Graphical abstract
Keywords: RNA therapeutics, SARS-CoV-2, Mutation, COVID-19, Drug safety and drug efficacy
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a global pandemic. Three viral proteins, the spike protein (S) for attachment of virus to host cells, 3-chymotrypsin-like cysteine protease (Mpro) for digestion of viral polyproteins to functional proteins, and RNA-dependent-RNA-polymerase (RdRp) for RNA synthesis are the most critical proteins for virus infection and replication, rendering them the most important drug targets for both antibody and chemical drugs. Due to its low-fidelity polymerase, the virus is subject to frequent mutations. To date, the sequence data from tens of thousands of virus isolates have revealed hundreds of mutations. Although most mutations have a minimum consequence, a small number of non-synonymous mutations may alter the virulence and antigenicity of the mutants. To evaluate the effects of viral mutations on drug safety and efficacy, we reviewed the biochemical features of the three main proteins and their potentials as drug targets, and analyzed the mutation profiles and their impacts on RNA therapeutics. We believe that monitoring and predicting mutation-introduced protein conformational changes in the three key viral proteins and evaluating their binding affinities and enzymatic activities with the U.S. Food and Drug Administration (FDA) regulated drugs by using computational modeling and machine learning processes can provide valuable information for the consideration of drug efficacy and drug safety for drug developers and drug reviewers. Finally, we propose an interactive database for drug developers and reviewers to use in evaluating the safety and efficacy of U.S. FDA regulated drugs with regard to viral mutations.
1. Introduction
COVID-19, the 2019 novel coronavirus disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in December of 2019, and has rapidly spread to more than 210 countries. As of November 30, 2020, more than 62,363,500 COVID-19 cases with more than 1,456,600 deaths have been documented worldwide (https://covid19.who.int/). Among the most-affected countries by SARS-CoV-2, the United States had almost 13,143,000 confirmed cases and 265,200 total deaths by the end of November 2020 (https://covid.cdc.gov/covid-data-tracker/). Worldwide, the unemployment rate increased dramatically, and trillions of dollars were spent in an effort to reduce infection and provide health care, including virus testing and patient treatment, to fight the COVID-19 pandemic. Considering the health and economic impacts of the COVID-19 pandemic, scientists, and clinicians have been acting rapidly to investigate the nature (such as genome structure, evolution, and mutations) of SARS-CoV-2 and the pathological processes (such as viral infection and host responses) of the COVID-19 disease, in order to discover potential drug targets, effective therapeutics, and vaccines to counter the evolving pandemic.
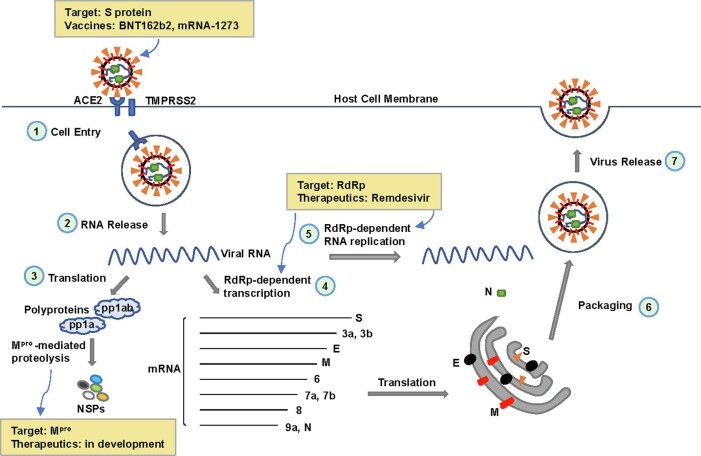
The SARS-CoV-2 virus, a novel member of the Betacoronavirus genus in the Coronaviridae family, is a positive-sense, single-stranded RNA virus, with approximately 29,900 nucleotides in its entire genome [1]. Structural studies and biochemical experiments have revealed many notable features of SARS-CoV-2. For example, several vital proteins are key factors at different stages of the viral infectious cycle (Fig. 1 ), including virus entry, replication, survival, and further infection: 1) the spike (S) protein of SARS-CoV-2 appears to have a high affinity to human receptor angiotensin-converting enzyme 2 (ACE2), which facilitates viral entry to human cells; 2) the RNA-dependent RNA polymerase (RdRp) catalyzes RNA-template dependent RNA synthesis, which is responsible for virus replication; and 3) the main protease (Mpro) digests polyproteins that are translated from viral RNA, resulting in individual functional viral proteins. These features help us better understand the biology, immunology, and pathology of the virus, which make these key proteins actionable drug targets.
Fig. 1.
The Infectious Cycle of SARS-CoV-2 and Therapeutic Targets to Key Proteins. 1) The spike (S) protein of SARS-CoV-2 binds to the angiotensin-converting enzyme 2 (ACE2) receptor on the host cell membrane, with TMPRSS2 in close proximity to aid viral fusion. The virus then enters host cells via endocytosis. As of December 2020, two mRNA vaccines targeting the S protein (BNT162b2 and mRNA-1273) have been authorized for emergency use by the U.S. Food and Drug Administration (FDA) for the prevention of COVID-19. 2) Upon cell entry, the viral RNA of SARS-CoV-2 is released into the cytoplasm. 3) Translation of the viral RNA produces polyproteins (pp1a and pp1ab), which are cleaved by the main protease (Mpro) to form non-structural proteins (NSPs). 4) Transcription of the viral RNA results in several mRNAs that are translated into essential viral proteins, including S, M (membrane), E (envelope), and N (nucleocapsid). 5) RNA-dependent RNA polymerase (RdRp) catalyzes viral RNA replication for new viral particle assembly. Remdesivir acts as an RdRp inhibitor and is the first drug approved by the U.S. FDA to treat COVID-19. 6) Newly synthesized viral RNA is packaged into new viral particles with the essential viral proteins. 7) Vesicles carrying new viral particles fuse with the cell membrane, releasing the new virus from host cells.
As the central dogma of molecular biology, RNAs primarily convert coding information from DNA into proteins; however, with many noncoding characteristics, RNAs are a group of essential molecules involved in almost all biological processes. RNA molecules and their biological features are used for diagnosis (e.g. testing kits), prevention (e.g. vaccines), and treatment (e.g. therapeutics) of human diseases. Particularly, as emerging drug targets, RNAs have been developed for innovative and promising therapeutics. RNA therapeutics refer to RNA-based therapeutic technologies and delivery strategies, including 1) biological interventions through RNA molecules, such as small interfering RNAs (siRNAs), microRNAs (miRNAs), noncoding RNAs, antisense oligonucleotides (ASOs), guide RNAs, mRNAs, and other types of RNAs; and 2) biochemical approaches involving mRNA degradation, inhibition of RNA or protein synthesis, and blockage of cellular functions. Therefore, most therapeutics for the treatment of COVID-19 can be considered RNA therapeutics.
It is worth noting that the incidence of nucleotide mutations is significantly high among RNA viruses, due to the lack of proofreading capability of their RNA polymerases. Specifically, the mutation rate of SARS-CoV-2 could be hundreds of thousands of times higher than that in host cells, which benefits viral evolution and hinders disease prevention and treatment. This review aims to address the importance of monitoring mutations in SARS-CoV-2 and their impacts on RNA therapeutics used for the treatment of COVID-19.
2. Genome structure of SARS-CoV-2 and key drug targets
With ~29,900 nucleotides in the positive strand of its RNA, SARS-CoV-2 has 12 fictional open reading frames (ORFs), by which 16 proteins are encoded [2]. Some of these proteins are expressed as polyproteins that require protease cleavage to produce functional individual proteins. Specifically, 16 non-structural proteins (NSPs) are composed of 7096 amino acids. From its 5′-cap to 3′-tail, the viral genome is arranged primarily by ORF1a and ORF1ab for NSPs, followed by other ORFs for structural proteins, including S, ORF3a, E, M, ORF6, ORF7a, ORF7b, ORF8, N, and ORF10 [1]. Briefly, the SARS-CoV-2 coded proteins can be categorized as replicase (R), protease (Pro), spike protein (S), membrane protein (M), nucleoproteins (N) including RdRp, and envelop protein (E). These proteins are important players in viral entry, replication, and survival in host cells, making them the major targets for therapeutics and vaccines [1].
2.1. The spike (S) protein
The coronavirus S protein is a transmembrane glycoprotein that is usually arranged as a trimer, and is the main protein that contributes to viral particle attachment, viral-host fusion, and virus entry into host cells [3], [4]. The monomer of the S protein is approximately 180 kD (1273 amino acids), containing an S1 subunit (671 amino acids) and an S2 subunit (587 amino acids) [5]. The S1 subunit contains two major domains, the N-terminal domain (NTD) and the C-terminal domain (CTD). At the CTD, the receptor-binding domain (RBD) is responsible for the attachment of a viral particle to the surface of host cells. The interacting counterpart molecule on the host cell surface is ACE2. ACE2 is a membrane protein, which primarily functions as a hormone that regulates blood vessel constriction and blood pressure; however, it works as a receptor protein in the host cell surface during coronavirus infection. Upon binding to the receptor ACE2, the polyprotein S undergoes conformational changes, which triggers cleavage of the S protein by the serine protease TMPRSS2 at the S1/S2 site to produce S1 and S2 subunits. The S2 subunit then undergoes a series of conformational changes, resulting in viral-host cellular fusion, followed by viral entry into host cells [6], [7], [8].
Biochemical characteristics of the S protein suggest that the S protein and its receptor protein ACE2 are valuable targets for developing therapeutics to block SARS-CoV-2 from integrating with host cells. Indeed, a battery of neutralizing antibodies (nAbs) and small molecules working as inhibitors has been investigated. Several nAbs have been cloned or isolated with binding affinities towards the S protein and/or ACE2 receptor. As summarized by Huang et al. [5], antibodies 4A8 (binding to NTD), 47D11 (binding to S1), n3130 (binding to RBD), n3088 (binding to both S1 and RBD), S309 (inhibiting ACE2/RBD binding), 311mab-31B5 (binding to RBD), 311mab-31D4 (biding to RBD), P2C-1F11 (biding to RBD), P2B-2F6 (binding to RBD), B38 (binding to RBD) and H4 (binding to RBD) are idea candidates for neutralizing the coronavirus. It is worth noting that pharmaceutical companies, such as Regeneron Pharmaceuticals and Eli Lilly and Company, have developed antibodies against the SARS-CoV-2 S protein to treat COVID-19, for which we will provide more detail later in the article. In addition, small molecules have been screened to determine the possibility of their interaction with the S protein. Although some compounds showed a high binding affinity with the S protein, such as rescinnamine, iloprost, and prazosin (drugs for the treatment of hypertension); sulfasalazine, azlocillin, penicillin, and cefsulodin (drugs for anti-bacterial infection); and posaconazole and itraconazole (drugs for anti-fungal infection), only a few compounds were able to interfere with the binding between the S protein and ACE2 receptor. For example, hesperidin was predicted to be able of preventing the interaction between RBD of the S protein and ACE2, thus inhibiting viral entry [9]. In SARS-CoV, the heptad repeat 1 domain (HR1) and heptad repeat 2 (HR2) domain of the SARS-CoV S2 subunit work together to facilitate virus–cell fusion and virus entry to host cells [10]. Owing to the high similarity in amino acid sequences between SARS-CoV and SARS-CoV-2, peptides were designed and developed to block the interaction of HR1/HR2 in SARS-CoV-2 with host cell membrane protein. For example, a lipopeptide EK1C4 was developed to block SARS-CoV-2 S-mediated virus–cell fusion [11]. Likewise, the lipopeptide IPB02 was found to be a potent virus-cell fusion inhibitor to prevent SARS-CoV-2 entry [12]. Small molecule drugs were screened to inhibit virus-cell fusion. For example, Viracept (nelfinavir mesylate), the only anti-HIV (human immunodeficiency virus) protease inhibitor approved by the U.S. Food and Drug Administration (FDA), is able to suppress S protein mediated fusion in SARS-CoV-2 infection with a high potency [13]. Proteolysis of the S protein at the S1/S2 and S2 sites by TMPRSS2, cathepsin B and L or furin is an essential step for SARS-CoV-2 virus entry. Protease inhibitors that can suppress the activities of TMPRSS2, cathepsin B and L, and furin are considered as potential drugs that block SAES-CoV-2 infection. Several inhibitors, such as camostat mesylate, E-64d, α-1-PDX, and hexa-D-arginine(D6R) are in this category [5].
2.2. The RNA-dependent RNA polymerase (RdRp)
The RdRp is an enzyme complex used by SARS-CoV-2 to replicate its genome and transcribe its viral genes. This complex contains three subunits which are encoded by three non-structural protein (nsp) genes, nsp12, nsp8, and nsp7 [14], [15]. The catalytic subunit nsp12 plays a critical role in viral RNA synthesis and transcription, while subunits nps8 and nps7 are cofactors responsible for de novo initiation and primer extension during RNA synthesis [16]. The nsp12 subunit possesses a nucleotidyltransferase domain in the N-terminus and an RdRp domain in the C-terminus [15], [17]. The binding site in the N-terminus of nsp12 is responsible for interacting with nucleoside triphosphate (NTP) substrate, in which amino acids N691, S682, and D623 are pivotal for recognizing the 2′–OH group of the NTP to distinguish RNA synthesis from DNA synthesis, resulting in the enzyme specificity of RdRp [14]. Briefly, upon binding to the RNA template and NTPs, the active site of nsp12 catalyzes the addition of a new nucleotide residue to the RNA chain, a step by which the newly synthesized viral RNA is extended. One copy of subunit nsp7 and one copy of nsp8 bind to different domains of nsp12 to form the RdRp complex while another copy of nsp8 may also provide polymerase activity. In general, structural analysis has indicated that the subunit nsp12 is responsible for template recognition, nucleotide binding, and catalysis; and the heterodimer consisting of nsp7 and nsp8 serves as the stabilizer of the complex; while the second subunit of nsp8 may be critical for extending the RNA template-binding surface [15], [17].
Amino acid sequence analysis revealed that the RdRp does not show sequence homology with human analog proteins, which provides an opportunity for the development of anti-viral drugs to inhibit the enzyme with minimum risk of affecting hosts. In addition, because RdRp lacks proofreading endonuclease activity, low-fidelity RNA synthesis occurs during replication [18], opening a door for the development of chain terminators and nucleoside analog inhibitors as potential drugs to block viral replication through inhibition of RdRp. RdRps in many viruses can be targeted by several drugs, including favipiravir, ribavirin, sofosbuvir, baloxavir, dasabuvir, remdesivir, galidesivir, pimorivir and beclabuvir [19]. Advances in structural analysis of SARS-CoV-2 RdRp are critical in facilitating the design of new drug candidates, while molecular modeling is a powerful tool to predict potential inhibitors of the enzyme. The recently published crystal structure of SARS-CoV-2 RdRp [14], [15] has been used to screen potential inhibitors as drugs for the treatment of COVID-19 using in silico approaches. Molecular docking is an important approach to evaluate potential interactions between molecules. Molecular data on structures and functional mechanisms can provide insights into conformational changes and molecular interactions based on the alteration of energy and binding affinities, which increase the possibility of identifying a battery of real compounds that can inhibit the activity of this protein. Using this comprehensive computational approach, Parvez et al predicted a group of drugs that may potentially inhibit the activity of SARS-CoV-2 RdRp, and revealed amino acids involved in the interaction between drugs and the RdRp protein [20]. Ranked by their binding affinity (from the highest to the lowest) and binding energy (from the lowest to the highest), these drugs are listed as rifabutin, rifapentine, fidaxomicin, 7-methyl-guanosine-5′-triphosphate-5′-guanosine, ivermectin, remdesivir, and favipiravir, of which many are U.S. FDA-approved drugs [20].
Approved by the Japanese and Chinese governments and evaluated by a Phase-3 clinical trial in the United States, favipiravir is a guanine analog targeting RdRp to treat influenza virus infection. With a high specificity (no interaction with human DNA polymerase and RNA transcriptase), its metabolite ribofuranosyl-5′-triphosphate (F-RTP) is a suicide substrate that inhibits RdRp enzyme activity, thus terminating viral RNA synthesis. Owing to a relatively high conservation of catalytic domains of RdRps among RNA viruses, it is speculated that favipiravir should have a broad spectrum of activity in inhibition of RNA virus replication; therefore, clinical trials to evaluate safety and efficacy of favipiravir in the treatment of COVID-19 patents have been conducted or are ongoing in multiple countries including China, Japan, the United States, Russia, Saudi Arabia, and India [21]. The preliminary conclusion is that favipiravir may be a potential drug to treat mild to moderate cases of COVID-19; however, larger and more randomized clinical trials are warranted to ensure its effectiveness and safety [21]. It is worth noting that remdesivir is the only drug approved by the U.S. FDA to treat COVID-19 patients, which will be discussed in detail later in the article.
2.3. The main protease (Mpro)
After translation, the two overlapping polyproteins (pp1a and pp1ab) encoded by the ORF1ab of SARS-CoV-2 are digested through a complicated multistep process by the Mpro (nsp5), also known as 3-chymotrypsin-like cysteine protease (3CLpro), resulting in 16 functional proteins, in which the Mpro has an autocleavage property that “frees” itself (nsp5) from nsp4 and nsp6. While the monomer of Mpro is considered inactive, two subunits of Mpro, each with three domains, work together as a homodimer to catalyze the hydrolysis of polypeptides by an acid-base mechanism. The Mpro possesses a unique dyad (Cys145 and His41) in the active site, which is critical for the enzyme to maintain its substrate specificity. The substrate specificity of the protease is relatively high: a glutamine must be present in the P1 position and a leucine residue is preferred in the P2 position (other hydrophobic residues are tolerated), for which the substrate is arranged as -P4-P3-P2-P1↓P1′-P2′-P3′- (from the N terminus to the C terminus). No homologous protein has been found in the human genome, limiting off-target effects introduced by inhibition of the enzyme; genetic conservation among various coronaviruses is relatively high, and crystal structures of the main proteases exhibited a high similarity among SARS-CoV, MERS-CoV and SARS-CoV-2, rendering similar substrate specificity among different coronaviruses [22], [23]. These features make Mpro an ideal antiviral target and facilitate drug repurposing based on previous efforts towards developing anti-coronavirus drugs.
Information of the biochemical characteristics and X-ray crystal structure of Mpro is utilized to design new drugs against SARS-CoV-2. In one study, two compounds (11a and 11b) targeting Mpro were designed and synthesized, which demonstrated an excellent inhibitory effect on Mpro enzyme activity and a significant inhibition potent towards viral infection by SARS-CoV-2. The interaction of each compound with the active site of Mpro was confirmed by X-ray crystal structures at a very high resolution (1.5 Ã…), while pharmacokinetic properties of both compounds showed low toxicity, demonstrating that these promising compounds have a great potential to be anti-SARS-CoV-2 drugs [23]. Similarly, with a series of synthesized Mpro inhibitors, Rathnayake et al. [24] demonstrated that two optimized compounds (6j and 6 h) showed effective inhibitory activity against several coronaviruses including SARS-CoV-2, using enzyme assay and cell-based assays. Furthermore, these compounds exhibited significant inhibition against MERS-CoV infection in a mouse model. These data suggested that Mpro inhibitors could be developed to drugs to treat patients infected by human coronaviruses.
On the other hand, many compounds have been identified that can potentially inhibit the catalytic activity of Mpro, using in silico screening, virtual drug repurposing, and wet-lab approaches. Utilizing deep learning technology, Ton et al developed a deep docking platform to provide rapid prediction of docking scores, which enabled the analysis of 1.3 billion compounds that led to the identification of 1,000 highly possible inhibitors of Mpro protein [25]. Based on analyses of structure characteristics of Mpro and the free energy of binding between the active site and potential targeting compounds, in silico approaches including molecular docking can provide insights that lead to discovering new drug candidates or repurposing preexisting drugs. In silico studies suggested that many compounds exhibit inhibitory effects against Mpro, and that these compounds include flavonoids, peptides, terpenes, quinolines, nucleoside or nucleotide analogues, conventional protease inhibitors, phenalene, antibiotic derivatives, imidazole, and indoles [26]. Biochemical high throughput screening (HTS) strategies have also been applied to screen synthetic or natural compounds. For example, a pioneer study was conducted by Jin et al. [27], in which they integrated structure-assisted drug design, virtual drug screening and HTS to identify drugs that may target SARS-CoV-2 Mpro. First, they found a mechanism-based inhibitor (N3) of Mpro through computer-assisted drug design, and then revealed the crystal structure of a complex in which Mpro and N3 interact. Using structure-based virtual screening in combination with biochemical high-throughput screening, they analyzed more than 10,000 compounds, including drugs, drug candidates and other compounds. Finally, they discovered that six compounds can inhibit Mpro significantly with IC50 values between 0.67 and 21.4 μM, of which the compound ebselen showed strong antiviral activity in cell-based assays [27]. In another study, with a recombinant Mpro, Coelho et al conducted HTS using a fluorescent assay to identify potential inhibitors from drugs, small molecules, and natural products, leading to the discovery of 13 Mpro inhibitors (e.g. thimerosal, phenylmercuric acetate, and benzophenone derivatives) with a low IC50 values (0.2–23 μM) [28].
3. Genetic mutations of SARS-CoV-2
The limited proofreading activity of SARS-CoV-2 RdRp introduces a substantial mutation rate (approximately 10−4) in the process of virus replication; some of these mutations will be “selected” in the progeny of the viral population due to pressure from the host immunological system and other environmental contributors [29], resulting in an accumulation of two mutants per month in the genome of SARS-CoV-2 [30]. After analyzing sequence data from 7666 viral genomes, Dorp et al suggested that mutations were very diversified and occurred in all countries – “everything is everywhere”, although the main phylogenetic clades were formed by virus strains sampled from the same geological area [31]. Cumulatively, as of September 2020, more than 10,000 single mutations in the SARS-CoV-2 genome have been recorded, in comparison to the first reported sequence reference published on January 5, 2020 [32], although many of the same mutations may be repeated in different virus isolates. Even though the majority of viral mutations is harmless with minimal impact on viral virulence, some of these mutations may change infectivity, survival capability, pathologic property, or immunogenicity and antigenicity of the virus [30], [32]. For example, the common mutation D614G on the S protein may increase binding affinity between the S protein and host ACE2 receptor, thus enhancing virus loads which lead to increased infectivity. Although the mutation does not change the ability of antibody recognition, the G-form variant is more readily neutralized by a human antibody [33]. By investigating 80 mutants and 26 modifications of the glycosylation site on the S protein, Li et al. [34] found that several mutants, including A475V, L452R, V483A, and F490L increased their resistance to neutralizing antibodies and deletion of glycosylation sites N331 and N343 decreased viral infectivity. Notably, the N234Q mutant was significantly resistant towards neutralization by antibodies, whereas the N165Q mutant increased sensitivity towards neutralization therapeutics. Likewise, mutations were identified in the SARS-CoV-2 RdRp. In an earlier study with limited viral sequence data, Pachetti found several mutations in RdRp. Interestingly, it was reported that viral strains with RdRp mutations exhibited a mutation rate three times higher than that of viral strains without a RdRp mutation [35]. Using in silico approaches, Shannon et al. proposed that two mutants, F480L and V557L, may introduce structural and functional changes on RdRp, resulting in patient’ resistance against remdesivir therapy [36]. D722Y, a missense mutation located in the catalytic site of RdRp, along with mutations V472D and L469S, may change the hydrophobicity of the binding pocket of RdRp, leading to a negative impact on the efficacy of remdesivir [37]. Key amino acids in the active site of an enzyme are critical for the function of the enzyme. For example, the Gly-11 located in the dimer interface is essential for Mpro dimerization. The G11A mutation has been shown to completely prevent Mpro from dimerization, leading to an abolishment of Mpro activity of coronaviruses [38]. With many recorded mutations of Mpro, Amamuddy et al used homology modeling and molecular dynamic analyses to reveal the biological impacts of these mutants. The D48E variant introduced a conformational change at the substrate binding site, resulting in an alteration towards the efficiency and substrate specificity of Mpro [39].
3.1. Identification of gene mutation hotspots in the three key proteins
To identify mutation hotspots that are potentially important to COVID-19, we analyzed mutations on the SARS-CoV-2 genome by focusing on the five genes that encode the RdRp (consisting of Nsp12, Nsp7, Nsp8), S, and Mpro proteins. The SARS-CoV-2 reference genome, deposited by Wu et al. in January 2020, was downloaded from the National Center for Biotechnology Information (NCBI) database (under accession number NC_045512). Complete SARS-CoV-2 genome sequences and their metadata were downloaded from the Gisaid EpiCovTM database https://db.cngb.org/gisaid/, which were accrued before Nov 19th, 2020. We filtered the genomes and selected those that were collected from May17th, 2020 to Nov 19th, 2020, resulting in a total of 102,478 genomes for subsequent analysis. Next, we disregarded sequences that contained “N” (indicating unidentified nucleotides), and thus retained 55,443 analysis-ready sequences for nsp12, 56,723 for nsp7, 56,391 for nsp8, 48,134 for S, and 54,897 for Mpro. The number of analysis-ready sequences for each gene was then used to normalize mutation frequencies. The MAFFT program [40] was used to conduct multiple sequence alignments. Statistical analysis was conducted using Python with Pandas and Numpy for data cleaning and Biopython for sequence manipulation.
We found a combination of 3750 point-mutations for the five genes (Fig. 2 ). A total of 236 mutations occurred in more than 0.01% of valid sequences; 138 out of the 236 mutations resulted in amino acid changes. We confirmed the occurrence of mutations located at positions 23,403 and 14,408 which have been reported previously. A23403G that leads to D614G in the S protein has been shown to significantly increase the infectivity of SARS-CoV-2 virus by reducing S1 protein shedding [41], [42]. In our analysis, A23403G was present in 40,201 sequences for the S protein and showed the highest occurrence percentage (83.5%). Another highly frequent mutation is C14408T, which occurred in 80.4% of the sequences for nsp12 in our results.
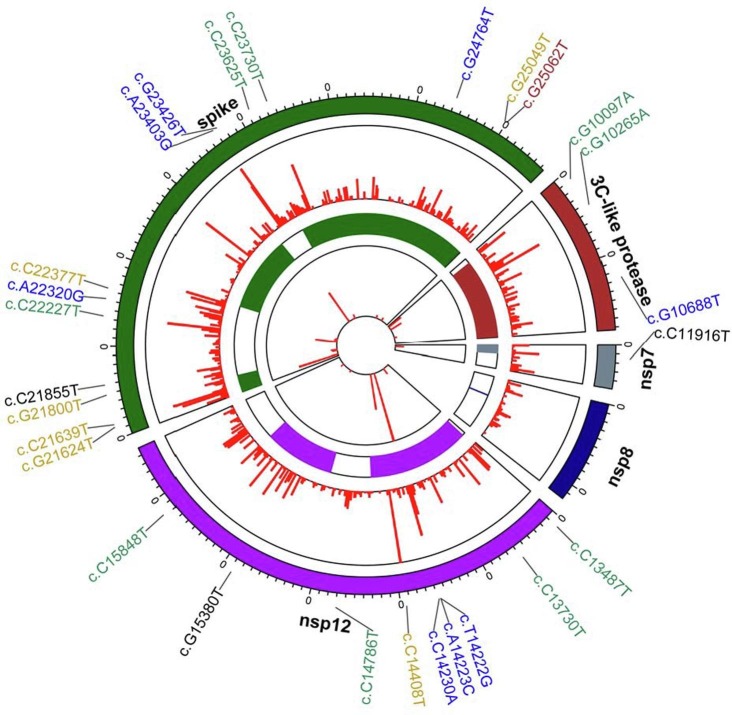
Fig. 2.
Mutation Spectrum of Three Key Proteins. The three key proteins are encoded by 5 genes of the SARS-CoV-2 genome: spike for S protein; 3-chymotrypsin-like for Mpro; and nsp7, nsp8 and nsp12 for RdRp. Twenty-seven non-synonymous mutations with high frequency occurrence are indicated in the figure for their positions (first circle, counted from the outer cycle to the inner cycle) and densities (second circle). Protein structures are color coded (third circle), the mutation frequency (log10 scale) of each amino acid is indicated at a specific position with the red line (fourth circle).
We then used the “BLOSUM90” matrix [43] to select high-impact protein mutations. “BLOSUM90” is one of the substitution matrices used for amino acid sequence alignment; it measures how likely a certain amino acid substitution is to occur in a protein based on empirical data. “BLOSUM90” also predicts how significant the impact of an amino acid substitution is to a protein’s structure; a low “BLOSUM90” score indicates a great impact. The thresholds we used to filter mutations were 1) at least 0.1% for occurrence percentage, 2) causing amino acid changes, and 3) less than 0 for “BLOSUM90” scoring. Our results revealed high-occurrence mutations that have not been previously reported, such as G10097A for Mpro (G15S, 3.02%, 1659 sequences), C11916T for nsp7 (S25L, 0.96%, 546 sequences), C13730T for nsp12 (A97V, 0.67%, 369 sequences), and G24368T for the S protein (D936Y, 0.2%, 94 sequences). Based on “BLOSUM90” scoring, these non-synonymous mutations which cause amino acid changes may significantly affect protein structures, and thus potentially influence the binding affinity of drugs and their target viral proteins. We next assessed the 27 high-ranking mutations, of which 12 occurred in nsp12, 2 in nsp7, 10 in S, and 3 in Mpro. No high-ranking mutation was found in nsp8, even though its protein has a larger sequence (198 aa) than nsp7 (83 aa). This suggests that nsp8 may be more stable than the other four viral genes.
3.2. Identify mutation hotspot patterns in different geographic areas
To analyze the mutation hotspot patterns in different geographic areas, we divided our dataset based on six geographic areas: North America, Oceania, Europe, Asia, Africa, and South America. Some mutations showed clearly different patterns for different geographic areas. The G15598A (nsp12, V720I), G13993T (nsp12, A185S), G15766T (nsp12, V776L), and G14202T (nsp12, E254D) mutations only occurred in Europe, but not in other areas (at the time of our analysis). In addition, G15406T (nsp12, A656T), G12067T (nsp7, M75I), C22227T (S, A222V), A22879C (S, N439K), and five other mutations showed an occurrence percentage of >90% in Europe, while T14222G (nsp12, L261C) and C10319T (Mpro, L89F) occurred in >90% of sequences from North America.
We then normalized mutation occurrence over the total number of mutations from each geographic area (Europe: 58602, North America: 22175, Oceania: 15778, Asia: 4409, Africa: 720, and South America: 795). G15594T (nsp12, K718N) and G22992A (S, S477N) showed much higher occurrence in Oceania, whereas C13730T (nsp12, A97V) was more likely to occur in Asia. No prevalent mutations were found in Africa and South America due to the relatively small numbers of mutations detected.
4. RNA-based therapeutics for infectious diseases
Efforts to develop RNA therapies were initiated in the 1990s. Recent advances in RNA biology have broadened the scope of therapeutic designs and actionable drug targets for human diseases. RNA therapies can be categorized based on their mechanisms of activity: those that target RNAs (e.g., ASOs, siRNAs, and miRNAs), those that bind to proteins (e.g., aptamers), and those that encode proteins (e.g., mRNAs), and catalytically active RNA molecules (e.g. ribozymes).
Therapeutic agents that target RNAs include ASOs, siRNAs, and short hairpin RNAs (shRNAs). ASOs are single-stranded oligonucleotides (13–25 nt long); they can target mRNAs to modulate their splicing, degradation, and translation, or inactivate miRNAs [e.g. miravirsen for hepatitis C virus (HCV) infection] [44], [45], [46], [47]. siRNAs are natural, double-stranded RNA molecules, whereas shRNAs are artificial with a hairpin structure that can give rise to siRNAs; both siRNAs and shRNAs regulate post-transcriptional gene silencing through RNAi (RNA interference) (e.g. patisiran for amyloidosis treatment) [48], [49]. RNAi-based therapies for infectious diseases gained momentum in the early 2000s. In 2002, McCaffrey et al effectively targeted a sequence from HCV via RNAi in mice, demonstrating the potential of RNAi therapy in silencing viruses [50]. In 2003, Song et al suppressed HIV replication in primary macrophages by targeting CCR5, a major HIV-1 coreceptor, and p24, a viral structural gene, via siRNA-mediated RNAi [51]. The most recent rising stars in RNA therapies are miRNAs, which also modulate gene silencing through RNAi. miRNAs may serve as both therapeutic agents and targets [52]. For example, miravirsen, an investigational ASO drug, treats HCV patients by targeting and functionally inhibiting host miR-122 in the liver to reduce HCV RNA levels without causing viral resistance [47].
RNA aptamers are single-stranded RNA oligonucleotides; they can bind to their cognate targets with high specificity and affinity to modulate their functions [53]. While other RNA-based therapeutics target intracellular components, RNA aptamers are able to target both intracellular and extracellular targets [54]. In addition, by binding to cell surface proteins, RNA aptamers serve as a powerful tool for targeted delivery of conjugated therapeutic agents (e.g., siRNAs and small molecules) into specific cell types [55]. RNA aptamers may exert antiviral functions in various mechanisms at different stages of the viral lifecycle [56]. First, aptamers can inhibit virus entry into host cells. For example, RNA aptamer AS1411 targets nucleolin, a cellular protein involved in viral entry, which disrupts the binding of the dengue virus [57]. Second, aptamers can inhibit viral replication. For instance, a 24-mer RNA aptamer targets the Japanese encephalitis virus (JEV) methyltransferase to suppress methylation activity that is required for JEV production in host cells [58]. Another mechanism is through aptamer-mediated delivery in chimera therapies. For example, an aptamer-siRNA chimera specifically targets the envelope glycoprotein GP120 of HIV-1 to inhibit viral replication; the aptamer transfers the siRNA to reach HIV-infected cells and mediate GP120 suppression [59]. RNA aptamers show immense potential in treating infectious diseases considering their broad targeting ability and versatile mechanisms of action.
mRNA-based therapies are a transformative platform for prevention and intervention of infectious diseases. mRNAs from in vitro transcription (IVT) have been developed for protein replacement therapies and vaccines for cancer and infectious diseases since the 1990s [60]. Synthetic mRNAs resemble natural mRNAs and can be delivered in vivo (e.g. direct injection) and ex vivo (e.g. transfection and electroporation of human cells) for protein replacement or supplementation purposes. mRNA vaccines are designed to deliver the sequences of chosen antigens from pathogens for antigen expression in host cells. A proof of concept for RNA-based vaccines emerged in 1993 with the injection of an mRNA from the influenza virus into mice, which induced a virus-specific T cell response [61]. There are two major types of mRNA vaccines: conventional and self-amplifying. Conventional mRNAs encode viral antigens and are immediately translated upon cell entry and endosomal release; the antigen levels rely on successfully delivered conventional transcripts during vaccination [62]. In self-amplifying mRNAs, the sequences of viral antigens and NSPs enable intracellular RNA amplification which results in high levels of antigens [63]. Extensive investigation has been conducted for mRNA vaccines, leading to numerous mRNA vaccines under preclinical and clinical developments. Several excellent reviews have detailed mRNA vaccines for infectious diseases [62], [63], [64], [65].
Ribozymes – RNA enzymes – are another group of RNA agents under exploration for gene therapy of various diseases. For infectious diseases, ribozymes are particularly suitable for inhibiting replication of RNA viruses including HIV-1, HCV, and hepatitis B virus [66], [67], [68], [69], [70], [71], [72]. For examples, hammerhead and hairpin ribozymes have been shown to impede HIV-1 infection in cell culture. The major cleavage sites for ribozymes reside in the 5′ leader, the packaging sequence (Ψ) and the env, gag and tat genes of HIV-1; most of these sites show little evidence of natural variation among HIV-1 strains [73], [74], [75].
RNA therapies show promising potential with their unique advantages but also face great challenges. RNA agents are fast acting compared to DNA therapeutics that need to enter the nucleus for transcription. RNA therapies are reversible due to natural degradation. Also, there is little risk of exogenous RNAs incorporating into the host genome and creating host genome instability. Lastly, RNA manufacturing is rapid, cost-efficient and easily scalable. The two main obstacles that hamper clinical application of RNA therapies are stability and delivery [76]. Unmodified RNAs are notoriously prone to rapid degradation and clearance from circulation, which reduces their bioavailability and half-life. Delivery of RNA agents to the right action sites of target cells with high enough concentration is critical to therapeutic efficacy. Although RNAs may diffuse into or be actively endocytosed by host cells, the cell entry efficiency may be significantly low, partly due to the negative charge of RNAs and cell membranes. Additionally, activation of innate immune nucleic acid sensors and off-target effects remain to be addressed to improve the safety of RNA therapies [77]. Efforts on overcoming these issues will propel the development and qualification of RNA therapeutics.
5. Promising RNA therapeutics targeting the three key proteins
Several promising RNA therapeutics have been completed or are in ongoing clinical trials, of which a few were approved or under Emergency Use Authorization (EAU) by the U.S. FDA. Remdesivir has drawn great attention as it is the first and only small molecule drug approved by the U.S. FDA to specifically treat COVID-19 patients. As an adenosine analog, remdesivir was initially developed to treat infection of another coronavirus, Ebola [78], [79], [80]. Remdesivir binds to the binding pocket of RdRp and terminates RdRp-dependent viral RNA synthesis [81], [82]. In vitro experiments showed that remdesivir was highly effective in inhibiting Ebola viral replication in several human cell lines, and in vivo studies proved that remdesivir significantly diminished Ebola virus in nonhuman primates [19]. Likewise, its effectiveness towards inhibition of SARS-CoV-2 was supported by results from in vitro experiments and preclinical in vivo animal studies [83]. Upon entry into host cells, the prodrug remdesivir is converted to remdesivir triphosphate (TP), an active form of the metabolite, which is a competitive substrate of adenosine triphosphate (ATP) for the viral RdRp. The competition between remdesivir-TP and ATP through RdRp activity causes a chain termination in synthesizing nascent RNA strand, thus inhibiting virus replication [84]. The mechanism of remdesivir in interacting with coronavirus RdRp and its inhibitory effects from both in vivo and in vitro experiments inspired many finished or ongoing clinical trials to test the safety and efficacy of remdesivir in the treatment of COVID-19 patients. For example, a randomized clinical trial with 596 patients reported that the administration of remdesivir could benefit severe COVID-19 patients, compared to patients with placebo [85]. In another randomized, open-label multi-center clinical trial with 394 participants, researchers found that both 5-day and 10-day courses of remdesivir treatment can improve symptoms of severe COVID-19 patients although there was no significant difference between 5-day and 10-day treatments [86]. Importantly, a double-blind, randomized, placebo-controlled trial (ACTT-1) was conducted with 1062 patients, of which 541 patients were treated with remdesivir while 521 patients were assigned a placebo. The ACTT-1 Study Group stated in its final reports that “remdesivir was superior to placebo in shortening the time to recovery in adults who were hospitalized with Covid-19 and had evidence of lower respiratory tract infection” [87]. On October 22, 2020, the U.S. FDA approved remdesivir (brand name Veklury) as the first treatment for hospitalized COVID-19 patients that are 12 years of age or older and weigh >40 kg [88].
RNA-based therapies targeting the S proteins have achieved great accomplishments as well. Moderna Inc.’s mRNA-1273 is an mRNA-based vaccine that encodes the S protein of SARS-CoV-2 and is delivered by lipid nanoparticles (LPN); the nucleoside-modified mRNA stabilizes the S protein in its prefusion conformation, which prevents the virus from entering host cells [89]. Vaccination with mRNA-1273 has shown neutralizing activities in nonhuman primates and human subjects in a Phase I study with no trial-limiting safety concerns [89], [90]. As of Oct 22, 2020, Moderna Inc. has completed enrollment of 30,000 participants for a Phase III clinical trial of mRNA-1273 [91]. Moderna Inc. announced with an early data release on Nov 16, 2020 that the mRNA-1273 vaccine candidate showed an efficacy of 94.5% (p < 0.0001) [92]. Another mRNA-based vaccine candidate currently in Phase III clinical testing is BNT162b2 from Pfizer and BioNTech. BNT162b2 is engineered similarly to mRNA-1273 (i.e. nucleoside modification and LPN encapsulation) and operates under the same mechanism by capturing the S protein for perfusion conformation [93]. BNT162b2 vaccination in mice and rhesus macaques both induced strong T-cell responses and fully protected the lungs of rhesus macaques from SARS-CoV-2 infection challenge. In a press release on Nov 18, 2020, Pfizer reported that BNT162b2 was 95% effective in their completed Phase III study [94]. Additionally, Regeneron Pharmaceuticals Inc. characterized a collection of antibodies against the SARS-CoV-2 S protein using sera from humanized mice and convalescent patients, and selected a pair of antibodies (R10933 and R10987) with high potency to bind the RBD and block viral entry to host cells, resulting in a potential therapeutic for the treatment of COVID-19 [95], [96]. Currently, an ongoing randomized, double-blind clinical trial has shown that Regeneron’s cocktail can decrease viral load and improve symptoms of patients, suggesting that the cocktail could be an effective strategy in the standard-of-care for COVID-19 patients. LY-CoV555 is an S protein antibody developed by Eli Lilly and Company, which exhibited neutralizing property towards SARS-CoV-2 and resulted in the protection of non-human primates from SARS-CoV-2 infection [97]. Furthermore, preliminary data analysis indicated that the combination therapy of LY-CoV555 and LY-CoV016 (namely bamlanivimab and etesevimab) decreased viral load, improved symptoms of COVID-19 patients, and reduced COVID-related hospitalization and emergency room visits, in a randomized, double-blinded clinical trial BLAZE-1 (NCT04427501) (https://investor.lilly.com/news-releases/news-release-details/lilly-provides-comprehensive-update-progress-sars-cov-2). As of Nov 21, 2020, the U.S. FDA has granted Emergency Use Authorizations for both bamlanivimab and casirivimab-imdevimab cocktails to treat COVID-19 patients who are 12 years of age or older, more than 40 kg of weight, and at high risk for progressing to severe COVID-19 [98], [99].
The main proteinase The Mpro plays an important role in viral replication and is another primary target for the development of antiviral therapy against COVID-19 [22]. The Mpro plays an important role in viral replication and is another primary target for the development of antiviral therapy against COVID-19 [22]. The Mpro of SARS-CoV-2 shares high levels of similarity in sequence and structure with other betacoronaviruses; thus, previously developed compounds that target the main proteases of other coronaviruses may be repurposed for combating the current pandemic [22]. Several compounds that inhibited the main proteases of SARS-CoV or MERS-CoV have been shown to co-crystallize with that of SARS-CoV-2 [23], [27], [100]. Recently, boceprevir, GC-376, and calpain inhibitors II and XII were identified to inhibit SARS-CoV-2 viral production by targeting the Mpro with high potency [101]. Specifically, these inhibitors showed EC50 values ranging from 0.49 to 3.37 µM in suppressing viral production in cell culture. In addition, lopinavir and ritonavir are two HIV-1 proteinase inhibitors that are being tested in clinical trials for treatment of COVID-19. In a randomized, controlled, open-label trial with a 199 subjects, lopinavir–Ritonavir treatment showed no benefit beyond standard care to hospitalized adult patients with severe COVID-19 [102]. An open-label, randomized, Phase 2 trial tested a triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin for the treatment of hospitalized adult patients with COVID-19 [103]. Early combination therapy was found to be safe and performed better than lopinavir–ritonavir alone in symptom alleviation and recovery time for patients with mild to moderate COVID-19 [103]. Early intervention and combinatory treatment may have individually or together helped achieve positive results in this latter trial. More investigation is needed to identify effective treatments against this appealing target.
6. The impact of mutations on drug safety and efficacy
Mutations in coronaviruses may reduce drug efficacy in several ways: 1) by decreasing the binding affinity of a drug-target complex; 2) by making the drug target constitutively active; and 3) by turning an antagonist drug to an agonist. The history of anti-HIV treatment has illustrated the impact of viral mutation rates on drug resistance [104]. The HIV-1 virus produces a possible single-nucleotide mutation per patient per day, which has led to resistance to the first approved anti-HIV drug, azidothymidine [105]. Similarly, multiple resistances have been observed for treatment of HCV, another fast-mutating RNA virus [5]. Moreover, resistance to protease and polymerase inhibitors exists in treatment-naïve patients, i.e. without selection that favors the mutations [106]. Combining multiple drugs, instead of increasing monotherapy potency, has proven to be a successful antiviral regimen that minimizes resistant mutation appearance and improves effectiveness [104].
The rapid infection of SARS-CoV-2 worldwide has drawn immense attention to its mutations and strains. Analysis of 220 SARS-CoV-2 genomic sequences derived from patients worldwide revealed different mutations in RdRp that emerged at different times (December 2019 – mid-Mach 2020) and locations, suggesting that SARS-CoV-2 is evolving and that multiple strains with their specific mutation patterns may coexist [35]. Among all newly identified mutations is P14408L, which resides near a hydrophobic cleft of RdRp and is associated with an overall increased mutation rate [35]. Previous studies have shown that naturally emerging mutations in RdRp may lead to drug resistance or reduce treatment efficacy due to weakened drug-RdRp binding, particularly near the mutation sites [35], [107], [108], [109]. In a recently published case report, a point mutation in RdRp (D484Y) was associated with the failure of remdesivir treatment in a 76-year-old female COVID-19 patient with post-rituximab B-cell immunodeficiency [110]. It is suspected that resistant mutations emerge under remdesivir treatment pressure which leads to treatment failure [110]. Therefore, it is crucial to identify resistant mutations and evaluate their impact on the conformation and activity of RdRp and its susceptibility to antiviral treatment. Meanwhile, new antiviral drugs for different targets along the viral lifecycle and cocktail therapies are planned for further testing to improve COVID-19 treatment.
Host-directed therapy design may resolve drug resistance stemming from fast-emerging viral mutations and strains. A recent study established an interaction map that consisted of 332 protein–protein interactions between SARS-CoV-2 and humans using affinity-purification mass spectrometry [111]. Through these interactions, the authors identified 66 host proteins or factors that were targeted by 69 approved or investigational compounds. The findings may facilitate drug repurposing for host-directed intervention towards mutation-associated drug resistance and pan-viral therapies as new treatment regimens.
7. Perspective
Advances in next-generation sequencing technology has made it possible to obtain a large amount of SARS-CoV-2 genome sequence data. Up to the end of November 2020, the GISAID database (https://www.gisaid.org) collected viral genome sequence data from approximately 224,000 isolates. On the other hand, studies in protein structural identification, biochemical characterization, and pharmacological investigation provide valuable information to understand how variants may influence drug efficacy/toxicity, and influence patients’ prognosis. Based on this information, an online interactive database, COVID-3D (http://biosig.unimelb.edu.au/covid3d/), was developed to monitor viral mutations; to visualize conformational changes of proteins in mutants; and to evaluate impacts of mutants on viral infectivity, host immunity, and drug-protein interaction [30].
To evaluate the effects of viral mutations on drug safety and efficacy, we propose to monitor and predict mutation-introduced protein conformational changes in three key viral proteins, and then evaluate their binding affinities and enzymatic activities with U.S. FDA regulated drugs, using computational modeling and machine learning processes. Finally, an interactive database will be provided and updated to evaluate viral mutation associated changes in drug safety and efficacy for U.S. FDA regulated drugs. As shown in Fig. 3 , the database SARS-CoV-2 Variome Structure-Substrate Database is proposed as 1) to accrue and monitor all mutations for three key viral proteins (the S protein, RdRp and Mpro) in the SARS-CoV-2 viral genome weekly, using sequencing data from the GISAID database and the nextstrain database (https://nextstrain.org/sars-cov-2/); 2) to predict protein conformational changes due to mutations, and to evaluate changes of enzyme activities or antigen-antibody affinities of mutants for three key viral proteins, using computational modeling and machine learning approaches with publicly available 3D structures for the three proteins; and 3) to develop a SARS-CoV-2 Variome Structure-Substrate Database and machine learning tools for evaluating potential effects of mutants on U.S. FDA regulated chemical and antibody drugs. The database contains mutants and their conformational changes, and predicted enzyme activities or antigen-antibody affinities. This is an interactive database that can be used by healthcare providers, drug developers, and drug reviewers to evaluate the potential effects of mutants on U.S. FDA regulated chemical and antibody drugs using machine learning tools. We believe that the SARS-CoV-2 Variome Structure-Substrate Database will provide strategic information concerning functional differences in mutant viral proteins and their potential effects on U.S. FDA regulated antibody and chemical drugs.
Fig. 3.
Scheme of the SARS-CoV-2 Variome Structure-Substrate Database. This is an interactive database. The database will collect and update all mutations for three key viral proteins in the SARS-CoV-2 genome. Users can explore and predict protein conformational changes of mutants, and to visualize changes in enzyme activities or antigen–antibody affinities of mutants for three key viral proteins, using computational modeling and machine learning approaches. Finally, users can evaluate potential effects of mutants on U.S. FDA regulated antibody and chemical drugs.
When this manuscript was under the initial editorial assessment, a novel phylogenetic cluster of SARS-CoV-2 mutations, known as the lineage B.1.1.7, was identified in the United Kingdom (https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563). This set of spike mutations has been escalating rapidly over the past several weeks and has been further spreading to dozens of countries, including Australia, Brazil, China, Germany, India and the United States. The B.1.1.7 lineage contains multiple genetic mutations in the S protein, of which several mutations are likely to exhibit biological effects. Notably, the mutation N501Y occurred at key interacting amino acids within the RBD, which increases the binding affinity between mutated virus and human ACE2 molecule; the deletion of 6 bases at the viral genome results in amino acid deletion at the positions 69 and 70 (69-70del) of the S protein, which is also associated with RBD conformational changes, thus making the virus more transmissible; the mutation P681H is closely adjacent to the furin cleavage site, which may affect the viral entry to human host cells. The biological significance of these mutations suggests that enhanced genomic and epidemiological surveillances are urgently needed for the control of current pandemic worldwide. More importantly, further laboratorial and clinical studies of antigenicity, infectivity, vaccine and treatment strategies to mitigate impacts of the mutants are warranted.
8. Disclaimer
The views presented in this article do not necessarily reflect those of the U.S. Food and Drug Administration. Any mention of commercial products is for clarification and is not intended as an endorsement.
CRediT authorship contribution statement
Conceptualization: Tong, Shi, Ning. Data Curation: Zeng, Li. Writing: Zeng, Li, Ning. Editing: TS, WT.
References
- 1.Zhang Y.Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181(2):223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djomkam A.L.Z., Olwal C.O., Sala T.B., Paemka L. Commentary: SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Front. Oncol. 2020;10:1448. doi: 10.3389/fonc.2020.01448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280 e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P., Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363(9413):938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Y., Yu D., Yan H., Chong H., He Y. Design of potent membrane fusion inhibitors against SARS-CoV-2, an emerging coronavirus with high fusogenic activity. J. Virol. 2020;94(14) doi: 10.1128/JVI.00635-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musarrat F., Chouljenko V., Dahal A., Nabi R., Chouljenko T., Jois S.D., Kousoulas K.G. The anti-HIV drug nelfinavir mesylate (Viracept) is a potent inhibitor of cell fusion caused by the SARSCoV-2 spike (S) glycoprotein warranting further evaluation as an antiviral against COVID-19 infections. J. Med. Virol. 2020 doi: 10.1002/jmv.25985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584(7819):154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 15.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.te Velthuis A.J., van den Worm S.H., Snijder E.J. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40(4):1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10(1):2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elena S.F., Sanjuan R. Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J. Virol. 2005;79(18):11555–11558. doi: 10.1128/JVI.79.18.11555-11558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu W., Chen C.Z., Gorshkov K., Xu M., Lo D.C., Zheng W. RNA-dependent RNA polymerase as a target for COVID-19 drug discovery. SLAS Discov. 2020;25(10):1141–1151. doi: 10.1177/2472555220942123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parvez M.S.A., Karim M.A., Hasan M., Jaman J., Karim Z., Tahsin T., Hasan M.N., Hosen M.J. Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach. Int. J. Biol. Macromol. 2020;163:1787–1797. doi: 10.1016/j.ijbiomac.2020.09.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agrawal U., Raju R., Udwadia Z.F. Favipiravir: a new and emerging antiviral option in COVID-19. Med. J. Armed Forces India. 2020;76(4):370–376. doi: 10.1016/j.mjafi.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 2020;30(17) doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368(6497):1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rathnayake A.D., Zheng J., Kim Y., Perera K.D., Mackin S., Meyerholz D.K., Kashipathy M.M., Battaile K.P., Lovell S., Perlman S., Groutas W.C., Chang K.O. 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice. Sci. Transl. Med. 2020;12(557) doi: 10.1126/scitranslmed.abc5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ton A.T., Gentile F., Hsing M., Ban F., Cherkasov A. rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Mol. Inform. 2020;39(8) doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh E., Khan R.J., Jha R.K., Amera G.M., Jain M., Singh R.P., Muthukumaran J., Singh A.K. A comprehensive review on promising anti-viral therapeutic candidates identified against main protease from SARS-CoV-2 through various computational methods. J. Genet. Eng. Biotechnol. 2020;18(1):69. doi: 10.1186/s43141-020-00085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 28.Coelho C., Gallo G., Campos C.B., Hardy L., Wurtele M. Biochemical screening for SARS-CoV-2 main protease inhibitors. PLoS ONE. 2020;15(10) doi: 10.1371/journal.pone.0240079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkataraman S., Prasad B., Selvarajan R. RNA Dependent RNA Polymerases: Insights from Structure, Function and Evolution. Viruses. 2018;10(2) doi: 10.3390/v10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Portelli S., Olshansky M., Rodrigues C.H.M., D'Souza E.N., Myung Y., Silk M., Alavi A., Pires D.E.V., Ascher D.B. Exploring the structural distribution of genetic variation in SARS-CoV-2 with the COVID-3D online resource. Nat. Genet. 2020;52(10):999–1001. doi: 10.1038/s41588-020-0693-3. [DOI] [PubMed] [Google Scholar]
- 31.van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., Owen C.J., Pang J., Tan C.C.S., Boshier F.A.T., Ortiz A.T., Balloux F. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect Genet. Evol. 2020;83 doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J., Wang R., Wang M., Wei G.W. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020;432(19):5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Groves D.C., Rowland-Jones S.L., Angyal A. The D614G mutations in the SARS-CoV-2 spike protein: implications for viral infectivity, disease severity and vaccine design. Biochem. Biophys. Res. Commun. 2020 doi: 10.1016/j.bbrc.2020.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Zhang L., Li X., Huang W., Wang Y. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284–1294 e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C., Zella D., Ippodrino R. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18(1):179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon A., Le N.T., Selisko B., Eydoux C., Alvarez K., Guillemot J.C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Badua C., Baldo K.A.T., Medina P.M.B. Genomic and proteomic mutation landscapes of SARS-CoV-2. J. Med. Virol. 2020 doi: 10.1002/jmv.26548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen S., Hu T., Zhang J., Chen J., Chen K., Ding J., Jiang H., Shen X. Mutation of Gly-11 on the dimer interface results in the complete crystallographic dimer dissociation of severe acute respiratory syndrome coronavirus 3C-like protease: crystal structure with molecular dynamics simulations. J. Biol. Chem. 2008;283(1):554–564. doi: 10.1074/jbc.M705240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheik Amamuddy O., Verkhivker G.M., Tastan Bishop O. Impact of early pandemic stage mutations on molecular dynamics of SARS-CoV-2 M(pro) J. Chem. Inf. Model. 2020;60(10):5080–5102. doi: 10.1021/acs.jcim.0c00634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.L. Zhang, C.B. Jackson, H. Mou, A. Ojha, E.S. Rangarajan, T. Izard, M. Farzan, H. Choe, The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity, bioRxiv (2020).
- 42.B. Korber, W.M. Fischer, S. Gnanakaran, H. Yoon, J. Theiler, W. Abfalterer, N. Hengartner, E.E. Giorgi, T. Bhattacharya, B. Foley, K.M. Hastie, M.D. Parker, D.G. Partridge, C.M. Evans, T.M. Freeman, T.I. de Silva, C.-G.G. Sheffield, C. McDanal, L.G. Perez, H. Tang, A. Moon-Walker, S.P. Whelan, C.C. LaBranche, E.O. Saphire, D.C. Montefiori, Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus, Cell 182(4) (2020) 812-827 e19. [DOI] [PMC free article] [PubMed]
- 43.Henikoff S., Henikoff J.G. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. U.S.A. 1992;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dias N., Stein C.A. Antisense oligonucleotides: basic concepts and mechanisms. Mol. Cancer Ther. 2002;1(5):347–355. [PubMed] [Google Scholar]
- 45.Benson M.D., Waddington-Cruz M., Berk J.L., Polydefkis M., Dyck P.J., Wang A.K., Plante-Bordeneuve V., Barroso F.A., Merlini G., Obici L., Scheinberg M., Brannagan T.H., 3rd, Litchy W.J., Whelan C., Drachman B.M., Adams D., Heitner S.B., Conceicao I., Schmidt H.H., Vita G., Campistol J.M., Gamez J., Gorevic P.D., Gane E., Shah A.M., Solomon S.D., Monia B.P., Hughes S.G., Kwoh T.J., McEvoy B.W., Jung S.W., Baker B.F., Ackermann E.J., Gertz M.A., Coelho T. Inotersen treatment for patients with hereditary transthyretin amyloidosis. N. Engl. J. Med. 2018;379(1):22–31. doi: 10.1056/NEJMoa1716793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roehr B. Fomivirsen approved for CMV retinitis. J. Int. Assoc. Physicians AIDS Care. 1998;4(10):14–16. [PubMed] [Google Scholar]
- 47.Janssen H.L., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y., Persson R., King B.D., Kauppinen S., Levin A.A., Hodges M.R. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368(18):1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 48.Kristen A.V., Ajroud-Driss S., Conceicao I., Gorevic P., Kyriakides T., Obici L. Patisiran, an RNAi therapeutic for the treatment of hereditary transthyretin-mediated amyloidosis. Neurodegener Dis. Manage. 2019;9(1):5–23. doi: 10.2217/nmt-2018-0033. [DOI] [PubMed] [Google Scholar]
- 49.Rao D.D., Vorhies J.S., Senzer N., Nemunaitis J. siRNA vs. shRNA: similarities and differences. Adv. Drug Deliv. Rev. 2009;61(9):746–759. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 50.McCaffrey A.P., Meuse L., Pham T.T., Conklin D.S., Hannon G.J., Kay M.A. RNA interference in adult mice. Nature. 2002;418(6893):38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 51.Song E., Lee S.K., Dykxhoorn D.M., Novina C., Zhang D., Crawford K., Cerny J., Sharp P.A., Lieberman J., Manjunath N., Shankar P. Sustained small interfering RNA-mediated human immunodeficiency virus type 1 inhibition in primary macrophages. J. Virol. 2003;77(13):7174–7181. doi: 10.1128/JVI.77.13.7174-7181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drury R.E., O'Connor D., Pollard A.J. The clinical application of MicroRNAs in infectious disease. Front. Immunol. 2017;8:1182. doi: 10.3389/fimmu.2017.01182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou J., Bobbin M.L., Burnett J.C., Rossi J.J. Current progress of RNA aptamer-based therapeutics. Front. Genet. 2012;3:234. doi: 10.3389/fgene.2012.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Germer K., Leonard M., Zhang X. RNA aptamers and their therapeutic and diagnostic applications. Int. J. Biochem. Mol. Biol. 2013;4(1):27–40. [PMC free article] [PubMed] [Google Scholar]
- 55.Chu T.C., Twu K.Y., Ellington A.D., Levy M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006;34(10) doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zou X., Wu J., Gu J., Shen L., Mao L. Application of aptamers in virus detection and antiviral therapy. Front. Microbiol. 2019;10:1462. doi: 10.3389/fmicb.2019.01462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Balinsky C.A., Schmeisser H., Ganesan S., Singh K., Pierson T.C., Zoon K.C. Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J. Virol. 2013;87(24):13094–13106. doi: 10.1128/JVI.00704-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han S.R., Lee S.W. Inhibition of Japanese encephalitis virus (JEV) replication by specific RNA aptamer against JEV methyltransferase. Biochem. Biophys. Res. Commun. 2017;483(1):687–693. doi: 10.1016/j.bbrc.2016.12.081. [DOI] [PubMed] [Google Scholar]
- 59.Zhou J., Li H., Li S., Zaia J., Rossi J.J. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol. Ther. 2008;16(8):1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sahin U., Kariko K., Tureci O. mRNA-based therapeutics–developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13(10):759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 61.Martinon F., Krishnan S., Lenzen G., Magne R., Gomard E., Guillet J.G., Levy J.P., Meulien P. Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur. J. Immunol. 1993;23(7):1719–1722. doi: 10.1002/eji.1830230749. [DOI] [PubMed] [Google Scholar]
- 62.Maruggi G., Zhang C., Li J., Ulmer J.B., Yu D. mRNA as a transformative technology for vaccine development to control infectious diseases. Mol. Ther. 2019;27(4):757–772. doi: 10.1016/j.ymthe.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bloom K., van den Berg F., Arbuthnot P. Self-amplifying RNA vaccines for infectious diseases. Gene Ther. 2020 doi: 10.1038/s41434-020-00204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang C., Maruggi G., Shan H., Li J. Advances in mRNA vaccines for infectious diseases. Front. Immunol. 2019;10:594. doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines – a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17(4):261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ojwang J.O., Hampel A., Looney D.J., Wong-Staal F., Rappaport J. Inhibition of human immunodeficiency virus type 1 expression by a hairpin ribozyme. Proc. Natl. Acad. Sci. U.S.A. 1992;89(22):10802–10806. doi: 10.1073/pnas.89.22.10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li M., Li H., Rossi J.J. RNAi in combination with a ribozyme and TAR decoy for treatment of HIV infection in hematopoietic cell gene therapy. Ann. N. Y. Acad. Sci. 2006;1082:172–179. doi: 10.1196/annals.1348.006. [DOI] [PubMed] [Google Scholar]
- 68.Feng Y., Leavitt M., Tritz R., Duarte E., Kang D., Mamounas M., Gilles P., Wong-Staal F., Kennedy S., Merson J., Yu M., Barber J.R. Inhibition of CCR5-dependent HIV-1 infection by hairpin ribozyme gene therapy against CC-chemokine receptor 5. Virology. 2000;276(2):271–278. doi: 10.1006/viro.2000.0536. [DOI] [PubMed] [Google Scholar]
- 69.Rossi J.J. Ribozyme therapy for HIV infection. Adv. Drug Deliv. Rev. 2000;44(1):71–78. doi: 10.1016/s0169-409x(00)00085-5. [DOI] [PubMed] [Google Scholar]
- 70.Wong-Staal F. Development of ribozyme gene therapy for HIV infection. Antibiot. Chemother. 1971;48(1996):226–232. doi: 10.1159/000425181. [DOI] [PubMed] [Google Scholar]
- 71.Lieber A., He C.Y., Polyak S.J., Gretch D.R., Barr D., Kay M.A. Elimination of hepatitis C virus RNA in infected human hepatocytes by adenovirus-mediated expression of ribozymes. J. Virol. 1996;70(12):8782–8791. doi: 10.1128/jvi.70.12.8782-8791.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.J. zu Putlitz, Q. Yu, J.M. Burke, J.R. Wands, Combinatorial screening and intracellular antiviral activity of hairpin ribozymes directed against hepatitis B virus, J Virol 73(7) (1999) 5381-7. [DOI] [PMC free article] [PubMed]
- 73.Li X., Gervaix A., Kang D., Law P., Spector S.A., Ho A.D., Wong-Staal F. Gene therapy targeting cord blood-derived CD34+ cells from HIV-exposed infants: preclinical studies. Gene Ther. 1998;5(2):233–239. doi: 10.1038/sj.gt.3300582. [DOI] [PubMed] [Google Scholar]
- 74.Sun L.Q., Pyati J., Smythe J., Wang L., Macpherson J., Gerlach W., Symonds G. Resistance to human immunodeficiency virus type 1 infection conferred by transduction of human peripheral blood lymphocytes with ribozyme, antisense, or polymeric trans-activation response element constructs. Proc. Natl. Acad. Sci. U.S.A. 1995;92(16):7272–7276. doi: 10.1073/pnas.92.16.7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lewin A.S., Hauswirth W.W. Ribozyme gene therapy: applications for molecular medicine. Trends Mol. Med. 2001;7(5):221–228. doi: 10.1016/s1471-4914(01)01965-7. [DOI] [PubMed] [Google Scholar]
- 76.Dammes N., Peer D. Paving the road for RNA therapeutics. Trends Pharmacol. Sci. 2020;41(10):755–775. doi: 10.1016/j.tips.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lieberman J. Tapping the RNA world for therapeutics. Nat. Struct. Mol. Biol. 2018;25(5):357–364. doi: 10.1038/s41594-018-0054-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chang W.T., Liu P.Y., Gao Z.H., Lee S.W., Lee W.K., Wu S.N. Evidence for the Effectiveness of Remdesivir (GS-5734), a Nucleoside-Analog Antiviral Drug in the Inhibition of I K(M) or I K(DR) and in the Stimulation of I MEP. Front. Pharmacol. 2020;11:1091. doi: 10.3389/fphar.2020.01091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lo M.K., Jordan R., Arvey A., Sudhamsu J., Shrivastava-Ranjan P., Hotard A.L., Flint M., McMullan L.K., Siegel D., Clarke M.O., Mackman R.L., Hui H.C., Perron M., Ray A.S., Cihlar T., Nichol S.T., Spiropoulou C.F. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017;7:43395. doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tchesnokov E.P., Feng J.Y., Porter D.P., Gotte M. Mechanism of inhibition of ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11(4) doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Al-Tawfiq J.A., Al-Homoud A.H., Memish Z.A. Remdesivir as a possible therapeutic option for the COVID-19. Travel Med. Infect. Dis. 2020;34 doi: 10.1016/j.tmaid.2020.101615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang J., Song W., Huang H., Sun Q. Pharmacological therapeutics targeting RNA-dependent RNA polymerase, proteinase and spike protein: from mechanistic studies to clinical trials for COVID-19. J. Clin. Med. 2020;9(4) doi: 10.3390/jcm9041131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eastman R.T., Roth J.S., Brimacombe K.R., Simeonov A., Shen M., Patnaik S., Hall M.D. Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent. Sci. 2020;6(5):672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Gotte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295(15):4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Spinner C.D., Gottlieb R.L., Criner G.J., Arribas Lopez J.R., Cattelan A.M., Soriano Viladomiu A., Ogbuagu O., Malhotra P., Mullane K.M., Castagna A., Chai L.Y.A., Roestenberg M., Tsang O.T.Y., Bernasconi E., Le Turnier P., Chang S.C., SenGupta D., Hyland R.H., Osinusi A.O., Cao H., Blair C., Wang H., Gaggar A., Brainard D.M., McPhail M.J., Bhagani S., Ahn M.Y., Sanyal A.J., Huhn G., Marty F.M., Investigators G.-U-. Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19: a randomized clinical trial. JAMA. 2020;324(11):1048–1057. doi: 10.1001/jama.2020.16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goldman J.D., Lye D.C.B., Hui D.S., Marks K.M., Bruno R., Montejano R., Spinner C.D., Galli M., Ahn M.Y., Nahass R.G., Chen Y.S., SenGupta D., Hyland R.H., Osinusi A.O., Cao H., Blair C., Wei X., Gaggar A., Brainard D.M., Towner W.J., Munoz J., Mullane K.M., Marty F.M., Tashima K.T., Diaz G., Subramanian A., Investigators G.-U-. Remdesivir for 5 or 10 days in patients with severe Covid-19. N. Engl. J. Med. 2020;383(19):1827–1837. doi: 10.1056/NEJMoa2015301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.D., Ruiz-Palacios G.M., Benfield T., Fatkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C., Members A.-S.G. Remdesivir for the Treatment of Covid-19 - Final Report. N. Engl. J. Med. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.US Food and Drug Administration, FDA Approves First Treatment for COVID-19, 2020.
- 89.L.A. Jackson, E.J. Anderson, N.G. Rouphael, P.C. Roberts, M. Makhene, R.N. Coler, M.P. McCullough, J.D. Chappell, M.R. Denison, L.J. Stevens, A.J. Pruijssers, A. McDermott, B. Flach, N.A. Doria-Rose, K.S. Corbett, K.M. Morabito, S. O'Dell, S.D. Schmidt, P.A. Swanson, 2nd, M. Padilla, J.R. Mascola, K.M. Neuzil, H. Bennett, W. Sun, E. Peters, M. Makowski, J. Albert, K. Cross, W. Buchanan, R. Pikaart-Tautges, J.E. Ledgerwood, B.S. Graham, J.H. Beigel, R.N.A.S.G. m, An mRNA Vaccine against SARS-CoV-2 - Preliminary Report, N Engl J Med 383(20) (2020) 1920-1931. [DOI] [PMC free article] [PubMed]
- 90.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O'Connell S., Bock K.W., Minai M., Nagata B.M., Andersen H., Martinez D.R., Noe A.T., Douek N., Donaldson M.M., Nji N.N., Alvarado G.S., Edwards D.K., Flebbe D.R., Lamb E., Doria-Rose N.A., Lin B.C., Louder M.K., O'Dell S., Schmidt S.D., Phung E., Chang L.A., Yap C., Todd J.M., Pessaint L., Van Ry A., Browne S., Greenhouse J., Putman-Taylor T., Strasbaugh A., Campbell T.A., Cook A., Dodson A., Steingrebe K., Shi W., Zhang Y., Abiona O.M., Wang L., Pegu A., Yang E.S., Leung K., Zhou T., Teng I.T., Widge A., Gordon I., Novik L., Gillespie R.A., Loomis R.J., Moliva J.I., Stewart-Jones G., Himansu S., Kong W.P., Nason M.C., Morabito K.M., Ruckwardt T.J., Ledgerwood J.E., Gaudinski M.R., Kwong P.D., Mascola J.R., Carfi A., Lewis M.G., Baric R.S., McDermott A., Moore I.N., Sullivan N.J., Roederer M., Seder R.A., Graham B.S. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020;383(16):1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moderna Inc, Moderna Completes Enrollment of Phase 3 COVE Study of mRNA Vaccine Against COVID-19 (mRNA-1273), 2020.
- 92.Moderna Inc, Moderna’s COVID-19 Vaccine Candidate Meets its Primary Efficacy Endpoint in the First Interim Analysis of the Phase 3 COVE Study, 2020.
- 93.Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., Kranz L.M., Walzer K.C., Hein S., Güler A., Loschko J., Maddur M.S., Tompkins K., Cole J., Lui B.G., Ziegenhals T., Plaschke A., Eisel D., Dany S.C., Fesser S., Erbar S., Bates F., Schneider D., Jesionek B., Sänger B., Wallisch A.-K., Feuchter Y., Junginger H., Krumm S.A., Heinen A.P., Adams-Quack P., Schlereth J., Kröner C., Hall-Ursone S., Brasky K., Griffor M.C., Han S., Lees J.A., Mashalidis E.H., Sahasrabudhe P.V., Tan C.Y., Pavliakova D., Singh G., Fontes-Garfias C., Pride M., Scully I.L., Ciolino T., Obregon J., Gazi M., Carrion R., Alfson K.J., Kalina W.V., Kaushal D., Shi P.-Y., Klamp T., Rosenbaum C., Kuhn A.N., Türeci Ö., Dormitzer P.R., Jansen K.U., Sahin U. A prefusion SARS-CoV-2 spike RNA vaccine is highly immunogenic and prevents lung infection in non-human primates. bioRxiv. 2020;2020(09) [Google Scholar]
- 94.Pfizer, pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-announce-vaccine-candidate-against, (2020).
- 95.Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., Wei Y., Atwal G.S., Murphy A.J., Stahl N., Yancopoulos G.D., Kyratsous C.A. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369(6506):1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., Chung K.M., Hermann A., Ullman E., Cruz J., Rafique A., Huang T., Fairhurst J., Libertiny C., Malbec M., Lee W.Y., Welsh R., Farr G., Pennington S., Deshpande D., Cheng J., Watty A., Bouffard P., Babb R., Levenkova N., Chen C., Zhang B., Romero Hernandez A., Saotome K., Zhou Y., Franklin M., Sivapalasingam S., Lye D.C., Weston S., Logue J., Haupt R., Frieman M., Chen G., Olson W., Murphy A.J., Stahl N., Yancopoulos G.D., Kyratsous C.A. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369(6506):1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.B.E. Jones, P.L. Brown-Augsburger, K.S. Corbett, K. Westendorf, J. Davies, T.P. Cujec, C.M. Wiethoff, J.L. Blackbourne, B.A. Heinz, D. Foster, R.E. Higgs, D. Balasubramaniam, L. Wang, R. Bidshahri, L. Kraft, Y. Hwang, S. Zentelis, K.R. Jepson, R. Goya, M.A. Smith, D.W. Collins, S.J. Hinshaw, S.A. Tycho, D. Pellacani, P. Xiang, K. Muthuraman, S. Sobhanifar, M.H. Piper, F.J. Triana, J. Hendle, A. Pustilnik, A.C. Adams, S.J. Berens, R.S. Baric, D.R. Martinez, R.W. Cross, T.W. Geisbert, V. Borisevich, O. Abiona, H.M. Belli, M. de Vries, A. Mohamed, M. Dittmann, M. Samanovic, M.J. Mulligan, J.A. Goldsmith, C.L. Hsieh, N.V. Johnson, D. Wrapp, J.S. McLellan, B.C. Barnhart, B.S. Graham, J.R. Mascola, C.L. Hansen, E. Falconer, LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection, bioRxiv (2020).
- 98.US Food and Drug Administration, Coronavirus (COVID-19) Update: FDA Authorizes Monoclonal Antibody for Treatment of COVID-19, (2020).
- 99.US Food and Drug Administration, Coronavirus (COVID-19) Update: FDA Authorizes Monoclonal Antibodies for Treatment of COVID-19, (2020).
- 100.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30(8):678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hung I.F., Lung K.C., Tso E.Y., Liu R., Chung T.W., Chu M.Y., Ng Y.Y., Lo J., Chan J., Tam A.R., Shum H.P., Chan V., Wu A.K., Sin K.M., Leung W.S., Law W.L., Lung D.C., Sin S., Yeung P., Yip C.C., Zhang R.R., Fung A.Y., Yan E.Y., Leung K.H., Ip J.D., Chu A.W., Chan W.M., Ng A.C., Lee R., Fung K., Yeung A., Wu T.C., Chan J.W., Yan W.W., Chan W.M., Chan J.F., Lie A.K., Tsang O.T., Cheng V.C., Que T.L., Lau C.S., Chan K.H., To K.K., Yuen K.Y. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395(10238):1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sanjuan R., Domingo-Calap P. Mechanisms of viral mutation. Cell. Mol. Life Sci. 2016;73(23):4433–4448. doi: 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Perelson A.S. Modelling viral and immune system dynamics. Nat. Rev. Immunol. 2002;2(1):28–36. doi: 10.1038/nri700. [DOI] [PubMed] [Google Scholar]
- 106.Cao Y., Bao Y., Xia W., Wu H., Wei F., Zhang Y., Zhang R., Xu X. Resistance-associated mutations to HCV protease inhibitors naturally pre-existed in HIV/HCV coinfected, treatment-naive patients. Clin Res Hepatol Gastroenterol. 2016;40(5):597–604. doi: 10.1016/j.clinre.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 107.Goldhill D.H., Te Velthuis A.J.W., Fletcher R.A., Langat P., Zambon M., Lackenby A., Barclay W.S. The mechanism of resistance to favipiravir in influenza. Proc. Natl. Acad. Sci. U.S.A. 2018;115(45):11613–11618. doi: 10.1073/pnas.1811345115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., Ray A.S., Cihlar T., Siegel D., Mackman R.L., Clarke M.O., Baric R.S., Denison M.R. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio. 2018;9(2) doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Young K.C., Lindsay K.L., Lee K.J., Liu W.C., He J.W., Milstein S.L., Lai M.M. Identification of a ribavirin-resistant NS5B mutation of hepatitis C virus during ribavirin monotherapy. Hepatology. 2003;38(4):869–878. doi: 10.1053/jhep.2003.50445. [DOI] [PubMed] [Google Scholar]
- 110.Martinot M., Jary A., Fafi-Kremer S., Leducq V., Delagreverie H., Garnier M., Pacanowski J., Mekinian A., Pirenne F., Tiberghien P., Calvez V., Humbrecht C., Marcelin A.G., Lacombe K. Remdesivir failure with SARS-CoV-2 RNA-dependent RNA-polymerase mutation in a B-cell immunodeficient patient with protracted Covid-19. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., Garcia-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]