated effector functions by influenza virus hemagglutinin antibodies requires two points of contact. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E5944–E5951. doi: 10.1073/pnas.1613225113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Corti D., Voss J., Gamblin S.J., Codoni G., Macagno A., Jarrossay D., Vachieri S.G., Pinna D., Minola A., Vanzetta F., Silacci C., Fernandez-Rodriguez B.M., Agatic G., Bianchi S., Giacchetto-Sasselli I., Calder L., Sallusto F., Collins P., Haire L.F., Temperton N., Langedijk J.P., Skehel J.J., Lanzavecchia A. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza a hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 276.Mallajosyula V.V., Citron M., Ferrara F., Lu X., Callahan C., Heidecker G.J., Sarma S.P., Flynn J.A., Temperton N.J., Liang X., Varadarajan R. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E2514–E2523. doi: 10.1073/pnas.1402766111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Hai R., Krammer F., Tan G.S., Pica N., Eggink D., Maamary J., Margine I., Albrecht R.A., Palese P. Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J. Virol. 2012;86:5774–5781. doi: 10.1128/JVI.00137-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Omer S.B., Yildirim I., Forman H.P. Herd immunity and implications for SARS-CoV-2 control. JAMA. 2020;324:2095–2096. doi: 10.1001/jama.2020.20892. [DOI] [PubMed] [Google Scholar]
- 279.V.C. Project 2020. https://www.vaccineconfidence.org/covid-19
- 280.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L, Hinsley W.R., Laydon D.J., Dabrera G., O’Toole A., Amato R., Ragonnet-Cronin M., Harrison I., Jackson B., Ariani C.V., Boyd O., Loman N.J., McCrone J.T., Gonçalves S., Jorgensen D., Myers R., Hill V., Jackson D.K., Gaythorpe K., Groves N., Sillitoe J., Kwiatkowski D.P., Flaxman S., Ratmann O., Bhatt S., Hopkins S., Gandy A., Rambaut A., Ferguson N.M. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data. medRxiv. 2021 doi: 10.1101/2020.12.30.20249034. [DOI] [Google Scholar]
- 281.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhar J., Doolabh D., Pillay S., San E.J., Msom N., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A.J., Engelbrecht S., Van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao M., Korsman S., Davies M., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkord A., Godzik A., Wibmer C.K., Sewell B.T., Lourenço J., Alcantara L.C.J., Pond S.L.K., Weaver S., Martin D., Lessells R.J., Bhiman J.N., Williamson C., de Oliveira T. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- 282.Voloch C.M., Silva F R.D., de Almeida L.G.P., Cardoso C.C., Brustolin O.J., Gerber A.L., Guimarães A.P.D.C., Mariani D., Costa R.M.D., Ferreira O.C., Cavalcanti A.C., Frauches T.S., de Mello C.M.B., Galliez R.M., Faffe D.S., Castiñeiras T.M.P.P., Tanuri A., de Vasconcelos A.T.R. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. medRxiv. 2020 doi: 10.1101/2020.12.23.20248598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Lambson B.E., Vermeulen M., van den Berg K., Rossouw T., Boswell M., Ueckermann V., Meiring S., von Gottberg A., Cohen C., Morris L., Bhiman J.N., Moore P.L. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv. 2021 doi: 10.1101/2021.01.18.427166. [DOI] [PubMed] [Google Scholar]
- 284.Xie X., Zou J., Fontes-Garfias C.R., Xia H., Swanson K.A., Cutler M., Cooper D., Menachery V.D., Weaver S., Dormitzer P.R., Shi P. Neutralization of N501Y mutant SARS-CoV-2 by BNT162b2 vaccine-elicited sera. bioRxiv. 2021 doi: 10.1101/2021.01.07.425740. [DOI] [PubMed] [Google Scholar]
- 285.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., Yang Z., Abernathy M.E., Huey-Tubman K.E., Hurley A., Turroja M., West K.A., Gordon K., Millard K.G., Ramos V., Da Silva J., Xu J., Colbert R.A., Patel R., Dizon J., Unson-O’Brien C., Shimeliovich I., Gazumyan A., Caskey M., Bjorkman P.J., Casellas R., Hatziioannou T., Bieniasz P.D., Nussenzweig M.C. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. bioRxiv. 2021 doi: 10.1101/2021.01.15.426911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.China COVID vaccine reports mixed results - what does that mean for the pandemic? Nature. 2021 doi: 10.1038/d41586-021-00094-z. [DOI] [PubMed] [Google Scholar]