Thaller et al. demonstrate that direct binding between phosphatidic acid (PA) and the ESCRT Chm7 is required for nuclear envelope surveillance; PA also accumulates at nuclear envelope herniations. Thus, tight control of PA metabolism is required for nuclear envelope homeostasis.
Abstract
Mechanisms that control nuclear membrane remodeling are essential to maintain the integrity of the nucleus but remain to be fully defined. Here, we identify a phosphatidic acid (PA)–binding capacity in the nuclear envelope (NE)–specific ESCRT, Chm7, in budding yeast. Chm7’s interaction with PA-rich membranes is mediated through a conserved hydrophobic stretch of amino acids, which confers recruitment to the NE in a manner that is independent of but required for Chm7’s interaction with the LAP2-emerin-MAN1 (LEM) domain protein Heh1 (LEM2). Consistent with the functional importance of PA binding, mutation of this region abrogates recruitment of Chm7 to membranes and abolishes Chm7 function in the context of NE herniations that form during defective nuclear pore complex (NPC) biogenesis. In fact, we show that a PA sensor specifically accumulates within these NE herniations. We suggest that local control of PA metabolism is important for ensuring productive NE remodeling and that its dysregulation may contribute to pathologies associated with defective NPC assembly.
Introduction
The nuclear envelope (NE) provides a selective barrier that ensures the integrity of nuclear-cytoplasmic compartmentalization. Disruption of NE function, either by perturbation of the nuclear membranes or by inhibition of the assembly of nuclear pore complexes (NPCs), is detrimental to cell viability, can result in losses in genome integrity, and can cause disease (Hatch and Hetzer, 2014; Lusk and King, 2017). Cells have evolved protective mechanisms that surveil and ameliorate disruptions of the nuclear compartment, including defective de novo NPC biogenesis (Thaller and Lusk, 2018; Lord and Wente, 2020).
NPC biogenesis during interphase occurs through an inside-out mechanism that begins at the inner nuclear membrane (INM; Otsuka et al., 2016). Although the molecular steps in NPC assembly remain obscure, a key event is the fusion of the INM and outer nuclear membrane (ONM). The mechanism of INM–ONM fusion is unknown, but local changes in lipid metabolism may play a pivotal role (Schneiter et al., 1996; Siniossoglou et al., 1998; Scarcelli et al., 2007; Hodge et al., 2010; Lone et al., 2015; Zhang et al., 2018). For example, the formation of aberrant NE blebs or herniations over malformed NPCs (Wente and Blobel, 1993; Scarcelli et al., 2007; Onischenko et al., 2017; Zhang et al., 2018; Thaller et al., 2019; Rampello et al., 2020; Allegretti et al., 2020) has been associated with perturbations in lipid metabolism (Schneiter et al., 1996; Hodge et al., 2010; Grillet et al., 2016) and is likely caused by defects in INM–ONM fusion and/or triggering of an NE surveillance pathway that monitors the integrity of nuclear-cytoplasmic compartmentalization (Webster et al., 2014, 2016; Thaller et al., 2019).
The principal components of the NE surveillance pathway are the endosomal sorting complexes required for transport (ESCRT) and integral INM proteins of the LAP2-emerin-MAN1 (LEM) family, including Heh1 in budding yeast (also known as Src1; LEM2 in higher eukaryotes; Lusk and Ader, 2020). The ESCRT Chm7 (orthologue of CHMP7) has nuclear export sequences (NESs) that restrict its access to the nucleus and Heh1/LEM2 (Thaller et al., 2019; Vietri et al., 2020). Perturbations to the nuclear transport system or nuclear membranes are predicted to disrupt the spatial segregation of Chm7 and Heh1, which leads to their binding and the subsequent activation of an ESCRT-dependent NE repair pathway (Thaller et al., 2019). The molecular mechanisms of NE repair remain obscure, but as ESCRTs also seal the NE at the end of an open mitosis (Olmos et al., 2015; Vietri et al., 2015; Gu et al., 2017; Ventimiglia et al., 2018; Willan et al., 2019; Warecki et al., 2020; Pieper et al., 2020), it seems likely that a Chm7–Heh1 partnership leads to the closure of nuclear pores through an ill-defined annular fusion event. As the hyperactivation of this pathway with gain-of-function mutants of Chm7 leads to aberrant membrane remodeling and expansion at the INM (Thaller et al., 2019; Vietri et al., 2020), there is an interesting relationship between changes in local lipid synthesis and membrane repair that needs to be mined.
Indeed, in Caenorhabditis elegans, there are functional interactions between ESCRT genes and regulators of phosphatidic acid (PA) metabolism (Penfield et al., 2020). As PA is a major precursor of both phospholipids and triacylglycerol, this pathway decides whether to generate phospholipids for membrane growth or to store fatty acids in lipid droplets (Carman and Han, 2011), some of which have been shown to grow from the INM (Romanauska and Köhler, 2018). Further, PA can destabilize membranes (Kooijman et al., 2003; Frey et al., 2006; Kwolek et al., 2015; Dymond, 2016), which can be deleterious but can also be leveraged to promote membrane fusion reactions (Zhukovsky et al., 2019). As such, PA levels are tightly controlled; whether changes in local PA can directly impact NE remodeling events remains unknown.
In our efforts to determine the mechanism of NE surveillance, we discovered that Chm7 directly binds membranes rich in PA. The data are consistent with a model in which PA binding is upstream of and required for Chm7’s interaction with Heh1. Mutation of a hydrophobic stretch that can be modeled as an amphipathic helix disrupts PA binding and Chm7’s function in the context of NPC misassembly-induced NE herniations. Further, we demonstrate that these herniations contain high levels of a PA sensor, suggesting the local accumulation of PA is both a physiological signal for Chm7 recruitment and a potential pathological signature of NPC misassembly. Thus, local control of PA at the NE is a key feature of essential NE remodeling events.
Results and discussion
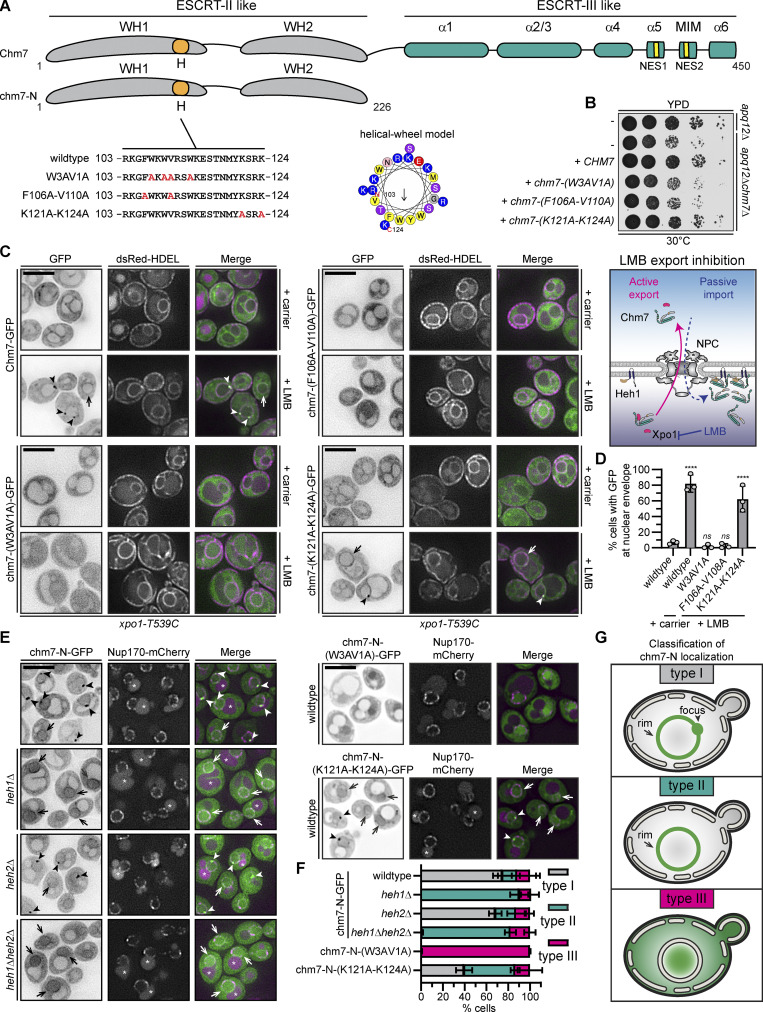
A conserved hydrophobic stretch is required for Chm7 function
Several functional domains have been identified within Chm7 that contribute to its localization at steady state and when NE surveillance is triggered (Olmos et al., 2016; Webster et al., 2016; Gu et al., 2017; Thaller et al., 2019; von Appen et al., 2020; Vietri et al., 2020; Gatta et al., 2020 Preprint). Of interest is a hydrophobic stretch (“H” in Fig. 1 A) first identified in the human protein, where it imparts binding to the ER (Olmos et al., 2016). This sequence is conserved in Chm7 but is less hydrophobic than its human counterpart (Fig. S1, A and B); it can be modeled as an amphipathic helix (Fig. 1 A and Fig. S1 B). To test its function, we generated three alleles (Fig. 1 A) where codons for either hydrophobic [chm7-(W3AV1A) and chm7-(F106A-V110A)] or charged [chm7-(K121A-K124A)] amino acid residues were mutated to encode alanine. We assessed the function of these alleles in apq12Δchm7Δ strains in which NE surveillance is required for growth in the context of NPC misassembly-induced NE herniations (Webster et al., 2016). Consistent with the conclusion that the hydrophobic amino acids conferred an important function, neither chm7-(W3AV1A) nor chm7-(F106A-V110A) could rescue apq12Δchm7Δ growth comparably to CHM7 or chm7-(K121A-K124A) (Fig. 1 B). Thus, the hydrophobic character of this stretch of amino acids is critical for Chm7 function.
Figure 1.
Chm7 interacts with the INM through a conserved hydrophobic stretch of amino acids. (A) Schematic of Chm7 and chm7-N with predicted winged helices (WHs), hydrophobic stretch (H), α helices (α), MIT-interacting motif (MIM), and NESs. Amino acid sequences encoded by alleles (with names at left) of CHM7 with amino acid changes indicated in red in the H sequence. In the middle is a helical wheel model of H generated with HeliQuest (Gautier et al., 2008). Numbers are amino acid positions of N- and C-terminal ends of the helix, and arrow indicates the hydrophobic moment. (B) The hydrophobic stretch is required for Chm7 function in the context of NPC misassembly. 10-fold serial dilutions of the indicated strains with indicated CHM7 alleles plated and grown on YPD. (C) The hydrophobic stretch is required for the focal accumulation of Chm7 at the INM. Deconvolved fluorescence micrographs of Chm7-GFP and indicated chm7-GFP proteins (fluorescence inverted), dsRED-HDEL (demarking the NE/ER), and merged images in the LMB-sensitive strain (xpo1-T539C) after treatment with carrier (MeOH) or LMB. Arrowheads and arrows point to focal and evenly distributed nuclear rim GFP fluorescence, respectively, at the NE. Scale bar, 5 µm. On the right is a cartoon interpretation of Chm7 localization upon LMB-induced inhibition of Xpo1. The top of the diagram is the cytosol. (D) Plot of the percentage of cells where Chm7-GFP or the indicated chm7-GFP mutants accumulate at the NE after treatment with carrier (only wild type shown) and LMB. Average and SD are shown from three independent experiments where >50 cells were counted per replicate. P values are from a one-way ANOVA with multiple comparisons and Dunnett’s post hoc test calculated compared against controls treated with carrier alone, where ns (not significant) is P > 0.05; ****, P ≤ 0.0001. (E) The hydrophobic stretch is required for the Heh1/2-independent association of chm7-N with the NE. Deconvolved fluorescence micrographs of chm7-N-GFP, chm7-N-(W3AV1A)-GFP, or chm7-N-(K121A-K124A)-GFP (GFP signal inverted) in the indicated strains with Nup170-mCherry and merged images shown. Arrowheads and arrows point to focal and evenly distributed nuclear rim GFP fluorescence, respectively, at the NE. Asterisks indicate vacuolar autofluorescence. Scale bar, 5 µm. (F) Plot of the percentage of cells with mean and SD from three independent experiments with 50 cells/experiment counted with type I, type II, or type III chm7-N-GFP distributions as diagrammed in the classification scheme in G. (G) Cartoons of a phenotypic classification scheme to help interpret chm7-N-GFP subcellular distributions in E.
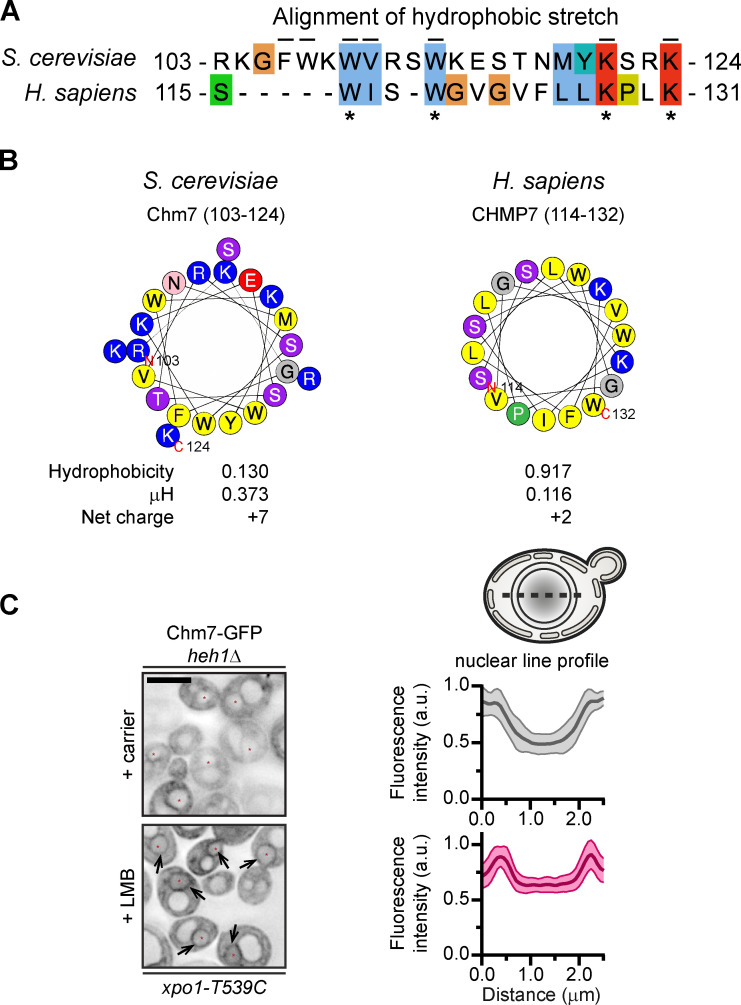
Figure S1.
Conserved hydrophobic stretch within Chm7 can be modeled as an amphipathic helix, and Chm7 accumulates at the INM without Heh1 (supplemental to Fig. 1). (A) Alignment of hydrophobic stretches in budding yeast and human. Color coding of properties of amino acids with conserved attributes as in ClustalW (Larkin et al., 2007), where hydrophobic residues are blue, positive-charged residues are red, polar residues are green, glycines are orange, prolines are yellow, and aromatic residues are cyan. Numbers are amino acids. Asterisks denote conserved residues; black lines are residues altered in mutational analysis in this study for Chm7 (top). (B) Helical wheel diagrams in which the letters are amino acids. Physicochemical properties were calculated by using HeliQuest (Gautier et al., 2008). Residues are color coded where hydrophobic residues are yellow, basic residues are blue, serine and threonine are purple, acidic residues are red, asparagine is pink, glycine is gray, and proline is green. (C) Chm7 accumulates at the INM in the absence of Heh1. Deconvolved inverted fluorescence micrographs of overexpressed Chm7-GFP in an heh1Δ LMB-sensitive strain (xpo1-T539C) after treatment with carrier (MeOH) or LMB. Arrows point out Chm7-GFP that accumulates at the nuclear rim specifically under conditions of LMB treatment. Scale bar, 5 µm. At right are line profiles bisecting the nucleus of carrier and LMB-treated cells where the mean Chm7-GFP fluorescence intensity (in a.u.) is plotted. The mean (normalized to max) is shown as thick lines and SD as thin lines from 20 randomly selected cells. Gray is the + carrier (control) treatment, and magenta is the +LMB treatment.
We next tested whether mutations of the hydrophobic stretch impacted Chm7’s recruitment to membranes. As Chm7 is predominantly cytosolic, we tested membrane association by inhibiting its nuclear export, which allows Chm7 access to the INM, where it binds to Heh1 (Fig. 1 C, schematic; Thaller et al., 2019). Specifically, we assessed the localization of Chm7-GFP and chm7-GFP mutants after inhibition of Xpo1 with leptomycin B (LMB; Neville and Rosbash, 1999); dsRED-HDEL was used as an ER marker (Madrid et al., 2006). Consistent with our prior work (Thaller et al., 2019), in 80% of cells, LMB treatment results in the relocalization of Chm7-GFP from the cytosol to the nuclear rim (Fig. 1 C, arrows), where it often accumulates in foci (Fig. 1 C, arrowheads; and Fig. 1 D). In striking contrast, chm7-(W3AV1A)-GFP and chm7-(F106A-V110A)-GFP accessed the nucleoplasm but failed to accumulate at the INM (Fig. 1, C and D). This effect was specific for the hydrophobic mutations, as chm7-(K121A-K124A)-GFP accumulated at the INM, albeit in fewer cells compared with Chm7-GFP (∼60%; Fig. 1 D). Thus, hydrophobic amino acids in the hydrophobic stretch are required for Chm7 to bind the INM.
The hydrophobic stretch confers binding to the NE independently of Heh1
Mutation of the hydrophobic residues could disrupt interactions with Heh1 or a previously undiscovered membrane-binding mechanism. We favored the latter idea, as Chm7-GFP can still accumulate at the NE in heh1Δ cells treated with LMB (Fig. S1 C). To evaluate this more directly, we visualized chm7-N-GFP (Fig. 1 A), which lacks the ESCRT-III domain and NESs and constitutively accesses the nucleus and INM. We observed two distinct localizations of chm7-N-GFP at the NE: a focal accumulation (Fig. 1 E, arrowheads) and a dispersed nuclear rim fluorescence (Fig. 1 E, arrows). We asked if these two distributions were dependent on Heh1 or its paralogue Heh2. We categorized the localizations as type I (nuclear rim and foci), type II (nuclear rim), and type III (no membrane association; Fig. 1 G). Interestingly, deletion of HEH1 shifted the distribution from type I to type II (Fig. 1, E and F). This result was specific for heh1Δ, as deletion of HEH2 had no impact on either distribution with ∼70% of cells classified as type I (Fig. 1, E and F). Interestingly, this remaining NE binding was disrupted by mutating the hydrophobic stretch, as chm7-N-(W3AV1A)-GFP failed to accumulate at the NE, with virtually all cells classified as type III (Fig. 1, E and F). Thus, the hydrophobic stretch contributes to a specific NE-binding activity of Chm7 that is independent of the LEM proteins.
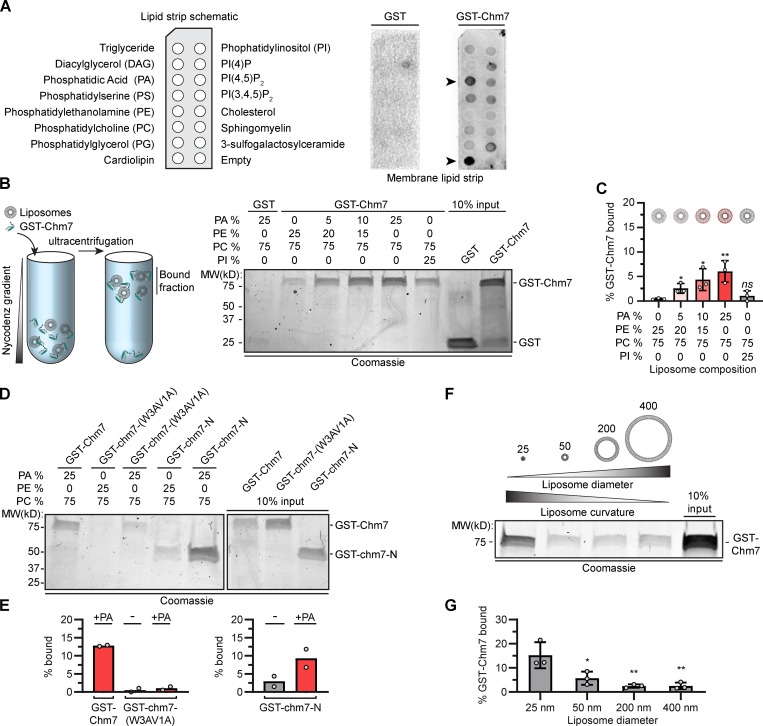
Chm7 binds directly to PA-rich lipid bilayers
We considered a model in which Chm7 directly binds to membrane lipids to explain the remaining NE-binding activity. We generated recombinant GST-Chm7 and evaluated binding to a lipid strip containing a spectrum of phospholipids (Fig. 2 A). Compared with GST alone, GST-Chm7 detected anionic lipids, but most noticeably PA and cardiolipin (Fig. 2 A). As cardiolipin is found in mitochondria, we focused on the possibility that Chm7 might directly bind to PA. To test lipid binding in a more physiological environment, we incubated GST-Chm7 with liposomes generated with increasing concentrations of PA, from 5% to 25%. These liposome–protein mixtures were overlaid with a Nycodenz gradient and subjected to ultracentrifugation (Fig. 2 B). Under these conditions, liposomes with bound proteins float; floated fractions are then separated by SDS-PAGE and visualized with Coomassie staining. Gratifyingly, as we increased the amount of PA in the liposomes, we observed a commensurate increase of GST-Chm7 in bound fractions, suggesting that Chm7 binds directly to PA or at least to PA-rich membranes (Fig. 2, B and C). This conclusion is further supported by the observation that Chm7 shows a weaker preference for PI-rich liposomes (that share a similar charge as PA) and for PE-rich liposomes (that share a similar cone shape; Fig. 2, B and C; van Meer et al., 2008). Consistent with the idea that these membrane interactions are mediated by the hydrophobic stretch, GST-chm7-N also shows perhaps more efficient binding to PA-rich liposomes, and GST-chm7-(W3AV1A) binding is clearly reduced (Fig. 2, D and E).
Figure 2.
Chm7 binds directly to PA-rich lipid membranes. (A) GST-Chm7 preferentially binds to PA and cardiolipin. At left is a schematic showing the location of lipids immobilized on membranes (lipid strip). These lipid strips were probed with either GST (middle) or GST-Chm7 (right). Specific binding was detected with anti-GST primary antibodies followed by HRP-conjugated secondary antibodies and ECL. Position of PA and cardiolipin is indicated with arrowheads. (B) GST-Chm7 specifically binds PA-rich liposomes. At left is a schematic of liposome flotation assay in which proteins and liposomes of defined lipid compositions are mixed, overlaid with a Nycodenz gradient, and then subjected to ultracentrifugation. Fractions that float (bound) are separated by SDS-PAGE, and proteins are detected with Coomassie staining; all liposomes are composed of 75% phosphatidylcholine (PC) with the indicated percentages of PA, phosphatidylethanolamine (PE), and phosphatidylinositol (PI). Numbers indicate the position of molecular weight (MW) standards in kD. (C) Plot of the percentage (relative to input) of GST-Chm7 bound to liposomes with increasing PA concentration as indicated. Error bars are SD from three independent experiments. P values were calculated from a one-way ANOVA with Tukey’s post hoc test where ns (not significant) is P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01. (D) Mutations in the hydrophobic stretch of Chm7 diminish binding to PA-rich membranes. Coomassie-stained gel of liposome flotation as in B but incorporating GST-chm7-N and GST-chm7-(W3AV1A). (E) Plot of the percentage of GST-Chm7, GST-chm7-(W3AV1A), and GST-chm7-N (relative to input) bound to liposomes with (+) or without (-) PA as in D. Average from two independent replicates (denoted as circles) is plotted. (F) Chm7 prefers high curvature. Coomassie-stained gel of liposome flotation and analysis as in B but changing liposome diameter as indicated above the liposome diagrams (in nm). All liposomes in this experiment are composed of 5% PA, 20% PE, and 75% PC. (G) Plot of the percentage (relative to input) of GST-Chm7 from experiments as in F. Error bars are SD from three independent experiments. P values were calculated from a one-way ANOVA with Tukey’s post hoc test where *, P ≤ 0.05; **, P ≤ 0.01.
We next assessed whether membrane curvature impacted Chm7’s interaction with lipids. We generated liposomes with different diameters (from ∼400 nm to ∼25 nm) and tested their ability to float GST-Chm7. We chose a low (5%) PA concentration to favor the specific evaluation of curvature sensitivity over PA binding. As shown in Fig. 2, F and G, GST-Chm7 preferred ∼25-nm diameter liposomes over those that were essentially flat (i.e., ∼200 and ∼400 nm). Thus, taken together, both lipid composition and curvature contribute to the efficiency by which Chm7 is recruited to membranes in vitro.
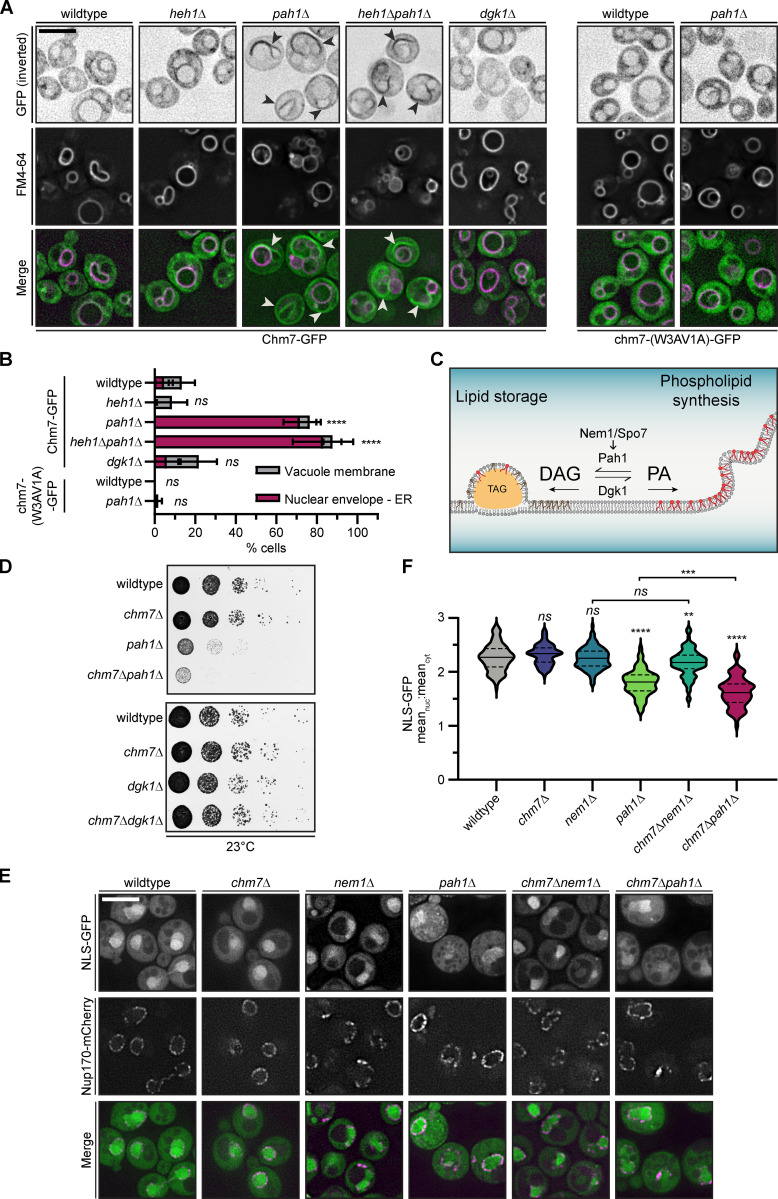
Cellular changes to PA levels alter Chm7 localization and disrupt nuclear-cytoplasmic compartmentalization
As Chm7 bound to membranes rich in PA in vitro, we tested how changing cellular PA levels impacted Chm7 distribution in vivo. We moderately overexpressed Chm7-GFP in strains where PA levels were elevated approximately twofold (pah1Δ; Han et al., 2006) or reduced (dgk1Δ; Han et al., 2008; Adeyo et al., 2011). pah1Δ cells also have elevated PA specifically at the INM (Romanauska and Köhler, 2018) and aberrant nuclear morphology (Santos-Rosa et al., 2005). Consistent with the in vitro binding data, only increasing PA levels led to a redistribution of Chm7-GFP from the cytosol (with a minor vacuole membrane localization; Fig. 3, A and B) to the NE/ER (arrowheads in Fig. 3 A; colocalization with dsRED-HDEL in Fig. S2 A) in ∼70% of cells (Fig. 3, A and B). Interestingly, these Chm7-rich membranes were closely apposed to vacuoles labeled with FM4-64 (Fig. 3 A). To ensure that the recruitment of Chm7-GFP was not a result of triggering NE surveillance and/or the mislocalization of Heh1, we visualized Chm7-GFP in heh1Δpah1Δ cells. As shown in Fig. 3, A and B and Fig. S2 A, loss of HEH1 had no impact on Chm7-GFP, with ∼80% of cells retaining NE/ER localization. These data thus echo chm7-N localization and lend support to the idea that Chm7 retains a membrane-binding activity distinct from its interaction with Heh1 through the hydrophobic stretch. Consistent with the latter conclusion, chm7-(W3AV1A)-GFP did not associate with any membranes in the context of elevated PA in the pah1Δ strain (Fig. 3, A and B; and S2 A).
Figure 3.
Elevating cellular PA levels leads to the recruitment of Chm7 to NE/ER membranes independently of Heh1. (A) The recruitment of Chm7 to NE/ER membranes in the context of elevated cellular PA is dependent on the hydrophobic stretch and not Heh1. Deconvolved fluorescence micrographs of Chm7-GFP (top left panels) or chm7-(W3AV1A)-GFP (top right panels) with vacuole membranes stained with FM4-64 and merged images. GFP fluorescence is inverted. Arrowheads point to Chm7-GFP at NE/ER membranes as confirmed in Fig. S2 A. Scale bar, 5 µm. (B) Plot of the percentage of cells from experiments as in A showing the relative distribution of Chm7-GFP or chm7-W3AV1A-GFP between NE/ER and vacuole membranes in the indicated genetic backgrounds. The mean and SD from three independent experiments of >50 cells is shown. P values were calculated from a two-way ANOVA with Dunnett’s post hoc test where ns (not significant) is P > 0.05; ****, P < 0.0001. (C) Cartoon of the PA metabolism pathway, which can be modulated to control both lipid storage and membrane growth as shown. Nem1/Spo7 dephosphorylate and activate Pah1, which in turn dephosphorylates PA to produce DAG. Thus, deletion of PAH1 and NEM1 elevates cellular PA levels. Dgk1 phosphorylates DAG to produce PA; therefore, there is less PA in dgk1Δ cells. TAG is triacylglycerol lipids stored in lipid droplets. Brown lipids represent DAG, and red lipids represent PA. (D) CHM7 is required to maintain fitness of pah1Δ cells. 10-fold serial dilutions of the indicated strains grown at 23°C for at least 48 h before imaging. (E) Nuclear-cytoplasmic compartmentalization is perturbed in pah1Δ and chm7Δpah1Δ cells. Deconvolved fluorescence micrographs of NLS-GFP, Nup170-mCherry, and merged images in the indicated strains cultured at 23°C. Scale bar, 5 µm. (F) Violin plot of the distribution of the mean nuclear (nuc) to cytosolic (cyt) fluorescence intensity of individual cells with median (solid line) quartiles (dotted line) from three independent experiments (50 cells/strain/experiment) of an NLS-GFP reporter from cells of the indicated genotype. The width of each plot represents the relative frequency distribution of cells. P values were calculated from a one-way ANOVA with Tukey’s post hoc test where ns is P > 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Figure S2.
Elevation of cellular PA redistributes Chm7-GFP to envelope/ER membranes (supplemental to Fig. 4). (A) Elevation of cellular PA levels leads to Chm7 recruitment to NE/ER membranes dependent on the hydrophobic stretch. Localization of Chm7-GFP or chm7-W3A1VA-GFP in the indicated strains with the NE and ER membranes visualized with dsRED-HDEL. GFP (inverted fluorescence), dsRED, and merged images are shown. Arrowheads point to colocalization of Chm7-GFP with NE/ER membranes. Scale bar, 2 µm. (B) CHM7 is required for fitness of pah1Δ but not dgk1Δ strains. 10-fold serial dilutions of the indicated strains grown on YPD plates at 30°C or 37°C for 48 h before imaging. (C) CHM7 is dispensable for the fitness of nem1Δ strains. 10-fold serial dilutions of the indicated strains grown on yeast extract peptone dextrose plates at the indicated temperatures for 48 h before imaging. (D) The NLS-DAG mCherry sensor does not focally accumulate at the NE in wild-type strains. Deconvolved inverted fluorescence micrographs of NLS-DAG sensor-mCherry at 23°C or at 37°C. Red asterisks demark nuclei. Numbers are mean percentage and SD of cells from three replicates of >50 cells per replicate with the appearance of NLS-DAG sensor-mCherry foci at the NE. Scale bar, 5 µm.
We next tested epistasis between PA metabolism genes (PAH1, DGK1, and NEM1; Fig. 3 C) and CHM7. As shown in Figs. 3 D and S2 B, while pah1Δ cells grew slower than wild-type cells at all temperatures, we observed an additional loss of fitness when CHM7 was deleted. These results were specific for PAH1 nulls, as CHM7 deletion did not impact dgk1Δ or nem1Δ growth (Fig. 3 D; and Fig. S2, B and C). We note that deletion of NEM1 would be predicted to elevate PA levels (Fig. 3 C), but this may not be the case (Bircham et al., 2020 Preprint). To gain insight into the underlying cause of the loss of fitness of chm7Δpah1Δ cells, we tested the distribution of a nuclear localization signal (NLS)–GFP reporter. We observed that pah1Δ cells, and in particular chm7Δpah1Δ cells, were unable to effectively accumulate NLS-GFP in the nucleus (Fig. 3, E and F). This was visible by relating the mean nuclear (N) and cytosolic (C) fluorescence as an N:C ratio, which was 2.3 in wild type, 1.8 in pah1Δ, and 1.6 in chm7Δpah1Δ cells (Fig. 3 F). A similar but less significant trend was observed in the N:C ratios of nem1Δ and chm7Δnem1Δ cells (Fig. 3 F). Therefore, Chm7 is protective of nuclear-cytoplasmic compartmentalization in the context of elevated PA. As increasing PA can disrupt membrane integrity (Kooijman et al., 2003; Kwolek et al., 2015; Dymond, 2016), we suggest that these results cumulatively point to an instability in the nuclear membranes, which fits well with data indicating that changes to lipid metabolism negatively impact NE integrity (Kinugasa et al., 2019; Barbosa et al., 2019; Lee et al., 2020).
Chm7 hyperactivation drives local PA sensor accumulation at the INM
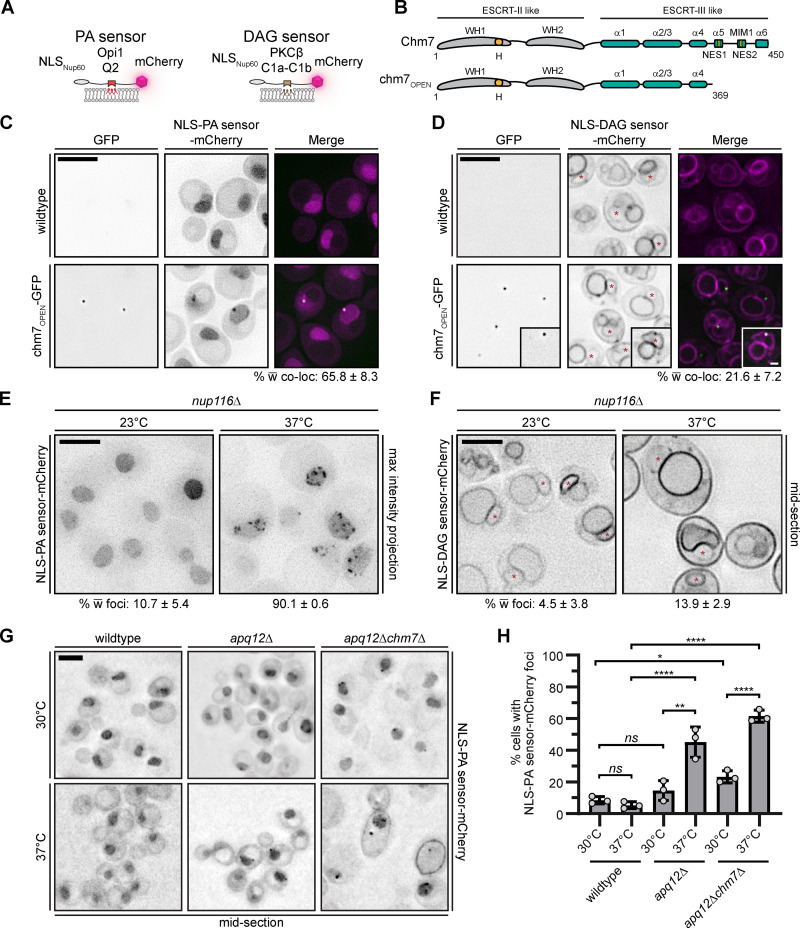
To further explore the relationship between Chm7 and PA, we tested whether PA accumulated at sites of Chm7 activation. We took advantage of fluorescent PA and diacylglycerol (DAG) sensors with an NLS, which allows monitoring of these lipids at the INM (Fig. 4 A; Romanauska and Köhler, 2018). As there is little PA at the INM under normal conditions, the PA sensor localizes throughout the nucleoplasm (Fig. 4 C, top panels; Romanauska and Köhler, 2018). In contrast, the DAG sensor localizes to the INM and can also be visualized at other membranes throughout the cell (Fig. 4 D, top panels; Romanauska and Köhler, 2018).
Figure 4.
Chm7 hyperactivation and NPC misassembly lead to local PA sensor accumulation at the INM. (A) Diagram of the PA and DAG sensors used to visualize the localization of PA and DAG in cells. (B) chm7OPEN is a dominant “activated” form of Chm7 that accumulates at the INM (Thaller et al., 2019). Schematic of Chm7 and chm7OPEN with predicted winged helices (WHs), hydrophobic stretch (H), α helices (α), MIT-interacting motif type 1 (MIM1), and NESs. Numbers denote amino acids. (C) The PA sensor specifically accumulates at sites of Chm7 hyperactivation. Deconvolved fluorescence micrographs of cells with wild-type CHM7 (no GFP) or chm7OPEN-GFP expressed with an NLS-PA sensor-mCherry. GFP (inverted), mCherry (inverted), and merged images are shown. Numbers are the mean percentage and SD of cells with colocalization (co-loc) of chm7OPEN-GFP with either lipid sensor from three independent replicates of 100 cells per replicate. (D) As in C, but with the NLS-DAG sensor-mCherry. Inset is representative of the few cells where there is colocalization between NLS-DAG sensor-mCherry and chm7OPEN-GFP. Note that as DAG sensor is constitutively at the NE, we only scored cells where there was a discernable enrichment of the DAG sensor at the chm7OPEN-GFP site. Red asterisks demark the nucleus. (E) The PA sensor specifically accumulates at the NE in strains with NPC misassembly-associated herniations. Deconvolved inverted fluorescence images of NLS-PA sensor-mCherry (maximum-intensity projections from a z-series) in an nup116Δ strain at 23°C or after 3 h at 37°C, which triggers herniation formation. Numbers at bottom are the mean percentage and SD of cells with NLS-PA sensor-mCherry at foci at the NE where 300 total cells were counted from three independent experiments. (F) As in E, but with the NLS-DAG sensor-mCherry and only a middle focal section shown. Asterisks mark the nucleus. (G) Deconvolved fluorescence micrographs of NLS-PA sensor-mCherry in the indicated strains after 3 h at 30°C (upper panels) or 37°C (lower panels). (H) Plot of the mean and SD of the percentage of cells (from three independent experiments) where the NLS-PA sensor-mCherry accumulates at foci at the NE. >50 cells were quantified per experiment. P values were calculated from a one-way ANOVA with Tukey’s post hoc test where ns (not significant) is P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001. Scale bars, 5 µm.
We first tested whether either sensor’s distribution was impacted by the expression of a truncation of Chm7, chm7OPEN (Fig. 4 B), which lacks the inhibitory helices in its ESCRT-III domain, resulting in an “open” form that is constitutively active at the INM (Webster et al., 2016; Thaller et al., 2019). At sites of chm7OPEN accumulation, there is an expansion of a fenestrated network of membrane emanating from the INM (Thaller et al., 2019). Under these conditions, the PA sensor localized to a focus at the nuclear periphery in 65% of cells, all of which colocalized with chm7OPEN-GFP (Fig. 4 C). In contrast, the DAG sensor was observed in a focal accumulation that colocalized with chm7OPEN in only ∼22% of cells (Fig. 4 D, inset). Thus, we suggest that chm7OPEN specifically impacts PA levels in this expanded INM domain either by directly recruiting PA or by inducing local changes to PA metabolism.
We next investigated lipid sensor distribution under more physiological conditions. For example, Chm7 accumulates at the NE upon perturbation of de novo NPC assembly, which is correlated with the presence of NE herniations in both apq12Δ and nup116Δ cells (Webster et al., 2016). We first examined PA and DAG sensor distribution in nup116Δ cells grown at 23°C or at herniation-forming (37°C) temperatures (Wente and Blobel, 1993). At 23°C, the PA sensor localized in the nucleus with no visible accumulation at the INM (Fig. 4 E), while the DAG sensor localized at the NE and vacuole (Fig. 4 F). Upon shifting the culture to 37°C, there was a striking redistribution of the PA sensor, which accumulated along the nuclear periphery in distinct foci in 90% of cells (Fig. 4 E); wild-type cells grown at 37°C did not exhibit this phenotype (Fig. 4 G). Similar results were observed in apq12Δ cells, although less cells (∼40%) showed a focal accumulation of the PA sensor at the nuclear periphery (Fig. 4, G and H). These changes were again specific for the PA sensor, as we only observed a DAG sensor focus in ∼14% of nup116Δ cells (Fig. 4 F), which was not significantly different from those seen in ∼8% of wild-type cells (Fig. S2 D).
We next asked whether Chm7 contributed to the focal PA sensor localization in these NE herniation-forming strains. Chm7 was dispensable for PA sensor accumulation, as ∼60% of apq12Δchm7Δ cells retained PA sensor foci at the NE (Fig. 4, G and H). Thus, the data indicate that local PA concentration likely precedes Chm7 recruitment under the conditions of NPC misassembly. Although these results might seem contradictory to the ability of chm7OPEN to drive the PA sensor to the INM (Fig. 4 C), it is important to consider that, in the chm7OPEN case, virtually all of the cellular chm7OPEN is in this dominant “active” form. We suggest, then, that the resulting high local concentration of this PA-binding activity may be capable of directly recruiting the PA sensor. This contrasts with the productive Chm7 recruitment that occurs in the context of NPC misassembly, which triggers a physiological NPC assembly surveillance pathway that requires local PA to help recruit Chm7. Cumulatively, we conclude that the changes in local PA sensor distribution in nup116Δ and apq12Δ cells are not caused by Chm7, but instead by the upstream insult that leads to Chm7 recruitment.
The PA sensor localizes at NE herniations
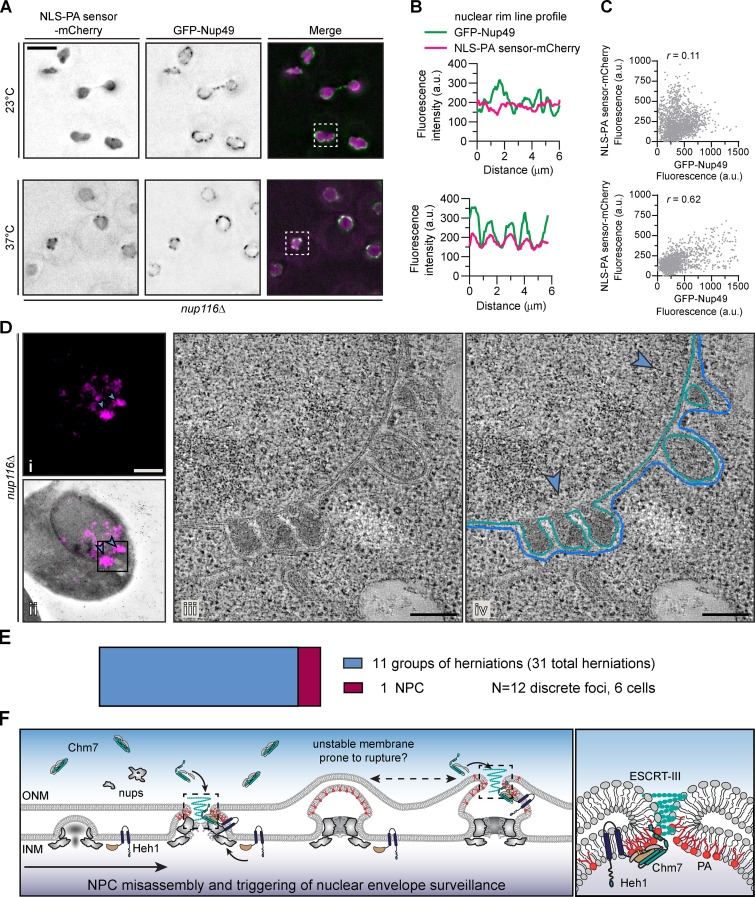
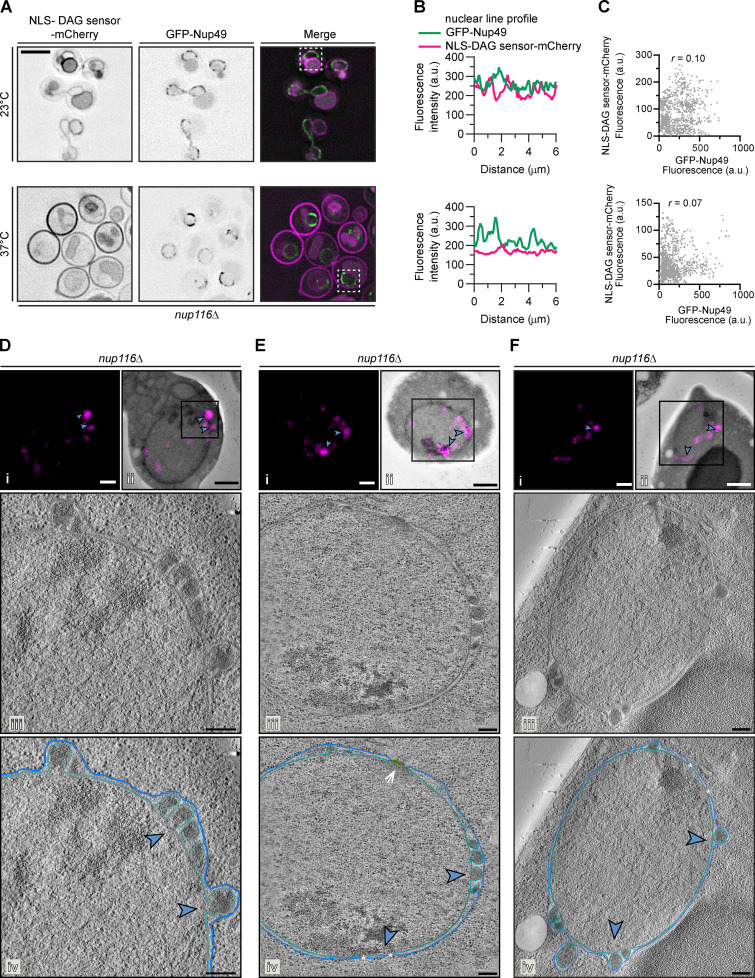
We therefore hypothesized that, in nup116Δ cells, PA accumulated at sites of NPC misassembly, perhaps within the herniations. To explore this possibility, we visualized the PA sensor alongside an NPC marker (GFP-Nup49) in nup116Δ cells by fluorescence microscopy. As expected, there was limited coincidence between PA sensor-mCherry and GFP-Nup49 fluorescence at 23°C (Fig. 5 A, top panels), as evident in line profiles along the NE of a single cell (boxed, Fig. 5 B, top) and in a scatterplot showing little correlation (r = 0.11) between the mCherry and GFP signal from many cells (Fig. 5 C, top). However, when herniations formed (37°C), we observed colocalization of NLS-PA sensor-mCherry and GFP-Nup49 (Fig. 5, A and B, bottom) with a positive correlation (r = 0.62) of the mCherry and GFP fluorescence (Fig. 5 C, bottom). In contrast, we did not observe obvious colocalization between the NLS-DAG sensor-mCherry and GFP-Nup49 at either temperature (Fig. S3, A–C).
Figure 5.
A PA sensor accumulates at NE herniations associated with NPC assembly defects. (A) The PA sensor colocalizes with NPCs under conditions of NE herniation formation. Deconvolved fluorescence micrographs of nup116Δ cells expressing NLS-PA sensor-mCherry and GFP-Nup49 at the indicated temperatures. In the GFP and mCherry images, the fluorescence is inverted, whereas the merge shows the GFP signal in green and mCherry in magenta. Scale bar is 5 µm. (B) Line profiles of the fluorescence intensities (in a.u.) along the nuclear rim of the boxed cells from corresponding top and bottom panels in A. (C) Correlation of the fluorescence intensity (in a.u.) of GFP-Nup49 and NLS-PA sensor-mCherry from line profiles drawn around the NE of 15 cells; r is the linear correlation coefficient (Pearson’s). Top and bottom panels correspond to top and bottom panels in A. (D) NLS-PA sensor-mCherry accumulates at NE herniations. Correlative light and electron microscopy of an nup116Δ strain grown at 37°C for 3 h before processing for EM. Fluorescence micrograph of NLS-PA sensor-mCherry alone (i) or overlaid on the corresponding low-magnification electron micrograph (ii) after correlation. Blue arrowheads point out focal accumulation of NLS-PA sensor-mCherry that are magnified in iv. Scale bars, 1 µm. (iii) Virtual slices from electron tomograms of the region indicated by the box in ii with 200-nm scale bar. (iv) Annotation of virtual slices from iii, with the ONM drawn in blue and the INM in teal and with the location of mCherry signal denoted with blue arrowheads (see Video 1). Scale bars, 200 nm. (E) Plot of the number of NE structures correlated with a total of 12 NLS-PA sensor-mCherry foci acquired from six unique cells/tomograms (other examples in Fig. S3). Area of colored rectangles is proportional to the total number of groups of herniations (31 individual herniations) and NPCs (1) identified. (F) Cartoon model incorporating PA into an NE/NPC assembly surveillance mechanism. From left to right: NPC assembly proceeds through the evagination of the INM and ultimate fusion with the ONM. Local PA (shown as red lipids) or DAG (not shown) might contribute to these events, likely through regulation of Pah1 (not shown). In contexts in which the NPC cannot be fully assembled (e.g., nup116Δ), Chm7 is recruited to the INM–ONM fusion site by a local accumulation of PA likely driven by its natural affinity for highly curved membranes. This local PA and membrane curvature would contribute to Chm7 recruitment. Chm7 activation is triggered by Heh1. The resulting local polymerization of Chm7 and other ESCRT-III subunits (teal spiral) would lead to membrane sealing. It is possible that the high local concentration of Chm7 during polymerization might also locally recruit PA and help to destabilize the nuclear membranes to promote fusion. Under conditions (like in nup116Δ) where NPCs cannot assemble properly, PA would continually build up, which might lead to the expansion of the herniations and likely instability in the herniation itself, leading to additional rupture (and repair) events. At right is a magnification of the boxed regions.
Figure S3.
The PA sensor accumulates in NE herniations (supplemental to Fig. 5). (A) The NLS-DAG mCherry sensor does not substantially colocalize with NPCs under conditions of herniation formation in nup116Δ cells. Deconvolved fluorescence micrographs of nup116Δ cells expressing NLS-DAG sensor-mCherry and GFP-Nup49 at the indicated temperatures. In the GFP and mCherry images, the fluorescence is inverted, whereas the merge shows the GFP signal in green and mCherry in magenta. Scale bar, 5 µm. (B) Line profiles of the fluorescence intensities (in a.u.) along the nuclear rim of the boxed cells from corresponding top and bottom panels in A. (C) Correlation of the fluorescence intensity (in a.u.) of GFP-Nup49 and NLS-DAG sensor-mCherry from line profiles drawn around the NE of 15 cells; r is the linear correlation coefficient (Pearson’s). (D–F) Examples of correlative light and electron tomography of NLS-PA mCherry-sensor localization in nup116Δ cells. Fluorescence micrographs of NLS-PA sensor-mCherry alone (i) or overlaid on the corresponding low-magnification electron micrograph (ii) after correlation. Blue arrowheads point out focal accumulation of NLS-PA sensor-mCherry that are magnified in iv. Scale bars, 1 µm. (iii) Virtual slices from electron tomograms of the region indicated by the box in ii with a 200-nm scale bar. (iv) Annotation of virtual slices from iii, with the ONM drawn in blue and the INM in teal and with the location of mCherry signal denoted with blue arrowheads. Asterisks demark locations of nuclear pores, and arrow points to a spindle pole body. Scale bars, 200 nm.
As light microscopy cannot resolve the herniations, we examined the distribution of the PA sensor in the context of nup116Δ NE ultrastructure using correlative light and electron tomography. The PA sensor fluorescence was visible as fluorescent puncta (Fig. 5 D, i, blue arrowheads), which were correlated with the corresponding electron micrograph at the edge of the nucleus (Fig. 5 D, ii). Higher-magnification views of the region under the PA sensor puncta (Fig. 5 D, ii, box) unambiguously identified the presence of NE herniations (Fig. 5 D, iii). In Fig. 5 D, iv, the INM and ONM are traced to allow for 3D reconstruction of the NE through the entire volume of the tomogram (see Video 1). In all, we examined six cells, which encompassed 12 discrete PA sensor foci (see also Fig. S3, D–F). With the exception of a single NPC (Fig. S3 E), the majority (11 out of 12) NLS-PA sensor-mCherry foci corresponded to groups of NE herniations (Fig. 5 E). Thus, these data provide compelling evidence that the NPC misassembly-associated NE herniations can recruit a PA sensor and support the conclusion that they contain high levels of PA.
Video 1.
The PA sensor accumulates at NE herniations (related to Fig. 5 D). Segmentation and isosurface rendering of correlative light and electron tomography of the NLS-PA sensor-mCherry (magenta) in an nup116Δ cell grown at 37°C for 3 h before processing for EM. The ONM is drawn in blue and the INM in teal. Scale bar, 200 nm. Playback speed, 10 frames per second.
Model and implications
We previously proposed a model whereby NPC assembly is surveilled by Chm7 at the step of INM–ONM fusion (Fig. 5 F; Webster et al., 2016; Thaller et al., 2019). When NPC assembly cannot be completed, for example in apq12Δ or nup116Δ cells (at 37°C), there is the formation of NE blebs or herniations. We now introduce PA as another component of this pathway (Fig. 5 F). We suggest that PA would have a natural affinity for sites of INM–ONM fusion because of their high curvature. As our in vitro data suggest, high curvature and PA are sufficient to recruit Chm7 to lipid bilayers (Fig. 2). In vivo, the data support that Chm7 membrane binding precedes and is in fact required for binding to Heh1. Thus, we suggest that an Heh1–Chm7 interaction reinforces the recruitment signal while simultaneously activating Chm7 polymerization, a likely prerequisite for membrane sealing (Fig. 5 F). Furthermore, as a putative Chm7–Heh1 polymer forms (predicated on that formed by CHMP7–LEM2; von Appen et al., 2020), it may also be possible to directly recruit additional PA (as suggested from chm7OPEN data in Fig. 4 C); this may have the added benefit of promoting the ultimate annular fusion event necessary for NE sealing in the context of NPC misassembly, but also likely in scenarios of NE rupture or postmitotic NE sealing, by making the membranes more fusogenic by destabilizing them.
Control of local PA metabolism may also be highly relevant in the context of normal NPC biogenesis as well. The observation that the NE herniations that arise during NPC misassembly are enriched for PA has several important implications. First, it suggests PA may directly contribute to herniation growth and be part of the pathology associated with these structures. Such an idea fits with work exploring the biogenesis of similar herniations in human cells with a loss of function of the AAA+ ATPase TorsinA (Goodchild et al., 2005; Laudermilch et al., 2016). As data grow linking these herniations to defective NPC assembly (Laudermilch et al., 2016; Pappas et al., 2018; Rampello et al., 2020), so does evidence that they are a product of defective lipid metabolism; in fact, recent work suggests that TorsinA may inhibit Lipin (Grillet et al., 2016; Cascalho et al., 2020; Jacquemyn et al., 2020 Preprint). Second, and as follows, while ultimately too much PA may be deleterious (Fig. 3, D–F; also see Zhukovsky et al., 2019), it seems probable that a tightly controlled system of PA generation and/or subsequent conversion to DAG may actually promote INM–ONM fusion during interphase NPC assembly. Definitively testing such a model requires more effective tools to precisely toggle PA levels (Tei and Baskin, 2020). Further, there is a need to develop additional tools to more fully explore how lipid synthesis and lipid flow across the nuclear pore membrane contribute to the lipid composition of the INM and ONM at steady state and during membrane remodeling.
Materials and methods
Saccharomyces cerevisiae culturing methods and strain generation
For all experiments, cells were grown to midlog phase in yeast extract peptone (1% Bacto-yeast extract [BD Biosciences], 2% Bacto-peptone [BD Biosciences], 0.025% adenine hemisulfate [Sigma-Aldrich]) supplemented with 2% raffinose (YPR; BD Biosciences), 2% D-galactose (YPG; Alfa Aesar), or 2% D-glucose (YPD; Sigma-Aldrich) or in synthetic complete medium (SC) lacking the indicated amino acids. The majority of experiments were performed at 30°C, unless otherwise indicated. Standard protocols were followed for transformation, mating, sporulation, and dissection as described in Amberg et al. (2005).
All yeast strains used in this study are listed in Table S1. Genetic modifications to generate gene deletions or the inframe incorporation of fluorescent protein genes were accomplished using a PCR-based approach using the pFA6a plasmid series (Table S2) as templates (Longtine et al., 1998). All oligonucleotides used in this study are listed in Table S3.
To genomically integrate the coding sequence for the NLS-PA sensor-mCherry or NLS-DAG sensor-mCherry, either pDT29 or pNA18 was linearized with StuI (New England Biolabs) and transformed into W303, apq12Δ (PCCPL249), chm7Δapq12Δ (CPL1323), and nup116Δ (Y1499) strains.
To generate strains to test complementation of chm7Δapq12Δ growth with chm7 alleles, a chm7Δapq12Δ (CPL1323) strain was transformed with StuI (New England Biolabs)–linearized pDT30, pDT31, pDT32, pDT33, pDT40, pDT41, or pDT44.
Plasmid generation
All plasmids used in this study are listed in Table S2. To generate pDT30, an ORF encoding CHM7-3xHA-GFP was PCR amplified from genomic DNA isolated from DTCPL1240 and assembled into pRS406-GAL1 digested with EcoRI/XbaI (New England Biolabs) using the Gibson Assembly Master Mix (New England Biolabs).
To generate pNA13, an ORF encoding GAL1-chm7-N was amplified from pDT30 with oligonucleotide primers containing SacI or PacI restriction sites. The PCR product was digested with SacI and PacI and gel purified (Qiagen) before ligation (T4 ligase; Invitrogen) into pDT30 digested with SacI/PacI (New England Biolabs).
Site-directed mutagenesis of the hydrophobic stretch coding sequences was performed using the Q5 site-directed mutagenesis kit (New England Biolabs) with either pCPLJJ50, pDT30, pDT40, or pNA13 as a template. Mutations were confirmed by sequencing.
pDT29 and pNA14 were generated by amplifying either NLS-Q2-mCherry from pRS316-CYC1-NLS-Q2-mCherry (gift from A. Köhler, Max Perutz Labs, Vienna, Austria) or the NLS-c1a-c1b-mCherry from pRS316-CYC1-NLS-c1a-c2b-mCherry (gift from A. Köhler) using Q5 polymerase. The PCR product was subsequently gel purified and assembled using Gibson Assembly (New England Biolabs) into pRS406-ADH1 digested with EcoRI/XbaI (New England Biolabs).
For generation of pDEST15-Chm7, used for bacterial expression of GST-Chm7 in lipid membrane strip binding experiments, the YJL049W cDNA coding for Chm7 (clone ScCD00011477) was supplied in pDONR201 by the Plasmid repository at Harvard Medical School (Hu et al., 2007; https://plasmid.med.harvard.edu/). The construct was shuttled into the pDEST15 gateway plasmid (Thermo Fisher Scientific) carrying an N-terminal GST tag for bacterial protein expression and purification.
Plate growth assays
To test for synthetic genetic interactions between CHM7 and PAH1 (DTCPL1634, 1659–60), NEM1 (DTCPL2010-13), or DGK1 (CPL1441, PMCPL379), or for growth complementation of chm7Δapq12Δ strains (PCCPL249, CPL1323, DTCPL1838, DTCPL1884-85, DTCPL1888, DTCPL1997-98), cells were cultured overnight, diluted to an OD600 of 0.4, and spotted onto YPD plates in 10-fold serial dilutions. Plates were wrapped in parafilm and incubated at either 23°C, 30°C, or 37°C for 36–48 h before imaging.
Microscopy
Cells were prepared for imaging as follows. Midlog-phase cells were collected by centrifugation at 375 g in a benchtop centrifuge for 1 min. Cells were resuspended in SC + 2% glucose and imaged immediately directly on the cover glass. Images were acquired on an Applied Precision DeltaVision microscope (GE Healthcare) using a 100×, 1.4 NA oil immersion objective (Olympus) and a CoolSnapHQ2 charge-coupled device camera (Photometrics).
To assess the localization of GFP fusions under the control of the GAL1 promoter, strains (DTCPL515-16, DTCPL2030, DTCPL2056-60, DTCPL2066-67) were grown in YPR to midlog phase. Expression of GFP fusions was induced upon addition of 1% galactose for 90 min before imaging.
To visualize NLS-PA sensor-mCherry or NLS-DAG sensor-mCherry in the apq12Δ and nup116Δ strains (DTCPL1979, DTCPL2035, DTCPL2039, DTCPL2050, PMCPL441, PMCPL450), cells were grown at RT and shifted to 37°C for 3 h before imaging. Similarly, to assess colocalization between NPCs and the lipid sensors, cells expressing GFP-Nup49 from pUN100-GFP-Nup49 (Belgareh and Doye, 1997) were grown in SC-leucine at 23°C and then shifted to 37°C for 3 h.
To assess the localization of Chm7-GFP (DTCPL1800, PMCPL381-384) and chm7-W3AV1A-GFP (DTCPL1873, PMCPL385), cells were grown to midlog phase in YPR, and expression of chm7-GFP fusions was induced upon the addition of 1% galactose for 45 min before imaging. For the additional staining of vacuole membranes, 1 ml of cultures was transferred to foil-wrapped tubes and incubated with the FM4-64 (1 µM in DMSO; Molecular Probes) for 1 h before washing once with SC + glucose before imaging.
Image processing/analysis
With the exception of those used for correlating fluorescent images to electron micrographs, all fluorescent micrographs presented were deconvolved using an iterative algorithm in softWoRx (version 6.5.1; Applied Precision/GE Healthcare). Micrographs and gel images were further processed in Fiji/ImageJ (Schindelin et al., 2012) and Adobe Photoshop. Deconvolved images were used for line profiles and colocalization analyses, but for all quantification of fluorescence intensity, unprocessed images were used.
To calculate N:C ratios of the NLS-GFP reporter (PLPC18), the average fluorescence of regions of the cytoplasm and nucleus within a middle z-section of individual cells was measured and related.
To evaluate the colocalization of GFP-Nup49 and the PA sensor or DAG sensor at the NE, the integrated density of GFP-Nup49 and NLS-PA sensor-mCherry fluorescence was measured from a freehand 4-pixel-wide line profile drawn around the NE of 15 cells using the Plot Profile plugin in Fiji (Schindelin et al., 2012). Corresponding values were plotted as a scatterplot, and the linear correlation coefficient (Pearson’s r) and linear regression were calculated in Prism (version 8.4; GraphPad Software).
LMB treatment
To inhibit Xpo1-mediated nuclear export, experiments were performed as previously described (Thaller et al., 2019). In brief, KWY175 (gift from B. Montpetit, University of California, Davis, Davis, CA)-expressing GFP fusions of Chm7 (DTCPL1815, DTCPL2052-2053, PMCPL436) were grown to midlog phase in YPR at 30°C. 20% galactose was added to the culture medium to a final concentration of 1%. After 45 min, glucose (final concentration, 2%) was added to the culture medium to arrest expression of GFP fusions. 2 ml of the culture were then treated with either 5 µl MeOH or LMB dissolved in 7:3 MeOH:H2O solution (Roche) at a final concentration of 50 ng/ml for 45 min before imaging.
Correlative light and electron tomography
Correlative microscopy of resin-embedded cells was performed as described previously (Kukulski et al., 2012). In brief, cells (DTCPL2050) were grown in YPD at RT for 5 h before shifting to the restrictive temperature (37°C) for 3 h. Cells were then pelleted by centrifugation for 2 min at 350 g and high-pressure frozen in the 200-µm recess of aluminum platelets (Engineering Office M. Wohlwend) using an HPM100 (Leica Microsystems). Samples were then freeze substituted in 0.1% uranyl acetate in acetone and embedded in Lowicryl HM20 (Polysciences) with the automated temperature control of an EM-AFS2 (Leica Microsystems) and manual agitation and solution exchange following published protocol (Kukulski et al., 2012). Resin-embedded cells were sectioned 250 nm thick using a Leica Artos 3D ultramicrotome equipped with a diamond knife (Diatome) and collected on 200 mesh copper grids with carbon support (01840; Ted Pella).
Fluorescence micrographs were acquired using the same imaging setup as described above. For each grid square of interest, 10 z-sections were acquired every 250 nm with a 3-s exposure with mCherry imaging conditions at 32% transmission. 15-nm protein A–coated gold beads (Cell Microscopy Core, University Medical Center Utrecht) were adhered to grids selected for EM.
Dual-axis tilt series were collected on an electron microscope (TF20; FEI) operated at 200 kV using a high-tilt tomography holder (model 2020; Fischione Instruments) from approximately −65 to +65 degrees (one-degree increments) at a binned pixel size of 1.242 nm on a 4k × 4k Eagle charge-coupled device (FEI) using a 150-µm C2 aperture and a 100-µm objective aperture in an automated fashion using SerialEM (Mastronarde, 2005). Reconstruction and segmentation were performed using IMOD (Kremer et al., 1996) in an automated fashion (Mastronarde and Held, 2017). For better visibility in the shown virtual slices, noise was reduced using a Gaussian filter in IMOD. Fluorescence images of resin sections have been rotated to match the orientation of electron tomogram virtual slices and have been adjusted for contrast individually.
Correlation of fluorescence microscopy and EM data was performed using the eC-CLEM plugin (Paul-Gilloteaux et al., 2017) in Icy (de Chaumont et al., 2012). The “anchor” map used by SerialEM during automated tilt series question, at 2.88-nm pixel size, was used for correlation with the fluorescence microscopy data. Correlation was performed by picking approximately six to eight corresponding points in both images based on morphological features of interest.
Statistical methods
All graphs were generated using Prism (version 8.4; GraphPad Software). Appropriate statistical significance tests were selected as indicated in the figure legends, and all values were calculated using Prism (version 8.4). P values are indicated on the graphs or in figure legends as follows: ns (not significant), P > 0.5; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. Error bars indicate the standard deviation from the mean. Individual replicate values are also plotted.
Recombinant GST fusion protein production and purification
To produce recombinant fusion proteins, Escherichia coli BL21 containing expression plasmids encoding for GST, GST-Chm7, GST-chm7-(W3AV1A), and GST-chm7-N were grown in Terrific Broth (2.4% Bacto-yeast extract [BD Biosciences], 1.2% Bacto-tryptone [BD Biosciences], 0.4% glycerol [Sigma-Aldrich]) overnight at 37°C, diluted to an OD600 of 0.1, and grown at 37°C until reaching an OD600 between 0.6 and 0.8. Subsequently, IPTG was added to a final concentration of 0.5 mM, and cells were cultured at 23°C for 6 h. The bacteria were collected by centrifugation for 10 min at 10,000 rpm at 4°C, washed with ice-cold water, and pelleted again at 4,000 rpm for 10 min at 4°C. Cell pellets were stored at −80°C.
Frozen cell pellets were resuspended in 15 ml of lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 2 mM MgCl2, 10% glycerol) with the addition of 150 mM NaCl, 1 mM DTT, Roche protease inhibitors, and 10 mg of lysozyme. The suspension was incubated on ice for 5–10 min and then lysed by sonication (Branson Sonifier 450). Lysates were pelleted at 11,700 rpm for 20 min at 4°C to remove cell debris. The supernatant was collected and incubated with 450 µl of glutathione Sepharose 4B (GE Healthcare) for 1 h at 4°C. Beads were then collected by centrifugation and washed three times in lysis buffer. GST fusion proteins were bound to the beads and were eluted upon the addition of glutathione (10 mM in 50 mM Tris buffer, pH 8.0), and the subsequent mixture was incubated at 4°C under constant rotation for 20 min. Beads were pelleted, and the supernatant was loaded into dialysis cassettes (3.5K, Slide-A-Lyzer; Thermo Fisher Scientific) that were then suspended in lysis buffer and dialyzed overnight to remove glutathione.
Protein binding to lipid strip membranes
GST-Chm7 binding to lipid strips was performed following the recommended manufacturer protocol (Membrane Lipid Strips P-6002; Echelon Biosciences Inc.). Briefly, membrane lipid strips with 100 pmol of 15 different membrane lipids were incubated in blocking buffer (PBS-T: 50 mM Na2HPO4, 150 mM NaCl, pH 7.4, 0.1% Tween 20, 3% BSA fatty acid free and globulin free) overnight at 4°C. Recombinantly produced GST-Chm7 or GST only was added to a membrane at a final concentration of ∼68 µg/ml in PBS-T and incubated overnight at 4°C. After discarding the protein solution, the strips were washed three times in PBS-T before incubation in primary antibody (GST tag mouse anti-tag, clone 8-326; Invitrogen) for 1 h at RT. The membrane was washed three times in PBS-T to remove any unbound primary antibody before incubating with the secondary antibody (antimouse IgG, HRP-linked antibody 7076; Cell Signaling Technology) for 1 h at room temperature. The membranes were washed in PBS-T before chemiluminescence detection.
Liposome generation
Liposomes were prepared as follows. Glycerophospholipids dissolved in chloroform of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate (840857C), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol (850142C), 1-oleoyl-2-palmitoyl-sn-glycero-3-phosphocholine (850457C, and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (850757C) were purchased from Avanti Polar Lipids. Lipid solutions were mixed in the desired ratios in a glass tube and were dried with N2 under a vacuum for 1 h. Dried lipid mixes were resuspended in SN buffer (20 mM Tris, pH 7.6, 100 mM NaCl, 5 mM MgCl2) to a final concentration of 10 mM. The suspensions were transferred to Eppendorf tubes and subjected to seven freeze–thaw cycles between liquid nitrogen and a 50°C water bath. Lipid mixtures were extruded 21 times through a double-membrane filter paper with pores of the desired liposome diameter (50 nm, 200 nm, 400 nm). The collected liposomes were stored on ice at 4°C until use in binding/flotation assays. 25-nm liposomes were generated by extrusion through a 100-nm membrane followed by three consecutive 1-min sonications (Branson Sonifier 450).
Liposome binding and flotation assays
For all experiments, 2 mM of prepared liposomes and 0.4 µM of selected GST fusions were mixed with SN buffer in a total volume of 150 µl in a Beckman 5 × 41–mm ultracentrifuge tube at 30°C for 1 h. Samples were adjusted to 40% Nycodenz by gently mixing with 150 µl of 80% Nycodenz in SN buffer. The mixture was layered with 250 µl of 30% Nycodenz in SN buffer and a top layer of 50 µl SN buffer. Samples were spun at 48,000 rpm at 4°C for 4 h in a Beckman SW Ti swinging-bucket rotor. 60 µl of floated sample (top layer) was collected using low-retention pipette tips. Laemmli SDS-PAGE sample buffer was added, and samples were denatured at 95°C for 5 min. Proteins were resolved on an SDS-PAGE gradient gel (4–20%; Bio-Rad Laboratories) and visualized by using SimplyBlue SafeStain (Thermo Fisher Scientific).
Online supplemental material
Fig. S1 compares the sequences and helical wheel models of the hydrophobic stretch between S. cerevisiae and Homo sapiens and shows Chm7-GFP at the nuclear rim in heh1Δ cells after LMB treatment. Fig. S2 shows that Chm7-GFP localizes to the NE/ER in pah1Δ cells, shows growth assays assessing epistasis between CHM7 and PAH1, DGK1, and NEM1, and shows micrographs and quantification of NLS-DAG sensor-mCherry NE foci in wild-type and nup116Δ cells. Fig. S3 shows micrographs of the DAG sensor in nup116Δ cells and examples of correlative light and electron tomography where PA sensor localizes to NE herniations in nup116Δ cells. Video 1 shows an isosurface rendering of the NE and fluorescence microscopy from correlative light and electron tomographs of NLS-PA sensor-mCherry in nup116Δ in Fig. 5 D. Table S1 lists the genotypes and origin of all S. cerevisiae strains used in this study. Table S2 lists all plasmids used in this study. Table S3 lists all oligonucleotides used in this study.
Supplementary Material
lists genotypes and origins of all S. cerevisiae strains used in this study.
lists all plasmids used in this study.
lists all oligonucleotides used in this study.
Acknowledgments
We are grateful to A. Köhler for the lipid sensors and M. Graham and X. Liu for EM expertise. We extend our thanks to T. Melia and L. Luan for help with liposome experiments.
This work was supported by the National Institutes of Health (R01 GM105672 to C.P. Lusk; R21 CA231269 to C.P. Lusk and M.C. King; F32 GM139285 to N.R. Ader; and T32 GM007223) and Engineering and Physical Sciences Research Council grant EP/M027821/1 (to B. Ciani).
The authors declare no competing financial interests.
Author contributions: Conceptualization: D.J. Thaller, B. Ciani, C.P. Lusk; methodology: D.J. Thaller, D. Tong, C.J. Marklew, N.R. Ader, S. Borah, B. Ciani, C.P. Lusk; investigation: D.J. Thaller, D. Tong, C.J. Marklew, N.R. Ader, P.J. Mannino, S. Borah, C.P. Lusk; validation: D.J. Thaller, P.J. Mannino; funding acquisition: B. Ciani, M.C. King, N.R. Ader, C.P. Lusk; project administration: B. Ciani, C.P. Lusk; supervision: B. Ciani, M.C. King, C.P. Lusk; visualization: D.J. Thaller, D. Tong, C.J. Marklew, N.R. Ader, P.J. Mannino; writing – original draft: D.J. Thaller, C.P. Lusk; writing – review and editing: all authors.
References
- Adeyo, O., Horn P.J., Lee S., Binns D.D., Chandrahas A., Chapman K.D., and Goodman J.M.. 2011. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192:1043–1055. 10.1083/jcb.201010111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegretti, M., Zimmerli C.E., Rantos V., Wilfling F., Ronchi P., Fung H.K.H., Lee C.W., Hagen W., Turonová B., Karius K., et al. 2020. In-cell architecture of the nuclear pore and snapshots of its turnover. Nature. 586:796–800. 10.1038/s41586-020-2670-5 [DOI] [PubMed] [Google Scholar]
- Amberg, D.C., Burke D., and Strathern J.N.. 2005. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 256 pp. [Google Scholar]
- Barbosa, A.D., Lim K., Mari M., Edgar J.R., Gal L., Sterk P., Jenkins B.J., Koulman A., Savage D.B., Schuldiner M., et al. 2019. Compartmentalized synthesis of triacylglycerol at the inner nuclear membrane regulates nuclear organization. Dev. Cell. 50:755–766.e6. 10.1016/j.devcel.2019.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh, N., and Doye V.. 1997. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J. Cell Biol. 136:747–759. 10.1083/jcb.136.4.747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bircham, P., Papagiannidis D., Lüchtenborg C., Ruffini G., Brügger B., and Schuck S.. 2020. Control of endoplasmic reticulum membrane biogenesis by regulators of lipid metabolism. bioRxiv. (Preprint posted February 24, 2020) 10.1101/2020.02.23.961722 [DOI]
- Carman, G.M., and Han G.-S.. 2011. Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem. 80:859–883. 10.1146/annurev-biochem-060409-092229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascalho, A., Foroozandeh J., Hennebel L., Swerts J., Klein C., Rous S., Dominguez Gonzalez B., Pisani A., Meringolo M., Gallego S.F., et al. 2020. Excess Lipin enzyme activity contributes to TOR1A recessive disease and DYT-TOR1A dystonia. Brain. 143:1746–1765. 10.1093/brain/awaa139 [DOI] [PubMed] [Google Scholar]
- de Chaumont, F., Dallongeville S., Chenouard N., Hervé N., Pop S., Provoost T., Meas-Yedid V., Pankajakshan P., Lecomte T., Le Montagner Y., et al. 2012. Icy: an open bioimage informatics platform for extended reproducible research. Nat. Methods. 9:690–696. 10.1038/nmeth.2075 [DOI] [PubMed] [Google Scholar]
- Dymond, M.K. 2016. Mammalian phospholipid homeostasis: evidence that membrane curvature elastic stress drives homeoviscous adaptation in vivo. J. R. Soc. Interface. 13:20160228. 10.1098/rsif.2016.0228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey, S., Richter R.P., and Görlich D.. 2006. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 314:815–817. 10.1126/science.1132516 [DOI] [PubMed] [Google Scholar]
- Gatta, A.T., Olmos Y., Stoten C.L., and Carlton J.G.. 2020. CDK1 controls CHMP7-dependent nuclear envelope reformation. bioRxiv. (Preprint posted April 21, 2020) 10.1101/2020.04.20.050674 [DOI] [PMC free article] [PubMed]
- Gautier, R., Douguet D., Antonny B., and Drin G.. 2008. HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics. 24:2101–2102. 10.1093/bioinformatics/btn392 [DOI] [PubMed] [Google Scholar]
- Goodchild, R.E., Kim C.E., and Dauer W.T.. 2005. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron. 48:923–932. 10.1016/j.neuron.2005.11.010 [DOI] [PubMed] [Google Scholar]
- Grillet, M., Dominguez Gonzalez B., Sicart A., Pöttler M., Cascalho A., Billion K., Hernandez Diaz S., Swerts J., Naismith T.V., Gounko N.V., et al. 2016. Torsins are essential regulators of cellular lipid metabolism. Dev. Cell. 38:235–247. 10.1016/j.devcel.2016.06.017 [DOI] [PubMed] [Google Scholar]
- Gu, M., LaJoie D., Chen O.S., von Appen A., Ladinsky M.S., Redd M.J., Nikolova L., Bjorkman P.J., Sundquist W.I., Ullman K.S., et al. 2017. LEM2 recruits CHMP7 for ESCRT-mediated nuclear envelope closure in fission yeast and human cells. Proc. Natl. Acad. Sci. USA. 114:E2166–E2175. 10.1073/pnas.1613916114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, G.S., Wu W.I., and Carman G.M.. 2006. The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281:9210–9218. 10.1074/jbc.M600425200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, G.-S., O’Hara L., Carman G.M., and Siniossoglou S.. 2008. An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J. Biol. Chem. 283:20433–20442. 10.1074/jbc.M802903200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch, E., and Hetzer M.. 2014. Breaching the nuclear envelope in development and disease. J. Cell Biol. 205:133–141. 10.1083/jcb.201402003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge, C.A., Choudhary V., Wolyniak M.J., Scarcelli J.J., Schneiter R., and Cole C.N.. 2010. Integral membrane proteins Brr6 and Apq12 link assembly of the nuclear pore complex to lipid homeostasis in the endoplasmic reticulum. J. Cell Sci. 123:141–151. 10.1242/jcs.055046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y., Rolfs A., Bhullar B., Murthy T.V.S., Zhu C., Berger M.F., Camargo A.A., Kelley F., McCarron S., Jepson D., et al. 2007. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Res. 17:536–543. 10.1101/gr.6037607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemyn, J., Foroozandeh J., Vints K., Swerts J., Verstreken P., Gounko N., Gallego S., and Goodchild R.. 2020. The Torsin/NEP1R1-CTDNEP1/Lipin axis regulates nuclear envelope lipid metabolism for nuclear pore complex insertion. bioRxiv. (Preprint posted July 5, 2020) 10.1101/2020.07.05.188599 [DOI]
- Kinugasa, Y., Hirano Y., Sawai M., Ohno Y., Shindo T., Asakawa H., Chikashige Y., Shibata S., Kihara A., Haraguchi T., et al. 2019. The very-long-chain fatty acid elongase Elo2 rescues lethal defects associated with loss of the nuclear barrier function in fission yeast cells. J. Cell Sci. 132:jcs229021. 10.1242/jcs.229021 [DOI] [PubMed] [Google Scholar]
- Kooijman, E.E., Chupin V., de Kruijff B., and Burger K.N.J.. 2003. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic. 4:162–174. 10.1034/j.1600-0854.2003.00086.x [DOI] [PubMed] [Google Scholar]
- Kremer, J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Kukulski, W., Schorb M., Welsch S., Picco A., Kaksonen M., and Briggs J.A.G.. 2012. Precise, correlated fluorescence microscopy and electron tomography of lowicryl sections using fluorescent fiducial markers. Methods Cell Biol. 111:235–257. 10.1016/B978-0-12-416026-2.00013-3 [DOI] [PubMed] [Google Scholar]
- Kwolek, U., Kulig W., Wydro P., Nowakowska M., Róg T., and Kepczynski M.. 2015. Effect of phosphatidic acid on biomembrane: experimental and molecular dynamics simulations study. J. Phys. Chem. B. 119:10042–10051. 10.1021/acs.jpcb.5b03604 [DOI] [PubMed] [Google Scholar]
- Larkin, M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Laudermilch, E., Tsai P.-L., Graham M., Turner E., Zhao C., and Schlieker C.. 2016. Dissecting Torsin/cofactor function at the nuclear envelope: a genetic study. Mol. Biol. Cell. 27:3964–3971. 10.1091/mbc.E16-07-0511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I.-J., Stokasimov E., Dempsey N., Varberg J.M., Jacob E., Jaspersen S.L., and Pellman D.. 2020. Factors promoting nuclear envelope assembly independent of the canonical ESCRT pathway. J. Cell Biol. 219:e201908232. 10.1083/jcb.201908232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lone, M.A., Atkinson A.E., Hodge C.A., Cottier S., Martínez-Montañés F., Maithel S., Mène-Saffrané L., Cole C.N., and Schneiter R.. 2015. Yeast integral membrane proteins Apq12, Brl1, and Brr6 form a complex important for regulation of membrane homeostasis and nuclear pore complex biogenesis. Eukaryot. Cell. 14:1217–1227. 10.1128/EC.00101-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Lord, C.L., and Wente S.R.. 2020. Nuclear envelope-vacuole contacts mitigate nuclear pore complex assembly stress. J. Cell Biol. 219:e202001165. 10.1083/jcb.202001165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk, C.P., and Ader N.R.. 2020. CHMPions of repair: emerging perspectives on sensing and repairing the nuclear envelope barrier. Curr. Opin. Cell Biol. 64:25–33. 10.1016/j.ceb.2020.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk, C.P., and King M.C.. 2017. The nucleus: keeping it together by keeping it apart. Curr. Opin. Cell Biol. 44:44–50. 10.1016/j.ceb.2017.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid, A.S., Mancuso J., Cande W.Z., and Weis K.. 2006. The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. J. Cell Biol. 173:361–371. 10.1083/jcb.200506199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde, D.N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51. 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Mastronarde, D.N., and Held S.R.. 2017. Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 197:102–113. 10.1016/j.jsb.2016.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville, M., and Rosbash M.. 1999. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 18:3746–3756. 10.1093/emboj/18.13.3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos, Y., Hodgson L., Mantell J., Verkade P., and Carlton J.G.. 2015. ESCRT-III controls nuclear envelope reformation. Nature. 522:236–239. 10.1038/nature14503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos, Y., Perdrix-Rosell A., and Carlton J.G.. 2016. Membrane binding by CHMP7 coordinates ESCRT-III-dependent nuclear envelope reformation. Curr. Biol. 26:2635–2641. 10.1016/j.cub.2016.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko, E., Tang J.H., Andersen K.R., Knockenhauer K.E., Vallotton P., Derrer C.P., Kralt A., Mugler C.F., Chan L.Y., Schwartz T.U., et al. 2017. Natively unfolded FG repeats stabilize the structure of the nuclear pore complex. Cell. 171:904–917.e19. 10.1016/j.cell.2017.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka, S., Bui K.H., Schorb M., Hossain M.J., Politi A.Z., Koch B., Eltsov M., Beck M., and Ellenberg J.. 2016. Nuclear pore assembly proceeds by an inside-out extrusion of the nuclear envelope. eLife. 5:e19071. 10.7554/eLife.19071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas, S.S., Liang C.-C., Kim S., Rivera C.O., and Dauer W.T.. 2018. TorsinA dysfunction causes persistent neuronal nuclear pore defects. Hum. Mol. Genet. 27:407–420. 10.1093/hmg/ddx405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul-Gilloteaux, P., Heiligenstein X., Belle M., Domart M.-C., Larijani B., Collinson L., Raposo G., and Salamero J.. 2017. eC-CLEM: flexible multidimensional registration software for correlative microscopies. Nat. Methods. 14:102–103. 10.1038/nmeth.4170 [DOI] [PubMed] [Google Scholar]
- Penfield, L., Shankar R., Szentgyörgyi E., Laffitte A., Mauro M.S., Audhya A., Müller-Reichert T., and Bahmanyar S.. 2020. Regulated lipid synthesis and LEM2/CHMP7 jointly control nuclear envelope closure. J. Cell Biol. 219:e201908179. 10.1083/jcb.201908179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper, G.H., Sprenger S., Teis D., and Oliferenko S.. 2020. ESCRT-III/Vps4 controls heterochromatin-nuclear envelope attachments. Dev. Cell. 53:27–41.e6. 10.1016/j.devcel.2020.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampello, A.J., Laudermilch E., Vishnoi N., Prophet S.M., Shao L., Zhao C., Lusk C.P., and Schlieker C.. 2020. Torsin ATPase deficiency leads to defects in nuclear pore biogenesis and sequestration of MLF2. J. Cell Biol. 219:e201910185. 10.1083/jcb.201910185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanauska, A., and Köhler A.. 2018. The inner nuclear membrane is a metabolically active territory that generates nuclear lipid droplets. Cell. 174:700–715.e18. 10.1016/j.cell.2018.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa, H., Leung J., Grimsey N., Peak-Chew S., and Siniossoglou S.. 2005. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24:1931–1941. 10.1038/sj.emboj.7600672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli, J.J., Hodge C.A., and Cole C.N.. 2007. The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes. J. Cell Biol. 178:799–812. 10.1083/jcb.200702120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter, R., Hitomi M., Ivessa A.S., Fasch E.V., Kohlwein S.D., and Tartakoff A.M.. 1996. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol. Cell. Biol. 16:7161–7172. 10.1128/MCB.16.12.7161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou, S., Santos-Rosa H., Rappsilber J., Mann M., and Hurt E.. 1998. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17:6449–6464. 10.1093/emboj/17.22.6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tei, R., and Baskin J.M.. 2020. Spatiotemporal control of phosphatidic acid signaling with optogenetic, engineered phospholipase Ds. J. Cell Biol. 219:e201907013. 10.1083/jcb.201907013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaller, D.J., and Lusk C.P.. 2018. Fantastic nuclear envelope herniations and where to find them. Biochem. Soc. Trans. 46:877–889. 10.1042/BST20170442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaller, D.J., Allegretti M., Borah S., Ronchi P., Beck M., and Lusk C.P.. 2019. An ESCRT-LEM protein surveillance system is poised to directly monitor the nuclear envelope and nuclear transport system. eLife. 8:e45284. 10.7554/eLife.45284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer, G., Voelker D.R., and Feigenson G.W.. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9:112–124. 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventimiglia, L.N., Cuesta-Geijo M.A., Martinelli N., Caballe A., Macheboeuf P., Miguet N., Parnham I.M., Olmos Y., Carlton J.G., Weissenhorn W., et al. 2018. CC2D1B coordinates ESCRT-III activity during the mitotic reformation of the nuclear envelope. Dev. Cell. 47:547–563.e6. 10.1016/j.devcel.2018.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri, M., Schink K.O., Campsteijn C., Wegner C.S., Schultz S.W., Christ L., Thoresen S.B., Brech A., Raiborg C., and Stenmark H.. 2015. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature. 522:231–235. 10.1038/nature14408 [DOI] [PubMed] [Google Scholar]
- Vietri, M., Schultz S.W., Bellanger A., Jones C.M., Petersen L.I., Raiborg C., Skarpen E., Pedurupillay C.R.J., Kjos I., Kip E., et al. 2020. Unrestrained ESCRT-III drives micronuclear catastrophe and chromosome fragmentation. Nat. Cell Biol. 22:856–867. 10.1038/s41556-020-0537-5 [DOI] [PubMed] [Google Scholar]
- von Appen, A., LaJoie D., Johnson I.E., Trnka M.J., Pick S.M., Burlingame A.L., Ullman K.S., and Frost A.. 2020. LEM2 phase separation promotes ESCRT-mediated nuclear envelope reformation. Nature. 582:115–118. 10.1038/s41586-020-2232-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warecki, B., Ling X., Bast I., and Sullivan W.. 2020. ESCRT-III-mediated membrane fusion drives chromosome fragments through nuclear envelope channels. J. Cell Biol. 219:e201905091. 10.1083/jcb.201905091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, B.M., Colombi P., Jäger J., and Lusk C.P.. 2014. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell. 159:388–401. 10.1016/j.cell.2014.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, B.M., Thaller D.J., Jäger J., Ochmann S.E., Borah S., and Lusk C.P.. 2016. Chm7 and Heh1 collaborate to link nuclear pore complex quality control with nuclear envelope sealing. EMBO J. 35:2447–2467. 10.15252/embj.201694574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente, S.R., and Blobel G.. 1993. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J. Cell Biol. 123:275–284. 10.1083/jcb.123.2.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willan, J., Cleasby A.J., Flores-Rodriguez N., Stefani F., Rinaldo C., Pisciottani A., Grant E., Woodman P., Bryant H.E., and Ciani B.. 2019. ESCRT-III is necessary for the integrity of the nuclear envelope in micronuclei but is aberrant at ruptured micronuclear envelopes generating damage. Oncogenesis. 8:29. 10.1038/s41389-019-0136-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Neuner A., Rüthnick D., Sachsenheimer T., Lüchtenborg C., Brügger B., and Schiebel E.. 2018. Brr6 and Brl1 locate to nuclear pore complex assembly sites to promote their biogenesis. J. Cell Biol. 217:877–894. 10.1083/jcb.201706024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhukovsky, M.A., Filograna A., Luini A., Corda D., and Valente C.. 2019. Phosphatidic acid in membrane rearrangements. FEBS Lett. 593:2428–2451. 10.1002/1873-3468.13563 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists genotypes and origins of all S. cerevisiae strains used in this study.
lists all plasmids used in this study.
lists all oligonucleotides used in this study.