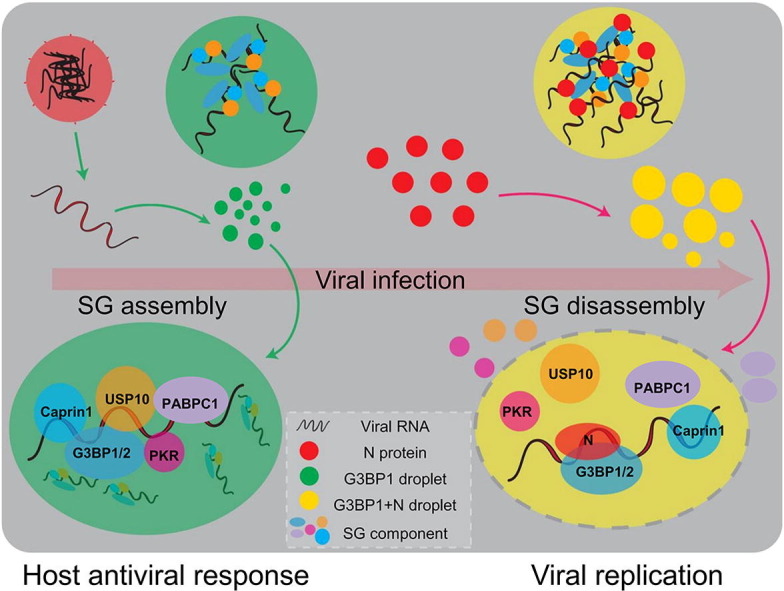
Graphical abstract
Keywords: SARS-CoV-2, Nucleocapsid protein, Stress granules, Liquid-liquid phase separation, G3BP, Viral production
Abstract
A key to tackling the coronavirus disease 2019 (COVID-19) pandemic is to understand how severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) manages to outsmart host antiviral defense mechanisms. Stress granules (SGs), which are assembled during viral infection and function to sequester host and viral mRNAs and proteins, are part of the antiviral responses. Here, we show that the SARS-CoV-2 nucleocapsid (N) protein, an RNA binding protein essential for viral production, interacted with Ras-GTPase-activating protein SH3-domain-binding protein (G3BP) and disrupted SG assembly, both of which require intrinsically disordered region1 (IDR1) in N protein. The N protein partitioned into SGs through liquid-liquid phase separation with G3BP, and blocked the interaction of G3BP1 with other SG-related proteins. Moreover, the N protein domains important for phase separation with G3BP and SG disassembly were required for SARS-CoV-2 viral production. We propose that N protein-mediated SG disassembly is crucial for SARS-CoV-2 production.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), responsible for the coronavirus disease 2019 (COVID-19) pandemic, has resulted in > 23,000,000 infections and > 800,000 deaths [1], [2]. SARS-CoV-2 is highly contagious and induces serious multi-organ damages [3], [4], [5], [6]. Despite tremendous efforts, how this deadly virus breaks the host defense remains poorly understood.
Stress granules (SGs) are dynamic large cytoplasmic mRNA-protein aggregates, formed in response to various stresses, including heat shock, oxidative stress, or viral infection [7], [8], [9]. SG formation can inhibit protein synthesis, restrict energy consumption and repair stress-induced damage [10], which facilitates cell survival and represents one of the defense mechanisms of host cells [11], [12]. In particular, SGs can counter viral infection by insulating invaders and limiting viral mRNA translation, but viruses have also evolved mechanisms to inhibit SG formation and to escape the host antiviral response [13], [14]. For example, foot-and-mouth disease virus and poliovirus inhibit SG formation by cleaving Ras GTPase-activating protein-binding protein (G3BP) 1 and 2, the key SG-nucleating proteins [15], [16]. Similar to SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV), the SARS-CoV-2 genome encodes four essential structural proteins, namely spike (S) glycoprotein, small envelope (E) protein, matrix (M) protein, and nucleocapsid (N) protein [17], [18], [19], with the N protein being the most abundant viral structure protein in infected cells. The N protein not only packages viral RNA and proteins into virions, but also promotes viral RNA transcription and replication, and impairs the host cell immune response [20], [21], [22]. Importantly, N protein interacts with the SG core component G3BP1/2 [23], [24], suggesting a potential impact on SG formation.
Here we report a novel mechanism whereby the N protein promotes viral production. First, the SARS-CoV-2 N protein indeed colocalized with G3BP and almost completely disrupted SG formation. Furthermore, the N protein underwent liquid-liquid phase separation (LLPS) and interacted with G3BP in an RNA-dependent manner, which conferred the N protein to partition into the SG component. Intrinsically disordered regions (IDRs)-mediated interaction and phase separation with G3BP is required for SG colocalization and disassembly. Importantly, the SARS-CoV-2 N protein impaired the interaction between G3BP1 and other SG-related proteins and led to SG reconstitution through phase separating with G3BP to occupy protein–protein interaction position of G3BP. In addition, we used an N protein-based transcomplementation SARS-CoV-2 cell culture model, and concluded that deletion of domains regulating the N protein to phase separate with G3BP and disassemble SG, disrupted viral production. Together, these data provide a novel function and mechanism by which the N protein regulates SG assembly and viral production.
2. Materials and methods
2.1. Plasmid construction
SARS-CoV-2 N protein-coding fragments were polymerase chain reaction (PCR)-amplified from the N protein expression construct in the PUC57 vector (Tsingke Biotech, Hangzhou, China) and subcloned into the pCS2 vector, while the coding region for SARS-CoV-2 N protein deletions (residues 1–175, residues 1–210, residues 51–C, residues 161–C, residues 211–C, and ΔSR) were amplified by PCR from a plasmid containing full-length SARS-CoV-2 N protein with appropriate sets of primers and was confirmed by sequencing. DNA fragments encoding human ubiquitin-specific peptidase 10 (USP10), double-stranded RNA-dependent protein kinase (PKR), and G3BP2 were PCR-amplified from cDNA that was reverse transcribed from HEK293T total RNA with reverse transcriptase (Vazyme, Nanjing, China) according to the manufacturer’s instructions. Exnase (Vazyme, Nanjing, China) was used to insert these sequences into the CMV-Flag or CMV-Myc backbone.
2.2. Cell culture and transfection
HeLa, Caco-2, and HEK293T cells were obtained from American type culture collection (ATCC, https://www.atcc.org/). Human bronchial epithelial cells (16HBE) were purchased from Sigma (St. Louis, USA). The 16HBE cells, HEK293T cells, Caco-2 cells, and HeLa cells were maintained in Dulbecco modified Eagle medium (DMEM; Gibco, Waltham, USA) supplemented with 10% fetal bovine serum (Gemini, Woodland, USA) and 1% penicillin/streptomycin (Gibco,) under standard tissue-culture conditions (37 °C, 5% CO2). The cells were seeded in 6-well plates and transfected with 2.5 μg plasmids at approximately 70% density using Lipofectamine 3000 Transfection Kit (Invitrogen, Carlsbad, USA). After transfection for 6 h, the cells were moved to fresh DMEM and cultivated for 24 h.
2.3. Stress treatment
Poly(I:C) (Sigma) was dissolved in RNase-free water containing 0.98% NaCl to make 5 mg/mL stock solution. Before use, poly(I:C) was incubated at 50 °C for 20 min followed by slow cooling to room temperature for annealing. To mimic stress induced by viruses, 2 μg poly(I:C) was transfected into the cells with lipofectamine 3000 for 7 h or the indicated time. To induce oxidative stress, the cells were treated with 200 μmol/L sodium arsenite for 45 min. To induce endoplasmic reticulum (ER) stress, the cells were treated with 2 mmol/L dithiothreitol (DTT) for 1 h. For cycloeximide treatment, the cells were incubated with 10 μg/mL cycloheximide and 200 μmol/L sodium arsenite simultaneously for 45 min.
2.4. Immunofluorescence
HeLa cells or 16HBE cells were seeded to reach 40%–60% confluence in 24-well glass bottom plates (Cellvis, Mountain View, USA) and transfected with plasmids for 24 h. Then, the cells were stressed with 200 μmol/L sodium arsenite for 45 min, poly(I:C) for 7 h or DTT for 1 h to induce SGs. After treatment, the cells were fixed in 4% paraformaldehyde for 15 min at room temperature and washed with phosphate buffered saline (PBS) for three times. Next, the fixed cells were incubated in blocking solution containing 5% (v/v) normal goat serum (Bioss Antibodies, Boston, USA), 0.3% (v/v) Triton X-100 in PBS for 2 h at room temperature. After that, the cells were incubated with primary antibodies against G3BP1 (Proteintech, Rosemont, USA), TIA1 (Proteintech), and Flag (Proteintech) overnight at 4 °C. Next, the cells were washed three times in PBS and then incubated with secondary antibodies conjugated to Alexa Fluor 488 (Cell Signaling Technology, Danvers, USA), and Alexa Fluor 647 (Cell Signaling Technology) at a 1:1000 dilution for 2 h at room temperature. During this period, a 24-well plate was wrapped in foil to keep it in a dark environment. Cell nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI; Yeasen, Shanghai, China). Cells were then washed three times in PBS for 10 min each time. Images were acquired with a Nikon spinning disk microscope with a 60× oil immersion objective. Fluorescence intensity was obtained using Fiji (National Institutes of Health, Bethesda, USA).
2.5. Western blotting
HeLa cells or HEK293T cells were rinsed with PBS (pH 7.4) and lysed in radio immunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China) supplemented with a protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, Waltham, USA) on ice for 30 min. Cell lysates were centrifuged for 10 min (12,000 × g, 4 °C) and the protein concentration was measured by a bicinchoninic acid (BCA) assay (Thermo Fisher Scientific). Equal amounts of protein were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Darmstadt, Germany). The membranes were blocked for 1 h at room temperature in Tris-buffered saline and 0.1% Tween 20 (TBST) containing 5% (w/v) nonfat milk and then incubated with primary antibodies at a 1:2000 dilution in the same buffer at 4 °C overnight. The protein bands were detected with horseradish peroxidase-conjugated secondary antibodies and immobilon western enhanced chemiluminescent solution (Millipore). The protein levels were analyzed using Western blottings with the corresponding antibodies. The protein levels were normalized by probing the same blots with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody.
2.6. Coimmunoprecipitation
HeLa cells or HEK293T cells were transfected with plasmids. Then, the cells were washed 3 times with PBS and harvested in 200 μL lysis buffer containing 10 mmol/L Tris HCl, pH 7.5, 150 mmol/L NaCl, 0.5 mmol/L ethylene diamine tetraacetie acid (EDTA), 0.5% NP-40, and a protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). The Cells were incubated for 30 min on ice and centrifuged for 10 min at 16,000 × g at 4 °C. Next, 25 μL agarose beads pre-equilibrated with dilution buffer (10 mmol/L Tris/Cl, pH 7.5, 150 mmol/L NaCl, 0.5 mmol/L EDTA) was added, and binding was performed for 2 h at 4 °C on a rotating wheel. Agarose beads were then washed 3 times using dilution buffer. The bound proteins were eluted by boiling the samples with 1× SDS loading buffer, and the supernatants were used for SDS-PAGE analysis.
2.7. Protein expression and purification
Protein-coding fragments were synthesized by GenScript Biotech Corporation (Nanjing, China), and deletions were generated by PCR and confirmed by sequencing. Full-length SARS-CoV-2 N protein was expressed and purified from E. coli BL21 (DE3) (TransGen Biotech, Beijing, China). E. coli were induced with 1.0 mmol/L isopropyl-β-D-thiogalactoside (IPTG) until the A 600nm reached 0.6 and grown at 16 °C for 15 h. Cells were harvested by centrifugation and resuspended in buffer A (20 mmol/L Tris-HCl, pH 8.0, 500 mmol/L NaCl, 10% (v/v) glycerol, 1 mmol/L phenylmethanesulfonyl fluoride (PMSF)). After lysis by EmulsiFlex-C3 (Avestin, Ottawa, Canada) at 4 °C, the lysates were cleared by centrifugation at 40,000 × g for 60 min. DNase and RNAase were added to the supernatant to the final concentration of 4 μg/mL and incubated at 20 °C for 3 h. The supernatant was then loaded on a polypropylene column (QIAGEN, Hilden, Germany) containing 4 mL His60-Ni-Superflow-Resin (TaKaRa, Kusatsu, Japan), which was pre-equilibrated by 10 mL buffer A. Proteins were eluted by 15 mL buffer B (20 mmol/L Tris-HCl, pH 8.0, 500 mmol/L NaCl, 10% (v/v) glycerol, 500 mmol/L imidazole). After that, proteins were further purified by size exclusion with a Superdex-200 column on an AEKTA purifier (GE Healthcare Life Sciences, Boston, USA). The fractions were analyzed by SDS-PAGE, concentrated and stored at −80 °C. All recombinant mCherry fusion proteins (mCherry-N protein FL, mCherry-N protein 1–175, mCherry-N protein 1–210, mCherry-N protein ΔSR, and mCherry-N protein 51–C) and mEGFP-G3BP1 protein were purified in the same manner.
2.8. In vitro droplet assay
For Ficoll-dependent droplet-formation experiments. Ficoll (0%, 1%, 2%, 5%, 10% (v/v)), a commonly used crowding agent, was added to 0.5, 1, 2, 7.5, or 15 μmol/L purified SARS-CoV-2 N protein. After incubation at room temperature for 5 min, reactions were then transferred in 96-well glass bottom plate (Cellvis) and observed under a Nikon spinning disk microscope equipped with a 60 × oil immersion objective. For RNA-dependent droplet-formation experiments. Total HeLa cellular RNA (0, 20, 60, 120, and 150 ng/μL), isolated by Trizol (TaKaRa), was coincubated with 0.5, 1, 4, 15, or 30 μmol/L purified SARS-CoV-2 N protein. Droplets were also visualized with Nikon spinning disk microscope. To confirm which domain is required for phase separation of N protein/G3BP in vitro, mCherry-N protein 1–175, mCherry-N protein 1–210, mCherry-N protein ΔSR, mCherry-N protein 51-C, or mCherry-N protein FL was coincubated with mEGFP-G3BP1 in the presence or absence of HeLa cellular RNA. LLPS droplets formation was observed under a Nikon spinning disk microscope. All the experiments were conducted in 150 mmol/L NaCl at pH 8.0.
2.9. Turbidity assay
Phase separation of the N protein with or without G3BP1 in the absence or presence of 50 ng/μL RNA, was induced as described above for the droplet assay. Turbidity measurements were conducted at 600 nm in 96-well plates with 40 μL samples using a plate reader (PerkinElmer, Enspire, USA). All experiments were performed in triplicate.
2.10. Fluorescence recovery after photobleaching (FRAP) analysis
The experiment was performed using a Nikon spinning disk microscope equipped with two laser systems. A region of the indicated protein droplets was bleached by a 488/561 nm wavelength laser with a light intensity of 80%. Only the center of the droplets was bleached. Fluorescence intensity recovery data were recorded at a given time interval. Fluorescence intensity was obtained using Fiji (National Institutes of Health). Fluorescence intensities of regions of interest (ROIs) were subtracted by background intensity and then normalized by prebleached intensities of the ROIs. The FRAP recovery curve was fit to the formula described previously [25].
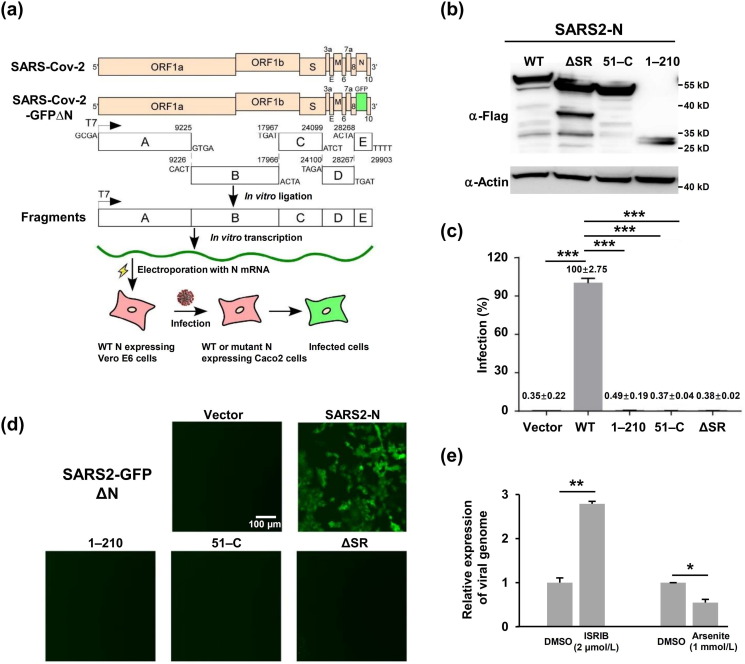
2.11. SARS-CoV-2 GFPΔN cell culture system
The ORF of N gene in the SARS-CoV-2 (Wuhan-Hu-1, NC_045512) genome was replaced with GFP (SARS-CoV-2 GFPΔN), and the entire genome was divided into A, B, C, D, and E fragments, which were synthesized by Genescript. PCR was performed to amplify each fragment with type IIS restriction endonuclease sites (BsaI or BsmBI) flanked at both ends of each DNA fragment and the T7 promoter was introduced upstream of fragment A. The DNA fragments were digested with BsaI (A, B, C, or E) or BsmBI (D) and then ligated as the template for T7 polymerase based on in vitro transcription (mMESSAGE mMACHINE T7 Transcription Kit, Thermo Fisher Scientific) to generate the SARS-CoV-2 GFPΔN viral genomic RNA. RNA was transfected into Caco-2 cells expressing the SARS-CoV-2 N protein by lentiviral transduction to produce the recombinant SARS-CoV-2 GFPΔN virus.
2.12. Real-time quantitative PCR (RT-qPCR)
Caco-2 cells were pretreated with 1 mmol/L sodium arsenite for 1 h prior to SARS-CoV-2 GFP ΔN virus infection at a multiplicity of infection (MOI) of 0.05. Caco-2 cells were treated with 2 μmol/L ISRIB and infected with SARS-CoV-2 GFP ΔN virus at an MOI of 0.05. Twenty-four hours post-infection, whole-cell RNAs were purified for RT-qPCR to analyze viral genome abundance. Viral genomic RNA expression was normalized to that of the DMSO-treated control. Then, qPCR primers targeting GAPDH (forward: 5'-GAAGGTGAAGGTCGGAGTC-3' and reverse: 5'-GAAGATGGTGATGGGATTTC-3'), viral genome (forward: 5'-CGAAAGGTAAGATGGAGAGCC-3' and reverse: 5'-TGTTGACGTGCCTCTGATAAG-3') were used.
2.13. Statistical analysis
All experiments were repeated, and Microsoft Excel (Professional 2019, Microsoft Corporation, Redmond, USA) or OriginPro (2019b, OriginLab, Northampton, USA) software was used for statistical analysis. All statistical values are displayed as the mean ± standard deviation (SD) or standard error of the mean (SEM), and the details are included in the corresponding figure legends. Differences between two groups were analyzed using a Student’s t-test. Multiple group comparisons were performed using a one-way analysis of variance (ANOVA) followed by post-hoc tests.
3. Results
3.1. N protein Interacts with G3BP and suppresses SG assembly
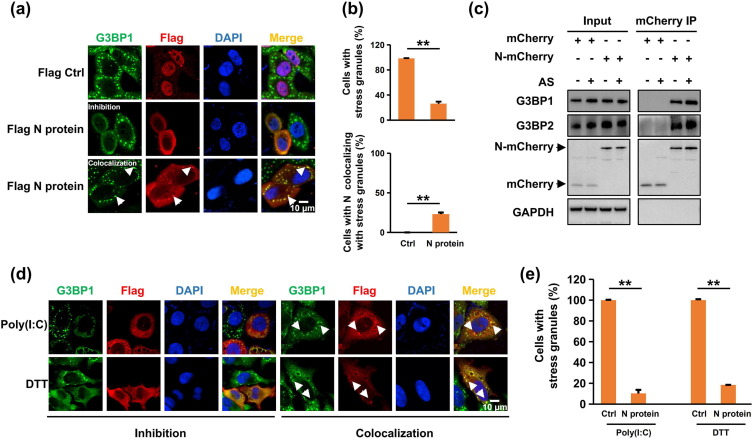
To investigate the roles of SARS-CoV-2 proteins in SG assembly, we expressed the S protein, N protein, RdRp, or PL protease in human bronchial epithelial (16HBE) cells and HeLa cells before inducing SG assembly using sodium arsenite (AS). SGs were visualized by immunostaining for G3BP1, an SG marker [26]. We found that SG assembly was markedly inhibited in the majority of N protein-transfected cells (74.0% in 16HBE cells and 89.4% in HeLa cells, Fig. 1 a, b, and Fig. S1a–c online). Moreover, the N protein interacted with the SG proteins G3BP1 and G3BP2, which was enhanced by SG induction (Fig. 1c), indicating that the N protein might enter SGs and promote SG disassembly by targeting G3BP1/2.
Fig. 1.
SARS-CoV-2 N protein interacted with G3BP and impaired SG assembly. (a) N protein colocalized with SGs and impaired SG assembly in 16HBE cells. The 16HBE cells transfected with Flag control or Flag-tagged N protein were treated with sodium arsenite (AS) for 45 min to induce SGs, followed by immunostaining for N protein (red) and G3BP1 (green). (b) Statistical analysis of the SG-assembly index and the SG-colocalization index of the N protein shown in panel A. Three independent experiments were performed in each case with similar outcomes, and representative data are shown as the mean ± SEM. Statistics: two-tailed t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (c) SG formation enhanced the N protein interaction with G3BP protein. HeLa cells transfected with mCherry control or mCherry-tagged N protein were subjected to immunoprecipitation with G3BP1 and G3BP2 antibodies. The presence of the N protein in the immunoprecipitates was assessed by Western blotting against anti-mCherry antibody. (d) N protein disrupted SG assembly under diverse stress conditions. HeLa cells transfected with the N protein were treated with poly(I:C) for 7 h or DTT for 1 h to induce SGs, followed by immunostaining for the N protein (red) and G3BP1 (green). (e) Statistical analysis of the SG-assembly index of the N protein is shown in panel D. Data are shown as the mean ± SEM. Statistics: two-tailed t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
The interaction with G3BP1/2 seemed specific, as the N protein did not (strongly) associate with other SG-related proteins tested (Fig. S1d online), including fragile X mental retardation 1 (FMR1), cell cycle associated protein 1 (Caprin1), eukaryotic translation initiation factor 4E (EIF4E), TIA1 cytotoxic granule associated RNA binding protein (TIA1), DEAD-box RNA helicase 3 (DDX3), and poly (A) binding protein cytoplasmic 1 (PABPC1). The N protein also disrupted the SGs induced by poly(I:C) (stressor mimicking viral infection) and DTT (ER stressor) (Fig. 1d, e, and Fig. S1e, f online). In addition, the N protein also colocalized with TIA1, another well-known SG marker [27] (Fig. S2a online), and cycloheximide (CHX), capable of disassembling AS-induced SGs [28], disrupted N protein/G3BP foci (Fig. S2b online). Of note, the N protein did not cause G3BP1/2 degradation (Fig. S2c online). We conclude that the N protein blocked SG assembly via interactions with G3BP1/2.
3.2. N protein undergoes RNA-dependent LLPS and partitions into the G3BP condensate
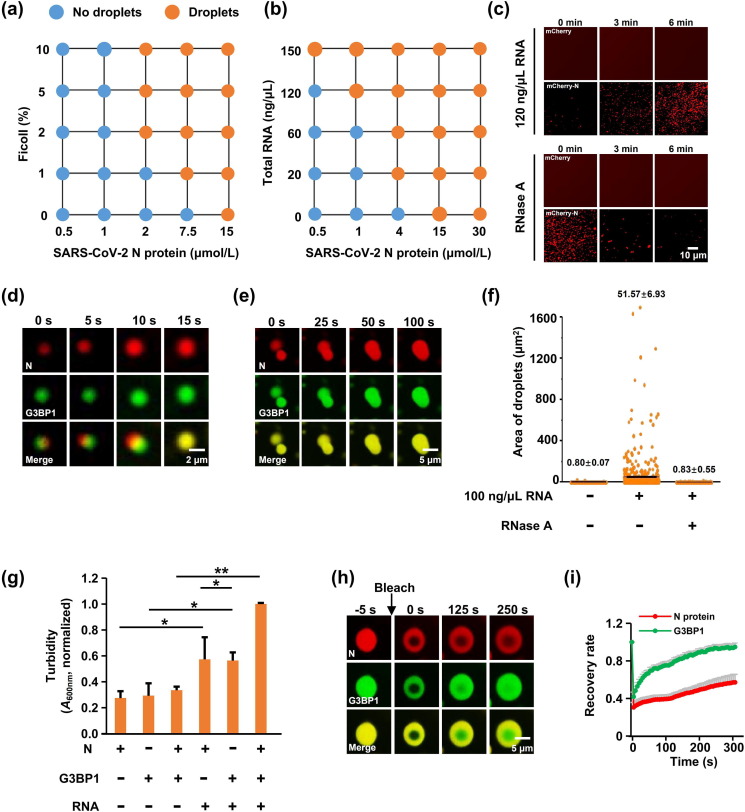
Coronaviral N protein binds the viral RNA to form a helical ribonucleoprotein (RNP) complex, known as the nucleocapsid, which usually exists as RNA granules, and assembles RNA and protein into condensates through LLPS [25, 29, 30]. We thus investigated whether the N protein exhibits features of liquid condensates. Notably, at physiological salt concentration (150 mmol/L NaCl), N protein formed droplets when the protein concentration reached 15 μmol/L (Fig. 2 a). Furthermore, the N protein did exhibit concentration-dependent LLPS upon the addition of the molecular crowding agent Ficoll (Fig. 2a and Fig. S3a online). Because the N protein binds to genomic RNA to participate in viral genome replication and subgenomic mRNA transcription [31], we next characterized the effect of RNA on LLPS of the N protein, found that HeLa RNA strongly promoted the N protein LLPS without impacting mCherry (Fig. 2b, c, and Fig. S3b online), with the effect abolished by RNase A as expected (Fig. 2c).
Fig. 2.
SARS-CoV-2 N protein phase separated with G3BP1 in an RNA-dependent manner. (a) Summary of the phase separation behaviors of the N protein with increasing concentrations of Ficoll (crowding agent). Corresponding images are shown in Fig S3a (online). (b) LLPS of the N protein was dependent on the RNA concentration. Corresponding images are shown in Fig. S3b (online). (c) LLPS of N protein-mCherry or mCherry protein with 120 ng/μL RNA, with or without the addition of 1 μg/mL RNase A. The concentration of relative proteins was 15 μmol/L, respectively. (d) N protein partitioned into G3BP droplets to form new heterotypic droplets in the presence of RNA. The concentration of relative proteins was 15 μmol/L, respectively. (e) Time-lapse micrographs of merging droplets. The concentration of relative proteins was 15 μmol/L, respectively. (f) Column scatter charts display the droplet area from Fig. S4c (online) for at least 500 droplets. (g) Turbidity of N protein ± G3BP1 with or without the addition of 50 ng/μL RNA. A600nm normalized to measurement of N protein-G3BP1-RNA. Data are shown as the mean ± SEM (n = 3). Statistics: two-tailed t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (h) FRAP of N protein-G3BP1-RNA liquid droplets. Bleaching was performed at the indicated time points. Time 0 indicates the start of recovery after photobleaching. (i) Plot showing the time course of recovery after photobleaching N protein-G3BP1-RNA droplets. Data are shown as the mean ± SD (n = 10).
SGs assemble via LLPS which occurs across a core protein-RNA and protein–protein interaction network, and G3BP1/2, the central node of this network, function as a molecular switch to trigger RNA-dependent LLPS [32], [33], [34]. A previous study using affinity-purification mass spectrometry (AP-MS) reported that the SARS-CoV-2 N protein binds G3BP1/2 proteins [24], which was confirmed in our study. Furthermore, SG formation is able to enhance the interaction between the N protein and G3BP1/2, raising the possibility of co-LLPS between the N protein and G3BP1/2. To test this possibility, we purified recombinant mEGFP-G3BP1 and mCherry-N fusion proteins, and mixed them at physiological salt concentrations. Consistent with a previous study [32], the G3BP1 protein alone almost did not form liquid-like droplets. When the N protein and G3BP1 were mixed, the two proteins formed heterotypic droplets that were much larger and more abundant than the droplets formed by N protein alone at 15 μmol/L (Fig. S4a online). Given that SGs are large cytoplasmic mRNA-protein aggregates and N protein associated with SGs through interaction with G3BP, we examined whether RNA binding enhances the phase separation of N protein/G3BP1. We incubated N protein and G3BP1 with RNA. Upon mixing, the N protein partitioned into G3BP1 droplets and small liquid droplets of N protein/G3BP1 fused into larger ones over time (Fig. 2d, e, Fig. S4b, and Supplementary Video 1 online). This effect was abolished by RNase A (Fig. 2f and Fig. S4c online). Next, we monitored the RNA-driven LLPS of N protein/G3BP1 in a turbidity assay based on the optical density of the protein solution. Consistent with the above results, the solution of N protein/G3BP1 became turbid upon RNA addition (Fig. 2g), revealing that the N protein partitioned into SGs through phase separation with G3BP1 in RNA-dependent manner.
To further characterize the LLPS of the N protein and G3BP1, we assessed the mobility of N protein/G3BP1 droplets by FRAP measurements. G3BP1-GFP molecules diffused rapidly within droplets, and exchanged between droplets and the surrounding solution. Surprisingly, N protein-mCherry displayed minimal recovery and less dynamic, indicating that N protein condensates are more stable than G3BP1 condensates (Fig. 2h, i). The N protein assembles nucleocapsid by encapsulating RNA, which may be facilitated by the stability of the N protein condensate.
3.3. IDR1 is required for RNA-dependent N protein phase separation with G3BP1
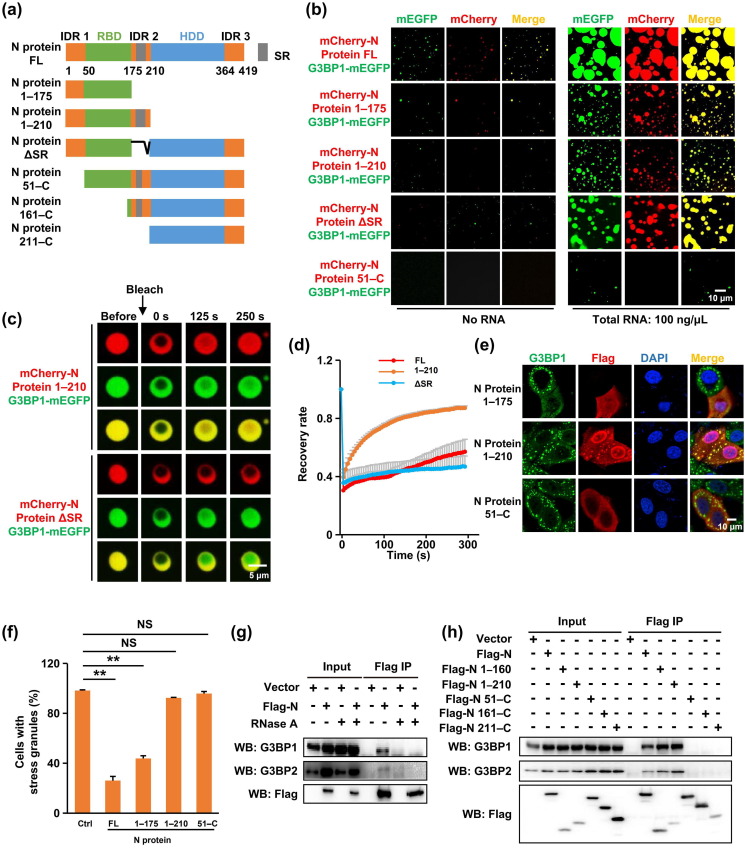
The N protein consists of three intrinsically disordered regions (IDRs) that are separated by two distinct but highly conserved domains: the RNA binding domain (RBD) and homodimerization domain (HDD) (Fig. 3 a). The CoV RBD binds RNA through electrostatic interactions, and the RNA binding capacity of the N protein is crucial for viral infectivity [35], [36]. The HDD of the SARS-CoV N protein folds into a β-sheet involved in homodimerization and is conferred with RNA binding activity [31], [37]. The Ser/Arg (SR)-rich domain, within IDR2, plays a pivotal role in SARS-CoV N protein multimerization [38]. To dissect the contribution of the various SARS-CoV-2 N domains to LLPS, we tested various N protein fragments, including residues 1–175 (containing IDR1 and RNA binding domain), 1–210 (containing IDR1, RNA binding domain, and SR region), 51–C (N protein lacking IDR1), 161–C (SR region, dimerization domain and IDR3), 211–C (dimerization domain and IDR3) and SR deletion (Fig. 3a). We found that aa 1–175 (containing IDR 1 and RNA binding domain) is necessary and sufficient for RNA-stimulated LLPS (Fig. 3b), with IDR1 being absolutely required (Fig. 3b, fragment 51–C), while deletion of HDD and IDR3 or the SR domain only attenuated LLPS (Fig. 3b).
Fig. 3.
IDR1-mediated interaction and RNA-dependent LLPS with G3BP were necessary for N protein-regulated SG localization and SG disassembly. (a) Schematic domain structure of the protein used in the study. (b) LLPS of individual domain constructs of N protein with G3BP1 in the absence or presence of RNA. The concentration of relative proteins was 15 μmol/L, respectively. (c) FRAP of N protein mutants-G3BP1-RNA liquid droplets. Bleaching was performed at the indicated time points. Time 0 indicates the start of recovery after photobleaching. (d) Plot showing the time course of recovery after photobleaching N protein mutants-G3BP1-RNA droplets. Data are shown as the mean ± SD (n = 10). (e) Characterization of N protein mutants required for SG-localization and SG-inhibition. The 16HBE cells transfected with N protein mutants were treated with sodium arsenite for 45 min to induce SGs, followed by immunostaining for N protein (red) and G3BP1 (green). Three independent experiments were performed in each case with similar outcomes, and representative data are shown. (f) Statistical analysis of the SG-assembly index of N protein mutants shown in panel e. Data are shown as the mean ± SEM (n = 3). Statistics: two-tailed t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant). (g) N protein interaction with G3BP was dependent on RNA binding. HEK293T cells transfected with Flag-tagged N protein were subjected to immunoprecipitation with anti-Flag antibody in the presence or absence of 100 μg/mL RNase A. The presence of G3BP1 and G3BP2 proteins in the immunoprecipitates was assessed by Western blotting against anti-G3BP1 and G3BP2 antibodies. (h) The interaction of N protein mutants with G3BP1 and G3BP2. HEK293T cells transfected with Flag-tagged N protein truncations were subjected to immunoprecipitation with anti-Flag antibody. The presence of G3BP1 and G3BP2 proteins in the immunoprecipitates was assessed by Western blotting against anti-G3BP1 and G3BP2 antibodies.
Surprisingly, deleting the C-terminus (containing HDD and IDR3), markedly facilitated N protein diffusion within droplets (Fig. 3c, d). We conclude that IDR1 triggers the N protein LLPS, while the C-terminus stabilizes the condensate presumably via mediating protein homodimerization.
3.4. N protein colocalizes with and dissembles SG via IDR1-mediated interaction with G3BP1
The essential role of IDR1 in LLPS predicts that IDR1 is also necessary for N protein interactions with SGs. Indeed, the N protein mutant lacking IDR1 (51–C) was unable to colocalize with G3BP1 or to disassemble SGs in vivo (Fig. 3e, f, and Fig. S5a online). Importantly, interaction of the N protein with G3BP was RNA-dependent (Fig. 3g) and required DR1 (Fig. 3h). These data suggest that the N protein phase separated with G3BP via IDR1 in an RNA-dependent manner, which was required for the N protein to enter and dissemble SGs.
Of note, compared with the full length N protein, residues 1–175 was more effectively colocalized with SG and less potent in dissembling SG (Fig. 3e, f), and further studies are needed to clarify the underlying mechanism.
3.5. N protein-mediated SG disassembly occurs through blocking the interaction between G3BP1 and other SG-related proteins and inducing SG reconstitution
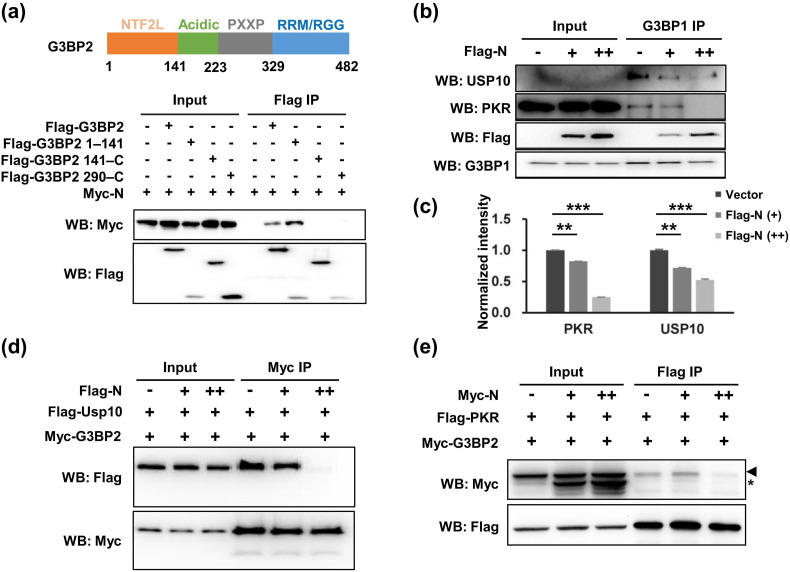
We next sought to reveal how the N protein disassembled SG. We found that the N protein bound N-terminal NTF2L in G3BP2 (Fig. 4 a). This domain is actively engaged in protein–protein interactions between G3BP1/2 and core SG components including Caprin1, USP10, and PKR, and is thus required for SG assembly [25], [32], [39], [40]. The N protein compromised G3BP1-USP10 and G3BP1-PKR interactions (Fig. 4b, c) and disrupted G3BP2-USP10 and G3BP2-PKR interactions (Fig. 4d, e). We propose that the N protein was recruited to SGs via phase separation with G3BP1/2 and outcompeted G3BP1/2 binding partners via interaction with the NTF2L domain, thus disrupting SGs.
Fig. 4.
SARS-CoV-2 N protein blocked the G3BP interaction with other SG-related proteins, thus inducing SG reconstitution. (a) The N protein-binding domain resides in the N-terminal NTF2L domain of the G3BP protein. HEK293T cells transfected with Myc-tagged N protein and Flag-tagged G3BP2 truncations were subjected to immunoprecipitation with anti-Flag antibody. The presence of N protein in the immunoprecipitates was assessed by Western blotting with an anti-Myc antibody. (b) N protein disrupted G3BP1-USP10 and G3BP1-PKR interactions. HEK293T cells transfected with increasing amounts of Flag-tagged N protein were subjected to immunoprecipitation with an anti-G3BP1 antibody. The presence of USP10 and PKR in the immunoprecipitates was assessed by Western blotting. (c) Quantification of immunoblot intensities in (b). Bars indicate mean ± SEM (n = 3). The intensity is the ratio of the coIP band to the IP band. Statistical analysis was performed with unpaired t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (d) N protein disrupted G3BP2-USP10 interaction. HEK293T cells transfected with Flag-tagged USP10, Myc-tagged G3BP2 and increasing amounts of Flag-tagged N protein were subjected to immunoprecipitation with anti-Myc antibody. The presence of USP10 in the immunoprecipitates was assessed by Western blotting against anti-Flag antibody. (e) N protein disrupted G3BP2-PKR interaction. HEK293T cells transfected with Flag-tagged PKR, Myc-tagged G3BP2, and increasing amount of Myc-tagged N protein were subjected to immunoprecipitation with anti-Flag antibody. The presence of G3BP2 in the immunoprecipitates was assessed by Western blotting against anti-Myc antibody. Arrowheads and asterisks indicate the bands of the G3BP2 and N protein, respectively.
3.6. The integrity of the N protein is required for SARS-CoV-2 viral production
Finally, we examined the effect of N protein mutations on SARS-CoV-2 production using an N protein-based transcomplementation model [41] (Fig. 5 a, b). Specifically, Caco-2 cells, a human epithelial colorectal adenocarcinoma cell line permissive to SARS-CoV-2 infection, were transduced with lentivirus expressing N protein or its derivatives before inoculation with a defective SARS-CoV-2 virus lacking the N protein but expressing GFP (GFPΔN virus, MOI = 0.05). The GFP signal became detectable within 48 h, and intensified within 72 h in Caco-2 cells expressing the N protein, but remained undetectable in cells expressing N protein mutants including residues 1–210, 51–C, or SR (Fig. S6 online). As expected, cell culture supernatant from Caco-2 cells expressing N protein but not the various mutants was able to infect Caco-2 cells expressing N protein and to produce a GFP signal (Fig. 5c, d). The failure of the N protein mutants to support viral production might result in part from their failure to encapsulate RNA. Indeed, chemical compounds that dissembled SGs increased SARS-CoV-2 production and vice versa, as evidenced by the alterations of viral genomic RNA abundance in each condition (Fig. 5e), reinforcing the notion that N protein-mediated SG disassembly contributes to SARS-CoV-2 production and infection.
Fig. 5.
The SARS-CoV-2 N protein-based genetic complementation system is utilized to assess WT or mutant N protein function in the viral life cycle. (a) Schematic diagram illuminating the SARS-CoV-2 N protein-based genetic complementation system for the assessment of N or N mutant function in the viral life cycle. (b) Immunoblotting assays were performed to detect the expression of different versions of the N proteins in Caco-2 cells transduced by lentivirus. (c) Flow cytometry quantified the percentage of GFP positive cells. Data are shown as the mean ± SEM (n = 3). Statistical analysis was performed with unpaired t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (d) Live-cell fluorescence microscopy of Caco-2-N cells infected by the cell culture supernatant from Caco-2 cells expressing different versions of the N protein. (e) SG assembly regulated SARS-CoV-2 infection. Caco-2 cells were pretreated with 1 mmol/L arsenite for 1 h prior to SARS-CoV-2 GFP ΔN virus infection at an MOI of 0.05. Caco-2 cells were treated with 2 μmol/L ISRIB and infected with SARS-CoV-2 GFP ΔN virus at MOI of 0.05. Viral genomic RNA expression was normalized to that of the DMSO treated control. Data are shown as the mean ± SEM (n = 3). Statistical analysis was performed with unpaired t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
4. Discussion and conclusion
The current study shows that the N protein phase separates with G3BP to block the interaction of G3BP and other SG-related proteins and thereby disassembles SGs, which may facilitate viral assembly and production. Furthermore, IDR1 in the N protein responsible for the interaction, and phase separation with G3BP is necessary for SG colocalization, SG disassembly, and viral production. Our discovery provides new insights into a previously unappreciated role of LLPS in SARS-CoV-2 pathogenesis.
Upon infection, viruses from diverse families can provoke stress response and SG formation in the host cells, but can subsequently block the SG assembly [14]. Although SG assembly as an innate defense mechanism can hamper viral proteins translation by sequestering mRNAs, the viruses might also exploit the high concentration of RNAs and RNA binding proteins in SG to package the virion. After completing virion assembly, viruses utilize viral proteins to dissemble SG, thereby promoting viral replication and production [23]. Viral proteins suppress the SG formation mainly by (1) cleaving SGs factors, (2) regulation of PKR activity, and (3) sequestering SGs proteins [9]. We found that the N protein acted via sequestering G3BP, but contrary to some viral proteins that sequester G3BP into viral replication complexes, inclusion bodies, or novel cytoplasmic foci, which are different from the SG foci [42], [43], [44]. The SARS-CoV-2 N protein localized with G3BP in the SG foci.
Viral replication compartments have the properties of liquid-like condensates [45], [46]. The biomolecular condensation activity of the N protein is important for viral assembly and replication [23]. For example, the measles virus N protein undergoes LLPS in vitro, which regulates the assembly of capsid-like particles [47]. Influenza A virus ribonucleoprotein forms liquid-like organelles at sites along the ER during Influenza A infection [48]. The N protein of numerous retroviruses shows the phase-separation property in vitro, and the HIV-1 N protein forms dynamic foci in mammalian cells, similar to biomolecular condensation [49]. In addition, the SARS-CoV N protein is involved in SARS-CoV replication [50]. Similarly, our study found that the SARS-CoV-2 N protein underwent RNA-dependent LLPS, supporting the concept that the liquid-like property of the N protein may facilitate the formation of replication compartments and viral assembly.
LLPS is often driven by IDRs responsible for oligomerization and/or contributing multivalent interactions [51], [52]. We found that for the N protein, the SR domain within IDR2 displayed the highest phase separation score, followed by the IDR1 within RBD domain [23]. However, IDR1 instead of IDR2 (or IDR3) was required for phase separation with G3BP. In addition, while SR domain in the SARS-CoV N protein is essential for SG regulation [53], the IDR1 in the SARS-CoV-2 N protein is essential for G3BP binding, SG localization, and SG disassembly.
The SARS-CoV-2 N protein enters SGs through phase separation with G3BP, leading to SG reconstitution and disassembly. Semliki Forest Virus (SFV) nsP3, a nonstructural protein, blocks SG assembly by hijacking G3BP1 into viral replication complexes [54], whereas Chikungunya virus nsP3 disrupts SGs by recruiting G3BP1 to novel cytoplasmic foci [42]. Thus, the mechanism whereby the SARS-CoV-2 N protein disrupts SGs is divergent from those used by several other viruses.
Importantly, our results showed that the domains required for the N protein to phase separate with G3BP and disassemble SGs were also required for SARS-CoV-2 viral production. Presumably, these domains could facilitate viral RNA encapsulation via inducing RNA-dependent LLPS. Consistent with this, chemically disrupting SGs also facilitated the escape of SARS-CoV-2 from host antiviral response. However, further studies are needed to directly demonstrate that the N protein regulates viral production via LLPS.
Altogether, our data indicate that via IDR1-mediated interaction with G3BP and RNA-dependent LLPS, the N protein enters SGs and outcompetes G3BP binding partners to dissemble SGs, which is required for efficient SARS-CoV-2 production. Our study provides a novel mechanism whereby SARS-CoV-2 outsmarts the host antiviral response and should aid in the development of COVID-19 therapy.
Conflict of interest
Qiang Ding and Xiaohui Ju have filed a patent application on the use of the SARS-CoV-2 transcomplementation system and its use for anti-SARS-CoV-2 drug screening (2020109679884). Other authors declare that they have no conflict of interest.
Acknowledgments
We thank all members of Huang lab for helpful discussions, Molecular Imaging Core Facility and Molecular and Cellular Biology Core Facility for support with imaging and cellular biology experiments, and Dr. Yichang Jia at Tsinghua University for providing arsenite. This work was supported by the National Natural Science Foundation of China (81830004, 31970755, and 31970173) and the Local Grant (608285568031).
Author contributions
Yu Zhang, Xingxu Huang, Cheng Huang, and Zhigang Jin conceived and supervised the project; Yu Zhang, Lingling Luo, Zhean Li, Tiejun Zhao, Boxing Jin, Yulin Zhou, Su He, and Jinhua Huang designed and performed the bulk of the experiments and analyzed nearly all experimental data with the assistances of Yan Zou, Peixiang Ma, Aibin Liang, Ping Li, Jia Liu, and Tian Chi; Xiaohui Ju, Xun Xu, and Qiang Ding designed and performed the experiment of viral production; Yu Zhang, Lingling Luo, and Zhean Li wrote the primary draft of manuscript, and all authors contributed to the final version.
Biographies

Lingling Luo is a graduate student under the supervision of Prof. Cheng Huang at School of Pharmacy, Shanghai University of Traditional Chinese Medicine. She is interested in the pathogenesis of SARS-CoV-2 and anti-SARS-CoV-2 drug screening.

Zhean Li is a postdoctoral researcher at the Shanghai Institute for Advanced Immunochemical Studies, ShanghaiTech University. His research interest focuses on the biological function of intracellular phase transitions and developing physical tools to manipulate the phase transitions process in vivo.

Tiejun Zhao serves as a principal investigator at the College of Chemistry and Life Sciences, Zhejiang Normal University. His research interest focuses on the pathological and oncogenic function of viral protein and signalling pathway in adult T-cell leukemia and virus-associated diseases.

Xiaohui Ju is a Ph.D. candidate under the supervision of Dr. Qiang Ding at School of Medicine, Tsinghua University. He is interested in the pathogenesis of Hepatitis E virus, SARS-CoV-2, and developing antivirals.

Xingxu Huang is a professor at School of Life Science and Technology, ShanghaiTech University. His research interest focuses on developing genome editing and base editing approaches for linkage of the genotype to phenotype and therapeutic purpose.

Qiang Ding is an assistant professor at Tsinghua University. He is interested in the establishment of novel experimental systems to dissect virus life cycle and virus-host interaction, which is critical for determining virus host range and discovery of novel antiviral targets.

Zhigang Jin serves as a principal investigator at the College of Chemistry and Life Sciences, Zhejiang Normal University. His research interest focuses on the physiological and pathological significance of phase separation in embryonic development, tumorigenesis, and viral diseases.

Cheng Huang is a full professor at School of Pharmacy, Shanghai University of Traditional Chinese medicine. He received his Ph.D. degree from Shanghai University of Traditional Chinese Medicine in 1997 and had two sessions of postdoc trainings from the Chinese Academy of Sciences and Baylor College of Medicine. He devotes his research to pharmacology and drug discovery.

Yu Zhang is a assistant professor at School of Life Science and Technology, ShanghaiTech University. Her research interest focuses on the role of phase separation in physiology and disease.
Footnotes
Supplementary materials to this article can be found online at https://doi.org/10.1016/j.scib.2021.01.013.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Wu F., Zhao S., Yu B. A new coronavirus associated with human respiratory disease in china. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang C., Horby P.W., Hayden F.G. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao B., Ni C., Gao R. Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell. 2020;11:771–775. doi: 10.1007/s13238-020-00718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas M.G., Loschi M., Desbats M.A. RNA granules: the good, the bad and the ugly. Cell Signal. 2011;23:324–334. doi: 10.1016/j.cellsig.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchan J.R., Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lloyd R.E. Regulation of stress granules and p-bodies during RNA virus infection. Wiley Interdiscip Rev RNA. 2013;4:317–331. doi: 10.1002/wrna.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson P., Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 11.Onomoto K., Yoneyama M., Fung G. Antiviral innate immunity and stress granule responses. Trends Immunol. 2014;35:420–428. doi: 10.1016/j.it.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCormick C., Khaperskyy D.A. Translation inhibition and stress granules in the antiviral immune response. Nat Rev Immunol. 2017;17:647–660. doi: 10.1038/nri.2017.63. [DOI] [PubMed] [Google Scholar]
- 13.Lloyd R.E. How do viruses interact with stress-associated RNA granules? PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White J.P., Lloyd R.E. Regulation of stress granules in virus systems. Trends Microbiol. 2012;20:175–183. doi: 10.1016/j.tim.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Visser L.J., Medina G.N., Rabouw H.H. Foot-and-mouth disease virus leader protease cleaves G3BP1 and G3BP2 and inhibits stress granule formation. J Virol. 2019;93:e00922–18. doi: 10.1128/JVI.00922-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White J.P., Cardenas A.M., Marissen W.E. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe. 2007;2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr Top Microbiol Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang S., Yang M., Hong Z. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm Sin B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagawa K., Lokugamage K.G., Makino S. Viral and cellular mRNA translation in coronavirus-infected cells. Adv Virus Res. 2016;96:165–192. doi: 10.1016/bs.aivir.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wurm T., Chen H., Hodgson T. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J Virol. 2001;75:9345–9356. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.You J., Dove B.K., Enjuanes L. Subcellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein. J Gen Virol. 2005;86:3303–3310. doi: 10.1099/vir.0.81076-0. [DOI] [PubMed] [Google Scholar]
- 23.Cascarina S.M., Ross E.D. A proposed role for the SARS-CoV-2 nucleocapsid protein in the formation and regulation of biomolecular condensates. FASEB J. 2020;34:9832–9842. doi: 10.1096/fj.202001351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon D.E., Jang G.M., Bouhaddou M. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Y., Protter D.S., Rosen M.K. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kedersha, N., Panas M.D., Achorn C.A. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol. 2016;212:845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilks N., Kedersha N., Ayodele M. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson P., Kedersha N. Stress granules. Curr Biol. 2009;19:R397–398. doi: 10.1016/j.cub.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Molliex A., Temirov J., Lee J. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin S.Y., Liu C.L., Chang Y.M. Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target. J Med Chem. 2014;57:2247–2257. doi: 10.1021/jm500089r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C.Y., Chang C.K., Chang Y.W. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J Mol Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang P., Mathieu C., Kolaitis R.M. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell. 2020;181:325–345.e28. doi: 10.1016/j.cell.2020.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guillen-Boixet J., Kopach A., Holehouse A.S. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell. 2020;181:346–361.e17. doi: 10.1016/j.cell.2020.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanders D.W., Kedersha N., Lee D.S.W. Competing protein-RNA interaction networks control multiphase intracellular organization. Cell. 2020;181:306–324.e28. doi: 10.1016/j.cell.2020.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Q., Yu L., Petros A.M. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 36.Fan H., Ooi A., Tan Y.W. The nucleocapsid protein of coronavirus infectious bronchitis virus: crystal structure of its N-terminal domain and multimerization properties. Structure. 2005;13:1859–1868. doi: 10.1016/j.str.2005.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan Y.W., Fang S., Fan H. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006;34:4816–4825. doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo H., Ye F., Chen K. SR-rich motif plays a pivotal role in recombinant SARS coronavirus nucleocapsid protein multimerization. Biochemistry. 2005;44:15351–15358. doi: 10.1021/bi051122c. [DOI] [PubMed] [Google Scholar]
- 39.Reineke L.C., Kedersha N., Langereis M.A. Stress granules regulate double-stranded RNA-dependent protein kinase activation through a complex containing G3BP1 and Caprin1. mBio. 2015;6 doi: 10.1128/mBio.02486-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solomon S., Xu Y., Wang B. Distinct structural features of Caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2α, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol Cell Biol. 2007;27:2324–2342. doi: 10.1128/MCB.02300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ju X., Zhu Y., Wang Y. A novel cell culture system modeling the SARS-CoV-2 life cycle. PLoS Pathog. 2021;17:e1009439. doi: 10.1371/journal.ppat.1009439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fros J.J., Domeradzka N.E., Baggen J. Chikungunya virus nsP3 blocks stress granule assembly by recruitment of G3BP into cytoplasmic foci. J Virol. 2012;86:10873–10879. doi: 10.1128/JVI.01506-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frolova E., Gorchakov R., Garmashova N. Formation of nsP3-specific protein complexes during sindbis virus replication. J Virol. 2006;80:4122–4134. doi: 10.1128/JVI.80.8.4122-4134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu Z., Wang Y., Tang Q. Inclusion bodies of human parainfluenza virus type 3 inhibit antiviral stress granule formation by shielding viral RNAs. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng Q., Wang L., Qin Z. Phase separation of Epstein-Barr virus EBNA2 and its coactivator EBNALP controls gene expression. J Virol. 2020;94:e01771–19. doi: 10.1128/JVI.01771-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heinrich B.S., Maliga Z., Stein D.A. Phase transitions drive the formation of vesicular stomatitis virus replication compartments. mBio. 2018;9:e02290–17. doi: 10.1128/mBio.02290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guseva S., Milles S., Jensen M.R. Measles virus nucleo- and phosphoproteins form liquid-like phase-separated compartments that promote nucleocapsid assembly. Sci Adv. 2020;6:eaaz7095. doi: 10.1126/sciadv.aaz7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alenquer M., Vale-Costa S., Etibor T.A. Influenza A virus ribonucleoproteins form liquid organelles at endoplasmic reticulum exit sites. Nat Commun. 2019;10:1629. doi: 10.1038/s41467-019-09549-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monette A., Niu M., Chen L. Pan-retroviral nucleocapsid-mediated phase separation regulates genomic RNA positioning and trafficking. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.03.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fang X., Wang L., Ishikawa R. Arabidopsis FLL2 promotes liquid-liquid phase separation of polyadenylation complexes. Nature. 2019;569:265–269. doi: 10.1038/s41586-019-1165-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hnisz D., Shrinivas K., Young R.A. A phase separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng T.Y., Lee K.R., Tarn W.Y. Phosphorylation of the arginine/serine dipeptide-rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. FEBS J. 2008;275:4152–4163. doi: 10.1111/j.1742-4658.2008.06564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Panas M.D., Varjak M., Lulla A. Sequestration of G3BP coupled with efficient translation inhibits stress granules in Semliki Forest virus infection. Mol Biol Cell. 2012;23:4701–4712. doi: 10.1091/mbc.E12-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.