Abstract
Without approved vaccines and specific treatments, COVID-19 is spreading around the world with above 26 million cases and approximately 864 thousand deaths until now. An efficacious and affordable vaccine is urgently needed. The Val308 – Gly548 of spike protein of SARS-CoV-2 linked with Gln830 – Glu843 of Tetanus toxoid (TT peptide) (designated as S1-4) and without TT peptide (designated as S1-5) were expressed and renatured. The antigenicity and immunogenicity of S1-4 were evaluated by Western Blotting (WB) in vitro and immune responses in mice, respectively. The protective efficiency was measured preliminarily by microneutralization assay (MN50). The soluble S1-4 and S1-5 protein was prepared to high homogeneity and purity. Adjuvanted with Alum, S1-4 protein stimulated a strong antibody response in immunized mice and caused a major Th2-type cellular immunity supplemented with Th1-type immunity. Furthermore, the immunized sera could protect the Vero E6 cells from SARS-CoV-2 infection with neutralizing antibody titer 256. Recombinant SARS-CoV-2 RBD with a built in T helper epitope could stimulate both strong humoral immunity supplemented with cellular immunity in mice, demonstrating that it could be a promising subunit vaccine candidate.
Keywords: SARS-CoV-2, RBD, TT peptide, Subunit vaccine
1. Introduction
Coronavirus disease (COVID-19) was claimed a pandemic by World Health Organization (WHO) and was initially reported in China [1]. Until 12 August 2020, 26,121,999 COVID-19 cases were reported with 864,618 deaths globally [2].
COVID-19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [3], [4]. Coronaviruses named after their morphology as spherical virions with a core shell and surface projections resembling a solar corona, are enveloped, positive single-stranded large RNA viruses that infect humans, but also a wide range of animals [5]. SARS-CoV-2 closely related to the SARS-CoV virus [6], belongs to the B lineage of the beta-coronaviruses [7]. There are more than 100 candidate vaccines in development worldwide, while among the vaccine technologies under evaluation are inactivated virus vaccines [8], recombinant protein subunit vaccines [9], [10], and nucleic acid vaccines [11], [12]. Based on previous experience with SARS-CoV vaccines, SARS-CoV-2 vaccines should refrain from immunopotentiation (i.e. antibody-dependent enhancement) that could lead to increased infectivity or eosinophilic infiltration [11]. Therefore, subunit vaccine might be one of the best candidate vaccines for its simpler in composition and less uncontrollable risk factors [13]. There are four major structural proteins including the nucleocapsid protein (N), the spike protein (S), a small membrane protein (SM) and the membrane glycoprotein (M) with an additional membrane glycoprotein (HE) in the HCoV-OC43 and HKU1 beta-coronaviruses [14]. Like other coronaviruses, SARS-CoV-2 infects lung alveolar epithelial cells by receptor-mediated endocytosis with angiotensin-converting enzyme II (ACE2) as the cell receptor [6], [8]. SARS-CoV-2 binds ACE2 with the receptor binding domain (RBD) of S protein [15], [16]. Full-length S (S1 and S2) or S1 which contains RBD could induce neutralization antibodies that prevent viral infection [17]. RBD is structurally and functionally different from the other S1 parts and highly stable and compact [13]. Numerous researchers have demonstrated that RBD and has the potential quality as a subunit vaccine [13], [18], [19], [20], containing abundant T cell and B cell epitopes including neutralization epitopes. Subunit vaccines possessed high safety profile, consistent production and could only induce humoral immune response, more importantly need appropriate adjuvants to induce high level neutralization antibody and giving effective protection [17], [21].
Tetanus toxoid (TT) as a T-helper-epitope-rich antigen could enhance effectively humoral and cellular immune responses of antigenic epitopes [22]. TT exerts the requisite immunological enhancement and promotes production of high levels of antibodies by achieving effective T-B cell reaction [22]. TT has been considered because it is safe and also has been commercially applied in making human vaccines [23]. Since generally most people have an immune response to TT, the resulting antibodies could function as immune modulation in the immune response to new antigens through antibody-antigen complex form [24]. Gln830 – Glu843 of TT (TT peptide) as a promiscuous T-helper epitope has a strong binding capacity to DR3 allele [25], [26] and is frequently used as T cell stimulator to enhance the immunogenicity of exogenous epitopes and induce cellular immunity for subunit vaccines [25], [27], [28]. TT peptide has only 14 amino acids, was easily expressed in the form of recombinant protein, and minimized the interference with epitope recognition and/or binding theoretically.
Without specific prevention measures, current effective management mainly focus on the enforcement of quarantine, isolation and physical distancing (i.e. negative pressure isolation) [9], [29], [30], [31]. An efficacious and affordable vaccine is urgently needed. In this study, we prepared a novel molecule of RBD linked TT peptide (designated as S1-4) and evaluated prelimarily its antigenicity, immunogenicity and protective efficiency as potential subunit vaccine.
2. Materials and methods
2.1. Preparation of SARS-CoV-2 subunit vaccine
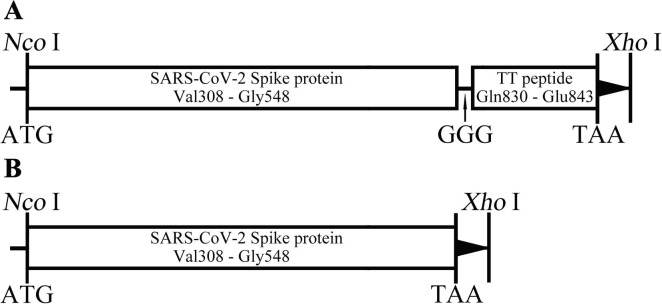
The SARS-CoV subunit antigen, designated as S1-4, consisted of the truncated spike protein Val308 – Gly548 (GenBank No. YP009724390) and TT peptide Gln830 – Glu843 with GGG as the linker (Fig. 1 A), while S1-5 only contained Val308 – Gly548 of spike protein (Fig. 1B). The genes codon-optimized encoding S1-4 and S1-5 protein were artificially synthesized (General Biol, Chuzhou, China) and sub-cloned into pET28a (+) vector using restriction endonucleases (NcoI and XhoI). The expected Molecular Weight (MW) of S1-4 and S1-5 protein were 29.0 kDa and 27.4 kDa, respectively. The positive plasmids were confirmed by restriction endonuclease analysis and sequencing, while the positive colonies of freshly transformed BL21(DE3) were chosen depending on the SDS-PAGE analysis of small-scale expression.
Fig. 1.
Schematic diagram of the gene of S1-4 (A) and S1-5 (B) in the pET28a vector.
Small-scale expression cultures inoculated with BL21(DE3) carrying S1-4 plasmids were induced by IPTG (1 mmol/L) at 37 °C for 2 h. A large-scale preparation was performed in 4-L LB medium induced by IPTG (1 mmol/L) at 32 °C for 4 h. The cell harvested by centrifugation (4000g, 10 min, 4 °C) were re-suspended with Buffer I (10 mmol/L Tris-HCl, 1 mmol/L EDTA, 0.1% Triton X-100, pH8.0) followed by sonic disruption (300 W, 20 s, 20 s, and 30 times). The pellets retained by centrifugation (17,300g, 20 min, 4 °C) were washed twice with Buffer I. After the pellets were dissolved with Buffer II (10 mmol/L Tris-HCl, 8 mol/L urea, pH8.0), S1-4 protein in the denatured solution was purified by DEAE ion-exchange chromatography (IEX) then followed by Octyl Sepharose 4 Fast Flow hydrophobic interaction chromatography (HIC). After being appropriately diluted in Buffer III (8 mol/L Urea, 0.3 mol/L Arginine, 1 mmol/L EDTA, 1 mmol/L Reduced glutathione, 10% Glycerol, pH8.0), S1-4 protein was renatured by gradually reducing the urea content with Buffer IV (15 mmol/L Na2CO3, 35 mmol/L NaHCO3, 1 mmol/L EDTA, 10% Glycerol) at 4 °C for at least 6 h each cycle. After refolding, S1-4/S1-5 protein solution was centrifuged to remove the precipitate and sterile filtered by 0.2 μm filters.
Protein analysis was conducted by SDS polyacrylamide gel electrophoresis (SDS-PAGE), Size Distribution analysis (Zetasizer Nano ZS90, Malvern, Worcestershire, UK) and Western Blotting (WB) using convalescent serum of COVID-19 patients, rabbit monoclonal antibody (MAb) against RBD, Rat polyclonal antibody (PAb) against SARS-CoV-2 and related HRP –conjugated secondary antibodies. Molecular weight (MW) and aggregation state of S1-4 protein complex in solution was detected according to Guerra’s method [32]. Briefly, size-exclusion chromatography (SEC) was performed equipped with Superdex 75 HR 10/300 GL column. SEC calibration curves were constructed based on linear regression analysis of peak elution volume (Ve) versus the logarithm of the MW of reference proteins (human gamma-globulin, 158 kDa; ovalbumin, 44 kDa; horse myoglobin, 17 kDa; B12 vitamin, 1.35 kDa). In parallel, the Ve of purified S1-4 protein was interpolated in the calibration plot to calculate MW and the aggregation state. Approximate tertiary structures of homo-trimer S1-4 (PDB: 6zgf.1.A) by homology modeling were constructed with SWISS-MODEL [32].
After being concentrated with protein concentrators (Pierce Biotechnology, Rockford. USA), S1-4 protein was mixed with aluminum hydroxide (Alum) at a ratio of 2:1 (v/v) for adsorption, and the antigen concentration in the final adsorption product was adjusted to 800 μg/mL. The mixture (S1-4/Alum) is designated as SARS-CoV-2 subunit vaccine.
The prokaryotic expression, chromatography purification and renaturation of S1-5 protein were performed according to those of S1-4 protein. Similarly, S1-5 protein was adjuvanted with Alum as a control (S1-5/Alum, 800 μg/mL).
2.2. Evaluation of humoral immunity of SARS-CoV-2 subunit vaccine in mice
All animal procedures were reviewed and approved by the Animal Care and Welfare Committee at the National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention (No. 20200201003). The neutralization tests were performed in a BSL-3 laboratory. Ten specific pathogen-free, female 4–6 weeks old BALB/c mice (Vital River Laboratories, Beijing, China) each group were immunized intramuscularly with 50 μL of S1-4/Alum (40 μg antigen/mg Alum), S1-5/Alum (40 μg antigen/mg Alum) or Alum alone (1 mg) respectively, with two booster shots on Day 7 and 14. Orbital venous blood was sampled at regular intervals (Day 0, 14, 28 and 35) and sera was collected by centrifugation (1000g, 10 min, 4 °C) after coagulation at room temperature for 2 h.
Enzyme linked immunosorbent assay (ELISA) was applied to evaluate the antibody response according to the previous study [33]. Anti-SARS-CoV-2 antibody titers were determined by an endpoint dilution ELISA on micro-well plates coated with inactivated SARS-CoV-2 (30 ng/well). Briefly, sera serially diluted in phosphate-buffered saline (PBS) were added into micro-well plates (100 μL/well) and incubated at 37 °C for 1 h. Goat anti-mouse IgG antibody (HRP conjugate) (Sigma-Aldrich, SL, USA) was used as the detection antibody (100 μL/well). Color reaction was developed with 3, 3′, 5, 5′-tetramethylbenzidine (TMB) and terminated with H2SO4 solution. The absorbance was read at 450 nm with an ELISA reader (Thermo Fisher, Mannheim, Finland). Antibody titers were determined as the highest dilution at which the mean absorbance of the sample was 2.1-fold greater than that of the control serum.
The neutralization antibody titers were detected by a traditional virus microneutralization assay (MN50) using SARS-CoV-2 (CN1 strain) [34], [35]. The assay was performed in quadruplicate, and a series of eight two-fold serial dilutions of the serum were assessed. Briefly, 100 tissue culture infective dose 50 (TCID50) units of SARS-CoV-2 were added to two-fold serial dilutions of heat-inactivated serum (60 °C, 30 min), and incubated for 1 h at 37 °C. The virus-serum complexes were added to Vero E6 cells monolayers (3 × 105/well) grown in an 96-well plates and incubated for 72 h. Record the results by visual inspection of the cytopathogenic effect (CPE) using an inverted microscope. The neutralizing titers (MN50) were calculated as the highest serum dilution at which half of four wells had intact cells and no CPE [36].
2.3. Investigation of immune response type by IFN-γ/IL-4 ELISpot and antibody isotypes assay
Mouse IFN-γ and IL-4 enzyme linked immunospot assay (ELISpot) assay were conducted according to the manufacturer’s guidelines (DaKeWe, Shenzhen, China). Briefly, splenocytes (3 × 105 per well, in triplicate) of immunized mice at Day 35 were plated into ELISpot plate and stimulated with PHA (Positive control) or S1-4/S1-5 protein. After incubation for 16 h in a 37 °C humidified incubator with 5% CO2, the cells were removed before adding the detection antibody (biotin conjugate). Two hours later at room temperature (RT), Streptavidin-HRP was added after washing plate. One hour later at RT, TMB substrate was applied until distinct spots emerged. Count spots in an ELISpot reader after stopping color development with deionized water.
In mice, IgG1 versus IgG2a reflected the differential Th2-Th1 reactivity [37]. Therefore, IgG1 and IgG2a responses SARS-CoV-2 were measured for sera obtained at Day35 by ELISA. Briefly, serial dilutions of mouse sera on Day 35 were incubated in the plates coated with inactivated SARS-CoV-2 (30 ng/well). Following washes, anti-mouse IgG1 and IgG2a-HRP conjugates (ThermoFisher, MA, USA) were applied as secondary antibody with TMB as the substrate. Antibody titers were defined as the dilution at which the mean absorbance of the sample was 2.1-fold greater than that of the control serum.
2.4. Statistical analysis
Antibody titers were log-transformed to establish geometric mean titers (GMT) for analysis. The number of specific IFN-γ- or IL-4-secreting T cells was expressed as spots forming cells (SFCs) per 3 × 105 cells. The related data were statistically analyzed by Mann-Whitney U test for non-parametric data and the paired t test for parametric data. SPSS software (22.0, Chicago, IL, USA) and GraphPad Prism software (8.00, San Diego, CA, USA) was applied to analyze the data and draw graphics, respectively. Significance was indicated when value of P < 0.05.
3. Results
3.1. S1-4 protein was prepared to high homogeneity and purity by prokaryotic expression and chromatography purification
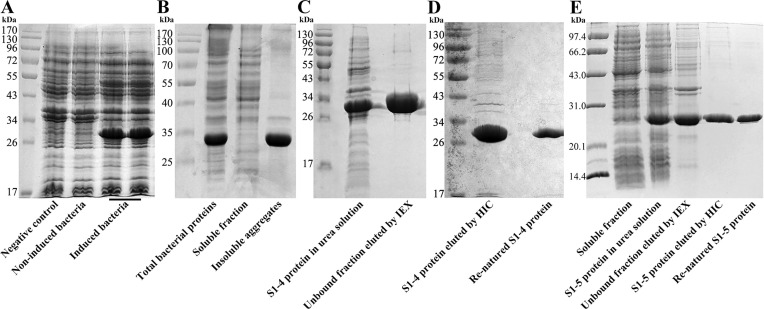
Upon induction, in small-expression, the expected S1-4 protein of 29.0 kDa was observed (Fig. 2 A) suggesting that S1-4 protein could be appropriately expressed in E. coli. Through large-scale expression (4 L LB medium), S1-4 protein accounted for approximately 26.38% in the total cell lysates, and increased up to 70.13% in the total proteins from the inclusion bodies (IBs) (Fig. 2B). Therefore, S1-4 protein expressed and existed mainly in the IBs form in E. coli. S1-4 protein treated with 8 mol/L urea (pH 8.0) and then purified by IEX existed mainly in the unbound fraction (Fig. 2C). The purity of S1-4 protein can reach 96.91% at 2.64 mg/mL through HIC (Fig. 2D). Renaturated by stepwise dialysis, the concentration and purity of S1-4 protein were 0.84 mg/mL and 98.71%, respectively (Fig. 2D). Correspondingly, the concentration and purity of S1-5 protein were 0.81 mg/mL and 99.10% respectively after the same procedure as S1-4 protein (Fig. 2E).
Fig. 2.
The preparation of S1-4 and S1-5 protein. (A). Small-scale expression of S1-4 protein. (B). Large-scale expression of S1-4 protein. (C). S1-4 protein purified by IEX. (D). S1-4 protein purified by HIC and renaturation. (E). S1-5 protein purified by IEX and HIC.
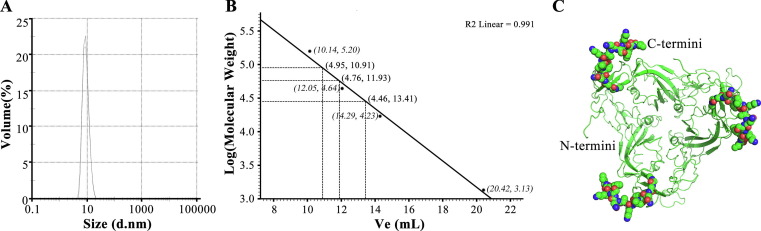
According to size distribution report by volume, the diameter of S1-4 protein was about 9.133 nm (Fig. 3 A). A calibration curve of log MW versus Ve was generated (log MW = −0.195 × Ve + 7.077, R2 = 0.991, P < 0.001; Fig. 3B). Three kinds of MW (28,986 Da, 57,544 Da and 89,146 Da) were found, which suggested the presence of monomeric (29.0 kDa), dimeric (59.0 kDa) and trimeric (87.0 kDa) forms of S1-4 protein. According to the predictive structure of homo-trimer S1-4 protein (Fig. 3C) and area under the peak curve on SEC, we speculated that most of S1-4 protein in solution has a trimeric structure with a diameter 9.133 nm and a MW 89.1 kDa.
Fig. 3.
Size distribution by volume (A), calibration curve for the standard protein and S1-4 protein on SEC column (B) and the predictive trimeric topology of S1-4 protein (C).
After diluted with normal saline, S1-4 and S1-5 protein were mixed respectively with aluminum adjuvant and adjusted antigen concentration in the final adsorption product was 800 μg/mL.
3.2. S1-4 protein possessed antigenicity in vitro
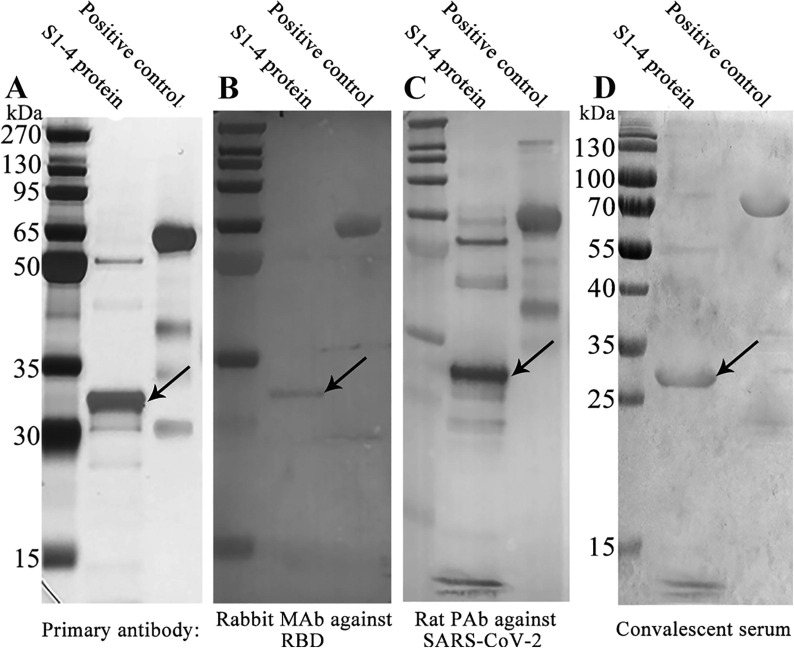
The antigenicity of S1-4 protein was demonstrated by WB analysis with a commercial recombinant RBD protein (Val16 - Arg685 of spike protein; 76.45 kDa) expressed in baculovirus-insect cells by Sino Biological (Beijing, China) as positive control (Fig. 4 ). Ten micrograms of S1-4 protein and commercial recombinant RBD protein were loaded onto the first three gels (Fig. 4A – 4C), with the exception of 30 µg of the two proteins in the fourth gel (Fig. 4D).
Fig. 4.
Western Blotting analysis of S1-4 protein.
Whether it was Rabbit monoclonal antibody against RBD (Fig. 4B), Rat polyclonal antibody against inactivated SARS-CoV-2 (Fig. 4C), or convalescent serum of COVID-19 patients (Fig. 4D) as the primary antibody, there was an obvious band at about 30 kDa, suggesting the high antigenicity of S1-4 protein. It was noteworthy that there was also an obvious band at about 59 kDa, which was probably a dimer (Fig. 4). There was no band at about 87 kDa, which was the MW of a trimer.
3.3. SARS-CoV-2 subunit vaccine displayed strong immunogenicity in mice and immunized sera could protect the Vero E6 cells from SARS-CoV-2 infection
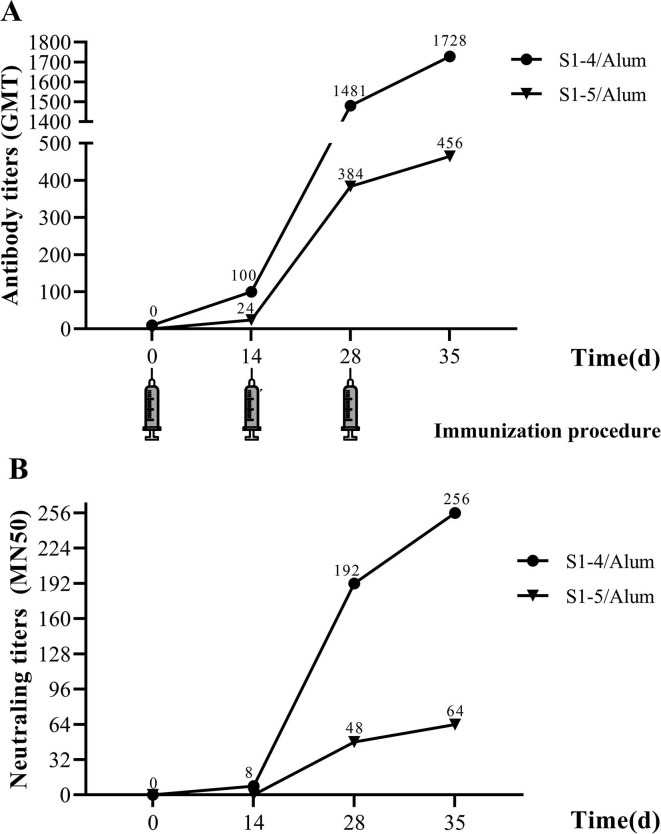
To analyze the immunogenicity of SARS-CoV-2 candidate subunit vaccine, we determined the anti-SARS-CoV-2 antibody titers in sera of immunized mice by ELISA based on inactivated SARS-CoV-2 (Fig. 5 A). The antibody titer (GMT) in mice immunized with S1-4/Alum came to 100 one week after the first injection, while it climbed to 1481 one week after the second injection. However, the antibody titer (GMT) in mice immunized with S1-5/Alum was only 24 after the first injection and 192 after the second injection. Finally, one week after the third injection, the antibody titer (GMT) in mice immunized with S1-4/Alum was 1728, while that of S1-5/Alum was 456. Obviously, S1-4 linked with TT peptide tail were more immunogenic than S1-5 without TT peptide. Similarly, the antibody titer increased rapidly after the second immunization between S1-4 and S1-5 protein. Interestingly, at the same sampling time, the antibody titer of mice immunized with S1-4/Alum was almost four times that of mice immunized with S1-5/Alum, suggesting the strong immune modulation of TT peptide.
Fig. 5.
Kinetics of serum anti-SARS-CoV-2 antibody in mice by ELISA (A) and serum neutralizing antibody in mice by microneutralization assay (B).
The virus neutralizing activity of SARS-CoV-2 subunit vaccine in immunized mice was examined using live SARS-CoV-2 (CN1 stain) operating within Biosafety Level 3 (BSL-3) Laboratory (Fig. 5B). The neutralizing antibody titer (MN50) were 192 on Day28 (one week after second immunization) and reached 256 on Day35 (one week after third immunization) in the sera of mice immunized with S1-4/Alum. However, the neutralizing antibody titers (MN50) of sera of mice immunized with S1-5/Alum were only 48 and 64 on Day28 and Day35, respectively. Furthermore, neutralizing antibody titers increased along with total antibody titers, whether in S1-4 or S1-5 protein immunized mice (P < 0.05). The anti-SARS-CoV-2 antibody titer at the same sampling time was approximately seven times higher than that of neutralizing antibody (P < 0.05). Those results indicated that TT peptide enhanced the neutralizing antibody level by stimulating the produce of more total antibody.
3.4. SARS-CoV-2 subunit vaccine caused a major Th2-type immunity supplemented with Th1-type immunity
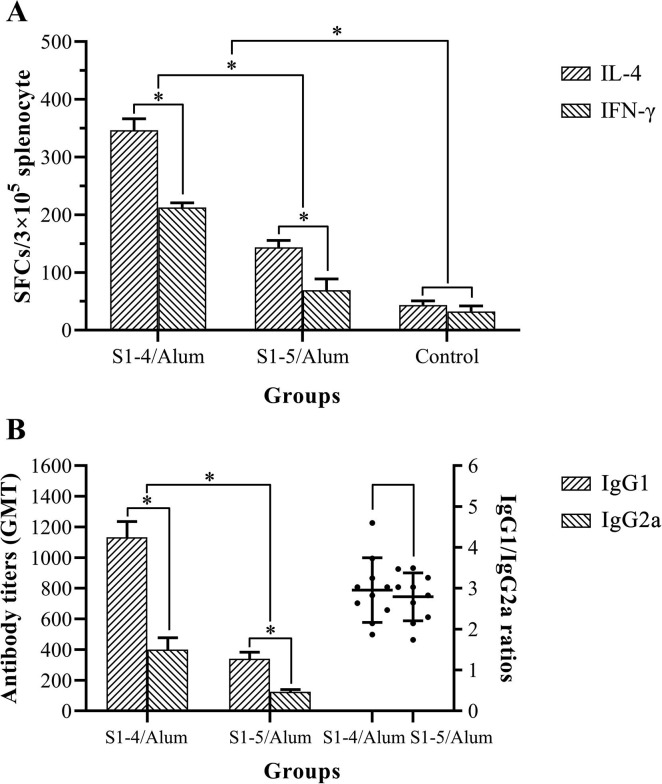
ELISpot assay was applied to enumerate Spot-forming cells (SFCs) for IL-4 and IFN-γ in splenocytes of immunized Balb/c mice (Fig. 6 A). The SFCs for IL-4 in S1-4/Alum were 346.7 ± 19.6 (Mean ± SD), while SFCs for IFN-γ were 212.7 ± 7.8. The SFCs for IL-4 in S1-5/Alum were 143.4 ± 11.7, while SFCs for IFN-γ were 69.3 ± 19.4. Whether in the S1-4/Alum or S1-5/Alum group, the SFCs for IL-4 were more than those for IFN-γ, suggesting a major Th2-type immunity in mice. Furthermore, a considerable number of IFN-γ SFCs existed in the two groups. These results indicated that not only Th2 (IL-4) but also Th1 (IFN-γ) SFCs were involved in the immune response against S1-4 and S1-5 protein. Remarkably, both IL-4 and IFN-γ SFCs were more in S1-4/Alum group than those in S1-5/Alum group (P < 0.05). The immune response type caused by S1-4 or S1-5 protein mainly based on Th2-type immunity supplemented with Th1-type immunity.
Fig. 6.
IL-4 & IFN-γ spot-forming cells (SFCs) in splenocytes of immunized mice (A) and antibody isotype analysis of immunized serum (B).
In Balb/c mice, IgG1 versus IgG2a also reflected the differential Th2-Th1 immunity, while the IgG1/IgG2a ratio was an indirect reflection of immune function in vivo [37]. Therefore, the antibody titers (GMT) of IgG1 and IgG2a against SARS-CoV-2 and IgG1/IgG2a ratios were measured by ELISA (Fig. 6B). The IgG1 titer in S1-4/Alum group was 1132.6 ± 103.4 (Mean ± SD), while IgG2a titer was 400.5 ± 77.4. Obviously, IgG1 titers were more than IgG2a titers in S1-4/Alum group (P < 0.05). IgG1 titer in S1-5/Alum group was 340.5 ± 42.8, while IgG2a titer was 124.4 ± 14.9. Obviously, IgG1 titers were also more than IgG2a titers in S1-5/Alum group (P < 0.05). The IgG1/IgG2a ratio in S1-4/Alum group was 2.95 ± 0.75, while that in S1-5/Alum was 2.79 ± 0.56. There was no significant difference in the IgG1/IgG2a ratio between the two groups (P > 0.05, Fig. 6B). Therefore, IgG1 response was stronger than IgG2a response, suggesting a major Th2-type immunity in mice.
4. Discussion
The independence of structure and function made RBD the ideal choice of SARS-CoV-2 subunit vaccine [21]. Several expression systems such as CHO-, baculovirus-, and yeast had been used to express recombinant RBD protein [16], [38], [39], [40]. Considering its efficiency, time-cost saving and the safety, nothing else could match the E. coli expression system, especially in the circumstance that SARS-CoV-2 vaccine is urgently needed worldwide. Xia had developed successfully the Hepatitis E virus (HEV) vaccine [41] and Human Papillomavirus (HPV) vaccine [42] by E. coli expression system. Furthermore, both of the vaccines had been licenced by National Medical Products Administration of China. By the approach of prokaryotic expression system, the annual productivity could reach billions of vaccine dosages if the candidate vaccine be approved, which would make it possible to prevent and eliminate COVID-19 on a global scale.
However, three key questions, glycosylation and renaturation, bear the brunt, if prokaryotic expression system is applied. First, there are only two predictive N-linked (Asn331 and Asn343) and O-linked (Thr323 and Thr385) glycosylation sites within Val308 – Gly548[43]. Practically, Val308 – Gly548 expressed in baculovirus expression system possessed 3N-glycosylation sites and 10O-glycosylation sites [39]. However, all the sites were distant from the bound ACE2[39]. It was presumed that glycosylations at those sites of RBD were not crucial for maintance of antigenicity and immunogenicity [13]. In this study, S1-4 protein without glycosylation was antigenic (Fig. 4) and immunogenic (Fig. 5A) enough to stimulate protective immunity (Fig. 5B), although it appeared that S1-4 protein reacted no better with the polyclonal sera than the commercial RBD (Val16 - Arg685 of spike protein) expressed in baculovirus expression system (Fig. 4). Surprisingly, we found that anti-S1-4 protein antibody titers were much higher than anti-SARS-CoV-2 antibody titers (P < 0.05). Secondly, the existence of four disulfide bonds (Cys336-Cys361, Cys379-Cys432, Cys480-Cys488, Cys391-Cys525) brings great difficulties to the reconstruction of natural structure of RBD [13]. Detailed dialysate buffer composition had been published in this study and its effectiveness had also been confirmed by our previous work of recombinant SARS-CoV RBD renaturation [44]. Finally, three kinds of aggregation states, monomeric (29.0 kDa), dimeric (59.0 kDa) and trimeric (87.0 kDa) forms of S1-4 protein, were demonstrated by SEC chromatography (Fig. 3B). Moreover, momomeric and dimeric forms was also found in the WB gels (Fig. 4). However, trimeric structure of S1-4 protein with a diameter 9.133 nm and a MW 89.1 kDa was the most convincing according to the predictive structure and area under the peak curve on SEC. The further study on the structure of S1-4 protein would be performed.
The Th1 cells produce cytokines that promote cell-mediated immune responses such as IFN-γ whilst the Th2 cells induce cytokines such as IL-4, IL-5 and IL-13, which will activate humoral immune responses [45]. Current evidences strongly indicated that Th1 type response is a key for successful control of SARS-CoV and MERS-CoV and probably is true for SARS-CoV-2 as well [17]. Stimulation of IgG2a (Th1-type) has been associated with increased efficacy of vaccination in mice [46]. On the whole, both humoral (Th1-type) and cellular immunity (Th2-type) were necessary to control SARS-CoV-2 infection. According to the ELISpot assay, S1-4/Alum could stimulate Th1- and Th2- type immune responses simultaneously in mice (Fig. 4B), although the latter was dominant. Anti-SARS-CoV-2 antibody isotype analysis also demonstrated that IgG1 response was dominant supplemented with IgG2a response, with an IgG1/IgG2a ratio 2.95. Furthermore, TT peptide played an important role in promoting cytokine secretion including Th1 and Th2-type cytokines (Fig. 4B). The presence of TT peptide in the construct as a strong T cell epitope ensured the T-B cell reaction in the process of antigen presentation. Despite the similar IL-4/IFN-γ ratio caused by both S1-4 and S1-5, the absolute values were different. S1-5 possessed the same T cell epitopes except TT peptide. The effect of TT peptide in immune response enhancement was obvious, especially Th2-type immunity (Fig. 6A). Almost all people have a pre-existing immunity to TT peptide, which would be helpful for immune modulation [24]. The immune efficacy of TT peptide, as a built in T helper epitope of S1-4 protein, would be evaluated in ongoing clinical trials.
Protective efficacy is crucial for a vaccine. Humoral immune response, especially the neutralization antibody response, plays an important role by limiting infection at later phase and prevents reinfection in the future [17]. Neutralization activity of immune serum is often the key to judge whether a candidate vaccine will be successful or not. Wu, et al. found that Ad5-nCoV expressing SARS-CoV-2 full-length spike protein could induce strong humoral immune responses in mice by intramuscular route with high titer of anti-recombinant SARS-CoV-2 S protein (Sino Biological, Beijing, China) antibody and neutralization antibody [47]. In this study, S1-4 protein could also induce high titer of anti-SARS-CoV-2 antibody and neutralization antobody in mice (Fig. 5). One week after the third immunization, anti-SARS-CoV-2 antibody titer detected by ELISA reached 1728 (Fig. 4A) while neutralizing antibody titer came to 256 measured by MN50 (Fig. 5). In contrast, S1-5 protein without TT peptide was barely satisfactory. Neutralizing activity elicited by SARS-CoV-2 mRNA vaccine developed by Corbett reached 819 reciprocal IC50 GMT in mice based on pseudo-virus neutralization assay [12]. Neutralization activity elicited by Ad5-nCoV vaccine reached about 28 (256) in mice based on SARS-CoV-2 neutralization assay [47]. In this study, the immune serum could protect Vero E6 cells from SARS-CoV-2 infection with neutralizing antibody titer 256 on Day35 (Fig. 4A).
5. Conclusion
Recombinant SARS-CoV-2 RBD with a built in T helper epitope could be prepared by prokaryotic system and chromatography purification with an ideal antigenicity and immunogenicity. Th2-type immunity was dominant supplemented with Th1-type immunity. The immune serum could protect Vero E6 cells from SARS-CoV-2 infection. This would make this subunit protein ideal vaccine candidate.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work is supported by a project funded by Denogen (Beijing) Biotechnology Co., Ltd. We thank Mrs Jian-rong Cao especially for the funding. The funder has no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Madabhavi I, Sarkar M, Kadakol N. COVID-19: a review. Monaldi archives for chest disease = Archivio Monaldi per le malattie del torace 2020;90(2). http://doi.org/10.4081/monaldi.2020.1298. [DOI] [PubMed]
- 2.World Health Organization. Weekly operational update on Coronavirus disease (COVID-19), https://www.who.int/docs/default-source/coronaviruse/situation-reports/wou-4-september-2020-approved.pdf?sfvrsn=91215c78_2 [accessed 4 September 2020].
- 3.Zhu N., Zhang D., Wang W., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. New England J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan W.J., Zhao X., Ma X.J., et al. A novel coronavirus genome identified in a cluster of pneumonia cases-Wuhan, China 2019–2020. China CDC Weekly. 2020;2(4):61–62. doi: 10.46234/ccdcw2020.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Velavan T.P., Meyer C.G. The COVID-19 epidemic. Tropical Med Int Health: TM & IH. 2020;25(3):278–280. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou P., Yang X.L., Wang X.G., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu R., Zhao X., Li J., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet (London, England) 2020;395(10224):565–574. doi: 10.1016/s0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H., Zhang Y., Huang B., et al. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. Cell. 2020;182(3):713–721.e719. doi: 10.1016/j.cell.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu F.C., Li Y.H., Guan X.H., et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet (London, England) 2020;395(10240):1845–1854. doi: 10.1016/s0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalita P., Padhi A.K., Zhang K.Y.J., et al. Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microb Pathog. 2020;145104236 doi: 10.1016/j.micpath.2020.104236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W.H., Strych U., Hotez P.J., et al. The SARS-CoV-2 Vaccine Pipeline: an Overview. Current Tropical Med Reports. 2020:1–4. doi: 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corbett K.S., Edwards D.K., Leist S.R., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020 doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su Q.D., Yi Y., Zou Y.N., et al. The biological characteristics of SARS-CoV-2 spike protein Pro330-Leu650. Vaccine. 2020;38(32):5071–5075. doi: 10.1016/j.vaccine.2020.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludwig S., Zarbock A. Coronaviruses and SARS-CoV-2: A Brief Overview. Anesth Analg. 2020;131(1):93–96. doi: 10.1213/ane.0000000000004845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–280.e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang J., Ye G., Shi K., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38(1):1–9. doi: 10.12932/ap-200220-0772. [DOI] [PubMed] [Google Scholar]
- 18.Sarkar B., Ullah M.A., Johora F.T., et al. Immunoinformatics-guided designing of epitope-based subunit vaccines against the SARS Coronavirus-2 (SARS-CoV-2) Immunobiology. 2020;225(3) doi: 10.1016/j.imbio.2020.151955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang D., Mai J., Zhou W., et al. Immunoinformatic Analysis of T- and B-Cell Epitopes for SARS-CoV-2 Vaccine Design. Vaccines. 2020;8(3) doi: 10.3390/vaccines8030355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noorimotlagh Z., Karami C., Mirzaee S.A., et al. Immune and bioinformatics identification of T cell and B cell epitopes in the protein structure of SARS-CoV-2: A systematic review. Int Immunopharmacol. 2020;86106738 doi: 10.1016/j.intimp.2020.106738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai L., Zheng T., Xu K., et al. A Universal Design of Betacoronavirus Vaccines against COVID-19, MERS, and SARS. Cell. 2020;182(3):722–733.e711. doi: 10.1016/j.cell.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He Q., Gao H., Gao M., et al. Immunogenicity and safety of a novel tetanus toxoid-conjugated anti-gastrin vaccine in BALB/c mice. Vaccine. 2018;36(6):847–852. doi: 10.1016/j.vaccine.2017.12.054. [DOI] [PubMed] [Google Scholar]
- 23.Lise L.D., Mazier D., Jolivet M., et al. Enhanced epitopic response to a synthetic human malarial peptide by preimmunization with tetanus toxoid carrier. Infect Immun. 1987;55(11):2658–2661. doi: 10.1128/iai.55.11.2658-2661.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mangsbo S.M., Fletcher E.A.K., van Maren W.W.C., et al. Linking T cell epitopes to a common linear B cell epitope: A targeting and adjuvant strategy to improve T cell responses. Mol Immunol. 2018:93115–93124. doi: 10.1016/j.molimm.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Moriyama H., Wen L., Abiru N., et al. Induction and acceleration of insulitis/diabetes in mice with a viral mimic (polyinosinic-polycytidylic acid) and an insulin self-peptide. PNAS. 2002;99(8):5539–5544. doi: 10.1073/pnas.082120099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai H., Chen M.S., Sun Z.Y., et al. Self-adjuvanting synthetic antitumor vaccines from MUC1 glycopeptides conjugated to T-cell epitopes from tetanus toxoid. Angew Chem Int Ed Engl. 2013;52(23):6106–6110. doi: 10.1002/anie.201300390. [DOI] [PubMed] [Google Scholar]
- 27.Abu-Zeid Y.A., Alwash R., Shaheen H.M., et al. Seroprevalence of antibodies to repetitive domains of Plasmodium vivax circumsporozoite protein in United Arab Emirates children. Trans R Soc Trop Med Hyg. 2002;96(5):560–564. doi: 10.1016/s0035-9203(02)90443-8. [DOI] [PubMed] [Google Scholar]
- 28.Boitel B, Blank U, Mège D, et al. Strong similarities in antigen fine specificity among DRB1* 1302-restricted tetanus toxin tt830-843-specific TCRs in spite of highly heterogeneous CDR3. J Immunol (Baltimore, Md: 1950) 1995;154(7):3245–55. [PubMed]
- 29.Li Z., Chen Q., Feng L., et al. Active case finding with case management: the key to tackling the COVID-19 pandemic. Lancet (London, England) 2020;396(10243):63–70. doi: 10.1016/s0140-6736(20)31278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou L., Wu Z., Li Z., et al. 100 Days of COVID-19 Prevention and Control in China. Clin Infect Dis: Off Publ Infect Dis Soc Am. 2020 doi: 10.1093/cid/ciaa725. [DOI] [Google Scholar]
- 31.Zhu F.C., Guan X.H., Li Y.H., et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet (London, England) 2020;396(10249):479–488. doi: 10.1016/s0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guerra L.L., Faccinetti N.I., Trabucchi A., et al. Novel prokaryotic expression of thioredoxin-fused insulinoma associated protein tyrosine phosphatase 2 (IA-2), its characterization and immunodiagnostic application. BMC Biotech. 2016;16(1):84. doi: 10.1186/s12896-016-0309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su Q.D., He S.H., Yi Y., et al. Intranasal vaccination with ebola virus GP amino acids 258–601 protects mice against lethal challenge. Vaccine. 2018;36(41):6053–6060. doi: 10.1016/j.vaccine.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Algaissi A, Hashem AM. Evaluation of MERS-CoV Neutralizing Antibodies in Sera Using Live Virus Microneutralization Assay. Methods Mol Biol (Clifton, NJ) 2020; 2099107-116. http://doi.org/10.1007/978-1-0716-0211-9_9. [DOI] [PMC free article] [PubMed]
- 35.Gao Q., Bao L., Mao H., et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science (New York, NY) 2020;369(6499):77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He Y., Zhou Y., Liu S., et al. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem Biophys Res Commun. 2004;324(2):773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maassen C.B., Boersma W.J., van Holten-Neelen C., et al. Growth phase of orally administered Lactobacillus strains differentially affects IgG1/IgG2a ratio for soluble antigens: implications for vaccine development. Vaccine. 2003;21(21–22):2751–2757. doi: 10.1016/s0264-410x(03)00220-2. [DOI] [PubMed] [Google Scholar]
- 38.Chen WH, Tao X, Agrawal A, et al. Yeast-Expressed SARS-CoV Recombinant Receptor-Binding Domain (RBD219-N1) Formulated with Alum Induces Protective Immunity and Reduces Immune Enhancement. bioRxiv: the preprint server for biology; 2020. http://doi.org/10.1101/2020.05.15.098079. [DOI] [PMC free article] [PubMed]
- 39.Yang J., Wang W., Chen Z., et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020 doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 40.Yuan M., Wu N.C., Zhu X., et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science (New York, NY) 2020;368(6491):630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J., Zhang X.F., Huang S.J., et al. Long-term efficacy of a hepatitis E vaccine. New England J Med. 2015;372(10):914–922. doi: 10.1056/NEJMoa1406011. [DOI] [PubMed] [Google Scholar]
- 42.Hu Y.M., Huang S.J., Chu K., et al. Safety of an Escherichia coli-expressed bivalent human papillomavirus (types 16 and 18) L1 virus-like particle vaccine: an open-label phase I clinical trial. Human Vacc Immunotherap. 2014;10(2):469–475. doi: 10.4161/hv.26846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vankadari N., Wilce J.A. Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerging Microbes Infect. 2020;9(1):601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi Y., Duan S.M., Zhang M.C., et al. Cloning and expression of SARS coronavirus spike gene fragment and its application. Chinese J Virol. 2003;19(3):267–268. [Google Scholar]
- 45.Mosmann T.R., Coffman R.L. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989:7145–7173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 46.Qi M., Zhang X.E., Sun X., et al. Intranasal Nanovaccine Confers Homo- and Hetero-Subtypic Influenza Protection. Small. 2018;14(13) doi: 10.1002/smll.201703207. [DOI] [PubMed] [Google Scholar]
- 47.Wu S., Zhong G., Zhang J., et al. A single dose of an adenovirus-vectored vaccine provides protection against SARS-CoV-2 challenge. Nat Commun. 2020;11(1):4081. doi: 10.1038/s41467-020-17972-1. [DOI] [PMC free article] [PubMed] [Google Scholar]